Abstract
Background
Aspergillus fumigatus (AFE) is a well-adapted, opportunistic fungus that causes a severe and commonly fatal disease, wherein IFN-γ is one of the most important protective cytokines. The aim of this study was to investigate the microRNA expression profile and explore the underlying mechanism during infection with AFE.
Methods
CD4+ T cells were activated by co-culturing with dendritic cells (DCs), which were pre-treated with AFE. Next, we performed microRNA microarray expression profiles of activated and control T cells, following which, miRNA-142-3P was selected. To explore the effect of miR-142-3P on T cell activation, miRNA-142-3P expression was disrupted by transient transfection with miR-142-3P mimic or inhibitor. Then, levels of RICTOR, phosphorylated AKT and IFN-γ were detected via Western blotting and qPCR respectively. We further used siRNA to decrease RICTOR expression and determined the role played by RICTOR in miR-142-3P mediated-IFN-γ expression by qPCR following AFE-mediated T cell activation.
Results
The heat-map of miRNA expression profiles showed that 54 microRNAs (miRNAs) were filtered, the levels of which, were significantly different between CD4+ T cells activated by AFE and control T cells, in which microRNA-142-3 was involved. Forced expression of miRNA-142-3P dramatically suppressed RICTOR levels, phosphorylated AKT and IFN-γ in AFE activated T cells. Conversely, loss of miRNA-142-3P elevated RICTOR levels, phosphorylated AKT and IFN-γ. Notably, RICTOR deficiency decreased AKT phosphorylation levels and IFN-γ secretion.
Conclusions
Observations indicated that down-regulation of microRNA-142-3p enhanced IFN-γ expression, and did so by promoting RICTOR expression in CD4+ T cells activated by AFE.
Keywords: miRNA-142-3p, Aspergillus fumigatus (AFE), T cells, gastric cancer, IFN-γ
Introduction
Invasive aspergillosis is one of the most severe pulmonary infections that provokes aspergillosis in patients with compromised immunity, and an associated heightened mortality rate of 50–95%, indicating a relatively poor outcome from the clinical management of this disease (1,2). Aspergillus fumigatus (AFE), identified as an opportunistic fungal pathogen that is quite abundant in external environments, is the main pathogenic fungus of invasive aspergillosis. It has been reported that AFE not only induces fungal infection in immune suppressed individuals but also exacerbates the inflammatory response to invasive aspergillosis.
Adaptive immunity is considered a critical consideration in AFE infectious diseases. Additionally, it is well-known that conventional myeloid dendritic cells (DCs) are highly potent professional antigen presenting cells (APCs), which exert important effects on adaptive immunity that is stimulated by Aspergillus. Once the invading microorganism is inhaled in the context of respirable hydrophobic conidia, resident phagocytic cells are activated and attempt to eliminate it. However, if the spores are not effectively removed, they will germinate into germ tube and/or hyphal morpho types, in which macrophages and DCs produce inflammatory factors and trigger adaptive immunity. DCs can capture aspergillus antigens, and present fungal peptides to CD4+ T cells, following which, the production of inflammatory cytokines and chemokines ensues, which contributes to T cell activation. These activated CD4+ T cells are both protective (Th1 and Th17) and pathological (Th2) (3-5).
It has been suggested that Th1-mediated adaptive immunity significantly promotes protective immune responses in invasive aspergillosis (6-8). Previous studies showed that the expression of IFN-γ is detected in early infectious stages of lungs that have been challenged with the microorganism (9,10). Furthermore, CD4 T cells have been identified as the main source of IFN-γ during late stages of infection, and in immunized mice (6,11,12), whereas IFN-γ is dominantly secreted by NK cells in the early infectious stages of involved lungs with neutropenic invasive aspergillosis (13). The results from prior clinical trials have illustrated the importance of adjunctive immunotherapy, which is based on the protective role of IFN-γ, which can dramatically restore immune functionality of patients affected by fungal sepsis caused by chronic granulomatous disease, renal transplant, and so on, in which the outcome is not significantly affected or improved by anti-fungal therapy (14-16).
MicroRNAs (miRNAs) are small non-coding RNAs of about 20–22 nucleotides, which have been reported to serve as functional regulators in different diseases, and do so mainly by promoting mRNA destabilization with the aim of inhibiting translation. Currently, miRNA plays a multitude of roles in various biological processes including immune responses. Prior studies showed that miRNAs have unique expression profiles in innate and adaptive immunity. In adaptive immunity, miRNAs engage in diverse processes including T cell activation, differentiation, proliferation, survival and TCR signaling pathway (17). Further, it has been reported that miRNAs have critical roles in infectious diseases, and do so by regulating the function of Th1 cells (18-21). However, the potential function of miRNAs in AFE activated CD4+ T cells, including IFN-γ secretion, and determining the underlying mechanisms remain unclear in the setting of aspergillosis.
The aim of this study was to identify differentially expressed miRNAs in human CD4+ T cells that were stimulated by an AFE extract, and then to identify the function of differentially-expressed miRNAs including IFN-γ secretion. Finally, we also sought to identify the regulatory mechanism of those identified miRNAs. Our study might provide novel insights into AFE promoting an infectious diseases, which might find clinical utility in the future management of aspergillosis.
Methods
Ethics statement
All the specimens in this study were used after obtaining written informed consent from all patients studied. The Ethics Committee of Changzheng Hospital had granted approval for this specific research protocol and study. It is confirmed that all the necessary consents, including the consent to participate in the study, and the consent to publish this work objectively and with the expected integrity, were obtained from all patients involved in this study.
Preparation of AFE antigen and specific antigen-activated T cells
The specific AFE antigen, as the mycelial extract, were prepared as described previously (22,23). The obtained protein lysate was concentrated using an endotoxin-free dialysis membrane with a 5-kDa pore size (pierce) to achieve a final protein concentration of approximately 800–1,000 mg/mL. Mononuclear cells were isolated from healthy peripheral blood by density, gradient centrifugation, from which, CD4+ T cells and CD14+ monocytic cells were respectively sorted by immuno-magnetic beads. Purified cells were then seeded in sterile 6-well plates (Thermo) at a density of 1×106 cells/mL. Cells were cultured for 5 days in RIPM-1640 medium (GIBCO Invitrogen Corp.) that was supplemented with 10% fetal calf serum (GIBCO Invitrogen Corp), 50 ng/mL of GM-CSF and 5 ng/mL IL-4 to generate immature DCs. The AFE antigen was added on the fifth day of DC generation, and after 24 h, antigen-primed DCs were harvested and mature DC cell markers were detected by flow cytometry.
The AFE antigen-treated DCs were co-cultured with the above purified CD4+ T cells for 24 h and the magnetic beads were then used to isolate CD4+ T cells. Isolated CD4+ T cells were marked by antibody PE-anti-IFN-Y, PE-anti-IL-4, PE-anti-IL-17 and PE-anti-IL-22 (BD Biosciences, New Jersey, USA) and analyzed by flow cytometry (FACScanW; BD Biosciences) equipped with Cell Quest software (BD Biosciences).
miRNA array
Prepared RNA samples from12 healthy donors (age- and gender-matched) were labeled using the cyanine 3 (Cy3)-PCP Agilent miRNA Complete Labeling kit and hybridized to SureHyb chambers (Agilent). Then, the slides were scanned using the Agilent Microarray Scanner (Agilent technologies, Santa Clara, CA, USA), and data were extracted from scanned images using the Feature Extraction software v.10.7 and normalized using Gene Spring Software v.11.0 (both software programs from Agilent technologies, Santa Clara).
Quantitative real-time polymerase chain reaction (qRT-PCR)
According to the manufacturer’s protocol, cDNA was reverse transcribed from total RNA samples, with a miRNA-specific stem-loop primer using a miRNA reverse transcription kit (Takara, Japan). The process of quantitative real-time PCR was performed in triplicate on an ABI 7500 system using the SYBR Green PCR Master mix (both from (ABI, Foster City, CA, USA). MiRNA expression levels were normalized to U6 or beta-actin as internal control.
Construction of RICTOR-specific siRNAs
RICTOR-specific siRNAs were designed with the BLOCK-IT RNAi Designer (https://rnaidesigner.invitrogen.com/rnaiexpress/), and synthesized using the chemical synthesis method by the RiboBio Gene Chem Co. (Guangzhou, China). The pGSci vector contains the H1 promoter driving the siRNA cassette, together with a CMV-driven coral EGFP (EGFP) cDNA and neomycin-resistance gene. The complementary sequences of the siRNA species were as follows:
S1 forwardc5' ACUUGUGAAGAAUCGUAUC dTdT 3';
S1 reverse 3'dTdTUGAACACUUCUUAGCAUAG 5';
S2 forward 5' GGCCAGACCUCAUGGAUAA dTdT 3';
S2 reverse 3'dTdT CCGGUCUGGAGUACCUAUU 5';
S3 forward 5'CAAACAAGGCUGUGAUAUU dTdT 3';
S3 reverse 3'dTdT GUUUGUUCCGACACUAUAA 5'.
A non-silencing siRNA was used as a negative control.
Western immunoblotting
Cell protein lysates were separated in 10% sodium dodecyl sulfate polyacrylamide gels that were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Roche Diagnostics, Mannheim, Germany), which were blocked by 5% blocking buffer and then detected with an anti-RICTOR, p-473 AKT and GAPDH specific antibodies. Protein loading was estimated using a mouse anti-actin monoclonal antibody. Lab Works Image Acquisition and Analysis Software (UVP, Upland, CA, USA) quantified specific protein band intensities. Antibodies were purchased from Abcam.
Flow-cytometric analysis
Approximately 1×106 cells were counted and stained with the following monoclonal antibodies: FITC-anti-CD4, PE-anti-IFN-Y, and PE-anti-IL-4. Subsequently, the cells were washed, fixed, and permeabilized with CvtofiX/Cytoperm buffer (BD Bioecience) and then analyzed by flow cytometry.
Statistical analysis
Data were analyzed by the Student’s t-test using the SPSS 15.0 statistical package (SPSS Inc., Chicago, IL, USA). An alpha value of P<0.05 was considered to be statistically significant.
Results
Elevated expression of IFN-γ in AFE treated CD4+ T cells
To obtain matured DCs activated by AFE antigen, CD14+ monocytes were sorted from peripheral blood mononuclear cells (PBMC) and induced into immature DCs that were further treated with the AFE extract. The expression of CD80 and CD83, which are recognized as markers of mature DCs, were analyzed to identify whether or not mature DCs had indeed been generated successfully. The result showed that the levels of CD80 and CD83 were significantly upregulated (Figure 1A,B). Next, to investigate the ability of AFE activated DC to stimulate activation of CD4+ T cells, a co-culture assay was performed and the relative markers of CD4+ T cells were then analyzed. Our data showed that AFE-treated DCs dramatically elevated CD69 expression in CD4+ T cells (Figure 1C). Notably, the inflammatory cytokines IL-17, IL-22, and IL-4, and especially IFN-γ were increased in CD4+ T cells that had been co-cultured with AFE-treated DCs (Figure 1D). Thus, the data suggested that AFE antigen promotes the maturity and activation of CD4+ T cells, in which IFN-γ expression is profoundly increased.
Figure 1.
Elevated expression of IFN-γ in CD4+ T cells treated with AFE. CD14+ PBMCs were isolated, induced to immature DCs and then incubated with/without AFE for 24 h. DCs markers, CD83 (A) and CD80 (B), were detected by flow-cytometric analysis; (C) CD4+ T cells were co-cultured with mature DCs (AFE) or immature DCs (control) for 24 h. Expression of CD4 and CD69 were detected by flow-cytometric analysis; (D) CD4+ T cells were sorted from PBMC and co-cultured with mature DCs (AFE) for 0, 24, 48 h. The mRNA levels of inflammatory cytokines (IFN-γ, IL-4, IL-17 and IL-22) (normalized for Actin) were measured by qPCR. *, P<0.05; **, P<0.001. PBMC, peripheral blood mononuclear cell; DC, dendritic cell; AFE, Aspergillus fumigatus.
MiRNA expression profiles in CD4+ T cells activated by AFE
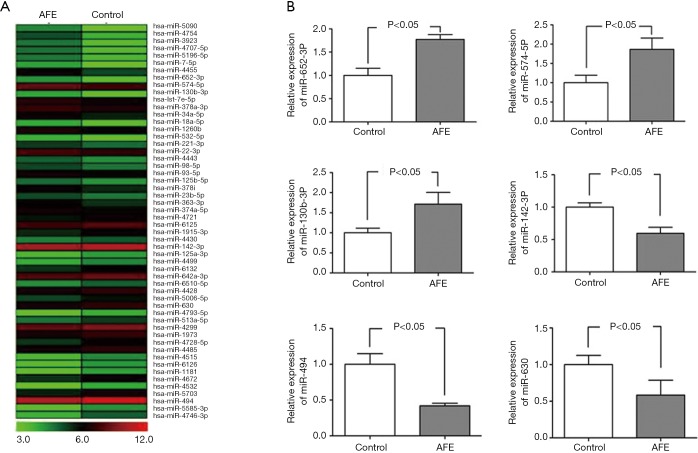
Prior studies suggested that in adaptive immunity, miRNAs engage diverse processes including T cell activation and differentiation, TCR signaling, and T cell proliferation and survival (17). Thus, to explore the underlying mechanism of AFE-induced CD4+ T cell maturity and activation, miRNA microarray analysis was used to identify the miRNA profiles of CD4+ T cells that were co-cultured with/without AFE antigen. A heat-map showed that a total of 54 miRNAs were significantly different when comparing activated CD4+ with non-activated T cells (Figure 2A) in which 26 miRNAs were up-regulated and 28 miRNAs were down-regulated (data not shown).
Figure 2.
miRNA expression profiles in CD4+ T cells activated by AFE. (A) RNAs of AFE activated CD4+ T cells and inactivated CD4+ T cells were extracted and subjected to miRNA microarray analysis. Heat map represented miRNAs dramatically changed, with ≥ 2-fold differences and P<0.05 between activated CD4+ T cells and inactivated CD4+ T cells. (B) the levels of altered miRNAs (normalized for U6) by miRNA microarray analysis in CD4+ T cells were measured by qPCR. AFE, Aspergillus fumigatus; miRNA, microRNAs.
To confirm the results of the microarray analyses, we performed quantitative RT-PCR analysis using RNA samples of 12 healthy donors. From this analysis, 6 from 54 of the differentially expressed miRNAs, which included miR-630, miR-494, miR-130b-3P, miR-652-3P, miR-574-5P and miR-142-3P, were subsequently selected. The expression data obtained by qRT-PCR were comparable to the results observed by microarray analysis (Figure 2B). Altogether, differentially expressed miRNAs, including miR-630, miR-494, miR-130b-3P, miR-652-3P, miR-574-5P and miR-142-3P might exhibit critical roles in AFE-induced CD4+ T cell maturation and activation.
MiR-142-3p inhibitor enhances RICTOR, phosphorylated AKT and IFN-γ expression
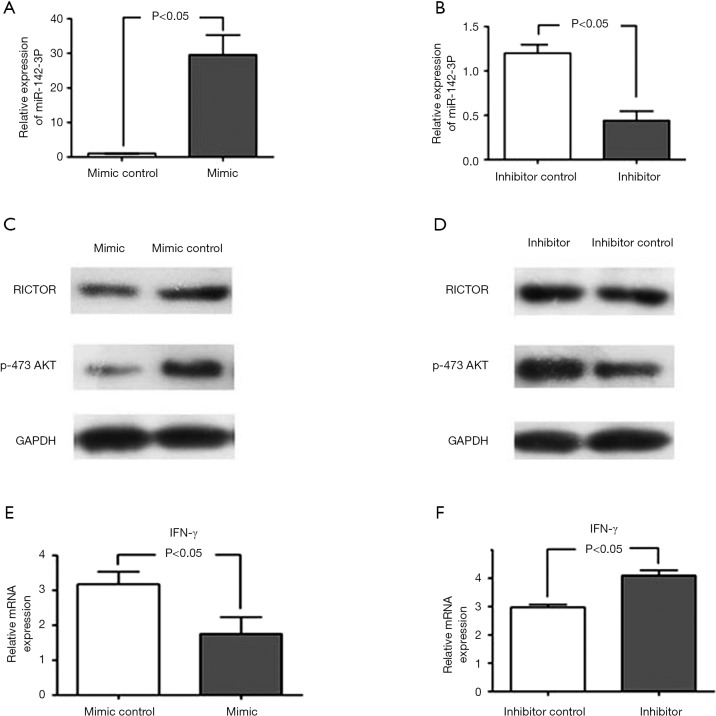
Previous data has shown that miR-142-3p is involved in the regulation of T cells (24,25), which led us to hypothesize whether miR-142-3p contributed to AFE-induced CD4+ T cell maturation and activation. Thus, to explore the potential effect of miR-142-3p, we disrupted the levels of expression of miR142-3p in AFE-induced CD4+ T cell by employing the miRNA-142-3P precursor or inhibitor. qPCR assays were used to detect miR-142-3p levels, and the results showed that levels of miR-142-3p were successfully disrupted in CD4+ T cells that had been activated by AFE (Figure 3A,B).
Figure 3.
The miR-142-3p inhibitor enhanced the expression of RICTOR, and of phosphorylated AKT and IFN-γ. AFE activated CD4+ T cells were transfected with miR-142-3p mimic (A), miR-142-3p inhibitor (B) and corresponding control regents for 24 h. Levels of miR-142-3p were detected by qPCR; western blotting analyses were used to measure RICTOR and p-AKT expression in AFE activated CD4+ T cells with miR-142-3p mimic (C) and miR-142-3p inhibitor (D) treatment; IFN-γ mRNA levels in AFE activated CD4+ T cells with miR-142-3p mimic (E) and miR-142-3p inhibitor (F) treatment were detected by qPCR. AFE, Aspergillus fumigatus.
It has been reported that RICTOR, which is a critical regulator of mTORC2 signaling, promotes Th1 cell activation, and does so by increasing the levels of phosphorylated AKT in vivo (26). Additionally, RICTOR was identified as a target of miR-142-3p in endothelial cells (27). Therefore, Western blotting was used to measure the expression of RICTOR and phosphorylated AKT in AFE-induced CD4+ T cells that had been transfected with/without the miR-142-3p precursor or inhibitor respectively. The data suggested that miR-142-3p inhibited the levels of RICTOR and phosphorylated AKT (Figure 3C,D). Importantly, IFN-γ levels were also detected and we found that miR-142-3p negatively regulated IFN-γ expression (Figure 3E,F). Collectively, our data illustrated that miR-142-3p inhibited AFE-induced CD4+ Th1 activation.
MiRNA-142-3p suppresses IFN-γ expression by boosting RICTOR/AKT signaling
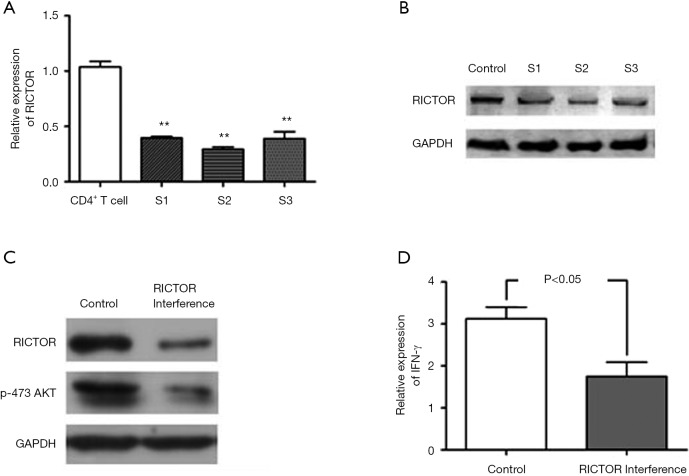
To further determine whether RICTOR mediates the inhibition of IFN-γ expression that was induced by miR-142-3p in AFE activated CD4+ T cells, we implemented RNAi to silence RICTOR. QRT-PCR (Figure 4A) and Weston blot (Figure 4B) assays confirmed the efficiency of RICTOR knockdown. The results suggested that RANi S2 exhibited optimal efficiency. Next, we investigated the role of RICTOR in CD4+ Th1 cell activation. Data showed that a deficiency in the functional expression of RICTOR notably decreased phosphorylated AKT and IFN-γ expression in AFE activated CD4+ T cells (Figure 4C,D). Taken together, the data demonstrated that RICTOR/AKT might mediate the miR-142-3p inhibited expression of IFN-γ in AFE activated CD4+ T cells.
Figure 4.
miRNA-142-3p suppresses IFN-γ expression and did so by stimulating RICTOR/AKT signaling AFE activated CD4+ T cells were transfected with control siRNA or RICTOR siRNAs for 24 h. The efficacy of RICTOR knockdown was measured by qPCR (A) and Western blotting (B) analysis; (C) p-473 AKT was detected by Western blotting; (D) the qPCR assay was implemented to measure IFN-γ expression in AFE activated CD4+ T cells that were transfected with control siRNA or RICTOR siRNAs. **, P<0.001 as compared with control. miRNA, microRNAs; AFE, Aspergillus fumigatus.
Discussion
In our study, the miRNA profile of human AFE-stimulated CD4+ T cells was investigated, following which, a total of 54 miRNA were identified as being significantly different between activated and inactivated CD4+ T cells including miR-630b, miR-494, miR-130b-3p, miR-652-3p, miR-574-5p and miR-142-3p. In particular, miR-142-3p was verified as being an important effector of IFN-γ expression in AFE-activated CD4+ T cells. Mechanically, we confirmed that miR-142-3p repressed IFN-γ expression in a RICTOR /AKT-dependent pathway.
IFN-γ, is one of the key cytokines that is secreted by CD4+ Th1 cells, which represent the major source of IFN-γ in pathogenic-mediated and infection-induced inflammation (28). In addition, IFN-γ can dramatically restore immune function of patients that present with fungal sepsis (14-16). In our study, the results from our mRNA analysis showed that the inflammatory cytokines IL-17, IL-22, and IL-4, and especially IFN-γ, were increased in CD4+ T cells that were co-cultured with AFE-treated DCs (Figure 1D), which demonstrated that as with other infections, AFE induces inflammatory pathways that can also activate T cells by enhancing IFN-γ expression.
Given the differentially expressed profiles of miRNA in activated and non-activated CD4+ T cells, we hypothesized that the significantly altered pattern of miRNA expression might have critical roles in the regulation of CD4+ T cell function in AFE-induced pathogenic infections. Notably, it was reported that miR-142-3p plays an important role in the maturation and migration of T cells (24,25). However, the correlation between miR-142-3p in AFE activated CD4+ T cells and IFN-γ generation has not been previously reported, which led us to explore the potential effects of miR-142-3p in this study. Our results showed that implementation of the miRNA-142-3p precursor or inhibitor, upregulated or inhibited the level of miR142-3p, and could negatively regulate IFN-γ expression in AFE-activated CD4+ T cells (Figure 3E,F).
As mentioned above, it is recognized that IFN-γ is one of the most important cytokines in Th1-mediated immunological responses. Further, miR-142-3p inhibits AFE-induced Th1 activation by repressing IFN-γ expression.
It has been reported that activation of mTORC2 signaling promotes activation of CD4+ Th1 cells and that RICTOR is a direct target of miR-142-3p in endothelial cell (26-27). Hence, RICTOR and the downstream regulator, phosphorylated AKT, were detected in AFE-induced CD4+ T cell that were transfected with/without the miR-142-3p precursor or inhibitor. Moreover, our results showed that miR-142-3p significantly inhibited the levels of RICTOR and phosphorylated AKT, which led us to determine whether miR-142-3p-mediated inhibition of IFN-γ production was mediated by RICTOR in AFE-activated CD4+ T cells. Therefore, we implemented RNAi to silence RICTOR in attempt to clarify this hypothesis. Our data showed that the miRNA-142-3p inhibitor enhanced the expression of RICTOR, AKT and IFN-γ in CD4+ T cells that had been activated by AFE. By contrast, phosphorylated AKT and IFN-γ expression was notably decreased when RICTOR was silenced (Figure 4C,D).
Overall, our studies demonstrated that miRNA-142-3p inhibited IFN-γ expression, and did so by targeting RICTOR in AFE-activated CD4+ T cells. This observation suggested that miR-142-3p might serve as a potential therapeutic target following infection with AFE.
Acknowledgments
Funding: This work was supported in part by a grant from the National Basic Research Program of China (973 Program) (Grant NO. 2013CB531603), and National Natural Science Foundation of China (NSFC No. 81471608) to Renqian Zhong and Changning district medical and health research projects of Shanghai (CNKW2018Y35) to Ning Ma.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Shanghai Changzheng Hospital Ethics Committee and the study was performed in accordance with the Declaration of Helsinki.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Cui N, Su LX, Wang H, et al. mTOR Modulates Lymphocyte Differentiation through T-bet and Eomesodermin in Response to Invasive Pulmonary Aspergillosis in Rats. Chin Med J (Engl) 2016;129:1704-10. 10.4103/0366-6999.185858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balloy V, Chignard M. The innate immune response to Aspergillus fumigatus. Microbes Infect 2009;11:919-27. 10.1016/j.micinf.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 3.Wang R, Wan Z, Li R. Th and Treg response induced by Aspergillus fumigatus pulsed dendritic cells in vitro. Chin Med J (Engl) 2014;127:3616-22. [PubMed] [Google Scholar]
- 4.Sun L, Chen C, Wu J, et al. TSLP-activated dendritic cells induce T helper type 2 inflammation in Aspergillus fumigatus keratitis. Exp Eye Res 2018;171:120-30. 10.1016/j.exer.2018.03.014 [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Zhang C, Jiang Y, et al. Innate and adaptive immune response to chronic pulmonary infection of hyphae of Aspergillus fumigatus in a new murine model. J Med Microbiol 2017;66:1400-8. 10.1099/jmm.0.000590 [DOI] [PubMed] [Google Scholar]
- 6.Cenci E, Perito S, Enssle KH, et al. Th1 and Th2 cytokines in mice with invasive aspergillosis. Infect Immun 1997;65:564-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cenci E, Mencacci A, Fe d'Ostiani C, et al. Cytokine- and T helper-dependent lung mucosal immunity in mice with invasive pulmonary aspergillosis. J Infect Dis 1998;178:1750-60. 10.1086/314493 [DOI] [PubMed] [Google Scholar]
- 8.Cenci E, Mencacci A, Del Sero G, et al. Interleukin-4 causes susceptibility to invasive pulmonary aspergillosis through suppression of protective type I responses. J Infect Dis 1999;180:1957-68. 10.1086/315142 [DOI] [PubMed] [Google Scholar]
- 9.Brieland JK, Jackson C, Menzel F, et al. Cytokine networking in lungs of immunocompetent mice in response to inhaled Aspergillusfumigatus. Infect Immun 2001;69:1554-60. 10.1128/IAI.69.3.1554-1560.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehrad B, Wiekowski M, Morrison BE, et al. Transient lung-specific expression of the chemokine KC improves outcome in invasive aspergillosis. Am J Respir Crit Care Med 2002;166:1263-8. 10.1164/rccm.200204-367OC [DOI] [PubMed] [Google Scholar]
- 11.Rivera A, Ro G, Van Epps HL, et al. Innate immune activation and CD4+ T cell priming during respiratory fungal infection. Immunity 2006;25:665-75. 10.1016/j.immuni.2006.08.016 [DOI] [PubMed] [Google Scholar]
- 12.Rivera A, Van Epps HL, Hohl TM, et al. Distinct CD4+-T-cell responses to live and heat-inactivated Aspergillusfumigatus conidia. Infect Immun 2005;73:7170-9. 10.1128/IAI.73.11.7170-7179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SJ, Hughes MA, Burdick M, et al. Early NK cell-derived IFN-{gamma} is essential to host defense in neutropenic invasive aspergillosis. J Immunol 2009;182:4306-12. 10.4049/jimmunol.0803462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrada C, Desai AG, Chirch LM, et al. Invasive easpergillosis in a renal transplant recipient successfully treated with interferon-gamma. Case Rep Transplant 2012;2012:493758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamashita K, Miyoshi T, Arai Y, et al. Enhanced generation of reactive oxygen species by interferon-γ may have contributed to successful treatment of invasive pulmonary aspergillosis in a patient with chronic granulomatous disease. Int J Hematol 2013;97:505-10. 10.1007/s12185-013-1315-y [DOI] [PubMed] [Google Scholar]
- 16.Delsing CE, Gresnigt MS, Leentjens J, et al. Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: a case series. BMC Infect Dis 2014;14:166. 10.1186/1471-2334-14-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun 2009;32:189-94. 10.1016/j.jaut.2009.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corral-Fernández NE, Cortes-García JD, Bruno RS, et al. Analysis of transcription factors, microRNAs and cytokines involved in T lymphocyte differentiation in patients with tuberculosis after directly observed treatment short-course. Tuberculosis (Edinb) 2017;105:1-8. 10.1016/j.tube.2017.03.007 [DOI] [PubMed] [Google Scholar]
- 19.Sánchez-Del Cojo M, López-Huertas MR, Díez-Fuertes F, et al. Changes in the cellular microRNA profile by the intracellular expression of HIV-1 Tat regulator: A potential mechanism for resistance to apoptosis and impaired proliferation in HIV-1 infected CD4+ T cells. PLoS One 2017;12:e0185677. 10.1371/journal.pone.0185677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Naqvi RA, Ali R, et al. FoxP3 provides competitive fitness to CD4+CD25+ T cells in leprosy patients via transcriptional regulation. Eur J Immunol 2014;44:431-39. 10.1002/eji.201343649 [DOI] [PubMed] [Google Scholar]
- 21.Zhao W, Liu C, Shi C, et al. Role of miR-124a in T cell activation and immunity in AIDS patients. Exp Ther Med 2017;14:4807-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corry DB, Grunig G, Hadeiba H, et al. Requirements for allergen-induced airway hyperreactivity in T and B cell-deficient mice. Mol Med 1998;4:344-55. 10.1007/BF03401741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koth LL, Rodriguez MW, Bernstein XL, et al. Aspergillus antigen induces robust Th2 cytokine production, inflammation, airway hyperreactivity and fibrosis in the absence of MCP-1 or CCR2. Respir Res 2004;5:12. 10.1186/1465-9921-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Oravecz-Wilson K, Mathewson N, et al. Mature T cell responses are controlled by microRNA-142. J Clin Invest 2015;125:2825-40. 10.1172/JCI78753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Li W, Wang S, et al. MiR-142-3p attenuates the migration of CD4+ T cells through regulating actin cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans. PLoS One 2014;9:e95514. 10.1371/journal.pone.0095514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen HH, Huang WT, Yang LW, et al. The PTEN-AKT-mTOR/RICTOR Pathway in Nasal Natural Killer Cell Lymphoma Is Activated by miR-494-3p via PTEN But Inhibited by miR-142-3p via RICTOR. Am J Pathol 2015;185:1487-99. 10.1016/j.ajpath.2015.01.025 [DOI] [PubMed] [Google Scholar]
- 27.Qin B, Shu Y, Long L, et al. MicroRNA-142-3p Induces Atherosclerosis-Associated Endothelial Cell Apoptosis by Directly Targeting Rictor. Cell Physiol Biochem 2018;47:1589-603. 10.1159/000490932 [DOI] [PubMed] [Google Scholar]
- 28.Ramstein J, Broos CE, Simpson LJ, et al. IFN-γ-Producing T-Helper 17.1 Cells Are Increased in Sarcoidosis and Are More Prevalent than T-Helper Type 1 Cells. Am J Respir Crit Care Med 2016;193:1281-91. 10.1164/rccm.201507-1499OC [DOI] [PMC free article] [PubMed] [Google Scholar]