Abstract
Background
This study was to establish nomogram models for prognostic evaluation of early-onset gastric cancer (EOGC) in both overall survival (OS) and cancer-specific survival (CSS).
Methods
EOGC patients from 2004 to 2015 were retrieved from the surveillance, epidemiology and end results (SEER) and further randomly assigned to training and validation sets. Univariate and multivariate cox analysis was used to screen out significant variables for construction of nomogram. Nomogram models were assessed by concordance index (C-index), calibration plot, receiver operating characteristics (ROCs) curve and decision curve analysis (DCA).
Results
A total of 549 EOGC were selected in this process. OS nomogram was constructed based on tumor size and tumor site. CSS nomogram was constructed based on tumor size, SEER stage and tumor site. In training set, C-index for the OS nomogram was 0.688 [95% confidence intervals (95% CI): 0.629–0.747], CSS nomogram 0.785 (95% CI: 0.735–0.835). In the external validation, the C-index for the OS nomogram was 0.633 (95% CI: 0.579–0.687), while for the CSS nomogram 0.733 (95% CI: 0.686–0.780). High quality of calibration plots both in OS and OS nomogram models was noticed. Nomograms displayed a comparable result to tumor-node-metastasis (TNM) stage and SEER stage for EOGC based on DCA.
Conclusions
The nomogram models provided an insightful and applicable tool to evaluate the prognosis of EOGC both in OS and CSS.
Keywords: Gastric cancer (GC), nomogram, overall survival (OS), cancer-specific survival (CSS), early-onset
Introduction
Gastric cancer (GC) remains one of the major malignancies in Asia, particularly in China, Japan and Korea (1-4). Both incidence and mortality of GC have been significantly reduced by solid medical interventions and systematic screening techniques (4). Although curative surgical resection is the primary therapy for most advanced GC cases, exact surgical strategies remain largely controversial between eastern and western countries (5,6). Meanwhile, postoperative recurrence is also one of the major challenges disturbing the outcome of GC. Previously, 194 out of 417 Chinese GC patients underwent curative resection (46.5%) suffered from recurrence (7). In fact, the mean recurrence time was less than 3 years (7).
Early-onset gastric cancer (EOGC) is defined as GC patients with age of young than 45 (8,9). EOGC is featured by diffuse histology, closer association in genetic factors and higher metastatic risk (8,10). In fact, the incidence of EOGC in ages 25 to 39 has been increasing in US whereas the incidence of GC in older group dramatically declines (11).
Rona et al. reported that GC patients aged <45 years had a higher proportion of poorly-differentiated histology, signet-cell type and advanced stage (12). Of note, surgical intervention was not significantly associated with better outcome (12). Meanwhile, Takatsu et al. also reported that lymph node metastasis was identified as a strong recurrence-indicator in young patient group, even they were featured by significantly fewer comorbidities and postoperative complications (13). However, two studies both indicated that no significant difference for stage-specific survival was identified between early-onset and control groups. Moreover, in country specific, US young GC patients were featured by proximal tumor and preoperative chemotherapy while Chinese young GC patients were featured by distal position and more advanced stage (14). However, both OS and disease-specific survival (DSS) was not significantly different between China and US (14).
Nomogram-based clinical modeling has been one of the most widely used statistic methods in clinical investigations. Featured by visual and mathematical advantages, nomogram facilitates the clinical implementation and probability calculation of risk factor or other predictor variables.
Generally, in the real world, the age-specific risk factors correlated to prognosis remain largely unnoticed. In fact, prognostic nomogram for EOGC is yet to be fully developed and validated. Given the comparable unsatisfied prognosis of EOGC, we believe that actual clinical outcome of EOGC patients requires a more solid, specific and statistical-power enhanced clinical model rather a general one for all GC. In fact, surveillance, epidemiology and end results (SEER) program enables the establishment of the nomogram for EOGC with sufficient registered cases.
Based on these premises, an EOGC-specific nomogram model is developed and validated to aid the prognosis evaluation for each EOGC individual using the data retrieved from SEER.
Methods
Data retrieved from SEER
Specific clinicopathological data and prognostic outcome of EOGC patients from 2004 to 2015 were retrieved from the SEER using reference number 14622-Nov2017 (15,16). This study did not require a local ethics approval or a statement. Because all the data used in this study were retrieved from the SEER database with public available approach. The identification of GC was based on the histological code and the cancer staging scheme (version 0204). The inclusion criteria included: (I) age <45 years old; (II) with complete tumor-node-metastasis (TNM) stage information; (III) only one primary tumor lesion (GC); (IV) with surgery performed; (V) complete survival data; (VI) without missing data in SEER cause-specific death classification; (VII) without unknown tumor size; (VIII) without unknown grade and race. All included EOGC were randomly assigned into training set and validation set. The median follow-up length was 20 [0–71] months. The training set included 276 EOGC patients and median follow-up length was 18 [0–71] months. The median follow-up length of validation set was 22 [0–71] months.
Clinical variables of EOGC
Clinical variables included sex, age, race, cancer staging scheme, grade, tumor size, American Joint Committee on Cancer (AJCC) TNM stage, SEER stage, tumor site, SEER cause-specific death classification, survival related information. Overall survival (OS) was the primary endpoint whereas cancer-specific survival (CSS) was the secondary endpoint. Age and tumor size were categorically divided based on the optimal cut-off value generated by X-tile software version 3.6.1 (Yale University School of Medicine, US).
Construction and validation of nomogram model
The Kaplan-Meier method and log-rank test were used for survival analysis while chi-square test was used for the comparison of categorical variables. Univariate cox analysis was used to screen out significant variables (P<0.2) for further multivariate analysis and construction of nomogram. In validation, the concordance index (C-index) was used for the measurement of nomogram between performance and predicted results. Calibration plots were used for the comparison between nomogram-predicted and actual outcome using a 45-degree line as an optimal model. Receiver operating characteristics (ROCs) curve was used for the sensitivity and specificity of nomogram. Furthermore, decision curve analysis (DCA) was used for the threshold probabilities range of nomogram in association with TNM stage and SEER stage. In addition, the nomogram was also compared to the TNM stage, SEER stage in terms of C-index. R software version 3.3.0 (R foundation for Statistical Computing, Vienna, Austria) was used for all analysis. Statistically significant cutoff value was set up as P<0.05. However, P<0.2 was selected as filter value for univariate to multivariate analysis.
Results
Input data from SEER
A total of 549 EOGC were selected in this process, of which 276 were randomly assigned to the training set while 273 cases were into validation set (Figure 1). For all patients, 280 (51.0%) were male and 363 (66.1%) were White. Based on the optimal cutoff value in age (age <33, 33–42, age >42), 309 (56.3%) were between 33–42 years old. Meanwhile, based on the optimal cutoff value in tumor size (tumor size <3.7, 3.7–6.8, >6.8 cm), 229 (41.7%) were less than 3.7 cm. The majority of grade is III (78.7%) while 83.8% were in M0 stage. The most common tumor site for EOGC was gastric antrum (including pylorus) (30.1%), followed by cardia (21.1%) and body of stomach (12.4%). Moreover, 59.4% of all patients were regional in SEER stage classification (Table S1).
Figure 1.
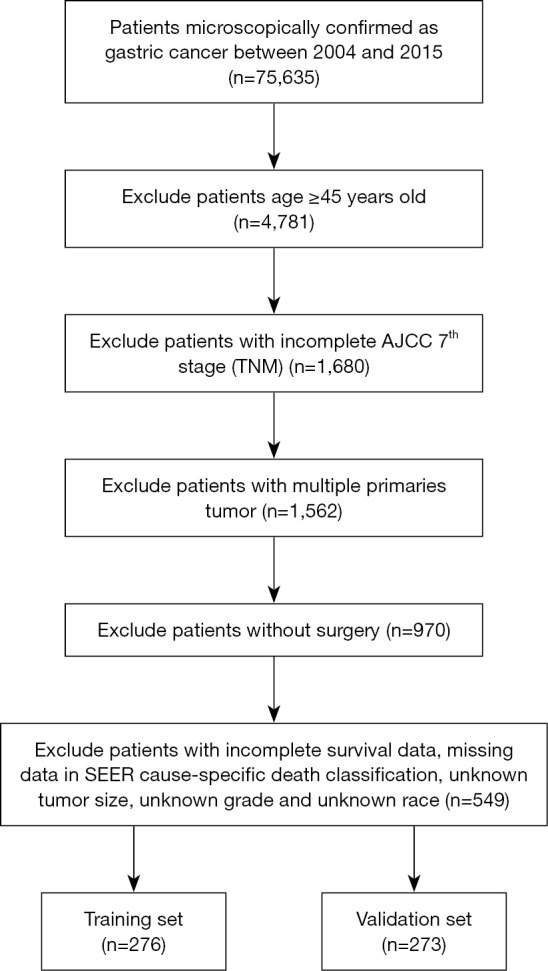
Flow diagram of the EOGC patients with training and validation sets. EOGC, early-onset gastric cancer; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; SEER, surveillance, epidemiology and end results.
Table S1. Patients’ demographics and clinicopathological characteristics.
| Variables | All patients, n (%) | Training set, n (%) | Validation set, n (%) | P |
|---|---|---|---|---|
| Total | 549 (100.0) | 276 (50.3) | 273 (49.7) | |
| Sex | 0.160 | |||
| Male | 280 (51.0) | 149 (54.0) | 131 (48.0) | |
| Female | 269 (49.0) | 127 (46.0) | 142 (52.0) | |
| Age, years | 0.081 | |||
| <33 | 100 (18.2) | 47 (17.0) | 53 (19.4) | |
| 33–42 | 309 (56.3) | 168 (60.9) | 141 (51.6) | |
| >42 | 140 (25.5) | 61 (22.1) | 79 (28.9) | |
| Race | 0.679 | |||
| White | 363 (66.1) | 181 (65.6) | 182 (66.7) | |
| Black | 74 (13.5) | 35 (12.7) | 39 (14.3) | |
| Other* | 112 (20.4) | 60 (21.7) | 52 (19.0) | |
| Grade | 0.513 | |||
| I | 18 (3.3) | 9 (3.3) | 9 (3.3) | |
| II | 80 (14.6) | 35 (12.7) | 45 (16.5) | |
| III | 432 (78.7) | 224 (81.2) | 208 (76.2) | |
| IV | 19 (3.5) | 8 (2.9) | 11 (4.0) | |
| Tumor size, cm | 0.699 | |||
| <3.7 | 229 (41.7) | 120 (43.5) | 109 (39.9) | |
| 3.7–6.8 | 184 (33.5) | 90 (32.6) | 94 (34.4) | |
| >6.8 | 136 (24.8) | 66 (23.9) | 70 (25.6) | |
| AJCC TNM stage (7th) | 0.592 | |||
| I | 94 (17.1) | 53 (19.2) | 41 (15.0) | |
| II | 127 (23.1) | 64 (23.2) | 63 (23.1) | |
| III | 239 (43.5) | 117 (42.4) | 122 (44.7) | |
| IV | 89 (16.2) | 42 (15.2) | 47 (17.2) | |
| AJCC T stage (7th) | 0.491 | |||
| T1 | 80 (14.6) | 39 (14.1) | 41 (15.0) | |
| T2 | 64 (11.7) | 37 (13.4) | 27 (9.9) | |
| T3 | 220 (40.1) | 113 (40.9) | 107 (39.2) | |
| T4 | 185 (33.7) | 87 (31.5) | 98 (35.9) | |
| AJCC N stage (7th) | 0.234 | |||
| N0 | 164 (29.9) | 92 (33.3) | 72 (26.4) | |
| N1 | 124 (22.6) | 59 (21.4) | 65 (23.8) | |
| N2 | 100 (18.2) | 52 (18.8) | 48 (17.6) | |
| N3 | 161 (29.3) | 73 (26.4) | 88 (32.2) | |
| AJCC M stage (7th) | 0.525 | |||
| M0 | 460 (83.8) | 234 (84.8) | 226 (82.8) | |
| M1 | 89 (16.2) | 42 (15.2) | 47 (17.2) | |
| Tumor site | 0.081 | |||
| C16.0—cardia, NOS | 116 (21.1) | 67 (24.3) | 49 (17.9) | |
| C16.1—fundus of stomach | 16 (2.9) | 10 (3.6) | 6 (2.2) | |
| C16.2—body of stomach | 68 (12.4) | 40 (14.5) | 28 (10.3) | |
| Gastric antrum (including pylorus) | 165 (30.1) | 76 (27.5) | 89 (32.6) | |
| C16.5—lesser curvature of stomach, NOS | 66 (12.0) | 28 (10.1) | 38 (13.9) | |
| C16.6—greater curvature of stomach, NOS | 25 (4.6) | 15 (5.4) | 10 (3.7) | |
| Stomach, NOS | 93 (17.0) | 40 (14.5) | 53 (19.4) | |
| SEER stage | 0.299 | |||
| Localized | 120 (21.9) | 67 (24.3) | 53 (19.4) | |
| Regional | 326 (59.4) | 162 (58.7) | 164 (60.1) | |
| Distant | 103 (18.8) | 47 (17.0) | 56 (20.5) |
*, American Indian/AK Native, Asian/Pacific Islander. AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; NOS, not otherwise specified; SEER, surveillance, epidemiology and end results.
Construction of nomogram
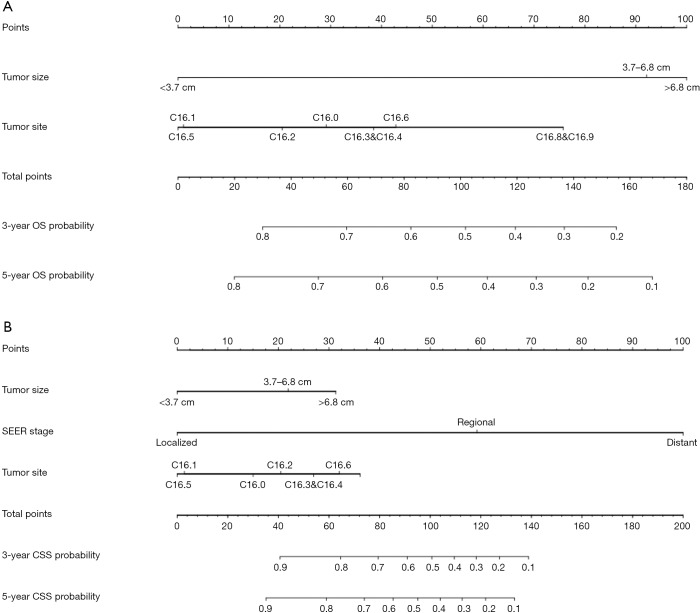
Tumor size, TNM stage, tumor site and SEER stage were significantly identified in univariate analysis in the training set (Table 1). However, only tumor size and tumor site were identified as significantly associated with OS in multivariate analysis. Next, the OS nomogram was constructed based on these two independent factors (Figure 2A). Moreover, in CSS analysis, tumor size, TNM stage, tumor site and SEER stage were identified by univariate analysis. However, only tumor size, SEER stage and tumor site were significantly identified in multivariate analysis and further subject to a CSS nomogram (Table 2, Figure 2B).
Table 1. Univariate and multivariate analysis of OS in the training set (n=276).
| Variables | No. of patients | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| P value | HR (95% CI) | P value | |||
| Sex | 0.752 | ||||
| Male | 149 | – | – | ||
| Female | 127 | – | – | ||
| Age, years | 0.976 | ||||
| <33 | 47 | – | – | ||
| 33–42 | 168 | – | – | ||
| >42 | 61 | – | – | ||
| Race | 0.411 | ||||
| White | 181 | – | – | ||
| Black | 35 | – | – | ||
| Other | 60 | – | – | ||
| Grade | 0.616 | ||||
| I | 9 | – | – | ||
| II | 35 | – | – | ||
| III | 224 | – | – | ||
| IV | 8 | – | – | ||
| Tumor size, cm | <0.001* | ||||
| <3.7 | 120 | Reference | |||
| 3.7–6.8 | 90 | 2.22 (1.20–4.13) | 0.0113* | ||
| >6.8 | 66 | 2.33 (1.22–4.46) | 0.0107* | ||
| AJCC TNM stage (7th) | <0.001* | ||||
| I | 53 | – | – | ||
| II | 64 | – | – | ||
| III | 117 | – | – | ||
| IV | 42 | – | – | ||
| AJCC T stage (7th) | <0.001* | ||||
| T1 | 39 | Reference | |||
| T2 | 37 | 2.45 (0.46–12.99) | 0.293 | ||
| T3 | 113 | 2.07 (0.40–10.65) | 0.3839 | ||
| T4 | 87 | 3.47 (0.66–18.43) | 0.1435 | ||
| AJCC N stage (7th) | <0.001* | ||||
| N0 | 92 | Reference | |||
| N1 | 59 | 0.70 (0.31–1.57) | 0.3882 | ||
| N2 | 52 | 0.93 (0.43–2.01) | 0.8443 | ||
| N3 | 73 | 1.25 (0.61–2.53) | 0.5414 | ||
| AJCC M stage (7th) | |||||
| M0 | 234 | Reference | |||
| M1 | 42 | 2.56 (0.56–11.74) | 0.226 | ||
| Tumor site | 0.062 | ||||
| C16.0—cardia, NOS | 67 | Reference | |||
| C16.1—fundus of stomach | 10 | 0.60 (0.17–2.10) | 0.4258 | ||
| C16.2—body of stomach | 40 | 0.75 (0.34–1.65) | 0.4768 | ||
| Gastric antrum (including pylorus) | 76 | 0.91 (0.48–1.73) | 0.7669 | ||
| C16.5—lesser curvature of stomach, NOS | 28 | 0.37 (0.14–0.98) | 0.0445* | ||
| C16.6—greater curvature of stomach, NOS | 15 | 1.10 (0.41–2.92) | 0.8523 | ||
| Stomach, NOS | 40 | 1.17 (0.54–2.49) | 0.6928 | ||
| SEER stage | <0.001* | ||||
| Localized | 67 | Reference | |||
| Regional | 162 | 2.49 (0.71–8.72) | 0.1533 | ||
| Distant | 47 | 3.53 (0.50–25.05) | 0.2076 | ||
*, Two-sided P values <0.05. OS, overall survival; HR, hazard ratio; CI, confidence intervals; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; NOS, not otherwise specified; SEER, surveillance, epidemiology and end results.
Figure 2.
OS and CSS associated nomograms for EOGC patients. (A) OS nomograms for EOGC in 3- and 5-year; (B) CSS nomograms for EOGC in 3- and 5-year. OS, overall survival; CSS, cancer-specific survival; EOGC, early-onset gastric cancer; SEER, surveillance, epidemiology and end results.
Table 2. Univariate and multivariate analysis of CSS in the training set (n=276).
| Variables | No. of patients | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| P value | HR (95% CI) | P value | |||
| Sex | 0.84 | ||||
| Male | 149 | – | – | ||
| Female | 127 | – | – | ||
| Age, years | 0.947 | ||||
| <33 | 47 | – | – | ||
| 33–42 | 168 | – | – | ||
| >42 | 61 | – | – | ||
| Race | 0.501 | ||||
| White | 181 | – | – | ||
| Black | 35 | – | – | ||
| Other | 60 | – | – | ||
| Grade | 0.457 | ||||
| I | 9 | – | – | ||
| II | 35 | – | – | ||
| III | 224 | – | – | ||
| IV | 8 | – | – | ||
| Tumor size, cm | <0.001* | ||||
| <3.7 | 120 | Reference | |||
| 3.7–6.8 | 90 | 1.92 (1.03–3.56) | 0.0396* | ||
| >6.8 | 66 | 2.23 (1.17–4.26) | 0.0151* | ||
| AJCC TNM stage (7th) | <0.001* | ||||
| I | 53 | – | – | ||
| II | 64 | – | – | ||
| III | 117 | – | – | ||
| IV | 42 | – | – | ||
| AJCC T stage (7th) | <0.001* | ||||
| T1 | 39 | Reference | |||
| T2 | 37 | 1.50 (0.26–8.52) | 0.6463 | ||
| T3 | 113 | 1.33 (0.25–7.04) | 0.7389 | ||
| T4 | 87 | 2.15 (0.40–11.72) | 0.3749 | ||
| AJCC N stage (7th) | <0.001* | ||||
| N0 | 92 | Reference | |||
| N1 | 59 | 0.65 (0.29–1.48) | 0.3043 | ||
| N2 | 52 | 0.93 (0.43–2.02) | 0.8533 | ||
| N3 | 73 | 1.26 (0.62–2.56) | 0.5281 | ||
| AJCC M stage (7th) | |||||
| M0 | 234 | Reference | |||
| M1 | 42 | 2.48 (0.54–11.4) | 0.2439 | ||
| Tumor site | 0.024* | ||||
| C16.0—cardia, NOS | 67 | Reference | |||
| C16.1—fundus of stomach | 10 | 0.40 (0.09–1.78) | 0.2285 | ||
| C16.2—body of stomach | 40 | 0.71 (0.32–1.59) | 0.4072 | ||
| Gastric antrum (including pylorus) | 76 | 0.88 (0.46–1.71) | 0.7122 | ||
| C16.5—lesser curvature of stomach, NOS | 28 | 0.38 (0.14–1.00) | 0.0506* | ||
| C16.6—greater curvature of stomach, NOS | 15 | 1.06 (0.40–2.84) | 0.9023 | ||
| Stomach, NOS | 40 | 1.15 (0.53–2.47) | 0.7294 | ||
| SEER stage | <0.001* | ||||
| Localized | 67 | Reference | |||
| Regional | 162 | 5.12 (1.14–23.11) | 0.0336* | ||
| Distant | 47 | 7.59 (0.89–64.81) | 0.064 | ||
*, two-sided P values <0.05. CSS, cancer-specific survival; HR, hazard ratio; CI, confidence intervals; AJCC, American Joint Committee on Cancer; TNM, tumor-node-metastasis; NOS, not otherwise specified; SEER, surveillance, epidemiology and end results.
Nomogram validation
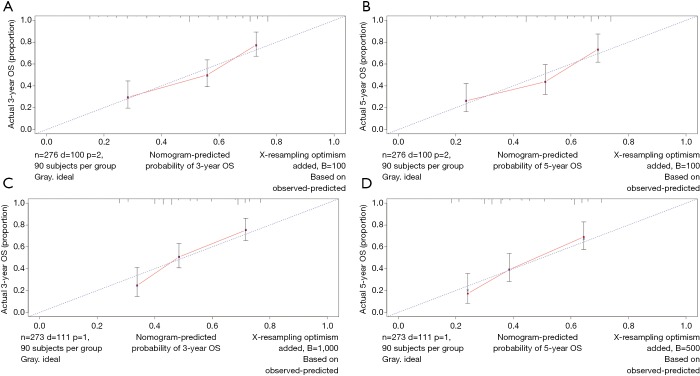
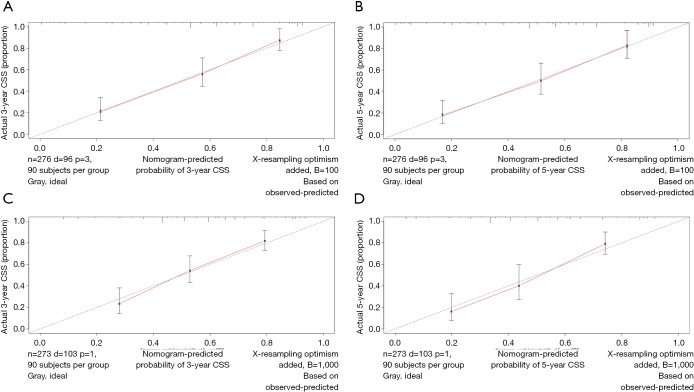
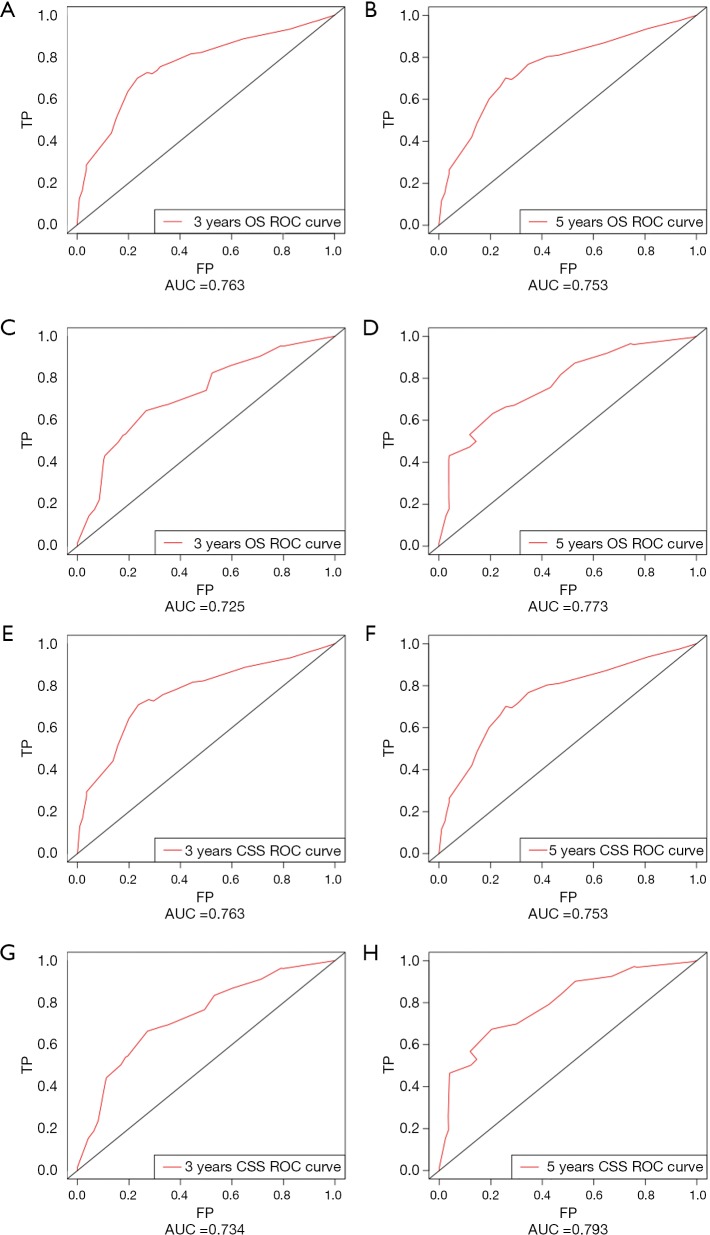
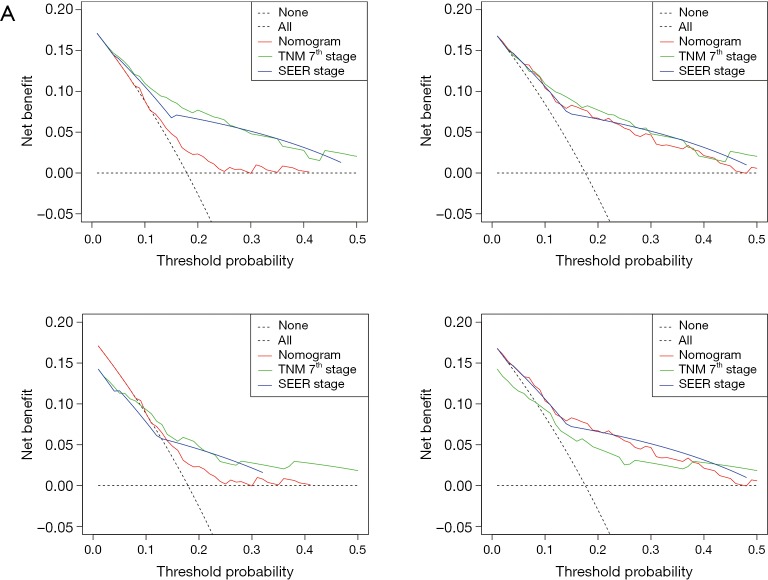
The OS and CSS nomograms were validated both internally and externally. In the internal validation, the C-index for the OS nomogram was 0.688 (95% CI: 0.629–0.747), while for the CSS nomogram 0.785 (95% CI: 0.735–0.835) (Table S2). In the external validation, the C-index for the OS nomogram was 0.633 (95% CI: 0.579–0.687), while for the CSS nomogram 0.733 (95% CI: 0.686–0.780) (Table S2). Meanwhile, high quality of calibration plots both in OS and OS nomogram models had been identified (Figures 3,4). In addition, high area under ROC curve (AUC) was noticed for both training and validation sets, respectively (Figure S1). The DCA results also indicated that nomograms showed a comparable clinical applicability similar to TNM stage and SEER stage (Figure S2). Specifically, both in training and validation sets, OS nomogram displayed a significantly better performance than TNM stage while CSS nomogram displayed better than SEER stage (Table S2). In addition, the etiology of non-cancer-related death in this study has been displayed. There are six types of non-cancer-related death that have been mentioned in the SEER concerning this study, including (I) accidents and adverse effects; (II) complications of pregnancy, childbirth, puerperium; (III) diseases of heart; (IV) nephritis, nephrotic syndrome and nephrosis; (V) other cause of death (COD); (VI) septicemia (Table S3).
Table S2. Comparison of C-indexes between the nomograms, TNM and SEER stages in EOGC patients.
| Survival types | Tumor stage types | Training set | Validation set | |||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |||
| OS | Nomogram | 0.688 | 0.629–0.747 | 0.633 | 0.579–0.687 | |||
| SEER stage | 0.708 | 0.659–0.757 | 0.267 | 0.685 | 0.642–0.728 | 0.198 | ||
| TNM 7th stage | 0.769 | 0.717–0.821 | 0.007 | 0.750 | 0.707–0.793 | <0.001 | ||
| CSS | Nomogram | 0.785 | 0.735–0.835 | 0.733 | 0.686–0.780 | |||
| SEER stage | 0.726 | 0.680–0.772 | 0.035 | 0.690 | 0.646–0.734 | 0.033 | ||
| TNM 7th stage | 0.782 | 0.732–0.832 | 0.457 | 0.762 | 0.719–0.805 | 0.366 | ||
TNM, tumor-node-metastasis; SEER, surveillance, epidemiology and end results; EOGC, early-onset gastric cancer; C-index, concordance index; HR, hazard ratio; CI, confidence intervals; OS, overall survival; CSS, cancer-specific survival.
Figure 3.
Calibration plots of OS associated nomograms in both training and validation sets. (A,B) Calibration plots of 3- and 5-year OS in training set; (C,D) calibration plots of 3- and 5-year OS in validation set. OS, overall survival.
Figure 4.
Calibration plots of CSS associated nomograms in both training and validation sets. (A,B) Calibration plots of 3- and 5-year CSS in training set; (C,D) calibration plots of 3- and 5-year CSS in validation set. CSS, cancer-specific survival.
Figure S1.
ROCs curve for the nomograms. (A) The ROC curve of nomogram with 3-year OS in training set; (B) the ROC curve of nomogram with 5-year OS in training set; (C) the ROC curve of nomogram with 3-year OS in validation set; (D) the ROC curve of nomogram with 5-year OS in validation set; (E) the ROC curve of nomogram with 3-year CSS in training set; (F) the ROC curve of nomogram with 5-year CSS in training set; (G) the ROC curve of nomogram with 3-year CSS in validation set; (H) the ROC curve of nomogram with 5-year CSS in validation set. ROC, receiver operating characteristic; OS, overall survival; CSS, cancer-specific survival; AUC, area under ROC curve; FP, false positive; TP, true positive.
Figure S2.
DCA of the nomograms for OS and CSS in both training and validation sets. (A,B) The DCA of nomogram in training set for both OS and CSS; (C,D) the DCA of nomogram in validation set for both OS and CSS. DCA, decision curve analysis; OS, overall survival; CSS, cancer-specific survival; SEER, surveillance, epidemiology and end results; TNM, tumor-node-metastasis.
Table S3. The etiology of non-cancer-related death.
| COD to site recode | SEER other COD classification | OS | CSS |
|---|---|---|---|
| Accidents and adverse effects | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
| Complications of pregnancy, childbirth, puerperium | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
| Diseases of heart | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
| Nephritis, nephrotic syndrome and nephrosis | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
| Other COD | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
| Septicemia | Dead (attributable to causes other than this cancer dx) | 1 | 0 |
COD, cause of death; SEER, surveillance, epidemiology and end results; OS, overall survival; CSS, cancer-specific survival.
Discussion
This study developed and validated prognostic nomogram models for both OS and CSS EOGC based on the public database SEER. By both internal and external validation, the nomograms used displayed comparably outcome to the TNM stage and SEER stage. The prognostic nomograms could facilitate the clinical prognostic evaluation and personalized treatment.
There are two reasons why this study focused on the nomogram of EOGC. Firstly, given the epidemiological facts, the incidence of EOGC remains largely unsatisfactory, particularly in patients between 25 and 39. However, it remains unclear whether the entire range of EOGC could be of high prognostic risk. Therefore, to develop a more specific nomogram that directly targets EOGC, instead of general patients’ group, could be of greater clinical value.
Secondly, given the fact that older GC patients are characterized by significantly declined incidence as well as potential reduced mortality rate, it is possible that those facts could contribute to the confounding bias of general prognostic indicator, particularly when focusing on the EOGC. In fact, the nomogram models in this study also reflected an individualized therapeutic management.
However, we admit that numerous variables, including sex and race, were not identified as prognostic significant. It is reasonable that in EOGC, potential prognostic significant factors may be diverse from general GC patients.
Moreover, the prognostic nomograms in this study may not display drastic different to older GC patients. However, it is also reasonable that the difference, whether or not existed between the EOGC-nomogram and older GC nomogram, did not lower the power of the nomogram developed in this study.
Of note, the nomogram used in OS included tumor size and tumor site while the nomogram used in CSS included tumor size, tumor site and SEER stage. Given the fact that optimal-cutoff categorized tumor size had been identified as significant independent factors in univariate/multivariate analysis, it is reasonably to presume the potential impact of tumor size in EOGC. In fact, conventional categorization based on 5 and 10 cm did not fully reflect the prognostic significance and clinical implications of tumor size in EOGC. Tumor size could be of great value to demonstrate the OS and CSS prognostic risk in this situation. In fact, based on our result, risk significantly increased in tumor size between 3.7–6.8 cm compared to tumor size <3.7 cm. Moreover, tumor size >6.8 cm demonstrated the highest risk compared to tumor size <3.7 cm. Previously, Saito et al. discovered that large size group (tumor size ≥8 cm) had been identified as an independent prognostic factor with worse prognosis (17). Of note, compared to small size group, large size group was featured by cases with undifferentiated histological type and lymphatic and venous invasion (17). Of note, tumor size >6.8 cm was a strong prognostic indicator both in OS and CSS. It may be potentially correlated with more advanced AJCC stage and occult disseminated tumor cells. Moreover, enhanced tumor cell proliferation and angiogenesis capability may also contribute to the biological features of larger tumors as well as extracellular matrix and stromal. In fact, intrinsic features beneath tumor size remain largely unexplored. Simple cutoff of tumor size, such as >5 or 10 cm, is not sufficient to fully characterize the prognostic and other clinical values. Thus, “bedside to bench” investigation is further warranted.
Up to now, SEER provides the largest sample size across various types of cancers. Moreover, similar nomogram models on several tumors by SEER have been explored (18,19). Although SEER does not fully provide numerous prognostic factors, for examples, surgical details (lymphadenectomy extent, D1, D2 or D2+), it remains one of the widely used datasets for nomogram modeling.
Meanwhile, there remains some limitation. Larger sample sizes are warranted for further independent validation and prognostic stratification analysis. The majority race of this data was from white, therefore could be with potential race heterogeneity.
Conclusions
The nomogram models provided an insightful and applicable tool to evaluate the prognosis of EOGC both in OS and CSS.
Acknowledgments
None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer 2007;10:75-83. 10.1007/s10120-007-0420-0 [DOI] [PubMed] [Google Scholar]
- 4.Ajani JA, D'Amico TA, Almhanna K, et al. Gastric cancer, version 3.2016, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2016;14:1286-312. 10.6004/jnccn.2016.0137 [DOI] [PubMed] [Google Scholar]
- 5.Songun I, Putter H, Kranenbarg EM, et al. Surgical treatment of gastric cancer: 15-year follow-up results of the randomised nationwide Dutch D1D2 trial. Lancet Oncol 2010;11:439-49. 10.1016/S1470-2045(10)70070-X [DOI] [PubMed] [Google Scholar]
- 6.Ajani JA, Bentrem DJ, Besh S, et al. Gastric cancer, version 2.2013: featured updates to the NCCN guidelines. J Natl Compr Canc Netw 2013;11:531-46. 10.6004/jnccn.2013.0070 [DOI] [PubMed] [Google Scholar]
- 7.Kang WM, Meng QB, Yu JC, et al. Factors associated with early recurrence after curative surgery for gastric cancer. World J Gastroenterol 2015;21:5934-40. 10.3748/wjg.v21.i19.5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milne AN, Offerhaus GJ. Early-onset gastric cancer: learning lessons from the young. World J Gastrointest Oncol 2010;2:59-64. 10.4251/wjgo.v2.i2.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milne AN, Carvalho R, Morsink FM, et al. Early-onset gastric cancers have a different molecular expression profile than conventional gastric cancers. Mod Pathol 2006;19:564-72. 10.1038/modpathol.3800563 [DOI] [PubMed] [Google Scholar]
- 10.Bautista MC, Jiang SF, Armstrong MA, et al. Impact of age on clinicopathological features and survival of patients with noncardia gastric adenocarcinoma. J Gastric Cancer 2014;14:238-45. 10.5230/jgc.2014.14.4.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson WF, Camargo MC, Fraumeni JF, Jr, et al. Age-specific trends in incidence of noncardia gastric cancer in US adults. JAMA 2010;303:1723-8. 10.1001/jama.2010.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rona KA, Schwameis K, Zehetner J, et al. Gastric cancer in the young: an advanced disease with poor prognostic features. J Surg Oncol 2017;115:371-5. 10.1002/jso.24533 [DOI] [PubMed] [Google Scholar]
- 13.Takatsu Y, Hiki N, Nunobe S, et al. Clinicopathological features of gastric cancer in young patients. Gastric Cancer 2016;19:472-8. 10.1007/s10120-015-0484-1 [DOI] [PubMed] [Google Scholar]
- 14.Strong VE, Russo A, Yoon SS, et al. Comparison of young patients with gastric cancer in the United States and China. Ann Surg Oncol 2017;24:3964-71. 10.1245/s10434-017-6073-2 [DOI] [PubMed] [Google Scholar]
- 15.Hankey BF, Ries LA, Edwards BK. The surveillance, epidemiology, and end results program: a national resource. Cancer Epidemiol Biomarkers Prev 1999;8:1117-21. [PubMed] [Google Scholar]
- 16.Hayat MJ, Howlader N, Reichman ME, et al. Cancer statistics, trends, and multiple primary cancer analyses from the surveillance, epidemiology, and end results (SEER) program. Oncologist 2007;12:20-37. 10.1634/theoncologist.12-1-20 [DOI] [PubMed] [Google Scholar]
- 17.Saito H, Osaki T, Murakami D, et al. Macroscopic tumor size as a simple prognostic indicator in patients with gastric cancer. Am J Surg 2006;192:296-300. 10.1016/j.amjsurg.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 18.Song W, Lv CG, Miao DL, et al. Development and validation of a nomogram for predicting survival in patients with gastrointestinal stromal tumours. Eur J Surg Oncol 2018;44:1657-65. 10.1016/j.ejso.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 19.Zhuang Y, Liu Y, Zhao H, et al. Age-related disparity in patients with colorectal cancer: a population-based study from SEER program. Int J Clin Exp Med 2017;10:3305-12. [Google Scholar]