Abstract
Background
Circulating cell-free DNA (cfDNA), which is present in the blood, is related to the apoptosis and necrosis of cancer cells; inflammation also influences the total plasma level of cfDNA. However, the total plasma cfDNA level has not been investigated in patients with hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) who experience cancer and HBV infection at the same time. The aim of the study was to investigate total plasma cfDNA in patients with HBV-related HCC.
Methods
HBV-related HCC patients were included from January 2018 to May 2019. All patients underwent hepatectomy and were diagnosed with HCC by histopathology. Peripheral blood samples were obtained preoperatively, and the levels of total plasma cfDNA were quantitated by a fluorometric double-stranded DNA (dsDNA) assay. We examined the correlation between cfDNA and clinical parameters, and recurrence-free survival was evaluated using Kaplan-Meier curves.
Results
Forty-eight HBV-related HCC patients were included. The average age in years was 50.90±13.15, and the mean albumin level was 41.63±5.38 g/L. HBV-DNA, Child-Turcotte-Pugh (CTP) class, TNM stage, tumor number and vascular invasion showed a relationship with total plasma cfDNA (P<0.05), and albumin, prothrombin time (PT) and tumor diameter had linear correlation with plasma cfDNA. Based on multivariate analysis, tumor diameter, vascular invasion, and CTP class (P<0.05) were independent risk factors of total plasma cfDNA. Median recurrence times for low-cfDNA and high-cfDNA groups were 14.729±0.712 and 9.264±1.22 months (P=0.026).
Conclusions
In addition to tumor diameter and vascular invasion, CTP class can influence total plasma cfDNA in HBV-related HCC patients, and the total plasma cfDNA level can be used as a biomarker to predict early recurrence in HBV-related HCC patients.
Keywords: Circulating cell-free DNA (circulating cfDNA), HBV-related hepatocellular carcinoma, hepatectomy
Introduction
Hepatocellular carcinoma (HCC), one of the most common digestive system tumors, is the 4th most common cancer worldwide; moreover, its incidence continues to rise, resulting in a significant morbidity and mortality burden in recent years (1-4). Hepatitis B virus (HBV) is a major etiological agent of chronic diseases and HCC, especially in Asia and Africa. HBV affects nearly 2 billion people worldwide, and approximately 350 million will become chronic carriers (5). Furthermore, HBV is one of the leading causes of chronic liver disease and increases the risk of developing fibrosis, cirrhosis and HCC (6). Hepatectomy, liver transplantation, and radiofrequency ablation (RFA) are the first-line treatments for early HCC (7,8). However, many patients diagnosed with advanced HCC or poor liver function reserves are not eligible for potentially curative therapies (9). In addition, a high recurrence rate after surgery affects the overall survival (OS) of patients with HBV-related HCC (10).
Tumor biopsy represents the standard diagnosis for HCC patients, providing both a diagnosis for HCC and tissue for molecular testing to guide the selection of precision therapies (11). Nonetheless, for patients with HBV-related HCC, especially those with poor liver function, the adverse consequences of tissue biopsy, such as bleeding, needle way transfer, increase mortality rates. Thus, it is important to identify a noninvasive diagnostic method for HBV-related HCC, and this is particularly important for patients with advanced disease or poor liver function. Liquid biopsies, especially those involving cell-free DNA (cfDNA) from plasma, are rapidly emerging as an important and minimally invasive adjunct to standard tumor biopsies.
Circulating cfDNA is short fragment of double-stranded DNA that can be detected in both healthy individuals and patients with disease (12). In the past decade, increasing research has focused on cell-free nucleic acid (cfNA), including DNA, mRNA and microRNA, which show high concentrations in the blood circulation of cancer patients (13). Accumulating evidence has confirmed that mircroRNAs are involved in HCC progression and can serve as biomarkers for diagnostics and monitoring therapies for HCCs (14), such as microRNA-622 (15) and microRNA-206 (16). cfDNA originates mainly from apoptotic or necrotic cells and fetal cells and can also be detected in patients with tumors (17) and diseases driven by inflammation. Furthermore, many studies demonstrate that cfDNA exacerbates autoimmune disease (18), trauma (19) and other diseases (20). In current cancer research, cfDNA is one of the most powerful tools for identifying tumor-specific mutations, providing new insight into tumor heterogeneity and the detection and monitoring of cancers (12).
Patients with HBV-related HCC present increased total plasma cfDNA, and chronic infection with HBV results in inflammation and oxidative stress, which can also increase the total plasma cfDNA level. However, the clinical value of total plasma cfDNA in HBV-related HCC patients is controversial. Early biomarkers for HBV-related HCC are mostly lacking, particularly when those common to both chronic HBV infection and HCC. In our study, we for the first time examined the level of total plasma cfDNA as a novel biomarker and evaluated the clinical value of cfDNA in patients with HBV-related HCC.
Methods
Patient section
The design of this study was approved by the XiangYa Hospital Central South University ethics committee (approval number 201703377). All clinical data involved in this study were obtained with patient consent. A total of 48 HCC patients were included from January 2018 to May 2019. All patients underwent hepatectomy and were diagnosed with HCC by postoperative pathology. Patients with infectious diseases, autoimmune diseases, pregnancy, metastatic liver cancer, and recurrent HCC, HCC patients who previously underwent transarterial chemoembolization (TACE), RFA or other treatments, and patients with incomplete data were excluded.
cfDNA assay
We collected blood samples into Streck tubes from HCC patients (10 mL) before surgery (Streck Inc., USA); the tubes contain stabilizing agents to prevent contamination of cfDNA with genomic DNA from white blood cells (18). Plasma was purified within 2 hours of blood collection by centrifugation at 1,600 ×g for 10 min at 4 °C. To generate a pure plasma fraction, the plasma was centrifuged at 16,000 ×g for 10 min, and DNA was extracted using QIAamp Circulating Nucleic Acid Kits (Qiagen, USA). The concentration of cfDNA was measured using a fluorometric Qubit dsDNA BR Assay Kit (Thermo Fischer Scientific, USA), and the amount is expressed as ng/mL of plasma.
Data collection
Patient clinical data, including age, sex, tumor diameter, TNM stage, Barcelona Clinic Liver Cancer (BCLC) stage, Child-Turcotte-Pugh (CTP) class, alpha-fetoprotein (AFP), albumin (ALB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and blood platelet (PLT), among others, were collected. All patients were followed up by telephone and out-patient interviews after the initial treatment. All cases were followed up until May 30, 2019. The prognostic endpoint of this study was recurrence-free survival (RFS). The endpoint of RFS was defined as recurrence or death from any cause.
Statistical methods
Pearson’s χ2 test or Fisher’s exact test was employed to compare categorical variables. A Wilcoxon rank sum test or Student’s t-test was used to evaluate continuous variables. Univariate and multivariate Cox proportional hazards regression analyses of cfDNA as an independent risk factor were also performed. The cutoff value of the cfDNA concentration was analyzed based on receiver operating characteristic (ROC) curves. RFS was estimated by the Kaplan-Meier method, and the results were compared with those of the log-rank test. Statistical analyses were performed using Prism software (GraphPad Prism Software, La Jolla, CA, USA) and SPSS 21.0 (SPSS Company, Chicago, IL, USA) for Windows. Statistical significance was set at 0.05.
Results
Clinicopathological characteristics of patients with HBV-related HCC
Forty-eight HBV-related HCC patients satisfied the study criteria and were included. The mean age of the HCC patients was 50.90±13.15 years, and the mean cfDNA concentration of these patients was 18.71±16.18 ng/mL. The mean prothrombin time (PT) was 13.71±1.05 s, and the mean tumor diameter was 6.08±4.10 cm. The majority of patients were male (91.67%, n=44). The cirrhosis prevalence was 71.08% (n=37), the CTP class A liver function percentage was 91.67% (n=44), the vascular invasion percentage was 16.67% (n=8), and the TNM stage I/II and III percentages were 77.08% (n=37) and 22.92% (n=11) (Table 1).
Table 1. The clinicopathological characteristics in patients with HCC (n=48).
| Variables | mean ± SD or N (n=48) |
|---|---|
| Age (years) | 50.90±13.15 |
| Albumin (g/L) | 41.63±5.38 |
| PLT (109/L) | 159.96±56.50 |
| cfDNA (ng/mL) | 18.71±16.18 |
| Total bilirubin (μmol/L) | 13.32±5.95 |
| Direct bilirubin (μmol/L) | 6.48±2.85 |
| ALT (U/L) | 40.76±37.73 |
| AST (U/L) | 43.62±32.22 |
| PT (s) | 13.71±1.05 |
| Tumor diameter (cm) | 6.08±4.10 |
| MELD score | 5.58±0.52 |
| Gender | |
| Male | 44 |
| Female | 4 |
| HBV-DNA | |
| <106 IU/mL | 44 |
| ≥106 IU/mL | 4 |
| Tumor capsular | |
| Absence | 39 |
| Presence | 9 |
| AFP | |
| ≤20 ng/mL | 17 |
| >20 ng/mL | 31 |
| Edmondson grade | |
| I–II | 35 |
| III–IV | 13 |
| Tumor number | |
| Single | 36 |
| Multiple | 12 |
| Satellite nodules | |
| Absence | 41 |
| Presence | 7 |
| Cirrhosis | |
| Absence | 11 |
| Presence | 37 |
| Vascular invasion | |
| Absence | 40 |
| Presence | 8 |
| CTP class | |
| A | 44 |
| B | 4 |
| BCLC stage | |
| 0/A | 34 |
| B | 6 |
| C | 8 |
| TNM stage | |
| I/II | 37 |
| III | 11 |
HCC, hepatocellular carcinoma; PLT, platelet; ALT, alanine transaminase; AST, glutamic oxaloacetic transaminase; PT, prothrombin time; MELD, model for end-stage liver disease; HBV, hepatitis B virus; AFP, alpha-fetoprotein; BCLC, Barcelona Clinic Liver Cancer score; CTP, Child-Turcotte-Pugh; TNM stage, Tumor, Lymph Node and Metastasis stage.
Relationship between plasma cfDNA and clinicopathological characteristics
Plasma cfDNA is released by both malignant and nonmalignant cells. Liver cells infected by HBV can release cfDNA into the bloodstream of HBV-related HCC patients, though the total plasma cfDNA in these patients is influenced by many factors. To clearly understand the relationship between total plasma cfDNA and clinicopathological characteristics, we analyzed the relationship between the cfDNA concentration and HBV-DNA, TNM stage, cirrhosis status and other clinical parameters.
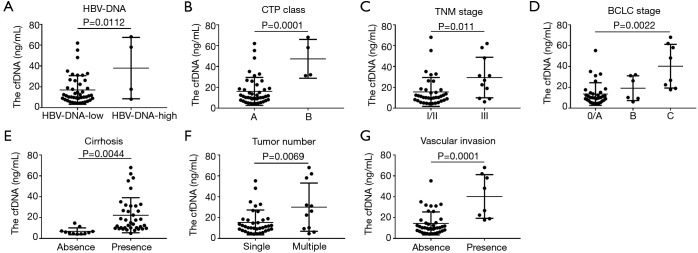
We divided the HBV-related HCC patients into two groups on the basis of the level of HBV-DNA: low-HBV-DNA group (HBV-DNA <106 IU/mL, n=44) and high-HBV-DNA group (HBV-DNA ≥106 IU/mL, n=4). The concentrations of total plasma cfDNA in the low-HBV-DNA and high-HBV-DNA groups were 16.96±2.07 ng/mL and 37.99±14.74 ng/mL, respectively (P=0.0112) (Figure 1A). Thus, HBV replication affects the total plasma cfDNA level, which may correlate with the level of HBV-DNA replication.
Figure 1.
The relationship between total plasma cfDNA and clinicopathological characteristics in HBV-related HCC patients. (A) The relationship between HBV-DNA and cfDNA; total plasma cfDNA in the low-HBV-DNA and high-HBV-DNA groups was 16.96±2.07 and 37.99±14.74 ng/mL (P=0.0112), respectively; (B) the relationship between CTP class and cfDNA; total plasma cfDNA in patients with CTP class A and class B was 16.11±2.02 and 47.33±9.30 ng/mL (P=0.0001), respectively; (C) the relationship between TNM stage and cfDNA; total plasma cfDNA in the patients with TNM stages I/II and III were 15.54±2.28 and 29.39±5.85 ng/mL (P=0.011); (D) the relationship between BCLC stage and cfDNA; total plasma cfDNA in the patients with BCLC 0/A stage, B stage and C stage were 13.56±1.87, 19.19±4.84 and 40.27±7.42 ng/mL (P=0.0022), respectively; (E) the relationship between liver cirrhosis and cfDNA; cfDNA concentrations in the absence and presence of liver cirrhosis were 6.84±0.99 and 22.24±2.77 ng/mL (P=0.0044), respectively; (F) the relationship between tumor number and cfDNA; the total plasma cfDNA concentrations in patients with single and multiple tumors were 15.35±1.96 and 30.03±6.98 ng/mL (P=0.0069), respectively; (G) the relationship between vascular invasion and cfDNA; total plasma cfDNA in HCC patients with and without vascular invasion was 14.40±1.75 and 40.27±7.42 ng/mL (P=0.0001), respectively. HBV, hepatitis B virus; CTP, Child-Turcotte-Pugh; BCLC, Barcelona Clinic Liver Cancer score; cfDNA, circulating cell-free DNA; HCC, hepatocellular carcinoma.
Total plasma cfDNA in CTP class A and class B patients was 16.11±2.02 ng/mL and 47.33±9.30 ng/mL, respectively (P=0.0001) (Figure 1B). Moreover, HBV-related HCC patients who had poor liver function had increased levels of total plasma cfDNA. CTP class can affect total plasma cfDNA, whereby an advanced CTP class is associated with increases. In clinical practice, CTP class can indicate the reserve function of the liver. We found that the CTP class to be strongly associated with the plasma cfDNA concentration, possibly because an advanced CTP class indicates poor liver reserve function and serious liver damage, promoting the release of cfDNA.
Regarding the correlation between tumor stage and total plasma cfDNA in HBV-related HCC patients, we found that total plasma cfDNA levels in TNM stages I/II and III were 15.54±2.28 and 29.39±5.85 ng/mL (P=0.011), respectively (Figure 1C). Additionally, the total plasma cfDNA levels in patients with BCLC 0/A stage, B stage and C stage were 13.56±1.87, 19.19±4.84 and 40.27±7.42 ng/mL, respectively, and differences between the three groups were statistically significant by the ANOVA test (P=0.0022) (Figure 1D). Furthermore, tumor stage, including the TNM classification and BCLC stage, in HBV-related HCC patients with an advanced tumor stage was associated with increased plasma cfDNA concentrations, indicating that tumor stage can influence total plasma cfDNA.
The cfDNA concentrations in the absence and presence of liver cirrhosis were 6.84±0.99 and 22.24±2.77 ng/mL, respectively. HBV-related HCC patients with liver cirrhosis had higher levels of total plasma cfDNA than did those without cirrhosis, and the difference between the two groups was statistically significant (P=0.0044) (Figure 1E).
Liver cirrhosis is known to be an independent risk factor for the occurrence of HCC. The high level of total plasma cfDNA in HBV-related HCC patients with cirrhosis may be caused by the following reasons: (I) liver cirrhosis enhances the degree of liver cell damage, resulting in cell death or apoptosis, which releases cfDNA into the blood; (II) cirrhotic nodules have significant cellular atypia and can be considered a precancerous form of HCC, and atypical cells can release increased amounts of cfDNA into the blood; (III) HCC patients often present portal hypertension, which is a typical hemodynamic change, and hemodynamic change may lead to damage of liver, endothelial and other cells, ultimately promoting cfDNA release.
To determine whether the tumor number can increase total plasma cfDNA, we divided the HCC patients into single and multiple tumor groups. The total plasma cfDNA concentration in the single and multiple tumor groups was 15.35±1.96 and 30.03±6.98 ng/mL (P=0.0069), respectively (Figure 1F), and we found a higher total plasma cfDNA concentration with a higher tumor number. Therefore, increased tumor number may enhance the release of cfDNA.
The total plasma cfDNA concentration in HCC patients with and without vascular invasion was 14.40±1.75 and 40.27±7.42 ng/mL (P=0.0001), respectively (Figure 1G). Accordingly, in HBV-related HCC, patients with vascular invasion have a high level of total plasma cfDNA.
Linear correlations between cfDNA and clinical parameters
We next analyzed linear correlations between cfDNA and other clinical parameters and found linear correlations with serum ALB, tumor diameter and PT (P<0.05) but none with PLT, total bilirubin, AST or other parameters (Table 2).
Table 2. The linear correlations of cfDNA and clinical parameters.
| Variables | cfDNA (μg/mL) | |
|---|---|---|
| Pearson correlation coefficient | P value | |
| PLT (109/L) | −0.18 | 0.227 |
| Albumin (g/L) | −0.41 | 0.004 |
| Total bilirubin (μmol/L) | 0.07 | 0.629 |
| Direct bilirubin (μmol/L) | 0.11 | 0.447 |
| ALT (U/L) | 0.13 | 0.382 |
| AST (U/L) | 0.23 | 0.121 |
| PT (s) | 0.35 | 0.015 |
| Tumor diameter (cm) | 0.51 | 0.0002 |
| MELD score | 0.18 | 0.220 |
cfDNA, circulating cell-free DNA; PLT, platelet; ALT, Alanine transaminase; AST, glutamic oxaloacetic transaminase; PT, prothrombin time; MELD, Model for end-stage liver disease.
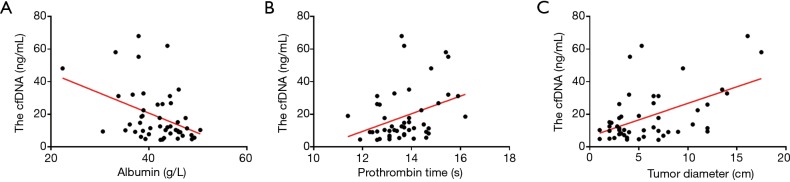
In addition, we detected a negative linear correlation between ALB and total plasma cfDNA (P=0.0042, R=−0.41); the equation was Y =−1.221*X +69.56 (Figure 2A), indicating that the higher was the serum ALB concentration, the lower was the total plasma cfDNA. Thus, an increased ALB level can indicate increased liver function in HBV-related HCC patients.
Figure 2.
Linear correlations between total plasma cfDNA and clinical parameters. (A) The linear correlation between ALB and total plasma cfDNA (P=0.0042, R=−0.41); (B) the correlation between prothrombin time and total plasma cfDNA (P=0.015, R=0.35); (C) the linear correlation between tumor diameter and total plasma cfDNA (P=0.0002, R=0.51). cfDNA, circulating cell-free DNA; ALB, albumin.
However, there was a positive linear correlation between total plasma cfDNA and tumor diameter and PT. For PT and total plasma cfDNA (P=0.015, R=0.35), the equation was Y=5.423*X −55.62 (Figure 2B). For the correlation between tumor diameter and total plasma cfDNA (P=0.0002, R=0.51), the equation was Y =2.032*X +6.35 (Figure 2C).
Independent risk factors associated with total plasma cfDNA in HBV-related HCC patients
Univariate and multivariate analyses were performed using age, serum ALB, ALT, AST, tumor diameter and other clinicopathologic variables to determine their associations with total plasma cfDNA in HBV-related HCC patients with or without recurrence. Serum ALB, tumor diameter, tumor number, vascular invasion, CTP class, and TNM stage were significant according to univariate analysis (P<0.05) (Table 3). These variables were then evaluated by multivariate analysis, which revealed tumor diameter [hazard ratio (HR), 1.48; 95% CI, 1.06–2.09; P=0.023], vascular invasion (HR, 0.04; 95% CI, 0.002–0.82; P=0.036), and CTP class (HR, 0.03; 95% CI, 0.001–0.75; P=0.033) to be independent factors for elevated total plasma cfDNA in HBV-related HCC patients (Table 3).
Table 3. Univariate and multivariate analyses of independent risk factors of the total plasm cfDNA in HBV-related HCC patients (n=48).
| Variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (years) | 1.02 (0.97–1.07) | 0.536 | – | – | |
| Albumin (g/L) | 0.84 (0.73–0.97) | 0.020 | 1.03 (0.84–1.27) | 0.780 | |
| PLT (109/L) | 0.99 (0.98–1.004) | 0.204 | – | – | |
| Total bilirubin (μmol/L) | 1.02 (0.92–1.13) | 0.690 | – | – | |
| Direct bilirubin (μmol/L) | 1.05 (0.85–1.31) | 0.630 | – | – | |
| ALT (U/L) | 1.004 (0.99–1.02) | 0.608 | – | – | |
| AST (U/L) | 1.02 (0.99–1.04) | 0.148 | – | – | |
| PT (s) | 1.77 (0.93–3.36) | 0.081 | – | – | |
| Tumor diameter (cm) | 1.38 (1.13–1.68) | 0.002 | 1.48 (1.06–2.09) | 0.023 | |
| MELD score | 1.93 (0.44–8.43) | 0.381 | – | – | |
| HBV-DNA (≥106 vs. <106 IU/mL) | 2.68 (0.34–21.12) | 0.353 | – | – | |
| Tumor capsular (absence vs. presence) | 4.0 (0.45–35.49) | 0.213 | – | – | |
| AFP (>20 vs. ≤20 ng/mL) | 2.57 (0.60–10.92) | 0.202 | – | – | |
| Edmondson grade (III–IV vs. I–II) | 2.89 (0.75–11.11) | 0.122 | – | – | |
| Satellite nodules (presence vs. absence) | 1.03 (0.18–6.09) | 0.970 | – | – | |
| Cirrhosis (presence vs. absence) | 1.11 (0.28–4.45) | 0.882 | – | – | |
| Tumor number (single vs. multiple) | 0.17 (0.04–0.71) | 0.015 | 0.35 (0.04–3.57) | 0.377 | |
| Vascular invasion (absence vs. presence) | 0.03 (0.003–0.29) | 0.002 | 0.04 (0.002–0.82) | 0.036 | |
| CTP class (A vs. B) | 0.08 (0.008–0.76) | 0.028 | 0.03 (0.001–0.75) | 0.033 | |
| TNM stage (I/II vs. III) | 0.07 (0.015–0.36) | 0.001 | 0.31 (0.03–3.28) | 0.329 | |
PLT, platelet; ALT, Alanine transaminase; AST, glutamic oxaloacetic transaminase; PT, prothrombin time; MELD, Model for end-stage liver disease; AFP, alpha-fetoprotein; CTP, Child-Turcotte-Pugh; TNM stage, Tumor, Lymph Node and Metastasis stage.
Total plasma cfDNA predicted HBV-related HCC patient prognosis
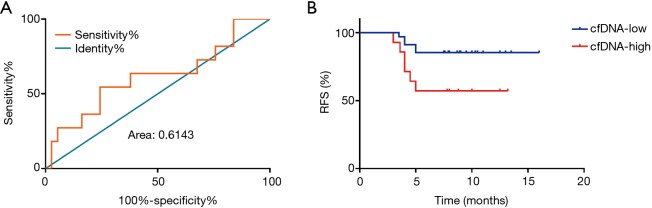
To determine whether the level of total plasma cfDNA can predict HBV-related HCC patient prognosis, patients who underwent liver resection were divided into recurrence and no recurrence groups on the basis of early recurrence. We then established the cutoff values of total plasma cfDNA in HBV-related HCC patients as 18.71 ng/mL by ROC curve analysis. The ROC area under the curve was 0.6143, and the 95% CI ranged 0.41 to 0.82 (Figure 3A).
Figure 3.
The ROC curve and RFS rate according to total plasma cfDNA in HBV-related HCC patients. (A) The ROC curve of total plasma cfDNA in HBV-related HCC patients. The ROC area under the curve was 0.6143, and the 95% CI was 0.41 to 0.82; (B) RFS in HCC patients according to the Kaplan-Meier method. The median recurrence times in the low-cfDNA group and the high-cfDNA group were 14.729±0.712 and 9.264±1.22 months (P=0.026), respectively. RFS, recurrence-free survival; cfDNA, cfDNA, circulating cell-free DNA; ROC, receiver operating characteristic; HBV, hepatitis B virus; HCC, hepatocellular carcinoma.
HBV-related HCC patients were divided into two groups based on the cutoff value of total plasma cfDNA: low-cfDNA (cfDNA ≤18.71 ng/mL) and the high-cfDNA (cfDNA >18.71 ng/mL) groups. Next, we analyzed RFS in HCC patients using the Kaplan-Meier method. The median recurrence times of the low-cfDNA group and the high-cfDNA group were 14.729±0.712 and 9.264±1.22 months (P=0.026), respectively (Figure 3B). Therefore, preoperative total plasma cfDNA can predict early recurrence in patients with HBV-related HCC.
Discussion
There is extensive research on plasma cfDNA, and many studies have verified associations between cancer, infectious diseases, inflammatory, normal cells and total plasma cfDNA. Indeed, cfDNA can be regarded as a diagnostic and prognostic biomarker for breast cancer (21) and for the detection of minor injuries in the field of forensic biomechanics (19). cfDNA can also assist in personalized therapy for lung cancer (22) and bladder cancer (23) patients. Overall, plasma cfDNA has a significant role in the early diagnosis and prognosis prediction of a variety of tumors (21,24,25).
Currently, there are no effective biomarkers for the early diagnosis of HCC, especially within the context of HBV infection. Although α-fetoprotein (AFP) clearly plays a role in the diagnosis of HCC and in surveillance after surgery, there are serious disadvantages to this marker, such as low sensitivity and false positive results. Nonetheless, AFP can be employed in HBV infection, cirrhosis, pregnancy, embryonic gonad adenocarcinoma and other cancers. Many genes and microRNAs have important roles in HCC diagnosis, yet genes and microRNAs cannot meet the requirements of clinical scale in terms of specificity and sensitivity. cfDNA can serve as a noninvasive approach for early diagnosis, which can improve the clinical outcomes of HCC patients (26). Regardless, the total plasma concentration of cfDNA in HBV-related HCC patients has not previously been assessed.
In this study, we mainly explored the clinical significance of the total plasma cfDNA in patients with HBV-related HCC and found that the concentration in patients with HBV-related HCC correlated with HBV infection, liver function, and tumor factors (e.g., tumor size, tumor stage, tumor number). However, there has been no research to date on liver function and total plasma cfDNA. In our study, we found that serum ALB and cfDNA had negative correlations, but PT and tumor diameter had a positive correlation. Furthermore, HCC patients with cirrhosis had higher total plasma cfDNA concentrations than did patients without cirrhosis. Multivariate analysis also showed that the CTP class was an independent factor for elevated total plasma cfDNA. Therefore, HBV infection and liver function status should be considered when using total plasma cfDNA to diagnose and monitor HBV-related HCC patients.
CTP class can accurately and sensitively reflect liver function in HCC patients, and serum ALB and PT are the main indices of liver function. The possible reasons why liver function affects total plasma cfDNA in HBV-related HCC patients are as follows: (I) HBV replication causes damage to liver cells and alters inflammation in the body. (II) HBV infection leads to liver cirrhosis, and liver cirrhosis is an independent risk factor for the development of HCC. Liver cirrhosis nodules exhibit cellular dysplasia, and liver cellular dysplasia may affect total plasma cfDNA. (III) HBV-related HCC patients usually present portal hypertension, which can alter hemodynamics and lead to death in liver cells, vascular endothelial cells and other cells, affecting the total plasma cfDNA level. Therefore, when analyzing the total plasma cfDNA of patients with HBV-related HCC, it is not comprehensive to consider only the effect of tumors on cfDNA concentrations; HBV and liver function should also be considered.
In addition, we found that the HCC patients who had increased preoperative total plasma cfDNA had poor prognoses and were prone to early recurrence. Accordingly, total plasma cfDNA can be used as a predictor for early recurrence in HBV-related HCC patients. Therefore, we speculate that hepatic artery perfusion chemotherapy with a targeted drug may be needed for HBV-related HCC patients with high preoperative cfDNA concentrations to achieve improved clinical survival, but further research and analyses are needed.
Conclusions
In summary, the level of total plasma cfDNA in HBV-related HCC patients can be influenced not only by tumor factors but also by HBV infection and liver function. Early recurrence in HBV-related HCC patients can be predicted by cfDNA.
Acknowledgments
Funding: This study was supported by grants from the National Nature Science Foundation of China (No. 81771932) and the Geneplus-Beijing Institute.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The design of this study was approved by the Xiangya Hospital Central South University ethics committee (approval number 201703377). All clinical data involved in this study were obtained with patient consent.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155-61. 10.1038/nmat4997 [DOI] [PubMed] [Google Scholar]
- 2.Kanwal F, Singal AG. Surveillance for Hepatocellular Carcinoma: Current Best Practice and Future Direction. Gastroenterology 2019;157:54-64. 10.1053/j.gastro.2019.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu K, Wang ZM, Li JN, et al. CLEC1B Expression and PD-L1 Expression Predict Clinical Outcome in Hepatocellular Carcinoma with Tumor Hemorrhage. Transl Oncol 2018;11:552-8. 10.1016/j.tranon.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai N, Peng E, Qiu X, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res 2018;37:172. 10.1186/s13046-018-0838-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007;132:2557-76. 10.1053/j.gastro.2007.04.061 [DOI] [PubMed] [Google Scholar]
- 6.Roberts LR, Gores GJ. Hepatocellular carcinoma: molecular pathways and new therapeutic targets. Semin Liver Dis 2005;25:212-25. 10.1055/s-2005-871200 [DOI] [PubMed] [Google Scholar]
- 7.Marasco G, Colecchia A, Colli A, et al. Role of liver and spleen stiffness in predicting the recurrence of hepatocellular carcinoma after resection. J Hepatol 2019;70:440-8. 10.1016/j.jhep.2018.10.022 [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Kang TW, Cha DI, et al. Radiofrequency ablation vs. surgery for perivascular hepatocellular carcinoma: Propensity score analyses of long-term outcomes. J Hepatol 2018;69:70-8. 10.1016/j.jhep.2018.02.026 [DOI] [PubMed] [Google Scholar]
- 9.Hyun MH, Lee YS, Kim JH, et al. Hepatic resection compared to chemoembolization in intermediate- to advanced-stage hepatocellular carcinoma: A meta-analysis of high-quality studies. Hepatology 2018;68:977-93. 10.1002/hep.29883 [DOI] [PubMed] [Google Scholar]
- 10.Tao YM, Huang JL, Zeng S, et al. BTB/POZ domain-containing protein 7: epithelial-mesenchymal transition promoter and prognostic biomarker of hepatocellular carcinoma. Hepatology 2013;57:2326-37. 10.1002/hep.26268 [DOI] [PubMed] [Google Scholar]
- 11.Harding JJ, Nandakumar S, Armenia J, et al. Prospective Genotyping of Hepatocellular Carcinoma: Clinical Implications of Next-Generation Sequencing for Matching Patients to Targeted and Immune Therapies. Clin Cancer Res 2019;25:2116-26. 10.1158/1078-0432.CCR-18-2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med 2018;379:1754-65. 10.1056/NEJMra1706174 [DOI] [PubMed] [Google Scholar]
- 13.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37. 10.1038/nrc3066 [DOI] [PubMed] [Google Scholar]
- 14.Yang N, Ekanem NR, Sakyi CA, et al. Hepatocellular carcinoma and microRNA: new perspectives on therapeutics and diagnostics. Adv Drug Deliv Rev 2015;81:62-74. 10.1016/j.addr.2014.10.029 [DOI] [PubMed] [Google Scholar]
- 15.Dietrich P, Koch A, Fritz V, et al. Wild type Kirsten rat sarcoma is a novel microRNA-622-regulated therapeutic target for hepatocellular carcinoma and contributes to sorafenib resistance. Gut 2018;67:1328-41. 10.1136/gutjnl-2017-315402 [DOI] [PubMed] [Google Scholar]
- 16.Wu H, Tao J, Li X, et al. MicroRNA-206 prevents the pathogenesis of hepatocellular carcinoma by modulating expression of met proto-oncogene and cyclin-dependent kinase 6 in mice. Hepatology 2017;66:1952-67. 10.1002/hep.29374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laufer-Geva S, Rozenblum AB, Twito T, et al. The Clinical Impact of Comprehensive Genomic Testing of Circulating Cell-Free DNA in Advanced Lung Cancer. J Thorac Oncol 2018;13:1705-16. 10.1016/j.jtho.2018.07.101 [DOI] [PubMed] [Google Scholar]
- 18.Xu Y, Song Y, Chang J, et al. High levels of circulating cell-free DNA are a biomarker of active SLE. Eur J Clin Invest 2018;48:e13015. 10.1111/eci.13015 [DOI] [PubMed] [Google Scholar]
- 19.Brodbeck K, Kern S, Schick S, et al. Quantitative analysis of individual cell-free DNA concentration before and after penetrating trauma. Int J Legal Med 2019;133:385-93. 10.1007/s00414-018-1945-y [DOI] [PubMed] [Google Scholar]
- 20.Ranucci R. Cell-Free DNA: Applications in Different Diseases. Methods Mol Biol 2019;1909:3-12. 10.1007/978-1-4939-8973-7_1 [DOI] [PubMed] [Google Scholar]
- 21.Hussein NA, Mohamed SN, Ahmed MA. Plasma ALU-247, ALU-115, and cfDNA Integrity as Diagnostic and Prognostic Biomarkers for Breast Cancer. Appl Biochem Biotechnol 2019;187:1028-45. 10.1007/s12010-018-2858-4 [DOI] [PubMed] [Google Scholar]
- 22.Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017;107:100-7. 10.1016/j.lungcan.2016.04.026 [DOI] [PubMed] [Google Scholar]
- 23.Riethdorf S, Soave A, Rink M. The current status and clinical value of circulating tumor cells and circulating cell-free tumor DNA in bladder cancer. Transl Androl Urol 2017;6:1090-110. 10.21037/tau.2017.09.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao W, Mao Y, Ge P, et al. Value of quantitative and qualitative analyses of circulating cell-free DNA as diagnostic tools for hepatocellular carcinoma: a meta-analysis. Medicine (Baltimore) 2015;94:e722. 10.1097/MD.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan L, Chen Y, Zhou J, et al. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis 2018;67:92-7. 10.1016/j.ijid.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 26.Cai J, Chen L, Zhang Z, et al. Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Gut 2019;68:2195-205. 10.1136/gutjnl-2019-318882 [DOI] [PMC free article] [PubMed] [Google Scholar]