Abstract
Adenosine deaminases acting on RNA (ADARs) catalyze the conversion of adenosine (A) to inosine (I) in double-stranded RNA, which can change the codons after transcription. Abnormal ADAR editing is present in a variety of cancers. However, the study of the biological effects of ADARs in cancer is not very deep. Here, we review current important ADAR-mediated editing events, related carcinogenic mechanisms and applications in clinical medicine. Further exploration in ADARs can provide a new direction for cancer treatment.
Keywords: Adenosine deaminases acting on RNA (ADARs), A to I RNA editing, carcinogenic mechanism, clinical utility
Introduction
Gene mutations have long been the focus of cancer occurrence and development. In recent years, epigenetic mechanisms and their abnormal function are also important in the development of neoplasia (1). RNA editing has been identified for the involvement in cancer (2), the most common type of which is ADAR-mediated RNA editing in higher eukaryotes, which may result in codon changes, cellular development defects and potential carcinogenic susceptibility. Thus, the imbalance in ADAR expression or activity may be a causative factor in a variety of diseases including cancer (3). This article reviews the structure and function of ADAR, ADAR-mediated RNA editing, the role of ADAR in cancer and related clinical application.
The ADAR family
ADAR proteins were first found in Xenopus embryos (4). ADAR genes are discovered in multicellular animals such as Drosophila melanogaster, Caenorhabditis elegans, birds, cuttlefish, and mammals including humans (5). Mammalian cells express 3 kinds of ADARs: ADAR1, ADAR2 and ADAR3. ADAR1 and ADAR2 form homodimers, whereas ADAR3 does not form dimers. ADAR3 can bind to ADAR1 and ADAR2 substrates, thereby inhibiting their activity (3). The ADAR proteins consist of two to three N-terminal dsRBDs and a conserved C-terminal catalytic deaminase domain (ADARcd) (6). The dsRBDs select and bind targets, and ADARcd catalyzes deamination (7). The structure of inosines is similar to that of guanosine (G), so cellular enzymes can recognize inosine as guanosine (7). In the N-terminal region, ADAR1 contains the Z-DNA binding domains Za and Zb, whose activity can influence neogenetic RNA, resulting in pre-splicing editing, which is absent in ADAR2 and ADAR3 (3). The ADAR1 protein has two isoforms, the p110 and p150 isoforms. P110-mediated editing is in the nucleus, while p150-mediated editing is in the nucleus and cytoplasm (8). ADAR2 is mainly in the nucleus, especially the nucleolus (9).
ADAR editing sites are currently estimated at over 100 million in the human genome (10), most of which occur in the short interspersed elements (SINEs) of introns and 3′ untranslated regions (3’-UTRs) in mature mRNAs and pre-mRNAs (11). Merely 0.4% of the editing takes place in the code sequences (12). ADAR editing occurs mainly in the Alu elements, which are about 300 bp long in mRNA. The Alus element is abundant in adenosine (11). ADAR can act on mRNA, miRNA, lncRNA and siRNA (13). ADAR editing can affect many biological processes, including increasing transcriptomic diversity and protecting cellular dsRNA from silencing and improper immune activation (14).
ADARs are critical for growth and development in animal models (6). ADAR1 is expressed at the highest level among the vertebrate enzymes (11). ADAR2 is expressed in almost all body parts and most abundantly in brain tissue (3). The ADAR2 transcript peaks in the CNS, particularly in the hippocampus, thalamus, subthalamic nuclei, globus pallidus, and cerebellar peduncles (11). Mice lacking ADAR1 die during embryonic stages with hemopoietic defects, widespread apoptosis, liver failure and IFN overexpression (15-18). ADAR2 knockout phenotypes are less severe than those of ADAR1 (19). ADAR2 edits the critical Q/R site in Gria2 transcript. ADAR2 knockout mice die of seizures caused by Gria2 under-editing. A knock-in editing-equivalent A-to-G mutation in the AMPA receptor subunit gene encoded by Gria2 transcript prevents seizures and death. The editing activity of ADAR2 is essential for normal development (20). ADAR expression imbalance results in many diseases including cancer, amyotrophic lateral sclerosis, dyschromatosis symmetrica hereditaria and Aicardi-Goutières syndrome (3,21).
ADAR3 is resembling to ADAR1 and ADAR2 in sequence and structure. However, ADAR3 does not display deaminase activity affecting physiological functions (22). The express of ADAR3 is only in the brain (23) and has negative correlation with the editing level of the brain, suggesting that ADAR3 is a negative regulator of ADAR2 editing (11). In glioblastoma multiforme (GBM), ADAR3 competes to Gria2 binding and editing with ADAR2 at the RNA-binding domain. ADAR3 might regulate tumorigenesis in cancer of the brain through regulating ADAR2/ADAR1-mediated editing (22).
ADARs have editing-independent effects on gene expression (24,25). For example, no significant correlation (r<0.05) between changes in gene expression and editing levels is observed after ADAR1 knockdown. The location and number of editing sites for each transcript are independent of changes in expression after ADAR1 knockout. These evidences suggest that ADAR1 can regulate gene expression independent of deamination activity (26). In-depth exploring functions of ADARs would create new ideas that may lead to the development of cancer treatments.
A-to-I RNA editing
RNA editing is a post-transcriptional mechanism that modifies RNA nucleotides without changing the template genomic DNA (27). Dysregulation of the editing process has serious consequences, and abnormal RNA editing within the transcriptome range detected in many kinds of tumors (21). A-to-I editing is the most common form in RNA base modification (10). The functional impact of A-to-I editing includes alteration of protein amino acid sequence, alterations in pre-mRNA splicing modes, alterations in miRNA seed sequences, alterations in gene regulation, alterations in target RNA stability, effects on lncRNA function, RNA interference and the formation of ribonucleoprotein (RNP) complexes, and transcript stability and subcellular localization (22,27,28). Furthermore, A-to-I editing in long double-stranded regions may prevent silencing of host RNAs and self-RNAs inappropriate activation of immune response (14).
Many A-to-I RNA editing events occur in cancers and are related to clinic (13,29,30). A-to-I editing can derive neoantigens (21) which immune system can recognize. Furthermore, RNA editing has temporal diversity, so there is no accumulation of mutations in the genome of the progeny cells in the tumor. The property helps promote the generation of chemotherapy-resistant cells (26). Identifying driver RNA editing sites could provide new therapeutic and diagnostic approaches.
ADAR in cancer
Growing evidence shows that ADAR-mediate RNA editing is the important contributor to pathogenesis. Bioinformatics analysis and experimental studies have shown that ADAR-mediated editing patterns and activities in tumors are different from normal tissues (31). The following summarizes the important targets and mechanisms of ADAR in cancer (Table 1).
Table 1. Known roles of ADAR in cancer.
| Targets | ADAR | Cancer types | Finding | References |
|---|---|---|---|---|
| AZINI | ADAR1 | HCC, ESCC, CRC | AZIN1 acts as an oncogene by inhibiting the tumor suppressor activities of antizyme | (13,32,33) |
| NSCLC, BC, PRAD | (34-36) | |||
| GLI1 | ADAR1 | MM | Edited GLI1 activates the Hedgehog signaling pathway and promotes malignant regeneration of MM and lenalidomide resistance in vitro | (37,38) |
| MB, BCC | Anti-tumorigenic effect | (37) | ||
| NEIL | ADAR1 | MM | Edited NEIL1 has reduced oxidative damage repair ability | (39) |
| BLCAP | ADAR1 | CC, HCC | Edited BLCAP increases cell proliferation by activating the Akt/mTOR signaling pathway or the pro-survival protein STAT3 | (40,41) |
| ADAR2 | Astrocytoma, CRC, bladder cancer | Decreased editing levels of BLCAP correlates with increased histological malignancy | (42) | |
| DHFR | ADAR1 | BC | Edited DHFR enhances cell proliferation and resistance to methotrexate | (43) |
| FAK | ADAR1 | LUAD | Edited FAK contributes to cell invasiveness and tumor recurrence in LUAD patients | (44) |
| Pri-let-7 | ADAR1 | CML | Reduction of let-7 levels enhances the self-renewal ability of leukemic stem cells | (45) |
| PU.1 | ADAR1 | CML | Increased hematopoietic differentiation toward the myeloid lineage | (46) |
| miR-200b | ADAR1 | HNSC, KIRP, THCA, UCEC | Edited miR-200b promotes cell invasion and migration | (47) |
| PCA3 | ADAR1 | Prostate cancer | Cancer cell proliferation, adhesion and migration | (48) |
| miR-455-5p | ADAR1 | Melanoma | Edited miR-455-5p reduces tumor growth and metastasis | (49) |
| miR-378a-3p | ADAR1 | Melanoma | Edited miR-378a-3p suppresses the malignant phenotype and melanoma metastasis | (50) |
| Gabra3 | ADAR1 | BC | Edited GABRA3 inhibits AKT activation and metastatic potential of cancer cells | (51) |
| CCNI | ADAR1 | Melanoma, OC, BC | Edited CCNI induces antigen-specific killing of tumor cells by CD8+ T cells | (52) |
| CDC14B | ADAR2 | GBM | Decreased CDC14B expression increases astrocytoma invasiveness and poor prognosis | (53,54) |
| GluA2 | ADAR2 | GBM | Edited GluA2 produces Ca2+ impermeable AMPA channels that inhibit cell migration and induces apoptosis | (55-58) |
| ADAR3 | astrocytoma | ADAR3 negatively regulates the editing of GluA2 at the Q/R site in astrocytoma | (59) | |
| miR221 | ADAR2 | GBM | MiR-221, miR-222, and miR-21 have oncogenic | (60,61) |
| miR222 | ADAR2 | GBM | Function by inhibiting tumor suppressor protein | |
| miR-21 | ADAR2 | GBM | p27kip1 (mir-221 and 222) and PDCD4 (miR-21) | |
| miR-376* | ADAR2 | GBM | Unedited miR-376* enhances invasion and migration whereas edited miR-376a* inhibits GBM progression | (62) |
| miR-589-3p | ADAR2 | GBM | Unedited miR589-3p inhibits the tumor suppressor protein PCDH9 | (63) |
| COPA | ADAR2 | HCC | The editing level of COPA is inversely correlated with the pathogenesis of HCC | (64) |
| CRC | Edited COPA is absent in invasive CRC with EMT phenotype, resulting in liver metastasis | (65) | ||
| miR-214 | ADAR2 | HCC | MiR-214 regulates proliferation, invasiveness and angiogenesis in HCC | (66-68) |
| miR-122 | ADAR2 | HCC | MiR-122 regulates the host gene and viral life cycle of HBV and HCV | (66,69) |
| IGFBP7 | ADAR2 | ESCC | Reduced editing and expression of IGFBP7 activates the Akt pathway, thereby inhibiting programmed cell death and promoting tumor growth | (70) |
| SLC22A3 | ADAR2 | Esophageal cancer | Edited SLC22A3 is increased in familial esophageal cancer and is associated with lymph node metastasis | (71) |
| miR-142 | ADAR1 and ADAR2 | AML | The miR-142s gene is located at a translocation site associated with aggressive B cell leukemia | (72) |
| HNRPLL | ADAR1 and ADAR2 | Kidney cancer, bladder cancer | Edited HNRPLL regulates the expression of cyclin D1 and TGFBR1 to promote cell proliferation | (73) |
| PODXL | ADAR1 and ADAR2 | GC | Edited PODXL provides a loss-of-function phenotype with reduced growth and invasiveness | (74,75) |
| miR-222 | ADAR1 | Melanoma | ADAR1 levels are decreased in melanoma. ADAR1 knockdown enhances miR-222 biogenesis and inhibits the expression of ICAM1 | (76) |
| COG3 I/V | ADAR3 | GBM | Excessive editing at the COG3 I/V site plays a pro-tumoral role in GBM and is associated with poor prognosis | (77) |
| PTPN6 | ADAR1 or ADAR2 | AML | PTPN6 plays an anti-cancer role by downregulating the broad spectrum growth-promoting receptors and cytokine receptors | (78) |
| RHOQ | ADAR1 or ADAR2 | CRC, GC, HCC, lung cancer | Edited RHOQ mediates dynamic actin cytoskeletal recombination, thus increasing the invasive potential | (29) |
HCC, hepatocellular carcinoma; ESCC, esophageal squamous cell carcinoma; NSCLC, non-small-cell lung cancer; CRC, colorectal cancer; BC, breast cancer; PRAD, prostate adenocarcinoma; MM, multiple myeloma; MB, medulloblastoma; BCC, basal cell carcinoma; CC, cervical cancer; LUAD, lung adenocarcinoma; CML, chronic myeloid leukemia; HNSC, head and neck squamous cell carcinoma; KIRP, kidney renal papillary cell carcinoma; THCA, thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; OC, ovarian cancer; GBM, glioblastoma multiforme; AML, acute myeloid leukemia; GC, gastric cancer.
RNA editing-dependent roles of ADAR1 in cancer
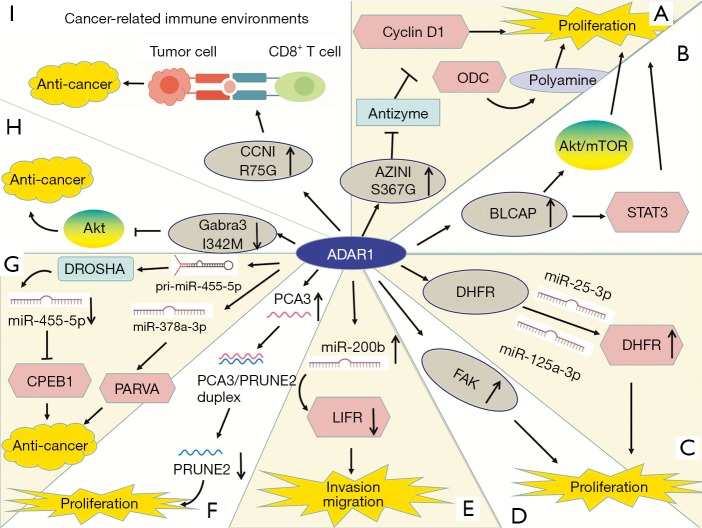
The mechanisms of ADAR1 in tumors are complex and it can promote or suppress cancer in different tumors (Figure 1). ADAR1-mediated editing of AZIN1 increases and promotes carcinogenesis in hepatocellular carcinoma (HCC). RNA-Seq shows that two ADAR1 transcript variants encoding the p110 and p150 isoforms are relatively abundant in liver tissue, among which p110 is the predominant form (32). Transcriptome sequencing of tumor tissue shows that ADAR1-mediated editing permutes serine to glycine at AZIN1 residue 367, inducing AZIN1 to transfer from the cytoplasm to the nucleus and tumorigenic phenotype, thereby enhancing the invasive capability (32,79). AZIN1 is homologous to ornithine decarboxylase (ODC), and its affinity for binding to enzymes is higher than that of ODC. Antizyme is a synthetic mammalian protein with ribosomal frameshift function. Antizyme binds to and promotes the degradation of ODC and cyclin D1 (CCND1) to adjust cell growth. Antizyme degrades ODC and inhibits polyamine uptake, thereby inhibiting cell proliferation, which suggests that it acts as a tumor suppressor gene (79). The affinity of edited AZIN1 protein for antizyme is stronger than wild-type form, and can neutralize the degradation of ODC and CCND1 mediated by antizyme, thereby increasing protein stability. ODC and CCND1 are important for regulating G1/S conversion, and their raised levels can increase protein synthesis, which is necessary to enter cell cycle (32). Accumulation of ODC and CCND1 leads to increase in cell proliferation, tumor initiation and metastasis (33). In summary, AZIN1 exerts carcinogenic effect by restraining antizyme activity. In addition, ADAR1 expression and AZIN1 over-editing are closely related to the occurrence, development, metastasis and recurrence of HCC (32). HCC occurrence involves other editing targets such as FLNB and COPA (64). In addition to HCC, AZIN1 levels are substantially elevated in non-small-cell lung cancer (NSCLC) (35), esophageal squamous cell carcinoma (ESCC) (34), colorectal cancer (CRC) (33), BC (13) and prostate adenocarcinoma (36). High-level AZIN1 editing is the prognostic indicator of population and tumor-free survival, and is the independent risk factor for lymph node and distant metastasis (33) (Figure 1A).
Figure 1.
The roles of ADAR1-mediated A-to-I RNA editing in cancer. (A,B,C,D,E,F) ADAR1 upregulation leads to increased editing of target mRNAs, such as AZINI, BLCAP, DHFR, FAK, leukemia inhibitory factor receptor (LIFR), and prostate cancer antigen 3 (PCA3) as well as miRNAs such as miR-200b, which contribute to cancer progression through various signaling pathways. (G,H) ADAR1 expression is downregulated in melanoma and BC, which decreases editing of miR-455-5p, miR-378a-3p, and Gabra3. Edited miR-455-5p, miR-378a-3p, and GABRA3 suppress cancer cell invasion and metastasis. (I) ADAR1 plays a role in cancer-related immune environments.
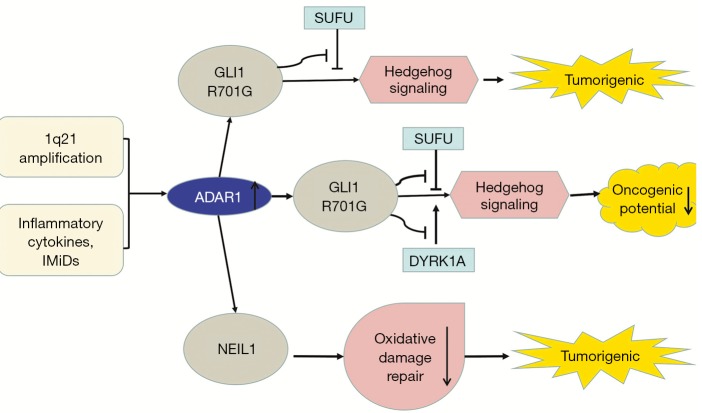
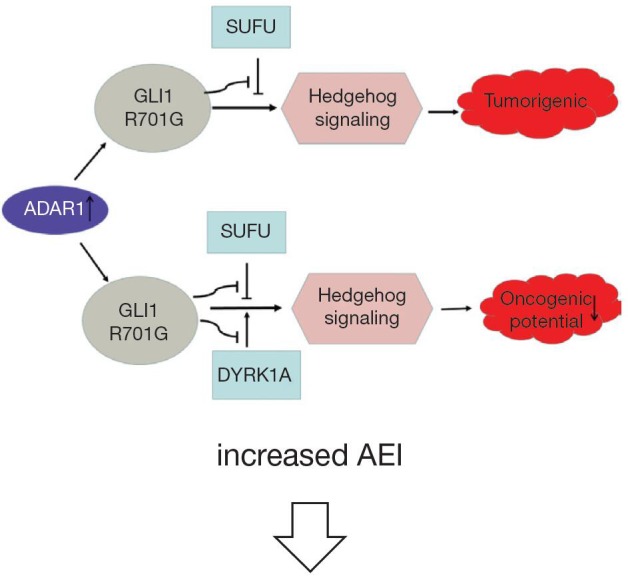
In RNA editing enzyme related to Multiple myeloma (MM), the expression of ADAR1 is the most abundant (37,80). Both 1q21 amplification and environmental factors such as inflammatory cytokines and immunomodulatory drugs (IMiDs) regulate A-to-I editing activity (38). Amplified ADAR1 edits glioma-associated oncogene 1 (GLI1), leading to an R/G amino acid change at residue 701 (38). This stabilizes GLI1 expression through blocking binding to negative regulator, suppressor of fused (SUFU) (37). Edited GLI1 has elevated transcriptional activity, and activates the Hedgehog signaling pathway, thereby promoting malignant regeneration of MM and lenalidomide resistance in vitro (81,82). Interestingly, in basal cell carcinoma and medulloblastoma, edited GLI1 exerts anti-tumorigenic effect. ADAR1-edited GLI1 (R701G) shows reduced oncogenic potential because edited GLI1 becomes less accostable to activator, dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) (37). Further research is needed to clarify the influencing factors of the tumorigenic and anti-tumor effects of ADAR1-mediate editing (Figure 2).
Figure 2.
The roles of ADAR1-mediated RNA editing in Multiple myeloma (MM), basal cell carcinoma (BCC) and medulloblastoma (MB). In MM, 1q21 amplification, inflammatory cytokines and immunomodulatory drugs (IMiDs) upregulate ADAR1 editing activity. Increased ADAR1 edits glioma-associated oncogene 1 (GLI1) and stabilizes GLI1 expression by preventing the binding of its negative regulator, suppressor of fused (SUFU). Edited GLI1 activates the Hedgehog signaling pathway and promote malignant regeneration of MM. In MB and BCC, edited GLI1 is less accessible to its activator, Dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) and shows reduced oncogenic potential. In addition, edited endonuclease 8-like 1 (NEIL1) in MM shows reduced oxidative damage repair ability.
Transcriptome analysis shows that endonuclease 8-like 1 (NEIL1) is also highly edited by ADAR1 in MM and is clinically important. NEIL1 is a basal excision repair protein involved in DNA damage repair. The edited NEIL1 shows reduced oxidative damage repair ability, enhancing cell sensitivity to combination treatment with agents inducing single-stranded and double-stranded DNA breaks (39) (Figure 2).
Bladder cancer-associated protein (BLCAP) was first identified in invasive bladder cancer (83). BLCAP inhibits tumorigenesis by stimulating cell apoptosis and inhibiting proliferation (84,85). ADAR1 edits many sites in BLCAP YXXQ domain to promote cervical cancer (CC) (40). Overexpression of BLCAP leads to growth inhibition, S-phase arrest, and apoptosis (84). The BLCAP transcript coding region is hyper-edited in CC (40) and in HCC (41). BLCAP editing activates Akt/mTOR signal pathway and the pro-survival protein STAT3 to promote cellular proliferation (40). By contrast, the editing levels of BLCAP are decreased in astrocytoma, bladder cancer, and CRC, and correlated with increased histological malignancy of the tumors, which is mediated by ADAR2 (42). ADAR1 and ADAR2 have different effects on BLCAP editing and further research is needed to clarify the reasons for this difference (Figure 1B).
ADAR1 expression is increased in BC (13,86,87). ADAR1 edits the pro-survival gene dihydrofolate reductase (DHFR) in 2 inverted Alu within the 3′-UTR of its transcript. DHFR is involved in folate metabolism. RNA editing prevents miRNA-mediated silencing of DHFR, thereby stabilizing its mRNA. DHFR is the target for methotrexate as well. ADAR1 binds to and edits miR-25-3p and miR-125a-3p to upregulate the expression of DHFR, thereby promoting cell proliferation and methotrexate resistance (43) (Figure 1C).
ADAR1-mediated editing occurs in focal adhesion kinase (FAK) at intron 26, which is a tumor metastasis promoting factor. In lung adenocarcinoma (LUAD), edited FAK stabilizes the FAK mRNA and protein, promoting invasion and recurrence (44) (Figure 1D).
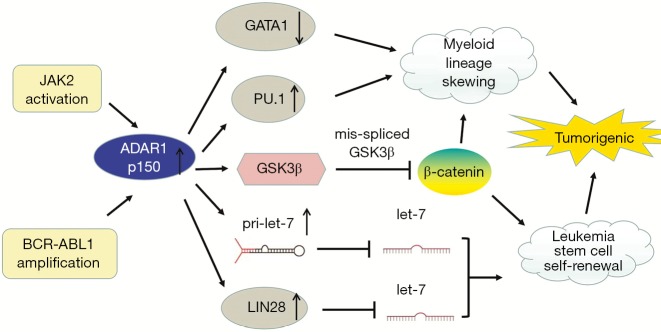
As previously stated, ADAR1 is involved in embryonic hematopoiesis (15,18). In blast crisis chronic myeloid leukemia (BC CML), BCR-ABL1 amplification upregulates expression of IFN-g pathway genes, and then inflammatory stimulation drives the expression of ADAR1 p150. In addition, JAK2 activation also promotes ADAR1 expression (46). The expression of ADAR1p110 remains unchanged (30). ADAR1 is required for leukemia cell survival (88). Overexpression of ADAR1 induces the pluripotency gene LIN28 and promotes pri-let-7 editing, causing a decrease in let-7 family (89). The decrease in let-7 increases the self-renewal ability of the leukemic stem cell (LSC) (45). In addition, increased levels of ADAR1 p150 are related to the upregulation of the myeloid transcription factor PU.1 and the downregulation of the erythroid transcription factor GATA1, which lead to myeloid lineage skewing. However, the effect of ADAR1 on PU.1 expression is not clear (46). ADAR1 p150 overexpression in cultured blood progenitor cells induces reprogramming of myeloid progenitor cells. This increases hematopoietic differentiation to myeloid lineage, possibly via the upregulation of PU.1 and generation of a mis-spliced version of glycogen synthase kinase 3β (GSK3β) mRNA. The identification of mis-spliced GSK3β as a negative regulator of β-catenin suggests that alternative splicing induced by ADAR1 promotes CML progression. β-catenin plays an important role in cell self-renewal, but mis-spliced GSK3β cannot downregulate β-catenin. ADAR1 was therefore identified, for the first time, as the driving factor for reprogramming malignant progenitor cells to self-renewing LSC (46) (Figure 3).
Figure 3.
The roles of ADAR1-mediated RNA editing in chronic myeloid leukemia. JAK2 activation and BCR-ABL1 amplification increase ADAR1 p150 expression. Increased ADAR1 p150 upregulates the myeloid transcription factor PU.1 and downregulates the erythroid transcription factor GATA1, which lead to myeloid lineage skewing. ADAR1 p150 overexpression induces reprogramming of myeloid progenitor cells, potentially through the generation of a mis-spliced version of GSK3β mRNA. The mis-spliced GSK3β cannot downregulate β-catenin, which is essential for cell self-renewal. ADAR1 overexpression induces the pluripotency gene LIN28 and promotes pri-let-7 editing, reducing let-7 production and further promoting the self-renewal ability of leukemic stem cells.
Over-editing of miR-200b can be found in a variety of cancers and is correlated with poor patient survival. Edited miR-200b by ADAR1 promotes cell invasion and migration mainly via its reduced ability to inhibit the epithelial-mesenchymal transition (EMT) regulators ZEB1 and ZEB2 and the concomitantly acquired ability to repress new targets, such as the metastasis suppressor leukemia inhibitory factor receptor (LIFR) (47) (Figure 1E).
The lncRNA prostate cancer antigen 3 (PCA3) is upregulated in prostate cancer (90). PCA3 is dominant-negative oncogene. ADAR1-mediated editing of PCA3 increases its stability and expression. PCA3 is antisense to tumor suppressor gene prune homolog 2 (PRUNE2), and forms a duplex with the PRUNE2 pre-mRNA. ADAR1-mediated A-to-I editing of the PCA3/PRUNE2 duplex at multiple sites downregulates PRUNE2 and upregulates PCA3 expression, thereby increasing cancer cell proliferation, adhesion, and migration (48) (Figure 1F).
The expression of ADAR1 is significantly lower in metastatic melanoma than in melanocytes (91). Activation of cyclic AMP-responsive element binding protein (CREB) downregulates ADAR1 expression (49,50). Knockdown of ADAR1 promotes the growth and metastasis of melanoma by controlling the biogenesis of oncogenic or tumor inhibiting miRNAs. Shoshan et al. found that editing pri-miR-455-5p by ADAR1 prevents cleavage by drosha ribonuclease III (DROSHA), resulting in a low level of mature miR-455-5p. Wild-type miR-455-5p inhibits tumor suppressor cytoplasmic polyadenylation element-binding protein 1 (CPEB1) to stimulate tumor proliferation and metastasis, whereas edited miR-455-5p has the opposite effect (49). In addition, ADAR1 mediated miR-378a-3p editing at +18 site is only in non-metastatic melanoma cells, whereas the wild-type form accumulates in metastatic cells. The oncogene PARVA promotes progress and metastasis of melanoma. Edited miR-378a-3p downregulates the expression of PARVA to prevent melanoma development and metastasis (50) (Figure 1G).
The expression of ADAR1 is lower in metastatic BC cells than in noninvasive cells (51). ADAR1 edits the conversion of isoleucine to methionine (I/M) in the GABAA receptor α-3 (Gabra3) transcript in the brain (92). The edited form of Gabra3 is expressed only in non-invasive BC tissues, and not in metastatic tumors (51,93). In BC, GABRA3 activates the AKT pathway, which promotes cell migration, invasion, and metastasis (51). The expression of edited GABRA3 decreases on the cell surface (94), thereby inhibiting AKT activation and BC metastasis (51) (Figure 1H).
ADAR1 also plays a role in cancer-related immune environments. In ovarian cancer, melanoma, and BC, human leukocyte antigen (HLA) molecules express increased ADAR1-edited peptides, including cyclin I (CCNI), which induce antigen-specific killing of oncocytes by CD8+ T cells. This suggests the possibility of exploring immunotherapeutic approaches based on RNA editing (52) (Figure 1I).
RNA editing-dependent roles of ADAR2 in cancer
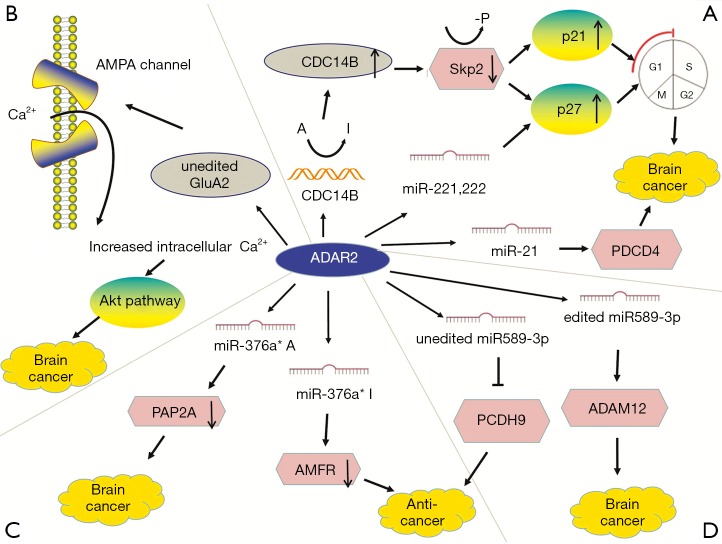
ADAR2 mediated editing can inhibit tumor occurrence and metastasis. ADAR2 is important in brain development and function (95). ADAR2 activity decreases in high-grade astrocytoma and GBM (31,60). Cells which overexpress ADAR2 are mainly concentrated in G1 phase (54). The CDC14B transcript is a novel candidate gene because it regulates S-phase entry through a process dependent on the E3 ligase S-phase kinase-associated protein 2 (skp2) (53). ADAR2 promotes the upregulation of CDC14B by editing multiple sites in the CDC14B phosphatase pre- mRNA intron 7, leading to degradation of SKP2 (53,54). ADAR2 silencing inhibits edits of entire recognized sites in CDC14B transcript and decreases the expression level of CDC14B mRNA. Decreased CDC14B expression is related to increased astrocytoma invasiveness and poor prognosis (54). The CDC14B phosphatase acts upstream of the Skp2/p21/p27 pathway and modulates GBM proliferation (53,54). In glioblastoma, Skp2 overexpression downregulates p21 and p27, which regulate G1/S cell cycle transition (96,97). The Skp2 protein is an E3-ligase and drives the ubiquitination and degradation of p27Kip1 and p21Cip1/Waf1/Sdi1 (98). CDC14B reduces Skp2 by Ser64 dephosphorylation, inhibiting the degradation of p21 and p27 (53). P21 and p27 are tumor suppressor genes. Active ADAR2 downregulates Skp2 and upregulates p21/p27 (54). In addition, the strong upregulation of active CDC14B further decreases the proliferation of U118 cells, possibly because the protein acts simultaneously at cell cycle checkpoints G1/S46 and G2/M, indicating that high ADAR2 expression significantly reduces cell multiplication (54,55). Overall, genes are rarely mutated in GBM, indicating that post-transcriptional regulation is critical in the pathology of GBM (97) (Figure 4A).
Figure 4.
The roles of ADAR2-mediated RNA editing in brain cancer development. ADAR2 expression or editing activity is often low in cancer. Editing levels of ADAR2 targets such as CDC14B, GluA2, miR-221, miR-222, miR-21, miR-376a* and miR-589-3p are decreased, contributing to brain cancer development, especially in glioblastoma multiforme.
In glioblastoma cells, GluA2 induces apoptosis (56), whereas in astrocytoma cells, ADAR2 overexpression completely edits the GluA2 Q/R site but does not have the same effect (55). Glutamate ionotropic receptor AMPA type subunit 2 (GluA2 or GRIA2) is a subunit of the AMPA glutamate receptor family, in which four subunits are combined in different ways to form a mature receptor. Activation of the mature receptor by the ligand glutamate converts it into neuronal excitatory ion channel. ADAR2 edits GluA2 at Q/R site, which makes the channel nearly impermeable to Ca2+ ions, thereby reducing intracellular calcium concentration (99). In glioblastoma, Q/R site editing reduces to 69–88% because of the significantly reduced editing activity of ADAR2 (57). High expression of the Ca2+ permeable AMPA receptor subunit in GBM indicates that the change of Ca2+ influx is associated with carcinogenesis. Edited GluA2 produces Ca2+ impermeable AMPA channels that inhibit cell migration and induce apoptosis, whereas unedited GluA2 promotes the malignant characteristics of cells (56). Unedited GluA2 and intracellular Ca2+ wave propagation induce the phosphorylation and activate Akt pathway in GBM (58) (Figure 4B).
In glioblastoma cells, ADAR2 regulates the expression of many miRNAs (100). ADAR2 editing prevents corresponding precursors from maturing to miR-221, miR-222 and miR-21 to restrict their expression (60). Decreased ADAR2 activity reduces the inhibition of the three oncomiRs, thereby promoting tumor growth and migration (101). ADAR2-mediated p27 downregulation is also related to miRNAs edited by ADAR2 in the brain (60). The three oncogenes inhibit the tumor suppressor protein p27kip1 and programmed cell death protein 4 (PDCD4) (61). In GBM, failure to edit miR-21 and miR-221/222 results in the downregulation of their respective targets PDCD4 and p27Kip1 (60) (Figure 4A).
MiR-376* is another miRNA that is under-edited in glioblastoma. Although the expression of miR-376* clusters does not change, ADAR2-mediated pri-miRNA editing is decreased in GBM, resulting in reduced seed sequence editing in mature miRNAs. The overexpression of unedited miR-376* promotes invasion and migration, whereas edited miR-376a* inhibits GBM progression. ADAR2 can induce a switch between the oncogenic and tumor-suppressive activities of miRNAs by changing target specificity. ADAR2-edited miR376a* binds to the 3′-UTR of autocrine motility factor receptor (AMFR), a receptor for the tumor motility-stimulating protein AMF, and downregulates it, thereby inhibiting tumor migration and invasion. Unedited miR376a* binds to the 3′-UTR of Ras-related protein 2a (RAP2A) mRNA to downregulate its expression, thereby promoting glioblastoma cell invasion. RAP2A acts on actin remodeling and exerts tumor suppressive effects. Invasion experiments show that AMFR upregulation has a greater effect on increasing the aggressiveness and migration of glioma than RAP2A downregulation. This study illustrated how RNA editing changes target specificity, leading to different effects between edited and unedited miRNAs (62) (Figure 4C).
In advanced GBM, miR-589-3p editing levels are significantly decreased. Unedited miR-589-3p inhibits protein protocadherin 9 (PCDH9) which suppress cancer development. In normal brain tissues, ADAR2 edits almost all miR-589-3p to diminish the expression of metalloproteinase domain-containing protein 12 (ADAM12), which can accelerate cancer metastasis (63) (Figure 4D).
ADAR2-mediated editing activity is generally low in GBM, particularly editing of GluA2 mRNA, although the expression of ADAR2 is not correspondingly downregulated (57). Both ADAR1 and ADAR3 are upregulated in brain tumors (59), however, decreased expression of all ADAR genes was reported in a different study (31). The low editing rate in glioblastoma may be due to inhibition of ADAR2 editing by overexpressed ADAR1, as suggested by in vitro experiments showing the dose-dependent inhibitory effect of ADAR1 overexpression on ADAR2 in vitro (55). Overexpression of ADAR3 may also be involved, as suggested by its effect on inhibiting GluA2 editing (59). It is also possible that the splicing variant of ADAR2 with reduced catalytic activity is the expressed form (57).
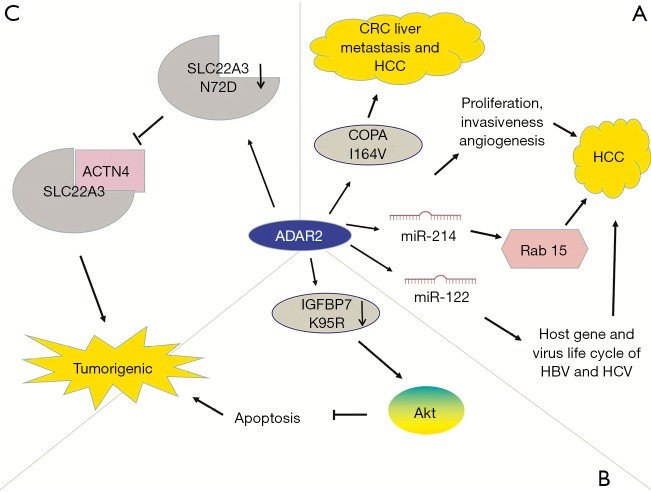
In addition to GBM, ADAR2 suppresses tumorigenicity in other cancers. ADAR2 downregulation occurs in approximately 50% probability in HCC, whereas increased ADAR2 expression in ADAR-deficient HCC weakens carcinogenic potential (64). The suppressive function of ADAR2 in HCC is mediated by edited coatomer protein complex subunit α (COPA) mRNA (I164V). Edited COPA is absent in invasive CRC with the EMT phenotype, resulting in liver metastasis (65). In addition, ADAR2 plays a role in regulating miR-214 and miR-122, which are reduced in HCC (66). MiR-214 promotes the occurrence of HCC by regulating proliferation, invasiveness, and angiogenesis (67,68). An editing event leads to a decrease in RNA transcription complementary to pri-miR-214, resulting in the decline in pri-miR-214 and miR-214 and an increase in target Rab15 protein (66). Rab15 is a Ras oncogene related to retinoic-acid-induced differentiation and receptor recycling, however, its specific role in HCC remains unclear (102,103). In HCC, miR-122 regulates the host gene and lifecycle of HBV and HCV (69). In summary, edited complementary antisense transcripts regulates the biogenesis of specific miRNAs involved in the pathogenesis of HCC, thus providing novel targets for the treatment of HCC (Figure 5A).
Figure 5.
The roles of ADAR2-mediated RNA editing in cancer development. ADAR2 downregulation has oncogenic effects. (A) The editing level of COPA is inversely correlated with the pathogenesis of hepatocellular carcinoma (HCC). Edited COPA is absent in CRC, leading to liver metastasis. ADAR2 regulates biogenesis of miR-214 and miR-122 in HCC. The RNA precursors of miR-214 and miR-122 are targets for ADAR2 editing. ADAR2 editing decreases miR-214 and its target gene Rab15. MiR-214 regulates HCC proliferation, invasiveness, and angiogenesis. MiR-122 regulates host gene and virus life cycle of HBV and HCV. (B) In esophageal squamous cell carcinoma (ESCC), decreased expression of ADAR2 reduces the editing frequency of IGFBP7, activates the Akt pathway and inhibits apoptosis, leading to malignant phenotype. (C) In familial esophageal cancer, SLC22A3 deregulation promotes metastasis and filopodia formation through decreasing direct binding to a-actinin-4 (ACTN4).
ADAR2 is downregulated in esophageal squamous cell carcinoma (ESCC) (70). Editing of insulin-like growth factor-binding protein 7 (IGFBP7) converts lysine residue into arginine (104,105). ADAR2-mediated editing stabilizes IGFBP7 by altering the protease recognition site of matriptase and inhibiting proteolytic cleavage, thereby inducing apoptosis. In ESCC, IGFBP7 is one of the pro-apoptotic genes showing decreased editing levels. Downregulation of ADAR2 in ESCC and decreased editing and expression of IGFBP7 (K95R) activate the Akt pathway, thereby inhibiting programmed cell death and promoting tumor growth (70) (Figure 5B).
ADAR2 also edits solute carrier family 22 member 3 (SLC22A3). Increased ADAR2 levels lead to editing and downregulation of SLC22A3 (N72D), which can inhibit the metastasis of esophageal cancer. SLC22A3 deregulation promotes metastasis and filopodia formation through decreasing direct binding to a-actinin-4 (ACTN4), an actin-binding protein (71) (Figure 5C).
RNA editing roles of ADAR1 and ADAR2 in cancer
The gene encoding miR-142s is located at the translocation site related to invasive B cell leukemia and is highly expressed in hematopoietic tissues, whereas rarely or not expressed in non-hematopoietic tissues (106). MiR-142-3p expression in patients with acute myeloid leukemia (AML) is significantly lower than that in controls (72). ADAR1 p110 and ADAR2 edit two specific pri-miR-142 sites (+4 and +5 sites), which suppresses pri-miR cleavage and significantly downregulates miR-142 expression (89).
ADAR1 edits the intron of heterogeneous nuclear ribonucleoprotein L-like (HNRPLL) to generate exon E12A, which reinforces the oncogenic splicing factor serine/arginine rich splicing factor 1, thereby promoting HNRPLL transcription. Edited HNRPLL can regulate the expression of cyclin D1 and transforming growth factor beta receptor 1 (TGFBR1) to stimulate cellular multiplication of renal and bladder carcinoma. ADAR2 can also edit HNRPLL, suggesting the existence of cross-talk between ADAR1 and ADAR2 (73).
In GC, ADAR2 edited podocalyxin-like (PODXL) codon H241R (histidine to arginine) results in reduced growth and invasiveness, which neutralizes the tumorigenic ability of unedited PODXL (74). PODXL is an oncogene that induces oncocytes migration and invasion via acting on actin-binding protein EZR (75). Unedited PODXL promotes the development of GC through adjusting cell adhesion mechanisms. In addition, ADAR1 and ADAR2 induce synonymous (ACA>ACG; T238T) and nonsynonymous (CAC>CGC; H241R) editing of PODXL, respectively. A high rate of ADAR2/ADAR1-mediated PODXL editing (H241R) can prevent carcinogenesis. In GC, the increase in ADAR1 and decrease in ADAR2 lead to an imbalance of PODXL editing and thus an increase in unedited PODXL, resulting in tumorigenesis. Inhibition of ADAR1 or recovery of potential ADAR2 activity is a potential strategy for the treatment of GC (75).
RNA editing-independent roles of ADAR
RNA editing enzymes can function independently of RNA editing capabilities, which is related to tumor proliferation, metastasis and immune escape (6). ADAR1 and ADAR2 can directly interact with miRNAs or affect miRNA biogenesis by regulating Dicer, Drosha, DiGeorge Syndrome Critical Region 8 (DGCR8), or other important factors to modulate the expression of miRNAs (25,100,107). The catalytically inactive forms of ADARs have RNA editing-independent functions.
ADAR1 levels are decreased in melanoma patients and cell lines, whereas ADAR2 levels remain unchanged. While ADAR1 level is changing, miRNA expression also changes, most of which do not experience RNA editing. ADAR1 silencing in vitro promotes malignant phenotype and metastasis, whereas ADAR1 overexpression inhibits melanoma cell proliferation independent of ADAR1 editing activity (91). MiR-17 and miR-432 overexpression induce downregulation of ADAR1 in melanoma (49,91). Galore-Haskel et al. found that ADAR1 knockdown promotes miR-222 biogenesis and inhibits the expression of the miR-222 target intercellular adhesion molecule 1 (ICAM1) (76). ICAM1 is the natural ligand of lymphocyte function associated antigen-1 (LFA-1), which acts as adhesion receptor of the immune system. Binding of LFA-1 and ICAM1 promotes the formation of the immune synapse (108) and activates T cells (109). In contrast, overexpression of ADAR1 upregulates ICAM1, increasing the sensitivity of tumors to TIL mediated killing. MiR-222 may be a potential marker for predicting the response of melanoma patients to ipilimumab, an immune checkpoint inhibitor (76).
The role of ADAR3 in cancer
ADAR3 is overexpressed in glioblastoma. ADAR3 negatively regulates GluA2 editing in astrocytoma. In addition, ADAR3 can directly bind to the GluA2 pre-mRNA to suppress ADAR2-mediated GluA2 editing (59). In GBM, over-editing of COG3 I/V promotes tumorigenesis and is related to poor prognosis. This suggests that COG3 I/V is a potential target of individualized therapy (77).
The exceptions
Some cancer-related editing targets are short of clear recognition to related editing enzymes, such as protein tyrosine phosphatase non-receptor type 6 (PTPN6) in AML and RAS homolog family member Q (RHOQ) in CRC.
AML is the first malignancy in which mRNA editing was shown to be associated with disease pathogenesis (78). PTPN6 is important for the development, proliferation and receptor-mediated mitogenic signaling of hematopoietic cells (110), and is identified as a tumor suppressor gene (111). PTPN6 exerts anti-cancer effect via reducing broad spectrum growth-promoting receptors and cytokine receptors. Edited PTPN6 produces a non-functional protein and suppresses its inhibitory action. Nevertheless, which editing enzyme editing PTPN6 remains to be further studied (78).
In CRC, edited RHOQ leads to the conversion of asparagine to serine, which increases protein activity and mediates dynamic actin cytoskeletal recombination, thus promoting invasive potential. Edited RHOQ is related to ascending relapse rate, which is higher while connected with mutant KRAS (G12D). RHOQ distorted editing is also common in GC, lung cancer and HCC (29).
Overall, these results reflect the mechanisms of ADAR in tumors and suggest potential large-scale clinical implications.
Clinical utility
Aberrant ADAR-mediated editing provides an underexplored mechanism for altering proteins and regulating dsRNA sequences which can act as driving factors and suggest biomarkers and cancer treatment targets. Monitoring ADAR expression levels or RNA editing activity is a useful early biomarker for detecting cancer even before clinical symptoms become apparent (Table 2).
Table 2. Clinical applications of ADAR in cancer treatment.
| Clinical utility | Marker (give an example) | Effect | References |
|---|---|---|---|
| Therapeutic targets | GLI1 | Drug 1: DYRK1A analogue | (37,38,81) |
| Drug 2: targeting edited GLI1 | |||
 |
|||
| increased AEI | |||
| Prognostic markers | Alu editing index (AEI) |  |
(30) |
| poor prognosis | |||
| DHFR is a target molecule of methotrexate | |||
| Predictive markers | DHFR |  |
(43) |
| Edited DHFR by ADAR1 is resistant to methotrexate in breast cancer |
DYRK1A, dual specificity tyrosine-phosphorylation-regulated kinase 1A; SUFU, suppressor of fused.
Therapeutic targets
ADAR1 is a promising therapeutic target because loss of activity leads to cell inherent fatality and IFN-I induction. ADAR1 repression directly kills cancer cells expressing high levels of ISGs through protein kinase R (PKR) activity (112). ADAR1 repression may enlarge IFN-I response by triggering IFN-β production by cells in the tumor microenvironment. These findings suggest that ADAR1 is a potential therapeutic target in cancer. Furthermore, ADAR1 inhibitors can synergize with cancer immunotherapy by stimulating cytotoxic T cells and natural killer cells (112). Similarly, ADAR2 is a promising target because it regulates multiple miRNAs and cellular pathways simultaneously in cancer cells. Compounds capable of regulating the expression of ADAR2 in glioblastoma could be developed as anti-cancer therapy.
The targets of ADAR are also therapeutic direction that can be explored. For example, FAK is a relatively mature biomarker in advanced solid malignancies. FAK inhibitors like VS-6063 (defactinib), GSK2256098 and VS-4718 are improved anticancer immunotherapy. (Content derives from https://clinicaltrials.gov).
In addition, molecular tools can be developed to selectively inhibit RNA editing. Site-selective editing inhibiting agent may treat cancers whose site is over-edited (113). In cancer cells with ADAR2 downregulation, SNAPtag technology can target specific RNA editing events, which may inhibit the development of cancer (114). The synthesis of RNA-guided editing enzymes for target-specific RNA editing is another potential direction. It is possible to target cancer-related genes and appropriately restore protein function through the CRISPR-Cas genome editing system (22).
Prognostic markers
The editing level of many RNA editing sites is related to survival time of patients, indicating that the level of editing at specific RNA sites can be used as a prognostic marker (30,47,86). For example, the Alu editing index (AEI) refers to weighted man editing level of all expressed Alu sequences. High index is related to poor prognosis of cancers (30). In addition, miR-122 is a liver-enriched miRNA. ‘Serum miR-122 may act as a real-time monitor marker of chemotherapy induced liver injury.’ (Content derives from https://clinicaltrials.gov).
Predictive markers
Particular ADAR editing events can influence reactions to cancer treatment, indicating that they are predictive biomarkers. For instance, edited COG3 and GRIA2 can improve the susceptibility to MEK inhibiting agents (86). In addition, hepatic and circulating miR-221 and miR-222 is of clinical meaning in HCC. (Content derives from https://clinicaltrials.gov) If assessment demonstrates that circulating miRNAs are relevant and reliable in diagnosis and follow-up of HCC, then treatments for the miRNAs and associated pathways may be used clinically rather than the necessary for pathology of liver biopsies.
In principle, regulating ADAR expression is a promising direction in the treatment of cancer. However, because many other diseases are related to altered A-to-I editing levels, reversing cancer by regulating RNA editing levels may have unintended consequences.
Conclusions
In recent years, high-throughput sequencing and TCGA have detected significant changes in ADAR-mediated editing in many cancers. RNA editing is the main biological function of ADARs. This review discussed current important ADAR-mediated editing events, related carcinogenic mechanism and the related clinical applications.
Taken together, research studies have revealed several issues that need to be addressed. The relationship among editing enzymes is complex, especially that between ADAR1 and ADAR2, which can cooperate or antagonize each other. For example, PODXL expression is controlled through rate between PODXL edited by ADAR1 and ADAR2. ADAR1 p150 and p110 show functional differences in cancer, however, studies often do not clearly distinguish the degree of involvement of each isoform. In addition, there are few studies addressing the role of ADAR3 in cancer. The mechanism by which ADARs activate or regulate oncogenic signaling remains unclear. The same editing event may result in a completely different function or phenotype, such as the aforementioned ADAR1-mediated GLI1 editing and ADAR2-mediated COPA editing. The extent to which RNA editing alterations affect protein modifications remains unclear. Some cancer-related editing sites lack distinct recognition of related editing enzymes, such as PTPN6 in AML and RHOQ in CRC. Furthermore, the interaction network between ADAR RNA editing and other tumorigenesis-related pathways and factors remains unclear.
In the future, potential biomarkers, cancer treatment targets, and molecular tools for regulation of RNA editing need to be identified and characterized for clinical application. ADAR RNA editing is also important for the accuracy and reference value of patient prognosis. Specific RNA editing events can selectively influence the response to cancer treatment. The application of RNA editing to improve the sensitivity to drugs and reduce the incidence of drug resistance is a key issue to be studied. Possible future directions in cancer treatment include the development of drugs targeting ADAR1, ADAR2, and editing sites; and exploring whether they can synergize with existing targeted drugs, immunotherapy, and other cancer treatments. Further elucidation of the mechanisms underlying ADAR functions will provide important insight for the development of anticancer treatments.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81572262), the Jiangsu Province’s Key Provincial Talents Program (ZDRCA2016028), 333 high class Talented Man Project (BRA2016516) and the Natural Science Foundation of the Jiangsu Higher Education Institution of China (18KJB320006).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27-36. 10.1093/carcin/bgp220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Federica Galeano ST, Franco Locatelli, Angela Gallo. A-to-I RNA editing: The “ADAR” side of human cancer. Seminars in Cell & Developmental Biology 2012;23:244-50. 10.1016/j.semcdb.2011.09.003 [DOI] [PubMed] [Google Scholar]
- 3.Wang C, Zou J, Ma X, et al. Mechanisms and implications of ADAR-mediated RNA editing in cancer. Cancer Lett 2017;411:27-34. 10.1016/j.canlet.2017.09.036 [DOI] [PubMed] [Google Scholar]
- 4.Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell 1987;48:607-13. 10.1016/0092-8674(87)90239-X [DOI] [PubMed] [Google Scholar]
- 5.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life 2009;61:572-8. 10.1002/iub.207 [DOI] [PubMed] [Google Scholar]
- 6.Qi L, Song Y, Chan THM, et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res 2017;45:10436-51. 10.1093/nar/gkx667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahman R, Xu W, Jin H, et al. Identification of RNA-binding protein targets with HyperTRIBE. Nat Protoc 2018;13:1829-49. 10.1038/s41596-018-0020-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckmann CR, Neunteufl A, Pfaffstetter L, et al. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol Biol Cell 2001;12:1911-24. 10.1091/mbc.12.7.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci U S A 2003;100:14018-23. 10.1073/pnas.2336131100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 2014;24:365-76. 10.1101/gr.164749.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinigaglia K, Wiatrek D, Khan A, et al. ADAR RNA editing in innate immune response phasing, in circadian clocks and in sleep. Biochim Biophys Acta Gene Regul Mech 2019;1862:356-69. 10.1016/j.bbagrm.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 12.Peng Z, Cheng Y, Tan BC, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 2012;30:253-60. 10.1038/nbt.2122 [DOI] [PubMed] [Google Scholar]
- 13.Fumagalli D, Gacquer D, Rothe F, et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell Rep 2015;13:277-89. 10.1016/j.celrep.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajendren S, Manning AC, Al-Awadi H, et al. A protein-protein interaction underlies the molecular basis for substrate recognition by an adenosine-to-inosine RNA-editing enzyme. Nucleic Acids Res 2018;46:9647-59. 10.1093/nar/gky800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Khillan J, Gadue P, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 2000;290:1765-8. 10.1126/science.290.5497.1765 [DOI] [PubMed] [Google Scholar]
- 16.Hartner JC, Schmittwolf C, Kispert A, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 2004;279:4894-902. 10.1074/jbc.M311347200 [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Miyakoda M, Yang W, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 2004;279:4952-61. 10.1074/jbc.M310162200 [DOI] [PubMed] [Google Scholar]
- 18.Hartner JC, Walkley CR, Lu J, et al. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 2009;10:109-15. 10.1038/ni.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritzell K, Xu LD, Otrocka M, et al. Sensitive ADAR editing reporter in cancer cells enables high-throughput screening of small molecule libraries. Nucleic Acids Res 2019;47:e22. 10.1093/nar/gky1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannion NM, Greenwood SM, Young R, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 2014;9:1482-94. 10.1016/j.celrep.2014.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth SH, Danan-Gotthold M, Ben-Izhak M, et al. Increased RNA Editing May Provide a Source for Autoantigens in Systemic Lupus Erythematosus. Cell Rep 2018;23:50-7. 10.1016/j.celrep.2018.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kung CP, Maggi LB, Weber JD. The Role of RNA Editing in Cancer Development and Metabolic Disorders. Front Endocrinol (Lausanne) 2018;9:762. 10.3389/fendo.2018.00762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan MH, Li Q, Shanmugam R, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 2017;550:249-54. 10.1038/nature24041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clerzius G, Gelinas JF, Daher A, et al. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol 2009;83:10119-28. 10.1128/JVI.02457-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heale BS, Keegan LP, McGurk L, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J 2009;28:3145-56. 10.1038/emboj.2009.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang IX, So E, Devlin JL, et al. ADAR regulates RNA editing, transcript stability, and gene expression. Cell Rep 2013;5:849-60. 10.1016/j.celrep.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Picardi E, D'Erchia AM, Lo Giudice C, et al. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res 2017;45:D750-D757. 10.1093/nar/gkw767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong H, An O, Chan THM, et al. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res 2018;46:7953-69. 10.1093/nar/gky396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han SW, Kim HP, Shin JY, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J Exp Med 2014;211:613-21. 10.1084/jem.20132209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz-Yaacov N, Bazak L, Buchumenski I, et al. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep 2015;13:267-76. 10.1016/j.celrep.2015.08.080 [DOI] [PubMed] [Google Scholar]
- 31.Paz N, Levanon EY, Amariglio N, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res 2007;17:1586-95. 10.1101/gr.6493107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Li Y, Lin CH, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 2013;19:209-16. 10.1038/nm.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shigeyasu K, Okugawa Y, Toden S, et al. AZIN1 RNA editing confers cancer stemness and enhances oncogenic potential in colorectal cancer. JCI Insight 2018. doi: . 10.1172/jci.insight.99976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin YR, Qiao JJ, Chan TH, et al. Adenosine-to-Inosine RNA Editing Mediated by ADARs in Esophageal Squamous Cell Carcinoma. Cancer Res 2014;74:840-51. 10.1158/0008-5472.CAN-13-2545 [DOI] [PubMed] [Google Scholar]
- 35.Hu X, Chen J, Shi X, et al. RNA editing of AZIN1 induces the malignant progression of non-small-cell lung cancers. Tumour Biol 2017;39:1010428317700001. 10.1177/1010428317700001 [DOI] [PubMed] [Google Scholar]
- 36.Olsen RR, Zetter BR. Evidence of a role for antizyme and antizyme inhibitor as regulators of human cancer. Mol Cancer Res 2011;9:1285-93. 10.1158/1541-7786.MCR-11-0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimokawa T, Rahman MF, Tostar U, et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol 2013;10:321-33. 10.4161/rna.23343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazzari E, Mondala PK, Santos ND, et al. Alu-dependent RNA editing of GLI1 promotes malignant regeneration in multiple myeloma. Nat Commun 2017;8:1922. 10.1038/s41467-017-01890-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teoh PJ, An O, Chung TH, et al. Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood 2018;132:1304-17. 10.1182/blood-2018-02-832576 [DOI] [PubMed] [Google Scholar]
- 40.Chen W, He W, Cai H, et al. A-to-I RNA editing of BLCAP lost the inhibition to STAT3 activation in cervical cancer. Oncotarget 2017;8:39417-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu X, Wan S, Ou Y, et al. RNA over-editing of BLCAP contributes to hepatocarcinogenesis identified by whole-genome and transcriptome sequencing. Cancer Lett 2015;357:510-9. 10.1016/j.canlet.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 42.Galeano F, Leroy A, Rossetti C, et al. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int J Cancer 2010;127:127-37. 10.1002/ijc.25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakano M, Fukami T, Gotoh S, et al. A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J Biol Chem 2017;292:4873-84. 10.1074/jbc.M117.775684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amin EM, Liu Y, Deng S, et al. The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK. Sci Signal 2017. doi: . 10.1126/scisignal.aah3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zipeto MA, Court AC, Sadarangani A, et al. ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis. Cell Stem Cell 2016;19:177-91. 10.1016/j.stem.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Q, Crews LA, Barrett CL, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 2013;110:1041-6. 10.1073/pnas.1213021110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Xu X, Yu S, et al. Systematic characterization of A-to-I RNA editing hotspots in microRNAs across human cancers. Genome Res 2017;27:1112-25. 10.1101/gr.219741.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salameh A, Lee AK, Cardo-Vila M, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci U S A 2015;112:8403-8. 10.1073/pnas.1507882112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shoshan E, Mobley AK, Braeuer RR, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol 2015;17:311-21. 10.1038/ncb3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velazquez-Torres G, Shoshan E, Ivan C, et al. A-to-I miR-378a-3p editing can prevent melanoma progression via regulation of PARVA expression. Nat Commun 2018;9:461. 10.1038/s41467-018-02851-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gumireddy K, Li A, Kossenkov AV, et al. The mRNA-edited form of GABRA3 suppresses GABRA3-mediated Akt activation and breast cancer metastasis. Nat Commun 2016;7:10715. 10.1038/ncomms10715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang M, Fritsche J, Roszik J, et al. RNA editing derived epitopes function as cancer antigens to elicit immune responses. Nat Commun 2018;9:3919. 10.1038/s41467-018-06405-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodier G, Coulombe P, Tanguay PL, et al. Phosphorylation of Skp2 regulated by CDK2 and Cdc14B protects it from degradation by APC(Cdh1) in G1 phase. EMBO J 2008;27:679-91. 10.1038/emboj.2008.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galeano F, Rossetti C, Tomaselli S, et al. ADAR2-editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene 2013;32:998-1009. 10.1038/onc.2012.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cenci C, Barzotti R, Galeano F, et al. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 2008;283:7251-60. 10.1074/jbc.M708316200 [DOI] [PubMed] [Google Scholar]
- 56.Ishiuchi S, Tsuzuki K, Yoshida Y, et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat Med 2002;8:971-8. 10.1038/nm746 [DOI] [PubMed] [Google Scholar]
- 57.Maas S, Patt S, Schrey M, et al. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A 2001;98:14687-92. 10.1073/pnas.251531398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishiuchi S, Yoshida Y, Sugawara K, et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J Neurosci 2007;27:7987-8001. 10.1523/JNEUROSCI.2180-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oakes E, Anderson A, Cohen-Gadol A, et al. Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma. J Biol Chem 2017;292:4326-35. 10.1074/jbc.M117.779868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomaselli S, Galeano F, Alon S, et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biology 2015;16:5. 10.1186/s13059-014-0575-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008;27:2128-36. 10.1038/sj.onc.1210856 [DOI] [PubMed] [Google Scholar]
- 62.Choudhury Y, Tay FC, Lam DH, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest 2012;122:4059-76. 10.1172/JCI62925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cesarini V, Silvestris DA, Tassinari V, et al. ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res 2018;46:2045-59. 10.1093/nar/gkx1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chan TH, Lin CH, Qi L, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut 2014;63:832-43. 10.1136/gutjnl-2012-304037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shelton PM, Duran A, Nakanishi Y, et al. The Secretion of miR-200s by a PKCzeta/ADAR2 Signaling Axis Promotes Liver Metastasis in Colorectal Cancer. Cell Rep 2018;23:1178-91. 10.1016/j.celrep.2018.03.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu WH, Chen CH, Yeh KH, et al. ADAR2-mediated editing of miR-214 and miR-122 precursor and antisense RNA transcripts in liver cancers. PLoS One 2013;8:e81922. 10.1371/journal.pone.0081922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shih TC, Tien YJ, Wen CJ, et al. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J Hepatol 2012;57:584-91. 10.1016/j.jhep.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 68.Li B, Han Q, Zhu Y, et al. Down-regulation of miR-214 contributes to intrahepatic cholangiocarcinoma metastasis by targeting Twist. Febs j 2012;279:2393-8. 10.1111/j.1742-4658.2012.08618.x [DOI] [PubMed] [Google Scholar]
- 69.Girard M, Jacquemin E, Munnich A, et al. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol 2008;48:648-56. 10.1016/j.jhep.2008.01.019 [DOI] [PubMed] [Google Scholar]
- 70.Chen YB, Liao XY, Zhang JB, et al. ADAR2 functions as a tumor suppressor via editing IGFBP7 in esophageal squamous cell carcinoma. Int J Oncol 2017;50:622-30. 10.3892/ijo.2016.3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu L, Qin YR, Ming XY, et al. RNA editing of SLC22A3 drives early tumor invasion and metastasis in familial esophageal cancer. Proc Natl Acad Sci U S A 2017;114:E4631-E4640. 10.1073/pnas.1703178114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang F, Wang XS, Yang GH, et al. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol Biol Rep 2012;39:2713-22. 10.1007/s11033-011-1026-5 [DOI] [PubMed] [Google Scholar]
- 73.Chen YT, Chang IY, Liu H, et al. Tumor-associated intronic editing of HNRPLL generates a novel splicing variant linked to cell proliferation. J Biol Chem 2018;293:10158-71. 10.1074/jbc.RA117.001197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan TH, Qamra A, Tan KT, et al. ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer. Gastroenterology 2016;151:637-650.e10. 10.1053/j.gastro.2016.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sizemore S, Cicek M, Sizemore N, et al. Podocalyxin increases the aggressive phenotype of breast and prostate cancer cells in vitro through its interaction with ezrin. Cancer Res 2007;67:6183-91. 10.1158/0008-5472.CAN-06-3575 [DOI] [PubMed] [Google Scholar]
- 76.Galore-Haskel G, Nemlich Y, Greenberg E, et al. A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme. Oncotarget 2015;6:28999-9015. 10.18632/oncotarget.4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silvestris DA, Picardi E, Cesarini V, et al. Dynamic inosinome profiles reveal novel patient stratification and gender-specific differences in glioblastoma. Genome Biology 2019;20:33. 10.1186/s13059-019-1647-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beghini A, Ripamonti CB, Peterlongo P, et al. RNA hyperediting and alternative splicing of hematopoietic cell phosphatase (PTPN6) gene in acute myeloid leukemia. Hum Mol Genet 2000;9:2297-304. 10.1093/oxfordjournals.hmg.a018921 [DOI] [PubMed] [Google Scholar]
- 79.Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun 2014;5:4726. 10.1038/ncomms5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eifler T, Pokharel S, Beal PA. RNA-Seq analysis identifies a novel set of editing substrates for human ADAR2 present in Saccharomyces cerevisiae. Biochemistry 2013;52:7857-69. 10.1021/bi4006539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasper M, Regl G, Frischauf AM, et al. GLI transcription factors: mediators of oncogenic Hedgehog signalling. Eur J Cancer 2006;42:437-45. 10.1016/j.ejca.2005.08.039 [DOI] [PubMed] [Google Scholar]
- 82.Xiao C, Ogle SA, Schumacher MA, et al. Hedgehog signaling regulates E-cadherin expression for the maintenance of the actin cytoskeleton and tight junctions. Am J Physiol Gastrointest Liver Physiol 2010;299:G1252-65. 10.1152/ajpgi.00512.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gromova I, Gromov P, Celis JE. bc10: A novel human bladder cancer-associated protein with a conserved genomic structure downregulated in invasive cancer. Int J Cancer 2002;98:539-46. 10.1002/ijc.10244 [DOI] [PubMed] [Google Scholar]
- 84.Yao J, Duan L, Fan M, et al. Overexpression of BLCAP induces S phase arrest and apoptosis independent of p53 and NF-kappaB in human tongue carcinoma: BLCAP overexpression induces S phase arrest and apoptosis. Mol Cell Biochem 2007;297:81-92. 10.1007/s11010-006-9332-2 [DOI] [PubMed] [Google Scholar]
- 85.Fan DG, Zhao F, Ding Y, et al. BLCAP induces apoptosis in human Ewing's sarcoma cells. Exp Biol Med (Maywood) 2011;236:1030-5. 10.1258/ebm.2011.010315 [DOI] [PubMed] [Google Scholar]
- 86.Han L, Diao L, Yu S, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015;28:515-28. 10.1016/j.ccell.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sagredo EA, Blanco A, Sagredo AI, et al. ADAR1-mediated RNA-editing of 3'UTRs in breast cancer. Biol Res 2018;51:36. 10.1186/s40659-018-0185-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Steinman RA, Yang Q, Gasparetto M, et al. Deletion of the RNA-editing enzyme ADAR1 causes regression of established chronic myelogenous leukemia in mice. Int J Cancer 2013;132:1741-50. 10.1002/ijc.27851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 2006;13:13-21. 10.1038/nsmb1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bussemakers MJ, van Bokhoven A, Verhaegh GW, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999;59:5975-9. [PubMed] [Google Scholar]
- 91.Nemlich Y, Greenberg E, Ortenberg R, et al. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest 2013;123:2703-18. 10.1172/JCI62980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohlson J, Pedersen JS, Haussler D, et al. Editing modifies the GABA(A) receptor subunit alpha3. Rna 2007;13:698-703. 10.1261/rna.349107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neman J, Termini J, Wilczynski S, et al. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc Natl Acad Sci U S A 2014;111:984-9. 10.1073/pnas.1322098111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Daniel C, Wahlstedt H, Ohlson J, et al. Adenosine-to-inosine RNA editing affects trafficking of the gamma-aminobutyric acid type A (GABA(A)) receptor. J Biol Chem 2011;286:2031-40. 10.1074/jbc.M110.130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wahlstedt H, Daniel C, Enstero M, et al. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res 2009;19:978-86. 10.1101/gr.089409.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coqueret O. New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol 2003;13:65-70. 10.1016/S0962-8924(02)00043-0 [DOI] [PubMed] [Google Scholar]
- 97.Abukhdeir AM, Park BH. P21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med 2008;10:e19. 10.1017/S1462399408000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kitagawa K, Kotake Y, Kitagawa M. Ubiquitin-mediated control of oncogene and tumor suppressor gene products. Cancer Sci 2009;100:1374-81. 10.1111/j.1349-7006.2009.01196.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fritzell K, Xu LD, Lagergren J, et al. ADARs and editing: The role of A-to-I RNA modification in cancer progression. Semin Cell Dev Biol 2018;79:123-30. 10.1016/j.semcdb.2017.11.018 [DOI] [PubMed] [Google Scholar]
- 100.Ota H, Sakurai M, Gupta R, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 2013;153:575-89. 10.1016/j.cell.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alon S, Mor E, Vigneault F, et al. Systematic identification of edited microRNAs in the human brain. Genome Res 2012;22:1533-40. 10.1101/gr.131573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nishimura N, Van Huyen Pham T, Hartomo TB, et al. Rab15 expression correlates with retinoic acid-induced differentiation of neuroblastoma cells. Oncol Rep 2011;26:145-51. [DOI] [PubMed] [Google Scholar]
- 103.Strick DJ, Elferink LA. Rab15 effector protein: a novel protein for receptor recycling from the endocytic recycling compartment. Mol Biol Cell 2005;16:5699-709. 10.1091/mbc.e05-03-0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gommans WM, Tatalias NE, Sie CP, et al. Screening of human SNP database identifies recoding sites of A-to-I RNA editing. Rna 2008;14:2074-85. 10.1261/rna.816908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Levanon EY, Hallegger M, Kinar Y, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res 2005;33:1162-8. 10.1093/nar/gki239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83-6. 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- 107.Bahn JH, Ahn J, Lin X, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat Commun 2015;6:6355. 10.1038/ncomms7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bachmann MF, McKall-Faienza K, Schmits R, et al. Distinct roles for LFA-1 and CD28 during activation of naive T cells: adhesion versus costimulation. Immunity 1997;7:549-57. 10.1016/S1074-7613(00)80376-3 [DOI] [PubMed] [Google Scholar]
- 109.Jenkins MR, Griffiths GM. The synapse and cytolytic machinery of cytotoxic T cells. Curr Opin Immunol 2010;22:308-13. 10.1016/j.coi.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dong Q, Siminovitch KA, Fialkow L, et al. Negative regulation of myeloid cell proliferation and function by the SH2 domain-containing tyrosine phosphatase-1. J Immunol 1999;162:3220-30. [PubMed] [Google Scholar]
- 111.Gauffin F, Diffner E, Gustafsson B, et al. Expression of PTEN and SHP1, investigated from tissue microarrays in pediatric acute lymphoblastic, leukemia. Pediatr Hematol Oncol 2009;26:48-56. 10.1080/08880010802625530 [DOI] [PubMed] [Google Scholar]
- 112.Gannon HS, Zou T, Kiessling MK, et al. Identification of ADAR1 adenosine deaminase dependency in a subset of cancer cells. Nat Commun 2018;9:5450. 10.1038/s41467-018-07824-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nakano M, Nakajima M. Significance of A-to-I RNA editing of transcripts modulating pharmacokinetics and pharmacodynamics. Pharmacol Ther 2018;181:13-21. 10.1016/j.pharmthera.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 114.Hanswillemenke A, Kuzdere T, Vogel P, et al. Site-Directed RNA Editing in Vivo Can Be Triggered by the Light-Driven Assembly of an Artificial Riboprotein. J Am Chem Soc 2015;137:15875-81. 10.1021/jacs.5b10216 [DOI] [PMC free article] [PubMed] [Google Scholar]