Abstract
Background
So far, there is a lack of reliable prognostic biomarkers for lung adenocarcinoma (ADC). Initially, we found that EF-hand and coiled-coil domain containing 1 (EFCC1) was a novel gene which was downregulated consistently with the progression of lung ADC in The Cancer Genome Atlas (TCGA) data through bioinformatics analysis. In this study, we aimed to evaluate the prognostic significance of EFCC1 in lung ADC in both TCGA data and clinical samples.
Methods
Firstly, the expression level and prognostic significance of EFCC1 in lung ADC were investigated in TCGA data. Then, the expression level of EFCC1 was validated by qPCR, Western blot, and immunohistochemistry (IHC) in five clinical lung ADC and matched adjacent non-tumor tissues. Finally, the association of EFCC1 expression with clinicopathological characteristics and overall survival (OS) in lung ADC patients was further evaluated in 130 clinical lung ADC samples with tissue microarray (TMA).
Results
In TCGA data, we found that decreased mRNA expression (P<0.001), elevated DNA methylation (P<0.001) of EFCC1 in lung ADC samples compared with normal lung samples, and low EFCC1 mRNA expression was associated with poor OS in lung ADC patients (HR =0.856, 95% CI: 0.754–0.970, P=0.015). In five clinical lung ADC and matched adjacent non-tumor tissues, both mRNA and protein levels of EFCC1 were lower in all lung ADC tissues than in their adjacent non-tumor counterparts. In 130 clinical lung ADC samples with TMA, EFCC1 expression was correlated with tumor-node-metastasis (TNM) stages (P=0.040) and lymph node metastasis status (P=0.001). The Kaplan-Meier survival curve revealed that low EFCC1 expression was significantly associated with poor OS in lung ADC patients (P=0.001) and multivariate Cox regression hazard model demonstrated that EFCC1 expression level was an independent prognostic factor for lung ADC patients (HR =0.557, 95% CI: 0.351–0.883, P=0.013).
Conclusions
Our findings suggested that decreased expression of EFCC1 was significantly associated with progression of lung ADC and could serve as a novel prognostic biomarker for lung ADC patients.
Keywords: EF-hand and coiled-coil domain containing 1 (EFCC1), lung adenocarcinoma (lung ADC), prognosis, biomarker
Introduction
Lung cancer is the most common cancer and the leading cause of cancer-related deaths worldwide (1). Lung adenocarcinoma (ADC), with increasing tendency annually, becomes the most frequent pathological subtype of lung cancer (2). Although the development of early diagnosis and introduction of targeted therapies have improved clinical outcomes of lung ADC, patients at advanced stages still have a high mortality rate (1). The tumor-node-metastasis (TNM) classification is the most important prognostic factor for lung ADC. Nevertheless, patients with the same TNM stage often have different clinical outcomes (3). Serum biomarkers which are routinely assessed in clinical practice, including carcinoembryonic antigen, cytokeratin 125, and neuro-specific enolase, have limited efficiency for prognostic evaluation (4). Therefore, it is necessary to identify novel biomarkers with clinicopathological significance and prognostic value for lung ADC.
During the past decade, the next-generation sequencing (NGS) technology has led a revolution in molecular biology and provided new insights into cancer research. Significant genetic and molecular events can be identified from NGS data through bioinformatic analysis (5). The Cancer Genome Atlas (TCGA) database has abundant genomic and clinicopathological data of various cancers. Initially, we identified genes which displayed consistent expression changes with the progression of lung ADC in TCGA data through bioinformatics analysis. Among the top ten upregulated and downregulated genes, the vast majority-such as B3GNT3, NME1, SMAD6, SPN, SRPK1and TUBB3-have been related to carcinogenesis, progression, or prognosis of lung cancer (6-12). However, the relationship between EF-hand and coiled-coil domain containing 1 (EFCC1) and lung cancer has not been reported.
EFCC1, a member of coiled-coil domain-containing (CCDC) family, is also termed coiled-coil domain containing 48 (CCDC48). Up to date, its expression patterns and bio-functions are still unclear. Previous studies have reported that CCDC family members CCDC6, CCDC8, CCDC88A, and CCDC67 were involved in the progression and have been closely related to the prognosis of lung cancer and other cancers (13-16). Based on the consistent downregulation trend of EFCC1 with the progression of lung ADC revealed by the bioinformatic analysis in TCGA data, we hypothesized that the expression of EFCC1 might also be related to the prognosis in lung ADC patients.
In this study, we first analyzed EFCC1 expression and its prognostic value in lung ADC in TCGA data. Then, both mRNA and protein expression levels of EFCC1 were validated in five clinical lung ADC and matched adjacent non-tumor tissue samples. Finally, the association of EFCC1 expression with clinicopathological characteristics and overall survival (OS) in lung ADC patients was further investigated in 130 clinical lung ADC samples with tissue microarray (TMA).
Methods
TCGA data and bioinformatic analysis
Matched genome and RNA sequencing data, DNA methylation data, and clinical data of 533 lung ADC patients were downloaded from TCGA Data Portal (https://cancergenome.nih.gov/). The mRNA expression level was expressed as log2-transformation of transcripts per million (TPM) +1. The DNA methylation level was expressed as a mean beta value. Data of 488 patients with survival information were used for survival analysis.
Bioinformatic analysis was performed using R software. Differences between groups were compared by the Wilcoxon rank-sum test. For survival analysis, a Cox proportional hazards model was used to determine whether the expression of EFCC1 was associated with OS. The survival curves were constructed according to the Kaplan-Meier method and compared with the log-rank test. P<0.05 was considered statistically significant.
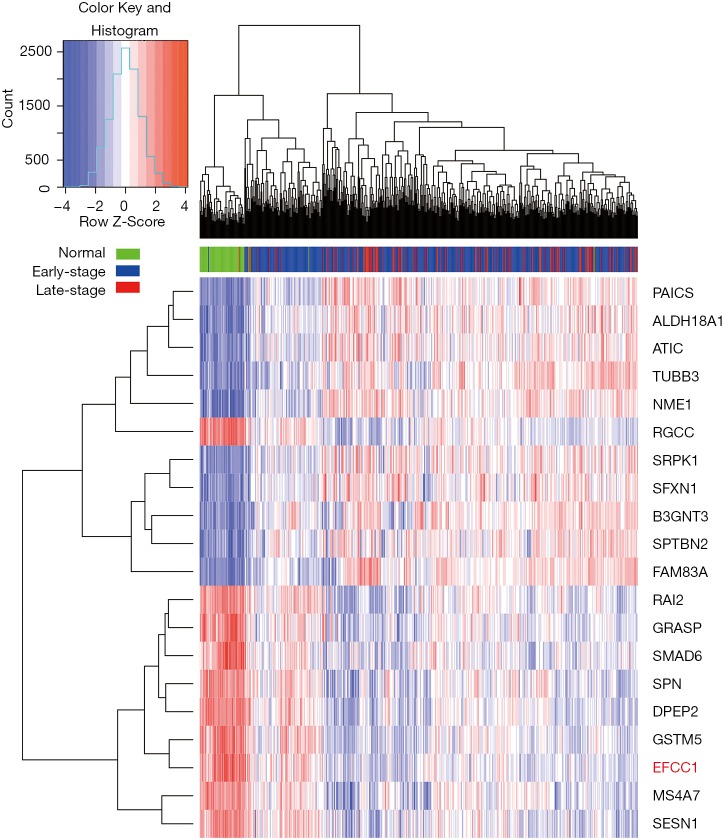
To identify genes that displayed consistent expression changes during cancer progression, we performed differential expression analysis for normal and early-stage (stage 0–II) samples (group 1), and early-stage and advanced-stage (stage III–IV) samples (group 2) using edgeR (17). The significant common genes (at the threshold of P<0.05 in both groups, q<0.05 in at least one group) with the same expression trend were considered for further analysis. And the top ten upregulated and downregulated genes (ordered by the P values in group 1) were shown by heatmap.
Patients and tissue samples
Fresh-frozen and paraffin-embedded lung ADC tissues with their matched adjacent non-tumor tissues of five patients were obtained from the Department of Thoracic Surgery, West China Hospital, Sichuan University from August 3, 2018, to August 14, 2018. The written informed consent of using biological samples and clinical data for scientific research was obtained from each patient. Fresh-frozen tissues were used for qPCR and Western blot, and paraffin-embedded tissue was used for immunohistochemistry (IHC) analysis.
To evaluate the correlation of EFCC1 expression with clinicopathological characteristics and prognosis in lung ADC patients, 130 lung ADC samples with TMA were obtained from Shanghai Outdo Biotech CO., Ltd. (China). The clinicopathological data included gender, age, tumor size, pathological grade, lymph node metastasis status, histological type, TNM stage, OS time, and survival status. OS was defined as the period from surgery to death or the last follow-up. This study was approved by the Ethics Committee of West China Hospital, Sichuan University.
qPCR
Total RNA was extracted from frozen tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Synthesis of cDNA was carried out using PrimerScriptTM RT reagent kit (Takara, Kumastu, Shiga, Japan). Then, the mRNA expression level of EFCC1 was qualified by qPCR using SYBR Premix Ex Taq TM II (Takara, Kumastu, Shiga, Japan). The GAPDH gene was used as the endogenous control. The signal was detected by a CFX Connect real-time detection system (Bio-Rad, Hercules, CA, USA). Primers for qPCR were as follows: EFCC1 (Forward primer: 5'-GCAAACCCGTCAAGAAAGGC-3'; Reverse primer: 5'-TCTAATAAGCCTGGACCGCTG-3'); GAPDH (Forward primer: 5'-GGAGCGATCCCTCCAAAAT-3'; Reverse primer: 5'-GGCTCTTCTCATACTTCTCATGG-3'). The relative expression of EFCC1 to GAPDH for each sample was calculated by ΔΔ Ct and expressed as 2–ΔΔ Ct.
Western blot
The fresh-frozen tissues were lysed with RIPA buffer (Solarbio, Beijing, China) in the presence of protease inhibitor PMSF (Solarbio, Beijing, China). Protein concentration was measured by BCA protein assay kit (Solarbio, Beijing, China). An equal amount of proteins (30 µg) were separated on 10% SDS-polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Merck Millipore, Cork, IRL). Then the membrane was blocked with defatted milk at room temperature for 1 hour and incubated with anti-EFCC1 rabbit polyclonal antibody (1:250; Abcam, #ab151108) at 4 °C overnight. After that, the membrane was incubated with fluorescence-labeled anti-rabbit IgG antibody (1:5,000; Li-Cor, #926-32211) at room temperature for 1 hour and then visualized using ODYSSEY CLX fluorescence developing system (Li-Cor, Lincoln, NE, USA). An anti-β-actin mouse monoclonal antibody was used as the loading control.
IHC and quantification analysis
The tissue sections were rewarmed at 65 °C for 3 hours and then deparaffinized and rehydrated with degraded alcohol. After that, heat-induced antigen retrieval was carried out with 0.01 M citrate salt buffer (ZSGB-BIO, Beijing, China) at 95 °C for 15 minutes. After being incubated with 0.3% H2O2 for 10 min and blocked with 10% fetal calf serum for 15 minutes, the tissue sections were incubated with anti-EFCC1 rabbit antibody (1:800; Abcam, #ab151108) at 4 °C overnight. Subsequently, these tissue sections were incubated with horseradish peroxidase-conjugated anti-rabbit antibody (1:1,000, ZSGB-BIO) at 37 °C for 15 minutes. Then, the sections were stained with DAB + substrate-chromogen solution (ZSGB-BIO, Beijing, China) at room temperature for 30 seconds and counterstained with hematoxylin. A sample incubated with PBS instead of antibodies served as the negative control. Two independent researchers assessed the immunostained result in a blinded fashion.
The expression level of EFCC1 was evaluated by both staining intensity and percentage of staining positive cells according to a semi-quantitative scoring system (18). Staining intensity was scored as 0 for negative staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining. Percentage of positive cells was quantified as 0 for ≤5% positive cells, 1 for 6–25%, 2 for 26–50%, 3 for 51–75% and 4 for ≥76%. The immunoreactivity score was then generated by multiplying score of staining intensity and percentage of positive cells. Based on immunoreactivity score of EFCC1, patients were divided into two subgroups: immunoreactivity score <6 into a subgroup of low EFCC1 expression while immunoreactivity score ≥6 into a subgroup of high EFCC1 expression.
Statistical analysis
Statistical analysis was performed using the SPSS 22.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 6 software (GraphPad Inc., La Jolla, CA, USA). The difference in EFCC1 expression between lung ADC and matched adjacent non-tumor tissues was compared using Student’s t-test. The relationship between EFCC1 and clinicopathological variables was analyzed by the Pearson chi-square test. The survival curve was constructed with the Kaplan-Meier method, and the significance of difference was examined by the log-rank test. The Cox proportional hazards regression model was used for univariate and multivariate analyses. The hazard ratio (HR) and 95% confidence interval (CI) was calculated to estimate the hazard risk of variables. P<0.05 was considered statistically significant.
Results
Expression patterns and prognostic significance of EFCC1 in lung ADC in TCGA data
Initially, we performed differential expression analysis for RNA sequencing data of normal lung, early-stage lung ADC, and advanced-stage lung ADC samples in TCGA data to identify genes that displayed consistent expression changes with lung ADC progression. Top ten upregulated and downregulated genes were presented in the heatmap (Figure 1), and EFCC1 was included in the top ten downregulated genes.
Figure 1.
The heatmap of top ten upregulated and downregulated genes with lung ADC progression based on RNA sequencing data in TCGA. Early-stage is for stage 0–II; late-stage is for stage III–IV. ADC, adenocarcinoma; TCGA, The Cancer Genome Atlas.
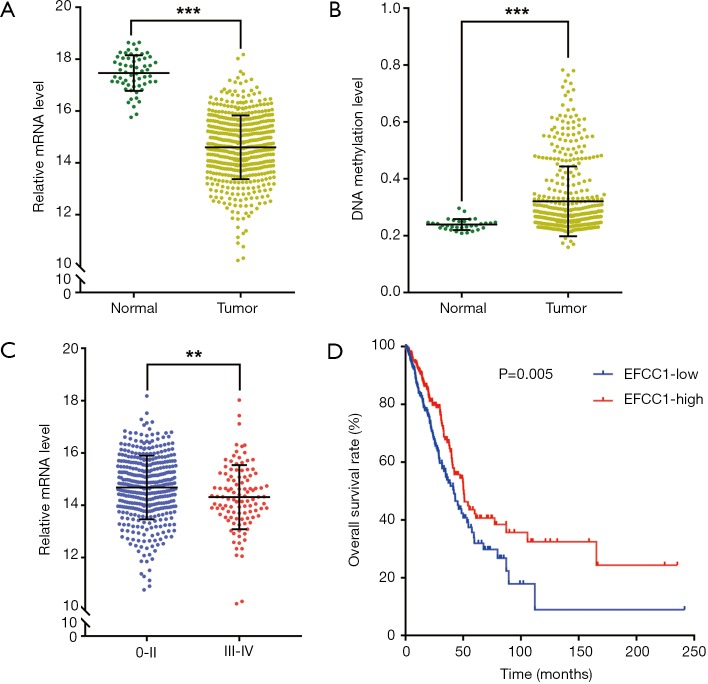
Then we analyzed EFCC1 expression level in lung ADC and normal lung samples in TCGA data. The results showed that the mRNA expression of EFCC1 was dramatically decreased in lung ADC samples compared with normal lung samples (P<0.001) (Figure 2A). The DNA methylation level of EFCC1 was significantly elevated in lung ADC samples compared with normal lung samples (P<0.001) (Figure 2B). Meanwhile, the mRNA expression of EFCC1 was lower in advanced-stage lung ADC samples than in early-stage lung ADC samples (P=0.005) (Figure 2C). Otherwise, no mutation or somatic copy number alteration of EFCC1 was found in lung ADC samples (data were not shown).
Figure 2.
EFCC1 expression and its prognostic value in lung ADC in TGCA data. (A) The mRNA level of EFCC1 was dramatically decreased in lung ADC samples compared with normal lung samples; (B) the DNA methylation level of EFCC1 was significantly higher in lung ADC samples compared with normal lung samples; (C) the mRNA level of EFCC1 was lower in stage III–IV than in stage 0–II lung ADC samples; (D) the Kaplan-Meier survival curve showed that lung ADC patients with low EFCC1 expression had an unfavorable OS (P=0.005). **P<0.01, ***P<0.001. EFCC1, EF-hand and coiled-coil domain containing 1; ADC, adenocarcinoma; TCGA, The Cancer Genome Atlas.
For survival analysis, 488 lung ADC patients with survival information in TCGA data were used. The patients were divided into groups with low or high EFCC1 expression according to the median value of EFCC1 mRNA expression. The Kaplan-Meier survival curve and log-rank test showed that lung ADC patients with low EFCC1 expression had an unfavorable OS (P=0.005) (Figure 2D). Multivariate Cox, proportional hazards analysis, demonstrated that EFCC1 expression was associated with OS of lung ADC patients (HR =0.856, 95% CI: 0.754–0.970; P=0.015), after adjustment for sex, age at diagnosis, TNM stage, history of other malignancies, history of neoadjuvant treatment and tobacco smoking history (Table 1).
Table 1. Univariate and multivariate Cox proportional hazards analysis of prognostic factor for lung ADC patients in TCGA data.
| Variable | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | ||
| EFCC1 expression | 0.828 | 0.736–0.932 | 0.002* | 0.856 | 0.754–0.970 | 0.015* | |
| Gender | 1.051 | 0.783–1.410 | 0.741 | ||||
| Age (year) | 1.009 | 0.998–1.020 | 0.109 | ||||
| History of smoking | 0.999 | 0.884–1.130 | 0.990 | ||||
| History of other malignancy | 1.537 | 1.047–2.257 | 0.028* | 2.018 | 1.358–2.999 | 0.001* | |
| Neoadjuvant treatment | 17.225 | 5.310–55.879 | <0.001* | 14.406 | 4.400–47.173 | <0.001* | |
| TNM stage | 1.638 | 1.425–1.882 | <0.001* | 1.650 | 1.428–1.906 | <0.001* | |
*P<0.05. ADC, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; CI, confidence interval; EFCC1, EF-hand and coiled-coil domain containing 1; TNM, tumor-node-metastasis.
Validation of EFCC1 expression in lung ADC and matched adjacent non-tumor tissues
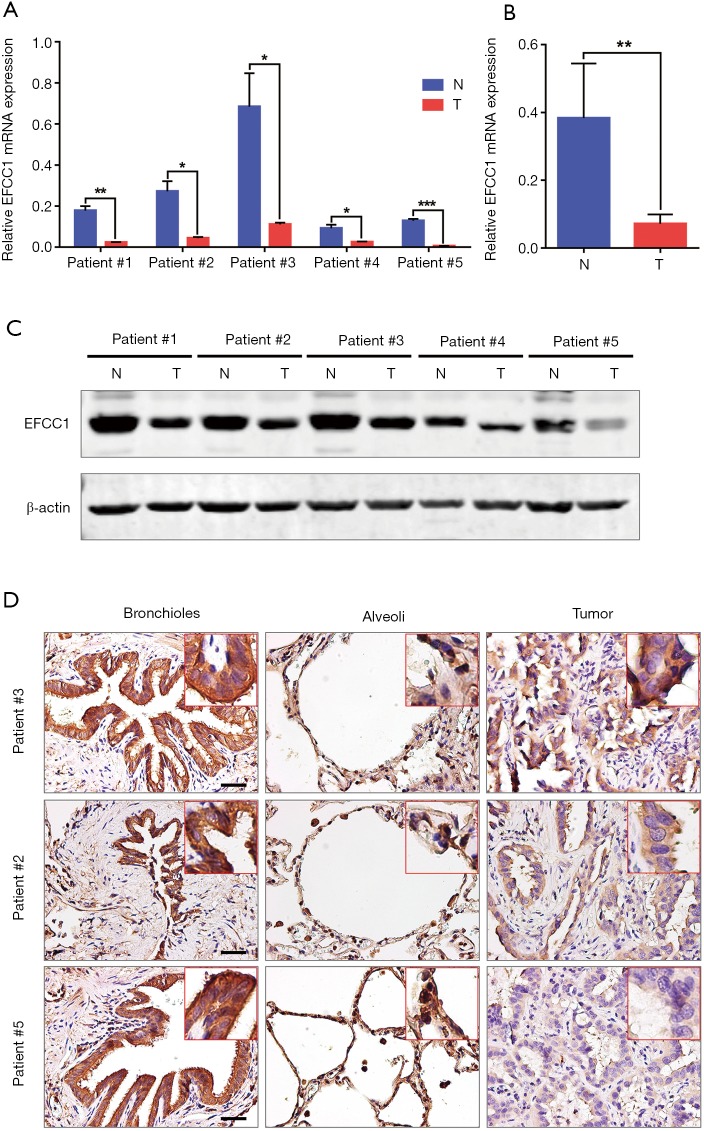
To further validate the decreased expression of EFCC1 in lung ADC, which was revealed in TCGA data, we examined the mRNA and protein expression levels of EFCC1 in five lung ADC and matched adjacent non-tumor tissues. Results of qPCR revealed that significantly lower expression of EFCC1 mRNA in all lung ADC tissues compared with their matched adjacent non-tumor tissues (all P<0.05) (Figure 3A,B). Western blot showed that EFCC1 protein expression was downregulated in all lung ADC tissues compared with their matched adjacent non-tumor tissues (Figure 3C). Also, IHC showed that the EFCC1 protein localized on the cytomembrane and cytoplasm. Strong staining of EFCC1 protein was found in all normal bronchial and alveolar epithelial cells, whereas the staining degree varied significantly in tumor cells and was lower than in normal bronchial and alveolar epithelial cells (Figure 3D).
Figure 3.
Decreased expression of EFCC1 in lung ADC tissues compared with matched adjacent non-tumor tissues. (A) and (B) qPCR showed significantly lower expression of EFCC1 mRNA in lung ADC tissues than in matched adjacent non-tumor tissues. *P<0.05, **P<0.01, ***P<0.001. (C) Western blot revealed that the protein expression of EFCC1 was decreased in lung ADC tissues compared with matched adjacent non-tumor tissues. (D) Representative immunohistochemical images of EFCC1 in lung ADC and matched adjacent non-tumor tissues (normal bronchioles and alveoli). Scale bar =20 µm. EFCC1, EF-hand and coiled-coil domain containing 1; ADC, adenocarcinoma; N, normal tissue; T, tumor tissue.
Correlation between EFCC1 expression and clinicopathological characteristics in lung ADC patients with TMA
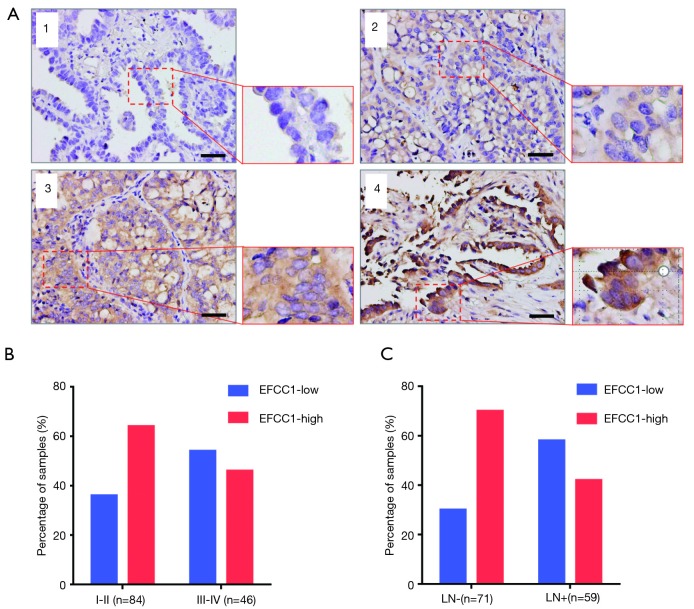
To evaluate the clinical significance of EFCC1 expression in lung ADC patients, immunohistochemical staining of 130 clinical lung ADC samples with TMA was performed, representative images of immunohistochemical staining were shown in Figure 4A. Overall, low EFCC1 expression was found in 42.3% (55/130) of samples and high EFCC1 expression found in 57.7% (75/130) of samples. The correlation between EFCC1 expression and clinicopathological characteristics were presented in Table 2. The results revealed that low EFCC1 expression was significantly associated with advanced TNM stages (stage III–IV) (P=0.040) and positive lymph node metastasis (P=0.001). For TNM stages, low EFCC1expression was found in 54.3% (25/46) of patients at advanced-stage (stage III–IV) whereas only in 35.7% (30/84) of patients at early-stage (stage I–II) (Figure 4B). And for lymph node metastasis status, 57.6% (34/59) of patients with lymph node metastasis while only 29.6% (21/71) of patients without lymph node metastasis had low EFCC1 expression (Figure 4C). Our results showed that EFCC1 expression had no significant correlation with gender, age, tumor location, tumor size, or pathological grade.
Figure 4.
EFCC1 expression in clinical lung ADC samples with TMA. (A) Representative images of different immunohistochemical staining intensities of EFCC1 in clinical lung ADC tissues. 1, negative staining; 2, weak staining; 3, moderate staining; 4, strong staining. Scale bar =20 µm. (B) The proportion of low EFCC1 expression was significantly higher in lung ADC patients at stage III–IV (54.3%) than those at stage I–II (35.7%) (P=0.040). (C) The proportion of low EFCC1 expression was significantly higher in lung ADC patients with lymph node metastasis (57.6%) than those without lymph node metastasis (29.6%) (P=0.001). EFCC1, EF-hand, and coiled-coil domain-containing; ADC, adenocarcinoma; TMA, tissue microarray; TNM, tumor-node-metastasis; LN, lymph node.
Table 2. Correlation between EFCC1 expression and clinicopathological variables in lung ADC patients with TMA.
| Clinicopathological variables | n | EFCC1 expression | χ2 value | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| All patients | 130 | 55 | 75 | ||
| Gender | 0.009 | 0.923 | |||
| Male | 75 | 32 | 43 | ||
| Female | 55 | 23 | 32 | ||
| Age (year) | 0.489 | 0.484 | |||
| ≤60 | 59 | 23 | 36 | ||
| >60 | 71 | 32 | 39 | ||
| Tumor location | 0.525 | 0.469 | |||
| Left | 52 | 20 | 32 | ||
| Right | 78 | 35 | 43 | ||
| Tumor size | 0.003 | 0.955 | |||
| ≤3 cm | 51 | 21 | 30 | ||
| >3 cm | 79 | 34 | 45 | ||
| Pathological grade | 1.028 | 0.311 | |||
| I–II | 89 | 35 | 54 | ||
| III | 41 | 20 | 21 | ||
| Lymph node metastasis | 10.387 | 0.001* | |||
| Negative | 71 | 21 | 50 | ||
| Positive | 59 | 34 | 25 | ||
| TNM stage | 4.228 | 0.040* | |||
| I–II | 84 | 30 | 54 | ||
| III–IV | 46 | 25 | 21 | ||
*P<0.05. EFCC1, EF-hand and coiled-coil domain containing 1; ADC, adenocarcinoma; TMA, tissue microarray; TNM, tumor-node-metastasis.
Decreased expression of EFCC1 predicted poor prognosis in lung ADC patients with TMA
Based on immunoreactivity scores of EFCC1 in clinical lung ADC samples with TMA, we further investigated the correlation between EFCC1 expression and survival in lung ADC patients. The Kaplan-Meier survival curve with log-rank test showed that patients with low EFCC1 expression had shorter OS time than those with high EFCC1 expression (P=0.001) (Figure 5).
Figure 5.
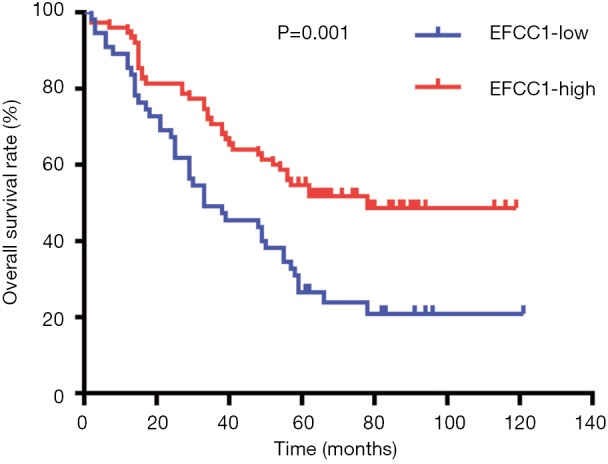
Low EFCC1 expression was associated with poor OS in lung ADC patients with TMA. EFCC1, EF-hand and coiled-coil domain containing 1; ADC, adenocarcinoma; TMA, tissue microarray; OS, overall survival.
In univariate Cox proportional hazards analysis, low EFCC1 expression was significantly associated with poor OS (HR =0.487, 95% CI: 0.321–0.759; P=0.001), besides of tumor size >3 cm (HR =1.620, 95% CI: 1.012–2.592; P=0.045), positive lymph node metastasis (HR =3.764, 95% CI: 2.360–6.003; P<0.001), and advanced TNM stages (HR =3.826, 95% CI: 2.422–6.045; P<0.001). The variables of significant difference were further analyzed by multivariate Cox proportional hazards regression models. The data showed that expression level of EFCC1 (HR =0.557, 95% CI: 0.351–0.883; P=0.013) could serve as an independent prognostic factor for lung ADC patients, like lymph node metastasis (HR =2.096, 95% CI: 1.154–3.805; P=0.015) and TNM stage (HR =2.127, 95% CI: 1.170–3.866; P=0.013). The details of univariate and multivariate Cox proportional hazards analysis were shown in Table 3.
Table 3. Univariate and multivariate Cox proportional hazards analysis of prognostic factor for lung ADC patients with TMA.
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | P value | ||
| EFCC1 expression (high vs. low) | 0.487 | 0.321–0.759 | 0.001* | 0.557 | 0.351–0.883 | 0.013* | |
| Gender (male vs. female) | 1.141 | 0.728–1.789 | 0.565 | ||||
| Age (year) (>60 vs. ≤60) | 1.446 | 0.918–2.278 | 0.112 | ||||
| Tumor location (right vs. left) | 1.098 | 0.700–1.723 | 0.684 | ||||
| Tumor size (>3 vs. ≤3 cm) | 1.620 | 1.012–2.592 | 0.045* | 1.284 | 0.779–2.116 | 0.327 | |
| Pathological grade (III vs. I–II) | 0.964 | 0.593–1.567 | 0.882 | ||||
| Lymph node metastasis (positive vs. negative) | 3.764 | 2.360–6.003 | <0.001* | 2.096 | 1.154–3.805 | 0.015* | |
| TNM stage (III–IV vs. I–II) | 3.826 | 2.422–6.045 | <0.001* | 2.127 | 1.170–3.866 | 0.013* | |
*P<0.05. ADC, adenocarcinoma; TMA, tissue microarray; CI, confidence interval; EFCC1, EF-hand and coiled-coil domain containing 1; TNM, tumor node metastasis.
Discussion
CCDC family is a group of genes coding coiled-coil domain-containing proteins, and more than 140 CCDC family members have been identified so far (16,19). Owing to the highly versatile folding motif, CCDC genes exhibit diverse functions (16). Previous studies have revealed that some CCDC genes were related to human diseases, including mitochondrial disease, diabetes, and other metabolic diseases, epigenetic disease, and cancers (16,20-23). Among cancer-related CCDC family members, some are upregulated and exhibit tumor-promoting functions, such as CCDC88A in pancreatic cancer, CCDC178 in hepatocellular carcinoma, and CCDC34 in bladder cancer and colorectal cancer (15,24-26). Some others are downregulated and exhibit tumor-suppressive activities, including CCDC6 and CCDC8 in lung cancer, and CCDC67 in gastric cancer (13,14,16). However, the functions and clinical significance of more than 60% of CCDC members, including EFCC1, remain unknown. Through differential expression analysis for RNA sequencing data in the TCGA database, we found that EFCC1 was included in genes which were significantly downregulated during lung ADC progression. This result prompts us to investigate the expression patterns and clinical significance of EFCC1 in lung ADC.
As the first study toward EFCC1, we initially analyzed EFCC1 expression and its clinical significance in TCGA data and then validated these results in clinical lung ADC samples to make reliable conclusions. In TCGA data, EFCC1 mRNA was lower in lung ADC samples than in normal lung samples. Likewise, both mRNA and protein of EFCC1 were significantly downregulated in lung ADC compared with their adjacent non-tumor counterparts in clinical tissue samples. Furthermore, EFCC1 mRNA was decreased form early-stage to advanced-stage lung ADC in the TCGA database. And low EFCC1 protein expression was associated with positive lymph node metastasis and advanced stages of lung ADC in clinical samples with TMA. All these results indicated that EFCC1 might be involved in carcinogenesis and progression of lung ADC, and EFCC1 expression level can be used to predict lymph node metastasis status and TNM stages for lung ADC patients. Also, we found that the methylation level of EFCC1 DNA was significantly higher in lung ADC samples than in normal lung samples in TCGA data. Since DNA methylation is considered a key mechanism for silencing of tumor suppressor genes in various cancers (27,28), our finding suggested that downregulation of EFCC1 in lung ADC may result from aberrant DNA methylation.
In survival analyses, the Kaplan-Meier survival curve with log-rank test showed that low EFCC1 expression was significantly associated with poor OS in lung ADC patients in both TCGA data and clinical samples. More importantly, multivariable Cox regression analysis further demonstrated that EFCC1 expression level was an independent prognostic factor for lung ADC in both sets of data. Despite specific mechanisms and involved signal pathways of EFCC1 in lung ADC need further investigations, our findings suggested that EFCC1 expression level can be served as an independent survival marker for lung ADC patients.
There are numerous genomic alterations accumulated during the pathological processes of cancers, some of them have been identified as oncogenic drivers, including KRAS, P53, PTEN, EGFR, BRAF, and ERBB2 mutation, and ALK, ROS1 and RET rearrangement (29,30). Those oncogenic drivers are important biomarkers and therapeutic targets for cancers (30). However, some other genes without mutation or rearrangement, which are only significantly upregulated or downregulated during the development of cancers, may not be the oncogenic drivers. In our study, we demonstrated these genes, such as EFCC1, could also serve as biomarkers for cancers.
Conclusions
In this study, we found aberrantly decreased expression of EFCC1 in lung ADC tissues compared with normal lung tissues. Low EFCC1 expression in lung ADC patients was significantly associated with positive lymph node metastasis and advanced TNM stages. Survival analyses indicated that low EFCC1 expression could serve as an independent factor for poor survival in lung ADC patients. Taken together, our findings suggest that EFCC1 could be a novel prognostic biomarker for lung ADC patients.
Acknowledgments
Funding: This study was supported by the National Natural Science Foundation of China (No. 81672311 to Dr. Lunxu Liu and No. 81602025 to Dr. Jiandong Mei).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the ethics committee of West China Hospital, Sichuan University (No. 2018-599).
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Patel MI, Cheng I, Gomez SL. US lung cancer trends by histologic type. Cancer 2015;121:1150-2. 10.1002/cncr.29180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rami-Porta R, Goldstraw P. Strength and weakness of the new TNM classification for lung cancer. Eur Respir J 2010;36:237-9. 10.1183/09031936.00016210 [DOI] [PubMed] [Google Scholar]
- 4.Li J, Lenferink AE, Deng Y, et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 2010;1:34. 10.1038/ncomms1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 2014;46:573-82. 10.1038/ng.2983 [DOI] [PubMed] [Google Scholar]
- 6.Gao L, Zhang H, Zhang B, et al. B3GNT3 overexpression is associated with unfavourable survival in non-small cell lung cancer. J Clin Pathol 2018;71:642-7. 10.1136/jclinpath-2017-204860 [DOI] [PubMed] [Google Scholar]
- 7.Sheng Y, Xu M, Li C, et al. Nm23-H1 is involved in the repair of ionizing radiation-induced DNA double-strand breaks in the A549 lung cancer cell line. BMC Cancer 2018;18:710. 10.1186/s12885-018-4592-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen XF, Zhang HT, Qi QY, et al. Expression of E-cadherin and nm23 is associated with the clinicopathological factors of human non-small cell lung cancer in China. Lung Cancer 2005;48:69-76. 10.1016/j.lungcan.2004.09.009 [DOI] [PubMed] [Google Scholar]
- 9.Jeon HS, Dracheva T, Yang SH, et al. SMAD6 contributes to patient survival in non-small cell lung cancer and its knockdown reestablishes TGF-beta homeostasis in lung cancer cells. Cancer Res 2008;68:9686-92. 10.1158/0008-5472.CAN-08-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Q, Cash SE, Andersen JJ, et al. CD43 in the nucleus and cytoplasm of lung cancer is a potential therapeutic target. Int J Cancer 2013;132:1761-70. 10.1002/ijc.27873 [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Hu X, Zhu Y, et al. Up-regulation of SRPK1 in non-small cell lung cancer promotes the growth and migration of cancer cells. Tumour Biol 2016;37:7287-93. 10.1007/s13277-015-4510-z [DOI] [PubMed] [Google Scholar]
- 12.McCarroll JA, Gan PP, Erlich RB, et al. TUBB3/betaIII-tubulin acts through the PTEN/AKT signaling axis to promote tumorigenesis and anoikis resistance in non-small cell lung cancer. Cancer Res 2015;75:415-25. 10.1158/0008-5472.CAN-14-2740 [DOI] [PubMed] [Google Scholar]
- 13.Morra F, Luise C, Visconti R, et al. New therapeutic perspectives in CCDC6 deficient lung cancer cells. Int J Cancer 2015;136:2146-57. 10.1002/ijc.29263 [DOI] [PubMed] [Google Scholar]
- 14.Jiang GY, Zhang XP, Zhang Y, et al. Coiled-coil domain-containing protein 8 inhibits the invasiveness and migration of non-small cell lung cancer cells. Hum Pathol 2016;56:64-73. 10.1016/j.humpath.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 15.Tanouchi A, Taniuchi K, Furihata M, et al. CCDC88A, a prognostic factor for human pancreatic cancers, promotes the motility and invasiveness of pancreatic cancer cells. J Exp Clin Cancer Res 2016;35:190. 10.1186/s13046-016-0466-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SJ, Jang HR, Kim M, et al. Epigenetic alteration of CCDC67 and its tumor suppressor function in gastric cancer. Carcinogenesis 2012;33:1494-501. 10.1093/carcin/bgs178 [DOI] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139-40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Detre S, Saclani Jotti G, Dowsett M. A. "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876-8. 10.1136/jcp.48.9.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 2001;11:82-8. 10.1016/S0962-8924(00)01898-5 [DOI] [PubMed] [Google Scholar]
- 20.Weraarpachai W, Antonicka H, Sasarman F, et al. Mutation in TACO1, encoding a translational activator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat Genet 2009;41:833-7. 10.1038/ng.390 [DOI] [PubMed] [Google Scholar]
- 21.Grassi MA, Tikhomirov A, Ramalingam S, et al. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet 2011;20:2472-81. 10.1093/hmg/ddr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi S, Fukuhara A, Taguchi T, et al. Identification of a new secretory factor, CCDC3/Favine, in adipocytes and endothelial cells. Biochem Biophys Res Commun 2010;392:29-35. 10.1016/j.bbrc.2009.12.142 [DOI] [PubMed] [Google Scholar]
- 23.Tuskan RG, Tsang S, Sun Z, et al. Real-time PCR analysis of candidate imprinted genes on mouse chromosome 11 shows balanced expression from the maternal and paternal chromosomes and strain-specific variation in expression levels. Epigenetics 2008;3:43-50. 10.4161/epi.3.1.5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Zhao Y, Wei L, et al. CCDC178 promotes hepatocellular carcinoma metastasis through modulation of anoikis. Oncogene 2017;36:4047-59. 10.1038/onc.2017.10 [DOI] [PubMed] [Google Scholar]
- 25.Gong Y, Qiu W, Ning X, et al. CCDC34 is up-regulated in bladder cancer and regulates bladder cancer cell proliferation, apoptosis and migration. Oncotarget 2015;6:25856-67. 10.18632/oncotarget.4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geng W, Liang W, Fan Y, et al. Overexpression of CCDC34 in colorectal cancer and its involvement in tumor growth, apoptosis and invasion. Mol Med Rep 2018;17:465-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer 2005;5:223-31. 10.1038/nrc1571 [DOI] [PubMed] [Google Scholar]
- 28.Lo PK, Lee JS, Liang X, et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res 2010;70:6047-58. 10.1158/0008-5472.CAN-10-1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito M, Shiraishi K, Kunitoh H, et al. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci 2016;107:713-20. 10.1111/cas.12941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015;16:e342-51. 10.1016/S1470-2045(15)00077-7 [DOI] [PubMed] [Google Scholar]