Abstract
Introduction
Herpes simplex virus type 1 (HSV1) in combination with genetic susceptibility has previously been implicated in Alzheimer's disease (AD) pathogenesis.
Methods
Plasma from 360 AD cases, obtained on average 9.6 years before diagnosis, and their age- and sex-matched controls, were analyzed for anti–HSV1 immunoglobulin (Ig) G with enzyme-linked immunosorbent assays (ELISAs). APOE genotype and nine other selected risk genes for AD were extracted from a genome-wide association study analysis by deCODE genetics, Reykjavik, Iceland.
Results
The interaction between APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) and anti–HSV1 IgG carriage increased the risk of AD (OR 4.55, P = .02). A genetic risk score based on the nine AD risk genes also interacted with anti–HSV1 IgG for the risk of developing AD (OR 2.35, P = .01).
Discussion
The present findings suggest that the APOEε4 allele and other AD genetic risk factors might potentiate the risk of HSV1-associated AD.
Keywords: Herpes simplex, HSV, Apolipoprotein E4, APOEε4, Alzheimer's disease, Dementia, Nested case-control study
1. Background
The pathogenesis of Alzheimer's disease (AD), the leading cause of major neurocognitive disorders, is still not fully understood. Sporadic AD is considered a multifactorial disease, triggered by environmental factors, in addition to genetic predisposition [1,2]. The genetic component in AD is complex, involving multiple susceptibility genes [3]; however, the ε4 allele of the apolipoprotein E gene (APOE) is the strongest genetic risk factor for AD. The APOE gene has three predominant allelic variants: APOEε2, ε3 and ε4. Possession of the APOEε4 allele increases the risk of AD with an odds ratio (OR) of 15 for homozygous carriers (APOEε4/ε4) and threefold for heterozygous carriers (APOEε2/ε4 or ε3/ε4) [4]. Although APOEε4 confers an increased risk of AD, it is not the single cause of the disease.
Large-scale genome-wide association studies (GWASs) have identified additional susceptibility genes for AD, including ATP-binding cassette subfamily A member 7 (ABCA7), bridging integrator 1 (BIN1), sialic acid binding Ig-like lectin 3 (CD33), Clusterin (CLU), complement receptor 1 (CR1), ephrin A1 (EPHA1), membrane-spanning 4-domain, subfamily A, member 4E (MS4A4E), nectin cell adhesion molecule 2 (NECTIN2), and phosphatidylinositol-binding clathrin assembly protein (PICALM). One emerging pattern is that many of these susceptibility genes associate with the complement system or immune mechanisms related to viral infections [[5], [6], [7]].
Infectious agents have been proposed as important environmental factors for AD development, and herpes simplex virus type 1 (HSV1) is the pathogen most strongly associated with AD [[8], [9], [10]]. Carriage of HSV1 is highly prevalent, reaching 80% in the adult population [11]. HSV1 DNA has been detected in the brains of patients with AD patients and colocalized with amyloid plaques in particular [[12], [13], [14], [15]]. More recently, transcriptomic studies also showed an increased abundance of HSV1 in the brains of patients with AD [16]. In epidemiological studies, both carriage of and reactivated HSV1 infection doubled the risk of developing AD [9,10,17]. Moreover, AD risk appears to decrease after treatment with antiherpetic medications in HSV1-infected patients [18]. The concept of HSV1-associated AD is further supported by the discovery that cultured cells and murine models infected with HSV1 display similar changes to AD pathology [[19], [20], [21], [22], [23]]. These observations are consistent with the finding that amyloid β has antimicrobial activity [[22], [23], [24]]. There are other suggested AD-associated pathogens, among which cytomegalovirus (CMV) [25,26], human herpes virus 6 (HHV6) [27], and Chlamydophila pneumoniae (C. pneumoniae) [28,29] are highlighted here because they have the common ability to establish latent or chronic infections within the central nervous system (CNS).
It has been hypothesized that concomitant carriage of several AD-related genes and their subsequent synergistic interactions might result in a genetic signature, which predisposes a person to HSV1-associated AD [7,30]. In postmortem studies, the combination of having APOEε4 and HSV1 in the brain was more highly associated with AD than having only one of these factors [15,31]. Previous research has connected the APOEε4 allele with HSV1 outcomes [[31], [32], [33], [34]], but no prospective epidemiological survey of AD has specifically investigated the HSV1-APOEε4 interaction. There are no studies that have examined the potential interaction between HSV1 and other AD-related genes. The aim of this study was to investigate interactions between HSV1, HSV2, CMV and C. pneumoniae; the APOEε4 allele; and nine additional AD risk genes, for the risk of subsequent AD development.
2. Methods
2.1. Participants and procedure
The nested case-control study was approved by the Regional Ethical Review Board in Umeå, Sweden (09-190M and 2017/17-31) and is based on the data from the Medical Biobank in Umeå (Northern Sweden Health and Disease Study [35]). The biobank contains plasma samples, previously donated during, for example, regular health checkups. From the biobank, samples from 360 individuals later diagnosed with AD and 360 matched controls were identified using a computerized procedure. Their plasma samples, obtained on average 9.6 years before diagnosis, were extracted for analysis. Controls were closely matched based on sampling date and age and exactly matched based on sex and cohort in the Medical Biobank.
2.2. AD diagnosis
Patients with AD were diagnosed at the Memory Clinic of the University Hospital of Northern Sweden in Umeå. The AD diagnoses were based on the criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th edition (DSM-IV) [36] and at least one brain imaging technique. The clinical diagnoses were also compatible with the NINCDS-ADRDA criteria [37]. Before final inclusion, an experienced specialist in psychogeriatric medicine verified the diagnoses. The controls were confirmed free of major neurocognitive disorder and alive at the time of diagnosis for their corresponding case. This procedure has been described extensively in a previous publication [10].
2.3. Plasma analyses
Plasma was analyzed for presence of anti–HSV IgG, anti–HSV1 and anti–HSV2 IgG, anti–CMV IgG, and anti–C. pneumoniae IgG with enzyme-linked immunosorbent assays (ELISAs). An in-house ELISA was used for the analyses of anti–HSV IgG and anti–CMV IgG as described in a previous publication [38]. Commercial ELISA kits were used to analyze anti–HSV1, anti–HSV2 (HerpeSelect 1, HerpeSelect 2, FOCUS Diagnostics), and anti–C. pneumoniae IgG (SeropCp™ Quant IgG).
To determine carriage of anti-HSV1, anti-HSV2, or anti-HSV1+anti-HSV2 IgG, each sample positive for anti–HSV IgG was further analyzed for anti–HSV2 IgG. If the sample is positive for anti–HSV2 IgG, additional analysis was performed for anti–HSV1 IgG, to separate individuals positive for anti–HSV1 IgG, anti–HSV2 IgG, or both.
2.4. Genotyping
Genotyping was performed at deCODE genetics (Reykjavik, Iceland) with Illumina genome-wide arrays (HumanOmniExpress-24). Variants with genotype yield <96 %, MAF <0.5 %, or failed Hardy-Weinberg test were excluded.
2.5. Statistical analyses
2.5.1. APOE genotype, infections, and the risk of AD
APOE genotype was classified ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4 based on single nucleotide polymorphisms (SNPs) rs429358 and rs7412. The impact of APOE genotype on AD risk was evaluated using conditional logistic regression with APOEε3/ε3 as reference. Owing to the small number (n = 3) of individuals with APOEε2/ε2, these three individuals were included in the group with APOEε2/ε3. To examine possible interactions between anti–HSV1, anti–HSV2, anti–CMV, and anti–C. pneumoniae IgG positivity, separately, and APOE genotype, conditional logistic regression for AD was repeated including the IgG positivity variables and interaction terms. For the analyses with interaction terms, APOE genotype was classified using variables for APOEε4 homozygosity (APOEε4/ε4), APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4), and APOEε2 carriage. Interactions with each of these three APOE genotype variables were analyzed separately, and significant interactions were included in the final models.
2.5.2. Selection of additional AD risk genes for a genetic risk score
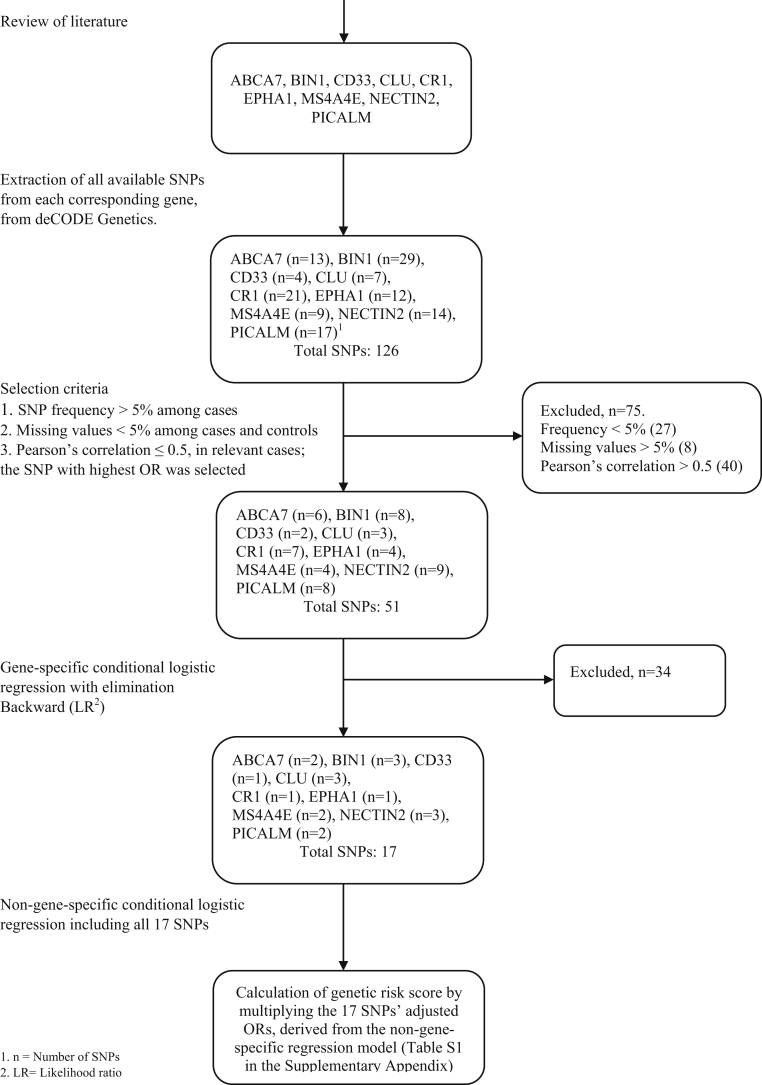
Nine different genes (ABCA7, BIN1, CD33, CLU, CR1, EPHA1, MS4A4E, NECTIN2, and PICALM) were selected after reviewing previous research [1,6,7,39]. All available SNPs from the selected genes were extracted from the genome-wide association study data files, resulting in a total of 126 SNPs (Fig. 1). Each individual SNP had 2 sets of alleles, “AA, AG, GG” or “AA, AC and CC.” The frequency of SNP variants was compared between AD cases and controls. The variants with a higher frequency among cases were given a value of 1 (“risk variant for AD”), and the variants with a lower frequency were subsequently given a value of 0 (“protective variant for AD”). Thus, every individual obtained a value of either 1 or 0 for each specific SNP. The resulting SNPs with a value of 1 or 0 were then analyzed for frequency and missing values in groups of cases and controls separately. Conditional logistic regression for each individual SNP was performed, with the outcome of AD.
Fig. 1.
Flow chart of gene selection and their corresponding single nucleotide polymorphisms.
The first selection of SNPs was made based on the following criteria: the rarest variant had to have a frequency >5% among cases, missing values should not exceed 10% among cases or controls, and finally, Pearson's correlation between two SNPs from the same gene should be <0.5 because of the risk of multicollinearity. If 2 SNPs originating from the same gene had a Pearson's correlation ≥0.5, the one with the highest OR for AD was selected. The SNPs which fulfilled the criteria for the first selection were included in a gene-specific multivariable conditional logistic regression for AD with backward elimination (likelihood ratio), that is separate models were made for each specific gene. The backward elimination (likelihood ratio) algorithm resulted in 1 to 3 SNPs per gene (Supplementary Table 1 in the Supplementary Appendix).
The final 17 SNPs identified by the gene-specific models were then integrated into the same non-gene-specific multivariable conditional logistic regression for AD and contributed to the genetic risk score (GRS). To achieve a weighted value for each individual SNP with regard to AD risk, carriers were assigned the value of the adjusted OR for AD derived from the non–gene-specific multivariable regression and value 1 for non-carriers. The GRS was calculated by the weighted value of each SNP being multiplied and then normalized. A squared normalized version of the GRS was also calculated to account for potential gene-gene interactions between different SNPs.
2.5.3. GRS of additional AD risk genes, infections and the risk of AD
Separate conditional logistic regression models were used to test the GRS and the squared GRS for the risk of AD. Analyses were then carried out to examine infection-gene interactions by including IgG positivity variables and interaction terms of GRS and squared GRS, respectively, with anti–HSV1 IgG, anti–HSV2 IgG, anti–CMV IgG, and anti–C. pneumoniae IgG positivity in separate conditional logistic models. The interaction between GRS and anti–C. pneumoniae IgG positivity for AD was tested with conditional logistic regression using GRS divided by its standard deviation instead of normalized deviation, to investigate the intercept for anti–C. pneumoniae IgG positivity at the lowest possible GRS.
To test if the interactions were independent of each other, we further specified a model including APOE genotype, GRS and anti–HSV1 IgG positivity as the main effects and two interaction terms: APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) x anti–HSV1 IgG and GRS x anti–HSV1 IgG. The same model was repeated including the squared GRS instead of the GRS.
SPSS Statistics, version 24 (IBM Corporation, Armonk, NY), was used. A two-sided P value <.05 was regarded as significant.
3. Results
Our study showed that individuals with the APOE genotypes ε2/ε3, ε3/ε4, and ε4/ε4 had ORs of 0.38 (P = .02), 3.22 (P < .001), and 19.15 (P < .001), respectively, for developing AD compared with those with APOEε3/ε3 (Table 1). Carrying APOEε2/ε4 did not significantly alter the risk of developing AD compared with carrying APOEε3/ε3 (OR 1.01, P = .99; Table 1).
Table 1.
Basic characteristics
| Group | Alzheimer's disease cases, n = 360 | Controls, n = 360 | Odds ratio for AD∗ (P value) |
|---|---|---|---|
| Age at plasma sampling, years mean ± SD | 61.2 ± 5.6 | 61.2 ± 5.6 | |
| Age at diagnosis, years mean ± SD | 70.8 ± 6.4 | ||
| Sex, female n (%) | 271 (75.3) | 271 (75.3) | |
| CT brain scans, n (%) | 309 (85.8) | ||
| MRIs of the brain, n (%) | 32 (8.9) | ||
| 99mTc SPECT/18F FDG-PET brain scans, n (%) | 172 (47.8) | ||
| Neuropsychological examinations, n (%) | 125 (34.7) | ||
| Analyses of biomarkers in cerebrospinal fluid, n (%) | 34 (9.4) | ||
| MMSE at diagnosis, mean ± SD | 21.9 ± 5.0 | ||
| APOE genotype | |||
| ε2/ε3†, n (%) | 12 (3.3) | 51 (14.2) | .38 (.02) |
| ε2/ε4, n (%) | 9 (2.5) | 13 (3.6) | 1.01 (.99) |
| ε3/ε3, n (%) | 116 (32.2) | 185 (51.4) | Ref |
| ε3/ε4, n (%) | 148 (41.1) | 74 (20.6) | 3.22 (<.001) |
| ε4/ε4, n (%) | 67 (18.6) | 6 (1.7) | 19.15 (<.001) |
| HSV1 IgG +, n (%) | 329 (91.4) | 317 (88.1) | 1.44 (.14) |
| HSV2 IgG +, n (%) | 52 (14.4) | 46 (12.8) | 1.15 (.53) |
| CMV IgG +, n (%) | 312 (86.7) | 318 (88.3) | .857 (.50) |
| C. pneumoniae IgG +, n (%) | 222 (61.7) | 220 (61.1) | 1.03 (.87) |
Abbreviations: SD, standard deviation; CT, computed tomography; MRI, magnetic resonance imaging; 99mTc SPECT/FDG-PET, technetium (99mTc) exametazime single-photon emission computed tomography/fludeoxyglucose (18F) positron emission tomography; MMSE, Mini-Mental State Examination; APOE, apolipoprotein E; HSV, herpes simplex virus; Ig, Immunoglobulin; CMV, cytomegalovirus; C. pneumoniae, Chlamydophila pneumoniae.
Simple conditional logistic regression. For APOE genotypes: with genotype APOEε3/ε3 as reference category.
APOEε2/ε2 was not analyzed separately because of the small numbers of individuals carrying this specific genotype (n = 3), and those individuals were included in the group APOEε2/ε3.
The interaction between APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) and anti–HSV1 IgG carriage was associated with increased risk of developing AD (OR 4.55, P = .02; Table 2), whereas the presence of only one factor was not (OR 0.83, P = .76 and OR 0.88 P = .73, respectively; Table 2). In the same model, the presence of the APOEε2 allele decreased the risk of AD (OR 0.36, P = .001; Table 2), and APOEε4 homozygosity (APOEε4/ε4) increased the risk (OR 20.48, P < .001; Table 2). There were no significant interactions between APOEε2 carriage and APOEε4 homozygosity (APOEε4/ε4) and anti–HSV1 IgG positivity (data not shown). There were no significant associations of anti–HSV2 IgG, anti–CMV IgG, or anti–C. pneumoniae positivity in the APOE models and no significant interactions with APOE variables (data not shown).
Table 2.
Conditional logistic regression of the interaction between APOEε4 heterozygosity/genetic risk score and herpes simplex virus type 1 carriage in the risk of Alzheimer's disease development
| Independent variables | Model 1, HSV1, and APOE variables |
Model 2, HSV1, and genetic risk score |
Model 3, HSV1, APOE variables, and genetic risk score |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Odds ratio | 95% confidence interval | P value | Odds ratio | 95% confidence interval | P value | Odds ratio | 95% confidence interval | P value | |
| Anti–HSV1 IgG+ | 0.88 | 0.43–1.85 | .73 | 1.91 | 1.09–3.37 | .03 | 1.31 | 0.56–3.03 | .53 |
| APOEε2 carriage | 0.36 | 0.19–0.67 | .001 | 0.35 | 17–0.75 | .007 | |||
| APOEε4 homozygosity (APOEε4/ε4) | 20.48 | 7.14–58.77 | <.001 | 20.80 | 5.98–72.40 | <.001 | |||
| APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) | 0.83 | 0.26–2.70 | .76 | 0.97 | 0.27–3.46 | .96 | |||
| APOEε4 heterozygosity x anti–HSV1 IgG+ | 4.55 | 1.29–16.06 | .02 | 3.75 | 0.95–14.82 | .06 | |||
| Genetic risk score | 1.42 | 0.82–2.44 | .21 | 1.26 | 0.55–2.84 | .59 | |||
| Genetic risk score x anti–HSV1 IgG+ | 2.35 | 1.21–4.56 | .01 | 2.21 | 0.87–5.63 | .10 | |||
Abbreviations: APOE, apolipoprotein E; HSV, herpes simplex virus; Ig, immunoglobulin.
17 different SNPs were identified (Fig. 1) and contributed to the GRSs (Supplementary Table 1 in the Supplementary Appendix). Missing values from the final SNPs ranged from 2.2 to 4.4% for cases and from 2.5 to 3.1% for controls and were excluded from the analyses. Thus, GRSs were calculated for 334 of the 360 AD cases and for 346 of the controls.
The GRS increased the risk of AD (OR 2.64 per standard deviation, P < .001), as did squared GRS (OR 5.61, P < .001). There was a significant interaction between the GRS and anti–HSV1 IgG positivity for the risk of developing AD (OR 2.35, P = .01; Table 2). In this model, GRS on its own did not significantly increase the risk (OR 1.42, P = .21; Table 2). The interaction between the squared GRS and anti–HSV1 IgG positivity with an outcome of AD was significant (OR 8.20, P = .005). There was also an interaction between both the GRS and squared GRS with anti–C. pneumoniae IgG for AD risk (OR 0.53, P = .04 and OR 0.13, P = .02). When using GRS divided by its standard deviation in the interaction model, anti–C. pneumoniae IgG was significantly associated with AD risk (OR 1.95 P = .04). There were no significant interactions between the GRS or the squared GRS and anti–HSV2 IgG and anti–CMV IgG positivity for AD risk (data not shown).
In the combined model with interaction terms of both APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) with anti–HSV1 IgG positivity and GRS with anti–HSV1 IgG positivity, the effect sizes of the interactions were nearly the same as models that included only one interaction term, although not significant (OR 3.75, P = .06 and OR 2.21, P = .10 respectively; Table 2). The effect sizes of the interactions between APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) and anti–HSV1 IgG positivity, and the squared GRS and anti–HSV1 IgG positivity, were also nearly the same but not significant in the combined model that included both interactions of APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) and squared GRS with anti–HSV1 IgG positivity (OR 3.85, P = .05 and OR 5.72, P = .08, respectively; Table 2).
4. Discussion
In this large nested case-control study, the APOEε4 allele and a GRS based on nine other AD-risk genes interacted with HSV1 for increased risk of developing AD. For the first time, the host genetic background can here be shown to interact with HSV1 carriage to increase the risk for developing AD in a prospective epidemiological material. The primary strengths of this study include a large number of cases with closely matched controls from the same population, combined with thorough clinical AD diagnosis.
The present results are in accordance with the recent findings of an interaction between APOEε4 and HSV for episodic memory decline [40]. Thus, an interaction between APOEε4 and HSV in AD development has been demonstrated in two large independent prospective epidemiological studies. The findings show that genetic background is important for the development of HSV1-associated AD. This corresponds with AD as a multifactorial disease, caused by genetic susceptibility in combination with environmental factors [1,2,7,16,30]. One plausible explanation could be that AD development with amyloid deposition is fueled by persistent and low-grade infection in the CNS over long periods of time. Host genetics might contribute to loss of immunological control over persistent infections, allowing the CNS entry and/or a shift from a protective (innate) immune response to neuropathological processes [16,23]. The squared GRS was associated with increased risk of AD, with a higher estimated risk effect than the non-squared GRS. This might indicate that there are gene-gene interactions between different risk genes, where concomitant carriage of many risk variants results in a genetic pattern which further increases the risk of HSV1-associated AD by multiplicative effects [7,30].
Interestingly, the GRS also interacted with anti–C. pneumoniae IgG in regard to the risk of developing AD, although the correlation had the opposite direction compared with HSV1, meaning that with a low GRS, C. pneumoniae carriage was associated with increased AD risk. This might indicate that C. pneumoniae could be contributing to AD risk in those individuals with the lowest genetic risk of HSV1-associated AD. This may also imply heterogeneity in AD pathogenesis and that the disease is multifactorial. In contrast, anti–HSV2 IgG or anti–CMV IgG did not interact with the APOEε4 allele, nor the GRS for AD risk.
For the present study, we selected nine genes consistently linked to the risk of AD from several studies for the calculation of the GRS [1,6,7,39]. This enabled investigation of their combined effect on AD. Nonetheless, this makes the contributing effects of individual genes indistinguishable from each other. This strategy was chosen because of the limited statistical power of the material. However, many other AD significant genes could also be worth investigating for their potential interactions with HSV1. The lack of desirable statistical power was also apparent when including the two interaction terms of APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) with HSV1 carriage and GRS with HSV1 carriage simultaneously in the models. While not significant, the ORs of the interactions were almost unaffected when included in the same model. Still, this could indicate that the effects of APOEε4 and other risk genes are independent of each other.
In conclusion, the interaction between APOEε4 heterozygosity (APOEε2/ε4 or ε3/ε4) and HSV1 carriage increased the risk of AD by approximately fivefold, whereas the presence of only one factor did not. A calculated GRS, based on nine additional risk genes, also interacted with anti–HSV1 IgG for increased risk of subsequent AD. The present findings suggest that the APOEε4 allele and other AD genetic risk factors might potentiate the risk of developing HSV1-associated AD. This could provide new insights into the possible mechanisms involved in the development of AD.
Research in Context.
-
1.
Systematic review: PubMed was used to search for previously published work. Alzheimer's disease (AD) is considered a multifactorial disease, triggered by genetic and environmental factors. A growing body of evidence indicates a potential role of herpes simplex virus type 1 (HSV1) in AD pathogenesis. Previous research has connected HSV1 outcomes with apolipoprotein Eε4 (APOEε4). However, no prospective epidemiological study has investigated the HSV1-APOEε4 interaction for AD risk.
-
2.
Interpretation: Our findings show that host genetic background interacts with HSV1 carriage to increase the risk of subsequent AD, consistent with our earlier findings concerning HSV and APOEε4 in episodic memory decline.
-
3.
Future directions: Interventional studies with antiviral agents are needed to prove the causal effect of herpes viruses in AD. Our results provide a foundation for future trials to target individuals carrying HSV1 in combination with certain genetic traits, thereby promoting a more individualized approach for treatment or prevention of AD.
Acknowledgments
The authors wish to express their gratitude to deCODE genetics for generously providing the genome-wide association study analyses. The study was conducted in the context of the CHANCES project, funded in the FP7 framework program of DG-RESEARCH in the European Commission. The Hellenic Health Foundation, Greece, coordinated the project. The study was further supported financially by grants from Västerbotten County Council, Knut and Alice Wallenberg Foundation, Umeå University, Kempe Foundations, Swedish Medical Association, Swedish Dementia Association, Erik and Anne-Marie Detlof foundation, Trolle-Wachtmeister Foundation, Dementia Fund in Västerbotten, Swedish Alzheimer Fund, Stohne Foundation, Bergvall Foundation and Umeå University Foundation for Medical Research.
Footnotes
Conflict of interest: The authors have no conflict of interest to report.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.trci.2019.09.014.
Supplementary data
References
- 1.Carter C.J. Susceptibility genes are enriched in those of the herpes simplex virus 1/host interactome in psychiatric and neurological disorders. Pathog Dis. 2013;69:240–261. doi: 10.1111/2049-632X.12077. [DOI] [PubMed] [Google Scholar]
- 2.Harris S.A., Harris E.A. Molecular mechanisms for herpes simplex virus type 1 pathogenesis in Alzheimer's disease. Front Aging Neurosci. 2018;10:48. doi: 10.3389/fnagi.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naj A.C., Schellenberg G.D. Genomic variants, genes, and pathways of Alzheimer's disease: an overview. Am J Med Genet B Neuropsychiatr Genet. 2017;174:5–26. doi: 10.1002/ajmg.b.32499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrer L.A., Cupples L.A., Haines J.L., Hyman B., Kukull W.A., Mayeux R. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 5.Carter C. Alzheimer's disease: APP, gamma secretase, APOE, CLU, CR1, PICALM, ABCA7, BIN1, CD2AP, CD33, EPHA1, and MS4A2, and their relationships with herpes simplex, C. Pneumoniae, other suspect pathogens, and the immune system. Int J Alzheimers Dis. 2011;2011:501862. doi: 10.4061/2011/501862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter C.J. APP, APOE, complement receptor 1, clusterin and PICALM and their involvement in the herpes simplex life cycle. Neurosci Lett. 2010;483:96–100. doi: 10.1016/j.neulet.2010.07.066. [DOI] [PubMed] [Google Scholar]
- 7.Licastro F., Carbone I., Ianni M., Porcellini E. Gene signature in Alzheimer's disease and environmental factors: the virus chronicle. J Alzheimers Dis. 2011;27:809–817. doi: 10.3233/JAD-2011-110755. [DOI] [PubMed] [Google Scholar]
- 8.Itzhaki R.F., Lathe R., Balin B.J., Ball M.J., Bearer E.L., Braak H. Microbes and Alzheimer's disease. J Alzheimers Dis. 2016;51:979–984. doi: 10.3233/JAD-160152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lovheim H., Gilthorpe J., Adolfsson R., Nilsson L.G., Elgh F. Reactivated herpes simplex infection increases the risk of Alzheimer's disease. Alzheimers Dement. 2015;11:593–599. doi: 10.1016/j.jalz.2014.04.522. [DOI] [PubMed] [Google Scholar]
- 10.Lovheim H., Gilthorpe J., Johansson A., Eriksson S., Hallmans G., Elgh F. Herpes simplex infection and the risk of Alzheimer's disease: A nested case-control study. Alzheimers Dement. 2015;11:587–592. doi: 10.1016/j.jalz.2014.07.157. [DOI] [PubMed] [Google Scholar]
- 11.Olsson J., Kok E., Adolfsson R., Lovheim H., Elgh F. Herpes virus seroepidemiology in the adult Swedish population. Immun Ageing. 2017;14:10. doi: 10.1186/s12979-017-0093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson G.A., Maitland N.J., Wilcock G.K., Craske J., Itzhaki R.F. Latent herpes simplex virus type 1 in normal and Alzheimer's disease brains. J Med Virol. 1991;33:224–227. doi: 10.1002/jmv.1890330403. [DOI] [PubMed] [Google Scholar]
- 13.Wozniak M.A., Shipley S.J., Combrinck M., Wilcock G.K., Itzhaki R.F. Productive herpes simplex virus in brain of elderly normal subjects and Alzheimer's disease patients. J Med Virol. 2005;75:300–306. doi: 10.1002/jmv.20271. [DOI] [PubMed] [Google Scholar]
- 14.Wozniak M.A., Mee A.P., Itzhaki R.F. Herpes simplex virus type 1 DNA is located within Alzheimer's disease amyloid plaques. J Pathol. 2009;217:131–138. doi: 10.1002/path.2449. [DOI] [PubMed] [Google Scholar]
- 15.Steel A.J., Eslick G.D. Herpes viruses increase the risk of Alzheimer's disease: a meta-analysis. J Alzheimers Dis. 2015;47:351–364. doi: 10.3233/JAD-140822. [DOI] [PubMed] [Google Scholar]
- 16.Readhead B., Haure-Mirande J.V., Funk C.C., Richards M.A., Shannon P., Haroutunian V. Multiscale analysis of independent Alzheimer's cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron. 2018;99:64–82.e7. doi: 10.1016/j.neuron.2018.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letenneur L., Peres K., Fleury H., Garrigue I., Barberger-Gateau P., Helmer C. Seropositivity to herpes simplex virus antibodies and risk of Alzheimer's disease: a population-based cohort study. PLoS One. 2008;3:e3637. doi: 10.1371/journal.pone.0003637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzeng N.S., Chung C.H., Lin F.H., Chiang C.P., Yeh C.B., Huang S.Y. Anti-herpetic medications and reduced risk of dementia in patients with herpes simplex virus infections-a nationwide, population-based cohort study in Taiwan. Neurotherapeutics. 2018;15:417–429. doi: 10.1007/s13311-018-0611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santana S., Recuero M., Bullido M.J., Valdivieso F., Aldudo J. Herpes simplex virus type I induces the accumulation of intracellular beta-amyloid in autophagic compartments and the inhibition of the non-amyloidogenic pathway in human neuroblastoma cells. Neurobiol Aging. 2012;33:430.e19–430.e33. doi: 10.1016/j.neurobiolaging.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 20.Wozniak M.A., Itzhaki R.F., Shipley S.J., Dobson C.B. Herpes simplex virus infection causes cellular beta-amyloid accumulation and secretase upregulation. Neurosci Lett. 2007;429:95–100. doi: 10.1016/j.neulet.2007.09.077. [DOI] [PubMed] [Google Scholar]
- 21.Piacentini R., Civitelli L., Ripoli C., Marcocci M.E., De Chiara G., Garaci E. HSV-1 promotes Ca2+ -mediated APP phosphorylation and Abeta accumulation in rat cortical neurons. Neurobiol Aging. 2011;32:2323.e13–2323.e26. doi: 10.1016/j.neurobiolaging.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Kumar D.K., Choi S.H., Washicosky K.J., Eimer W.A., Tucker S., Ghofrani J. Amyloid-beta peptide protects against microbial infection in mouse and worm models of Alzheimer's disease. Sci Transl Med. 2016;8:340ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eimer W.A., Vijaya Kumar D.K., Navalpur Shanmugam N.K., Rodriguez A.S., Mitchell T., Washicosky K.J. Alzheimer's disease-associated beta-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron. 2018;99:56–63.e3. doi: 10.1016/j.neuron.2018.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soscia S.J., Kirby J.E., Washicosky K.J., Tucker S.M., Ingelsson M., Hyman B. The Alzheimer's disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One. 2010;5:e9505. doi: 10.1371/journal.pone.0009505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lurain N.S., Hanson B.A., Martinson J., Leurgans S.E., Landay A.L., Bennett D.A. Virological and immunological characteristics of human cytomegalovirus infection associated with Alzheimer disease. J Infect Dis. 2013;208:564–572. doi: 10.1093/infdis/jit210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovheim H., Olsson J., Weidung B., Johansson A., Eriksson S., Hallmans G. Interaction between cytomegalovirus and herpes simplex virus type 1 associated with the risk of Alzheimer's disease development. J Alzheimers Dis. 2018;61:939–945. doi: 10.3233/JAD-161305. [DOI] [PubMed] [Google Scholar]
- 27.Carbone I., Lazzarotto T., Ianni M., Porcellini E., Forti P., Masliah E. Herpes virus in Alzheimer's disease: relation to progression of the disease. Neurobiol Aging. 2014;35:122–129. doi: 10.1016/j.neurobiolaging.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Balin B.J., Gerard H.C., Arking E.J., Appelt D.M., Branigan P.J., Abrams J.T. Identification and localization of Chlamydia pneumoniae in the Alzheimer's brain. Med Microbiol Immunol. 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- 29.Little C.S., Joyce T.A., Hammond C.J., Matta H., Cahn D., Appelt D.M. Detection of bacterial antigens and Alzheimer's disease-like pathology in the central nervous system of BALB/c mice following intranasal infection with a laboratory isolate of Chlamydia pneumoniae. Front Aging Neurosci. 2014;6:304. doi: 10.3389/fnagi.2014.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porcellini E., Carbone I., Ianni M., Licastro F. Alzheimer's disease gene signature says: beware of brain viral infections. Immun Ageing. 2010;7:16. doi: 10.1186/1742-4933-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itzhaki R.F., Lin W.R., Shang D., Wilcock G.K., Faragher B., Jamieson G.A. Herpes simplex virus type 1 in brain and risk of Alzheimer's disease. Lancet. 1997;349:241–244. doi: 10.1016/S0140-6736(96)10149-5. [DOI] [PubMed] [Google Scholar]
- 32.Koelle D.M., Magaret A., Warren T., Schellenberg G.D., Wald A. APOE genotype is associated with oral herpetic lesions but not genital or oral herpes simplex virus shedding. Sex Transm Infect. 2010;86:202–206. doi: 10.1136/sti.2009.039735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgos J.S., Ramirez C., Sastre I., Bullido M.J., Valdivieso F. ApoE4 is more efficient than E3 in brain access by herpes simplex virus type 1. Neuroreport. 2003;14:1825–1827. doi: 10.1097/00001756-200310060-00013. [DOI] [PubMed] [Google Scholar]
- 34.Burgos J.S., Ramirez C., Sastre I., Valdivieso F. Effect of apolipoprotein E on the cerebral load of latent herpes simplex virus type 1 DNA. J Virol. 2006;80:5383–5387. doi: 10.1128/JVI.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hallmans G., Agren A., Johansson G., Johansson A., Stegmayr B., Jansson J.H. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort–evaluation of risk factors and their interactions. Scand J Public Health Suppl. 2003;61:18–24. doi: 10.1080/14034950310001432. [DOI] [PubMed] [Google Scholar]
- 36.American Psychiatric Association . 4th ed. American Psychiatric Association; Washington, DC: 2000. Diagnostic and Statistical Manual of Mental Disorders. Text Revision. [Google Scholar]
- 37.McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 38.Sjostrom S., Hjalmars U., Juto P., Wadell G., Hallmans G., Tjonneland A. Human immunoglobulin G levels of viruses and associated glioma risk. Cancer Causes Control. 2011;22:1259–1266. doi: 10.1007/s10552-011-9799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.http://www.alzgene.org Available at:
- 40.Lovheim H., Norman T., Weidung B., Olsson J., Josefsson M., Adolfsson R. Herpes simplex virus, APOEvarepsilon4, and cognitive decline in old age: results from the Betula Cohort Study. J Alzheimers Dis. 2019;67:211–220. doi: 10.3233/JAD-171162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.