Abstract
Background
Resistance to thyroid hormone beta (RTHβ) is characterized by elevated thyroid hormone and unsuppressed thyroid-stimulating hormone (TSH), caused by thyroid hormone receptor beta gene (THRB) defects. Most mutations producing RTHβ phenotype are located in CG-rich regions of THRB, encoding the T3-binding and hinge domains of the receptor. However, a region encompassing codons 384–425 is virtually devoid of RTHβ-causing mutations, termed “cold region.”
Case
A 49-year-old woman was diagnosed with Hashimoto thyroiditis in her twenties, and levothyroxine (LT4) was initiated. During LT4 treatment she had slightly elevated free thyroxine and TSH levels, suggesting the possibility of RTHβ.
Results
Sequencing of THRB identified a heterozygous missense variant c.1154G>A producing p.G385E in the proband. Since this variant of unknown significance (VUS) has not been reported in RTHβ individuals and considering its location in the “cold region” of THRB, we questioned its relevance. In silico functional prediction algorithms showed conflicting results: PolyPhen-2 predicted this VUS to be probably damaging with a score of 1.000, while SIFT predicted it to be tolerated with a score of 0.07, thus making additional investigations necessary. Genotyping of family members revealed that the proband's mother and sister, without RTHβ phenotype, also harbored the same variant. This indicates that the THRB G385E variant is unlikely to manifest RTHβ phenotype and confirms its “cold” status.
Conclusions
This study illustrates that assigning causality of a THRB VUS for RTHβ based only on in silico prediction algorithms is not always fully reliable. Additional phenotype-genotype segregation in family members can assist in predicting functional consequences of missense mutations.
Keywords: Thyroid hormone receptor beta gene, Cold region, Resistance to thyroid hormone beta
What Is Known about This Topic?
Mutations in the thyroid hormone receptor beta gene (THRB) in the region encoding the T3-binding and the adjacent hinge domains of the receptor have been reported to cause resistance to thyroid hormone beta (RTHβ).
A region encompassing codons 384–425 of THRB is virtually devoid of RTHβ-causing mutations and was termed “cold region.”
What Does This Case Report Add?
Although the G385E variant was identified in an individual in whom the diagnosis of RTHβ was suspected, the lack of genotype-phenotype correlation in family members suggests that this variant is unlikely to produce RTHβ phenotype and assigns putative “cold” status.
This report highlights the importance of phenotype-genotype segregation in family members, in addition to the in silico prediction algorithms, in assessing functional relevance of a variant of unknown significance in THRB as a cause of RTHβ.
Introduction
Resistance to thyroid hormone beta (RTHβ) is caused by mutations in thyroid hormone receptor beta gene (THRB) leading to impaired tissue responsiveness to thyroid hormone. It is characterized by elevated thyroid hormone levels and unsuppressed thyroid-stimulating hormone (TSH). As with other genetic conditions, the majority of mutations causing RTHβ occur in CG-rich regions or CpG hot spots. THRB mutations are located in the T3-binding and the adjacent hinge domains of the thyroid hormone receptor [1, 2, 3]. Yet, there is a “cold region” that is virtually devoid of mutations, even though it contains CG-rich regions. This region was formerly believed to encompass codons 348–437 of THRB [4]. In an in vitro study, artificial missense mutations in CpG dinucleotides located in this region produced no or minimal impairment in T3 binding and had no or weak dominant negative effect, suggesting that their presence is unlikely to be identified because they would fail to produce abnormalities typical of RTHβ [4]. However, as more mutations were identified in individuals with RTHβ [2, 3], including mutations in the codons 383 [5, 6] and 426 [7], this region has been narrowed to codons 384–425. In this report, a variant located in the cold region of THRB was identified in a woman in whom the diagnosis of RTHβ was suspected. Whether this variant has clinical significance is an important question to address, especially since its location is in the so-called “cold region.” Therefore, further investigations to assess the clinical significance of this variant were performed.
Case Presentation
A 49-year-old woman was diagnosed as having Hashimoto thyroiditis in her twenties, and levothyroxine (LT4) treatment was initiated. Information regarding her thyroid function tests (TFTs) prior to starting LT4 treatment is not available. Thyroid ultrasonography showed diffusely atrophic gland with heterogeneous architecture consistent with Hashimoto thyroiditis. During LT4 treatment, she had normal to slightly elevated free thyroxine (FT4) and TSH levels and presented a mix of hypothyroid and hyperthyroid symptoms. TFT results while she was taking 137 µg of LT4 daily are shown in Figure 1. The proband had elevated serum total thyroxine (TT4), FT4 index (FT4I), total reverse triiodothyronine (TrT3), and TSH levels while serum total triiodothyronine (TT3) was normal. Positive thyroid peroxidase antibodies indicated the presence of autoimmune thyroid disease (AITD). An assay using polyethylene glycol precipitation to detect the presence of antibodies to T4 was performed and revealed that there was no excess binding of T4 to serum IgG, thus T4 antibodies were negative. Testing for heterophile antibodies (Mayo Clinic assay) was negative. While pituitary MRI showed a 3-mm lesion, possibly a small microadenoma, alpha subunit was low at 0.3 ng/mL, ruling out the possibility of a TSHoma. Further, high-dose liothyronine treatment of 100 μg for 5 days resulted in TSH suppression from 1.98 to 0.05 mIU/L. As RTHβ could not be ruled out, THRB sequencing was performed.
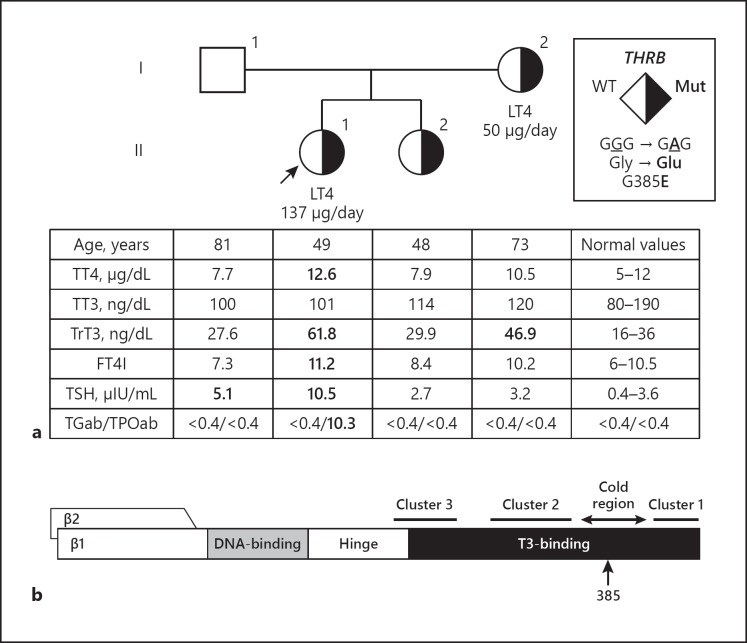
Fig. 1.
a Pedigree of the family with TFTs and THRB variant. TFTs are aligned below each symbol representing a family member. Abnormal values are shown in bold. The proband is indicated with an arrow. Square symbols indicate males, circles females. Roman numerals to the left of the pedigree indicate the generation and Arabic numerals to the right of each symbol individual family members. Half-filled symbols indicate subjects heterozygous for the novel THRB mutation, as indicated in the legend. b Schematic representation of the thyroid hormone receptor beta (TRβ) and its functional domains, showing the location of amino acid 385 within the cold region. FT4I, free thyroxine index; LT4, levothyroxine; TFTs, thyroid function tests; TGab, thyroglobulin antibodies; THRB, thyroid hormone receptor beta gene; TPOab, thyroperoxidase antibodies; TrT3, total reverse triiodothyronine; TSH, thyroid-stimulating hormone; TT3, total triiodothyronine; TT4, total thyroxine; WT, wild type.
Sequencing of the proband's THRB by Quest Diagnostics identified a heterozygous missense variant of unknown significance (VUS) c.1154G>A in exon 10 (transcript ID: ENST00000396671.7) that results in the substitution of the normal amino acid glycine at the position 385 with a glutamic acid (p. Gly385Glu; G385E). This variant has not been reported in individuals with RTHβ or in the gnomAD [8] database, but has been reported in ClinVar. Two commonly used in silico functional prediction algorithms showed conflicting results with PolyPhen-2 [9] predicting this VUS to be probably damaging with a score of 1.000, while SIFT [10] predicted it to be tolerated with a score of 0.07. Although such algorithms cannot fully capture the consequences of each VUS for the function of the protein and are not always in agreement in their prediction, they are the first line of analysis of novel variants. When their functional prediction is not similar, one cannot conclude on the relevance of the VUS. Therefore, considering that it is located in the “cold region” of THRB, we questioned whether this variant was related to the RTHβ phenotype and responsible for the proband's TFT abnormalities.
Blood samples were obtained from the proband's family members living in India, and TFTs as well as genotyping of all family members of the proband were performed. TFT results from her parents and sister were not consistent with the RTHβ phenotype (Fig. 1). The proband's father had only a slightly elevated TSH level, a condition which can occur with aging and could be considered as normal [11]. The proband's mother was treated with 50 µg of LT4 daily. She had negative thyroid antibodies and had only a slightly elevated TrT3 level. Information regarding the diagnosis and TFTs prior to treatment of the proband's mother could not be obtained. The proband's sister was not taking thyroid medication and her TFTs were normal. Sequencing of exon 10 of THRB of all family members revealed that the proband's mother and sister also harbored the heterozygous G385E variant, while the proband's father harbored the wild-type THRB (Fig. 1).
Follow-up of the proband revealed that an increase of the LT4 dose from 137 to 150 µg daily increased her FT4I to 13.8 and normalized TSH to 0.6 µIU/mL, while her TT3 remained in the normal range, at 129 ng/dL. The high total and free serum T4 with clearly normal T3 may represent irregular intake of LT4 or increased inactivation of T4 (see high rT3) seen in some individuals on LT4 replacement.
Although genetic defects in the deiodinases are not known, we also considered the possibility of a thyroid hormone metabolism defect in which a deficiency in one of the 5′-deiodinases (D) could result in this TFT pattern. Sequencing of D1 was negative, and for D2 the proband did not harbor the variant Thr92Ala previously shown to correlate with a higher LT4 dose/kg needed to normalize TSH in homozygous carriers [12].
Discussion and Conclusions
The G385E variant is located in a CG-rich area (CGCCCGGGGC) prone to mutations [1], but in the “cold region” of THRB. The cold region was formerly believed to encompass codons 348–437 of THRB. A previous in vitro study predicted that mutations in this region would fail detection by not giving rise to TFT abnormalities [4]. Indeed, artificial missense mutations created according to the hot spot mutagenesis rule (C to T and G to A substitutions in CpGs) showed either normal affinity to T3 (V376I, P384L, R410Q, V414M, R429Q, R429W) or affinity reduced to a level of 60–80% (A352T, R383C, R383H). In addition, all showed no or weak dominant negative effect, which could be due to the fact that this region appears to play an important role in the dimerization of the receptor [4].
More mutations were later identified in individuals with RTHβ [2, 3]. The R383H mutation, which was shown to have a T3-binding affinity 70% that of wild type [4], was identified in individuals with RTHβ [5, 6]. In vitro functional study demonstrated that R383H impairs corepressor release and transcriptional activity of negatively regulated genes [5]. In addition, a de novo T426I mutation was reported in an individual with RTHβ and facial dysmorphism [7]. Therefore, the cold region, which is devoid of reported cases of RTHβ, has been narrowed to a 42-amino-acid-long region encompassing codons 384–425. At the population level, 12 variants in this region (P384L, G385R, L386V, E390D, R391K, D397G, S398G, N408S, H413D, V414M, K420R, and V425L) were identified in 26 alleles among ∼251,400 alleles in the gnomAD database [8]. No information about the clinical correlation with the RTHβ phenotype is available in this large database. The majority of these variants were predicted by in silico tools to be benign, except for G385R, which is predicted to be probably damaging, and P384L, D397G, and K420R, which have conflicting predictions among algorithms.
The G385E variant in the cold region of THRB identified in this study has not been reported in individuals with RTHβ. Two in silico prediction tools reported conflicting results regarding the functional effect of the mutation. Thus, it was important to identify whether this variant has a clinical significance related to RTHβ phenotype or not. In order to do that, genotype-phenotype correlations in the family members were evaluated.
Considering that the G385E THRB variant was found not only in the proband but also in two relatives without the RTHβ phenotype, this variant is unlikely to be responsible for the proband's TFT abnormalities. As detailed in the results, antibodies to T4 were negative, the possibility of a TSHoma was ruled out, and sequencing abnormalities in the 5′-deiodinases were not found. Thus, the findings of elevated serum TT4, FT4I, and TrT3 levels with normal TT3 level were likely attributable to LT4 treatment for her AITD. A study in athyreotic patients with normal serum TSH levels on LT4 treatment showed higher serum FT4 and lower FT3 levels than in untreated euthyroid controls [13]. Furthermore, 7.2% of patients treated with LT4 had serum FT4 levels above the upper limit of normal [13]. Finally, we cannot exclude that the THRB variant might have some effect on the proband, but not on the mother and sister harboring the same mutation.
The importance of thorough investigations prior to the assignment of a VUS as a cause for the RTHβ phenotype was also demonstrated in a previous study, in which a THRB variant was incidentally identified in a family with AITD [14]. In that study, sequencing of THRB was performed in the proband and all family members because the proband was, at that time, incorrectly diagnosed with RTHβ. The G339S variant was present in three relatives who had normal TFTs, but was not found in the proband, confirming that the G339S variant was unlikely to manifest the RTHβ phenotype.
In conclusion, we report a novel VUS in the cold region of THRB identified in an individual who was suspected to have RTHβ. Genotyping the family members revealed that not only the proband, but also her mother and sister, who did not have the RTHβ phenotype, harbored the same variant. This finding suggested that the G385E variant in THRB is unlikely to cause RTHβ. This study illustrates that the assignment of a VUS in THRB as a cause of RTHβ in an individual based only on in silico prediction algorithms is not always fully reliable. Additional phenotype-genotype segregation in family members can assist in predicting functional consequences of missense mutations. In the future, more sophisticated computational tools taking into considerations protein-specific functional domains might be able to accurately predict the clinical significance of missense VUS.
Statement of Ethics
Signed informed consent approved by the institutional review board was obtained from all individuals.
Disclosure Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported in part by grants R01DK15070 and R01DK110322 from the National Institutes of Health to S. Refetoff and A.M. Dumitrescu, respectively. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
Author Contributions
M. Korwutthikulrangsri performed the genetic analysis of the patient and was the first author in writing the manuscript. C. Dosiou examined the patient and contributed to the acquisition of the clinical information. A.M. Dumitrescu and S. Refetoff contributed to the conception of the study. All authors revised and approved the final manuscript.
References
- 1.Weiss RE, Weinberg M, Refetoff S. Identical mutations in unrelated families with generalized resistance to thyroid hormone occur in cytosine-guanine-rich areas of the thyroid hormone receptor beta gene. Analysis of 15 families. J Clin Invest. 1993 Jun;91((6)):2408–15. doi: 10.1172/JCI116474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumitrescu AM, Refetoff S. The syndromes of reduced sensitivity to thyroid hormone. Biochim Biophys Acta. 2013 Jul;1830((7)):3987–4003. doi: 10.1016/j.bbagen.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pappa T, Refetoff S. Human genetics of thyroid hormone receptor beta: resistance to thyroid hormone beta (RTHβ) Methods Mol Biol. 2018;1801:225–40. doi: 10.1007/978-1-4939-7902-8_18. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi Y, Sunthornthepvarakul T, Refetoff S. Mutations of CpG dinucleotides located in the triiodothyronine (T3)-binding domain of the thyroid hormone receptor (TR) beta gene that appears to be devoid of natural mutations may not be detected because they are unlikely to produce the clinical phenotype of resistance to thyroid hormone. J Clin Invest. 1994 Aug;94((2)):607–15. doi: 10.1172/JCI117376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clifton-Bligh RJ, de Zegher F, Wagner RL, Collingwood TN, Francois I, Van Helvoirt M, et al. A novel TR beta mutation (R383H) in resistance to thyroid hormone syndrome predominantly impairs corepressor release and negative transcriptional regulation. Mol Endocrinol. 1998 May;12((5)):609–21. doi: 10.1210/mend.12.5.0113. [DOI] [PubMed] [Google Scholar]
- 6.Safer JD, O'Connor MG, Colan SD, Srinivasan S, Tollin SR, Wondisford FE. The thyroid hormone receptor-beta gene mutation R383H is associated with isolated central resistance to thyroid hormone. J Clin Endocrinol Metab. 1999 Sep;84((9)):3099–109. doi: 10.1210/jcem.84.9.5985. [DOI] [PubMed] [Google Scholar]
- 7.Menzaghi C, Di Paola R, Corrias A, Einaudi S, Trischitta V, De Sanctis C, et al. T426I a new mutation in the thyroid hormone receptor beta gene in a sporadic patient with resistance to thyroid hormone and dysmorphism. Mutations in brief no. 192. Online. Hum Mutat. 1998;12((4)):289. [PubMed] [Google Scholar]
- 8.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016 Aug;536((7616)):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7((4)):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012 Jul;40((Web Server issue)):W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Surks MI, Boucai L. Age- and race-based serum thyrotropin reference limits. J Clin Endocrinol Metab. 2010 Feb;95((2)):496–502. doi: 10.1210/jc.2009-1845. [DOI] [PubMed] [Google Scholar]
- 12.Torlontano M, Durante C, Torrente I, Crocetti U, Augello G, Ronga G, et al. Type 2 deiodinase polymorphism (threonine 92 alanine) predicts L-thyroxine dose to achieve target thyrotropin levels in thyroidectomized patients. J Clin Endocrinol Metab. 2008 Mar;93((3)):910–3. doi: 10.1210/jc.2007-1067. [DOI] [PubMed] [Google Scholar]
- 13.Gullo D, Latina A, Frasca F, Le Moli R, Pellegriti G, Vigneri R. Levothyroxine monotherapy cannot guarantee euthyroidism in all athyreotic patients. PLoS One. 2011;6((8)):e22552. doi: 10.1371/journal.pone.0022552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen CC, Dumitrescu A, Guerra-Argüero LM, Gállego-Suárez C, Vazquez-Mellado A, Vinogradova M, et al. Incidental identification of a thyroid hormone receptor beta (THRB) gene variant in a family with autoimmune thyroid disease. Thyroid. 2013 Dec;23((12)):1638–43. doi: 10.1089/thy.2013.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]