Abstract
Gene regulatory networks and their evolution are important in the study of animal development. In the nematode, Caenorhabditis elegans, the endoderm (gut) is generated from a single embryonic precursor, E. Gut is specified by the maternal factor SKN-1, which activates the MED → END-1,3 → ELT-2,7 cascade of GATA transcription factors. In this work, genome sequences from over two dozen species within the Caenorhabditis genus are used to identify MED and END-1,3 orthologs. Predictions are validated by comparison of gene structure, protein conservation, and putative cis-regulatory sites. All three factors occur together, but only within the Elegans supergroup, suggesting they originated at its base. The MED factors are the most diverse and exhibit an unexpectedly extensive gene amplification. In contrast, the highly conserved END-1 orthologs are unique in nearly all species and share extended regions of conservation. The END-1,3 proteins share a region upstream of their zinc finger and an unusual amino-terminal poly-serine domain exhibiting high codon bias. Compared with END-1, the END-3 proteins are otherwise less conserved as a group and are typically found as paralogous duplicates. Hence, all three factors are under different evolutionary constraints. Promoter comparisons identify motifs that suggest the SKN-1, MED, and END factors function in a similar gut specification network across the Elegans supergroup that has been conserved for tens of millions of years. A model is proposed to account for the rapid origin of this essential kernel in the gut specification network, by the upstream intercalation of duplicate genes into a simpler ancestral network.
Keywords: GATA factors, cell fate specification, gene regulatory network, developmental system drift, Caenorhabditis
Central to the development of a metazoan is the activation of tissue-specific gene regulatory networks (GRNs) that drive subdivision of progenitors and emergence of features of terminal differentiation (Davidson 2010). On evolutionary time scales, changes in such networks drive appearance of novel features, but these changes can also occur without changes in morphology or development (Peter and Davidson 2016). Such differences in GRNs that nonetheless drive homologous developmental processes exemplify Developmental System Drift (DSD) (True and Haag 2001). In the nematode genus Caenorhabditis, which includes the well-studied species C. elegans, examples of DSD include the gene networks that produce the derived character of hermaphroditism, which evolved at least three independent times in the genus, and vulval development (Haag et al. 2018; Félix 2007; Ellis and Lin 2014).
A relatively understudied area in Caenorhabditis is the evolutionary dynamics of GRNs that drive embryonic development. One reason may be that the close relatives to C. elegans exhibit indistinguishable embryogenesis, differing perhaps by the timing of some developmental milestones (Memar et al. 2019; Zhao et al. 2008; Levin et al. 2012). Another reason for the paucity of evo-devo studies in embryogenesis is that the dissection of a GRN requires cause-and-effect associations to be probed through experimental perturbations (Davidson et al. 2002). The powerful tools of forward and reverse genetics in C. elegans have only recently become available in related species, most notably C. briggsae, which like C. elegans is hermaphroditic and supports RNA-mediated interference (Zhao et al. 2010). A third, and more important limitation, is that very few embryonic GRNs are known at high resolution in C. elegans that could serve as a comparison.
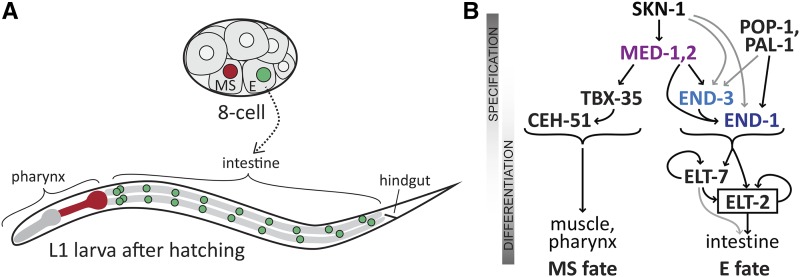
The gene regulatory network that specifies the C. elegans endoderm is an example of a set of interacting transcription factors that has been studied in great detail (Maduro 2017). In the early embryo, the founder cells E and MS are born (Figure 1A). The E cell generates the entire endoderm (intestine), while its sister cell MS generates many mesodermal cell types, including the part of the pharynx, and many body muscle cells (Sulston et al. 1983). Many components of the GRN underlying MS and E development are known with high precision, and in most of cases, regulatory inputs have been confirmed to be direct and cis-regulatory sites have even been identified in upstream regions (Maduro et al. 2001; Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2005; Wiesenfahrt et al. 2015; Du et al. 2016). This network is therefore a highly suitable system in which to examine questions of GRN evolution and developmental system drift.
Figure 1.
Embryonic origin of the E blastomere and simplified diagram of the gene regulatory network for endomesoderm specification in C. elegans. (A) The E cell and its sister cell MS are found ventrally in the 8-cell embryo (approximately 50 μm long). MS generates mesodermal cells including body muscles and the posterior portion of the pharynx, shown in red on the diagram of the larva (approximately 200 μm long). E generates the 20 cells of the intestine, whose nuclei are shown in green on the larva. (B) Specification of MS and E fates begins with the same SKN-1 and MED-1,2 factors, but then bifurcates into an MS pathway that includes the T-box factor TBX-35 and the homeobox factor CEH-51, while endoderm specification involves activation of END-3 and END-1. These upstream transient factors ultimately activate ELT-2 (and its paralogue ELT-7) which maintain intestinal fate. Additional input into E specification occurs by input from TCF/POP-1 and Caudal/PAL-1. All of MED-1,2, END-1,3 and ELT-2,7 are GATA type transcription factors. Arrows indicate transcriptional activation of the gene encoding a downstream factor.
The endomesoderm specification network works as follows. A simplified diagram is shown in Figure 1B. Specification of both MS and E begins with accumulation of maternal SKN-1 protein. SKN-1 is an unusual transcription factor that binds DNA as a monomer through a Skn domain consisting of a homeodomain-like amino half recognizing an A/T-rich sequence, and a bZIP-like carboxyl basic domain recognizing a TCAT sequence (Pal et al. 1997; Carroll et al. 1997; Blackwell et al. 1994; Lo et al. 1998). SKN-1 directly activates expression of med-1 and med-2, which encode nearly identical divergent GATA-type transcription factors that recognize an atypical AGTATAC core site (Broitman-Maduro et al. 2005; Lowry et al. 2009). SKN-1 and MED-1,2 are important for specification of both MS and E, as loss of activity of these genes results in a penetrant failure to specify MS, and an incompletely penetrant failure to specify E (Bowerman et al. 1992; Maduro et al. 2001). In MS, the MEDs specify mesodermal fate in part through activation of tbx-35 (Broitman-Maduro et al. 2006). In E, SKN-1 and MED-1,2 contribute to activation of the paralogous end-1 and end-3 genes. These encode similar GATA factors that are expressed in the early E lineage, with end-3 being activated slightly earlier than end-1 (Maduro et al. 2005a; Maduro et al. 2002; Zhu et al. 1997; Baugh et al. 2003). In turn, the END-3 and END-1 proteins activate elt-2, a GATA factor that sets and maintains, through positive autoregulation, the fate of intestinal cells and is the central regulator for all intestinal genes (McGhee et al. 2009; Fukushige et al. 1998; Fukushige et al. 1999). The elt-7 gene encodes a similar GATA factor that shares function and expression with elt-2, but which itself is not essential for normal development (Sommermann et al. 2010; Dineen et al. 2018). All of END-1, END-3, ELT-2 and ELT-7 have similar DNA-binding properties and interact with canonical GATA binding sites of the type HGATAR (Wiesenfahrt et al. 2015; Du et al. 2016). Many additional studies have revealed unexpected nuance and complexity to the myriad of factors in this network, confirming that the sum of upstream inputs into elt-2 activation is not merely additive. Upstream factors have distinguishable roles in establishment of robust cell divisions, gut morphogenesis and activation of genes important for metabolic function of the intestine (Dineen et al. 2018; Maduro et al. 2015; Boeck et al. 2011; Choi et al. 2017; Sawyer et al. 2011).
Integrated with the SKN-1 → MED-1,2 → END-1,3 feed-forward regulatory chain is the Wnt/β-catenin asymmetry pathway, which acts in the asymmetric MS vs. E fate decision through the nuclear effector TCF/POP-1 (Lin et al. 1995; Maduro et al. 2002; Owraghi et al. 2010; Rocheleau et al. 1997; Shetty et al. 2005; Thorpe et al. 1997). In MS, POP-1 represses gut fate by preventing activation of end-1 and end-3, while in E, POP-1 is an activator that contributes to activation of end-1 through its association with a divergent β-catenin, SYS-1 (Maduro et al. 2005b; Shetty et al. 2005). The POP-1 contribution to gut specification is not the major regulatory input, however, because loss of pop-1 still results in endoderm specification from E (Lin et al. 1995). The contribution of POP-1 is detectable when depletion of pop-1 is combined with loss of skn-1, med-1,2 (together) or end-3, which produces loss of gut specification in a majority of embryos (Maduro et al. 2005a; Maduro et al. 2005b; Shetty et al. 2005; Maduro et al. 2007; Maduro et al. 2015; Owraghi et al. 2010). An additional minor input into gut specification in C. elegans is through maternally provided PAL-1 protein, a Caudal-like factor whose primary role is specification of a different blastomere called C (Hunter and Kenyon 1996; Maduro et al. 2005b).
A small number of studies have investigated the evolutionary dynamics of gut specification in species closely related to C. elegans. In C. briggsae, the end-1 and end-3 orthologs (the latter of which is found as two nearby paralogues, end-3.1 and end-3.2) are expressed in the early E lineage, and simultaneous knockdown of C. briggsae end-1, end-3.1 and end-3.2 by RNAi results in a failure to specify gut (Lin et al. 2009; Maduro et al. 2005a). In C. briggsae and C. remanei, most orthologs of the med genes, when introduced individually as high-copy transgenes, can fully complement the embryonic lethality of C. elegans med-1,2(-) embryos (Coroian et al. 2006). Together these studies suggest that the med and end factors play similar roles in all three species, as might be expected. Somewhat unexpectedly, however, knockdown of skn-1 and pop-1 orthologs in C. briggsae was found to produce different phenotypes from C. elegans, suggesting that the way that SKN-1 and POP-1 interact with their downstream target genes is subject to evolutionary changes even among very closely related species, i.e., the hallmark of developmental system drift (Lin et al. 2009; Zhao et al. 2010). From these few studies, then, a model emerges of a core endoderm specification pathway, where some regulatory inputs into the pathway are subject to more rapid evolutionary change than others.
An important way that properties of a GRN can be studied on an evolutionary scale is to examine features of orthologous genes in related species (Peter and Davidson 2016). However, given the essential requirement for the gut specification network in C. elegans, a paradox became apparent when genome sequences outside of the genus were completed: No med or end orthologs could be identified in the related nematode Pristionchus pacificus, while putative orthologs of elt-2 and skn-1 can be found in Pristionchus and in even more divergent species (data not shown) (Dieterich et al. 2008; Schiffer et al. 2014; Couthier et al. 2004). In recent years, however, the number of known species within the Caenorhabditis genus has grown considerably, opening possibilities for studying evolution of development through sequence comparisons (Kiontke et al. 2011). In the past two years, new sequence assemblies have become available for over two dozen Caenorhabditis genomes both within and outside of the so-called “Elegans supergroup” of species that are most closely related to C. elegans (Félix et al. 2014; Stevens et al. 2019). Collectively, this powerful set of sequences captures tens of millions of years of genome evolution (Stein et al. 2003; Cutter 2008).
In this work, I have used a primarily in silico approach to identify orthologs of the med, end-3 and end-1 genes among the Caenorhabditis genome sequence assemblies (Haag and Thomas 2015). Patterns of conservation of gene structure, protein structure and putative cis-regulatory sites are revealed in the med and end genes that confirm known information from C. elegans and reveal new insights into the MED and END proteins and the evolutionary dynamics of the network. The results complement studies that identify genome-wide conserved putative cis-regulatory motifs among close relatives of C. elegans (Zhao et al. 2012; Siepel et al. 2005; Grishkevich et al. 2011). A surprising finding is that the endoderm network likely originated at the base of the Elegans supergroup, in a manner that can be hypothesized to have resulted from the rapid serial intercalation of successive duplications of an ancestral GATA factor, likely elt-2. Other unexpected findings are that the MED, END-3 and END-1 proteins are evolving at different rates, and that END-1 contains previously unrecognized, highly conserved domains that distinguish it from END-3. The resulting suite of MED/END-3/END-1 factors from 20 species forms a starting point for future studies on GRN evolution in Caenorhabditis.
Materials and Methods
Identification of putative med and end orthologs
Sequence scaffolds and predicted proteins were downloaded from the Caenorhabditis Genomes Project (CGP) website (http://download.caenorhabditis.org) in late 2017. Searches were performed using the NCBI Windows 64-bit BLAST 2.7.1+ executable (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/) on a 64-bit Core i7 PC running Microsoft Windows 10, complemented by searching on both the CGP site and WormBase (http://wormbase.org). FASTA files containing sequence scaffolds, and others containing protein predictions, were searched by TBLASTN and BLASTP respectively using the protein sequences of C. elegans MED-1, END-1 and END-3. The updated C. elegans VC2010 sequence was also searched to confirm the med and end genes (Yoshimura et al. 2019).
Putative orthologous genes were identified using recommended best practices (Haag and Thomas 2015). Genes were first predicted by matching high-scoring segment pairs from TBLASTN results with genomic sequence, predicting the gene structure by identifying consensus intron splice donor and acceptor sequences, and comparing with the predicted genes from the assembly projects (Spieth et al. 2014; Stevens et al. 2019). Identification of gene structure started with the coding region for the DNA-binding domains and progressed both upstream and downstream. As analysis progressed, conserved features of the med and end genes and their gene products, within and among closely related species, became apparent, and these were used to refine the gene predictions. Searching of representative orthologs from each species back to the C. elegans genome confirmed that the predictions were the best matches. In some cases, the gene predictions from the assembly projects included short (<50 bp) predicted introns that could also be read through as coding. For these, a case-by-case judgment was made as to whether to include such introns in favor of maximizing amino-acid level homology. Some of the predictions within less-conserved regions could be incorrect, but these would not be expected to dramatically affect the analysis presented here. Similar judgments were made when multiple in-frame start codons were possible at the 5′ end of a gene, or when open reading frames could be extended in the 3′ direction by splicing around a stop codon. While no molecular validation of predicted genes was made, the manual curation of gene predictions favoring maximal similarity of gene and protein structures provides a surrogate validation by conservation across related species. This is the approach taken computationally for gene predictions by algorithms such as TWINSCAN (Korf et al. 2001).
It is highly likely that the gene set described here includes artifactual duplicates, particularly among the MEDs. The quality and coverage of the genome assemblies, as well as the maintenance of heterozygosity in sequenced strains, are known to produce artifactual paralogues that are really alleles of one locus (Haag and Thomas 2015; Barriere et al. 2009). Some of these may still have been included as orthologs because they corresponded to a predicted gene from the sequence assembly. For example, the two end-1 genes in C. brenneri are nearly identical with one found on a small sequence scaffold, suggesting that there is only one end-1 ortholog in this species. The inclusion of such nearly identical duplicates is not expected to affect inter-species comparisons, for which a representative single gene/protein was chosen. Gene models categorized as pseudogenes were more straightforward to find because they were truncated, had in-frame stop codons or frame shifts in the DNA-binding domain, or were missing essential amino acids such as one of the four cysteines in the C4 zinc finger. These may be expressed genes but were deemed unlikely to result in a functional protein.
Comparison of my protein predictions to those of the various sequence projects validated the approach used to identify med and end orthologs. Of the genes identified and deemed not to be pseudogenes, 54% (94/174) were identical to a predicted coding sequence (CDS) from the assemblies, 32% (56/174) partially overlapped an existing CDS, and 14% (24/174) did not correspond to a predicted CDS. Differences from assembly project predictions often resulted from missing carboxyl and/or amino ends because of large introns, or extensions of open reading frames that maximized ORF length only. Completely missed predictions tended to be of the small intronless med genes that are often missed by gene-finding algorithms. Data from cDNA sequences were generally not found to be useful, likely because the transient expression of the med and end factors in the earliest stages of embryogenesis means that med and end RNAs are generally absent from mixed-stage cDNA preparations.
Predicted genes/proteins have been provisionally named med-1.n/MED-1.n, end-3.n/END-3.n, and end-1.n/END-1.n (where n = 1, 2, 3, etc.). Lower numbers correspond roughly to the rank order of identified high-scoring segment pairs from the TBLASTN search, which favors both stronger similarity with the C. elegans search sequence and scaffolds that contain multiple hits. Where a single ortholog was found in a species, it was named as med-1/MED-1, end-1/END-1 or end-3/END-3. For analyses where a single representative of a set of paralogues was used, it was the first numbered one, except for pseudogenes or one of the apparent two-fingered MEDs, in which case the next paralogue was used.
Identification of conserved regulatory motifs
A representative set of promoters, one per Elegans supergroup species per factor, was compiled to identify putative cis-regulatory motifs. This was done to reduce artifacts arising from overrepresentation of sets of very similar promoters resulting from intraspecific paralogs, which tended to have very similar promoters (data not shown). To identify sites starting with known binding sites, a JavaScript program was written to count occurrence of sites and compute p values assuming a Poisson distribution, following the approach used in a prior work (Maduro et al. 2015). To identify motifs ab initio by their conservation, MEME (http://meme-suite.org/tools/meme) was used with expected site distribution with any number of repetitions (anr), the number of motifs to be identified as 10, and a maximum motif width of 12. Alternative parameters generally retrieved the same highly represented sites, except that motifs with higher E-values (and hence less conserved) could be different. Searches of the end-1 and end-3 promoters as separate groups produced qualitatively similar results as those that used both together, except that MED-like sites became rare enough among the end-1 genes that they were not reported as significant by MEME. I did not consider sites whose E-values were greater than 1e-02 as these occurred among a small number of med and/or end genes. Some of these may represent less-conserved regulatory motifs, although they were not recognized as belonging to known factors from C. elegans. The site locations and promoter sequences are in Supplemental File S1.
Phylogenetic analysis
Alignments and simple Maximum-Likelihood trees were performed using MUSCLE as implemented in MEGA-X (Kumar et al. 2018; Edgar 2004). The tree for the DNA-binding domains was produced using RAxML as implemented in the RAxML-NG web service (https://raxml-ng.vital-it.ch) with default parameters, except that the BLOSUM62 substitution matrix was used and bootstrapping was activated (Kozlov et al. 2019; Stamatakis 2014). I note that construction of trees using the proteins described here results in disagreements with the more robust trees of Stevens et al. (2019), with only closely related species retaining the same relationship, such as the interfertile species C. briggsae and C. nigoni (Woodruff et al. 2010). This is what would be expected from rapidly evolving genes. Consistent with this, calculations of synonymous and non-synonymous substitutions rates did not produce interpretable information because of the high rates of molecular evolution in Caenorhabditis in general (Cutter 2008). Moreover, the fastest rates of evolution in Caenorhabditis occur in early zygotic regulators with transient expression, which accurately describes the MED and END factors (Cutter et al. 2019). Because fast-evolving proteins are being compared among 20 species (as opposed to only two or three), the major conclusions regarding conserved amino acids and stringency of selection are nonetheless self-evident from the alignments and topology of phylogenetic trees.
Additional software
Gene modeling, sequence alignments and other analyses were performed with Vector NTI 6 and the MEGA-X software package (Kumar et al. 2018). Generation of tables and drawing of to-scale diagrams in SVG format were aided by custom programs written by the author in JavaScript and Python. These scripts are available by request. Protein alignments were annotated using BoxShade (https://embnet.vital-it.ch/software/BOX_form.html) to generate EPS-formatted files. Data were compiled in Microsoft Excel and figures were assembled in Adobe Illustrator.
Data availability
Sequences identified in this work are available as Supplemental files. Supplemental material available at figshare: https://doi.org/10.25387/g3.9820622.
Results
Med, end-3 and end-1 are found together in the elegans supergroup
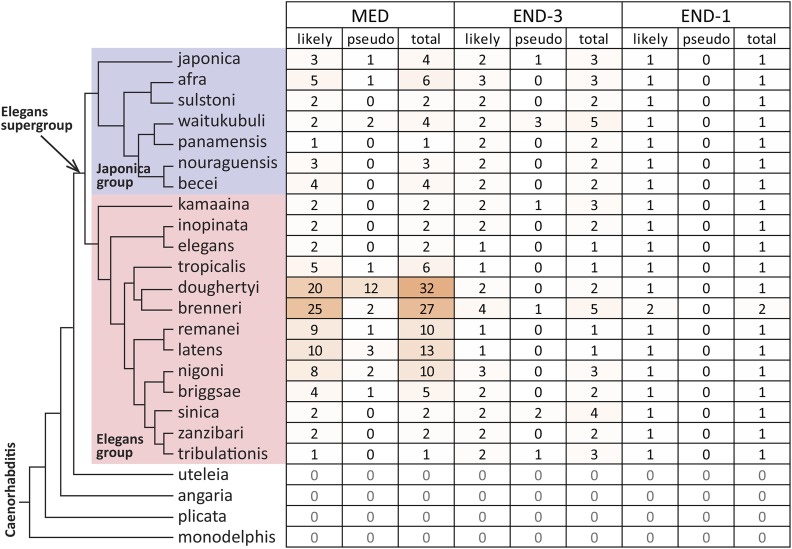
I searched sequence scaffolds from 27 species of the Caenorhabditis Genomes Project (http://caenorhabditis.org) with TBLASTN using the protein sequences of C. elegans MED-1, END-3 and END-1. C. elegans, C. briggsae and C. remanei were included as their sequences have been updated since earlier reports on med and end genes from these (Coroian et al. 2006; Maduro et al. 2005a; Yoshimura et al. 2019). As shown in Figure 2, at least one ortholog of each of the three genes was found in 20 species comprising the Elegans supergroup, a clade that includes the Japonica and Elegans groups (Stevens et al. 2019; Kiontke et al. 2011). Consistent with the absence of even more distant MED or END orthologs, the number of putative GATA factors in the genomes of species outside the Elegans supergroup was smaller, typically 5 or fewer, and putative orthologs were better matched to other C. elegans GATA factors like ELT-3 (data not shown). Across the 20 species searched in the Elegans supergroup, end-1 orthologs were unique in each genome except for C. brenneri (which may have two end-1 genes), while multiple paralogs within a species was the norm for the end-3 orthologs with an average of 2.0 copies per genome, and the med orthologs, found an average of 5.6 copies. The high average copy number of the med orthologs is driven by the 20 or more genes found in C. doughertyi and C. brenneri. Excluding these two species, the average number of med genes is 3.7 copies per genome. Of 208 genes identified for all three factors, 34 were deemed to be the result of unresolved heterozygosity or were likely pseudogenes (counted together under “pseudo” in Figure 2); these were eliminated from further study. It is still likely that some falsely identified med paralogues persist in the predicted gene set; hence, occurrence of nearly identical paralogues should be interpreted with caution (see Materials and Methods). In any event, the identification of false duplicates would not change the results of inter-species comparisons, for which a single representative gene was chosen for each factor. I note that because many comparisons were done with a single representative ortholog for each factor per species, it is possible that some species-specific evolutionary novelty will be missed.
Figure 2.
Orthologs of the MED, END-3 and END-1 factors among species whose sequences were searched. Species are shown after the most recent phylogeny (Stevens et al. 2019) with the Japonica group in light blue and the Elegans group in pink. The species C. parvicauda, C. castelli, C. quiockensis, and C. virilis, which contain no orthologs of the MED and END factors, have been omitted for simplicity. Table cells are colored by the number of orthologs.
Conserved linkage of end-1 and end-3 orthologs
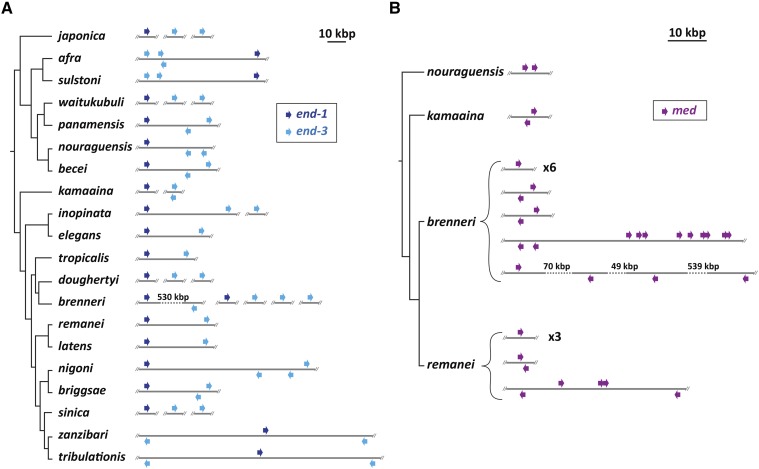
In C. elegans and C. briggsae the end-1 and end-3 genes are within ∼30 kbp of each other (Maduro et al. 2005a). Microsynteny of this type has been observed in other genes of these two species (Kent and Zahler 2000; Coghlan and Wolfe 2002). To see if microsynteny of end-1 and end-3 is common, I examined whether end-1 and end-3 orthologs in other species may be linked. As shown in Figure 3A, in 12/18 of the remaining Elegans supergroup species, end-1 and end-3 are found on the same scaffold with an average separation of ∼37 kbp and a range of 20-63 kbp. In C. brenneri, which has two end-1 and five end-3 orthologs, one scaffold carries both an end-1 and an end-3, however the distance between them is ∼530 kbp. In the remaining five species, the end-1 and end-3 genes are found on different scaffolds. Because it is possible for sequence scaffolds to break between two linked genes, there may be additional synteny among these. For example, in C. sinica the scaffold containing the end-1 ortholog is 32 kbp in size with the end-1 gene located 3 kbp from one end, raising the possibility that although its end-3 ortholog is on a different scaffold, end-1 and end-3 may be nearby in the genome. Closely related species have similar patterns of end-1 and end-3 synteny, for example between C. afra and C. sulstoni, and between C. zanzibari and C. tribulationis (Figure 3A). Although synteny is conserved, the relative orientation of linked end-1 and end-3 paralogues varies, with examples of all four possible linked arrangements. In C. elegans, end-1 and end-3 are encoded on the same strand with end-1 upstream of end-3. In C. sulstoni, two end-3 paralogs are upstream of end-1 with all three genes on the same strand. In C. zanzibari and C. tribulationis, end-1 is on one strand in between two end-3 paralogs on the other strand, hence in one end-1/3 pair the genes point toward each other, and in the other they are divergently transcribed. These differing arrangements are consistent with the high rate of intrachromosomal rearrangements previously noted for Caenorhabditis (Coghlan and Wolfe 2002).
Figure 3.
Synteny and relative orientation among med and end genes found on sequence scaffolds. Except where noted by a number, inter-gene distances are shown relative to the scale bar at the top of each panel. (A) Patterns of microsynteny among end-1 (dark blue) and end-3 (light blue) orthologs among the Elegans supergroup species. (B) Patterns of microsynteny among med orthologs for a subset of species in the Elegans supergroup.
Prevalance of linked med and linked end-3 duplications
In C. briggsae, two end-3 paralogues are found in an inverted orientation within several kbp, and in C. remanei, two clusters of closely linked med paralogues are found (Coroian et al. 2006; Maduro et al. 2005a). Similar linked duplications of these genes are found in other species. Among the end genes shown in Figure 3A, 7/10 species with at least two end-3 genes show two of them within 10 kbp. Among the 18 species with at least two med genes, linked pairs can be found in nine of them, in which at least two med genes occur within 5 kbp of each other. Examples of linked med duplications are shown for four of the Elegans supergroup species in Figure 3B. In the most extreme case, 9/25 C. brenneri med orthologs are clustered across a 23-kbp region, with an additional tandem pair located ∼22 kbp away. Linked duplications are therefore a common occurrence, particularly for the med genes.
Absence of a conserved intron in the Elegans group
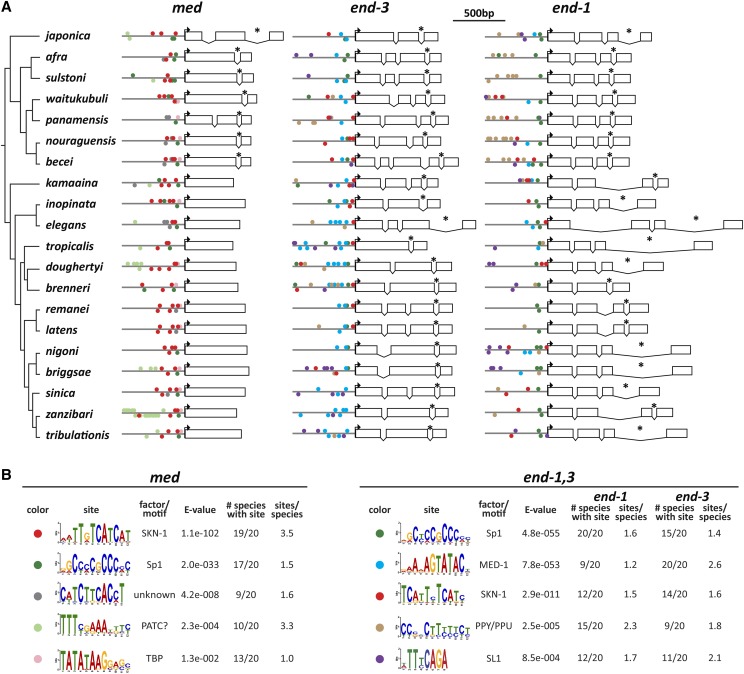
I next examined the evolutionary changes in med and end gene structures across the Elegans supergroup. For simplicity, a single representative med, end-3 and end-1 gene was used for each species because intraspecific paralogs generally showed identical splicing patterns. The gene structures are shown in scale diagrams in Figure 4A, depicting intron/exon structures arranged by the phylogeny of Stevens et al. (2019). Intron positions are also indicated on diagrams of the predicted proteins in Figure 8. Of particular significance, prior work found that the med genes of C. elegans, C. briggsae, and C. remanei have no introns, unlike all other GATA factors in these species including the end genes (Coroian et al. 2006; Gillis et al. 2008; Maduro et al. 2001). As shown in Figure 4A, while all representative med genes are found to be intronless across the Elegans group, the meds from the Japonica group share a common intron (indicated by an asterisk) within the C4 zinc finger coding region that is found in the same position in all end-1 and end-3 genes. In addition to this conserved intron, within the Japonica group, the C. japonica and C. panamensis med genes each have one more upstream intron at non-homologous positions.
Figure 4.
med and end gene structures and conserved promoter motifs. (A) Gene structures. 600bp of promoter are shown as a line, and the coding DNA sequence (CDS) predictions are shown relative to the scale bar at the top. Boxes are exons, and spaces joined by a ’V’ are introns. Bent arrows indicate the location of the predicted start codon. An asterisk denotes the intron conserved among all end genes and Japonica group med genes. (B) Motifs identified by MEME for the med and end-1,3 genes. The motifs are symbolized by a colored circle on the promoters in (A). Some of the motifs are shown in their reverse complement from the MEME output files in Supplemental Files S13 and S14.
Figure 8.
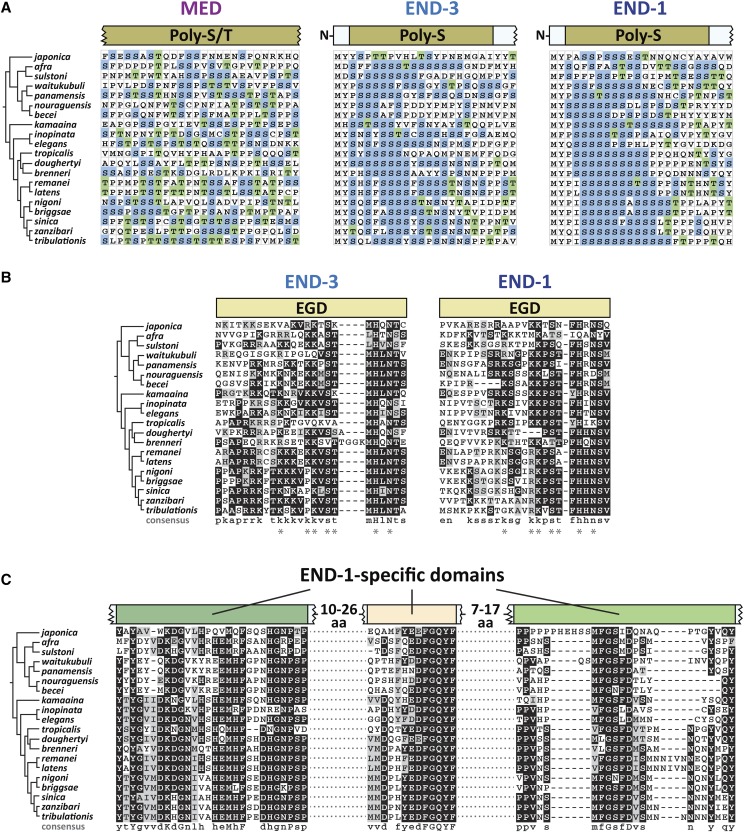
Conserved MED and END protein domains. The top part of the figure shows the MED, END-3 and END-1 protein structures with conserved domains in colored regions. Triangles represent the positions of introns in the coding regions as shown in the gene models in Fig. 4A. The bottom of the figure shows the names of the domains, which are shown at the amino acid level in Figs. 9 and 10. The MED orthologs have a variable region high in serine and threonine (Poly-S/T), while END-1 and END-3 share an amino-terminal polyserine domain (Poly-S) of variable length and an Endodermal GATA Domain (EGD). The END-1 orthologs share three additional regions not found in END-3. The species are arranged after the phylogeny in (Stevens et al. 2019).
Differences in introns among end-3 and end-1 genes
The conserved intron that interrupts the zinc finger is the only one shared between the end-3 and end-1 genes (Figure 4A). As a group, the end-3 orthologs show the highest variability in the number of introns, with C. tropicalis having only the one conserved intron, C. becei having four introns total, and the remaining species having two or three. The end-1 orthologs are far less diverse, sharing the same four exons with three introns, except for C. brenneri which is missing the second intron. In terms of size, the end-3 introns tend to be smaller overall, with introns larger than 100 bp most apparent within the Elegans group end-1 genes. Hence, the positions of introns in the end-1 orthologs appear to be under a greater constraint than those of the end-3 genes.
Identification of conserved promoter motifs
The occurrence of med and end genes in 20 related species affords the opportunity to identify conserved cis-regulatory sites and infer conservation of the structure of the gut specification network. The expectation is that conserved regulatory inputs found in C. elegans should be reflected in the occurrence of similar cis-regulatory sites mediating the same promoter-DNA interactions in the other species. I first searched for known binding sites for C. elegans factors among the Elegans supergroup med and end orthologs using methods previously used in C. elegans (Maduro et al. 2015). A size of 600bp upstream of the ATG was chosen for these and subsequent analyses, as the known regulatory interactions with the C. elegans med and end genes generally occur within a few hundred base pairs of the ATG (Broitman-Maduro et al. 2005; Maduro et al. 2001; Shetty et al. 2005; Bhambhani et al. 2014). Among the med upstream regions, I found widespread conservation of only SKN-1-like sites, and among the end-3 orthologs, only MED sites (Supplemental Material, Tables S1, S2 and S3). While these results support conservation of activation of med orthologs by a SKN-1-like factor, and activation of end-3 orthologs by MED-like factors, a complementary (and superior) approach is to search for over-represented motifs ab initio. I therefore searched 600bp upstream of representative med and end genes from all 20 species using the MEME discovery algorithm (Bailey and Elkan 1994). The results are summarized in Figure 4B, with the sites indicated by color-coded circles on the promoters in Figure 4A. The locations of the sites diagrammed in Figure 4 are listed in Supplemental File S1.
SKN-1 binding sites in the med and end genes
Among the med orthologs, a motif resembling two overlapping SKN-1 sites was identified in 19/20 species. The core of this motif, RTCATCAT, is found in two clusters in the C. elegans med genes and DNA fragments containing these sites are capable of binding recombinant SKN-1 DNA-binding domain in vitro (Maduro et al. 2001). The same core is found in SKN-1 binding sites in gcs-1, a known SKN-1 target gene in the fully developed intestine (An and Blackwell 2003). As in C. elegans, the SKN-1 sites in the med genes are found within 300 bp of the predicted start site in most of the other species, which is apparent from the diagram in Figure 4A. In C. panamensis, which contains only a single putative med gene, an RTCATCAT site was not identified by MEME although six ’core’ RTCAT sites were found by direct searching (P ≤ 0.05, Poission distribution). The low E-value of 1.1e-102 and presence of an average of 3.5 sites per species strongly suggest that activation of med orthologous genes likely occurs by SKN-1 in most Elegans supergroup species.
Among the end-1 and end-3 genes, a TCATTYTCATC site was identified by MEME in 12/20 end-1 genes and 14/20 end-3 genes (E-value 2.9e-11). Most of this site (underlined) overlaps with 8/9 bases of the WWWRTCATC site for SKN-1 (Etheve et al. 2016; Mathelier et al. 2014). Unlike the SKN-1 sites in the med genes, which occur an average of 3.5 times per gene, these putative SKN-1 sites in the end genes, when present, occur only 1.5 times per end-1 gene and 1.6 times per end-3 gene. I hypothesize that this site represents a degenerate (low-affinity) SKN-1 binding site. Prior evidence in C. elegans had suggested that SKN-1 contributes directly to end-1,3 activation independently of the MEDs, though the precise sites have not been reported (Maduro et al. 2015).
Sp1 binding sites
A motif resembling the binding site for Sp1 is found in the promoters of med (17/20 species, E-value of 2.0e-33), end-1 (20/20 species), and end-3 genes (15/20 species), with an E-value of 4.8e-55 for the two end genes. This same motif has been found among many C. elegans promoters, suggesting that regulation by Sp1 is not restricted to gut specification (Grishkevich et al. 2011). Reduction of function of sptf-3, a gene encoding an Sp1-like factor, causes a decrease in specification of E and a reduction in expression of end-1 and end-3 reporters (Sullivan-Brown et al. 2016). From the widespread conservation of the Sp1 binding sites, it is likely that Sp1 contributes to E specification across many species in the Elegans supergroup through direct binding of the med, end-1 and end-3 orthologous genes.
MED binding sites in the end-1 and end-3 genes
Prior work identified the binding sites for the MED factors in the end-1 and end-3 genes, defining a core sequence of AGTATAC that is distinct from the HGATAR site of canonical GATA factors (Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2005; Lowry et al. 2009). As anticipated by the results from searching for this site directly, MEME identified a highly conserved MED site motif in 9/20 end-1 genes and 20/20 end-3 genes (E-value 7.8e-53 across both end-1 and end-3). Across the nine species with MED sites identified in end-1, there are an average of 1.2 sites per gene, while for end-3, there are 2.6 sites on average. The location and spacing of the sites are consistent with results from C. elegans, with sites occurring within 200 bp of the predicted translation start site and showing a spacing (when multiple sites are present) of ∼50 bp (Broitman-Maduro et al. 2005).
Polypyrimidine motif
MEME identified a pyrimidine-rich motif in 15/20 end-1 genes and 9/20 end-3 genes (E-value 2.5e-05). This motif, consisting primarily of C and T, is most apparent among the Japonica group end-1 genes. The complement of the pyrimidine-rich motif is purine-rich, hence these motifs are called PPY/PPU (polypyrimidine/polypurine) tracts (Sawicka et al. 2008). This motif shows a strand bias by gene: 30/34 sites among the end-1 genes have the polypyrimidines on the top strand, while the sites are evenly distributed on either strand (9/16 on the top strand) in the end-3 genes. Polypyrimidine tracts are generally associated with messenger RNAs where they would be present as one strand, and interact with polypyrimidine-tract binding proteins (PTBs) (Sawicka et al. 2008). The human Pur-alpha protein (PURA) can bind to purine-rich motifs (Bergemann et al. 1992). A Pur-alpha-like protein in C. elegans, PLP-1, was previously identified as having a regulatory input into end-1 activation through a purine-rich site (Witze et al. 2009). However, the PPY/PPU motif identified by MEME was not found in either of the C. elegans end genes.
Additional overrepresented motifs
Three additional sites were found by MEME among the med genes. A motif containing a TCTKCAC core is found in 9/20 species med genes with an average of 1.6 sites per gene (E-value 4.2e-08). The motif sequence does not immediately suggest a putative regulatory factor, although it tends to be found among the SKN-1 sites, suggesting it is related to SKN-1 binding. A motif containing TTTNNAAA was found at a higher E-value of 2.3e-04 in 10/20 med genes with an occurrence of 3.3 sites per gene, with one species C. zanzibari, containing 16 of them. This site resembles previously identified periodic AT clusters (PATCs) suggesting it may be a more general motif (Frøkjær-Jensen et al. 2016). A motif resembling a TATA-box was found in 13/20 species’ med genes with an even higher E-value of 1.3e-02 (Grishkevich et al. 2011). This may be a bona fide basal promoter site, as it is found within tens of base pairs from the translation start in these 13 genes. Finally, among the end genes, an “SL1 motif” was found in 12/20 end-1 genes and 11/20 end-3 genes (E-value 8.5e-04) (Grishkevich et al. 2011). The SL1 sequence is typically found at the 5′ end of genes whose transcripts become trans-spliced to the SL1 spliced leader sequence (Allen et al. 2011). The motif was not found in the C. elegans end-1/3 genes, consistent with prior work that neither of these genes in C. elegans is known to be trans-spliced (Zhu et al. 1997; Allen et al. 2011). Its relevance as a motif is uncertain, as in most of the end promoters that contain it, the site is more than 300bp upstream of the predicted start site.
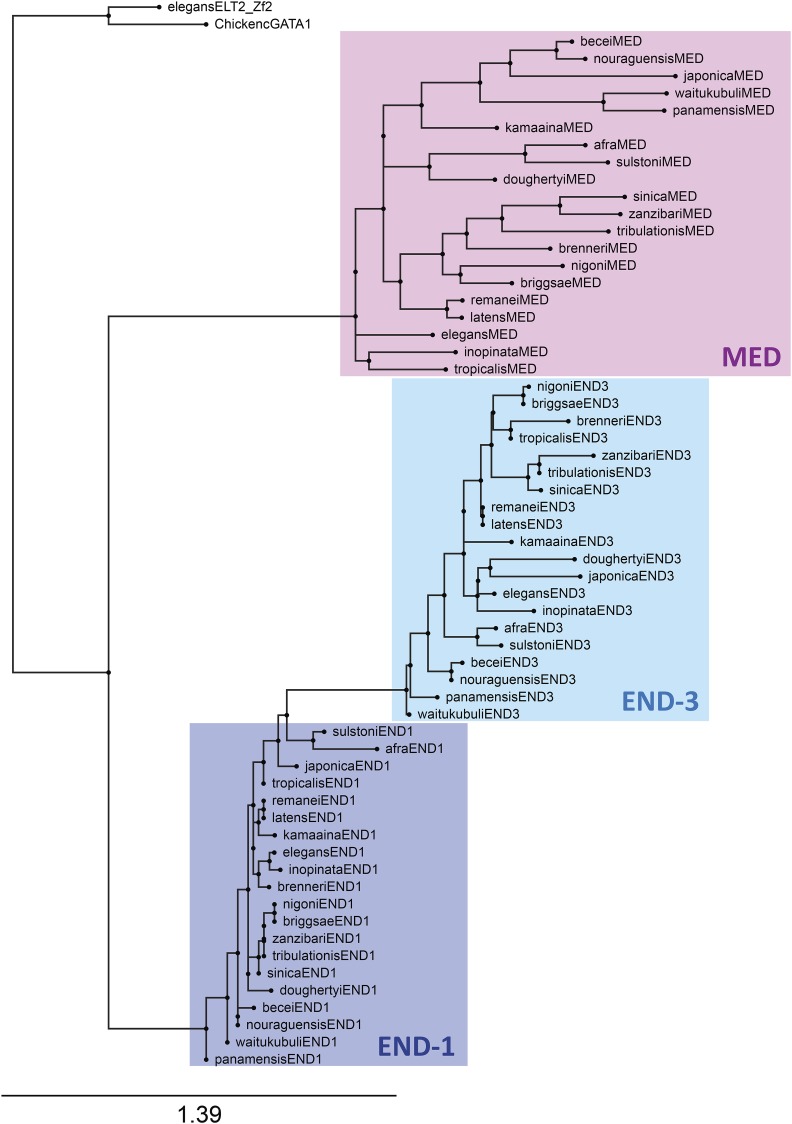
Phylogenetic analysis confirms that med, end-3 and end-1 form distinct clades
The gene structure and promoter motifs suggest that the med, end-3 and end-1 genes form distinct families among the 20 species of the Elegans supergroup. To confirm that this is reflected at the protein level, I aligned the DNA-binding domains (DBDs) among representative MED, END-3 and END-1 factors (one per species) and used this to construct a phylogenetic tree ab initio with the RAxML-NG method (Kozlov et al. 2019; Stamatakis 2014). As shown in Figure 5, MED, END-3 and END-1 form three broad clades, with the END-1 factors showing the highest similarity as a group, followed by the END-3 factors, and finally the more diverse MED factors. A high diversity of the MED factors was previously observed among the med genes from C. elegans, C. briggsae and C. remanei (Coroian et al. 2006). The grouping of the factors increases confidence that the correct orthologs have been assigned and shows that different rates of protein evolution have occurred among the three factors.
Figure 5.
Phylogenetic tree of representative MED, END-3 and END-1 DNA-binding domains. The DNA-binding domains of C. elegans ELT-2 and chicken GATA1 are shown as outgroups. Each of the three factors forms a distinct clade, with the END-1 factors showing the highest similarity, followed by END-3, then the MEDs as the most diverse group.
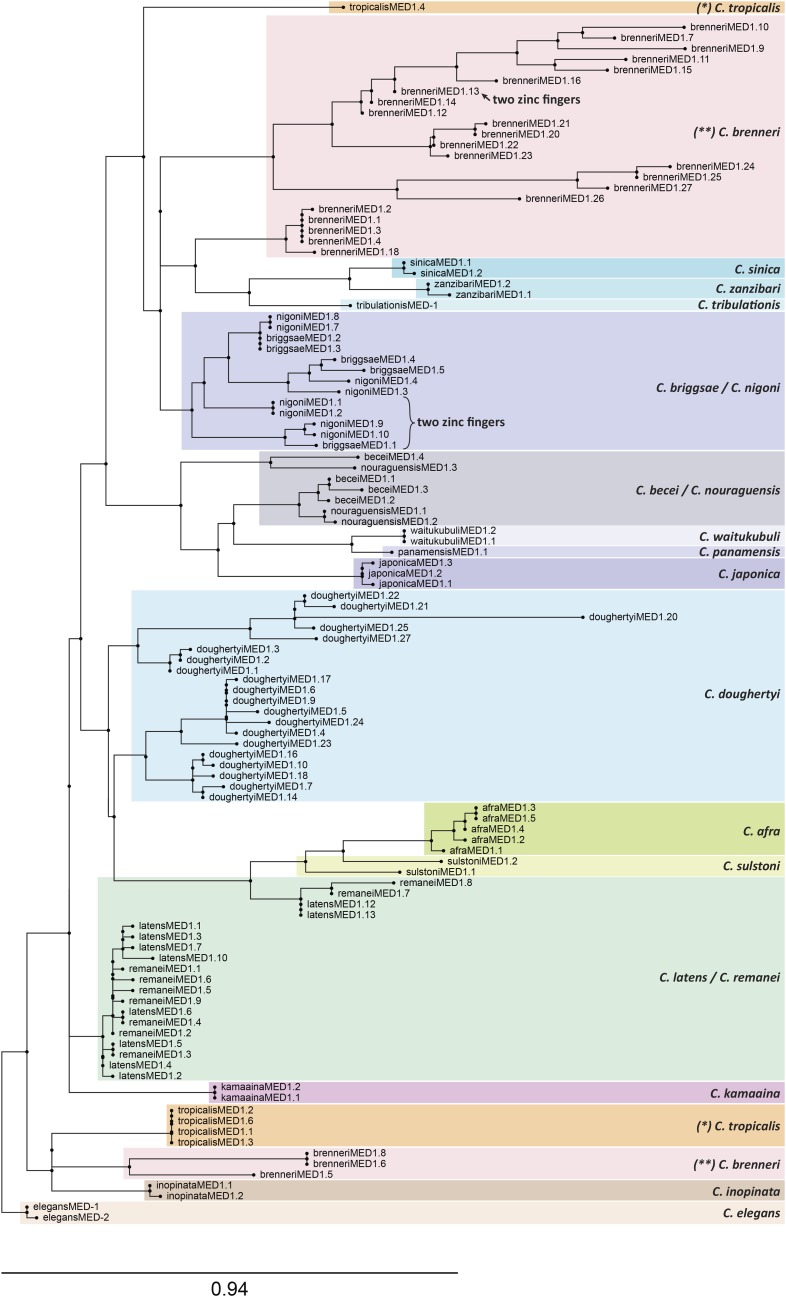
Gene amplification within and among species
While end-1 is represented by a unique ortholog among all species (except C. brenneri which may have two end-1 genes), med and end-3 orthologs are often found as two or more duplicate genes within a species. The two C. briggsae END-3 paralogues are highly similar, suggesting recent duplication, and the multiple med genes among C. elegans, C. briggsae and C. remanei are also much more alike within each species (Coroian et al. 2006; Maduro et al. 2005a). To test how general this phenomenon is, I aligned and constructed trees for all MED DBDs, and separately, the END DBDs. In the tree of MED factors shown in Figure 6, most med duplications have occurred post-speciation from a small number of founding genes. The 20 MED factors in C. doughertyi cluster in a way that suggests there may have been only one or two ancestral med genes that underwent multiple rounds of amplification. In the case of C. brenneri, the MEDs form two clusters of 22 and 3 genes each, suggesting there were only a few ancestral factors. A similar division occurs among the C. tropicalis MEDs, which suggests two ancestral med genes. There are three groups in which paralogous MED factors are clustered within species pairs: C. briggsae with C. nigoni, C. becei with C. nouraguensis, and C. latens with C. remanei. Within each cluster, the pattern suggests that both species inherited two or three med paralogues from a common ancestor, which then each underwent further amplification post-speciation. Among the remaining 9 species that have 2-5 med genes each, the paralogous MEDs clustered together as a single group, suggesting a single ancestral gene. This unusually widespread pattern of duplications both pre- and post-speciation, not seen in the end genes, shows that the med genes are under different evolutionary constraints.
Figure 6.
Phylogenetic tree of all MED factors, showing high prevalence of duplications across the Elegans supergroup. In most cases, paralogous duplicates likely arose post-speciation, although there are examples that suggest that some species each inherited two or three genes from a common ancestor that later underwent further duplications. The tree was generated by RAxML using the MED DNA-binding domains (Kozlov et al. 2019; Stamatakis 2014).
I note here that six genes were found that encode MED-like factors with two C4 zinc fingers, indicated on the tree in Figure 6. In each case, the two fingers are highly similar, so only one of the two fingers was used to generate the tree. Four of the “two-fingered” genes are present as two paralogous pairs in C. nigoni, one is found in C. briggsae, and another is found in C. brenneri (Figure 6). C. nigoni and C. briggsae are very closely related, suggesting they inherited the same two-fingered med gene from a common ancestor (Kiontke et al. 2011). The positions of the six two-fingered MED factors in the phylogeny are hence consistent with two-finger MED-type GATA factors having arisen twice, likely by an interstitial duplication, because the two fingers in each share a nearly identical amino acid sequence. The observation of two-fingered GATA factors is noteworthy because among vertebrates, GATA factors generally have two zinc fingers, and even within C. elegans, there is a two-fingered GATA factor, ELT-1 (Gillis et al. 2009; Lowry and Atchley 2000; Page et al. 1997).
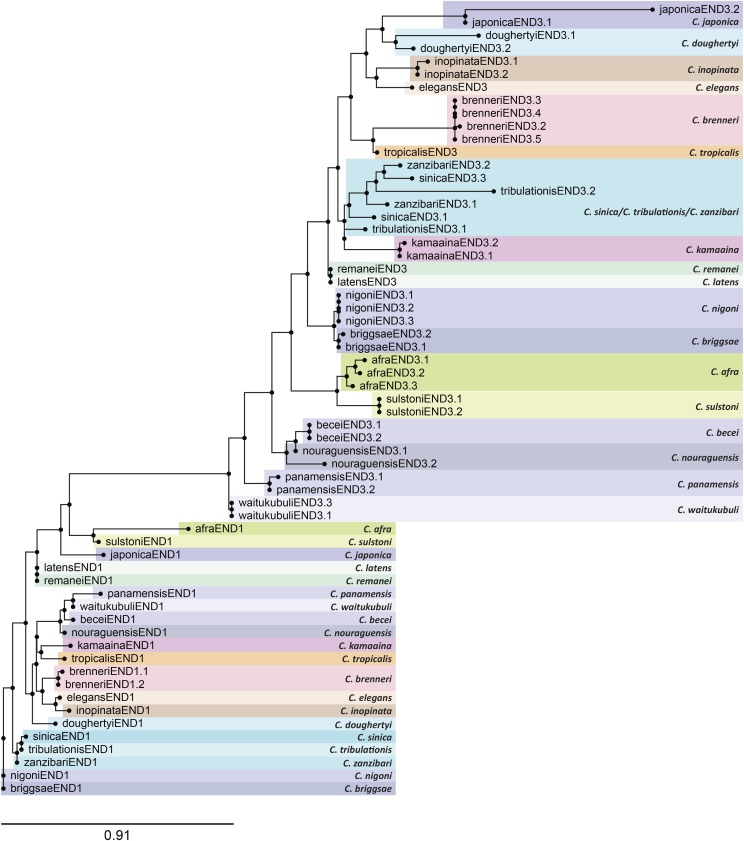
A tree of the DBDs of the END-1 and END-3 orthologs is shown in Figure 7. As mentioned earlier, all END-1 orthologs are unique in each species except for the two possible end-1 paralogues in C. brenneri. Among the END-3s, intraspecific amplification is implied for all species with two or more END-3s, except for a cluster containing END-3 paralogues from C. sinica, C. tribulationis, and C. zanzibari. This portion of the tree is most consistent with two paralogous end-3 genes having been present in the common ancestor of all three species. Hence, duplications do occur among the end-3 paralogues, but at a far lower frequency than with the med genes.
Figure 7.
Phylogenetic tree of all END-3 and END-1 factors, showing tendency for END-1 factors to be unique, and END-3 factors to have undergone some duplications. The tree was generated by RAxML using the END-3 and END-1 DNA-binding domains (Kozlov et al. 2019; Stamatakis 2014).
Conserved domains of MED, END-3 and END-1
Prior alignments of the ENDs from C. elegans and C. briggsae revealed three conserved domains: An amino-terminal polyserine (Poly-S) region, a short region immediately upstream of the zinc finger, called the Endodermal GATA Domain (EGD), and the GATA-type zinc finger and basic domains (Maduro et al. 2005a). Among the MEDs, only the latter two domains are conserved (Coroian et al. 2006). Taking advantage of the 20 Elegans supergroup species, I aligned representative MED and END proteins to both generalize these earlier findings and to identify other conserved domains that might have been missed. The alignments revealed both expected and previously unknown conserved regions, shown diagrammatically in Figure 8. On this figure, the corresponding positions of introns are also indicated to reveal patterns of conservation of the gene structure in relation to these conserved regions.
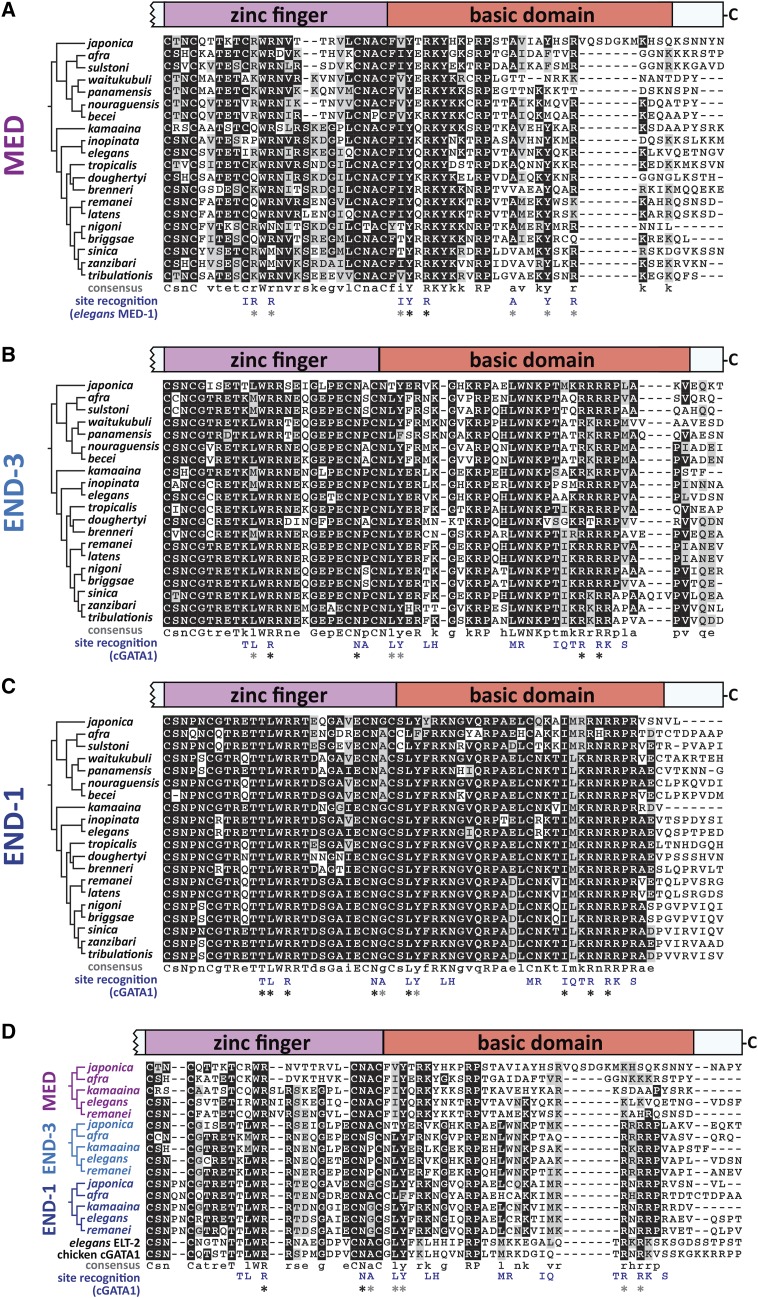
MED, END-3 and END-1 DNA-binding domains
An alignment of representative DBDs for the MED, END-3 and END-1 factors, one per species, is shown in Figure 9 (Edgar 2004). Consistent with their recognizing an atypical binding site, the MED DBDs share features that distinguish them from the END-3 and END-1 DBDs (Figure 9A). Among the Elegans group MED factors, the C4 zinc finger has 18 amino acids between the two pairs of cysteines, with a structure of CXXC-X18-CXXC, while the Japonica group members are diverged from this structure and have 16-17 amino acids, i.e., CXXC-X16-17-CXXC. A consensus sequence with 11 invariant amino acids is shown below the alignment in Figure 9A. While the group of MED factor DBDs appears to be diverse, the identification of a conserved MED-like motif among the end-3 promoters suggests that the MED factors have nonetheless coevolved to continue recognizing a similar binding site in each species. The solution structure of a C. elegans MED-1 DBD::binding site complex revealed that recognition of the MED binding site is mediated by 9 amino acids, indicated at the bottom of Figure 9A (Lowry et al. 2009). In comparing these with the corresponding amino acids in the other MED DBDs, there is evidence of conservation as shown by asterisks. Two of the 9 amino acids, a tyrosine (Y) and arginine (R) just after the zinc finger, are invariant. Five of the remaining amino acids are found in most of the MED DBDs. The remaining two are the isoleucine (I) and the first arginine in the zinc finger. The arginine is somewhat conserved, as in most MEDs it is an arginine or a lysine (K), both of which are basic. The isoleucine (I) is not conserved, and is replaced by a cysteine (C) in most other MEDs. This amino acid may not be critical for recognition of a MED binding site, however, as prior work showed that transgenes containing individual med genes from C. briggsae and C. remanei can fully complement the embryonic lethal phenotype of C. elegans med-1; med-2 double mutants; in the MED factors from both of these species, the corresponding amino acid is a cysteine. Overall, despite the higher divergence among the MEDs as a group, there appears to be selection for the 8/9 amino acids known to be involved in site recognition in C. elegans MED-1. Added to the apparent conservation of MED-like binding sites in the respective end-3 orthologs in every species, the data suggest maintenance of the DNA-binding specificity of the MEDs.
Figure 9.
DNA-binding domains (DBDs) and additional carboxyl amino acids aligned using MUSCLE (Edgar 2004). The zinc fingers and basic domains are shown for representative sequences of (A) MED, (B) END-3, (C) END-1, and (D) a representative subset of all three factors. Consensus sequences are shown below each alignment. The phylogeny of Stevens et al. (2019) is shown to the left of the species names for reference. Under the consensus sequences, the amino acids that mediate site recognition by the C. elegans MED-1 DBD for (A) and cGATA1 for (B), (C) and (D) are shown (Omichinski et al. 1993; Lowry et al. 2009). Asterisks show corresponding amino acids that are invariant (black) or are generally conserved (gray).
In contrast with the divergent MEDs, the DBDs of the END-3 and END-1 orthologs are more alike and share greater similarity to those of canonical GATA factors. The ENDs, ELT-2 and cGATA1 have an invariant CXXC-X17-CXXC zinc finger structure with 17 amino acids between the 2nd and 3rd cysteines. Consensus sequences for END-3 and END-1, shown below the alignments in Figures 9B and 9C, contain 23 invariant amino acids for END-3, and 31 for END-1, i.e., 2x and 3x more than the 11 invariant amino acids among the MED DBDs. A solution structure for END-1 or END-3 has not been reported, but as a surrogate I have shown, beneath both alignments, the 18 amino acids in the cGATA1 zinc finger known to mediate base contacts (Omichinski et al. 1993). END-3 is conserved at 7/18 of these positions with 4 amino acids being invariant, while END-1 has 10/18 positions conserved, of which 8 are invariant. Hence the END-1s are structurally more like cGATA1 than are the END-3s. Moreover, the END-1 orthologs are also invariant at more positions, indicating that they are under the most evolutionary constraint.
An amino acid in the END-3 DBD is worth further comment. The proline between the 3rd and 4th cysteines of the zinc finger, in sequence CNPC, was substituted by a leucine in the EMS-induced C. elegans mutant end-3(zu247) (Maduro et al. 2005a). This mutant has a phenotype indistinguishable from the null mutant end-3(ok1448) which lacks most of the DBD (Owraghi et al. 2010). While this position is also a proline in 12/20 species, among the other END-3s it is serine (S) or alanine (A). Serine has a short polar side chain, while alanine is short and hydrophobic, however leucine is also hydrophobic but longer, suggesting that the longer side chain at this position compromises the structure of the zinc finger. This position is variable among the MED and END-1 orthologs, where it is a proline (P), alanine (A), serine (S), or glycine (G), indicating this position is under relaxed selection.
Another difference between the END-3s and END-1s is the amino end of the C4 zinc finger between the 1st and 2nd cysteines. GATA factors in general, including the MEDs, END-3, ELT-2 and cGATA1, have two amino acids in the pattern CXXC. Most of the END-3s are CSNC, while the END-1s have either CSNPNC (12 species), CSNPSC (6 species), CSNQNC (C. afra) or CNPNC (C. becei). It is not known what effect the extra one or two amino acids have on the structure of the zinc finger, however this variation in structure is found only in the END-1 orthologs.
Finally, as a set, the DBDs from the MEDs and ENDs of a subset of the Elegans supergroup species are shown with ELT-2 and cGATA1 in Figure 9D, showing that all three factors share conserved amino acids with each other and with canonical GATA factors. Overall, 7/18 of the amino acids known to mediate DNA recognition in cGATA1 are broadly conserved (Omichinski et al. 1993).
Serine-rich domains in MEDs and ENDs
The MED and END factors share an upstream region of variable size enriched in serine, with or without threonine. Both are polar amino acids. These are shown diagrammatically in Figure 8, as the amino-most conserved domain among the MEDs and ENDs, and in amino acid sequence alignment in Figure 10A. Among the MEDs, the Poly-S/T region is variable in size, consists of both serines and threonines, and is the only other conserved feature upstream of the DNA-binding domain. Because of the size variability, the alignment in Figure 10A represents only part of an overlapping region among MEDs of all 20 species. Among the ENDs, a similar Poly-S domain, consisting almost exclusively of homopolymeric clusters of serines, is found at the amino terminus starting at the 3rd or 4th amino acid (Figure 10A). In one exception, the Poly-S domain is all but gone in C. japonica END-3. As noted earlier, the Poly-S region had been previously recognized in the C. elegans and C. briggsae end genes (Maduro et al. 2005a).
Figure 10.
Other conserved domains of unknown significance among the MED and END proteins. (A) A portion of the alignment of Poly-S/T domains (MED factors) and the Poly-S domains (END-3 and END-1). Serines are highlighted in blue and threonines in green. (B) Extended Endodermal GATA Domains (EGDs) immediately upstream of the zinc fingers of END-3 and END-1. A consensus sequence is shown beneath each alignment, with amino acids similar between END-3 and END-1 shown with an asterisk (*). (C) Highly conserved regions among the END-1 factors showing highly conserved amino acids and a consensus sequence beneath the alignment.
An unexpected feature of the Poly-S region in the end genes bears further description. Although serine is coded by six codons – TCT, TCC, TCA, TCG, AGT and AGC – the serines among the Poly-S regions in the end-3 and end-1 orthologs are coded almost exclusively (99%, 554/557) by TCN codons (N = any base). Moreover, two of the four TCN codons, TCT and TCC, are used 50% and 22% of the time. Among C. elegans genes, TCN represents 75% of serine codons, and among these, TCT and TCC occur only 28% and 18% of the time, respectively (https://www.genscript.com/tools/codon-frequency-table). This preferential use of TCT and TCC codons for serine in the Poly-S regions, among the TCN codons, is statistically significant (P < 10−40, χ2-test). The possible implications of this codon bias are discussed later.
Conservation of the end family gata domain (EGD)
Previous work identified the END family GATA Domain, or EGD, immediately upstream of the C. elegans and C. briggsae END-1 and END-3 DBDs (Maduro et al. 2005a). This domain does not occur among the other C. elegans GATA factors, suggesting it is uniquely important for function of END-1 and END-3. Among the 20 species in the Elegans supergroup, the END-1 and END-3 orthologs across 20 species do contain a conserved region immediately upstream of the zinc finger. This is shown diagrammatically in Figure 8, and by sequence alignment in Figure 10B. Whereas the original report had the domain consisting of 9 amino acids, an extended domain is apparent that consists of approximately 25 amino acids. Seven of these (shown by an asterisk in the figure) are highly conserved between the END-3 and END-1 factors, but there are additional conserved amino acids within each group of factors. Moreover, the domain is more conserved among the END-3 orthologs. While the EGDs tend to be enriched in basic amino acids, suggesting they may be involved in general DNA binding, their significance remains unknown.
END-1 specific domains
Among the END-3 orthologs, the region between the Poly-S and the EGD regions is variable in size and does not exhibit sequences with extensive conservation (Figure 8). In contrast, the END-1 orthologs display three additional domains that are highly conserved across all 20 species (Figures 8 and 10C). A consensus sequence shows high conservation with many invariant regions. These domains are apparently novel, as a BLAST search using this region of END-1 did not identify related proteins other than predicted orthologs of END-1 within Caenorhabditis. With the identification of these extended sequence similarities, the END-1 orthologs across the 20 species are highly conserved throughout their lengths, while the END-3 and MED orthologs are conserved only in parts.
Discussion
In this work I have identified and compared the gene and protein structures of the MED, END-3 and END-1 GATA transcription factors among 20 Caenorhabditis species of the Elegans supergroup. Predictions were made by manual curation, guided by known features of the network from C. elegans and informed by comparison of gene and protein structures together. The results confirm coevolution of cis-regulatory sites, gene structures and protein sequence over tens of millions of years of evolution. Many of the conserved features, including the DNA-binding domains, and binding sites for SKN-1, MED, and an Sp1-like factor, are consistent with known properties of the med and end genes in C. elegans (Maduro et al. 2001; Maduro et al. 2015; Sullivan-Brown et al. 2016; Broitman-Maduro et al. 2005). Prior work has also shown that orthologous meds and/or ends from a few of these species can function as transgenes in C. elegans (Coroian et al. 2006; Maduro et al. 2005a). Hence, I hypothesize that the med, end-3 and end-1 genes function in a core endoderm specification network across the Elegans supergroup that originated in a common ancestor.
High rates of med gene duplication
The med, end-3 and end-1 genes showed distinct patterns of gene duplication among species. Occurrence of duplicate med genes is disproportionately high, with an average of 5.6 med genes per species (or 3.7 if C. doughertyi and C. brenneri are not counted), compared with 2.0 end-3 genes and a single end-1 per species, except for C. brenneri which may have two end-1 genes (Figure 2). In most cases, sequence similarity was consistent with most med duplicates having arisen post-speciation, with exceptions resulting from likely inheritance of two or three med genes from a recent common ancestor (Figure 6).
The disproportionate amplification of the meds compared with the ends suggests that there is ongoing selective pressure for increased numbers of med genes. The high amplification of the meds is unusual, as redundancy of GATA factors in tissue specification is typically not more than twofold in other systems (Gillis et al. 2009; Tremblay et al. 2018; Murakami et al. 2005). Across the Elegans supergroup, the occurrence of MED binding sites in the end genes (particularly end-3) argues for positive selection for the presence of these sites, and hence the MED factors that bind them. Loss of MED binding sites in the C. elegans end genes results in aberrant intestinal lineage development, metabolic defects, and reduced viability (Choi et al. 2017; Maduro et al. 2015). Hence, duplications of med genes might select for increased med expression to make gut specification more robust. C. elegans has a high rate of segmental duplications compared with other species, with a higher gene dose generally leading to increased mRNA production (Konrad et al. 2018). Alternatively, it may be that MED factors in some species have become collectively reduced in their ability to be activated or to activate target genes, in a way that maintains multiple copies due to complementary degenerative mutations (Force et al. 1999). Protein degeneracy would be consistent with the lower degree of sequence conservation among the MED DNA-binding domains in C. brenneri, which has experienced an extreme amplification of med genes (Figure 9). However, this does not explain amplification of med genes in C. doughertyi, whose MED DNA-binding domains are more similar as a group, unless they are all collectively degenerate in some way. In C. elegans, which has two nearly identical med genes, either med gene is dispensable, although when med-1 is deleted, med-2 becomes haploinsufficient in 35% of embryos due to a failure to specify the MS blastomere (Maduro et al. 2007). Hence, maintenance of copies of med genes may be occurring by selection for robust specification of MS rather than E (Maduro et al. 2001). This still does not explain the extreme amplification, although it could explain why a driving force for duplications is not apparent from the structure of the end genes.
Rather than increase expression through gene duplication, it seems equally possible for a small number of mutations to increase expression or activity of any one med gene. Hence, some other constraint may select against a small number of med genes in some species. For example, a reduction in SKN-1 activity could limit the expression of individual med genes and select for med gene amplification as a compensatory mechanism. It is also likely that at least some duplicated med genes have acquired new essential functions. Consistent with this, not all med orthologs from C. remanei are able to rescue C. elegans med-1; med-2 double mutants, even as multicopy transgenes, which would be expected to overcome expression limitations (Coroian et al. 2006). Future work to quantify the contributions of individual med genes in other Elegans supergroup species, or to test expression of these when introduced into C. elegans as single-copy transgenes, may shed some light on what mechanisms may be driving increased med copy number.
Linkage of end orthologs
In most species, end-1 was found within ∼35 kbp of end-3 (Figure 3A). One possibility for maintenance of this synteny is that the two genes may be coregulated. Three lines of evidence argue against this possibility, at least for C. elegans. First, there is at least one unrelated gene between the ends, the neural gene ric-7 (Hao et al. 2012). Second, the end-1,3 genes are not precisely co-expressed as accumulation of end-3 mRNA precedes that of end-1 (Baugh et al. 2003; Maduro et al. 2007; Raj et al. 2010). Third, unlinked single-copy transgenes of wild-type end-1 and end-3 are able to completely replace function of the endogenous genes when introduced into an end-1,3(-) strain, suggesting that linkage is not a prerequisite for their expression (Maduro et al. 2015). It may be, therefore, that synteny of end-1 and end-3 merely reflects their origin as a tandem duplication of an ancestral end gene.
A pair of partially redundant developmental factors in C. elegans, LIN-12 and GLP-1, which encode highly similar Notch orthologs, are a good comparison for the END-1/3 pair (Rudel and Kimble 2002). These paralogous genes are similar in structure and have overlapping function in C. elegans development (Moskowitz and Rothman 1996). The two genes are approximately 30 kbp apart in the C. elegans genome with apparently unrelated intervening genes (http://wormbase.org). The lin-12/glp-1 pair is conserved in closely related species, and likely arose from the duplication of a progenitor gene at the base of the Elegans supergroup (Stevens et al. 2019; Rudel and Kimble 2002). A search of the Elegans supergroup genomes finds examples where lin-12 and glp-1 orthologs are found within tens of kbp on the same sequence scaffolds, suggesting microsynteny is conserved in at least some species (data not shown). The conservation of microsynteny for lin-12 and glp-1, like that of end-1 and end-3, then, likely results from the origin of the genes as a linked duplication, followed by the tendency for genomic segments tens of kbp in size to stay intact within the genus (Coghlan and Wolfe 2002).
Identification of known and previously unrecognized cis-regulatory sites
The MEME search recovered binding sites for regulators previously known to activate the med and end genes in C. elegans (Figure 4B). In the case of the med orthologs, these were binding sites for SKN-1, while for the end genes, these were binding sites for both SKN-1 and MED-1. The conservation of these sites supports the hypothesis that these genes have maintained the same regulatory hierarchy as in C. elegans, with SKN-1 activating the med genes, and both SKN-1 and the MED proteins activating the end genes. The MED sites in the Elegans supergroup end genes are found in all end-3 orthologs but only 9/20 end-1 orthologs. C. elegans end-3 has four MED sites and these are collectively essential for end-3 activation, although even a single MED site in a single-copy end-3 transgene is sufficient for activation (Maduro et al. 2015). In contrast, C. elegans end-1 has only two MED sites, and these are less important for end-1 expression due to the stronger parallel input by TCF/POP-1 and PAL-1 into end-1 as compared with end-3 (Maduro et al. 2015; Maduro et al. 2005b). Hence, the lower number of MED sites in the end-1 genes may reflect stronger input from other factors. The likely sites for SKN-1 in end-1 and end-3 were not previously known because they do not contain the same pattern of SKN-1 site core sequences as present in the med promoters. An intriguing hypothesis is that the SKN-1 sites in the end genes may be of lower affinity than those in the med genes. Because expression of the end genes is delayed by at least one cell cycle compared with med-1,2, lower-affinity SKN-1 sites could potentially allow for delayed activation, preventing expression of the ends before EMS has divided into MS and E. A similar affinity difference has been hypothesized for early- and late-acting binding sites of the pharynx regulator PHA-4 (Gaudet et al. 2004). As the SKN-1 sites in the end genes were not found in all species, it is possible that the input from SKN-1 directly into gut specification through the ends is lost or further weakened in some species. This might make the SKN-1 → MED → END-1,3 pathway more strictly linear, similar to the SKN-1 → MED → TBX-35 pathway that specifies MS in C. elegans (Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2009). In MS, loss of the MED factors results in the absence of MS-derived fates, consistent with an inability of SKN-1 to specify MS without the MED factors. Finally, an additional suspected regulatory input was from an Sp1-like factor, likely to be SPTF-3 (Sullivan-Brown et al. 2016). Most of the med, end-3 and end-1 orthologs have a consensus Sp1 binding site (Figure 4B). Together, the recovery of these sites from an ab initio search of their putative promoters lends strong support to the hypothesis of conservation of this gene network across the Elegans supergroup.
MEME-identified sites of lower significance, and not as broadly conserved, are either unknown or reflect putative core promoter elements. These include one with core sequence TCTKCAC, a polypyrimidine motif, putative PolyA/T cluster, a TATA-binding protein (TBP) site, and an SL1 motif. The latter two were previously found in many promoters in five Elegans supergroup species (Grishkevich et al. 2011). The putative PolyA/T cluster is associated with germline expression (Frøkjær-Jensen et al. 2016). The other two motifs are of unknown significance. The TCTKCAC motif is found in the C. elegans med genes, hence it is possible to test its significance directly. The site was found three times, and close to the previously identified SKN-1 sites, suggesting the site may play an accessory role to SKN-1 activation.
It is particularly conspicuous that sites for minor regulatory inputs known in C. elegans were not found to be widely conserved, either by a direct search or through MEME. This includes sites for TCF/POP-1 and the Caudal ortholog PAL-1, both of which are genetically known to contribute to end-1 and end-3 expression, and for which binding sites are known or suspected based on prior work (Bhambhani et al. 2014; Maduro et al. 2005b; Robertson et al. 2011; Shetty et al. 2005). In C. elegans, END-3 is also a suspected contributor to activation of end-1 based on reduction of end-1 mRNA in an end-3 mutant background (Maduro et al. 2007). The failure to recover sites for these regulators suggests that these inputs are poorly conserved or lie outside of the regions that were searched, or else the binding sites have changed among the various species. Given how easily SKN-1 and MED sites were found, it could also be that different species have evolved different sets of supportive regulatory inputs. The apparent qualitative differences in regulatory input of SKN-1 and POP-1 in C. briggsae, revealed through cryptically different reduction-of-function phenotypes between C. briggsae and C. elegans, suggests that reinforcing regulatory inputs may evolve rapidly (Lin et al. 2009). Even within C. elegans, widespread cryptic variation in input from SKN-1 and the Wnt pathway (which acts through POP-1) was observed among C. elegans wild isolates (Torres Cleuren et al. 2019). An emerging model seems to be that the core SKN-1 → MED → END-1,3 regulatory cascade is conserved, while additional regulatory inputs that reinforce this cascade evolve rapidly and would thus be expected to be species-specific. Putative cis-regulatory sites that mediate these supporting inputs might therefore occur in only a subset of species in the Elegans supergroup and would be missed in the analysis done here.
End-3 and end-1: The same but different
In C. elegans, end-1 and end-3 clearly have overlapping function. Complete loss of both genes has a fully penetrant failure to specify endoderm, while null alleles either for gene alone have either no effect (end-1) or a weak effect (end-3) on gut specification (Maduro et al. 2005a; Owraghi et al. 2010). A similar result was obtained using RNAi in C. briggsae (Maduro et al. 2005a). As well, overexpression of either end gene in C. elegans is sufficient to induce endoderm differentiation in non-endodermal lineages (Maduro et al. 2005a; Zhu et al. 1998). Within their DNA-binding domains, the END-3 and END-1 orthologs are clearly more similar to each other than they are to the MEDs (Figures 5, 9).
Despite these similarities, END-3 and END-1 differ in ways that suggest they have at least some unique functions. First, the END-1 DBDs are more highly conserved as a group, while those of END-3 are under slightly more relaxed selection. This is apparent in the way that the DBDs appear in a phylogenetic tree (Figure 7) and in the degree of invariant amino acids in an alignment (Figures 9B, 9C). Within their DBDs, the END-1s have twice as many similar amino acids in common with vertebrate cGATA1 than the END-3s have in common with cGATA1, notably in amino acid positions known to mediate sequence recognition (Figures 9B, 9C).
Additional evidence is consistent with both shared and divergent activity of END-3 and END-1 in C. elegans. Recent work inferred the binding sites for C. elegans END-1 and END-3 as RSHGATAASR and RKWGATAAGR, respectively, which are very similar though not identical (Weirauch et al. 2014; Lambert et al. 2019). Other work has shown that recombinant DNA-binding domains of C. elegans END-1 and END-3 can bind canonical GATA sites in the promoter of C. elegans elt-2, although END-1 has a higher affinity for such sites (Du et al. 2016; Wiesenfahrt et al. 2015). From this work, Endoderm GATA Domains (EGDs) immediately upstream of the DBDs show conserved amino acids between END-3s and END-1s but many more that are unique to either EGD (Figure 10B). Although the function of the EGDs remains unknown, their conservation and proximity to the DBDs suggest an accessory role in protein-DNA interaction that is unique to the ENDs among the Caenorhabditis GATA factors.
The Poly-S region of END-3 and END-1: protein domain or polypyrimidine tract?
END-3 and END-1 share an amino-terminal segment, far from the DNA-binding domain, that is enriched for homopolymers of serine (Figure 10A). Such a domain is not found in the other C. elegans GATA factors, nor is enrichment for serine found in vertebrate GATA factors (Kaneko et al. 2012; Yang et al. 1994). This suggests that the Poly-S domain plays some other function besides DNA binding and transactivation. The selection for TCT and TCC codons suggests that the Poly-S regions have been maintained for a reason other than a selection for what they contribute to the END-1 and END-3 proteins. Beyond transcriptional activation of the end-1 and end-3 genes, post-transcriptional regulatory mechanisms could potentially fine-tune END-1,3 protein levels. At the level of mRNA, the preference for these codons, as opposed to UCG and UCA, results in maintenance of a polypyrimidine tract in the mRNA. Support for a possible role of such a tract in the endoderm GRN is that in some species (e.g., C. latens and C. remanei), the med orthologs also have an apparent enrichment of T and C bases in the first part of their coding regions. In other systems, polypyrimidine tract binding proteins (PTBs) have various roles in RNA metabolism, including regulation of splicing and mRNA stability, though in these cases the tracts occur outside of coding regions (Sawicka et al. 2008). There is a C. elegans PTB gene, ptb-1, but its function has not been described (http://wormbase.org). At the level of translation, repeats of the same UCY serine codon could cause starvation for limiting amounts of a particular seryl-tRNASer, leading to ribosome pausing (Darnell et al. 2018). However, it is not clear why there would be selection to delay translation of end mRNA, particularly as given the rapid early cell divisions of the C. elegans embryo, it makes more sense to express the gene products as rapidly as possible. A more benign reason for the maintenance of the serine codon repeats is that they might be an artifact of a trinucleotide repeat expansion process (Koren and Trifonov 2011). Indeed, in that study, amino acid repeats in vertebrate proteins were most likely to be found in the first exon, i.e., at the amino end, consistent with their location in the end-3 and end-1 genes. Hence, the role of the Poly-S domain, if any, remains open for speculation until structure-function studies are performed.
END-1 orthologs are conserved throughout their lengths
An additional unexpected finding emerged from the alignment of END-1 orthologs that distinguishes them among the MED/END proteins. Between the Poly-S and EGD domains, the END-3 orthologs as a group are diverse in size and sequence, whereas the END-1 orthologs are more similar in size and show several regions of high conservation (Figure 10C). These END-1-specific domains can be grouped into three regions containing blocks of invariant amino acids. The most striking of these is the center domain which contains an invariant sequence of FGQYF across all species END-1s. None of these highly conserved domains is found in other proteins, apart from predicted END-1 orthologs. The high conservation is further supported by the conservation of introns in the end genes. The end-1 genes have four introns with only one of these absent in C. brenneri (Figure 4A). In contrast, the end-3 genes were more likely to experience intron gains and losses over the same evolutionary time period, with most of these occurring in the variable region between the amino-terminal Poly-S and EGD domains (Figure 8). A cursory examination of the amino acids in the END-1-specific domains suggests that these are on the outside of the protein, perhaps mediating protein-DNA or protein-protein interactions that do not occur with END-3 (data not shown).
Taken together, these data show that across the Elegans supergroup, the END-1s are highly conserved proteins with greater similarity to vertebrate GATA factors than the more diverse END-3s proteins. This predicts that END-1 has unique features in transcriptional activation, and that the target genes activated by each of these factors are likely to include both common and distinct targets.
Med othrologs: A divergent and diverse subclass of GATA factors
The MED orthologs among the 20 species were found to be divergent from the END-3/END-1 factors, and to comprise a more diverse group of proteins, even within the DNA-binding domain (Figures 5, 9). The divergence of the DBD from that of the ENDs, ELT-2 and cGATA1 is expected, because the C. elegans MEDs were recognized to be divergent GATA factors that recognize a different binding site with an AGTATAC core (Broitman-Maduro et al. 2005; Lowry et al. 2015). Despite the high divergence of the MED factors as a group, indicating relaxed selection, there appears to be maintenance of their binding site sequence over evolutionary time. This is supported by the conservation, across all 20 species, of most of the amino acids that were found to mediate protein-DNA recognition in C. elegans MED-1 (Figure 9A), and more importantly, by the MEME identification of AGTATAC binding sites among all end-3 orthologous genes and 9/20 end-1 genes (Figure 4). Furthermore, transgenes of most of the C. briggsae and C. remanei meds were individually able to complement C. elegans med-1,2 double mutants in both gut and mesoderm specification despite limited conservation, albeit in high copy number transgenes (Coroian et al. 2006). Selection is likely not acting solely on the MEDs for end gene activation, as there are other direct MED targets in C. elegans whose orthologs in the Elegans supergroup were not investigated here, including in the early MS lineage (Broitman-Maduro et al. 2006; Broitman-Maduro et al. 2005). The lower conservation suggests that the MED DBDs may simply be more accommodating of amino acid substitutions than are the DBDs of END-3 or END-1.
Outside of the DNA-binding domain, the MEDs as a group lack the type of conserved regions seen in the ENDs. The only other feature found is a variable enrichment for serine and threonine of unknown significance. This region does not resemble the homopolymeric serine regions at the amino end of the ENDs (Figure 10A). Rather, it is a higher prevalence for S/T that lacks a recognizable context. A serine-threonine rich motif was found to be important for nuclear localization of the mineralocorticoid receptor in vertebrates, suggesting that this region of the MED orthologs may play a similar role (Walther et al. 2005). Until structure-function analyses are done, the significance of the serine/threonine enrichment will remain unknown.
The MED/END cascade is a derived charachter
The existence of a gut precursor is a conserved lineage feature found in more distantly related nematode species (Schierenberg 2006; Houthoofd et al. 2003; Schulze and Schierenberg 2011; Boveri 1892). It must therefore be that species outside the Elegans supergroup specify the gut precursor without MED/END factors. The most upstream factor SKN-1, and the downstream gut identity factor ELT-2, are also more widely conserved than just the Elegans supergroup (Schiffer et al. 2014; Couthier et al. 2004). If SKN-1 still specifies MS and E outside of the Elegans supergroup, the simplest hypothesis is that specification of gut occurs by direct activation of an elt-2-like gene by SKN-1. An attempt to demonstrate bypass of the end-1 and end-3 genes was successful using an elt-2 transgene under regulatory control of the end-1 promoter in a C. elegans strain lacking end-1 and end-3 (Wiesenfahrt et al. 2015). However, this transgene worked best in a high copy-number array, and not in single-copy. Furthermore, expression of this transgene is likely to be at least partially dependent upon regulatory input by MED-1,2, based on studies with an end-1 promoter lacking MED binding sites (Maduro et al. 2015). As an alternative to direct SKN-1 → ELT-2 regulation, there could be one or more non-GATA regulators between them, analogous to the MED/END cascade. Regardless of how gut specification occurs outside of the Elegans supergroup, some set of evolutionary events must have set in motion a breakdown of the ancestral specification mechanism, favoring the evolution and fixation of the SKN-1/MED/END cascade as the dominant mode of E specification.
Evolutionary Origin Of the SKN-1 → MED → end-1,3 cascade
The co-occurrence of the MED and END factors suggests that these genes evolved within a short time at the base of the Elegans supergroup (Figure 11A). A preliminary search for orthologs of ELT-7 also found evidence that this factor likely originated at the same time, as 18/20 of the Elegans supergroup species have a clear elt-7 ortholog while species outside do not (data not shown). At the start of this work there was an expectation that there might have been one or more “transitional” species with only part of the network upstream of ELT-2, for example with only the end-3 and end-1 factors, or only one end-like factor. Since no such species were found apart from the two species that may lack elt-7 orthologs, it may be that for the med and end factors, a transitional species has not yet been sequenced, or is extinct, or that the orthologs are highly diverged. The reduced number of recognizable GATA factors in species outside of the Elegans supergroup argues against this possibility, however.
Figure 11.
Origin of the MED, END-3 and END-1 factors. (A) Origin of all three factors at the base of the Elegans supergroup, followed by loss of a conserved intron in an ancestral med gene at the base of the Elegans group. (B) Hypothetical microhomology-mediated end joining (MMEJ) event that could delete the conserved zinc finger intron at the base of the Elegans group, using a 6-bp identity in-frame microhomology in an extant C. japonica med gene. At top, the microhomology is shown for the top strand. In the bottom part, complementary strands are shown pairing across the microhomology, which if resolved could result in an in-frame deletion of the intron, after (van Schendel and Tijsterman 2013). This would also require maintenance of the AAC codon for asparagine immediately to the right of the homology. (C) Speculative model for generation of the SKN-1/MED/END regulatory cascade through intercalation by serial duplications of an ancestral autoregulating elt-2 gene. A bent arrow indicates the transcription start site, with the regulatory activity of the protein product of the gene shown as a colored line from the bent arrow. The promoter is to the left of the bent arrow. The positions in the promoters are only meant to qualitatively convey positive regulation and not indicate number or position of binding sites.
The data strongly suggest that the med and end genes might have been derived from the same ancestral gene. This hypothesis is supported by the existence of an intron in the zinc finger domain of all med and end genes, except for the Elegans group med genes where loss of this intron occurred. This intron is also found in elt-2 and elt-7 in C. elegans and at least some of the other species in the Elegans supergroup (Fukushige et al. 1998; Sommermann et al. 2010)(data not shown). Intron loss is common throughout the genus, and occurs more frequently than intron gain (Roy and Penny 2006). One mechanism by which this particular intron could have been lost in an ancestral med gene of the Elegans group is through germline gene conversion from a reverse-transcribed (spliced) mRNA (Roy and Gilbert 2005). An alternative mechanism could be through microhomology-mediated end joining, or MMEJ, of a double-stranded break in the gene (McVey and Lee 2008; van Schendel and Tijsterman 2013). Indeed, in one of the C. japonica med genes, a short stretch of six base pairs upstream of this intron recurs close to the 3′ splice site of the intron itself, such that a repair of a double-stranded chromosome break by MMEJ would result in an in-frame removal of the intron (Figure 11B). This would also require that the asparagine codon (AAC) is somehow maintained, which may be possible given the observed types of MMEJ repair of double-stranded breaks induced by Cas9 cleavage, e.g., (Taheri-Ghahfarokhi et al. 2018). Regardless of the mechanism, loss of this intron likely occurred only once in the last common ancestor to the Elegans group. I note in passing that the converse property, lack of intron gain in the Elegans group med genes, may be accounted for by selection for rapid gene expression through avoidance of mRNA splicing; most early zygotic Drosophila genes are intronless, for example (Guilgur et al. 2014). However, a small number of the med gene predictions in the Elegans supergroup do have introns (Supplemental File S1).
The structural conservation among the 20 Elegans supergroup MEDs and ENDs lead me to propose a model by which the MED/END cascade arose through a process of duplication and intercalation, from elt-2 upwards, as shown in Figure 11C. This model combines gene duplications, which shape Caenorhabditis genomes, and the mechanism of intercalation of factors into an ancestral regulatory network (Booth et al. 2010; Lipinski et al. 2011). I include duplication of elt-2 to produce elt-7 based on preliminary data suggesting that this gene also originated at the same time as the MEDs and ENDs. Indeed, a common origin of all these upstream factors is further supported by their similar size of 174-242 amino acids, while ELT-2 is approximately twice as large. One interpretation of this size difference could be that ELT-2, as the central regulator of intestinal fate, has additional structural features unique to this role (McGhee et al. 2009). In contrast, the upstream med and end factors are transiently expressed and seem to serve to robustly activate elt-2, while elt-7 plays an accessory role with elt-2 (Maduro et al. 2005a; Maduro et al. 2015; Sommermann et al. 2010; Wiesenfahrt et al. 2015; Zhu et al. 1997). Indeed, function of the ends and elt-7 can be replaced by early activation of just elt-2 alone, as mentioned earlier (Wiesenfahrt et al. 2015).
Patterns of structural similarity among the factors upstream of ELT-2 lead to hypotheses about their origin. The similarity of the END-3 and END-1 orthologs and their tendency to be <50 kbp apart in a species suggests that they originated from a common progenitor together, or that one was a duplicate of the other. Considering the stronger resemblance of the DNA-binding domain of END-1 with that of ELT-2 and vertebrate cGATA1, a reasonable hypothesis is that end-1 originated first, as a duplicate of an ancestral elt-2 gene that was both activated by SKN-1 and maintained its own expression through positive autoregulation. In parallel, elt-7 would be duplicated from elt-2 to become its paralogue. Positive autoregulation of ELT-2 and ELT-7 is known and for ELT-2 has even been visualized in vivo (Fukushige et al. 1999; Sommermann et al. 2010). Duplication of elt-2 has likely occurred to generate the extant paralogous (and likely inactive) C. elegans elt-4 gene, and more significantly, C. elegans elt-7, a paralogue of elt-2 that shares overlapping function, expression and autoregulation with elt-2 (Sommermann et al. 2010; Fukushige et al. 2003). Although not necessary at this step, if the SKN-1 sites in the elt-2 promoter became degenerate, the end-1 prototype would be stable because it would be necessary to relay input from SKN-1 into elt-2,7. A paralogous end-3 prototype gene might then have originated as a simple linked duplication of end-1. Lending support for elt-2 as a progenitor for the end genes is the presence of the conserved intron in the zinc finger coding region found in all end-1/3 orthologs and in C. elegans elt-2/7. The two end genes could be stabilized by the complete loss of SKN-1 sites in the elt-2 promoter, degeneracy of SKN-1 sites in the end-1 promoter, and coevolution of END-3 with binding sites in the end-1 promoter. In this state, END-1 also acts to amplify input from END-3 into elt-2.
A challenge is to account for the origin of a med-like progenitor, given the evidence that they form a structurally divergent set of regulators. In this work it was found that while the Elegans group species have intronless med genes, obscuring their origin, the putative Japonica group meds share a common intron in the zinc finger coding region that is in the same location as the aforementioned intron in all extant end-3 and end-1 genes. This leads to the hypothesis that a prototype med gene arose as a duplicate of one of these genes. The slightly higher structural similarity of the MED DBD with that of END-1 (Figure 5) suggests the prototype may have arisen from end-1, but it could also have been end-3. Co-evolution of the MED DNA-binding domain with cognate sites in end-1 and end-3 would reduce autoregulation of the end genes and fix the MED factor within the network, though END-3 could retain the ability to contribute to end-1 activation. Degeneration of the SKN-1 sites in end-3 would strengthen the requirement for the MED factors as they would become necessary to relay SKN-1 input to end-3. Further refinement of the network would strengthen regulatory input of the meds by SKN-1, activation of end-3 by the MEDs, and other regulatory inputs into end-1. Further selection on the END-1 coding region might have been enforced by protein-protein interactions with other factors that contribute to gut specification.
Although this model is highly speculative, there is supporting evidence for a similar model in evolution of the Bicoid (Bcd) gene in an ancestor to cyclorrhaphan flies, a group that includes Drosophila (Driever and Nusslein-Volhard 1989; Stauber et al. 1999). Bcd specifies anterior fates in early cyclorrhaphan embryos, while outside of this group bcd is not found, and other factors play an analogous role (Lynch et al. 2006; McGregor 2005). Bcd arose as a duplicate of the Hox gene Zen, and likely acquired derived DNA-binding characteristics primarily through two missense mutations in the DNA-binding domain (Liu et al. 2018; McGregor 2005). From studies in the flour beetle Tribolium, which lacks bcd, it is hypothesized that Bcd took over functions of some of its downstream gap gene targets, which it then became an activator of (McGregor 2005). Bcd is proposed to have originated ∼140 Mya at the base of the Cyclorrhapha, a longer time period than the estimated tens of millions of years since the common ancestor to the Elegans supergroup (Wiegmann et al. 2011; Coghlan and Wolfe 2002; Cutter 2008). Recruitment of Bcd into A/P specification in Drosophila likely required more steps than the MED/END cascade, because in my proposed model for C. elegans endoderm specification, the cascade originated through duplication and modification of a factors already in an ancestral version of the network. Hence, it is plausible that emergence of the MED/END network could have occurred at the base of the Elegans supergroup on a shorter evolutionary time scale. Furthermore, in analogy to Bcd, the initial evolution of the MED DBD that resulted in a change in its binding site to a non-GATA target site might have been driven by a small number (or even just one) change(s) in a key amino acid. With the sequences of med genes from 20 species, such structure-function correlations can now be examined.
Studies on the evolution of Bcd suggest a possible explanation as to why a more layered gene cascade might have evolved for embryonic gut specification within the Elegans supergroup. The emergence of Bcd may have conferred a more rapid specification of segment identity, allowing developmental time to become faster without sacrificing robustness (McGregor 2005). By extension to the Elegans supergroup, it is possible that the SKN-1 → MED → END-1,3 gene regulatory cascade coincided with an increase in developmental speed in Caenorhabditis, perhaps as part of the transition to very early and rapid cell fate specification (Schierenberg 2001; Laugsch and Schierenberg 2004). Elucidation of gut specification mechanisms in Caenorhabditis species outside of the Elegans supergroup, compared with their developmental speed, could provide evidence for this hypothesis, or alternatively identify non-GATA factors that play the same role as the MED/END cascade.
In the meanwhile, the identification of MED, END-3 and END-1 orthologs in 20 species sets the stage for studies to test hypotheses about evolution of gene regulatory networks, structure-function correlations in the evolution of novel DNA-binding domains, and features of developmental system drift. As the study of gene regulatory networks becomes more computational, the set of MED and END orthologs identified here will provide a basis for future studies integrating gene network architecture with transcriptomics data, for example (Omranian and Nikoloski 2017; Nomoto et al. 2019).
Acknowledgments
I am indebted to Mark Blaxter, Lewis Stephens and colleagues at the Caenorhabditis Genomes Project in Edinburgh for prepublication access to the genome sequences of Caenorhabditis species and for advice during this work. I also thank Eric Haag (University of Maryland, College Park) for helpful advice in interpretation of search results. I am similarly grateful to the anonymous reviewers for their very helpful insights and suggestions. Earlier versions of this work were completed under my NSF grant IOS#1258054. I also thank Christian Turner, a former UCR undergraduate supported by NIH Award T34GM062756 from the National Institute of General Medical Sciences (MARCU-STAR) program to UC Riverside, for having performed preliminary searches of available Caenorhabditis sequences in 2014.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.9820622.
Communicating editor: M.-A. Félix
Literature Cited
- Allen M. A., Hillier L. W., Waterston R. H., and Blumenthal T., 2011. A global analysis of C. elegans trans-splicing. Genome Res. 21: 255–264. 10.1101/gr.113811.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. H., and Blackwell T. K., 2003. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 17: 1882–1893. 10.1101/gad.1107803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., and Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36. [PubMed] [Google Scholar]
- Barriere A., Yang S. P., Pekarek E., Thomas C. G., Haag E. S. et al. , 2009. Detecting heterozygosity in shotgun genome assemblies: Lessons from obligately outcrossing nematodes. Genome Res. 19: 470–480. 10.1101/gr.081851.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., Hill A. A., Slonim D. K., Brown E. L., and Hunter C. P., 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900. 10.1242/dev.00302 [DOI] [PubMed] [Google Scholar]
- Bergemann A. D., Ma Z. W., and Johnson E. M., 1992. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded-DNA-binding properties of the encoded protein. Mol. Cell. Biol. 12: 5673–5682. 10.1128/MCB.12.12.5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhambhani C., Ravindranath A. J., Mentink R. A., Chang M. V., Betist M. C. et al. , 2014. Distinct DNA binding sites contribute to the TCF transcriptional switch in C. elegans and Drosophila. PLoS Genet. 10: e1004133 10.1371/journal.pgen.1004133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Bowerman B., Priess J. R., and Weintraub H., 1994. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266: 621–628. 10.1126/science.7939715 [DOI] [PubMed] [Google Scholar]
- Boeck M. E., Boyle T., Bao Z., Murray J., Mericle B. et al. , 2011. Specific roles for the GATA transcription factors end-1 and end-3 during C. elegans E-lineage development. Dev. Biol. 358: 345–355. 10.1016/j.ydbio.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth L. N., Tuch B. B., and Johnson A. D., 2010. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature 468: 959–963. 10.1038/nature09560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T., 1892. Ueber die Entstehung des Gegensatzes zwischen den Geschlechtszellen und den somatischen Zellen bei Ascaris megalocephala, nebst Bemerkungen zur Entwicklungsgeschichte der Nematoden. Sitzungsberichte der Gesellschaft für Morphologie und Physiologie in München 8: 114–125. [Google Scholar]
- Bowerman B., Eaton B. A., and Priess J. R., 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075. 10.1016/0092-8674(92)90078-Q [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G., Lin K. T.-H., Hung W., and Maduro M., 2006. Specification of the C. elegans MS blastomere by the T-box factor TBX-35. Development 133: 3097–3106. 10.1242/dev.02475 [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G., Maduro M. F., and Rothman J. H., 2005. The noncanonical binding site of the MED-1 GATA factor defines differentially regulated target genes in the C. elegans mesendoderm. Dev. Cell 8: 427–433. 10.1016/j.devcel.2005.01.014 [DOI] [PubMed] [Google Scholar]
- Broitman-Maduro G., Owraghi M., Hung W., Kuntz S., Sternberg P. W. et al. , 2009. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development 136: 2735–2746. 10.1242/dev.038307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A. S., Gilbert D. E., Liu X., Cheung J. W., Michnowicz J. E. et al. , 1997. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes Dev. 11: 2227–2238. 10.1101/gad.11.17.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Broitman-Maduro G., and Maduro M. F., 2017. Partially compromised specification causes stochastic effects on gut development in C. elegans. Dev. Biol. 427: 49–60. 10.1016/j.ydbio.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Coghlan A., and Wolfe K. H., 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res. 12: 857–867. 10.1101/gr.172702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coroian C., Broitman-Maduro G., and Maduro M. F., 2006. Med-type GATA factors and the evolution of mesendoderm specification in nematodes. Dev. Biol. 289: 444–455. 10.1016/j.ydbio.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Couthier A., Smith J., McGarr P., Craig B., and Gilleard J. S., 2004. Ectopic expression of a Haemonchus contortus GATA transcription factor in Caenorhabditis elegans reveals conserved function in spite of extensive sequence divergence. Mol. Biochem. Parasitol. 133: 241–253. 10.1016/j.molbiopara.2003.10.012 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., 2008. Divergence times in Caenorhabditis and Drosophila inferred from direct estimates of the neutral mutation rate. Mol. Biol. Evol. 25: 778–786. 10.1093/molbev/msn024 [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Garrett R. H., Mark S., Wang W., and Sun L., 2019. Molecular evolution across developmental time reveals rapid divergence in early embryogenesis. Evol Lett 3: 359–373. 10.1002/evl3.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell A.M., Subramaniam A.R., and O’Shea E.K., 2018. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol Cell 71: 229–243 e211. 10.1016/j.molcel.2018.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., 2010. Emerging properties of animal gene regulatory networks. Nature 468: 911–920. 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. H., Rast J. P., Oliveri P., Ransick A., Calestani C. et al. , 2002. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 246: 162–190. 10.1006/dbio.2002.0635 [DOI] [PubMed] [Google Scholar]
- Dieterich C., Clifton S. W., Schuster L. N., Chinwalla A., Delehaunty K. et al. , 2008. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat. Genet. 40: 1193–1198. 10.1038/ng.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dineen A., Osborne Nishimura E., Goszczynski B., Rothman J. H., and McGhee J. D., 2018. Quantitating transcription factor redundancy: The relative roles of the ELT-2 and ELT-7 GATA factors in the C. elegans endoderm. Dev. Biol. 435: 150–161. 10.1016/j.ydbio.2017.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., and Nusslein-Volhard C., 1989. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature 337: 138–143. 10.1038/337138a0 [DOI] [PubMed] [Google Scholar]
- Du L., Tracy S., and Rifkin S. A., 2016. Mutagenesis of GATA motifs controlling the endoderm regulator elt-2 reveals distinct dominant and secondary cis-regulatory elements. Dev. Biol. 412: 160–170. 10.1016/j.ydbio.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., and Lin S. Y., 2014. The evolutionary origins and consequences of self-fertility in nematodes. F1000Prime Rep. 6: 62 10.12703/P6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheve L., Martin J., and Lavery R., 2016. Dynamics and recognition within a protein-DNA complex: a molecular dynamics study of the SKN-1/DNA interaction. Nucleic Acids Res. 44: 1440–1448. 10.1093/nar/gkv1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M. A., 2007. Cryptic quantitative evolution of the vulva intercellular signaling network in Caenorhabditis. Curr. Biol. 17: 103–114. 10.1016/j.cub.2006.12.024 [DOI] [PubMed] [Google Scholar]
- Félix M. A., Braendle C., and Cutter A. D., 2014. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One 9: e94723 Erratum: 10: e0118327. 10.1371/journal.pone.0094723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L. et al. , 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Jain N., Hansen L., Davis M. W., Li Y. et al. , 2016. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 166: 343–357. 10.1016/j.cell.2016.05.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Goszczynski B., Tian H., and McGhee J. D., 2003. The Evolutionary Duplication and Probable Demise of an Endodermal GATA Factor in Caenorhabditis elegans. Genetics 165: 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T., Hawkins M. G., and McGhee J. D., 1998. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198: 286–302. [PubMed] [Google Scholar]
- Fukushige T., Hendzel M. J., Bazett-Jones D. P., and McGhee J. D., 1999. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc. Natl. Acad. Sci. USA 96: 11883–11888. 10.1073/pnas.96.21.11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J., Muttumu S., Horner M., and Mango S. E., 2004. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2: e352 10.1371/journal.pbio.0020352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis W. Q., Bowerman B. A., and Schneider S. Q., 2008. The evolution of protostome GATA factors: molecular phylogenetics, synteny, and intron/exon structure reveal orthologous relationships. BMC Evol. Biol. 8: 112 10.1186/1471-2148-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis W. Q., St John J., Bowerman B., and Schneider S. Q., 2009. Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family. BMC Evol. Biol. 9: 207 10.1186/1471-2148-9-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkevich V., Hashimshony T., and Yanai I., 2011. Core promoter T-blocks correlate with gene expression levels in C. elegans. Genome Res. 21: 707–717. 10.1101/gr.113381.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilgur L. G., Prudencio P., Sobral D., Liszekova D., Rosa A. et al. , 2014. Requirement for highly efficient pre-mRNA splicing during Drosophila early embryonic development. eLife 3: e02181 10.7554/eLife.02181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., Fitch D. H. A., and Delattre M., 2018. From “the Worm” to “the Worms” and Back Again: The Evolutionary Developmental Biology of Nematodes. Genetics 210: 397–433. 10.1534/genetics.118.300243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag E. S., and Thomas C. G., 2015. Fundamentals of Comparative Genome Analysis in Caenorhabditis Nematodes. Methods Mol. Biol. 1327: 11–21. 10.1007/978-1-4939-2842-2_2 [DOI] [PubMed] [Google Scholar]
- Hao Y., Hu Z., Sieburth D., and Kaplan J. M., 2012. RIC-7 promotes neuropeptide secretion. PLoS Genet. 8: e1002464 10.1371/journal.pgen.1002464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd W., Jacobsen K., Mertens C., Vangestel S., Coomans A. et al. , 2003. Embryonic cell lineage of the marine nematode Pellioditis marina. Dev. Biol. 258: 57–69. 10.1016/S0012-1606(03)00101-5 [DOI] [PubMed] [Google Scholar]
- Hunter C. P., and Kenyon C., 1996. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87: 217–226. 10.1016/S0092-8674(00)81340-9 [DOI] [PubMed] [Google Scholar]
- Kaneko H., Kobayashi E., Yamamoto M., and Shimizu R., 2012. N- and C-terminal transactivation domains of GATA1 protein coordinate hematopoietic program. J. Biol. Chem. 287: 21439–21449. 10.1074/jbc.M112.370437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., and Zahler A. M., 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 10: 1115–1125. 10.1101/gr.10.8.1115 [DOI] [PubMed] [Google Scholar]
- Kiontke K. C., Felix M. A., Ailion M., Rockman M. V., Braendle C. et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339 10.1186/1471-2148-11-339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A., Flibotte S., Taylor J., Waterston R. H., Moerman D. G. et al. , 2018. Mutational and transcriptional landscape of spontaneous gene duplications and deletions in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 115: 7386–7391. 10.1073/pnas.1801930115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren Z., and Trifonov E. N., 2011. Role of everlasting triplet expansions in protein evolution. J. Mol. Evol. 72: 232–239. 10.1007/s00239-010-9425-0 [DOI] [PubMed] [Google Scholar]
- Korf I., Flicek P., Duan D., and Brent M. R., 2001. Integrating genomic homology into gene structure prediction. Bioinformatics 17: S140–S148. 10.1093/bioinformatics/17.suppl_1.S140 [DOI] [PubMed] [Google Scholar]
- Kozlov A. M., Darriba D., Flouri T., Morel B., and Stamatakis A., 2019. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., and Tamura K., 2018. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35: 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S. A., Yang A. W. H., Sasse A., Cowley G., Albu M. et al. , 2019. Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51: 981–989. 10.1038/s41588-019-0411-1 [DOI] [PubMed] [Google Scholar]
- Laugsch M., and Schierenberg E., 2004. Differences in maternal supply and early development of closely related nematode species. Int. J. Dev. Biol. 48: 655–662. 10.1387/ijdb.031758ml [DOI] [PubMed] [Google Scholar]
- Levin M., Hashimshony T., Wagner F., and Yanai I., 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22: 1101–1108. 10.1016/j.devcel.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Lin K. T., Broitman-Maduro G., Hung W. W., Cervantes S., and Maduro M. F., 2009. Knockdown of SKN-1 and the Wnt effector TCF/POP-1 reveals differences in endomesoderm specification in C. briggsae as compared with C. elegans. Dev. Biol. 325: 296–306. 10.1016/j.ydbio.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Thompson S., and Priess J. R., 1995. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83: 599–609. 10.1016/0092-8674(95)90100-0 [DOI] [PubMed] [Google Scholar]
- Lipinski K. J., Farslow J. C., Fitzpatrick K. A., Lynch M., Katju V. et al. , 2011. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr. Biol. 21: 306–310. 10.1016/j.cub.2011.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Onal P., Datta R. R., Rogers J. M., Schmidt-Ott U. et al. , 2018. Ancient mechanisms for the evolution of the bicoid homeodomain’s function in fly development. eLife 7: e34594 10.7554/eLife.34594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M. C., Ha S., Pelczer I., Pal S., and Walker S., 1998. The solution structure of the DNA-binding domain of Skn-1. Proc. Natl. Acad. Sci. USA 95: 8455–8460. 10.1073/pnas.95.15.8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry J., Yochem J., Chuang C. H., Sugioka K., Connolly A. A. et al. , 2015. High-Throughput Cloning of Temperature-Sensitive Caenorhabditis elegans Mutants with Adult Syncytial Germline Membrane Architecture Defects. G3 (Bethesda) 5: 2241–2255. 10.1534/g3.115.021451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry J. A., and Atchley W. R., 2000. Molecular evolution of the GATA family of transcription factors: conservation within the DNA-binding domain. J. Mol. Evol. 50: 103–115. 10.1007/s002399910012 [DOI] [PubMed] [Google Scholar]
- Lowry J. A., Gamsjaeger R., Thong S. Y., Hung W., Kwan A. H. et al. , 2009. Structural analysis of MED-1 reveals unexpected diversity in the mechanism of DNA recognition by GATA-type zinc finger domains. J. Biol. Chem. 284: 5827–5835. 10.1074/jbc.M808712200 [DOI] [PubMed] [Google Scholar]
- Lynch J. A., Brent A. E., Leaf D. S., Pultz M. A., and Desplan C., 2006. Localized maternal orthodenticle patterns anterior and posterior in the long germ wasp Nasonia. Nature 439: 728–732. 10.1038/nature04445 [DOI] [PubMed] [Google Scholar]
- Maduro M., Hill R. J., Heid P. J., Newman-Smith E. D., Zhu J. et al. , 2005a Genetic redundancy in endoderm specification within the genus Caenorhabditis. Dev. Biol. 284: 509–522. 10.1016/j.ydbio.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., 2017. Gut Development in C. elegans. Semin. Cell Dev. Biol. 66: 3–11. 10.1016/j.semcdb.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Broitman-Maduro G., Choi H., Carranza F., Chia-Yi Wu A. et al. , 2015. MED GATA factors promote robust development of the C. elegans endoderm. Dev. Biol. 404: 66–79. 10.1016/j.ydbio.2015.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M. F., Broitman-Maduro G., Mengarelli I., and Rothman J. H., 2007. Maternal deployment of the embryonic SKN-1→MED-1,2 cell specification pathway in C. elegans. Dev. Biol. 301: 590–601. 10.1016/j.ydbio.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Kasmir J. J., Zhu J., and Rothman J. H., 2005b The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev. Biol. 285: 510–523. 10.1016/j.ydbio.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Lin R., and Rothman J. H., 2002. Dynamics of a developmental switch: recursive intracellular and intranuclear redistribution of Caenorhabditis elegans POP-1 parallels Wnt-inhibited transcriptional repression. Dev. Biol. 248: 128–142. 10.1006/dbio.2002.0721 [DOI] [PubMed] [Google Scholar]
- Maduro M. F., Meneghini M. D., Bowerman B., Broitman-Maduro G., and Rothman J. H., 2001. Restriction of mesendoderm to a single blastomere by the combined action of SKN-1 and a GSK-3beta homolog is mediated by MED-1 and -2 in C. elegans. Mol. Cell 7: 475–485. 10.1016/S1097-2765(01)00195-2 [DOI] [PubMed] [Google Scholar]
- Mathelier A., Zhao X., Zhang A. W., Parcy F., Worsley-Hunt R. et al. , 2014. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 42: D142–D147. 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Fukushige T., Krause M. W., Minnema S. E., Goszczynski B. et al. , 2009. ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 327: 551–565. 10.1016/j.ydbio.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor A. P., 2005. How to get ahead: the origin, evolution and function of bicoid. BioEssays 27: 904–913. 10.1002/bies.20285 [DOI] [PubMed] [Google Scholar]
- McVey M., and Lee S. E., 2008. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 24: 529–538. 10.1016/j.tig.2008.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memar N., Schiemann S., Hennig C., Findeis D., Conradt B. et al. , 2019. Twenty million years of evolution: The embryogenesis of four Caenorhabditis species are indistinguishable despite extensive genome divergence. Dev. Biol. 447: 182–199. 10.1016/j.ydbio.2018.12.022 [DOI] [PubMed] [Google Scholar]
- Moskowitz I. P., and Rothman J. H., 1996. lin-12 and glp-1 are required zygotically for early embryonic cellular interactions and are regulated by maternal GLP-1 signaling in Caenorhabditis elegans. Development 122: 4105–4117. [DOI] [PubMed] [Google Scholar]
- Murakami R., Okumura T., and Uchiyama H., 2005. GATA factors as key regulatory molecules in the development of Drosophila endoderm. Dev. Growth Differ. 47: 581–589. 10.1111/j.1440-169X.2005.00836.x [DOI] [PubMed] [Google Scholar]
- Nomoto Y., Kubota Y., Ohnishi Y., Kasahara K., Tomita A. et al. , 2019. Gene Cascade Finder: A tool for identification of gene cascades and its application in Caenorhabditis elegans. PLoS One 14: e0215187 10.1371/journal.pone.0215187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omichinski J. G., Clore G. M., Schaad O., Felsenfeld G., Trainor C. et al. , 1993. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261: 438–446. 10.1126/science.8332909 [DOI] [PubMed] [Google Scholar]
- Omranian N., and Nikoloski Z., 2017. Computational Approaches to Study Gene Regulatory Networks. Methods Mol. Biol. 1629: 283–295. 10.1007/978-1-4939-7125-1_18 [DOI] [PubMed] [Google Scholar]
- Owraghi M., Broitman-Maduro G., Luu T., Roberson H., and Maduro M. F., 2010. Roles of the Wnt effector POP-1/TCF in the C. elegans endomesoderm specification gene network. Dev. Biol. 340: 209–221. 10.1016/j.ydbio.2009.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page B. D., Zhang W., Steward K., Blumenthal T., and Priess J. R., 1997. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 11: 1651–1661. 10.1101/gad.11.13.1651 [DOI] [PubMed] [Google Scholar]
- Pal S., Lo M. C., Schmidt D., Pelczer I., Thurber S. et al. , 1997. Skn-1: evidence for a bipartite recognition helix in DNA binding. Proc. Natl. Acad. Sci. USA 94: 5556–5561. 10.1073/pnas.94.11.5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I. S., and Davidson E. H., 2016. Implications of Developmental Gene Regulatory Networks Inside and Outside Developmental Biology. Curr. Top. Dev. Biol. 117: 237–251. 10.1016/bs.ctdb.2015.12.014 [DOI] [PubMed] [Google Scholar]
- Raj A., Rifkin S. A., Andersen E., and van Oudenaarden A., 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918. 10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson S. M., Lo M. C., Odom R., Yang X. D., Medina J. et al. , 2011. Functional analyses of vertebrate TCF proteins in C. elegans embryos. Dev. Biol. 355: 115–123. 10.1016/j.ydbio.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau C. E., Downs W. D., Lin R., Wittmann C., Bei Y. et al. , 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90: 707–716. 10.1016/S0092-8674(00)80531-0 [DOI] [PubMed] [Google Scholar]
- Roy S. W., and Gilbert W., 2005. The pattern of intron loss. Proc. Natl. Acad. Sci. USA 102: 713–718. 10.1073/pnas.0408274102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. W., and Penny D., 2006. Smoke without fire: most reported cases of intron gain in nematodes instead reflect intron losses. Mol. Biol. Evol. 23: 2259–2262. 10.1093/molbev/msl098 [DOI] [PubMed] [Google Scholar]
- Rudel D., and Kimble J., 2002. Evolution of discrete Notch-like receptors from a distant gene duplication in Caenorhabditis. Evol. Dev. 4: 319–333. 10.1046/j.1525-142X.2002.02027.x [DOI] [PubMed] [Google Scholar]
- Sawicka K., Bushell M., Spriggs K. A., and Willis A. E., 2008. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 36: 641–647. 10.1042/BST0360641 [DOI] [PubMed] [Google Scholar]
- Sawyer J. M., Glass S., Li T., Shemer G., White N. D. et al. , 2011. Overcoming redundancy: an RNAi enhancer screen for morphogenesis genes in Caenorhabditis elegans. Genetics 188: 549–564. 10.1534/genetics.111.129486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierenberg E., 2001. Three sons of fortune: early embryogenesis, evolution and ecology of nematodes. BioEssays 23: 841–847. 10.1002/bies.1119 [DOI] [PubMed] [Google Scholar]
- Schierenberg E., 2006. Embryological variation during nematode development. (January 02, 2006), WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.55.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer P. H., Nsah N. A., Grotehusmann H., Kroiher M., Loer C. et al. , 2014. Developmental variations among Panagrolaimid nematodes indicate developmental system drift within a small taxonomic unit. Dev. Genes Evol. 224: 183–188. 10.1007/s00427-014-0471-2 [DOI] [PubMed] [Google Scholar]
- Schulze J., and Schierenberg E., 2011. Evolution of embryonic development in nematodes. Evodevo 2: 18 10.1186/2041-9139-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P., Lo M. C., Robertson S. M., and Lin R., 2005. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev. Biol. 285: 584–592. 10.1016/j.ydbio.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M. et al. , 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermann E. M., Strohmaier K. R., Maduro M. F., and Rothman J. H., 2010. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification→differentiation transition. Dev. Biol. 347: 154–166. 10.1016/j.ydbio.2010.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J., Lawson D., Davis P., Williams G., and Howe K., 2014. Overview of gene structure in C. elegans. (October 29, 2014), WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.65.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M., Jackle H., and Schmidt-Ott U., 1999. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc. Natl. Acad. Sci. USA 96: 3786–3789. 10.1073/pnas.96.7.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L. D., Bao Z., Blasiar D., Blumenthal T., Brent M. R. et al. , 2003. The Genome Sequence of Caenorhabditis briggsae: A Platform for Comparative Genomics. PLoS Biol. 1: E45 10.1371/journal.pbio.0000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens L., Felix M. A., Beltran T., Braendle C., Caurcel C. et al. , 2019. Comparative genomics of 10 new Caenorhabditis species. Evol Lett 3: 217–236. 10.1002/evl3.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan-Brown J. L., Tandon P., Bird K. E., Dickinson D. J., Tintori S. C. et al. , 2016. Identifying Regulators of Morphogenesis Common to Vertebrate Neural Tube Closure and Caenorhabditis elegans Gastrulation. Genetics 202: 123–139. 10.1534/genetics.115.183137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Schierenberg E., White J. G., and Thomson J. N., 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Taheri-Ghahfarokhi A., Taylor B. J. M., Nitsch R., Lundin A., Cavallo A. L. et al. , 2018. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 46: 8417–8434. 10.1093/nar/gky653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe C. J., Schlesinger A., Carter J. C., and Bowerman B., 1997. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90: 695–705. 10.1016/S0092-8674(00)80530-9 [DOI] [PubMed] [Google Scholar]
- Torres Cleuren Y. N., Ewe C. K., Chipman K. C., Mears E. R., Wood C. G. et al. , 2019. Extensive intraspecies cryptic variation in an ancient embryonic gene regulatory network. eLife 8: e48220 10.7554/eLife.48220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M., Sanchez-Ferras O., and Bouchard M., 2018. GATA transcription factors in development and disease. Development 145: dev164384 10.1242/dev.164384 [DOI] [PubMed] [Google Scholar]
- True J. R., and Haag E. S., 2001. Developmental system drift and flexibility in evolutionary trajectories. Evol. Dev. 3: 109–119. 10.1046/j.1525-142x.2001.003002109.x [DOI] [PubMed] [Google Scholar]
- van Schendel R., and Tijsterman M., 2013. Microhomology-mediated intron loss during metazoan evolution. Genome Biol. Evol. 5: 1212–1219. 10.1093/gbe/evt088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther R. F., Atlas E., Carrigan A., Rouleau Y., Edgecombe A. et al. , 2005. A serine/threonine-rich motif is one of three nuclear localization signals that determine unidirectional transport of the mineralocorticoid receptor to the nucleus. J. Biol. Chem. 280: 17549–17561. 10.1074/jbc.M501548200 [DOI] [PubMed] [Google Scholar]
- Weirauch M. T., Yang A., Albu M., Cote A. G., Montenegro-Montero A. et al. , 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443. 10.1016/j.cell.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann B. M., Trautwein M. D., Winkler I. S., Barr N. B., Kim J. W. et al. , 2011. Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. USA 108: 5690–5695. 10.1073/pnas.1012675108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenfahrt, T., J.Y. Berg, E.O. Nishimura, A.G. Robinson, B. Goszczynski et al., 2015 The Function and Regulation of the GATA Factor ELT-2 in the C. elegans Endoderm. Development. [DOI] [PMC free article] [PubMed]
- Witze E. S., Field E. D., Hunt D. F., and Rothman J. H., 2009. C. elegans pur alpha, an activator of end-1, synergizes with the Wnt pathway to specify endoderm. Dev. Biol. 327: 12–23. 10.1016/j.ydbio.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Felix M. A., and Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012. 10.1534/genetics.110.120550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Gu L., Romeo P. H., Bories D., Motohashi H. et al. , 1994. Human GATA-3 trans-activation, DNA-binding, and nuclear localization activities are organized into distinct structural domains. Mol. Cell. Biol. 14: 2201–2212. 10.1128/MCB.14.3.2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura J., Ichikawa K., Shoura M. J., Artiles K. L., Gabdank I. et al. , 2019. Recompleting the Caenorhabditis elegans genome. Genome Res. 29: 1009–1022. 10.1101/gr.244830.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Ihuegbu N., Lee M., Schriefer L., Wang T. et al. , 2012. Conserved Motifs and Prediction of Regulatory Modules in Caenorhabditis elegans. G3 (Bethesda) 2: 469–481. 10.1534/g3.111.001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Boyle T. J., Bao Z., Murray J. I., Mericle B. et al. , 2008. Comparative analysis of embryonic cell lineage between Caenorhabditis briggsae and Caenorhabditis elegans. Dev. Biol. 314: 93–99. 10.1016/j.ydbio.2007.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Flibotte S., Murray J. I., Blick D., Boyle T. J. et al. , 2010. New tools for investigating the comparative biology of Caenorhabditis briggsae and C. elegans. Genetics 184: 853–863. 10.1534/genetics.109.110270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Fukushige T., McGhee J. D., and Rothman J. H., 1998. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 12: 3809–3814. 10.1101/gad.12.24.3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Hill R. J., Heid P. J., Fukuyama M., Sugimoto A. et al. , 1997. end-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 11: 2883–2896. 10.1101/gad.11.21.2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences identified in this work are available as Supplemental files. Supplemental material available at figshare: https://doi.org/10.25387/g3.9820622.