Abstract
Tanning response and melanocyte differentiation are mediated by the central transcription factor MITF. This involves the rapid and selective induction of melanocyte maturation genes, while concomitantly the expression of other effector genes is maintained. In this study, using cell‐based and zebrafish model systems, we report on a pH‐mediated feed‐forward mechanism of epigenetic regulation that enables selective amplification of the melanocyte maturation program. We demonstrate that MITF activation directly elevates the expression of the enzyme carbonic anhydrase 14 (CA14). Nuclear localization of CA14 leads to an increase of the intracellular pH, resulting in the activation of the histone acetyl transferase p300/CBP. In turn, enhanced H3K27 histone acetylation at selected differentiation genes facilitates their amplified expression via MITF. CRISPR‐mediated targeted missense mutation of CA14 in zebrafish results in the formation of immature acidic melanocytes with decreased pigmentation, establishing a central role for this mechanism during melanocyte differentiation in vivo. Thus, we describe an epigenetic control system via pH modulation that reinforces cell fate determination by altering chromatin dynamics.
Keywords: epigenetics and zebrafish, histone acetylation, p300/CBP, pH regulation, pigmentation
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Post-translational Modifications, Proteolysis & Proteomics
Melanocyte differentiation is mediated by the transcription factor MITF. A feed‐forward loop involving MITF and the carbonic anhydrase 14 enables the rapid and selective expression of pigmentation genes via pH‐dependent epigenetic marks.

Introduction
Gene expression networks, as well as upstream pathways that govern their expression, are well‐studied for skin melanocytes 1. Precursors of these cells are neural crest derived and are present in several locations in skin such as hair follicles, dermis, and presumably the epidermis 2. In response to specific cues from the wnt and melanocortin pathways, these precursors are believed to migrate and rapidly mature into pigmented melanocytes in skin 3. Microphthalmia‐associated transcription factor (MITF), the central melanocyte specific transcription factor, is crucial for almost all aspects of development, maintenance, and survival of melanocytes across vertebrates 4.
To mediate these wide ranging effector functions, MITF target genes are selectively activated during specific melanocyte transitions 5. The level and activity of MITF are governed by cAMP/PKA and MAPK signaling pathways, respectively 6, 7, 8. M/E‐box‐mediated direct activation is observed for several of the MITF target genes. Physical interaction of MITF with factors such as YY1 and proximal interplay with Sox10 mediates activation of a subset of these effector genes, thereby adding another layer of complexity to this selective activation 9. Recruitment of transcriptional co‐regulators such as BRG1 and p300/CBP facilitates the dynamic interaction of MITF to a subset of promoters 10, 11. A key challenge still is to selectively activate certain gene modules dynamically, while maintaining other effector functions of MITF 9, 12. This is evident during tanning response and melanocyte differentiation where MITF activates pigmentation genes several fold, while maintaining the expression of proliferation and survival genes at normal levels 13. These recent findings indicate that multiple linked regulatory loops enable selective outcomes in pigmentation during the tanning response. The basis of this selectivity by MITF and the mechanistic understanding is just beginning to emerge, and epigenetic factors are thought to play a class‐specific activator role 11, 13, 14, 15. In this context, experiments by Laurette et al, followed by analysis by Malcov‐Brog et al, recently described increased H3K27 acetylation at several pigmentation genes downstream of MITF.
Epigenetic regulation is likely to be intricately linked to cellular cues that cooperate with external signals in modulating networks involved in determining cell fates. pH homeostasis is a crucial biological process that is linked to several cellular pathways and is a plausible candidate 16, 17, 18. While the importance of pH balance in cell homeostasis is well appreciated, emerging evidence indicates that pH could impact on cellular events by programmatically modulating existing networks 18, 19. An increase of the intracellular pH is, e.g., critical for the differentiation of mouse embryonic stem cells, as well as Drosophila adult follicle stem cells, highlighting a role of pH in controlling key cell fates 20.
A large family of carbon dioxide‐metabolizing enzymes, carbonic anhydrases (CA), regulate pH across life forms 21. Members of this family are ubiquitously expressed in many cell types and localize to distinct subcellular compartments 22, 23. CA14 is a type I transmembrane protein expressed in a variety of cell types, but has been studied mainly in retina, brain, kidney, smooth muscle, and cardiomyocytes 24, 25. Studies using knock‐out mice have established a role of CA14 in buffering alkaline shifts in brain 26. Intracellular expression of CA14 in the sarcolemma of smooth muscle cells is known to modulate impulse induced muscle contractions 27. Cells of the retinal pigmented epithelium (RPE) demonstrate high expression of CA14 on the apical region, and Ca14 knock‐out mice are deficient in eliciting a functional retinal light response 28.
There are several links between pH and melanin synthesis. The enzymes involved in melanogenesis, namely tyrosinase (Tyr), tyrosinase‐related protein (Tyrp1), and dopachrome tautomerase (Dct) that reside within the melanosomes, are modulated by luminal pH changes. V‐type ATPases are involved in maintaining an acidic pH of these lysosome‐related organelles. Optimal pH for melanogenesis has remained controversial, and marginal luminal alkalinization by protonophores is thought to promote melanin synthesis 29. pH changes in the endolysosomal compartments also alter the trafficking and maturation of melanosomal enzymes in addition to their activity, as well as the type of melanin being synthesized 30, 31, 32. Predictably, alterations of the cellular pH modulate the pigmentation in melanocytes, inducing a robust alteration in the net melanin synthesis. Therefore, we decided to systematically study the impact of pH on the entire melanogenic program at various levels.
In this study, we demonstrate that intracellular pH is a critical cue for amplifying the melanocyte maturation program. We trace genes that follow a concordant pattern of regulation with pigmentation, and identify CA14 to be a MITF regulated gene. CA14 acts as a feed‐forward activator of the MITF regulatory network and amplifies the expression of melanocyte maturation genes by altering histone acetylation marks via programmed intracellular pH changes. Using cell‐based and zebrafish model systems, we demonstrate that a feed‐forward loop involving CA14 is critical to mediate the melanocyte maturation program downstream of MITF.
Results
Alkaline intracellular pH induces pigmentation by enhancing the melanogenesis gene expression
When culturing B16 cells in DMEM, the CO2‐HCO3 − buffering system maintains a stable media pH. Hence, we resorted to establish a pH‐pigmentation link by modulating the prevailing CO2 levels. We set up progressive pigmentation using the B16 pigmentation oscillator model as described earlier 33, and described in Appendix Fig S1. In this setup, B16 cells progressively activate the melanogenesis gene expression program and increase pigmentation over a course of 8–12 days. To alter the pH, 10% CO2 levels were tested. We measured the extracellular pH (pHe) using the standard pH electrode under controlled conditions of temperature and CO2 saturation. While the cells grown in 5% CO2 showed an increase in pHe from around 7.4–7.8 on day 4 after induction of the pigmentation program, the 10% CO2 sustained a constant pH of around 7.4 (Fig EV1A). This trend was reflected in the intracellular pH, which is close to 7.9 on day 4 under the routine 5% CO2 condition. However, using 10% CO2 the pH remained close to 7.0, similar to day 0, where the cells are depigmented (Fig 1A). When cells were assessed for the cumulative accumulation of melanin on day 8, under the 10% CO2 condition we observed less pigmentation (Fig 1B). A melanin content assay performed using synthetic melanin standard confirmed that the levels of melanin are significantly low (Fig EV1B). Further electron microscopic evaluation of day 8 cells confirmed that indeed melanin laden stage III and IV melanosomes are dramatically reduced under these conditions (Fig 1C).
Figure EV1. 10% CO 2 leads to extracellular acidification leading to decreased melanin content and increased proliferation.
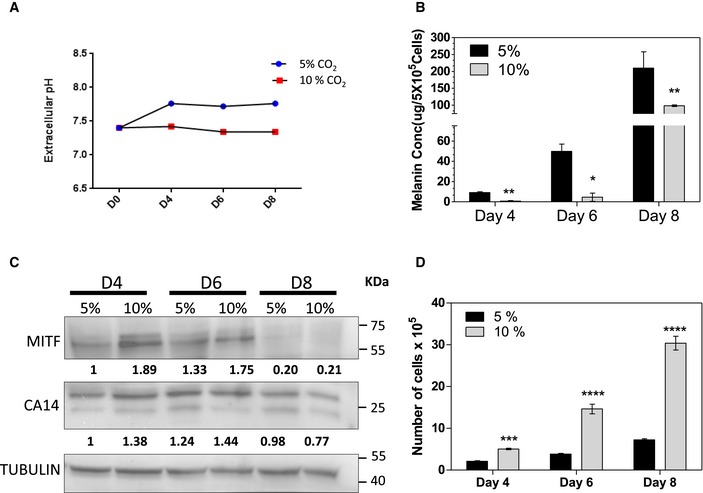
- Line graph showing extracellular pHe at various days of pigmentation in 5% and 10% CO2 cultured B16 cells.
- Bar graph depicts the colorimetric melanin estimation of B16 melanoma cells cultured under 5% CO2 and 10% CO2 growth conditions, on indicated days of pigmentation using synthetic melanin as a standard. Bars represent mean ± SEM independent biological replicate, n = 3.
- Western blot analysis of CA14 and MITF normalized to tubulin on days 4, 6, and 8 under 5 and 10% CO2.
- Alteration of cellular pH by the modulation of CO2 results in changes in cell proliferation in B16 cells as measured by cell count. Bars indicate mean ± SEM across three independent experiments.
Figure 1. Modulation of intracellular pH by CO 2 results in predictable changes in pigmentation via a transcriptional response.
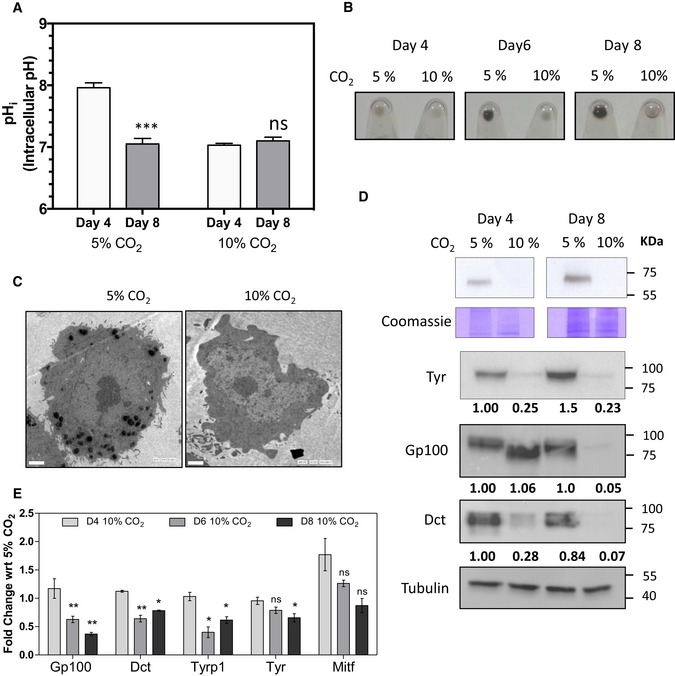
- B16 cells were maintained at 5 or 10% CO2 for indicated days, and intracellular pH was measured by ratiometric imaging using BCECF‐AM for the respective conditions. While the pHi of 10% CO2 grown cells on day 4 and day 8 remained close to 7.0, in 5% condition the pHi on day 4 was close to 7.9. Bars represent mean ± SEM, and the data are obtained from three biological replicates with around 60 cells each.
- Pellets of B16 cells at various days cultured under 5 or 10% CO2 conditions. While the initial day 4 pellets look comparable, but on day 6 and day 8, 5% CO2 grown cells accumulate melanin, while the 10% CO2 grown cells remain less pigmented.
- Transmission electron micrograph images of day 8 cells indicate that the 5% CO2 grown cells have many electron‐dense melanosomes, whereas the 10% CO2 grown cells are devoid of these structures. Scale bars: 2 μm.
- Top panel shows in‐gel tyrosinase activity developed using L‐DOPA as the substrate and below is part of the gel stained using Coomassie brilliant blue. (bottom panel) Western blot analysis of Tyr, Gp100, and Dct proteins normalized to tubulin. Numbers represent tubulin normalized fold changes corresponding to day 4 cells grown at 5% CO2. 10% CO2 reduces expression of pigmentation‐related markers.
- qRT‐PCR analysis of pigmentation‐related gene transcripts Gp100, Dct, Tyrp1, Tyr, and Mitf normalized to 18s rRNA on days 4, 6, and 8 during pigmentation. The fold changes are depicted for the corresponding days for cells grown at 5% CO2. Progressive reduction in the mRNA expression of pigmentation genes is observed at the transcript level. Bars represent mean ± SEM, and the data are obtained from four independent replicate experiments.
We then analyzed steady‐state protein levels of components of the melanosomal machinery. All the three pigmentation proteins, Tyr, Dct and Gp100, were drastically reduced under 10% CO2 (Fig 1D). However, the levels of MITF were not decreased; rather, we observed a mild increase in the level of this central transcription factor (Fig EV1C). In concordance with earlier studies that suggested a pH‐dependent alteration in protein stability, the activity of tyrosinase assessed by L‐DOPA based in‐gel assays, as well as Western blot analysis confirmed a dramatic reduction 30. However, a decrease in Dct and Gp100 levels was unanticipated. Gp100 showed a greater decrease on day 8 and had comparable expression levels on day 4, with alterations in the mobility suggesting processing differences 34.
Surprisingly, transcript levels of these downstream pigmentation genes were lower in 10% CO2 condition (Fig 1E). Thus, keeping cells in an acidic pHi delays pigmentation by the decreased expression of pigmentation genes, despite comparable levels of MITF. We also observed an increase in cell numbers at 10% CO2 (Fig EV1D). Therefore, there is seemingly an additional level of cellular programming induced by intracellular pH changes that governs melanogenesis beyond enzyme activity and protein stability, and involving transcriptional regulation.
A candidate effector CA14 follows a concordant expression pattern with pigmentation genes
We set out to identify the molecular mechanism behind this pH‐dependent transcriptional response. Previously, we have established an autonomous B16 cell model and demonstrated the utility of this model to identify underlying networks that govern pigmentation. A second model involves the growth of pigmented melanoma tumor derived from mice and grown in vitro for four consecutive passages. In the second model, pigmented cells derived from tumor progressively lose pigmentation and the underlying gene expression pattern when grown in vitro. Details of the models used and a set of genes that showed a concordant regulation across these two reversible models are provided in Ref. 33 (see also Appendix Fig S1). Among the common set of regulated genes, five of the fifty candidates, Tyr, Tyrp1, Mlana, Mcoln3, Si, and Rab27a are targets of the central melanocyte transcription factor MITF. These are well‐known pigmentation genes directly involved in the process of melanogenesis and melanosome maturation, which are directly linked to the process of melanocyte differentiation. From this analysis carbonic anhydrase 14 emerged as a putative candidate gene that could directly regulate pH, as it is regulated together with pigmentation in both the cellular model systems (Appendix Fig S1).
We then analyzed the pattern of expression of carbonic anhydrases from the data and observed that of the several CAs expressed in melanocytes, only CA14 showed a regulation pattern similar to the melanocyte differentiation genes tyr, dct, and tyrp1 (Fig 2A) (GSE54359) 33. Hence, we proceeded to characterize the regulation of CA14 with an aim to establish its role in melanocyte maturation.
Figure 2. CA14 expression is directly controlled by Mitf.
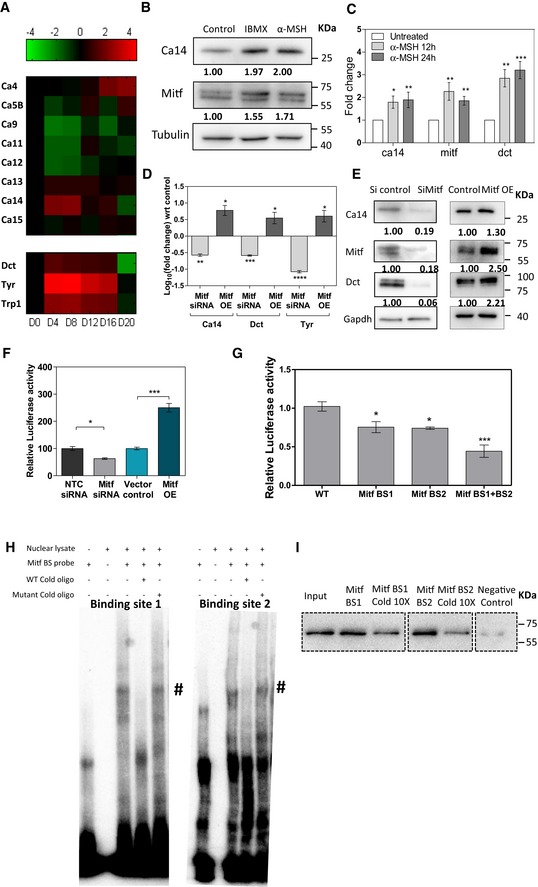
- (top) Panel of carbonic anhydrases expressed in B16 cells along with (bottom) pigmentation genes during the course of in vitro pigmentation oscillation. The fold change in the mRNA expression is represented as a heat‐map relative to the expression on day 0. Notably CA14 follows a concordant pattern of expression with the pigmentation genes.
- Western blot analysis of CA14, Mitf, and normalization by tubulin, on treatment with Mitf inducers 60 μM isobutyl methyl xanthine (IBMX) and 600 nM alpha‐melanocyte stimulating hormone (α‐MSH) for 72 h in Melan‐A cells. Fold change with respect to control untreated cells normalized to tubulin expression is depicted as numbers below each lane.
- qRT‐PCR for ca14, Mitf, and Dct transcripts upon treatment with α‐MSH for 12 and 24 h in Melan‐A cells. Fold change is depicted (mean ± SEM, n = 3, biological replicates) calculated using 18s rrna as the reference.
- qRT‐PCR for ca14, Dct, and Tyr transcripts upon Mitf knockdown and overexpression in B16 cells. Log10 fold change is depicted (mean ± SEM, n ≥ 3, biological replicates) calculated using Tubulin as the reference with respect to scrambled siRNA or pcDNA3 vector overexpression.
- Western blot analysis of CA14, Mitf, and Dct normalized to Gapdh upon knockdown and overexpression of Mitf with respect to scrambled siRNA or pcDNA3 vector overexpression as control.
- Dual‐luciferase assay performed on B16 cells transfected with luciferase construct (pGL4.23) containing 3 kb upstream region of ca14 promoter, upon Mitf knockdown and overexpression. Renilla luciferase driven by cytomegalovirus promoter (pGL4.75) was used for reference. Bars represent mean ± SEM across three independent experiments.
- Dual‐luciferase assay performed on B16 cells transfected with luciferase construct (pGL4.23) containing Mitf binding site mutations in the 3 kb upstream region of ca14 promoter upon Mitf overexpression. Renilla luciferase driven by cytomegalovirus promoter (pGL4.75) was used for reference. Bars represent mean ± SEM across three independent experiments.
- Electrophoretic Mobility Shift Assay (EMSA) performed on B16 nuclear lysate using labeled wild‐type (BS1 and BS2) probes encompassing E/M box sequence in ca14 promoter. When indicated, reactions were carried out in presence of an excess (10‐fold) of unlabeled homologous or mutated probe. # indicates the bands specific to the shift.
- Western blot analysis of biotinylated probes (ca14 promoter region containing Mitf binding sites) pull‐down probed with Mitf antibody. When indicated, reactions were carried out in presence of an excess (10‐fold) of unlabeled homologous probe. Negative control indicates pull‐down experiment performed with non‐biotinylated DNA. 10% input was taken as control.
CA14 expression is induced upon activation of the melanocortin pathway via MITF
As CA14 showed concordant expression with pigmentation genes and correlated with the activity of MITF, we set out to identify whether Ca14 is indeed a downstream gene and could play an important role in mediating the effects of MITF in melanocytes. Overexpression of Mitf, followed by global gene expression analysis, revealed a set of regulated genes 35. Chromatin immunoprecipitation (ChIP) studies have independently identified several promoters occupied by this transcription factor 36. Combined analysis of both these approaches resulted in a set of genes enriched in known targets of Mitf, along with several other putative candidates. In this set of genes CA14 features, and hence emerged as a promising downstream target of MITF. From publicly available “The Cancer Genome Atlas” (TCGA) datasets, we analyzed the expression of CA14 and MITF across 473 melanoma samples and observed a strong positive correlation between MITF and CA14 mRNA reads (Pearson's correlation coefficient 0.61), and this was comparable to that of other bona fide targets of MITF such as Tyr, Mcoln3, and Mlana (Fig EV2A), suggesting CA14 to be a MITF target gene.
Figure EV2. CA14 is a putative pigmentation regulator associated with MITF .
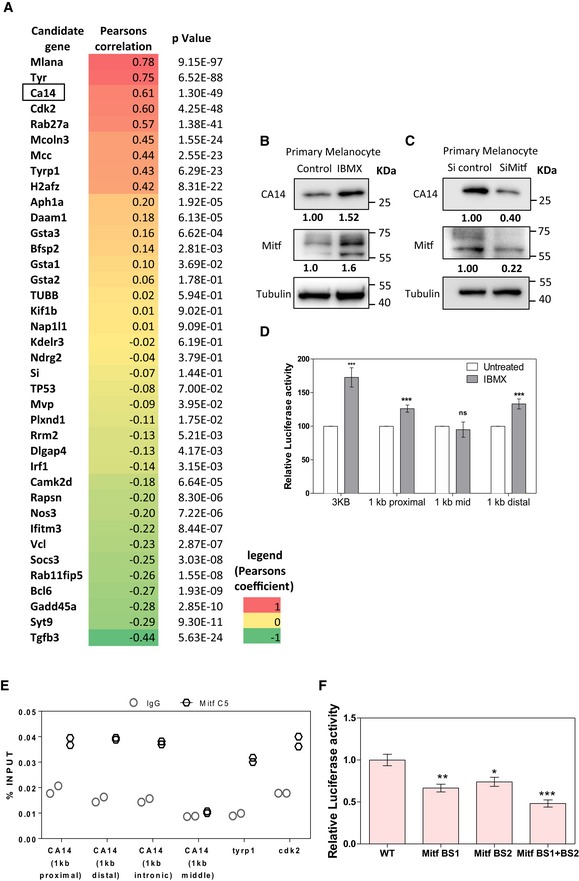
- Correlation analysis of the expression of the melanocyte maturation‐associated genes identified from the two reversible pigmentation models with the expression of MITF in the Cancer genome atlas (TCGA) data of melanoma samples (SKCM). Pearson's correlation coefficient and P‐value describing the chance of the correlation being random was calculated across 473 melanoma samples with the normalized gene expression using RNA sequencing data from TCGA and represented as a heat‐map. Expression of CA14 has a very high correlation with that MITF, suggesting that it may be a target gene like others such as Tyr, Cdk2, and Mcoln3.
- Western blot analysis of CA14 and MITF normalized to tubulin in primary human melanocytes upon 100 μM IBMX treatment.
- Western blot analysis of CA14 and MITF normalized to tubulin in primary human melanocytes upon silencing of MITF using siRNA.
- Luciferase reporter assay using ca14 promoter in B16 cells normalized to renilla luciferase upon 100 μM IBMX treatment. Indicated are the four constructs tested, 3kB upstream region, or 1 kb each of proximal, mid‐, and distal regions. Bars represent mean ± SEM across n = 3 biological replicates.
- Chromatin immunoprecipitation studies with C5 MITF antibody followed by qPCR analysis of select regions of the ca14 promoter. Circle represents percent input in IgG enrichment, and dark hexagons represent percent input in MITF ChIP. Two independent replicates are depicted.
- Luciferase reporter assay using the ca14 promoter in B16 cells followed by 100 μM IBMX treatment, normalized to renilla luciferase, upon mutation of E/M box sequence 1, 2, or both. Bars represent mean ± SEM across n = 3 biological replicates.
α‐Melanocyte‐stimulating hormone (α‐MSH) and iso‐butyl methyl xanthine (IBMX) are used as inducers of Mitf to stimulate melanocytes 37. We carried out similar experiments in mouse Melan‐A cells, as well as the primary human melanocytes. Treatment of Melan‐A cells with 60 μM IBMX or 1 μM α‐MSH for 48 h and 72 h resulted in the induction of Mitf and the downstream target gene Dct, confirming activation of the melanocortin pathway. The antibody used to detect CA14 was validated by Western blot, and we could identify the 25‐kDa form of CA14. The intensity of the band was reduced upon silencing with a pool of siRNA against Ca14 and increased upon expression of Ca14 ORF in an expression vector, confirming specificity of the antibody (Fig EV3D–F). With IBMX as well as α‐MSH treatments, we could observe elevation of the 25‐kDa form of CA14 protein, indicating that activation of Mitf mediates Ca14 induction (Fig 2B). We observed a similar induction of CA14 in primary human melanocytes treated with 60 μM IBMX for 48 h (Fig EV2B). A concomitant decrease was observed upon MITF silencing (Fig EV2C).
Figure EV3. Mitf regulates pH via CA14, and pigmentation changes associated with CA14 are observed in primary human melanocytes.
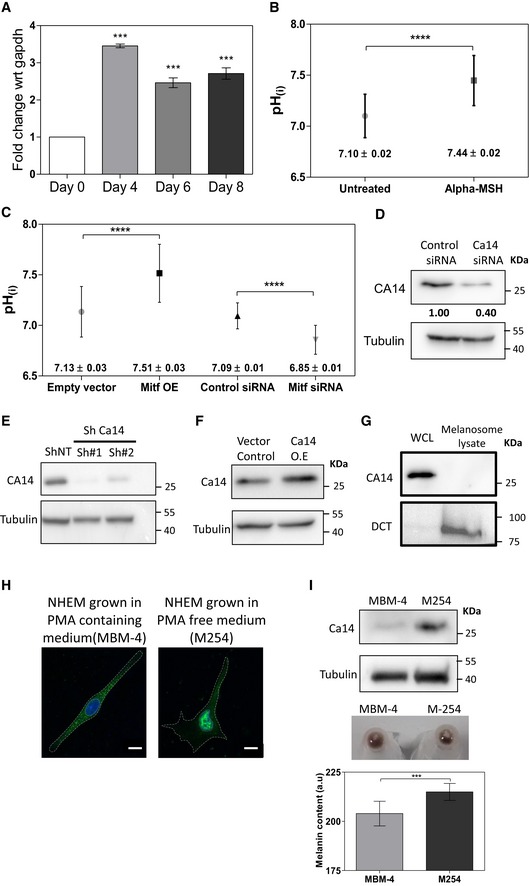
- Levels of ca14 mRNA measured by qRT PCR analysis across different days of pigmentation. Bars indicate mean ± SEM across three independent experiments.
- pHi upon activation of MITF using 1 μM α‐MSH in B16 cells. Data from 50 cells across two independent experiments. The solid symbol indicates mean, and error bars indicate SEM.
- pHi upon MITF overexpression and knockdown using siRNA in B16 cells. Data from 50 cells across two independent experiments. The solid symbol indicates mean, and error bars indicate SEM.
- Western blot analysis of CA14 normalized to tubulin upon silencing of Ca14 using siRNA.
- Western blot analysis of CA14 normalized to tubulin upon silencing of Ca14 using two different sequence independent shRNAs.
- Western blot analysis of CA14 normalized to tubulin upon overexpression of Ca14.
- Fractionation of melanosomes from B16 cells followed by Western blot analysis depicting the levels of CA14 and Dct to assess localization to melanosomes.
- Immunofluorescence study of CA14 in NHEM (normal human primary melanocytes) counter‐stained with DAPI, upon culturing of cells under two different media conditions as indicated. Scale bars: 10 μm.
- Western blot analysis of CA14 in NHEM cells grown under the two conditions. (bottom) Cell pellets and their melanin intensity quantitation. Bars represent mean ± SEM across n = 3 biological replicates.
Next, we performed qRT‐PCR analysis to monitor the mRNA levels in B16 cells treated with 1 μM α‐MSH for 12 and 24 h, as the transcript‐level changes are expected to occur earlier than protein‐level changes. We observed a robust elevation in the expression of pigmentation gene transcript of Dct, and the elevation in Mitf and Ca14 levels were comparable, modest but significant (Fig 2C). Silencing of Mitf using siRNA or its forced expression using a tagged construct, resulted in decrease and increase respectively, with levels of Ca14 similar to that of Dct and Tyr (Fig 2D), hence confirming MITF‐dependent CA14 expression in melanocytes.
MITF directly binds to Ca14 promoter and regulates its expression
To probe the induction of Ca14 by MITF further, we cloned the 3‐kb upstream region of Ca14 in a reporter vector and performed dual‐luciferase assays. Substantial elevation of reporter activity with IBMX was observed, confirming that the induction is a transcriptional response (Fig EV2D). To fine‐map the responsive sites, we adopted two strategies. In the first strategy, we created three clones of the mouse Ca14 promoter carrying the 1 kb transcription start site proximal region, and the mid or the distal 1 kb region. Analysis of MITF binding sites in the promoter of the Ca14 gene indicated multiple sites in both the human and mouse promoter regions (Appendix Fig S2). Luciferase assays confirmed that the proximal and the distal regions are inducible by IBMX, whereas the middle 1 kb promoter sequence was not IBMX inducible.
Further, chromatin immunoprecipitation using C5 monoclonal antibody to MITF demonstrated binding of the protein to a 3 kb as well as proximal and distal regions. Comparable binding of MITF to these Ca14 upstream regions to that present in the Tyrp1 and Cdk2 promoters suggested direct binding of MITF to the Ca14 promoter. However, the middle region did not show appreciable binding of MITF (Fig EV2E). We propose that there are multiple binding sites within the Ca14 promoter responsible for induction by MITF. Mitf dependence of Ca14 expression was probed by the downregulation of this transcription factor by siRNA and forced expression followed by Western blot analysis. The known downstream Mitf target gene Dct was dramatically downregulated, and both Mitf and Ca14 showed a comparable downregulation of around 80%. The opposite trend was observed when the levels of Mitf were elevated (Fig 2E). Similar observations were made using primary human melanocytes and a siRNA against human MITF, confirming Mitf‐dependent CA14 expression (Fig EV2C). The same trend was seen in a promoter assays using the 3 kb Ca14 promoter (Fig 2F). As the ChiP experiments suggested the MITF responsive site to be in the 1st and 3rd kb regions, we mutated the predicted E/M box sequences in the 3 kb promoter and performed promoter assays upon increased Mitf expression. We observed that the two binding sites contribute additively to MITF responsiveness (Fig 2G). Direct binding was evaluated using these two E/M box regions as a probe in Electrophoretic Mobility Shift Assays (EMSA). The shift was abrogated with an E/M box probe but not with the mutated probe (Fig 2H). The MITF monoclonal antibody C5 induced a shift ablation and not a super shift with the Ca14 promoter region (Appendix Fig S3). We therefore performed synthetic promoter pull‐down assays and observed binding of Mitf that could be abrogated with wild‐type E/M box DNA (Fig 2I). Thus, we establish that the melanocyte differentiating melanocortin signaling pathway controls Ca14 expression via direct binding of MITF to the Ca14 promoter.
CA14 is essential for melanocyte maturation in zebrafish
In cultured melanocytes, the pH is governed by prevailing CO2 concentrations, which may override intracellular pH programs. Therefore, to address the role of CA14 on melanocyte functions, we utilized a morpholino (MO)‐based transient silencing approach using the zebrafish model system. Herein, the pigment‐producing cells termed melanophores are ontologically equivalent to higher vertebrate melanocytes, and the underlying gene networks are highly conserved. During embryogenesis, melanophores rapidly mature between 48 and 72 hours post‐fertilization (hpf). Additionally, in the absence of melanosome transport, melanocyte maturation is easy to visualize and to monitor.
After titrating the dose of the CA14 MO based on viability of embryos, scoring of the phenotypes was carried out (Appendix Fig S4). Analysis of melanophores at 48 hpf indicated that the ca14 morphants were lightly pigmented compared to control non‐targeting MO‐injected embryos at the same concentration (Fig 3A–C). To assess melanophore numbers, we generated morphants in the transgenic zebrafish line Tg:ftyrp1‐GFP where the melanophores are fluorescently marked with GFP driven by a fugu Tyrp‐1 promoter 38. To prevent masking of GFP fluorescence by melanin, the embryos were treated with phenylthiourea (PTU), a potent tyrosinase inhibitor. Similar pattern of melanophore positioning and comparable number of melanophores were observed in the morphant fishes, suggesting that the melanophore numbers are unaltered, indicating melanocyte specification and survival are unaffected (Fig 3B and D). However, the mature heavily pigmented melanophores were drastically reduced in the ca14 morphants. Quantitation of the bright field images using ImageJ platform suggested that the morphant melanophores had a higher mean gray value, indicating that they were lighter with less melanin content (Fig 3C). These observations strongly suggest that CA14 plays an important role in the process of melanogenesis, a crucial event associated with melanocyte maturation. Furthermore, upstream events such as specification, migration, and patterning of melanocytes are seemingly unperturbed. We observed a progressive increase in the expression of pigmentation genes tyr, dct, and tyrp1b, concomitant with the known melanocyte maturation process however in the ca14 morphants that elevation was severely curtailed (Fig 3E–G). These data indicate that CA14 mediates melanocyte maturation by altering the pigmentation gene expression. We then went ahead to elucidate the mechanism of CA14‐mediated pigmentation using cultured cells as the model system.
Figure 3. Transient silencing of ca14 in zebrafish decreases melanocyte maturation.

-
ABrightfield images of the lateral view of control and ca14 morphant embryos at 48 h post‐fertilization (hpf). The black structures observed are melanophores, the melanin‐containing cells equivalent to mammalian melanocytes. These are pale in the morphants. Scale bars: 100 μm.
-
BRepresentative fluorescence images of control and ca14 morphant embryos at 48 hours post‐fertilization (hpf) from ftyrp1:GFP line, where the melanocytes are marked by the expression of GFP. Number of melanocytes remains unaffected in the morphants. Scale bars: 100 μm.
-
CMelanin quantitation from the bright field images of control and ca14 morphants carried out using ImageJ platform. The mean gray values are inversely linked to melanin content of the embryo and are represented as scatter plot across melanophores from n = 50 across 3 biological replicates animals.
-
DNumber of ftyrp1 promoter‐driven GFP‐positive melanophores from control and ca14 morphant embryos at 48 hpf remains unchanged (n = 3, biological replicates).
-
E–G(E) qRT‐PCR quantification of pigmentation genes tyrosinase (tyr), (F) dopachrome tautomerase (dct), and (G) tyrosinase‐related protein 1b (tyrp1b) between control and ca14 morphants at 36 and 48 hpf, compared to 24 hpf. Error bars represent SEM across three independent experiments normalized to corresponding control Morpholino set, with rps11 as the normalization control.
CA14 increases intracellular pH during pigmentation
Since Ca14 is a MITF‐dependent gene, expression changes during pigmentation and a downstream modulation of pH due to its carbonic anhydrase activity emerged as a possibility. To study this, we induced pigmentation in B16 cells using pigmentation oscillation (see Appendix Fig S1). On different days of the pigmentation oscillator the levels of the Ca14 mRNA and protein using qRT PCR and Western blot analysis, respectively, were quantified. CA14 expression at both levels was high during early phase (day 4) and progressively decreased at the later days of pigmentation (Figs 4A and EV3A).
Figure 4. CA14 causes increase in pH i observed during pigmentation program.
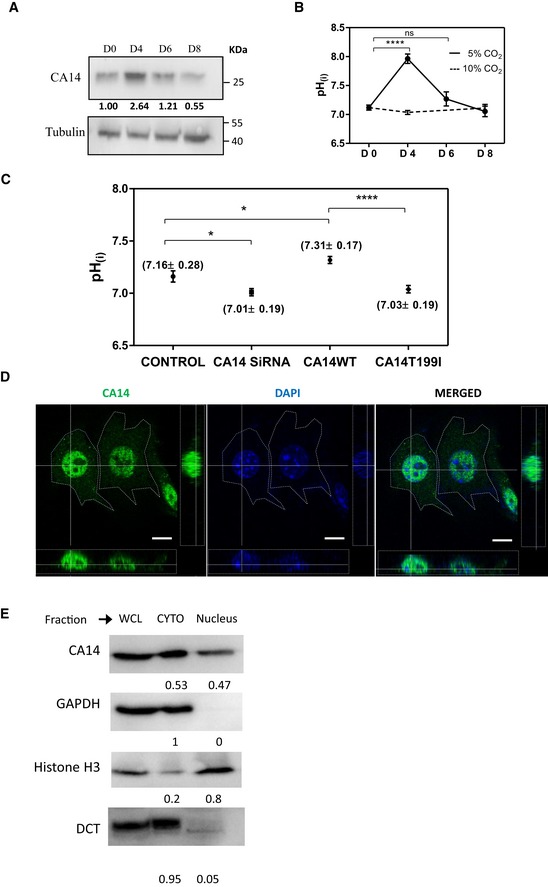
- Western blot analysis of CA14 normalized to tubulin during pigmentation. Indicated days (day 0, 4, 6, and 8) represent number of days after initiating the pigmentation program in the pigment oscillator model. Numbers indicate normalized fold change with respect to tubulin with day 0 as the reference.
- Intracellular pH probed by ratiometric pH‐sensitive fluorescent dye BCECF‐AM in B16‐melanoma cells during different days of pigmentation. The data represent mean ± SD of at least 100 randomly chosen cells across three experiments. Student's t‐test was performed to compare day 0 cells with day 4 and day 6 cells grown under 5% CO2 (day 0 and day 4), P value < 0.0001 and (day 0 and day 6) P value > 0.05 not significant. The trend in pHi follows Ca14 expression in 5% CO2 condition. While pigmentation‐conducive 5% CO2 shows an increase in pHi, the non‐conducive 10% CO2 does not show the elevation in pHi.
- pHi probed by BCECF‐AM in B16‐melanoma cells on siRNA knockdown of ca14 and on overexpression of wild type or the catalytically inactive form of CA14 (CA14T199I). The data represent mean ± SEM of at least 30 transfected cells across two experiments. Statistical analysis was performed using unpaired t‐test. *P ≤ 0.05, ****P ≤ 0.0001.
- Confocal image of immunocytochemistry using CA14 antibody (green) on permeabilized B16 melanoma cells, counterstained by DAPI. Orthogonal projections in both channels and the overlay are depicted. CA14 signal colocalizes in the nuclear compartment stained by DAPI. Scale bars: 10 μm.
- Western blot analysis of subcellular fractions enriched in nucleus and cytosol (post‐nuclear supernatant), using CA14 antibody. H3 antibody was used as a marker for nuclear, GAPDH, and DCT for cytosolic enrichments. Numbers represent relative expression in the indicated fraction.
Source data are available online for this figure.
As carbonic anhydrases mediate pH buffering, we measured pHi using the ratiometric pH sensitive dye (BCECF‐AM) that reports intracellular pH. We observed a consistent increase in pHi from around 7.0 on day 0 to 7.9 on day 4, which is subsequently restored to around 7.0 on day 8 of the oscillator at the pigmentation‐conducive 5% CO2 condition (Fig 4B). Strikingly, this trend followed changes in CA14 expression. Non‐permissive pigmentation conditions involving 10% CO2 did not induce a sharp rise in pHi on day 4 (Fig 4B). We observed an increase in CA14 protein levels at day 4 in both 5% and 10% conditions (Fig EV1C). We then proceeded to unequivocally establish the role of CA14 in altering pHi.
We transfected B16 cells with C‐terminal mCherry tagged CA14 to trace the transfected cells for pH measurements. There was a marginal but significant elevation of pHi, which could not be observed for the catalytically inactive mutant form (CA14T166I). Silencing ca14 using a cocktail of four independent siRNAs caused intracellular acidification and reduced pHi (Fig 4C). Together, these data confirm the role of CA14 in elevating intracellular pH. CA14‐mediated elevation of pHi in B16 cells raised the question about the localization of CA14 in melanocytes.
CA14 is localized to the nucleus in melanocytes
CA14 is an alpha‐type carbonic anhydrase domain containing type I transmembrane protein, containing a short signal sequence 39. Hence, the catalytic motif is predicted to function in buffering extracellular pH (pHe), alternately its localization to intracellular membranes would modulate organelle pH. Expression of CA14 is high in adult brain, retinal pigmented epithelium, liver, heart, and skeletal muscle, and its localization in various cell types differs 27, 40, 41, 42. While in smooth muscle cells its expression is detected in sarcoplasmic reticulum, in other cell types it has been found at the plasma membrane 27. Being a MITF‐inducible gene, CA14 like other targets could be localized to melanosomes, where pH regulation is known to be critical 43. However, as it controls pHi, it should be localized at the cytoplasm or a connected subcellular compartment.
Immunofluorescence studies were carried out on B16 cells to identify the localization of CA14 in melanocytes. Intracellular localization of CA14 could be detected in the nucleus of B16 cells, under endogenous as well as forced expression conditions (Fig 4D). Fractionation of cells to enrich the nuclear and cytoplasmic (post‐nuclear) fraction, followed by Western blot analysis, confirmed substantial localization of around 50% in the nucleus (Fig 4E). To confirm the nuclear localization we generated N‐ or C‐terminal tagged constructs. While the elevation in Ca14 was observed using the C‐terminal tagged construct, the tag could not be detected, suggesting cleavage of the C‐terminal region. The 26‐kDa form of CA14 was found to be localized to the nucleus and was similar to MITF localization detected by immunofluorescence staining of HA‐tagged MITF over‐expressed in B16 cells (Appendix Fig S5). Melanosomal fractions from B16 cells did not contain CA14 (Fig EV3G). The localization of CA14 to the nucleus is strikingly similar to another transmembrane paralog CA9 that also localizes to the nucleus and is involved in nucleolar gene expression 44. CA9 has been an important target for pH modulation in several cancers including melanoma 45.
While culturing primary human melanocytes in the laboratory, we had previously observed that the cells grown under MBM‐4 medium compared to M254 medium were more proliferative and had less melanin content (Fig EV3I, lower panel). Similar observations were also reported earlier 46. We observed that the expression of CA14 was higher in M254 medium and localization was primarily in the nucleus (Fig EV3I, upper panel). Whereas in the MBM‐4 media, cells had lower expression of CA14 and the immunolocalization was found to be diffuse (Fig EV3H). These observations further add credence to a possible role of CA14 in melanocyte maturation. To understand the mechanism, we proceeded to investigate the effect of Ca14 silencing on the expression of pigmentation genes in B16 cells.
Ca14 mediates pigmentation gene expression through a transcriptional response
As Ca14 is regulated by MITF, and considering its ability to control pHi and its role in sustaining melanin content of zebrafish melanophores, it is a likely candidate to mediate melanocyte maturation. Hence, we set out to address the effect of its downregulation on the expression of pigmentation genes. We transfected B16 cells with a pool of four siRNAs targeting Ca14 or a control non‐targeting siRNA pool and the cells were subjected to downstream experiments (Fig EV3D). Independently, shRNA mediated knockdown was also carried out (Fig EV3E). Western blot analysis was performed on the cell lysates and a reduction of around 50% was observed for CA14 (Figs 5A and EV3E). Expression of pigmentation genes was found out to be lower upon Ca14 silencing in both the approaches (Fig 5A, Appendix Fig S5). The mRNA levels of Dct and Tyr were found to be downregulated by qRT PCR analysis (Fig 5B). We further confirmed that this downregulation happens at the transcriptional level by performing luciferase assays using Dct and Tyr promoter driven firefly luciferase (Fig 5C and D). Both promoters were marginally but consistently downregulated by around 30% upon silencing of Ca14.
Figure 5. CA14 brings about transcriptional regulation of downstream pigmentation genes through histone acetylation.
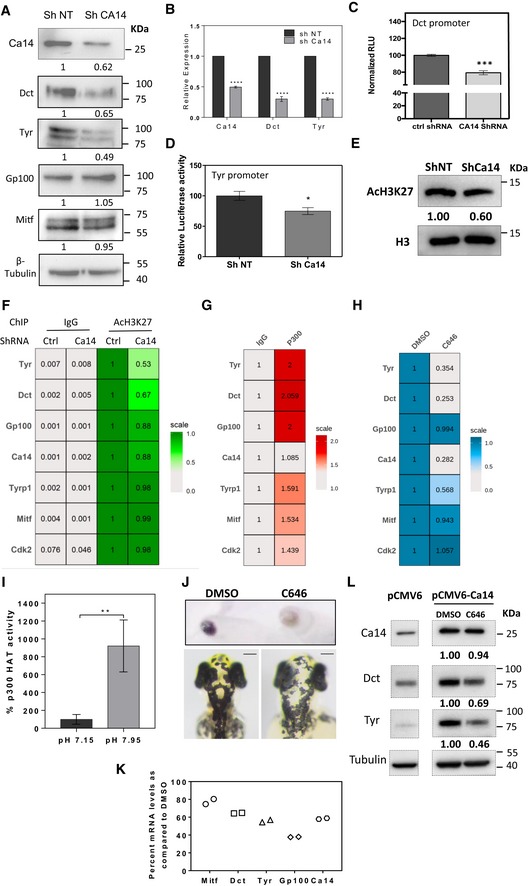
- Western blot analysis of Dct, Gp100, Tyr, and Mitf proteins along with CA14 and tubulin upon knockdown using ca14 shRNA in B16 cells. Numbers represent tubulin normalized fold changes with respect to control non‐targeting shRNA.
- Gene expression analysis using qRT‐PCR of ca14, Dct, and Tyr upon ca14 knockdown using shRNA in B16 cells. Fold change normalized to gapdh with respect to control non‐targeting shRNA. Bars represent mean ± SEM across four independent biological replicates.
- Luciferase assay of Dct promoter cloned downstream of firefly luciferase (pGL4.23), on knockdown using ca14 shRNA. Bars represent mean ± SEM across three independent biological replicates.
- Luciferase assay of Tyr promoter cloned downstream of firefly luciferase (pGL4.23), on knockdown using ca14 shRNA. Bars represent mean ± SEM across three independent biological replicates.
- Western blot analysis was carried out for H3K27 acetylation normalized to total H3, upon ca14 knockdown in B16 melanoma cells.
- Chromatin immunoprecipitation using acetylated H3K27 antibody or normal rabbit IgG as a control on day 4 pigmenting cells of the oscillator in control non‐targetting shRNA and ca14 shRNA transfected cells. qRT‐PCR was performed for select promoters in the immunoprecipitate and input DNA. Relative enrichment with respect to AcH3K27 in control cells under each promoter is depicted as a heat‐map. Statistics for ChIP enrichment is provided in Appendix Table S5.
- Chromatin immunoprecipitation using P300 antibody or normal rabbit IgG as a control on B16 cells. qRT‐PCR was performed for select promoters in the immunoprecipitate and input DNA. Relative fold enrichment of DNA compared to IgG under each promoter is depicted as a heat‐map. Statistics for ChIP enrichment is provided in Appendix Table S6.
- Chromatin immunoprecipitation using acetylated H3K27 antibody or normal rabbit IgG in DMSO‐ and C646‐treated B16 cells. qRT‐PCR was performed for select promoters in the immunoprecipitate and input DNA. Relative fold enrichment of DNA compared to DMSO under each promoter is depicted as a heat‐map. Statistics for ChIP enrichment is provided in Appendix Table S7.
- In vitro histone acetyl transferase (HAT) activity as a function of pH for recombinant HAT domain of p300 protein is depicted. Compared to the HAT activity at pH 7.15, the activity at pH 7.95 was almost 10‐fold.
- Cell pellets and representative bright field images of B16 cells and zebrafish embryos treated with DMSO and EP300/CBP inhibitor 10 μM C646. Decrease in pigmentation is observed in both the models. Scale bars: 100 μm.
- Real‐time qRT‐PCR levels of pigmentation transcripts Dct, Tyr, Gp100, ca14, and Mitf upon selective inhibition of p300/CBP HAT activity in B16 cells. Bars represent mean ± SD across two replicate experiments normalized to 18s rRNA. p300 HAT inhibition resulted in depigmented cells with a decrease in pigmentation gene transcripts.
- Western blot analysis of Dct, Tyr, and CA14 normalized to tubulin from B16 cells transfected with control empty vector or CA14 overexpression vector. A set of CA14 overexpressing cells was treated with 10 μM C646 for 24 h, and Western blot analysis was carried out. Numbers represent fold changes with respect to DMSO‐treated CA14 forced expressed cells. C646‐mediated p300/CBP inhibition reverses the induction of Dct and Tyr caused by forced expression of Ca14.
Given the nuclear expression of CA14, it is likely that local pH changes may culminate in specific transcriptional response. Since Tyr, Dct, and the other melanocyte differentiation genes are direct targets for MITF, it is surprising that CA14 is able to modulate gene expression without affecting MITF levels. It is therefore likely that CA14 could mediate transcriptional activation by facilitating chromatin alterations, which in turn could facilitate MITF‐mediated gene expression. This possibility also allows for promoter‐specific alterations and selectivity in downstream gene expression. Changes in histone acetylation upon intracellular pH changes have been reported before 19; hence, we set out to investigate this possibility.
Ca14 promotes H3K27 acetylation marks on select MITF target genes
Thus far we could establish that CA14 is present in the nucleus, increases intracellular pH, and enhances the expression of pigmentation genes. We went ahead and monitored global changes in histone acetylation using Western blot analysis and observed a modest but consistent decrease across AcH3K27 (acetylated histone 3 at Lys 27 position), but not in AcH3K18, or AcH3K9 (Figs 5E and EV4A). This provided an exciting possibility of local pH changes culminating in epigenetic marks that could in turn affect pigmentation gene expression profiles downstream of MITF. While the trend of decrease in AcH3K27 was consistent across three independent biological replicate experiments, the effect was marginal, leading to around 30–40% reduction in global acetylation levels (Fig 5E). We then proceeded to query the specific changes in a battery of promoters of MITF target genes using chromatin immunoprecipitation (ChIP).
Figure EV4. Histone acetylation changes upon ca14 silencing and C646 treatment.
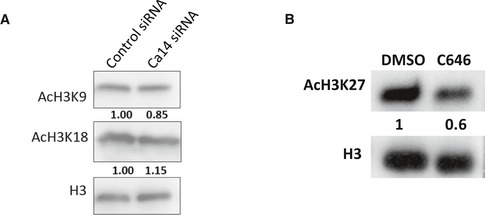
- Western blot analysis of control and ca14 siRNA‐treated cells for various acetylation modifications on histone H3. Quantitation with respect to to control cells is depicted below the blot images.
- Western blot analysis of acetylated H3K27 levels with respect to total H3 levels upon 10 μM C646 treatment.
Source data are available online for this figure.
We transfected B16 cells with control non‐targeting or Ca14‐targeting shRNA constructs and allowed cells to initiate pigmentation by setting up pigmentation oscillation. Cells were subjected to cross‐linking on day 5, a day after the peak in pHi is observed, and ChIP was carried out with antibodies against acetylated H3K27 as well as control IgG. Relative enrichment with respect to the input DNA was quantitated by qRT‐PCR analysis using promoter‐specific primers. The enrichment of AcH3K27 was decreased at the promoters of Tyr, Dct, and Gp100 (Fig 5F). We observed that AcH3K27 occupancy at the Tyr promoter was reduced by around 50%, at the Dct promoter by 33% and at Gp100 by a marginal but consistent decrease of around 12%. However, AcH3K27 levels at promoters of other MITF targets (Ca14, Tyrp1, and Cdk2), as well as the promoter of Mitf itself, remained unaltered upon CA14 silencing (Fig 5F). We further assessed the binding of p300 at these promoters using ChIP and observed binding at most of the promoters analyzed (Fig 5G). Further, inhibition by 10 μM C646, a p300/CBP selective inhibitor of the histone acetyl transferase (HAT) activity, demonstrated a decrease in acetylated H3K27 in the Tyr and Dct promoters (Fig 5H). This experiment confirmed that CA14 brings about the promoter‐specific changes in activation marks and provides a molecular basis of pigmentation gene expression control mediated by MITF. These data also reinforce earlier observations that the pigmentation gene targets of MITF have unusually high H3K27 acetylation profiles as compared to global average or other targets of MITF, and is functionally important 13.
Histone acetyl transferase activity of p300 is elevated at an alkaline pH
We then set out to identify the pH‐dependent mechanism of histone acetylation. p300/CBP emerged as a probable candidate as it mediates H3K27 acetylation. Additionally, a physical interaction of MITF with p300/CBP has already been established 10. In vitro activity of recombinant p300 HAT domain was assayed with a synthetic peptide of histone H3 and acetyl CoA, and the product CoA was measured by coupling with a fluorophore. Activity at around the neutral pH 7.15 was almost 1/8th of its activity at pH 7.95, suggesting that acetylation by p300 would be clearly high at alkaline pH (Fig 5I). Hence, this study provided a possible effector of pHi that could control pigmentation gene expression.
To confirm the involvement of p300/CBP in pigmentation, we incubated B16 cells with 10 μM C646 and measured pigmentation as well as expression of melanocyte maturation genes. Upon inhibition, B16 cells, despite being subjected to pigmentation permissive 5% CO2 condition, showed lesser pigmentation (Fig 5J). Incubation of zebrafish embryos between 20 and 48 hpf with 10 μM C646 resulted in decreased pigmentation of the melanophores (Fig 5J). The expression of melanocyte differentiation genes in B16 cells was downregulated with marginal changes in MITF transcript levels, confirming that p300/CBP facilitates the melanocyte maturation process by enhancing the expression of differentiation genes (Fig 5K). Further, C646 treatment abrogated CA14 overexpression mediated induction of Dct and Tyr (Fig 5L), confirming that the CA14 effects are mediated via H3K27 acetylation by p300/CBP.
Taken together, we demonstrate that MITF activates CA14 transcriptionally. CA14 increases pHi that in turn can activate p300 HAT activity, which acetylates histone H3 in the promoters of melanocyte differentiation genes, facilitating their transcriptional activation by MITF. Hence, this feed‐forward loop enables quick and heightened expression of differentiation genes under conditions where the cells require rapid pigmentation.
Targeted mutation of ca14 reiterates the role of feed‐forward loop on pigmentation
We chose to address the role of this feed‐forward loop mediated by CA14 in pigmentation under physiological conditions, where rapid and programmed induction of pigmentation genes is involved. We created a germline mutant in zebrafish by targeting the ca14 coding region using the CRISPR‐Cas9 system. We injected Cas9‐guide RNA complexes into fertilized zebrafish embryos at the single cell stage and observed a variety of phenotypes in F0, which include microcephaly, small eye size, mild enlargement of heart, and decreased pigmentation. These phenotypes recapitulate the known effects of high CA14 expression in brain, heart, and eye. After screening for mutants using the T7 endonuclease assay, siblings were grown to adulthood. Genotyping revealed a frame shift mutation at the third codon by the deletion of two bases. Thereby the mutant gene would encode a truncated protein lacking most the coding amino acids (Fig EV5). Further experiments were carried out using F2 embryos obtained by an in‐cross of a homozygous mutant male and a heterozygous female fish with the same mutation. We obtained around 50% of embryos with small eye size with decreased pigmentation, following a Mendelian pattern of inheritance. Mutant embryos showed the two base deletion and the non‐phenotypic siblings were heterozygous based on PCR confirmation of the mutation. Thereby, the pigmentation phenotype observed in the morphant embryos is recapitulated in the genetic mutants, confirming the regulatory role of CA14 in the maturation of melanocytes (Fig 6A).
Figure EV5. Schematic of zebrafish gene ca14 depicting that CRISPR target region and the observed mutation in ca14 fs003 .
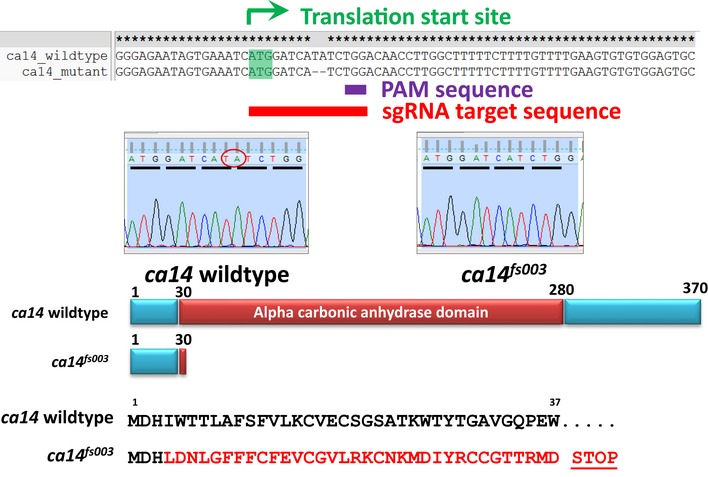
(Top) DNA sequence alignment of ca14 and ca14 fs003 mutants. Depicted are the PAM sequence and the region of the crispr guide RNA (sgRNA). Below is the chromatogram depicting the deletion. (Bottom) Diagrammatic representation of the mutant gene and the amino acid sequence encoded by ca14 fs003.
Figure 6. Targeted null mutation of ca14 by CRISPR demonstrates immature acidic melanocytes.
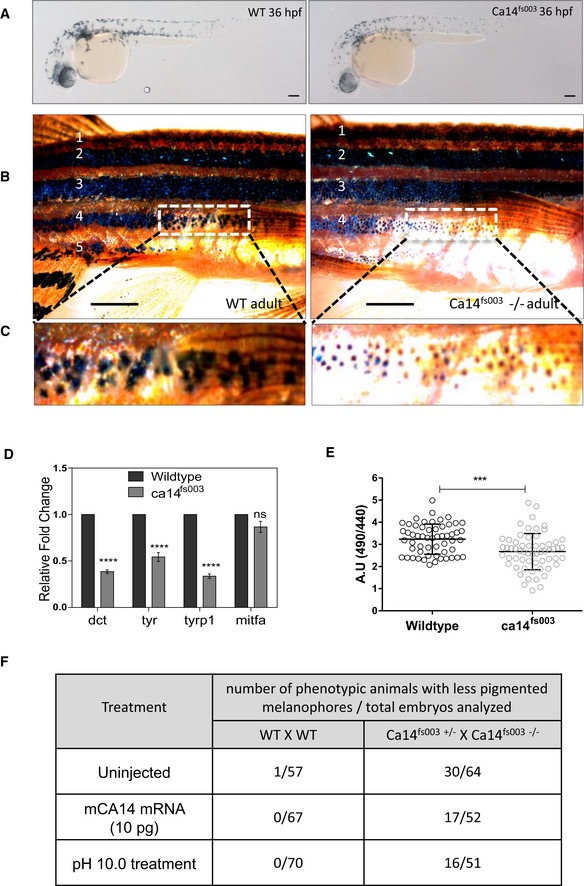
- Brightfield images of the lateral view of CRISPR targeted mutant ca14 fs003 and control embryos at 36 hours post‐fertilization (hpf) in F2 generation fishes. Scale bar 100 μm.
- Wild‐type and ca14 fs003−/− adult male animals were dark‐adapted for 24 h. Image of the lateral view was captured under identical lighting and image capture settings. Scale bar: 5 mm.
- Zoomed up portion from the fourth lateral line demonstrates melanophores to be laden with less melanin content in the ca14 mutant animal.
- qRT‐PCR quantification of pigmentation genes tyrosinase (tyr), dopachrome tautomerase (dct), tyrosinase‐related protein 1b (tyrp1b), and micropthalmia associated factor a (mitfa) between control and ca14 fs003 at 36 hpf, using rps11 gene as the normalization reference. While mitfa remains unaffected, all other pigmentation genes are downregulated in the absence of functional ca14. Bars represent mean ± SEM across n = 3 biological replicates.
- Intracellular pHi probed by BCECF‐AM in ca14 fs003 and sibling control zebrafish embryos. Readings were obtained from trunk region melanophores. Bars represent mean ± SEM across n = 3 biological replicates, at least 10 embryos each. A decrease in the ratio indicates acidification of melanocytes.
- Wild‐type as well as embryos obtained from the cross of ca14 fs003−/− and ca14 fs003+/− were left uninjected or treated with embyo water buffered at pH 10.0 (between 18 hpf till 36 hpf) or injected with 10 pg of in vitro transcribed mouse ca14 mRNA. Embryos were scored for pigmentation at 36 hpf. Ratios represent number of less pigmented phenotypic embroys to the total number of embryos scored. The proporition of less pigmented animals decreased upon mouse Ca14 mRNA injection as well as increased extracellular alkalinization.
In the adult stage, the CA14 mutation induced a visible decrease in pigmentation; however, high melanophore density in the lateral and dorsal region precluded assessment of melanin. We therefore subjected the wild‐type adult and the ca14 fs003−/− fishes to dark adaptation to disperse the existing melanin within the melanocyte so that the content could be easily assessed. We observed distinct non‐overlapping melanophores present in the fourth and fifth stripes near the caudal fin, and in this region, a substantial reduction in the melanin content could be observed in the mutant fish (Fig 6B and C).
Finally, we chose the 36 hpf time point to analyze the expression of differentiation genes when the pigment cells undergo migration and maturation. A decreased gene expression of tyr, tyrp1b, and dct could be observed in the mutant embryos (Fig 6D), confirming that the cells are in an immature less pigmented state and that pigmentation promoting gene expression is severely reduced in the absence of CA14.
Ca14 mutant melanophores are acidic with reduced pigmentation
In a final set of experiments, we subjected the wild type and the ca14 fs003−/− to intracellular pH measurements. Mutant embryos had lower ratiometric values of BCECF (490/440), suggesting an acidic intracellular pH in melanocytes (Fig 6E). We also attempted a chemical rescue approach by subjecting the mutant embryos to water buffered at various pH between 24 hpf and 36 hpf, when the melanocyte maturation commences. While an acidic pH of 5 did not show a rescue but rather showed a marginal increase in deformities, alkaline pH 10 rescued the phenotype marginally and animals with small eyes had now substantially high melanin content (Fig 6F). The extent of pH‐mediated rescue was comparable when the animals were injected with mouse ca14 mRNA. It is interesting to note that in the ca14 fs003−/− mutant animals, the number of melanocytes remained unaltered, but the expression of differentiation effectors was relatively low. Therefore, in the absence of ca14, melanocytes would still respond to external cues and activate Mitf; however, the extent of pigmentation would be severely curtailed. The mutant melanophores clearly showed decreased melanin content, thereby unequivocally establishing a role for this feed‐forward loop involving MITF, CA14, and the differentiation genes in ensuring heightened pigmentation.
Discussion
Several genes that alter melanin content and melanocyte functions have been identified using naturally occurring mutants and targeted gene disruptions 47. Recently, siRNA‐based genome‐wide screen revealed some of the important pigmentation genes 48. While these perturbation‐based approaches reveal the identity of components, biologically relevant regulators and the physiological context are often the most difficult to decipher. In our earlier work, the ca14 gene was observed to be co‐regulated with the pigmentation status in B16 melanoma cells 33. Based on this observation, we have decided to analyze if Ca14 is under the direct control of Mitf and to identifying the role of CA14 in melanocyte functions. In the current study, using cell as well as whole animal‐based approach using zebrafish, we demonstrate an important role for CA14 and downstream pH changes as an amplifier of the maturation process in melanocytes.
As a general theme, pH changes and cell fate decisions have attracted a lot of attention, but precise mechanistic understanding is often limited by the widespread changes brought about by pH 18. Ca14 was identified as a potential Mitf target gene based on promoter binding and upregulation of mRNA in melanocytes overexpressing Mitf 35, 36. However, the physiological implications of this observation were not readily apparent. Based on our study, we propose a model where Ca14 is an early gene induced by MITF, and together both accelerate gene expression of a subset of pigmentation genes resulting in a feed‐forward amplification. Similar modes of network interactions are seen often during cell differentiation programs. For instance, during chondrogenic differentiation two key transcription factors, Bmp2 and Shh, operate to regulate Sox9 in a positive feedback loop to stimulate cellular differentiation 49.
While a link between pH and cell differentiation is often observed across many systems, mechanisms that connect changes in pH to pigmentation gene expression were not immediately apparent. An increase in intracellular pH is necessary for the differentiation of follicle stem cells in Drosophila (FSCs) as well as mouse embryonic stem cells (mESCs) 20. The Drosophila Na+–H+ exchanger DNhe2 is involved in lowering the pHi in differentiating cells and impairs prefollicular cell differentiation, whereas increasing pHi promotes excess differentiation toward a polar/stalk cell fate through the suppression of the Hedgehog pathway. Together with our observations, it is now apparent that this metabolic link involving pH and cell differentiation may be a far more widespread mechanism that is operational in many cell types. Earlier studies indicate that Mitf regulates the differentiation of precursors to mature osteoclasts by the induction of carbonic anhydrase II, which is responsible for extracellular acidification 50, 51. It is likely that pH‐mediated induction of differentiation and its regulation by Mitf may be an underlying theme conserved across several cell types.
The role of p300/CBP in MITF‐mediated gene expression has emerged from multiple evidences. Activated MITF physically interacts with p300/CBP, and Mitf immunoprecipitates show HAT acitivity 10, 52. However, the physiological effect of this association remained enigmatic, as MITF mutants defective in binding to p300/CBP still activated transcription 53. The footprint of a SWI/SNF complex involving BRG1 on MITF targets has distinct clusters of genes with high as well as low H3K27 acetylation status 11. Thus, it is likely that distinct subsets of MITF targets show different levels of activation. This was also recently alluded to by 13. The authors demonstrate that H3K27 acetylation patterns are unusually high in pigmentation related MITF targets compared to proliferation and sustenance genes, making dynamic regulation of H3K27 acetylation a prerequisite for a subset of these genes, while others are less dependent on this activation. This is also evident in p300 ChIP and C646 inhibition studies, where effectors of MITF other than Tyr and Dct show differential responsiveness to HAT inhibition. The identification of CA14‐mediated p300 activation thus provides a plausible explanation for the context‐dependent target selectivity by MITF.
The p300 histone acetyl transferase is a central epigenetic regulator and its observed pH‐mediated activation is likely to be a general phenomenon. The crystal structure of the p300 HAT domain revealed an unusual hit and run “Theorell‐Chance” catalytic mechanism 54. Subsequent molecular dynamic simulations suggest a proton relay from the ε amino group of the acceptor lysine substrate, and it is predicted that the involved side chains have a high pKa 55. Standard enzymatic assays involving p300 are routinely carried out at an alkaline pH 7.8–8.0, which would favor deprotonation, providing a molecular mechanism of pH‐mediated HAT activation. The transient increase in pH mediated by CA14 facilitates H3K27 acetylation by p300, thereby promoting gene activation.
The CA14‐mediated feed‐forward loop described here would be operational under conditions where rapid melanocyte maturation is required. This is anticipated during the UV‐induced sun tanning response as well as in developmental states where melanocytes mature into high melanin‐containing cells. A recent study identified Ca14 to be downregulated in the lesional depigmented skin of vitiligo subjects 56. It is thus tempting to speculate that the depigmentation in vitiligo could be caused by the decrease in CA14 that could affect melanocyte maturation, in addition to the loss of mature melanocytes. Advances made in this study therefore significantly underline the importance of the biochemical milieu as an intermediate effector in determining cell fate decisions.
Materials and Methods
Cell line and culture
B16 mouse melanoma cell line was cultured in DMEM‐high glucose (Gibco, Life Technologies) medium supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies), and cells were maintained in 5% (or at 10% CO2 when indicated) in a 37°C incubator. The orthologous non‐cancerous murine melanocyte line Melan‐A was cultured at10% CO2 at 37°C in RPMI‐1640 medium (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS; Gibco, Life Technologies), 400 nM phorbol‐myrystyl‐13‐acetate (PMA; Sigma) and 0.003% phenylthiourea (PTU, Sigma). Primary human melanocytes were grown in proliferative conditions in PMA containing medium MBM4 (Lonza). For differentiation, cells were switched to M254 medium for 3‐4 population doublings (Thermofisher Scientific, Life Technologies).
Setup of pigmentation oscillation model in B16 cells
Detailed protocol described in ref. 33. Briefly, B16 cells were seeded at density of 100 cells/cm2 in DMEM‐high glucose medium supplemented with 10% FBS. Cells were cultured at 5% CO2 and were harvested at day 4, 6, and 8 for downstream experiments. The cells progressively pigmented and the expression of pigmentation genes were induced in this model system (Appendix Fig S1). To modulate the pH, cells were alternatively cultured in 10% CO2 and subsequent changes in pH, melanin content and gene expression were investigated. Treatment with 10 μM P300/CBP inhibitor C646 or vehicle control DMSO treatment was performed at day 1 of pigmentation oscillator and terminated on day 6 for downstream analysis.
Measurement of intracellular pH
For mammalian cells
Intracellular pH was measured by using the ratiometric dye BCECF‐AM (Molecular probes, Thermo scientific), as described in Ref. 33. Briefly, cells were plated in 35‐mm dishes (IBIDI), and on the day of pH, measurement cells were incubated with 0.2 μM of BCECF‐AM dye for 2 min at 37°C under indicated conditions. In order to generate a pH calibration curve, cells were incubated in pH calibration solution (in mM: 1glucose, 140 KCl, 1 MgSO4, 30 HEPES, 25 NaCl, 1 CaCl2, 1 NaH2PO4) with pH range of 6–8.5 and added 10 μM Nigericin (Thermofisher Scientific). Quantitative images were acquired, and fluorescence ratio was measured at dual excitation wavelength of 440 and 490 nm in Leica SP8 confocal microscope.
For zebrafish embryos
2 dpf zebrafish embryos were taken and dechorionated. Ten embryos were incubated with 10 mM BCECF‐AM (Thermofisher scientific) in embryo water at 28°C incubator for 15 min. After incubation, the embryos were washed, embedded in methylcellulose, and taken for quantitative imaging using Leica SP8 confocal microscopy.
Generation of wild‐type and mutant ca14 promoter constructs, Ca14 expression construct, and the inactive mutant
The ca14 3 kb promoter (3,000 bp upstream of transcription start site) was amplified from mouse genomic DNA and cloned upstream of luciferase cassette in KpnI/HindIII site of pGL4.23 (Promega) using primers listed in Appendix Table S1. Ca14 coding sequence was amplified from mouse B16 cDNA and cloned in mcherryN1 vector (Clontech) in KpnI/HindIII site. Site‐directed mutagenesis for Ca14 3 kb promoter (1 kb binding site and 3 kb binding site mutations) and CA14 (Thr199Ile) were carried out using SDM II kit (Agilent) using primers listed in Appendix Table S1. Truncated constructs of 1 kb (proximal, middle, distal from TSS) were generated from Ca14 3 kb promoter. Dct promoter used in this study is reported elsewhere 33. Tyr promoter used in this study was a kind gift from Dr Krishnamurthy Natarajan, Jawaharlal Nehru University, Delhi, India.
Plasmid and silencing RNA transfections
B16 cells were trypsinized and seeded at density of 1 × 105 cells/well in 6 well plate (BD Bioscience) and incubated overnight in antibiotic containing DMEM + 10% FBS. At the time of transfection, the cells were replaced with serum and antibiotic free media OptiMEM (Gibco, Life Technologies). Lipofectamine 2000 was used at a ratio of 1:2 with CA14 siRNA or Mitf siRNA or the scrambled control siRNA (Dharmacon) (details provided in Appendix Table S3). The cells were incubated for 6 hours with the transfection mixture containing Lipofectamine 2000, OptiMEM, and the siRNA. The media was then replaced with antibiotic containing DMEM + 10% FBS and incubated for 72 h, and downstream experiments were performed.
NHEM cells were trypsinized and seeded at density of 1 × 105 cells/well in 6‐well plate (BD Bioscience) and incubated overnight with antibiotic containing M254 (Thermofisher Scientific). Next day, cells were washed with DPBS (Gibco, Life Technologies) and were replaced with antibiotic‐free OptiMEM media (Gibco, Life Technologies). Cellfectin (Invitrogen) was used at a ratio of 1:2 with MITF siRNA and CA14 siRNA (Dharmacon) and incubated for 6 h. The transfection mixture was replaced with antibiotic containing M254 media and incubated for 72 h.
Treatment for the induction of MITF
Melan‐A cells were trypsinized and seeded at density of 1 × 105 cells/well in 6‐well plate (BD Bioscience) and incubated overnight with antibiotic containing RPMI 1640 + 10% FBS. Next day, fresh media was added to the cells and treatment was performed with α‐MSH (1 μM; Sigma) and IBMX (60 μM; Sigma). The cells were incubated at 37°C at 10% CO2 and were then harvested for protein isolation after 48 and 72 h.
RNA Isolation and cDNA synthesis
Cells were trypsinized with 0.1% trypsin, and the pellet was washed twice with 1X phosphate‐buffered saline (PBS; Gibco Life Technologies). To the pelleted cells, 1 ml of TriZol (Ambion, Invitrogen) was added and stored at −80°C overnight. The RNA was isolated using standard Trizol‐based method. The isolated RNA was subjected to DNAse (Qiagen) treatment for 20 min at room temperature. Column purification of the RNA was performed using RNAeasy mini kit (Qiagen, Cat 74104) according to manufacturer's protocol. 100–500 ng of RNA was taken for cDNA synthesis using Superscript III (Invitrogen, Life Technologies) according to manufacturer's protocol. Q‐RT PCR experiments were performed using TaqMAN assay probes (details provided in Appendix Table S2) according to manufacturer's protocol using Lightcycler 480 II (Roche).
Isolation of nuclear, cytoplasmic (post‐muclear supernatant fraction) and melanosomal fractions
Nuclear proteins were extracted using NE‐PER™ nuclear and cytoplasmic extraction reagents (Thermoscientific; 78833) according to manufacturer's protocol. Melanosomes were isolated using protocol previously described 57. The B16 tumor from mice was excised and washed in homogenization buffer and was homogenized with 120 strokes of dounce glass homogenizer. Post‐nuclear supernatant was isolated and then separated on a stepwise density gradient and centrifuged at 100,000 g for 1 h at 4°C in a swing out rotor. The stage III and IV melanosomes which preferentially localized to the highest density fraction (1.8–2.0) were collected, and protein was isolated using NP‐40 lysis buffer.
Ca14 promoter binding to Mitf
Biotinylated Ca14 BS1 and BS2 complementary sequences (500 ng) were annealed immobilized on streptavidin‐conjugated Dyna beads (Sigma) followed by incubation with B16 nuclear lysates for 4 hat 4°C on a rocker. The supernatant was discarded, and the DNA‐bead–bound proteins were washed three times with buffer (10 mM Hepes, 10 mM KCl, 0.1 M EDTA, 0.1 M EGTA, 1 × protease inhibitor mixture). The bound proteins were eluted by heating the samples with SDS sample dye at 100°C for 5 min and loaded onto SDS/PAGE gel followed by Western blot analysis with Mitf‐C5 antibody (Abcam). Corresponding non‐biotinylated DNA was used at 10X molar excess for cold competition in this assay.
Electrophoretic mobility shift assay
EMSA was carried out using BS1 (TGGTGAGGCTTAGGTCCACGTGCCTGTGTAGCCTGGG) and BS2 (CCCTCTTCTCCCCTTGCACCTGGCCCAAGGACCACCAA) probes annealed with complementary sequences. B16 nuclei were collected by hypotonic lysis and resuspended in 30 μl NE buffer (250 mM Tris pH 7.8, 60 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF) and lysed by 3 freeze–thaw cycles. Nuclear lysates were cleared by 14,000 g centrifugation and adjusted to 1 μg/μl total protein concentration. 2 μg of total nuclear protein was incubated at room temperature for 15 min with 0.1 pmoles of 32P‐labeled double‐stranded oligonucleotides in 3 μl binding buffer (10 mM Tris‐Cl pH 7.5, 50 mM NaCl, 10% glycerol, 1% NP‐40, 1 mM EDTA, 0.1 μg/μl poly‐dIdC). Complexes were resolved on a non‐denaturing gel and visualized using a Typhoon Scanner (GE Healthcare Life Sciences).
Protein Isolation and Western blot analysis
Cells were trypsinized with 0.1% trypsin and the pellet was washed twice with 1× phosphate‐buffered saline (PBS; Gibco Life Technologies). NP‐40 lysis buffer (Invitrogen) was added to the pellet and was incubated on ice for 30 min with pipetting at interval of 10 min. The cells were centrifuged down at 11,200 g for 30 min at 4°C (Eppendorf Centrifuge 5415 R). The supernatant was collected and transferred to a fresh microfuge tube. For Histone blots, lamelli lysis buffer was used. The protein was estimated using standard BCA protocol (Pierce BCA protein assay kit; Thermoscientific). Equal amount of protein from each sample were resolved in 10 or 12% % SDS gel in 1× Tris glycine buffer. The gel was blotted onto 0.45 μm PVDF membrane (Millipore) at 300 mA for 2 h. 5% Skim milk was used for blocking for 1 h at room temperature. Incubation with primary antibody was performed for overnight at 4°C. Primary antibody dilutions are provided in (Appendix Table S4). After washing the blot with 1× TBST, the blot was incubated with HRP‐conjugated secondary antibody for 1 h at room temperature. After washing with 1× TBST, the blot was developed using Immobilon Western (Millipore) in the Syngene GBOX Chemiluminescence instrument. Densitometry analysis was performed using ImageJ software.
Chromatin immunoprecipitation and qRT PCR
B16 Melanoma cells at 80% confluence was fixed with 10% formalin (Sigma Cat No HT501128) and incubated at 37°C for 10 min. 2.5 M Glycine was added to the cells and again incubated at 37°C for 10 min. Cells were washed with ice‐cold 1× PBS containing protease inhibitors. Cells were then scraped and centrifuged at 112 g for 5 min at 4°C. The cell pellet was lysed in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris (pH 8.1)) on ice for 30 min. The cells were then sonicated on bioruptor (DIAGENODE) in ice. The chromatin lysate was then estimated for protein content using BCA kit (Pierce). 5–10 μg of MITF C5 antibody (Abcam) or AcH3K27 antibody (Abcam) or P300 antibody (Cell Signaling) was taken and incubated with Protein G or Protein A Dynabeads (Thermoscientific) overnight at 4°C on a rotator. The next day, the sera was cleared and washed with ice‐cold dilution buffer (2 mM EDTA, 150 mM NaCl, 20 mM Tris HCl (pH 8)). 500 μg of chromatin lysate was added to the beads and the final volume was made up to 750 μl using the dilution buffer and incubated for 6 h at 4°C in a rotator. 10% of the lysate was kept separately as input. After incubation, the magnetic beads were washed with low salt buffer (0.1% SDS, 1% Triton X‐100, 2 mM EDTA, 20 mM Tris HCl (pH 8), 150 mM NaCl), high salt buffer (0.1% SDS, 1% Triton X‐100, 2 mM EDTA, 20 mM Tris HCl (pH 8), 500 mM NaCl), and LiCl buffer (0.25 M LiCl, 1% Igepal C‐630, 1 mM EDTA, 10 mM Tris HCl (pH 8), 1% deoxycholate). Finally, the magnetic beads were incubated overnight at 65°C in elution buffer (1% SDS, 0.75% sodium bicarbonate) and 1 μl of 20 mg/ml proteinase K (Sigma) for elution and subsequent reverse cross‐linking. The magnetic beads were separated from supernatant and column purified using QIAgen PCR purification kit, the input control was also included in the purification step. SYBR qRT‐PCR was setup using 5 μl of eluted DNA (primers in Appendix Table S1), and graphs were plotted as percentage input or fold enrichment.
Immunofluorescence
The cells plated on coverslips were washed twice with 1× PBS and then fixed with 4% PFA at 37°C for 20 min. The cells were again washed twice with 1× PBS and permeabilized with 0.01% Triton X‐100 (Sigma). The cells were blocked with 5% normal goat serum (Jackson Laboratories) overnight at 4°C. The cells were washed twice with PBST (1× PBS with 0.01% Tween‐20). The cells were then incubated with 1:50 dilution of CA14 antibody (Abcam) or HA tag antibody (Abcam ab9110) in a moist chamber for 1 h at room temperature. Incubation with secondary antibody (Alexafluor488; Molecular probes, Thermoscientific) was performed at room temperature for 1 h. The cells were again washed with PBST. The cells were then mounted on slides using Antifade slowfade DAPI (Molecular probes, Invitrogen) and visualized using Confocal Microscope (Zeiss LSM 510 or Leica SP8).
P300 HAT activity assay
HAT activity assay was performed using KAT3B/P300 inhibitor fluorometric assay screening kit (Abcam, ab196996) according to manufacturer's protocol with minor modification. Tris–Cl pH 7.15 and pH 7.95 was added at an additional concentration of 50 mM to buffer provided in the kit, and the assays were performed with the modified buffers.
Zebrafish ethics statement
Fish experiments were performed in strict accordance with the recommendations and guidelines laid down by the CSIR Institute of Genomics and Integrative Biology, India. The protocol was approved by the Institutional Animal Ethics Committee (IAEC) of the CSIR Institute of Genomics and Integrative Biology, India (Proposal No 45a). All efforts were made to minimize animal suffering.
Zebrafish strains and maintenance
The wild‐type strain Assam WT (ASWT) and Tyrp1 reporter, Tg(ftyrp1: GFP) zebrafish lines were used for morpholino injections and were maintained according to standard zebrafish husbandry protocols. ftyrp1:GFP plasmid was a kind gift from Dr Xiangyun Wei 38, University of Pittsburgh School of Medicine, and the transgenic line was created at CSIR‐IGIB zebrafish facility using tol2 transposase microinjections in ASWT line. Phenylthiourea (PTU; 0.003%) was added to embryo water before 24 hours post‐fertilization (hpf) to prevent melanin from masking the GFP fluorescence. For pH rescue experiments, the embryos were grown in embryo media buffered with 10 mM HEPES.
Morpholino knockdown of zebrafish Ca14
Antisense morpholino was synthesized from Gene Tools against Ca14 of zebrafish. The morpholino was designed to block the translation of car14 gene. Morpholino sequence with translation initiation site (initiator ATG codon) is underlined CCATGATTTCACTATTCTCCCTACA. Standard control was obtained from Genetools and injected at same dosage as ca14 morphlino.
ca14 CRISPR mutant generation
Zebrafish ca14 CRISPR was designed using ECRISP software (http://www.e-crisp.org/E-CRISP/). The CRISPR sgRNA sequence was in vitro transcribed using mMessage mMachine T7 ULTRA kit (Thermofisher scientific). 300 pg of sgRNA was injected along with 500 pg of spcas9 protein (kind gift from Dr Debojyoti Chakraborty, CSIR‐IGIB). The F0 embryos were screened using T7 endonuclease assay for mutations, and the putative mutant siblings were grown to adulthood and inbred to get F1 animals. Mutations in the F1 fishes were confirmed using sanger sequencing.
The cross involving the ca14fs003 reported herein involves a ca14 fs003−/− male with a ca14 fs003 +/− female that accounts for 50% of the embryos to be phenotypic. The genotypes of the phenotypic embryos and normally pigmented embryos arising from this cross were ascertained to be ca14 fs003−/− and ca14 fs003 +/− respectively using PCR based amplification.
C646 treatment
Zebrafish embryos were dechorionated at 20 hpf and treated with 10 μM of C646 in embryo water with DMSO as vehicle control. The embryos were subsequently imaged at 54 hpf.
Zebrafish imaging
The embryos were manually dechorinated and embedded in 2% methylcellulose. Control and Morphant embryos were imaged laterally and dorsally for melanophore quantitation. Brightfield Images were taken using Zeiss Stemi 2000‐C microscope; for fluorescence microscopy, Zeiss Axioscope A1 was used.
RNA Isolation from embryos
Total RNA was extracted from zebrafish embryos using TriZol (Invitrogen). cDNA was synthesized using Superscript III kit (Invitrogen). Real‐time quantitative PCR was performed using SYBR Green (Kapa Biosystems) or TAQMAN probes (Appendix Table S2), and data were generated in ROCHE Lightcycler 480 II.
mRNA injection in zebrafish embryos
For rescue experiments, the coding sequence of mouse Ca14 gene was cloned with kozak sequence inserted in front of translation start site (Appendix Table S1). The amplicon was cloned into TOPO‐Zero blunt vector (Thermofisher Scientific). RNA was made using in vitro transcription kit T7 Ultra mRNA synthesis kit (Ambion – Thermofisher Scientific). 10 pg of Ca14 WT or Ca14T199I RNA was injected into the cell at one cell stage Ca14fs003 embryos.
Estimation of melanin content
Dorsal images of Control and ca14 morphant embryos were taken at 2 dpf. The images were imported to ImageJ, and mean gray values were taken for melanophores of each embryo set. Mean gray values are inverse proportional to the melanin content of the cell. Corresponding values were then plotted using GraphPad Prism.
Statistical analysis and graphs
Student's t‐test was performed to obtain statistical significance in the data. Asterisk on the error bar corresponds to *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001 and ns P > 0.05. Graphs were plotted using GraphPad prism.
Author contributions
DAR, VG, RSG and VTN designed experiments. DAR, VG, SAB, FS, and YJS executed the experiments with cultured cells. AS performed experiments pertaining to electron microscopy. BS, SS, and AV were involved in cloning and HG in experiments with primary human melanocytes. DAR, YJS, SS, and VTN were involved in the design and execution of zebrafish experiments. DAR, SAB, YJS, VG along with SS, RSG and VTN were involved in data analysis, interpretations, and writing of the manuscript.
Conflict of interest
R.S.G. is the co‐founder of the board of Vyome Biosciences, a biopharmaceutical company in the area of dermatology unrelated to the work presented here. Other authors do not have any conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
This work was supported by the Council for Scientific and Industrial Research (CSIR), India, through grant (TOUCH‐BSC0302) and (GRAFT‐MLP1810) Department of Biotechnology through the grant (GAP0182). We acknowledge the infrastructural support of CSIR to the imaging facility (VISION‐BSC0403) and Manish Kumar for help with imaging. DAR acknowledges ICMR for Research Fellowship (BMS/FW/CMB/2015‐23210/Sep2015/09/DL/govt). R.S.G. is a J.C. Bose Fellow of the Department of Science and Technology, Government of India (SB/S2/ JCB‐038/2015). TNV acknowledges CSIR support through the Young Scientist Award (OLP1118). Dr Soumen Basak and Alvina Deka (National Institute of Immunology) for help with EMSA experiments. Dr Krishnamurthy Natarajan, and Shalini Yadav, Jawaharlal Nehru University for Dct and Tyr promoter constructs. Ayush Aggarwal for help with generation of figures.
EMBO Reports (2020) 21: e48333
References
- 1. Bennett DC (1983) Differentiation in mouse melanoma cells: initial reversibility and an on‐off stochastic model. Cell 34: 445–453 [DOI] [PubMed] [Google Scholar]
- 2. Mort RL, Jackson IJ, Patton EE (2015) The melanocyte lineage in development and disease. Development 142: 1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris ML, Erickson CA (2007) Lineage specification in neural crest cell pathfinding. Dev Dyn 236: 1–19 [DOI] [PubMed] [Google Scholar]
- 4. Levy C, Khaled M, Fisher DE (2006) MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12: 406–414 [DOI] [PubMed] [Google Scholar]
- 5. Levy C, Fisher DE (2011) Dual roles of lineage restricted transcription factors: the case of MITF in melanocytes. Transcription 2: 19–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Price ER, Horstmann MA, Wells AG, Weilbaecher KN, Takemoto CM, Landis MW, Fisher DE (1998) α‐Melanocyte‐stimulating hormone signaling regulates expression of microphthalmia, a gene deficient in Waardenburg syndrome. J Biol Chem 273: 33042–33047 [DOI] [PubMed] [Google Scholar]
- 7. Bertolotto C, Abbe P, Hemesath TJ, Bille K, Fisher DE, Ortonne J‐P, Ballotti R (1998) Microphthalmia gene product as a signal transducer in cAMP‐induced differentiation of melanocytes. J Cell Biol 142: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, Narayan R, Flaherty KT, Wargo JA, Root DE et al (2013) A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504: 138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Song JS, Bell RJ, Tran TN, Haq R, Liu H, Love KT, Langer R, Anderson DG, Larue L et al (2012) YY1 regulates melanocyte development and function by cooperating with MITF. PLoS Genet 8: e1002688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato S, Roberts K, Gambino G, Cook A, Kouzarides T, Goding CR (1997) CBP/p300 as a co‐factor for the Microphthalmia transcription factor. Oncogene 14: 3083–3092 [DOI] [PubMed] [Google Scholar]
- 11. Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, Van Otterloo E, Imrichova H, Siddaway R, Aerts S et al (2015) Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife 4: e06857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Praetorius C, Grill C, Stacey SN, Metcalf AM, Gorkin DU, Robinson KC, Van Otterloo E, Kim RS, Bergsteinsdottir K, Ogmundsdottir MH et al (2013) A polymorphism in IRF4 affects human pigmentation through a tyrosinase‐dependent MITF/TFAP2A pathway. Cell 155: 1022–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malcov‐Brog H, Alpert A, Golan T, Parikh S, Nordlinger A, Netti F, Sheinboim D, Dror I, Thomas L, Cosson C et al (2018) UV‐protection timer controls linkage between stress and pigmentation skin protection systems. Mol Cell 72: 444–456.e447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de la Serna IL, Ohkawa Y, Higashi C, Dutta C, Osias J, Kommajosyula N, Tachibana T, Imbalzano AN (2006) The microphthalmia‐associated transcription factor requires SWI/SNF enzymes to activate melanocyte‐specific genes. J Biol Chem 281: 20233–20241 [DOI] [PubMed] [Google Scholar]
- 15. Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL (2010) Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia‐associated transcription factor target genes in melanoma. Oncogene 29: 81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boron WF (2004) Regulation of intracellular pH. Adv Physiol Educ 28: 160–179 [DOI] [PubMed] [Google Scholar]
- 17. Simons M, Gault WJ, Gotthardt D, Rohatgi R, Klein TJ, Shao Y, Lee HJ, Wu AL, Fang Y, Satlin LM et al (2009) Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat Cell Biol 11: 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tatapudy S, Aloisio F, Barber D, Nystul T (2017) Cell fate decisions: emerging roles for metabolic signals and cell morphology. EMBO Rep 18: 2105–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB et al (2012) Histone acetylation regulates intracellular pH. Mol Cell 49: 310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ulmschneider B, Grillo‐Hill BK, Benitez M, Azimova DR, Barber DL, Nystul TG (2016) Increased intracellular pH is necessary for adult epithelial and embryonic stem cell differentiation. J Cell Biol 215: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindskog S (1997) Structure and mechanism of carbonic anhydrase. Pharmacol Ther 74: 1–20 [DOI] [PubMed] [Google Scholar]
- 22. Karler R, Woodbury DM (1960) Intracellular distribution of carbonic anhydrase. Biochem J 75: 538–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reibring CG, El Shahawy M, Hallberg K, Kannius‐Janson M, Nilsson J, Parkkila S, Sly WS, Waheed A, Linde A, Gritli‐Linde A (2014) Expression patterns and subcellular localization of carbonic anhydrases are developmentally regulated during tooth formation. PLoS One 9: e96007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujikawa‐Adachi K, Nishimori I, Taguchi T, Onishi S (1999) Human carbonic anhydrase XIV (CA14): cDNA cloning, mRNA expression, and mapping to chromosome 1. Genomics 61: 74–81 [DOI] [PubMed] [Google Scholar]
- 25. Kaunisto K, Parkkila S, Rajaniemi H, Waheed A, Grubb J, Sly WS (2002) Carbonic anhydrase XIV: luminal expression suggests key role in renal acidification. Kidney Int 61: 2111–2118 [DOI] [PubMed] [Google Scholar]
- 26. Shah GN, Ulmasov B, Waheed A, Becker T, Makani S, Svichar N, Chesler M, Sly WS (2005) Carbonic anhydrase IV and XIV knockout mice: roles of the respective carbonic anhydrases in buffering the extracellular space in brain. Proc Natl Acad Sci USA 102: 16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wetzel P, Scheibe RJ, Hellmann B, Hallerdei J, Shah GN, Waheed A, Gros G, Sly WS (2007) Carbonic anhydrase XIV in skeletal muscle: subcellular localization and function from wild‐type and knockout mice. Am J Physiol Cell Physiol 293: C358–C366 [DOI] [PubMed] [Google Scholar]
- 28. Ogilvie JM, Ohlemiller KK, Shah GN, Ulmasov B, Becker TA, Waheed A, Hennig AK, Lukasiewicz PD, Sly WS (2007) Carbonic anhydrase XIV deficiency produces a functional defect in the retinal light response. Proc Natl Acad Sci USA 104: 8514–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watabe H, Valencia JC, Yasumoto K, Kushimoto T, Ando H, Muller J, Vieira WD, Mizoguchi M, Appella E, Hearing VJ (2004) Regulation of tyrosinase processing and trafficking by organellar pH and by proteasome activity. J Biol Chem 279: 7971–7981 [DOI] [PubMed] [Google Scholar]
- 30. Halaban R, Patton RS, Cheng E, Svedine S, Trombetta ES, Wahl ML, Ariyan S, Hebert DN (2002) Abnormal acidification of melanoma cells induces tyrosinase retention in the early secretory pathway. J Biol Chem 277: 14821–14828 [DOI] [PubMed] [Google Scholar]
- 31. Bellono NW, Escobar IE, Lefkovith AJ, Marks MS, Oancea E (2014) An intracellular anion channel critical for pigmentation. Elife 3: e04543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wakamatsu K, Nagao A, Watanabe M, Nakao K, Ito S (2017) Pheomelanogenesis is promoted at a weakly acidic pH. Pigment Cell Melanoma Res 30: 372–377 [DOI] [PubMed] [Google Scholar]
- 33. Natarajan VT, Ganju P, Singh A, Vijayan V, Kirty K, Yadav S, Puntambekar S, Bajaj S, Dani PP, Kar HK et al (2014) IFN‐gamma signaling maintains skin pigmentation homeostasis through regulation of melanosome maturation. Proc Natl Acad Sci USA 111: 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoashi T, Tamaki K, Hearing VJ (2009) The secreted form of a melanocyte membrane‐bound glycoprotein (Pmel17/gp100) is released by ectodomain shedding. FASEB J 24: 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R et al (2008) Novel MITF targets identified using a two‐step DNA microarray strategy. Pigment Cell Melanoma Res 21: 665–676 [DOI] [PubMed] [Google Scholar]
- 36. Strub T, Giuliano S, Ye T, Bonet C, Keime C, Kobi D, Le Gras S, Cormont M, Ballotti R, Bertolotto C et al (2011) Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene 30: 2319–2332 [DOI] [PubMed] [Google Scholar]
- 37. Motiani RK, Tanwar J, Raja DA, Vashisht A, Khanna S, Sharma S, Srivastava S, Sivasubbu S, Natarajan VT, Gokhale RS (2018) STIM1 activation of adenylyl cyclase 6 connects Ca(2 + ) and cAMP signaling during melanogenesis. EMBO J 37: e97597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zou J, Beermann F, Wang J, Kawakami K, Wei X (2006) The Fugu tyrp1 promoter directs specific GFP expression in zebrafish: tools to study the RPE and the neural crest‐derived melanophores. Pigment Cell Res 19: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Whittington DA, Grubb JH, Waheed A, Shah GN, Sly WS, Christianson DW (2004) Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane‐associated isozymes. J Biol Chem 279: 7223–7228 [DOI] [PubMed] [Google Scholar]
- 40. Vargas LA, Alvarez BV (2012) Carbonic anhydrase XIV in the normal and hypertrophic myocardium. J Mol Cell Cardiol 52: 741–752 [DOI] [PubMed] [Google Scholar]
- 41. Purkerson JM, Schwartz GJ (2007) The role of carbonic anhydrases in renal physiology. Kidney Int 71: 103–115 [DOI] [PubMed] [Google Scholar]
- 42. Hallerdei J, Scheibe RJ, Parkkila S, Waheed A, Sly WS, Gros G, Wetzel P, Endeward V (2010) T tubules and surface membranes provide equally effective pathways of carbonic anhydrase‐facilitated lactic acid transport in skeletal muscle. PLoS One 5: e15137 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Ancans J, Tobin DJ, Hoogduijn MJ, Smit NP, Wakamatsu K, Thody AJ (2001) Melanosomal pH controls rate of melanogenesis, eumelanin/phaeomelanin ratio and melanosome maturation in melanocytes and melanoma cells. Exp Cell Res 268: 26–35 [DOI] [PubMed] [Google Scholar]
- 44. Sasso E, Vitale M, Monteleone F, Boffo FL, Santoriello M, Sarnataro D, Garbi C, Sabatella M, Crifo B, Paolella LA et al (2015) Binding of carbonic anhydrase IX to 45S rDNA genes is prevented by exportin‐1 in hypoxic cells. Biomed Res Int 2015: 674920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Supuran CT, Winum JY (2015) Carbonic anhydrase IX inhibitors in cancer therapy: an update. Future Med Chem 7: 1407–1414 [DOI] [PubMed] [Google Scholar]
- 46. Kormos B, Belso N, Bebes A, Szabad G, Bacsa S, Szell M, Kemeny L, Bata‐Csorgo Z (2011) In vitro dedifferentiation of melanocytes from adult epidermis. PLoS One 6: e17197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bennett DC, Lamoreux ML (2003) The color loci of mice–a genetic century. Pigment Cell Res 16: 333–344 [DOI] [PubMed] [Google Scholar]
- 48. Ganesan AK, Ho H, Bodemann B, Petersen S, Aruri J, Koshy S, Richardson Z, Le LQ, Krasieva T, Roth MG et al (2008) Genome‐wide siRNA‐based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet 4: e1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeng L, Kempf H, Murtaugh LC, Sato ME, Lassar AB (2002) Shh establishes an Nkx3.2/Sox9 autoregulatory loop that is maintained by BMP signals to induce somitic chondrogenesis. Genes Dev 16: 1990–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lehenkari P, Hentunen TA, Laitala‐Leinonen T, Tuukkanen J, Vaananen HK (1998) Carbonic anhydrase II plays a major role in osteoclast differentiation and bone resorption by effecting the steady state intracellular pH and Ca2+ . Exp Cell Res 242: 128–137 [DOI] [PubMed] [Google Scholar]
- 51. Lu SY, Li M, Lin YL (2010) Mitf induction by RANKL is critical for osteoclastogenesis. Mol Biol Cell 21: 1763–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Price ER, Ding HF, Badalian T, Bhattacharya S, Takemoto C, Yao TP, Hemesath TJ, Fisher DE (1998) Lineage‐specific signaling in melanocytes. C‐kit stimulation recruits p300/CBP to microphthalmia. J Biol Chem 273: 17983–17986 [DOI] [PubMed] [Google Scholar]
- 53. Vachtenheim J, Sestakova B, Tuhackova Z (2007) Inhibition of MITF transcriptional activity independent of targeting p300/CBP coactivators. Pigment Cell Res 20: 41–51 [DOI] [PubMed] [Google Scholar]
- 54. Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA (2008) The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451: 846–850 [DOI] [PubMed] [Google Scholar]
- 55. Zhang X, Ouyang S, Kong X, Liang Z, Lu J, Zhu K, Zhao D, Zheng M, Jiang H, Liu X et al (2014) Catalytic mechanism of histone acetyltransferase p300: from the proton transfer to acetylation reaction. J Phys Chem B 118: 2009–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu R, Broady R, Huang Y, Wang Y, Yu J, Gao M, Levings M, Wei S, Zhang S, Xu A et al (2012) Transcriptome analysis reveals markers of aberrantly activated innate immunity in vitiligo lesional and non‐lesional skin. PLoS One 7: e51040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Watabe H, Kushimoto T, Valencia JC, Hearing VJ (2005) Isolation of melanosomes. Curr Protoc Cell Biol Chapter 3: Unit 3 14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5


