Abstract
Triple-negative breast cancer (TNBC) is the most difficult subtype of breast cancer to treat due to a paucity of effective targeted therapies. Many studies have reported that breast cancer stem cells (BCSCs) are enriched in TNBC and are responsible for chemoresistance and metastasis. In this study, we identify LRP8 as a novel positive regulator of BCSCs in TNBC. LRP8 is highly expressed in TNBC compared to other breast cancer subtypes and its genomic locus is amplified in 24% of TNBC tumors. Knockdown of LRP8 in TNBC cell lines inhibits Wnt/β-catenin signaling, decreases BCSCs, and suppresses tumorigenic potential in xenograft models. LRP8 knockdown also induces a more differentiated, luminal-epithelial phenotype and thus sensitizes the TNBC cells to chemotherapy. Together, our study highlights LRP8 as a novel therapeutic target for TNBC as inhibition of LRP8 can attenuate Wnt/β-catenin signaling to suppress BCSCs.
Keywords: LRP8, Triple-negative breast cancer, Breast cancer stem cells, Tumorigenesis, Wnt/β-catenin signaling
1. Introduction
Triple-negative breast cancer (TNBC) is a heterogeneous disease diagnosed pathologically by the lack of expression of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) [1]. Although accounting for only 10–15% of all breast cancers, TNBC has the worst prognosis due to its high rate of relapse and metastasis and the lack of effective targeted therapies [1,2]. Currently, cytotoxic chemotherapy is the main therapeutic strategy for TNBC, irrespective of disease stage [3]. However, TNBC patients with residual disease after chemotherapy have significantly worse survival than non-TNBC patients with residual disease [4]. Therefore, recent research of TNBC focuses on identifying novel therapeutic targets and mechanisms of chemoresistance.
The aggressive nature of TNBC has been attributed to the presence of cancer stem cells, also termed “tumor-initiating cells”. These cells are characterized by their ability to self-renew and differentiate into non-stem cancer cells that form the tumor bulk [5]. Cancer stem cells have been identified as important targets for cancer treatment due to their enhanced metastatic capability and resistance to conventional chemotherapy. Increasing evidence has shown that breast cancer stem cells (BCSCs) are enriched in TNBC compared to other breast cancer subtypes [6–10], which is concordant with the inherently aggressive clinical behavior of TNBC. Thus, it is important to target BCSCs to achieve a better outcome in TNBC treatment.
We conducted siRNA screening in a TNBC cell line to identify important regulators of BCSCs in TNBC. From this screening, we identified low-density lipoprotein receptor-related protein 8 (LRP8, also known as apolipoprotein E receptor 2, apoER2) as a novel positive regulator of BCSCs in TNBC. The biological function of LRP8 has been studied in the developing brain where it has been shown to regulate neuronal migration. Upon binding to its ligand, Reelin, at the cell surface, LRP8 triggers phosphorylation of the cytoplasmic adaptor protein Disabled-1 (Dab-1) to activate the downstream signaling cascades [11–14]. A recent study reports that LRP8-Reelin signaling is also important for cognitive function [15]. However, the function of LRP8 in cancers and cancer stem cells is not well studied.
Here, we show that LRP8 expression is elevated in TNBC as compared to other breast cancer subtypes, which may be due to a higher rate of LRP8 copy number gain or amplification in TNBC. Furthermore, higher LRP8 expression is associated with poor patient survival. Knockdown of LRP8 decreases BCSCs, metastatic potential, and tumorigenesis of TNBC and sensitizes TNBC cells to chemotherapy. Mechanistically, we show that LRP8 knockdown leads to inhibition of Wnt/β-catenin signaling pathway and potentially extracellular-signal regulated kinase (ERK)/MAPK pathway. In conclusion, LRP8’s localization to the cell surface and its importance for maintaining BCSCs in TNBC position LRP8 as a valuable therapeutic target for the treatment of TNBC through eradication of BCSCs.
2. Materials and methods
2.1. Cell lines and chemicals
HCC1937 was grown in RPMI1640 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% FBS and 1× Antibiotic-Antimycotic (Invitrogen; Thermo Fisher Scientific, Inc.). SUM149 was grown in F12 (Invitrogen; Thermo Fisher Scientific, Inc.) containing 5% FBS, 1× Antibiotic-Antimycotic, 5 μg/mL of insulin (Gibco; Thermo Fisher Scientific, Inc.), and 1 μg/mL of hydrocortisone (Sigma-Aldrich, St. Louis, MO, USA). HEK293T was grown in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS and 1× Antibiotic-Antimycotic. Cells were cultured in a 5% CO2 incubator at 37 °C. Docetaxel and 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H tetrazolium-bromide (MTT) were purchased from Sigma-Aldrich.
2.2. siRNA knockdown of LRP8
Cells were transfected with Silencer Select siRNA (Thermo Fisher Scientific, Inc.) targeting mRNA of LRP8 (Cat. No. s15367) in parallel with AllStars Negative Control siRNA (Cat. No. SI03650318, Qiagen, Palo Alto, CA, USA). Transfection was conducted by using Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s instructions.
2.3. Knockdown of LRP8 by inducible shRNA
Oligonucleotides containing the shRNA sequence targeting mRNA of LRP8 (Supplementary Table 2) were ligated into AgeI and EcoRI digested Tet-pLKO-puro lentiviral vector. The LRP8 shRNA was designed to target a different mRNA sequence from those targeted by the LRP8 siRNA to confirm the effects of the siRNA. Tet-pLKO-puro was a gift from Dmitri Wiederschain (Addgene plasmid # 21915) and TetpLKO-puro-Scrambled shRNA was a gift from Charles Rudin (Addgene plasmid # 47541). To produce virus particles, the shRNA constructs were co-transfected with the packaging vectors psPAX2 and pMD2.G into HEK293T cells. The viral supernatants were collected 48 h after transfection and then added to cells in the presence of 4 μg/mL polybrene (Sigma-Aldrich). Twenty-four hours later, cells were selected with puromycin (1 μg/mL) for 5 days. To induce shRNA knockdown, doxycycline at a final concentration of 100 ng/mL was added to the culture media.
2.4. RNA-Seq and data analysis
SUM149 cells were transfected with control or LRP8 siRNA in quadruplicate. After 72 h, total RNA was extracted with Direct-zol kit (Zymo, Irvine, CA, USA), and mRNA libraries were prepared using TruSeq (Illumina, Hayward, CA). RNA-Seq was performed in the University of Michigan DNA Sequencing Core using Illumina Hi-Seq 4000 with 50 cycle single-end reads. The sequencing reads were mapped to human transcripts annotated in GENCODE [16] using Bowtie [17]. Only uniquely mapped reads were used for further analysis. Gene expression levels were estimated as reads/kilobase/million mapped reads (RPKM) [18] using rSeq [19]. Differentially expressed genes were detected using edgeR [20]. Genes with a FDR value < 0.05 and a fold change ≥2 folds were considered significant (Supplementary Table 1). Gene ontology analysis was conducted using genes with a |log2 fold change| (|log2FC|) ≥ 1.5 in DAVID [21]. Gene Set Enrichment Analysis (Broad Institute) was used to correlate gene functions and signaling pathways that are significantly affected in LRP8 knockdown cells. All METABRIC data were accessed from the cBioPortal for Cancer Genomics website [22,23].
2.5. Mammosphere formation assay
Mammosphere formation was conducted as previously reported [24]. Briefly, dissociated single cells were seeded at a density of 5000 cells/well in Mammocult (StemCell Technologies) in ultra-low attachment 6-well plates (Corning Inc., Corning, NY, USA). After culturing for 6 days, primary mammospheres greater than 50 μm in diameter were counted. Primary mammospheres were then dissociated to single cells to be seeded at the same density as the primary mammosphere culture for secondary mammosphere formation.
2.6. Invasion assay
In vitro cellular invasion and migration was evaluated by matrigel-based transwell assays following the manufacturer’s instruction (Corning Inc.). Seventy-two hours after siRNA transfection, cells were seeded at a density of 1 × 104 in serum-free medium in the top chamber of the transwell. Medium containing serum as attractant was added to the bottom chamber of the transwell. Twenty-four hours after seeding, the migrated/invaded cells were fixed with ice-cold methanol and stained with 0.05% crystal violet. Five pictures were taken at 20× magnification in random fields.
2.7. Immunoblotting
Cells were lysed in RIPA buffer containing 5 mM EDTA, 1× protease inhibitor cocktail (Thermo Fisher) and 1× phosphatase inhibitor cocktail (Millipore). Proteins were separated by SDS-PAGE and probed with antibodies. LRP8 and CK19 antibodies were purchased from Abcam (Cambridge, MA, USA). β-actin antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Non-phospho (active) β-catenin (Ser33/37/Thr41) antibody was purchased from Cell Signaling (Danvers, MA, USA). The detailed information of the antibodies was listed in Supplementary Table 3.
2.8. Flow cytometry
CD44 (BD Biosciences, San Jose, CA, USA) and CD24 (BioLegend, San Diego, CA, USA) staining was conducted 3–4 days after siRNA or shRNA knockdown of LRP8. The cells were analyzed on a MoFlo Astrios flow cytometer (Beckman Coulter, Indianapolis, IN, USA). Additional information of the antibodies was listed in Supplementary Table 3.
2.9. RNA preparation and qRT-PCR
Total RNA was extracted from siRNA treated cells with Direct-zol (Zymo) and then reverse transcribed using QuantiTect Reverse Transcription kit (Qiagen). The cDNA was then subjected to quantitative PCR on a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Inc.) using SYBR Green (Thermo Fisher Scientific, Inc.). YWHAZ was used to normalize and calculate relative gene expression. Primers used for qRT-PCR were listed in the Supplementary Table 2.
2.10. Mouse xenograft model
The animal studies were conducted following the protocols approved by the University Committee on the Use and Care of Animals at the University of Michigan. Female NOD/SCID mice (Jackson Laboratory, Bar Harbor, ME, USA) were used to evaluate the effects of LRP8 knockdown on tumor growth and tumorigenicity. Briefly, 5000 SUM149 cells carrying doxycycline-inducible control or LRP8 shRNA were injected into the inguinal mammary fat pad of 6–8-week-old mice. Doxycycline diet (625 mg/kg) (Envigo, Haslett, MI, USA) was given to mice starting from 5 weeks after implantation when palpable tumors were observed. At the end of tumor growth monitoring, tumors were harvested and dissociated by using Tumor Dissociation Kit, human (Miltenyi Biotec, Auburn, CA, USA), and then DAPI and H-2Kd (BD Biosciences) double-negative live human cancer cells were sorted by flow cytometry. Limiting dilution assay was conducted by inoculating the sorted and serially diluted cancer cells (2,500, 500 and 100 cells/inoculation) into the inguinal mammary fat pad of tumor-free mice. Tumor formation was monitored for 3 months. The frequency of tumor-initiating cells was calculated by using Extreme Limiting Dilution Analysis (ELDA) [25].
2.11. Immunohistochemistry
Tumors were isolated from mouse xenografts and fixed in 10% formalin (Thermo Fisher Scientific, Inc.) followed by paraffin embedding. Immunohistochemistry was performed in the University of Michigan In-Vivo Animal Core (IVAC). Briefly, slides were deparaffinized and subjected to heat-induced antigen retrieval using a commercial pressure chamber (Biocare Decloaker, Biocare Medical) and buffer (Biocare Diva). Immunostaining was performed using an automated immunohistochemical stainer (Biocare Intellipath FLX®, Biocare Medical, Concord, CA) and included blocking for endogenous peroxidases and non-specific binding (Biocare Rodent Block M). The primary antibody (anti-CK19, cat #ab7754, Abcam) was applied for 30 min at a concentration of 1:200. Detection was performed by a biotin-free polymer based commercial detection system (Biocare Univ HRP Polymer) with the chromogen diaminobenzidine (DAB) and a hematoxylin nuclear counterstain. Negative controls were performed with each run using a universal control (Biocare #NC498) in place of the primary antibody.
2.12. Immunofluorescence staining
FFPE sections were deparaffinized and rehydrated by dipping three times in xylene, two times in 100% ethanol and once each in 95% and 70% ethanol. Antigen retrieval was performed by heating slides in citrate buffer (pH = 6.0) in a microwave for 10 min. Samples were then treated with ice-cold 1:1 Methanol:Acetone for 1 min and washed with PBS. Blocking buffer consisting of 5% goat serum (Sigma Aldrich) diluted in PBS was applied at room temperature for 1 h to prevent non-specific adhesion. Primary antibodies targeting LRP8 (1:200), Cytokeratins 8/18 (1:200), and vimentin (5 μg/mL) were diluted in blocking buffer and applied overnight in a humidified chamber at 4 °C. Sections were washed three times for 5 min each with PBS. Secondary and direct-conjugated antibodies were diluted in blocking buffer and applied for 6–8 h in a humidified chamber at 4 °C: CD44-BV510 (2 μg/ml), CD24 phycoerythrin (2 μg/ml), goat anti-mouse immunoglobulin M (IgM) AF488 (2 μg/mL), goat anti-rabbit immunoglobulin G (IgG) AF647 (2 μg/mL), and goat anti-guinea pig immunoglobulin G (IgG) DL755 (2.5 μg/mL). Slides were again washed three times for 5 min each with PBS, then treated with DAPI (1 μg/mL, Thermo Fisher Scientific) to label nuclei. Finally, the tissue sections were mounted with coverslips using Prolong Diamond Antifade Mountant (Thermo Fisher Scientific) and imaged with an Olympus IX-83 microscope using six optical filter sets corresponding to each fluorophore.
2.13. Chemoresistance test
SUM149 cells were transfected with control siRNA or LRP8-targeted siRNA. At 48 h after transfection, cells were reseeded into a 96-well plate at a density of 1 × 104 cells/well. Twenty-four hours later, cells were treated with Docetaxel from 0 to 500 nM for 72 h and the cell viability was then measured by a MTT assay. Briefly, cells were incubated with 0.5 mg/mL of MTT solution at 37 °C for 2 h. The MTT solution was removed and then 50 μL of DMSO was added to each well. The absorbance at 570 nm was read by using a plate reader (BioTek, Winooski, VT, USA).
2.14. Statistical analysis
Two-tailed Student’s t-test was used to compare the statistical difference between two groups. One-way ANOVA was used if the comparison involved more than two groups. A P-value < 0.05 was considered significant.
3. Results
3.1. LRP8 is highly expressed in triple-negative breast cancer
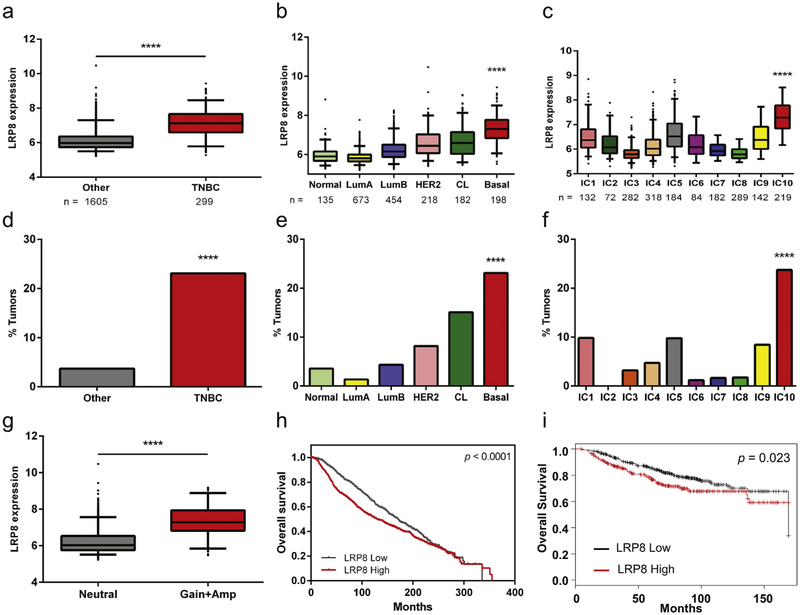
To determine if LRP8 is expressed differentially among the various breast cancer subtypes, we analyzed the METABRIC patient data set, which contains nearly 2,000 breast cancer cases with gene expression, copy number alteration and clinical data [26]. Pathologically, LRP8 expression was elevated in TNBC compared to other subtypes (Fig. 1a). Stratification of this patient cohort based on molecular classification [27] identified higher LRP8 expression in basal-like breast cancer (Fig. 1b), a subtype that constitutes ~80% of TNBC [28–30]. Using a genome-driven classification [26], elevated LRP8 expression was found associated with the IC10 subgroup of breast cancers that shows high concordance with the basal-like subtype (Fig. 1c).
Fig. 1.
LRP8 is highly expressed in TNBC. a-c Breast cancer samples in the METABRIC data set were stratified by different classification methods. LRP8 was found highly expressed in a TNBC (****p < 0.0001, Student’s t-Test), b basal-like subtype of the molecular subtypes (Normal: normal-like breast cancer, Lum A: luminal A subtype, Lum B: luminal B subtype, CL: claudin-low subtype, Basal: basal-like subtype; ****p < 0.0001, One-way ANOVA), and c IC10 of the integrative clusters (****p < 0.0001, One-way ANOVA). d-f Percentage of LRP8 copy number gain or amplification in breast cancer subtypes using different classifications (****p < 0.0001, χ2-test). g Comparison of LRP8 expression between breast cancer patients with LRP8 diploid (neutral) and those with LRP8 copy number gain or amplification (Amp) (****p < 0.0001, Student’s t-Test). h, i Kaplan-Meier survival analysis of breast cancer patients with low or high expression of LRP8. h data from the METABRIC data set and i data from Kaplan-Meier Plotter [32].
Further analysis of the METABRIC data set showed that elevated LRP8 expression in TNBC may be due to copy number alterations. Among all METABRIC breast cancer patients, copy number gain or amplification at the LRP8 genomic locus was identified in 142 cases (6.5%) (data not shown). Interestingly, the incidence of LRP8 copy number gain or amplification increased to 23–24% in TNBC, basal-like and IC10 breast cancers (Fig. 1d–f). The claudin-low subtype, which was common within TNBC [31], also showed a higher rate of LRP8 copy number gain or amplification compared to the luminal and HER2 subtypes (Fig. 1e). Furthermore, patients with LRP8 copy number gain or amplification had significantly higher mRNA expression, suggesting that copy number alterations could be the causal mechanism underlying elevated LRP8 expression in these patients (Fig. 1g). Lastly, patients with high expression of LRP8 had poorer survival than the rest of the cohort (Fig. 1h), which may be due to the strong correlation between LRP8 expression and TNBC. The survival analysis result of METABRIC was consistent with that of another data set [32] (Fig. 1i). Therefore, the LRP8 expression level could potentially be used as a prognosis marker.
3.2. Knockdown of LRP8 decreases BCSCs and invasiveness in TNBC
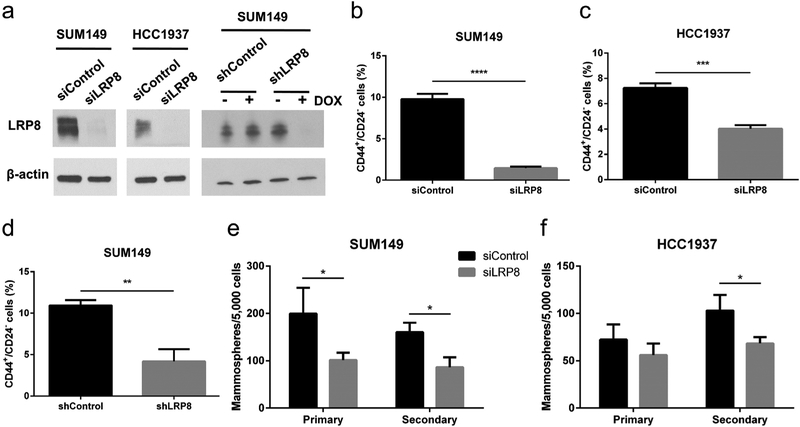
To study the function of LRP8 in TNBC, we performed siRNA or doxycycline (tetracycline)-inducible shRNA knockdown of LRP8 in two TNBC cell lines, SUM149 and HCC1937 (Fig. 2a), and examined the effect on CD44+/CD24− cells, a well-studied BCSC population with self-renewal and tumor-initiating ability [33,34]. We found that LRP8 knockdown significantly reduced the percentage of CD44+/CD24− cells (Fig. 2b–d). Consistent with the reduction of the CD44+/CD24− BCSC population, LRP8 knockdown decreased the mammosphere formation capacity of TNBC cells (Fig. 2e and f), indicating an impairment of the self-renewal ability of BCSCs.
Fig. 2.
LRP8 knockdown suppresses BCSCs in TNBC. a Western blot of LRP8 and β-actin (loading control) in LRP8 (siLRP8) or control (siControl) siRNA treated SUM149 and HCC1937 and in LRP8 (shLRP8) or control (shControl) shRNA treated SUM149. b-d Flow cytometry analysis of CD44+/CD24− BCSCs in SUM149 and HCC1937 with LRP8 knockdown by siRNA and in SUM149 with LRP8 knockdown by shRNA. e, f Primary and secondary mammosphere formation in SUM149 and HCC1937 with LRP8 knockdown by siRNA. The results of b-f were shown as mean ± S.D. (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, Student’s t-Test).
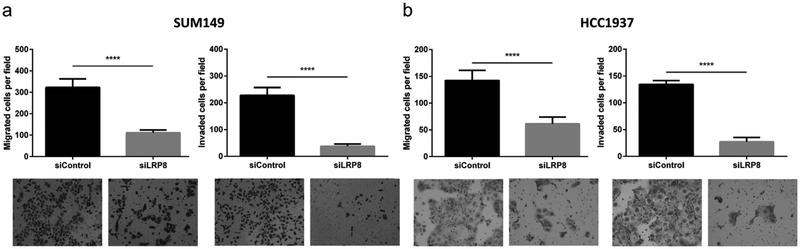
Epithelial cancer cells can acquire stemness and CD44+/CD24− expression status through epithelial-to-mesenchymal transition (EMT) [35], a process that also plays an important role in cancer metastasis [36]. To investigate whether silencing of LRP8 was able to inhibit metastasis of TNBC cells, we used a matrigel-based transwell assay to evaluate the effect of LRP8 knockdown on cellular migration and invasion. The results showed that LRP8 knockdown significantly decreased both migration and invasion of TNBC cells (Fig. 3a and b). Our data suggest that knockdown of LRP8 may decrease the metastatic potential of TNBC.
Fig. 3. LRP8 knockdown inhibits invasiveness of TNBC cells.
Matrigel-based migration and invasion transwell assay. Migrated and invaded cells in a SUM149 and b HCC1937 with LRP8 knockdown by siRNA. The results were shown as mean ± S.D. (n = 5, ****p < 0.0001, Student’s t-Test).
3.3. Knockdown of LRP8 reduces tumorigenicity of TNBC
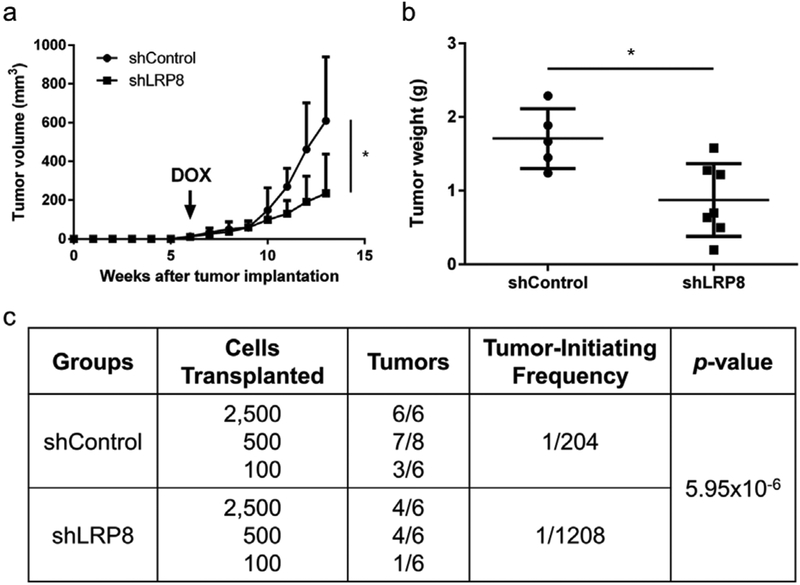
To investigate the impact of LRP8 knockdown on tumorigenesis, we used a doxycycline-inducible vector to generate stable SUM149 cell lines carrying LRP8 shRNA (shLRP8) or negative control shRNA (shControl). Upon shLRP8 induction with doxycycline in vitro, LRP8 protein was decreased to an almost undetectable level (Fig. 2a). These stable SUM149 cell lines were injected into the mammary fat pads of NOD-SCID mice and shRNA was induced by doxycycline treatment when the tumors were palpable in all the mice. As shown in Fig. 4a and b, LRP8 knockdown significantly suppressed tumor growth. Furthermore, the result of the secondary limiting dilution transplantation experiment revealed that silencing of LRP8 significantly reduced the frequency of tumor-initiating cells by 6-fold, from 1/204 (shControl) to 1/1208 (shLRP8) (Fig. 4c).
Fig. 4.
LRP8 knockdown inhibits tumorigenesis of TNBC. a Tumor growth of SUM149 carrying doxycycline-inducible shLRP8 (n = 7) or shControl (n = 5). The cells were injected into the mammary fat pad of NOD-SCID mice. Doxycycline diet (DOX) was given when palpable tumors were observed in all mice (6 weeks after implantation). The results were shown as mean + S.D. (*p < 0.05, Student’s t-Test). b Tumor weight at the end of tumor monitoring (13 weeks after implantation). *p < 0.05, Student’s t-Test. c Secondary limiting dilution transplantation of the primary tumors. The frequency of tumor-initiating cells was calculated by using Extreme Limiting Dilution Analysis (ELDA).
3.4. Signaling pathways altered by LRP8 knockdown in TNBC cells
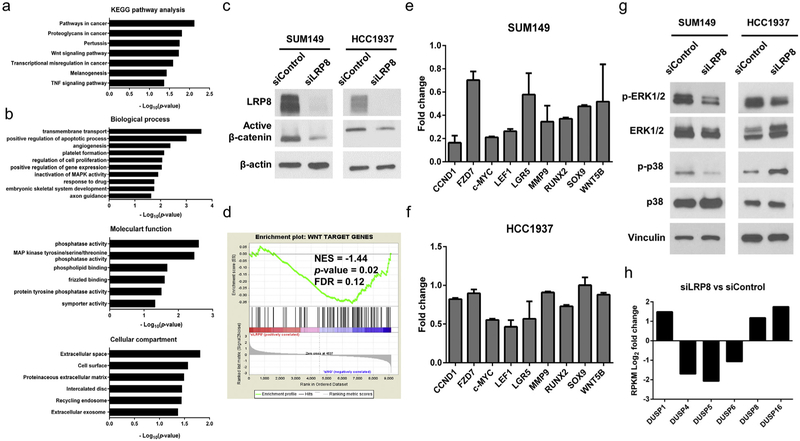
To study the molecular mechanism of BCSC inhibition mediated by LRP8 knockdown, we performed RNA sequencing (RNA-Seq) in SUM149 cells treated with control or LRP8 siRNA. Knockdown of LRP8 resulted in 366 genes upregulated and 416 genes downregulated at ≥ 2 folds (edgeR FDR < 0.05) (Supplementary Table 1). To identify signaling pathways and cellular functions affected by LRP8 knockdown, we conducted pathway analysis and gene ontology (GO) analysis of the RNA-Seq data using DAVID. In the pathways known to regulate self-renewal and stemness of cancer cells, canonical Wnt/β-catenin signaling pathway was the most significantly dysregulated signaling pathway in the LRP8 knockdown SUM149 cells (Fig. 5a). In addition, the GO analysis revealed that mitogen-activated protein kinase (MAPK) pathways were also interrupted by LRP8 knockdown (Fig. 5b). The alteration of the MAPK pathways might be due to the dysregulation of MAPK phosphatases caused by LRP8 knockdown (Fig. 5b). Furthermore, LRP8 knockdown significantly affected cell survival mechanisms such as proliferation and apoptosis (Fig. 5b), which might be the reason for the reduced tumor burden in the LRP8 knockdown xenografts.
Fig. 5.
Signaling pathways altered by LRP8 knockdown in TNBC cells. RNA-Seq was conducted for SUM149 transfected with control siRNA or LRP8-targeted siRNA. a Pathway analysis and b GO analysis of the differentially expressed genes from the RNA-Seq data were conducted using DAVID. Canonical Wnt/β-catenin and MAPK signaling pathways were enriched upon LRP8 knockdown. c Western blot of active (Non-phospho) β-catenin and β-actin (loading control) in SUM149 and HCC1937 with LRP8 knockdown by siRNA. d GSEA of the RNA-Seq data using a gene set of previously reported Wnt downstream target genes. The expression of Wnt target genes was negatively correlated with the LRP8 knockdown cells. ES: enrichment score. e, f qRT-PCR for the selected Wnt target genes in SUM149 and HCC1937. Gene expression fold change between siLRP8 and siControl was calculated using the 2−ΔΔCt method and YWHAZ was used for normalization. The results were shown as mean ± S.D. g Western blot of vinculin (loading control), phosphorylated and total protein of p38 and ERK1/2 in SUM149 and HCC1937 with LRP8 knockdown by siRNA. h Gene expression fold change of DUSPs between siLRP8 and siControl in SUM149 cells. RPKM: reads per kilobase million.
To determine the effect of LRP8 knockdown on the canonical Wnt/β-catenin pathway, we first examined the level of active (non-phosphorylated) β-catenin [37,38] by western blot and found that knockdown of LRP8 decreased the level of active β-catenin in SUM149 and HCC1937 cells (Fig. 5c). We next conducted gene set enrichment analysis (GSEA) of the RNA-Seq data using a Wnt signature gene set [39] and demonstrated that Wnt target genes were negatively correlated with the LRP8 knockdown SUM149 cells (Fig. 5d), indicating that loss of LRP8 downregulated Wnt downstream targets. The quantitative reverse transcription PCR (qRT-PCR) analysis of the selected Wnt target genes in the LRP8 knockdown SUM149 and HCC1937 further supported the inhibition of Wnt signaling in these cells (Fig. 5e and f).
We also performed Western blot to examine the effects of LRP8 knockdown on MAPK signaling pathways. We found that LRP8 knockdown decreased the phosphorylation of ERK1/2 in both SUM149 and HCC1937 cells (Fig. 5g). However, LRP8 knockdown led to opposite effects on the phosphorylation of p38 MAPK in SUM149 and HCC1937 cells (Fig. 5g). Given that silencing of LRP8 significantly altered the expression of MAPK phosphatases (Fig. 5b), we examined the RNA-Seq data and found that dual-specific phosphatases (DUSPs) were the most dysregulated MAPK phosphatases caused by LRP8 knockdown (Fig. 5h). We hypothesized that individual DUSPs might dominate the dephosphorylation of MAPKs in different cellular context. For instance, DUSP4–6 might dominate the dephosphorylation of p38 MAPK over DUSP1, DUSP8, and DUSP16 in HCC1937. Nevertheless, more studies will be needed to clarify how these DUSPs regulate specific MAPK pathways in different cellular context.
3.5. LRP8 knockdown shifts TNBC cells towards a more differentiated, epithelial cell state
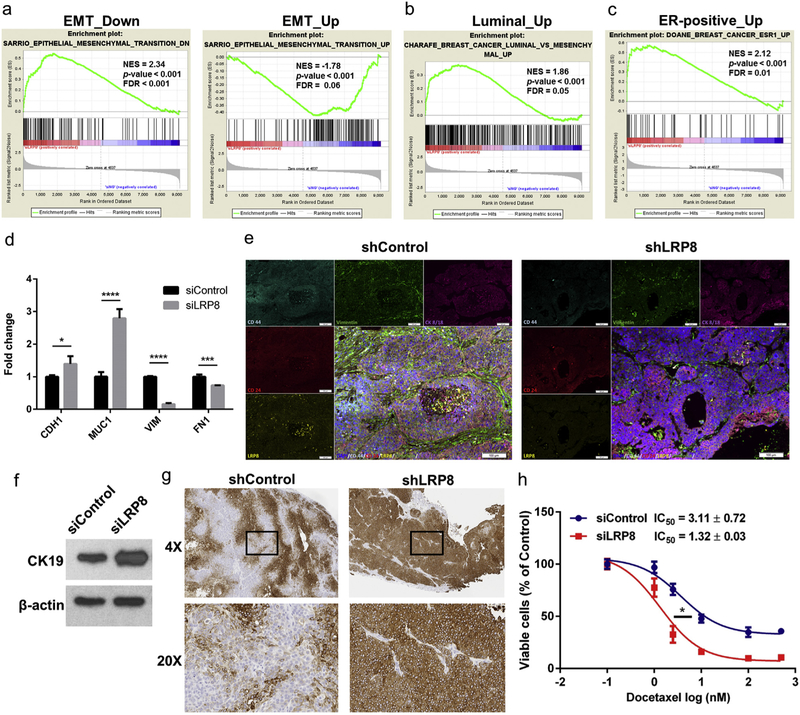
We next conducted a more comprehensive GSEA of the LRP8 knockdown RNA-Seq data. We found that LRP8 knockdown shifted the gene signature of SUM149 cells from a basal-mesenchymal state towards a more luminal-epithelial state (Fig. 6a–c). Specifically, gene sets from mammary cells undergoing EMT were negatively correlated with the LRP8 knockdown cells (Fig. 6a). In addition, the genes that were upregulated in luminal breast cancer and ER-positive breast cancer were positively correlated with the LRP8 knockdown cells (Fig. 6b and c).
Fig. 6.
LRP8 knockdown shifts TNBC cells from a basal-mesenchymal state to a more differentiated luminal-epithelial state. a-c GSEA enrichment plots of EMT and luminal breast cancer gene sets in the LRP8 knockdown RNA-Seq data. a (left panel) Genes downregulated in mammary epithelial cells undergoing EMT (epithelial gene signature) were positively correlated with the LRP8 knockdown cells; (right panel) Genes upregulated in cells undergoing EMT (mesenchymal gene signature) were negatively correlated with the LRP8 knockdown cells. Genes upregulated in luminal breast cancer (b) and ER-positive breast cancer (c) were positively correlated with the LRP8 knockdown cells. NES: normalized enrichment score, FDR: false discovery rate. d qRT-PCR for the selected epithelial (CDH1: E-cadherin, MUC1: mucin 1) and mesenchymal (VIM: vimentin, FN1: fibronectin) genes in SUM149 with siLRP8 or siControl. Fold change between siLRP8 and siControl was calculated using the 2−ΔΔCt method and YWHAZ was used for normalization. The results were shown as mean ± S.D. (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, Student’s t-Test). e Immunofluorescence staining of doxycycline-inducible control or LRP8 knockdown SUM149 xenograft tumors. f Western blot of CK19 in SUM149 cells with LRP8 knockdown by siRNA in vitro. g CK19 IHC in SUM149 tumors with control or LRP8 knockdown by doxycycline-inducible shRNA. h Relative cell survival of siControl or siLRP8 knockdown SUM149 cells treated with docetaxel (0, 1, 2.5, 10, 100, and 500 nM) for 72 h. The results were shown as mean ± S.D. (n = 3). IC50 of docetaxel was calculated and compared (*p < 0.05, Student’s t-Test).
We next tested the hypothesis that TNBC cells were shifted from a basal-mesenchymal state towards a more luminal-epithelial state upon LRP8 knockdown. We selected the classic mesenchymal genes vimentin (VIM) and fibronectin (FN1) and epithelial genes E-cdherin (CDH1) and mucin 1 (MUC1) and evaluated their expression by qRT-PCR. Knockdown of LRP8 decreased the expression of mesenchymal genes and concomitantly increased the expression of epithelial genes (Fig. 6d). This phenomenon was again confirmed by the immuno-fluorescence staining on tumor biopsies, which demonstrated a dramatic decrease of vimentin and CD44 and increase of CD24 expression in the LRP8 knockdown tumors (Fig. 6e). We also found that knockdown of LRP8 in cell culture and xenografts led to elevated expression of CK19 (Fig. 6f, g), a luminal-epithelial cytokeratin of breast cancer, whose negativity was correlated with poor prognosis [40,41]. Furthermore, LRP8 knockdown sensitized SUM149 cells to docetaxel treatment. The IC50 of docetaxel was significantly decreased by 2.4-fold in the LRP8 knockdown cells compared to control cells (Fig. 6h). Together, our results indicated that silencing of LRP8 could shift TNBC cells towards a more differentiated, epithelial cell state and sensitize them to chemotherapy.
4. Discussion
TNBC has poorer prognosis compared with other breast cancer subtypes due to its inherent aggressiveness and the lack of targeted therapies [3]. Current treatments for TNBC are limited to surgery and chemotherapy. The later has been shown to induce BCSCs [42], which are believed to be responsible for metastasis and drug-resistance. As a result, identification of therapeutic targets to eliminate BCSCs is necessary to achieve a better outcome for TNBC patients. By investigating the METABRIC data set, we have found that LRP8, a transmembrane protein, is highly expressed in TNBC in comparison to other breast cancer subtypes. Not only does LRP8 serve as a marker of poor prognosis in breast cancer, but our study also suggests that LRP8 is essential for the maintenance of BCSCs and tumorigenicity in TNBC.
In our pathway analysis to scrutinize LRP8’s mechanism of action, we find that silencing of LRP8 significantly inhibits Wnt signaling pathway. Our results support previous studies which link LRP8 to Wnt signaling regulation. In normal tissues, LRP8 regulates osteoblast differentiation through Wnt signaling [43]. In addition, LRP8 gene amplification and overexpression have been reported in lung cancer and its oncogenic role has been suggested to be linked to Wnt signaling [44]. In the canonical Wnt signaling pathway, LRP5/6 function as Wnt co-receptors that bind to Wnt ligands and then inactivate the destruction complex to prevent proteosomal degradation of β-catenin [45]. Like LRP5/6, LRP8 belongs to the low-density lipoprotein receptor-related protein (LRP) family and shares the conserved domains, such as LDL repeats and EGF receptor-like domains [46]. Furthermore, LRP8 has been reported to interact directly with Wnt3a, indicating that LRP8 might serve as a novel Wnt co-receptor [43]. Other studies have shown that ligation of LRP8 by activated protein C, a plasma protein with anticoagulant and cytoprotective activities, leads to Dab-1 phosphorylation and subsequent activation of PI3K and AKT and inactivation of GSK-3β by phosphorylation [47,48]. Phosphorylation of GSK-3β releases β-catenin from the destruction complex and thereby stabilizes β-catenin [49–51]. Therefore, LRP8 may activate Wnt signaling via different regulatory mechanisms. Our results and the previous studies suggest that LRP8 plays an important role in activating Wnt signaling in TNBC.
It is worth noting that LRP8 is functionally redundant to LRP5/6 in Wnt signaling activation. Therefore, one concern is that LRP5/6 may potentially compensate LRP8 knockdown-mediated Wnt signaling inhibition. In our RNA-Seq data, we do not find LRP8 knockdown elevate the expression of LRP5/6. Interestingly, LRP8 knockdown increase the expression of LRP1 by 2.3 folds (Supplementary Table 1). LRP1 has been reported to interact with Frizzled-1, which causes the disruption of Wnt receptor/co-receptor complex formation and thus represses the canonical Wnt signaling pathway [52]. Furthermore, LRP8 knockdown increase the expression of SFRP1, the secreted Wnt antagonist, by 3.1 folds (Supplementary Table 1). These data suggest that LRP8 knockdown can elevate the expression of Wnt pathway repressors which may compromise LRP5/6-mediated Wnt signaling activation. Such hypothesis is also supported by our mouse xenograft models, which demonstrate that long-term knockdown of LRP8 still significantly inhibits tumorigenesis.
Wnt signaling plays a critical role in regulating self-renewal of stem cells and cancer stem cells [53,54]. In a mouse model, Wnt3a is sufficient to sustain mouse mammary stem cells to form spheres in vitro and enhance the reconstitution of mammary gland in vivo [55]. Studies have shown that overexpression of Wnt1 in mice induces spontaneous mammary cancers [56,57]. Recent work elegantly demonstrates that human mammary cells undergoing EMT have increased expression of WNT5A and decreased expression or protein secretion of Wnt antagonists SFRP1 and DKK1 [58]. Restoration of SFRP1 expression in these EMT mammary cells reduces cell migration and mammosphere formation in vitro and inhibits tumorigenesis in xenograft models [58]. In addition, Wnt signaling has been reported to be particularly active in TNBC and is associated with metastasis and tumorigenesis in TNBC [59–61]. These studies reveal the profound effects of Wnt signaling in BCSCs and TNBC, hence highlighting the value of targeting LRP8 to inhibit Wnt signaling in TNBC.
Emerging evidence has demonstrated the importance of MAPK signaling pathways in regulating EMT and TICs. Previous studies have demonstrated that activation of MAPK pathways is required or can synergistically enhance TGF-β-induced EMT [62–65]. MAPKs can also phosphorylate TWIST1, an EMT transcription factor, and thus protect it from proteasome-mediated degradation [66]. In our study, silencing of LRP8 decreases the phosphorylation of ERK1/2, which may contribute to the inhibition of EMT according to previous studies [62,64–66]. Interestingly, LRP8 knockdown increases the phosphorylation of p38 in HCC1937 cells. Given that silencing of LRP8 significantly inhibits the expression of specific DUSPs, the elevated p38 phosphorylation in the LRP8 knockdown HCC1937 cells implies a potential resistant mechanism of targeting LRP8 as loss of DUSPs has been reported to activate MAPK pathways and induce both stem cell-like phenotypes and chemoresistance in TNBC [67,68]. Therefore, a more thorough study will be needed to elucidate the effects of targeting LRP8 on DUSPs and the MAPK pathways.
TNBC/basal-like breast cancer has worse prognosis compared to the luminal subtypes of breast cancer [69]. Also, TNBC has been linked to a less differentiated stem or progenitor cell state contrary to the luminal subtypes which recapitulate a more differentiated epithelial cell state in human mammary epithelial hierarchy [70]. As the emerging evidence proves the contribution of BCSCs to tumorigenesis and drug-resistance, promoting TNBC cell differentiation is a rational and potentially effective strategy for TNBC treatment. A recent study has demonstrated that EMT reversion triggers mammary cells to leave the mesenchymal tumor-initiating state and differentiate into a chemotherapy-sensitive epithelial (non-stem) cell state [71]. Like the aforementioned study, our results show that upon LRP8 knockdown, TNBC cells acquire the more differentiated luminal-epithelial gene signature and become more sensitive to the treatment with docetaxel.
In conclusion, our study shows that LRP8 is a novel oncogene in TNBC. The expression of LRP8 is highly associated with TNBC. Silencing of LRP8 can significantly suppress BCSCs and tumorigenesis in TNBC via Wnt signaling inhibition. Furthermore, knockdown of LRP8 shifts TNBC cells to a more differentiated, luminal-epithelial cell state and sensitizes them to chemotherapy. These results suggest LRP8 may serve as a therapeutic target to inhibit BCSCs in TNBC.
Supplementary Material
Acknowledgements
The flow cytometry work in this research was supported by the University of Michigan Cancer Center Support Grant (NCI (National Cancer Institute), United States Grant P30CA046592) from the National Institutes of Health.
Footnotes
Conflicts of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Declarations of interest
None.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.canlet.2018.09.022.
References
- [1].Malorni L, Shetty PB, De Angelis C, Hilsenbeck S, Rimawi MF, Elledge R, Osborne CK, De Placido S, Arpino G, Clinical and biologic features of triple-negative breast cancers in a large cohort of patients with long-term follow-up, Breast Canc. Res. Treat 136 (2012) 795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dawood S, Triple-negative breast cancer: epidemiology and management options, Drugs 70 (2010) 2247–2258. [DOI] [PubMed] [Google Scholar]
- [3].Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L, Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease, Nat. Rev. Clin. Oncol 13 (2016) 674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Liedtke C, Mazouni C, Hess KR, Andre F, Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B, Green M, Cristofanilli M, Hortobagyi GN, Pusztai L, Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer, J. Clin. Oncol 26 (2008) 1275–1281. [DOI] [PubMed] [Google Scholar]
- [5].Beck B, Blanpain C, Unravelling cancer stem cell potential, Nat. Rev. Canc 13 (2013) 727–738. [DOI] [PubMed] [Google Scholar]
- [6].Ma F, Li H, Wang H, Shi X, Fan Y, Ding X, Lin C, Zhan Q, Qian H, Xu B, Enriched CD44(+)/CD24(−) population drives the aggressive phenotypes presented in triple-negative breast cancer (TNBC), Canc. Lett 353 (2014) 153–159. [DOI] [PubMed] [Google Scholar]
- [7].Foulkes WD, Smith IE, Reis-Filho JS, Triple-negative breast cancer, N. Engl. J. Med 363 (2010) 1938–1948. [DOI] [PubMed] [Google Scholar]
- [8].Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, Grabau D, Ferno M, Borg A, Hegardt C, The CD44+/CD24− phenotype is enriched in basal-like breast tumors, Breast Cancer Res. 10 (2008) R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Korsching E, Jeffrey SS, Meinerz W, Decker T, Boecker W, Buerger H, Basal carcinoma of the breast revisited: an old entity with new interpretations, J. Clin. Pathol 61 (2008) 553–560. [DOI] [PubMed] [Google Scholar]
- [10].Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW, A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes, Canc. Cell 10 (2006) 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J, Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2, Cell 97 (1999) 689–701. [DOI] [PubMed] [Google Scholar]
- [12].Benhayon D, Magdaleno S, Curran T, Binding of purified Reelin to ApoER2 and VLDLR mediates tyrosine phosphorylation of Disabled-1, Brain Res Mol Brain Res 112 (2003) 33–45. [DOI] [PubMed] [Google Scholar]
- [13].Bock HH, Herz J, Reelin activates SRC family tyrosine kinases in neurons, Curr. Biol 13 (2003) 18–26. [DOI] [PubMed] [Google Scholar]
- [14].Arnaud L, Ballif BA, Forster E, Cooper JA, Fyn tyrosine kinase is a critical regulator of disabled-1 during brain development, Curr. Biol 13 (2003) 9–17. [DOI] [PubMed] [Google Scholar]
- [15].Telese F, Ma Q, Perez PM, Notani D, Oh S, Li W, Comoletti D, Ohgi KA, Taylor H, Rosenfeld MG, LRP8-Reelin-Regulated neuronal enhancer signature underlying learning and memory formation, Neuron 86 (2015) 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R, The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression, Genome Res. 22 (2012) 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Langmead B, Trapnell C, Pop M, Salzberg SL, Ultrafast and memory-efficient alignment of short DNA sequences to the human genome, Genome Biol. 10 (2009) R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B, Mapping and quantifying mammalian transcriptomes by RNA-Seq, Nat. Methods 5 (2008) 621–628. [DOI] [PubMed] [Google Scholar]
- [19].Jiang H, Wong WH, Statistical inferences for isoform expression in RNA-Seq, Bioinformatics 25 (2009) 1026–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data, Bioinformatics 26 (2010) 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc 4 (2009) 44–57. [DOI] [PubMed] [Google Scholar]
- [22].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Canc. Discov 2 (2012) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci. Signal 6 (2013) pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS, In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells, Gene Dev. 17 (2003) 1253–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hu Y, Smyth GK, ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays, J. Immunol. Meth 347 (2009) 70–78. [DOI] [PubMed] [Google Scholar]
- [26].Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A,Russell R, McKinney S, Group M, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups, Nature 486 (2012) 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D, Molecular portraits of human breast tumours, Nature 406 (2000) 747–752. [DOI] [PubMed] [Google Scholar]
- [28].Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D, How basal are triple-negative breast cancers? Int. J. Canc 123 (2008) 236–240. [DOI] [PubMed] [Google Scholar]
- [29].Prat A, Pineda E, Adamo B, Galvan P, Fernandez A, Gaba L, Diez M, Viladot M, Arance A, Munoz M, Clinical implications of the intrinsic molecular subtypes of breast cancer, Breast 24 (Suppl 2) (2015) S26–S35. [DOI] [PubMed] [Google Scholar]
- [30].Lehmann BD, Pietenpol JA, Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes, J. Pathol 232 (2014) 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM, Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer, Breast Cancer Res. 12 (2010) R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lanczky A, Nagy A, Bottai G, Munkacsy G, Szabo A, Santarpia L, Gyorffy B, miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients, Breast Canc. Res. Treat 160 (2016) 439–446. [DOI] [PubMed] [Google Scholar]
- [33].Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF, Prospective identification of tumorigenic breast cancer cells, Proc. Natl. Acad. Sci. U.S.A 100 (2003) 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liao MJ, Zhang CC, Zhou B, Zimonjic DB, Mani SA, Kaba M, Gifford A, Reinhardt F, Popescu NC, Guo W, Eaton EN, Lodish HF, Weinberg RA, Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity, Canc. Res 67 (2007) 8131–8138. [DOI] [PubMed] [Google Scholar]
- [35].Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA, The epithelial-mesenchymal transition generates cells with properties of stem cells, Cell 133 (2008) 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nieto MA, Huang RY, Jackson RA, Thiery JP, Emt 166 (2016) (2016) 21–45 Cell. [DOI] [PubMed] [Google Scholar]
- [37].Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT, The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3, Gene Dev. 10 (1996) 1443–1454. [DOI] [PubMed] [Google Scholar]
- [38].Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW, Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC, Science 275 (1997) 1787–1790. [DOI] [PubMed] [Google Scholar]
- [39].Van der Flier LG, Sabates-Bellver J, Oving I, Haegebarth A, De Palo M, Anti M, Van Gijn ME, Suijkerbuijk S, Van de Wetering M, Marra G, Clevers H, The intestinal Wnt/TCF signature, Gastroenterology 132 (2007) 628–632. [DOI] [PubMed] [Google Scholar]
- [40].Fujisue M, Nishimura R, Okumura Y, Tashima R, Nishiyama Y, Osako T, Toyozumi Y, Arima N, Clinical significance of CK19 negative breast cancer, Cancers 5 (2012) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Parikh RR, Yang Q, Higgins SA, Haffty BG, Outcomes in young women with breast cancer of triple-negative phenotype: the prognostic significance of CK19 expression, Int. J. Radiat. Oncol. Biol. Phys 70 (2008) 35–42. [DOI] [PubMed] [Google Scholar]
- [42].Samanta D, Gilkes DM, Chaturvedi P, Xiang L, Semenza GL, Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells, Proc. Natl. Acad. Sci. U.S.A 111 (2014) E5429–E5438. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [43].Zhang J, Zhang X, Zhang L, Zhou F, van Dinther M, Ten Dijke P, LRP8 mediates Wnt/beta-catenin signaling and controls osteoblast differentiation, J. Bone Miner. Res 27 (2012) 2065–2074. [DOI] [PubMed] [Google Scholar]
- [44].Garnis C, Campbell J, Davies JJ, Macaulay C, Lam S, Lam WL, Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis, Hum. Mol. Genet 14 (2005) 475–482. [DOI] [PubMed] [Google Scholar]
- [45].Komiya Y, Habas R, Wnt signal transduction pathways, Organogenesis 4 (2008) 68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rey JP, Ellies DL, Wnt modulators in the biotech pipeline, Dev. Dynam 239 (2010) 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang G, Assadi AH, McNeil RS, Beffert U, Wynshaw-Boris A, Herz J, Clark GD, D’Arcangelo G, The Pafah1b complex interacts with the reelin receptor VLDLR, PLoS One 2 (2007) e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yang XV, Banerjee Y, Fernandez JA, Deguchi H, Xu X, Mosnier LO, Urbanus RT, de Groot PG, White-Adams TC, McCarty OJ, Griffin JH, Activated protein C ligation of ApoER2 (LRP8) causes Dab1-dependent signaling in U937 cells, Proc. Natl. Acad. Sci. U.S.A 106 (2009) 274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kitagawa M, Hatakeyama S, Shirane M, Matsumoto M, Ishida N, Hattori K,Nakamichi I, Kikuchi A, Nakayama K, Nakayama K, An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin, EMBO J. 18 (1999) 2401–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Aberle H, Bauer A, Stappert J, Kispert A, Kemler R, beta-catenin is a target for the ubiquitin-proteasome pathway, EMBO J. 16 (1997) 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P, Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly, Science 272 (1996) 1023–1026. [DOI] [PubMed] [Google Scholar]
- [52].Zilberberg A, Yaniv A, Gazit A, The low density lipoprotein receptor-1, LRP1, interacts with the human frizzled-1 (HFz1) and down-regulates the canonical Wnt signaling pathway, J. Biol. Chem 279 (2004) 17535–17542. [DOI] [PubMed] [Google Scholar]
- [53].Nusse R, Clevers H, Wnt/beta-Catenin signaling, disease, and emerging therapeutic modalities, Cell 169 (2017) 985–999. [DOI] [PubMed] [Google Scholar]
- [54].Holland JD, Klaus A, Garratt AN, Birchmeier W, Wnt signaling in stem and cancer stem cells, Curr. Opin. Cell Biol 25 (2013) 254–264. [DOI] [PubMed] [Google Scholar]
- [55].Zeng YA, Nusse R, Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture, Cell Stem Cell 6 (2010) 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE, Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice, Cell 55 (1988) 619–625. [DOI] [PubMed] [Google Scholar]
- [57].Nusse R, van Ooyen A, Cox D, Fung YK, Varmus H, Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15, Nature 307 (1984) 131–136. [DOI] [PubMed] [Google Scholar]
- [58].Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA, Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast, Cell 145 (2011) 926–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, Deng X, Chen L, Kim CC, Lau S, Somlo G, Yen Y, FZD7 has a critical role in cell proliferation in triple negative breast cancer, Oncogene 30 (2011) 4437–4446. [DOI] [PubMed] [Google Scholar]
- [60].Xu J, Prosperi JR, Choudhury N, Olopade OI, Goss KH, beta-Catenin is required for the tumorigenic behavior of triple-negative breast cancer cells, PLoS One 10 (2015) e0117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH, Wnt/beta-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome, Am. J. Pathol 176 (2010) 2911–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL, Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro, Neoplasia 6 (2004) 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yu L, Hebert MC, Zhang YE, TGF-beta receptor-activated p38 MAP kinase mediates Smad-independent TGF-beta responses, EMBO J. 21 (2002) 3749–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Grande M, Franzen A, Karlsson JO, Ericson LE, Heldin NE, Nilsson M, Transforming growth factor-beta and epidermal growth factor synergistically stimulate epithelial to mesenchymal transition (EMT) through a MEK-dependent mechanism in primary cultured pig thyrocytes, J. Cell Sci 115 (2002) 4227–4236. [DOI] [PubMed] [Google Scholar]
- [65].Uttamsingh S, Bao X, Nguyen KT, Bhanot M, Gong J, Chan JL, Liu F, Chu TT, Wang LH, Synergistic effect between EGF and TGF-beta1 in inducing oncogenic properties of intestinal epithelial cells, Oncogene 27 (2008) 2626–2634. [DOI] [PubMed] [Google Scholar]
- [66].Hong J, Zhou J, Fu J, He T, Qin J, Wang L, Liao L, Xu J, Phosphorylation of serine 68 of Twist1 by MAPKs stabilizes Twist1 protein and promotes breast cancer cell invasiveness, Canc. Res 71 (2011) 3980–3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, Gomez H, Cook RS, Arteaga CL, Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer, Canc. Res 73 (2013) 6346–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lu H, Tran L, Park Y, Chen I, Lan J, Xie Y, Semenza GL, Reciprocal regulation of DUSP9 and DUSP16 expression by HIF1 controls ERK and p38 MAP kinase activity and mediates chemotherapy-induced breast cancer stem cell enrichment, Canc. Res 78 (2018) 4191–4202. [DOI] [PubMed] [Google Scholar]
- [69].Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H, Breast cancer subtypes and the risk of local and regional relapse, J. Clin. Oncol. : Off. J. Am. Soc. Clin. Oncol 28 (2010) 1684–1691. [DOI] [PubMed] [Google Scholar]
- [70].Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, Feleppa F, Huschtscha LI, Thorne HJ, kConFab SB Fox M Yan JD French MA Brown GK Smyth JE Visvader GJ Lindeman, Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers, Nat. Med 15 (2009) 907–913. [DOI] [PubMed] [Google Scholar]
- [71].Pattabiraman DR, Bierie B, Kober KI, Thiru P, Krall JA, Zill C, Reinhardt F, Tam WL, Weinberg RA, Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability, Science 351 (2016) aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.