Abstract
Accumulating evidence has suggested that microRNAs (miRNAs) play important roles in regulating the progression of cancerby acting as tumor suppressors or oncogenes. Here, our results demonstrated that miR-5582-5p was significantly down-regulated in non-small cell lung cancer (NSCLC) tissues and cell lines compared with normal controls. Overexpression of miR-5582-5p markedly inhibited the proliferation and migration of NSCLC cells. Consistently, the apoptosis of NSCLC cells was also significantly promoted by overexpressed miR-5582-5p. Functional study uncovered that miR-5582-5p bound the 3’-untranslated region (UTR) of fibroblastic growth factor-10 (FGF-10) and decreased the expression of FGF-10 in NSCLC cells. FGF-10 was up-regulated in NSCLC tissues and inversely correlated with the level of miR-5582-5p in NSCLC tissues. Overexpression of FGF-10 significantly reversed the inhibitory effect of miR-5582-5p on the proliferation of NSCLC cells. Taken together, our results demonstrated the functional mechanism of miR-5582-5p in suppressing malignant behaviors of NSCLC cells by targeting FGF-10. These findings demonstrated that miR-5582-5p might be a novel therapeutic target in the treatment of NSCLC.
Keywords: miR-5582-5p, NSCLC, FGF-10
Introduction
Lung cancer is one of the most common malignancies and the leading cause of cancer-related death worldwide [1-4]. Non-small cell lung cancer (NSCLC) is a major histologic type of lung cancer and accounts for approximately 80% of all primary cases [5-7]. Surgical resection plus chemotherapy and radiotherapy has improved the outcome of NSCLC patients; however, the prognosis of NSCLC patients remains unsatisfactory. Considering the serious side effects of chemotherapy and radiotherapy, specific molecularly targeted agents that might benefit the treatment of cancer patients have been the focus of cancer research. Therefore, exploring the underlying molecular mechanisms that are involved in regulating cancer progression is urgent.
MicroRNAs (miRNAs) are a class of single-stranded, non-coding RNAs with the length of approximately 19-25 nucleotides [8]. miRNAs act as post-transcriptional regulators of gene expression by complementarily binding to the 3’-untranslated region (3’-UTR) of the targeted mRNAs [9,10]. The biological consequence of the miRNA-miRNA binding leads to the degradation or translation inhibition of the mRNAs [11]. Due to the critical roles of miRNAs in modulating gene expression, diverse roles of miRNAs in regulating cell proliferation, differentiation and apoptosis have been reported [12]. Notably, accumulating evidence has suggested that dysregulation of miRNAs plays important roles in the initiation and progression of cancer [13-15]. For example, miR-1258 suppressed the progression of NSCLC by targeting the GRB2/Ras/Erk pathway [16]. A recent study showed that miR-1179 inhibited the growth and invasion of NSCLC by down-regulating the expression of sperm-associated antigen 5-mediated AKT pathway [17]. miR-5582-5p was a novel tumor suppressor that inducedapoptosis and cell cycle arrest in colon cancer cells [18]. However, the function of miR-5822-5p in NSCLC has not been characterized.
Epithelial mesenchymal transition (EMT) is a critical process related to the initiation and progression of cancer. EMT is activated by many cellular signals [19-21]. Fibroblastic growth factor-10 (FGF-10) is a well-defined regulator of type I EMT [22]. Overexpression of FGF-10 has been found in a variety of cancers and is correlated with the advanced development of cancer [23]. Interrupting the FGF-10 receptor suppressed the progressive behavior of cancer cells. Thus, blocking the FGF-10 signal or down-regulating the expression of FGF-10 might inhibit the progression of cancer. As miRNAs are a class of negative regulators of gene expression, those miRNAs that target FGF-10 in cancer cells deserve investigation.
In this study, we found that miR-5582-5p was down-regulated in NSCLC tissues and cell lines. Overexpressed miR-5582-5p inhibited the proliferation and induced apoptosis of NSCLC cells. Mechanistic study revealed that miR-5582-5p targeted FGF-10 and decreased the expression of FGF-10 in NSCLC cells. Our results demonstrated a tumor suppressive role of miR-5582-5p in NSCLC by modulating the level of FGF-10.
Materials and methods
Clinical samples and ethics statement
Sixty NSCLC patients were enrolled between Feb 2014 to August 2016 at PLA Army General Hospital. The NSCLC tissues and paired adjacent normal tissues were obtained from each participant by minimal invasive surgery. All the samples were maintained at -80°C before the experiments. This study was approved by the Ethics Committee of the PLA Army General Hospital. Written informed consent was received from all the patients.
Cell culture and transfection
The normal lung epithelial cell line HBE and human NSCLC cell lines including A549, H1299, SK-MES-1 and NCI-H460 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin. Cells were maintained at 37°C with 5% CO2. The miR-5582-5p mimics and corresponding negative control miRNA were purchased from Ribobio (Guangzhou, China). The oligo transfection was performed using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Real-time quantitative PCR
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The RNA concentration was determined with the NanoDrop-2000 (Bio-Rad, Hercules, USA). Reverse transcription was performed using the Transcript First Strand cDNA Synthesis Kit (Roche, USA) following the protocol of manufacturer. For the qPCR analysis, FastSTART Universal SYBR Green Master (Roche, USA) was used in a final volume of 20 μl reaction mixtures on the ABI PRISM 7500 Real-time PCR system (Applied Biosystems, Foster City, CA, USA). The expression of U6 RNA was detected as the endogenous control for the normalization. Relative level of miR-5582-5p was analyzed according to the 2-ΔΔCT method.
Cell proliferation
The proliferation of NSCLC cells was evaluated with the Cell Counting Kit-8 (CCK-8, Dojindao, Japan). Cells transfected with the indicated miRNA were seeded in the 96-well plate with the confluence of 1,000 cells per well. After cultured for 24 h, the CCK-8 solution was added into the medium at the final concentration of 0.5 mg/mL and incubated at 37°C for 2 h. Viable cells were measured by absorbance at 450 nm with the microplate reader (Bio-Rad, Hercules, USA).
Western blot
Total protein was extracted using the NP-40 lysis buffer (Beyotime, Shanghai, China). Equal amount of protein was separated with 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto the polyvinylidene fluoride membrane (Bio-Rad, Hercules, USA). After blocking with 5% non-fat milk for 1 h at room temperature (RT), the membrane was incubated with the primary antibody against FGF-10 (Cat#PA1-25518, Thermo Fisher Scientific, MA, USA) or GAPDH (#5174, Cell Signaling Technology, MA, USA) at 4°C overnight. After washing three times with TBST, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (1:5000; Bio-Rad, Hercules, USA) for 1 h at RT. The protein bands were developed with enhanced chemiluminescence (ECL) reagent (GE Healthcare, Little Chalfont, UK) according to the manufacturer’s instructions.
Dual luciferase report assay
The full length of wild-type (WT) or mutant 3’-UTR of FGF-10 was amplified and constructed into the pMIR-Report vector (Promega Corporation, Madison, WI, USA), respectively. A549 cells were seeded in the 96-well plate. After cultured for 24 h, cells were co-transfected with the WT or mutant FGF-10 3’-UTR luciferase reporter and miR-5582-5p mimic or control miRNA. Cells were harvested after transfection for 48 h and the luciferase activity was analyzed using the Dual Luciferase Assay Kit (Promega Corporation, Madison, WI, USA) following the manufacturer’s instructions.
Cell apoptosis assay
The apoptosis of A549 cells was assessed using the FITC Annexin V Apoptosis Detection Kit (BD Biosciences, La Jolla, CA, USA) according to the manufacturer’s instructions. Briefly, cells were cultured in the 6-well plate and transfected with miR-5582-5p mimics or corresponding control miRNA. After transfection for 48 h, cells were collected and stained with the FITC and propidium (PI) reagent according to the manufacturer’s instructions. The percentage of cell apoptosis was determined using the Becton Dickinson FACScan instrument (BD PharmingenTM, La Jolla, CA, USA).
Statistical analysis
Data are presented as mean ± standard deviation (SD) from three independent experiments. Statistical analysis was performed using the GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA). Significant difference was analyzed withStudent’s t test or one-way analysis of variance (ANOVA) followed by least-significant difference post-doc test. Statistical significance was defined as P<0.05.
Results
miR-5582-5p is down-regulated in NSCLC tissues and cell lines
To explore the function of miR-5582-5p in NSCLC, RT-qPCR was performed to detect the expression of miR-5582-5p in the NSCLC tissues. As shown in Figure 1A, the level of miR-5582-5p was significantly decreased in NSCLC tissues compared with that of the adjacent normal tissues. The expression of miR-5582-5p in normal lung epithelial cell line HBE and NSCLC cell lines including A549, SK-MES-1, H1299 and NCI-H460 was also measured. The result indicated that miR-5582-5p was remarkably down-regulated in NSCLC cell lines compared to control cells (Figure 1B). These results suggested the potential important roles of miR-5582-5p in NSCLC.
Figure 1.
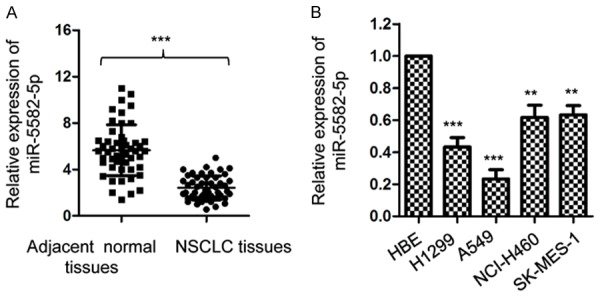
The expression of miR-5582-5p was decreased in NSCLC tissues. A. Expression level of miR-5582-5p in 60 paired NSCLC tissues and adjacent normal tissues was determined using RT-qPCR assay. ***P<0.001, Student’s t test. B. Relative expression of miR-5582-5p in NSCLC cells lines and normal cell line HBE was detected. **P<0.01, ***P<0.001 vs. control group, ANOVA followed by LSD post-hoc test.
miR-5582-5p suppressed the proliferation and induced apoptosis of NSCLC cells
As miR-5582-5p was down-regulated in NSCLC, A549 cells were transfected with miR-5582-5p mimics or control miRNA to investigate the biologic function of miR-5582-5p in regulating the growth of NSCLC cells. The transfection efficiency of miR-5582-5p was validated by RT-qPCR (Figure 2A). The proliferation of A549 cells with overexpressed miR-5582-5p or control vector was evaluated with CCK-8 analysis. As presented in Figure 2B, overexpression of miR-5582-5p significantly inhibited the proliferation of A549 cells. To further investigate the negative modulation of miR-5582-5p on the growth of NSCLC cells, the apoptosis of A549 cells with the ectopically expressed miR-5582-5p was detected. The result showed that transfection of miR-5582-5p significantly increased the apoptosis of A549 cells (Figure 2C). Consistent with these results, overexpression of miR-5582-5p inhibited the colony formation of A549 cells (Figure 2D). These results indicated the inhibitory effect of overexpressed miR-5582-5p on the growth of NSCLC cells.
Figure 2.
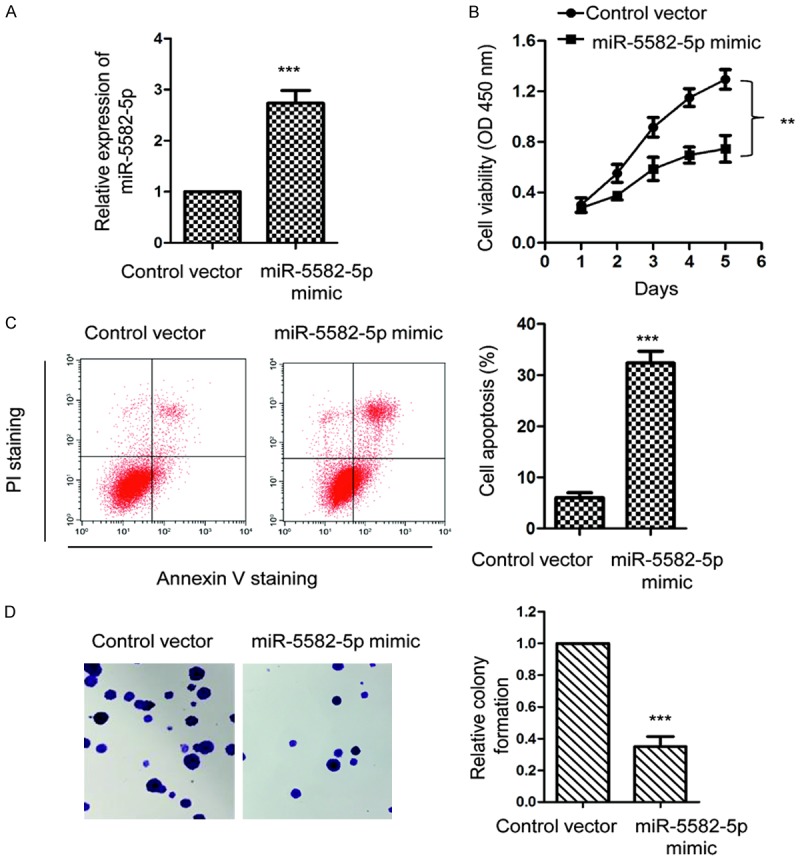
Overexpression of miR-5582-5p inhibits the growth of NSCLC cells. A. A549 cells were transfected with miR-5592-5p mimic or control vector and the expression level of miR-5582-5p was confirmed by the RT-qPCR. ***P<0.001, Student’s t test. B. Cell counting kit-8 assay was performed to analyze the regulation of miR-5582-5p overexpression on the proliferation of A549 cells. **P<0.01, vs. control group, ANOVA followed by LSD post-hoc test. C. FACS analysis was used to measure the apoptosis of A549 cells with the overexpression of miR-5582-5p. ***P<0.001, Student’s t test. D. Transfection of miR-5582-5p significantly suppressed the colony formation of A549 cells. ***P<0.001, Student’s t test.
miR-5582-5p targets FGF-10 in NSCLC cells
To explore the underlying molecular mechanism by which miR-5582-5p suppressed the proliferation of NSCLC cells, the potential targets of miR-5582-5p were predicted with the miRDB database. The output showed the potential binding sites of miR-5582-5p at the 3’-UTR of FGF-10 (Figure 3A). To further confirm this prediction, the full length wild-type or mutant 3’-UTR of FGF-10 was inserted into the luciferase reporter vector, respectively. As shown in Figure 3B, miR-5582-5p mimics markedly reduced the luciferase activity of WT but not the mutant 3’-UTR of FGF-10. This data suggested the binding between miR-5582-5p and the 3’-UTR of FGF-10. To investigate the consequence of the interaction between miR-5582-5p and the 3’-UTR of FGF-10, mRNA level of FGF-10 in A549 cells with overexpressed miR-5582-5p was evaluated by RT-qPCR. The result showed that the mRNA level of FGF-10 was significantly decreased with the transfection of miR-5582-5p in A549 cells (Figure 3C). Western blot analysis indicated the reduced the protein abundance of FGF-10 with the overexpression of miR-5582-5p (Figure 3D). Taken together, these results demonstrated that miR-5582-5p targeted FGF-10 and negatively modulated the expression of FGF-10 in NSCLC cells.
Figure 3.
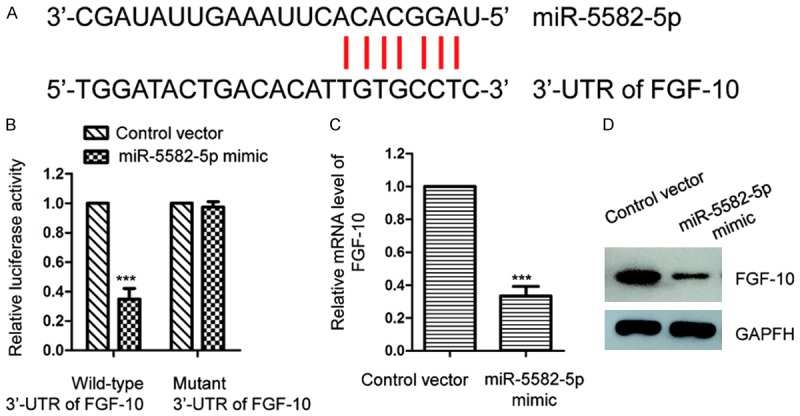
FGF-10 is a target of miR-5582-5p. A. The predicted binding sites of miR-5582-5p at the 3’-UTR region of FGF-10. B. A549 cells were transfected with the wild-type or mutant 3’-UTR of FGF-10 in the presence of miR-5582-5p mimic or control vector. Luciferase assay was performed to detect binding between miR-5582-5p to the 3’-UTR of FGF-10. ***P<0.001, Student’s t test. C. RT-qPCR analysis was carried out to check the mRNA level of FGF-10 in A549 cells with the transfection of miR-5582-5p or control vector. ***P<0.001, Student’s t test. D. The protein level of FGF-10 was detected with the overexpression of miR-5582-5p in A549 cells.
Overexpression of FGF-10 reversed the suppression effect of miR-5582-5p on the growth of NSCLC cells
To validate that the inhibitory influence of miR-5582-5p on the growth of NSCLC cells was through regulating FGF-10, A549 cells were co-transfected with Flag tagged FGF-10 and miR-5582-5p mimics or control vector. The restoration of FGF-10 was confirmed by western blot with anti-Flag antibody (Figure 4A). The CCK-8 assay showed that reintroduction of FGF-10 significantly abrogated the suppressive effect of miR-5582-5p on the proliferation of A549 cells (Figure 4B). Beyond this, the apoptotic percentage of A549 cells with the transfection of FGF-10 and miR-5582-5p mimics was also evaluated by FACS analysis. The data demonstrated that up-regulated apoptosis of A549 cells by miR-5582-5p was remarkably attenuated with the co-transfection of FGF-10 (Figure 4C). These results suggested that FGF-10 reversed the tumor suppressive function of miR-5582-5p in NSCLC cells.
Figure 4.
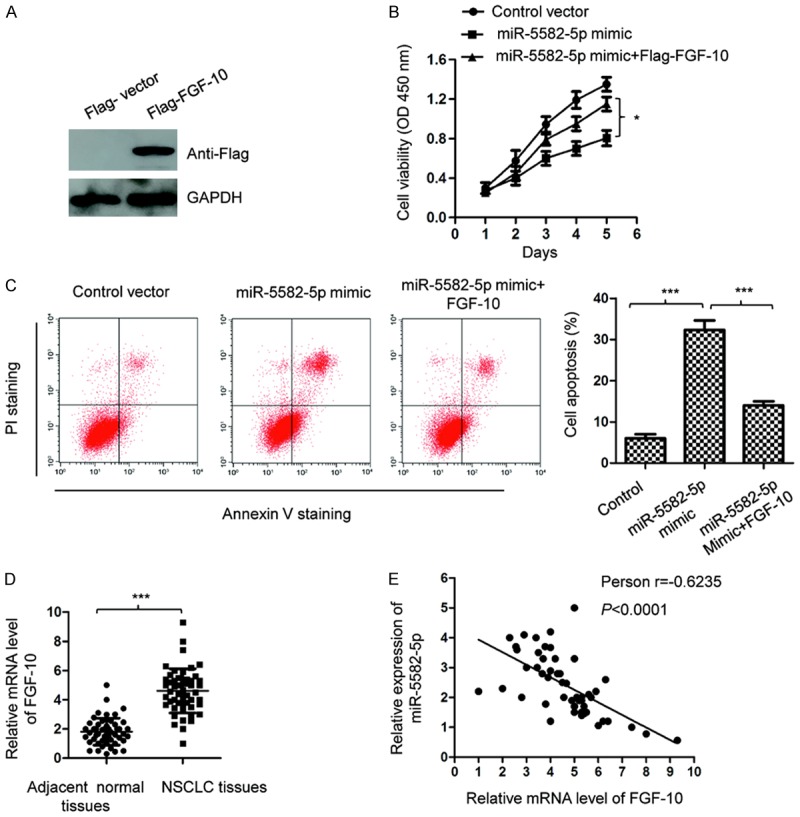
Restoration of FGF-10 reversed the inhibitory effect of miR-5582-5p on the growth of A549 cells. A. A549 cells were transfected with Flag vector or Flag-FGF-10, and the expression of FGF-10 was determined by western blot with anti-Flag antibody. B. A549 cells were co-transfected with miR-5582-5p and Flag-FGF-10. The cell proliferation was detected with the CCK-8 analysis. *P<0.05, ANOVA followed by LSD post-doc test. C. The apoptosis of A549 cells transfected with miR-5582-5p and FGF-10 was determined by the FACS assay. ***P<0.001, ANOVA followed by LSD post-doc test. D. The mRNA level of FGF-10 in NSCLC tissues and adjacent normal tissues was measured by RT-qPCR. ***P<0.001, Student’s t test. E. The expression of FGF-10 was inversely correlated with the level of miR-5582-5p in NSCLC tissues. P<0.0001, Spearman test.
To further support the negative regulatory relationship between FGF-10 and miR-5582-5p, the expression of FGF-10 in NSCLC tissues and adjacent normal tissues was detected. The mRNA abundance of FGF-10 was significantly increased in NSCLC tissues compared with that of the corresponding normal controls (Figure 4D). Spearman’s test suggested the significantly negative correlation between the expression of miR-5582-5p and FGF-10 in NSCLC tissues (Figure 4E). Those findings supported the conclusion that miR-5582-5p negatively modulated FGF-10 in NSCLC.
Discussion
Accumulating evidence has suggested critical roles of miRNAs in the development of cancer; thus, exploring the underlying molecular mechanisms by which miRNAs regulate the malignant behaviors of cancer cells might provide novel insights into the treatment. In the current study, the expression and function of miR-5582-5p in NSCLC was investigated. The results uncovered that miR-5582-5p was significantly down-regulated in NSCLC tissues and cell lines. Molecular study indicated that overexpression of miR-5582-5p inhibited the proliferation and increased the apoptosis of NSCLC cells. These results suggested the potential tumor suppressive role of miR-5582-5p in the progression of NSCLC.
miR-5582-5p was first identified as a novel tumor suppressive miRNA in colon cancer. Overexpression of miR-5582-5p led to proliferation defects and promoted the apoptosis of colon cancer cells both in vitro and in vivo [18]. Consistent with a previous report, our findings also demonstrated that miR-5582-5p inhibited the growth and trigged the apoptosis of NSCLC cells. To further confirm the tumor suppressive function of miR-5582-5p in cancers, the expression and functional mechanisms of miR-5582-5p in other types of cancers deserve further investigation. Considering the decreased level of miR-5582-5p in NSCLC tissues, it would be interesting to evaluate the correlation between the expression of miR-5582-5p with the clinical features of NSCLC patients. Additionally, given the significant inhibitory effect of miR-5582-5p on the proliferation of NSCLC cells, the potential anti-cancer function of miR-5582-5p calls for further validation by in vivo study.
To understand the molecular mechanism by which miR-5582-5p modulated the proliferation of NSCLC cells, the targets of miR-5582-5p were predicted with a bioinformatics database. FGF-10 was predicted as one of the possible binding partners of miR-5582-5p. miR-5582-5p bound the 3’-UTR of FGF-10 and decreased the expression of FGF-10 in NSCLC cells. Interestingly, a previous study demonstrated that miR-5582-5p targeted GAB1 and SHC2 to induce the apoptosis of cancer cells [18]. CDK2 was also identified as a target of miR-5582-5p and inhibited the cell cycle progression [18]. These results indicated that miR-5582-5p modulated the growth of cancer cells by regulating the expression of different targets. In the present study, FGF-10 was overexpressed in NSCLC tissues and inversely correlated with the expression of miR-5582-5p. Restoring the level of FGF-10 significantly reversed the inhibitory effect of miR-5582-5p on the proliferation of NSCLC cells. It would be of interest to see whether the regulatory axis of miR-5582-5p/FGF-10 also works in other types of cancer.
In conclusion, we found that miR-5582-5p was down-regulated in NSCLC tissues. Overexpression of miR-5582-5p significantly inhibited the proliferation of NSCLC cells by targeting FGF-10. Our results provided the possible mechanism by which miR-5582-5p modulated the malignant behaviors of NSCLC cells. These findings suggested miR-5582-5p as a novel target for anti-cancer therapy in NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Huang CY, Ju DT, Chang CF, Muralidhar Reddy P, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomedicine (Taipei) 2017;7:23. doi: 10.1051/bmdcn/2017070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moretti F, D’Antona P, Finardi E, Barbetta M, Dominioni L, Poli A, Gini E, Noonan DM, Imperatori A, Rotolo N, Cattoni M, Campomenosi P. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget. 2017;8:94980–94996. doi: 10.18632/oncotarget.21739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Revesz D, Engelhardt EG, Tamminga JJ, Schramel F, Onwuteaka-Philipsen BD, van de Garde EMW, Steyerberg EW, Jansma EP, De Vet HCW, Coupe VMH. Decision support systems for incurable non-small cell lung cancer: a systematic review. BMC Med Inform Decis Mak. 2017;17:144. doi: 10.1186/s12911-017-0542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vestergaard HH, Christensen MR, Lassen UN. A systematic review of targeted agents for non-small cell lung cancer. Acta Oncol. 2018;57:176–186. doi: 10.1080/0284186X.2017.1404634. [DOI] [PubMed] [Google Scholar]
- 5.Campbell AM, Decker RH. Mini-review of conventional and hypofractionated radiation therapy combined with immunotherapy for non-small cell lung cancer. Transl Lung Cancer Res. 2017;6:220–229. doi: 10.21037/tlcr.2017.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H, Senan S, Nossent EJ, Boldt RG, Warner A, Palma DA, Louie AV. Treatment-related toxicity in patients with early-stage non-small cell lung cancer and coexisting interstitial lung disease: a systematic review. Int J Radiat Oncol Biol Phys. 2017;98:622–631. doi: 10.1016/j.ijrobp.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Counago F, Rodriguez A, Calvo P, Luna J, Monroy JL, Taboada B, Diaz V, Rodriguez de Dios N. Targeted therapy combined with radiotherapy in non-small-cell lung cancer: a review of the oncologic group for the study of lung cancer (spanish radiation oncology society) Clin Transl Oncol. 2017;19:31–43. doi: 10.1007/s12094-016-1512-2. [DOI] [PubMed] [Google Scholar]
- 8.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 12.Gentilin E, Degli Uberti E, Zatelli MC. Strategies to use microRNAs as therapeutic targets. Best Pract Res Clin Endocrinol Metab. 2016;30:629–639. doi: 10.1016/j.beem.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–2315. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu H, Xu W, Huang Y, Yang S. Circulating miRNAs: promising biomarkers of human cancer. Asian Pac J Cancer Prev. 2011;12:1117–1125. [PubMed] [Google Scholar]
- 16.Jiang W, Wei K, Pan C, Li H, Cao J, Han X, Tang Y, Zhu S, Yuan W, He Y, Xia Y, Chen L, Chen Y. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif. 2018;51:e12502. doi: 10.1111/cpr.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song L, Dai Z, Zhang S, Zhang H, Liu C, Ma X, Liu D, Zan Y, Yin X. MicroRNA-1179 suppresses cell growth and invasion by targeting sperm-associated antigen 5-mediated akt signaling in human non-small cell lung cancer. Biochem Biophys Res Commun. 2018;504:164–170. doi: 10.1016/j.bbrc.2018.08.149. [DOI] [PubMed] [Google Scholar]
- 18.An HJ, Kwak SY, Yoo JO, Kim JS, Bae IH, Park MJ, Cho MY, Kim J, Han YH. Novel miR-5582-5p functions as a tumor suppressor by inducing apoptosis and cell cycle arrest in cancer cells through direct targeting of GAB1, SHC1, and CDK2. Biochim Biophys Acta. 2016;1862:1926–1937. doi: 10.1016/j.bbadis.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Sung WJ, Kim H, Park KK. The biological role of epithelial-mesenchymal transition in lung cancer (review) Oncol Rep. 2016;36:1199–1206. doi: 10.3892/or.2016.4964. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Huang S, Sun YL. Epithelial-mesenchymal transition in pancreatic cancer: a review. Biomed Res Int. 2017;2017:2646148. doi: 10.1155/2017/2646148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skovierova H, Okajcekova T, Strnadel J, Vidomanova E, Halasova E. Molecular regulation of epithelial-to-mesenchymal transition in tumorigenesis (review) Int J Mol Med. 2018;41:1187–1200. doi: 10.3892/ijmm.2017.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurban G, Ishiwata T, Lu YP, Fujii T, Kawahara K, Naito Z, Yamada N, Asano G. [Expression and intracytoplasmic signal transduction pathway of fibroblast growth factor (FGF)-10 in human cervical cancer cell lines] . J Nippon Med Sch. 2001;68:253–258. doi: 10.1272/jnms.68.253. [DOI] [PubMed] [Google Scholar]
- 23.Sugimoto K, Yoshida S, Mashio Y, Toyota N, Xing Y, Xu H, Fujita Y, Huang Z, Touma M, Wu Q. Role of FGF10 on tumorigenesis by MS-K. Genes Cells. 2014;19:112–125. doi: 10.1111/gtc.12118. [DOI] [PubMed] [Google Scholar]


