Abstract
Systemic lupus erythematosus (SLE) is a challenging disease caused by both genetic and environmental influences. Symptoms of SLE vary and they may come and go, therefore diagnosis and treatment of the disease is difficult. Serum metabolites can not only serve as biomarkers of the disease but also can reveal the pathogenesis. Thus, it is important to find reliable biomarkers for early diagnosis and treatment of the disease, which would greatly benefit SLE patients. Our purpose was to study the metabolite profiles in active systemic lupus erythematosus and to identify metabolites that are significantly altered. Serum samples from 34 participants (17 SLE and 17 healthy) were collected and analyzed. Untargeted lipidomics and metabolomics were used to study the metabolite profiles in serum by high-performance liquid chromatography-tandem mass spectrometry. Serum enzyme-linked immunosorbent assay was performed to validate differentially expressed metabolites. We identified differential expression of over 50 metabolites. These metabolites include several new SLE related metabolite species such as ceramide, trimethylamine N-oxide, xanthine, which were significantly elevated in the serum of active systemic lupus erythematosus patients. Some other metabolites include acylcarnitine, caffeine, hydrocortisone, itaconic acid and serotonin were down-regulated. Our study characterizes the circulating metabolites in active systemic lupus erythematosus and provides several candidate biomarkers for the diagnosis and potential therapeutic targets of the disease.
Keywords: Systemic lupus erythematosus, lipidomics, metabolomics
Introduction
Systemic lupus erythematosus (SLE) is a chronic inflammatory autoimmune disease in which healthy tissue in many parts of the body is mistakenly attacked by the immune system [1]. It can damage the joints, skin, kidneys, heart, lungs, blood vessels and brain. Most of the cases occur in young females with a nine times higher rate than men of childbearing age [1].
The cause of SLE is not clear. Genetics and environmental factors are thought to be involved [2]. Environmental triggers often act by cellular pathways containing disease-associated polymorphisms. Immunological abnormalities including T-cells, dendritic cells and B-cell hyperactivity are involved in the pathogenesis of SLE [3], causing organ damage in host tissues, but the precise cellular and molecular mechanisms leading to autoimmune disease remain poorly understood. Consequently, the sensitivity and specificity of the biomarkers used today to diagnose and to monitor disease activity are not perfect.
Diagnosis can be difficult because symptoms of SLE vary and they may come and go. Usually, a combination of symptoms and laboratory tests is used to diagnose it, which may take months or years for doctors. As a chronic inflammatory disease, early treatment of SLE can prevent irreversible organ damage and improve quality of life. Finding reliable biomarkers for early diagnosis of the disease would greatly benefit SLE patients and optimization of treatment. Lipidomic and metabolic profiling is one approach that could help find candidate biomarkers which are helpful in the diagnosis and monitoring of the disease.
In this study, we find several serum biomarkers that show high sensitivity and specificity by lipidomics and metabolomics in active SLE patients when compared with healthy controls. We identified several metabolites including ceramide, trimethylamine N-oxide, xanthine, and hydrocortisone that are changed in SLE, which has not been reported before and might contribute to the pathophysiology and serve as therapeutic target of SLE.
Materials and methods
Patients
Primary serum specimens from 32 SLE patients (17 active SLE patients, 15 inactive SLE patients) were obtained from Peking University Third Hospital (Table S1). The study was approved by the local ethics committee for clinical studies. The samples were obtained only from patients or healthy people who agreed to undergo the laboratory research. The control subjects were recruited from hospital and laboratory personnel, and were age and sex-matched with the study patients.
Sample preparation for metabolomics and lipidomics
For metabolite extraction, 100 μL serum was extracted according to polarity using liquid-liquid extraction. In brief, 400 μL chloroform: methanol (2:1) was added into the serum, followed by vortexing for 15 min at room temperature. Then the mixture was centrifuged at 13,000 rpm for 15 min. The upper aqueous phase (hydrophilic metabolites) and the lower organic phase (hydrophobic metabolites) were separately collected and evaporated under vacuum. The residue was stored at -80°C until further analysis. For LC-MS analysis, the aqueous phase was dissolved in 50 μL water. The organic phase was dissolved in 100 μL chloroform: methanol (1:1), and diluted by 300 μL isopropanol (IPA): acetonitrile (ACN): water (2:1:1). After centrifugation at 12000 rpm for 15 min, 6 μL of supernatant were injected for LC-MS/MS analysis.
High-performance liquid chromatography
Lipidomics and metabolomics were performed on an Ultimate 3000 UHPLC system coupled with Q-Exactive MS (Thermo Scientific). For the aqueous phase (metabolomics), an Xbridge amide column (100 × 4.6 mm i.d., 3.5 μm; Waters) was employed for compound separation at 30°C. The mobile phase A consisted of 5 mM ammonium acetate in water with 5% acetonitrile, and mobile phase B was acetonitrile. The flow rate was 0.5 mL/min with the following linear gradient: 0 min, 90% B; 3 min, 90% B; 15 min, 35% B, 18 min, 35% B; 19 min, 90% B and 24 min, 90% B.
For the lipidomics, chromatographic separation was performed on a reversed phase X-select CSH C18 column (4.6 mm × 100 mm, 2.5 μm, Waters, USA) at 50°C. Two solvents, containing 10 mM ammonium acetate and 0.1% formic acid, were used for gradient elution: (A) ACN/water (3:2, V/V), (B) IPA/ACN (9:1, V/V). The gradient program was: 0 min-40% B; 2 min-43% B; 2.1 min-50% B; 12 min 60% B; 12.1-75% B; 18 min-99% B; 19 min-99% B; 20 min-40% B; 25 min-40% B. The flow rate was set to 0.6 ml/min.
Mass spectrometry
Data-dependent acquisition (DDA) was performed using the Q-Exactive MS (Thermo Scientific). Acquisition was performed in positive ion mode and negative ion mode separately in profile type. Each acquisition cycle consists of 1 survey scan (MS1 scan) at 70,000 resolution from 50 to 750 m/z for the hydrophilic metabolites and mass range from m/z 200 to 1200 for the lipids, followed by 10 MS/MS scans in HCD mode at 17,500 resolution using stepped normalized collision energy (step-NCE) of 15, 30 and 45. The automatic gain control (AGC) target was set to 3e6 (maximum injection time 20 ms) and 2e5 (maximum injection time 100 ms) for the MS1 and MS/MS scan. The dynamic exclusion was set to 8 s. The parameters of ion source were: spray voltage 3.3 kV for positive ion mode and 3.0 kV for negative ion mode; Ion source sheath gas 40; aux gas 10; capillary temperature 320°C; probe heater temperature 300°C; S-lens RF level 55.
Data processing and statistical analysis
The acquired raw data were processed using MS-DIAL software according to the instructions in the software tutorial. Metabolites were identified using MS-DIAL by comparing the acquired MS/MS spectra to those in spectra library, which includes MS1 and MS/MS information of common metabolite species. The mass tolerance of identification for MS1 and MS/MS spectra were set to 0.01 Da and 0.05 Da. Datasets containing m/z values, retention time, metabolite name, and peak area were exported as an Excel file, and then the Excel file was imported into the MetaboAnalyst 3.0 Web service for multivariate analysis. Principal component analysis (PCA) with QC samples was used to determine whether there was any clustering, trend, or outlier. Further volcano plot analysis with fold-change (FC) > 2.0 and false discovery rate (FDR) < 0.05 by the Student’s t-test was used to identify significantly differential metabolites. Biomarker analysis with ROC curves were calculated to investigate whether the characteristics of the differential metabolites could be efficiently exploited as a sensitive biomarker of SLE.
ELISA analysis of serum ceramide
The serum derived from age- and gender-matched active SLE patients (n = 17), inactive SLE (n = 15) and healthy individuals (n = 32) was prepared as previously described. The ELISA kit for testing ceramide was obtained from J&L Biological (Shanghai, China). The experimental procedure follows ELISA protocol.
Results
Lipidomic profiling of features from SLE and healthy groups
We totally identified 613 and 153 nonredundant lipids in positive and negative ion mode after the filtration of the unidentified features. 15 classes of lipids were identified in positive mode, including cholesteryl ester, acylcarnitine, Ceramide (Cer) [NDS], Cer [NS], diacylglycerol (DG), HexCer [NS], lysoPC, lysophosphatidylethanolamine (lysoPE), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidyl serine (PS), EtherPC, EtherPE, sphingomyelin (SM), triglyceride (TG). 17 classes of lipids were identified in negative mode, including Cer [AP], Cer [NS], EtherPE, fatty acid (FA), branched fatty acid esters of hydroxy fatty acid (FAHFA), HexCer [NS], HexCer [AP], lysoPC, lysoPE, lysoPG, lysoPI, PC, PE, PG (phosphatidyl glycerol), PI (phosphatidyl inositol), PS (phosphatidyl serine) and SM. There were 36 lipids that were identified in both positive and negative mode and totally 730 unique identified lipids in combination. PCA with QC samples was performed to assess the experiment quality, which showed that the QC samples were clustered well in both positive and negative ion mode (Figure 1A and 1B). This indicates that our experiment met the required quality for subsequent difference analysis.
Figure 1.
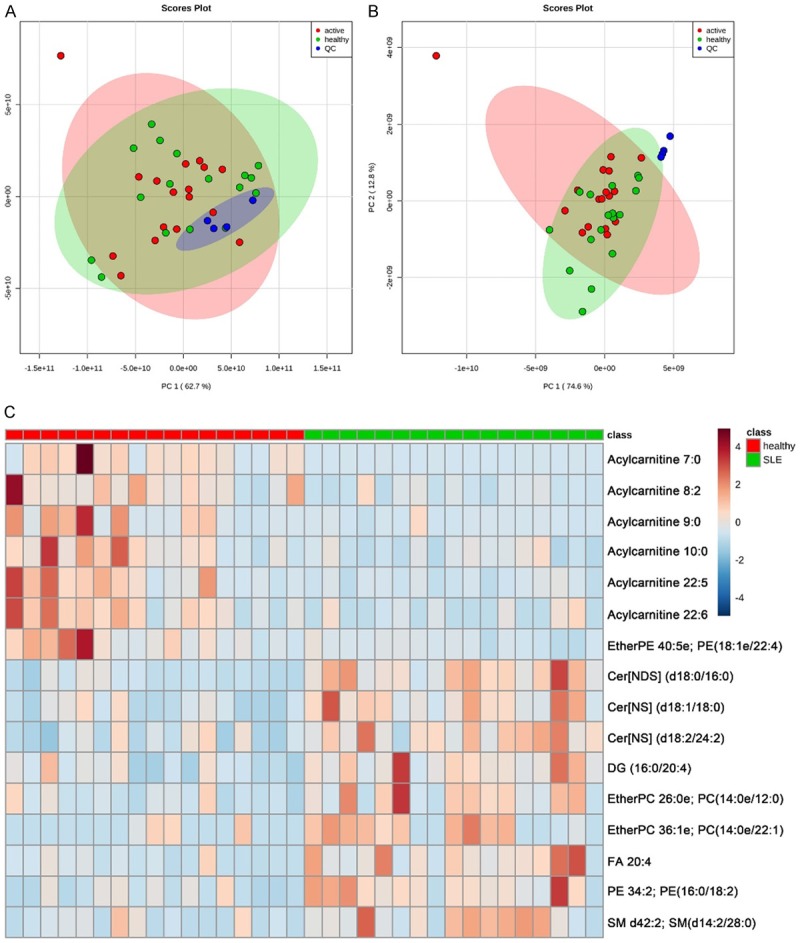
PCA score plots and heatmap of lipidomics. Overview of PCA score plots from all SLE (red), all healthy (green), and QC (blue) in positive mode (A) and negative mode (B). (C) Heatmap of the 16 significantly altered lipids in positive mode (+) and negative mode (-). The color is positively correlated with the intensity of change in metabolites, with red indicating up-regulation and green indicating down-regulation.
To find the dysregulated lipids between SLE and healthy groups, we performed volcano plot with fold change analysis and t-tests. FDR-adjusted p value of 0.05 and fold change of 2 were used as cutoffs. Finally, we identified 16 lipids that were differentially expressed between SLE and healthy people, in which 9 lipids were significantly upregulated and 7 downregulated (Table 1). A heat map was generated to show the differential expression of each lipid (Figure 1C). Among the 16 differential lipids, many are ceramides and acylcarnitines. Acylcarnitines are downregulated, while ceramides including Cer [NDS] [d18:0/16:0], Cer [NS] [d18:1/18:0] and Cer [NS] [d18:2/24:2] are significantly increased in SLE. In addition to ceramides, PE34:2, DG36:4, PE34:2, SM42:2, EtherPCs and FA 20:4 (also known as arachidonic acid, ARA) were up-regulated in SLE.
Table 1.
Differentially expressed metabolites between groups
| Compound Name | m/z | FC (SLE/healthy) | p-value | AUC |
|---|---|---|---|---|
| Lipids | ||||
| AC 7:0 | 274.2029 | 0.018102 | 0.023883 | 0.792 |
| AC 8:2 | 284.185 | 0.29371 | 0.023883 | 0.872 |
| AC 9:0 | 302.2325 | 0.23912 | 0.034984 | 0.82 |
| AC 10:0 | 316.2479 | 0.44434 | 0.02626 | 0.808 |
| AC 22:5 | 474.3582 | 0.27054 | 0.013191 | 0.803 |
| AC 22:6 | 472.3421 | 0.25971 | 0.015736 | 0.785 |
| EtherPE 40:5e; PE (18:1e/22:4) | 778.5778 | 0.3324 | 0.028642 | 0.818 |
| Cer [NDS] d34:0; Cer [NDS] (d18:0/16:0) | 540.5347 | 2.9874 | 0.000545 | 0.948 |
| Cer[NS] d36:1; Cer [NS] (d18:1/18:0) | 566.5503 | 2.0169 | 0.00626 | 0.862 |
| Cer[NS] d42:4; Cer [NS] (d18:2/24:2) | 644.5974 | 2.1661 | 0.004681 | 0.851 |
| DG 36:4; DG (16:0/20:4) | 634.5393 | 2.2814 | 0.023883 | 0.836 |
| EtherPC 26:0e; PC (14:0e/12:0) | 636.4952 | 2.1805 | 0.02626 | 0.785 |
| EtherPC 36:1e; PC (14:0e/22:1) | 774.6351 | 3.9421 | 0.02626 | 0.77 |
| FA 20:4 | 303.2345 | 2.8245 | 0.013191 | 0.869 |
| PE 34:2; PE (16:0/18:2) | 714.5118 | 2.0389 | 0.010404 | 0.841 |
| SM d42:2; SM (d14:2/28:0) | 871.7102 | 3.2379 | 0.02587 | 0.789 |
| Hydrophilic metabolites | ||||
| ADP | 426.021 | 0.012737 | 0.014476 | 0.983 |
| Caffeine | 195.0866 | 0.08511 | 0.012849 | 0.682 |
| Hydrocortisone | 363.216 | 0.12165 | 8.38E-05 | 0.913 |
| Itaconic acid | 129.0181 | 0.49143 | 0.007551 | 0.792 |
| Serotonin | 177.1033 | 0.13863 | 0.001925 | 0.917 |
| 2-Coumaric acid | 165.0535 | 2.0555 | 0.000917 | 0.837 |
| Acetylcholine | 146.1167 | 2.879 | 0.008465 | 0.761 |
| Beta-Guanidinopropionic acid | 132.0762 | 7.4822 | 4.36E-05 | 0.92 |
| D-(+)-Galacturonic acid | 193.0343 | 2.3402 | 0.02063 | 0.761 |
| Inosine | 269.0871 | 633.95 | 0.044327 | 0.941 |
| Rac-Glycerol 3-phosphoate | 170.9989 | 2.1369 | 0.020283 | 0.751 |
| S-3-Amino-4-phenylbutyric acid | 180.1058 | 617.59 | 0.000581 | 0.934 |
| S-3-Amino-5-methylhexanoic acid | 146.1168 | 3.006 | 0.007551 | 0.772 |
| Trimethylamine N-oxide | 76.076 | 2.6833 | 0.049358 | 0.692 |
| Xanthine | 151.0247 | 6.2 | 0.000318 | 0.941 |
| Arginine | 175.1184 | 2.2096 | 6.37E-06 | 0.948 |
| Asparagine | 133.06 | 3.1734 | 8.38E-05 | 0.938 |
| L-Glutamic acid | 148.0597 | 2.221 | 0.007512 | 0.837 |
| L-Glutamic acid (negative) | 146.0443 | 1.663 | 0.04 | 0.74 |
| L-Histidine | 156.0759 | 1.8883 | 3.89E-04 | 0.851 |
| L-Histidine (negative) | 154.0611 | 4.0105 | 3.00E-06 | 0.92 |
| Serine | 106.0497 | 2.9539 | 2.00E-05 | 0.903 |
| L-beta-Homoproline | 130.0859 | 615.67 | 0.000719 | 0.91 |
| L-beta-Homothreonine | 134.0804 | 135.95 | 0.000161 | 0.73 |
| L-beta-Homovaline | 132.1015 | 10.47 | 4.51E-05 | 0.919 |
| Allopurinol | 137.0452 | 7.9612 | 0.009513 | 0.893 |
| Dimefline | 324.1681 | 84.427 | 8.15E-05 | 0.905 |
| D-Sorbitol | 183.0847 | 17.663 | 0.002287 | 0.862 |
| Dulcitol | 181.0709 | 208.28 | 2.00E-05 | 0.889 |
| Flonicamid | 228.0331 | 19993 | 0.000191 | 0.964 |
| Leupeptin | 427.3008 | 3444 | 2.00E-05 | 0.894 |
| Maltitol | 343.1237 | 505.27 | 0.000191 | 0.92 |
| Mycophenolic acid | 321.1322 | 5392.5 | 0.044327 | 0.917 |
| Prednisolone | 361.1991 | 381.9 | 0.006423 | 0.779 |
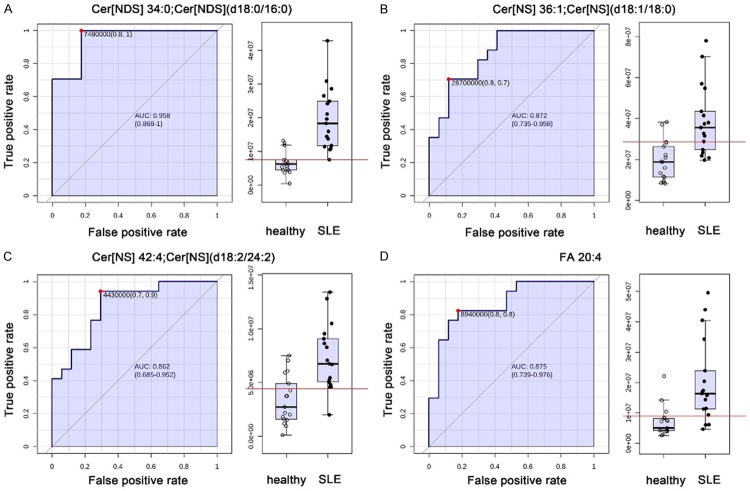
In order to find potential biomarkers to discriminate SLE from healthy people, we further performed classical univariate receiver operating characteristic (ROC) curve analyses to generate ROC curve, to calculate area under the curve (AUC) and their 95% confidence intervals. The ROC results showed that the AUC of these differential features are all above 0.75 with Cer [NDS] [d18:0/16:0] reaching up to 0.958 and FA 20:4 reaching up to 0.875 (Figure 2).
Figure 2.
ROC curves of the important altered lipids. (A) Cer[NDS] d34:0; Cer[NDS] [d18:0/16:0], (B) Cer[NS] d36:1; Cer[NS] [d18:1/18:0], (C) Cer[NS] d42:4; Cer[NS] [d18:2/24:2] and (D) FA 20:4.
Metabolomic profiling of SLE patients and healthy groups
We totally identified 110 and 83 nonredundant metabolites in positive and negative ion mode. 20 metabolites were detected in both positive and negative mode and 173 metabolites totally in combination. The QC samples were clustered well in both positive and negative ion modes in the PCA analysis (Figure 3A and 3B), which indicates that our experiment met the required quality for subsequent difference analysis.
Figure 3.
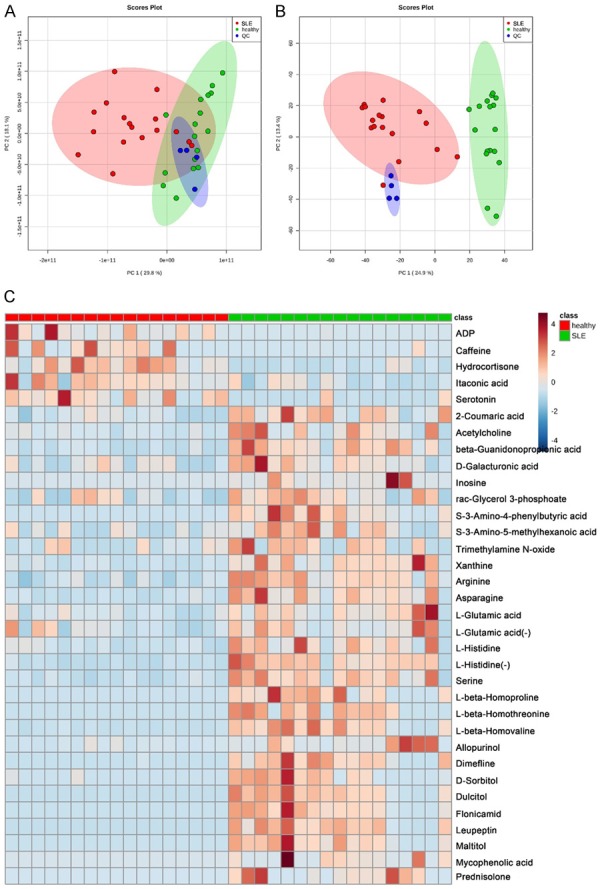
PCA score plots and heatmap of metabolomics. Overview of PCA score plots from all SLE (red), all healthy (green), and QC (blue) in positive mode (A) and negative mode (B). (C) Heatmap of the 34 significantly altered metabolites in positive mode (+) and negative mode (-). The color is positively correlated with the intensity of change in metabolites, with red indicating up-regulation and green indicating down-regulation.
The subsequent analysis was done by lipidomics. There were 34 features that were significantly changed between SLE and healthy group (Table 1). A heat map was generated to show the differential expression of each metabolite (Figure 3C). The result showed that ADP, caffeine, hydrocortisone, itaconic acid and serotonin were down-regulated, while others including several amino acids, drug metabolites, 2-coumaric acid, acetycholine, beta-guanidonopropionic acid, xanthine, inosine, galacturonic acid, rac-glycerol 3-phosphoate and trimethylamine N-oxide (TMAO) were up-regulated in the SLE group.
Classical univariate ROC curve analyses were performed to calculate AUC and their 95% confidence intervals. The AUC of arginine and asparagine were 0.958 and 0.943 respectively. The AUC of serotonin, hydrocortisone and histidine were all 0.924. The AUC of xanthine was 0.948 (Figure 4).
Figure 4.
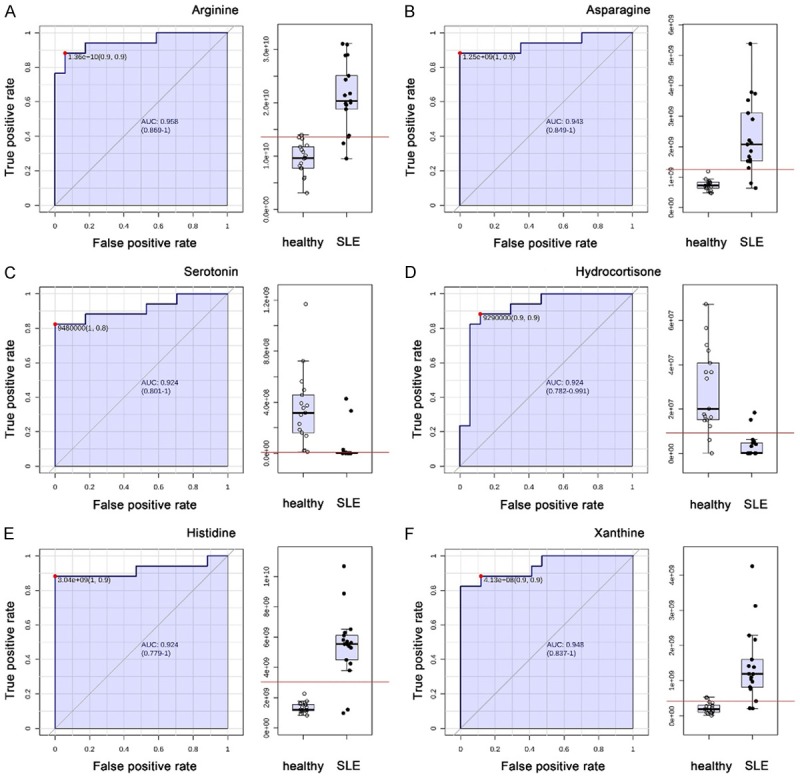
ROC curves of the important altered metabolites. A. Arginine; B. Asparagine; C. Serotonin; D. Hydrocortisone; E. Histidine; F. Xanthine.
Metabolic pathway analysis
A functional pathway analysis was performed to show the most relevant pathways that are affected as shown in Figure S1A. The most significant pathways that are dysregulated are aminoacyl-tRNA biosynthesis, sphingolipid metabolism, nitrogen metabolism, cyanoamino acid metabolism, caffeine metabolism, alanine/aspartate and glutamate metabolism, and methane metabolism.
Measurement of ceramide concentration in serum by ELISA
Several ceramides in the lipidomic profiling were elevated in SLE. To verify this and test if it has difference between active and inactive SLE, we performed an ELISA to measure its abundance in serum from active SLE (n = 17), inactive SLE (n = 15) and healthy groups (n = 32). The result showed that ceramides are dramatically increased in active and inactive SLE when compared to healthy ones, while there is no difference between active and inactive SLE (Figure S1B).
Discussion
Although previous studies have been performed to evaluate the metabolic disturbance of SLE in humans by metabolomics and different potential biomarkers were obtained [4-6], our studies revealed several significantly valuable novel biomarkers including ceramide, TMAO, xanthine and hydrocortisone. Despite this, SLE patients tends to be depressed partially because of lower a level of serotonin [7,8], which is in accordance with our results that serotonin is significantly decreased in SLE.
SLE was reported to be strongly associated with defects in apoptotic clearance, which has an important pathogenic effect [9]. Ceramide, one of the major sphingosine-based lipid second messengers, was reported to be related to oxidative stress, which is involved in apoptosis signaling [10]. The elevated level of serum ceramide may contribute to cell apoptosis, which can aggravate the disease progression [11]. Ceramide is a metabolite that come from the cleavage of sphingomyelin (SM), which is mediated by sphingomyelinase (SMase). ROS can promote the activation of SMase to release ceramide and ceramide in turn promotes the production of ROS as a positive feedback [10]. ROS is thought to play an important role in SLE [12], therefore inhibition of SMase may benefit SLE patients.
Rapid-onset cardiovascular disease (CVD) is a major side effect of many patients with SLE [13]. Traditional risk factors such as lipid alteration as well as autoimmunity are thought to contributes to accelerated atherosclerosis [13]. TMAO is a product of the oxidation of trimethylamine (TMA), a common metabolite derived from gut microbiota metabolism of choline [14]. Circulating TMAO levels are shown to be strongly associated with atherosclerosis. TMAO may contribute to atherosclerosis progression in part by increasing cholesterol accumulation within macrophages [14]. Here we show TMAO may be another risk factor that contributes to atherosclerosis in patients with SLE. However, the mechanism by which TMAO is elevated in SLE patients is unclear. A previous study shows that Flavin mono-oxygenase family members, FMO1 and FMO3 converts TMA to TMAO, in which the activity of FMO3 is much higher [15]. Serum TMAO level is significantly correlated to FMO3 expression [15]. We suspect that the expression level of FMO3 might be upregulated in SLE, which thereafter leads to the increased level of circulating TMAO. If this can be verified, the inhibition of FMO3 can be a method to alleviate atherosclerosis in SLE patients, especially in females because hepatic FMO3 expression is higher in females compared to males [15].
Our study shows xanthine increased in SLE patients. Xanthine is a product on the pathway of purine degradation by the xanthine oxidase enzyme. Xanthine oxidase catalyzes the oxidation of hypoxanthine to xanthine and further to uric acid, accompanying with the generation of reactive oxygen species [16]. An increase of xanthine may be due to the inhibition of xanthine oxidase by allopurinol since its metabolite caffeine decreases in SLE, while the inhibitor of xanthine oxidase, allopurinol, increases. Allopurinol is usually used to improve cardiovascular health by inhibition of xanthine oxidase [17].
It is interesting that hydrocortisone as an anti-inflammatory hormone decreases in SLE. This may be an influence of the use of prednisolone since it is about four times as strong in its anti-inflammatory effect and its serum level increases.
In conclusion, our untargeted lipidomics and metabolomics provide several novel biomarkers for SLE. We found that ceramide, TMAO, xanthine, and hydrocortisone were dramatically changed in SLE serum. However, the molecular mechanism how these factors increased remains to be elucidated, which may contribute to the pathophysiology and treatment of SLE.
Acknowledgements
We thank Dr. Juntuo Zhou for proof-reading the manuscript. This work was supported by the Fund for Fostering Young Scholars of Peking University Health Science Center (Grant No. BMU2018PY006), the National Natural Science Foundation of China (No. 81501390).
Written informed consent was obtained from all patients prior to sampling.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet. 2014;384:1878–1888. doi: 10.1016/S0140-6736(14)60128-8. [DOI] [PubMed] [Google Scholar]
- 2.Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, Chen L, Cunninghame Graham DS, Bentham J, Roberts AL, Chen R, Zuo X, Wang T, Wen L, Yang C, Liu L, Yang L, Li F, Huang Y, Yin X, Yang S, Rönnblom L, Fürnrohr BG, Voll RE, Schett G, Costedoat-Chalumeau N, Gaffney PM, Lau YL, Zhang X, Yang W, Cui Y, Vyse TJ. Genome-wide association meta-analysis in chinese and european individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet. 2016;48:940–946. doi: 10.1038/ng.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahren-Herlenius M, Dörner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet. 2013;382:819–831. doi: 10.1016/S0140-6736(13)60954-X. [DOI] [PubMed] [Google Scholar]
- 4.Bengtsson AA, Trygg J, Wuttge DM, Sturfelt G, Theander E, Donten M, Moritz T, Sennbro CJ, Torell F, Lood C, Surowiec I, Rännar S, Lundstedt T. Metabolic profiling of systemic lupus erythematosus and comparison with primary sjogren’s syndrome and systemic sclerosis. PLoS One. 2016;11:e0159384. doi: 10.1371/journal.pone.0159384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, Xie C, Han J, Ye Y, Weiel J, Li Q, Blanco I, Ahn C, Olsen N, Putterman C, Saxena R, Mohan C. Metabolic disturbances associated with systemic lupus erythematosus. PLoS One. 2012;7:e37210. doi: 10.1371/journal.pone.0037210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyanova S, Temu T, Cox J. The maxquant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 7.Bachen EA, Chesney MA, Criswell LA. Prevalence of mood and anxiety disorders in women with systemic lupus erythematosus. Arthritis Rheum. 2009;61:822–829. doi: 10.1002/art.24519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyerhoff J, Dorsch CA. Decreased platelet serotonin levels in systemic lupus erythematosus. Arthritis Rheum. 1981;24:1495–1500. doi: 10.1002/art.1780241207. [DOI] [PubMed] [Google Scholar]
- 9.Shao WH, Cohen PL. Disturbances of apoptotic cell clearance in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:202. doi: 10.1186/ar3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrieu-Abadie N, Gouaze V, Salvayre R, Levade T. Ceramide in apoptosis signaling relationship with oxidative stress. Free Radic Biol Med. 2001;31:717–728. doi: 10.1016/s0891-5849(01)00655-4. [DOI] [PubMed] [Google Scholar]
- 11.Tserng KY, Griffin RL. Ceramide metabolite, not intact ceramide molecule, may be responsible for cellular toxicity. Biochem J. 2004;380:715–722. doi: 10.1042/BJ20031733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat Rev Rheumatol. 2013;9:674–686. doi: 10.1038/nrrheum.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skaggs BJ, Hahn BH, McMahon M. Accelerated atherosclerosis in patients with SLE-mechanisms and management. Nat Rev Rheumatol. 2012;8:214–23. doi: 10.1038/nrrheum.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ardan T, Kovaceva J, Cejkova J. Comparative histochemical and immunohistochemical study on xanthine oxidoreductase/xanthine oxidase in mammalian corneal epithelium. Acta Histochem. 2004;106:69–75. doi: 10.1016/j.acthis.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Dierckx R, Cleland JG. Xanthine oxidase inhibition for the treatment of cardiovascular disease: a systematic review and meta-analysis. Cardiovasc Ther. 2014;32:57–58. doi: 10.1111/1755-5922.12059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.