Abstract
Background: The role of lipid metabolism played in cancer cell growth attracts more attention. SREBP1 is a common lipid regulatory factor. It has been reported that SREBP1 can promote tumor cell resistance. The aim of this study was to investigate its role in chemoresistance of colorectal cancer (CRC). Methods: The expression of SREBP1 in CRC tissues was analyzed by immunohistochemistry. Using a viability assay, the sensitivity to 5-fluorouracil in two colon cancer cell lines (HT-29 and SW620) was measured and its correlation with different expression levels of SREBP1 protein by western blot was investigated. Results: The protein expression of SREBP1 in CRC tissues was higher than that in normal colon tissues. We found that over-expression of SREBP1 through SREBP1 gene transfection enhances the resistant of CRC cell lines to 5-FU, and SREBP1 silencing through SREBP1 shRNA transfection can promote apoptosis in 5-FU treated SW620 cells. Further study indicated that SREBP1 could inhibit the expression of caspase7 and reduce PARP1 cleavage fragments. Conclusion: Our results suggest that SREBP1 protect the 5-FU treated CRC cells through caspase7 dependent PARP1 cleavage in apoptosis pathway and potentially provide a new target in the treatment of CRC.
Keywords: Colorectal cancer (CRC), sterol-regulatory element binding protein 1 (SREBP1), resistance, caspase7
Introduction
Recently, lipid metabolism alteration has been considered to be the initiating factor of tumor occurrence and progression [1,2]. SREBP1 is a major transcription factor that regulates lipid synthesis gene expression. EGFR signaling can activated SREBP1 by increasing glucose uptake, promotes N-glycosylation of SREBP cleavage-activating protein (SCAP) [3]. The expression of SREBP1 and its target gene in endometrial cancer cells was significantly increased compared with normal cells, and knockout of SREBP1 can inhibit cell growth [4]. The expression of SREBP1 is high in other cancers such as prostate cancer [5], breast cancer [6], colon cancer [7], liver cancer [8]. SREBP1 induces oxidative stress in prostate cancer by increasing the expression of ROS (reactive oxygen species) and NOX5 [9]. In colon cancer, it has been found that there is a high expression of FAS in colon cancer tissues which is related to the activation of fatty acid synthesis [10]. Later research on the role of SREBP in CRC has found that tumor cells can recognize and respond to up-regulation of SREBP1 and FAS in response to a deficiency of endogenous fatty acids, and demonstrates that SREBP1 is involved in the transcriptional regulation of lipogenic genes in CRC [11]. However, the mechanisms of SREBP1 have not yet been comprehensively identified during CRC progression.
We found SREBP1 expression in colorectal tissue was significantly increased compared with normal tissue, suggesting that SREBP1 may have a role in tumorigenesis. SREBP1 were over expressed in HT29 and silenced in SW620 cell lines. SREBP1 promoted cell proliferation, led to cells resistant to 5-FU. Further experiments showed that this proliferation and drug resistance is through the SREBP1 apoptosis signal pathway in the regulation of caspase7, while inhibiting the PARP1 cleavage, thereby inhibiting the apoptosis of colon cancer cells.
Methods
Tissues and patients
All cases were pathologically confirmed. Additionally, formalin-fixed, paraffin-embedded specimens, including primary carcinoma specimens (n=30) and corresponding non-tumor normal tissues specimens used for IHC were collected from CRC patients who underwent surgery from 2008-2010. Primary carcinomas were assessed according to the 7th edition of the American Joint Committee on Cancer (AJCC) staging system. All CRC patient data and tissue samples were collected from the Tumor Hospital of Harbin Medical University. No patients received preoperative radiotherapy or chemotherapy. The study was approved by the Ethics Committee of the Tumor Hospital of Harbin Medical University, Harbin, China.
Antibodies
Caspase-3 (8G10) Rabbit mAb, Caspase-7 (D2Q3L) Rabbit mAb, Caspase-9 (C9) Mouse mAb, PARP Antibody were purchased from Cell Signaling Technology (Danvers, MA,USA); Anti-SREBP1 antibody was purchased from Abcam (Cambridge, MA, USA); β-actin mouse mAb was purchased from (Genscript, JiangSu, China).
Cell culture
HT29 and SW620 cell lines were purchased from the American Type Culture Collection, and grown in RPMI or L15 medium supplemented with 10% fetal bovine serum in 37°C incubator with a humidified, 5% CO2 atmosphere.
Lentiviral vector construction
The SRRBP1 sequence was synthesized by Shanghai GenePharma Co. and cloned into pLVX vector. Four short hairpin RNAs (shRNAs) target sequence for SREBP1 gene were synthesized by Shanghai GenePharma Co, Ltd, to deplete the expression of SREBP1 in colon cancer cells. A scrambled shRNA was used as a negative control. shRNA oligos were cloned into the pLKO vector. Then recombinant lentivirus was generated by co-transfecting shRNA plasmids or over-expression plasmids and pHelper plasmids into HEK293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instruction.
Lentivirus infection
Viral titer of shRNA plasmids or over-expression plasmids was 2×10^9 ifu/ml which was determined by ELISA test. For cell infection, SW620 cells and HT29 cells were seeded in six-well plates and infected with the constructed lentivirus containing at a multiplicity of infection (MOI) of 60. At 24 h after infection, puromycin was added, the final concentration was 2 ug/ml, and stable transfected cell lines were screened. At 48 h after infection, the knockdown or over-expression efficiency was determined by protein level.
Immunoblot assay
In brief, cells were collected using a scraper and washed once with cold PBS. The cells were then lysed in lysis buffer (50 mM Tris-HCl, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 0.1% NP-40) supplemented with 1% protease inhibitor cocktail. Equal amounts of proteins were size-fractionated by 7.5-15% SDS-PAGE. At least three independent experiments were performed.
Evaluation of SREBP1 immunohistochemical results
Used the immunohistochemical test, and SREBP1 (ab191857, Abcam, USA) concentration was 1:300. IHC staining sections were observed by optical microscopy. Ten high power visual fields were selected for each section, and 100 tumor cells were observed in each visual field. The average positive cell proportion was calculated. The positive criterion was brown-yellow granules in nucleus or cytoplasm. The positive (+) criterion was positive cells >10%, the negative (-) criterion was positive cells <10% or no positive cells.
CCK-8 assay
5,000 cells per well were seeded into a 96-well plate 24 h before experimentation. Cells were treated with different compounds for 72 h. CCK-8 was added to the 96-well plate and incubated at 37°C for 1 h. Absorbance of each sample was read at 450 nm.
Flow cytometry
Cells were washed twice with cold PBS and then resuspended in 1× Binding Buffer at a concentration of 1×10^6 cells/ml. 100 µl of the solution (1×10^5 cells) was transferred to a 5 ml culture tube. 5 µl FITC Annexin 5 µl PI (BD) was added (BD). Gently vortex the cells and incubate for 15 min at RT (25°C) in the dark. Add 400 µl of 1× Binding Buffer to each tube. We analyzed by flow cytometry within 1 h.
Statistical analysis
Three independent experiments were performed prior to statistical analysis. The data represent the means ± S.D. P<0.05, by unpaired Student’s t-test, was considered statistically significant.
Results
SREBP1 is over-expressed in CRC
30 pairs of tumor and corresponding non-tumor normal tissues of colorectal cancer patients were collected at the Tumor Hospital of Harbin Medical University. Immunohistochemistry was used to detect the protein expression of SREBP1 in CRC tissues. We confirmed that SREBP1 expression appeared up-regulated in colorectal adenocarcinoma compared with normal colorectal tissues (Figure 1A). The positive rate of SREBP1 in tumor tissues was 86%, compared with 23.5% in normal tissues (P<0.05) (Figure 1B).
Figure 1.
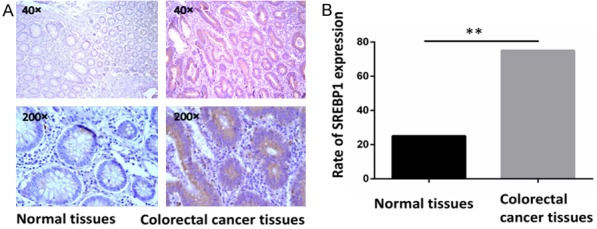
SREBP1 was highly expressed in colorectal adenocarcinoma tissues compared with normal colorectal tissues. A. SREBP1 had different expression levels in colorectal adenocarcinoma tissues and normal colorectal tissues. B. SREBP1 was expressed in different percentages. The positive rate in tumor tissues was 86%, compared with 23.5% in normal tissues (P<0.05).
Over-expression of SREBP1 enhances the resistant of colon cancer cell lines HT29 to 5-FU
In order to verify the effect of SREBP1 on CRC, we constructed a lentivirus over-expressing SREBP1 and infected HT29 cell line, which normally has low expression of SREBP1, to obtain HT29 cell line stably over-expressing SREBP1, called HT29+. In the 5-FU drug resistance experiment, we found that the resistance to 5-FU in HT29+ was significantly higher than that of control group HT29 (Figure 2A). IC50 of HT29 and HT29+ was 0.209 mg/ml vs 2.847 mg/ml, Flow cytometry analysis showed that the ratio of apoptosis cells in HT29 group was significantly higher than that in HT29+ group (Figure 2A, 2C).
Figure 2.
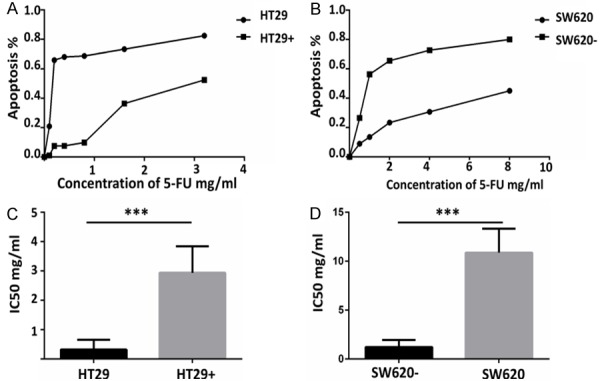
Different expression levels of SREBP1 may affect the sensitivity of colon cancer cells to 5-FU. A. SREBP1 overexpression HT29 cells are less sensitive to 5-FU than a control group by CCK-8 detection. B. SREBP1 knocked down SW620 cells by transfected with SREBP1 shRNA are more sensitive to 5-FU than a control group by CCK-8 detection. C, D. IC50 of SREBP1 over-expression HT29 cells and SREBP1 knocked down SW620 cells compared with the control group.
SREBP1 silencing can promote apoptosis of 5-FU in SW620 cells
In order to fully prove the function of SREBP1, we silenced SREBP1 by transfected SW620 cells with SREBP1 shRNA. We also used CCK-8 and flow cytometry to detect the 5-FU-induced apoptosis in SW620. The result showed that SREBP1 knockdown can promote 5-FU-induced apoptosis in SW620 cells (Figure 2B). The IC50 of SW620 and SREBP1 shRNA-treated SW620 required for 5-Fu-induced apoptosis were 10.69 mg/ml and 1.085 mg/ML respectively (Figure 2D). The flow cytometry analysis showed that the proportion of early apoptosis and late apoptosis in SREBP1 knockdown group was significantly higher than that in control group (Figure 3D-F).
Figure 3.
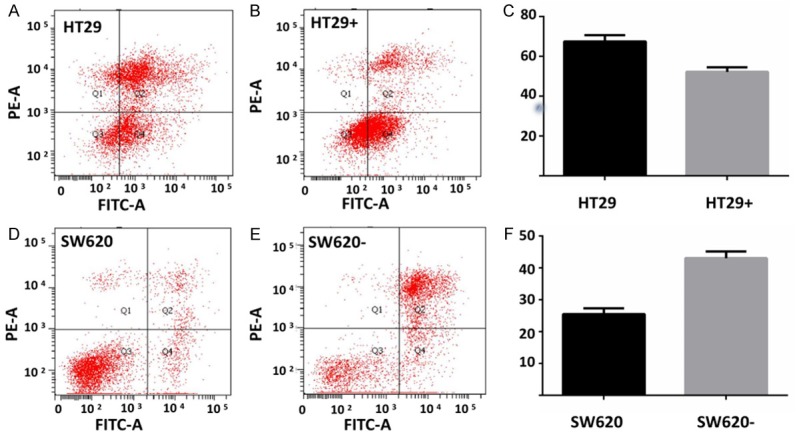
SREBP1 can promote intestinal cancer cells resistance to 5-FU. A-C. The number of apoptotic cells in SREBP1 over-expressing cells after 5-FU treatment for 72 hours was less than that in the control group. D-F. The number of apoptotic cells in SREBP1 knockdown SW620 cells after 5-FU treatment for 72 hours was less than that in the control group.
SREBP1 can reduce the expression of caspase7
We examined the expression of apoptosis related proteins in cell lines with over-expression or knockdown SREBP1, and their corresponding control groups with or without 5-FU treated. We found that the expression of caspase7 was reversely correlated with the expression of SREBP1. The expression of caspase7 was decreased in 5-FU-induced apoptosis in HT29+ and SW620 cells with highly expressed SREBP1 (Figure 4).
Figure 4.
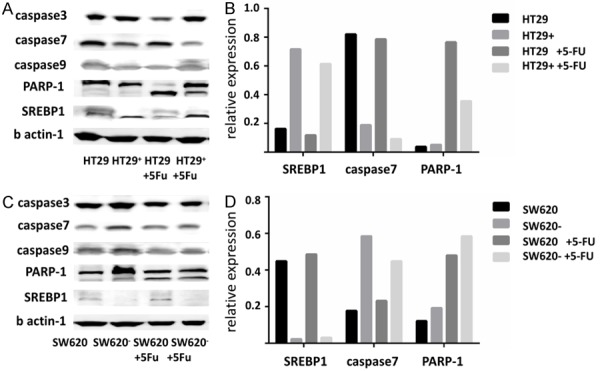
SREBP1 reduced the expression of caspase7 and inhibited the cleavage of PRAP-1. A, B. The relationship between expression of SREBP1 and the expression level of apoptosis-related proteins was tested by western blot. C, D. Relative expression of SREBP1, caspase7 and PARP-1.
SREBP1 can inhibit the cleavage of PRAP1
We detected the cleavage of PARP1 with western blot, and the cleavage of parp-1 associated with caspase7 was also reduced accordingly. PARP1, a downstream molecule of caspase7, can be cleaved by caspase7 and lost the ability to repair DNA damage, thereby promoting apoptosis (Figure 4).
Discussion and conclusion
CRC is a common malignant tumor. Tumor resistance to chemotherapeutic drugs is the main reason leading to the failure of chemotherapy [12]. The mechanism of resistance and the way to overcome resistance are a focus of research. SREBP1 is widely studied in lipid metabolism but less studied in cancer especially in CRC. According to the previous study, apoptosis of SREBP1 mainly focused on the lipid metabolism pathway [13]. SREBP1 acted as a downstream molecule, its deletion caused energy metabolism disorder and led to apoptosis [14].
In addition to FAS-related lipid metabolism associated with tumor proliferation, the previous study showed that SREBP1 knockdown can promote HeLa cell apoptosis through the caspase pathway, which was not caused by FASN, the downstream of SREBP1 [15]. Most recent studies had shown that SREBP1 mainly acted on downstream molecules, especially FAS, thereby controlling lipid metabolism in tumor cells and promoting tumor progression, and had been widely studied in lipid metabolism [7,16,17]. An early study showed that caspase3 can regulate the activity of SREBP1 [18]. However, the mechanism of the role in tumor formation, development, metastasis and drug resistance was still unclear, and tissue-specific regulation and function of SREBP1 have not yet been determined. SREBP1 has been less studied for tumor resistance.
Using clinical samples, we found that the expression of SREBP1 in colon cancer tissues was significantly higher than that in normal tissues. Further experiments found that SREBP1 over-expression can inhibit 5-Fu-induced apoptosis. By overexpression or knockdown of SREBP1, we confirmed that SREBP1 can indeed inhibit apoptosis, and inhibition of apoptosis through overexpression of SREBP1 inhibition. The down-regulated expression of caspase7 further inhibits the cleavage of PARP1, thereby inhibiting cell apoptosis. We found that SREBP1 inhibit apoptosis of CRC through SREBP1-caspase7-PARP1 axis possibly. This result indicates that SREBP1 may affect 5-FU-induced apoptosis in CRC cells through a non-lipid metabolic pathway. The next study needs to explore the mechanism of how SREBP1 regulates caspase7, and a way to reverse chemo-resistance both in vitro and in vivo.
SREBP1-caspase7-PARP1 is a novel pathway leading to drug resistance in colon cancer, which provides a new target of SREBP1 in the treatment of CRC.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (81401921). The funding bodies had no role in the design of the study or collection, analysis, and interpretation of the data; or writing the manuscript. We gratefully acknowledge Professor J. Wang and Professor B. Fang of MD Anderson Cancer Center for many valuable suggestions. The authors also gratefully appreciate Professor X.Liu for languages edition.
Harbin Tumor Hospital of Medical University is a teaching hospital. Each patient has signed an informed consent form for secondary use of medical history/biological specimen.
Disclosure of conflict of interest
None.
References
- 1.Li J, Condello S, Thomes-Pepin J, Ma X, Xia Y, Hurley TD, Matei D, Cheng JX. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell Stem Cell. 2017;20:303–314. doi: 10.1016/j.stem.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, He W, Luo M, Zhou Y, Chang G, Ren W, Wu K, Li X, Shen J, Zhao X, Hu Y. SREBP1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 2015;36:4133–4141. doi: 10.1007/s13277-015-3047-5. [DOI] [PubMed] [Google Scholar]
- 3.Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, Cheng X, Euthine V, Hu P, Guo JY, Lefai E, Kaur B, Nohturfft A, Ma J, Chakravarti A, Guo D. Glucose-mediated N-glycosylation of SCAP is essential for SREBP-1 activation and tumor growth. Cancer Cell. 2015;28:569–581. doi: 10.1016/j.ccell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W, Tai Y, Zhou J, Gu W, Bai Z, Zhou T, Zhong Z, McCue PA, Sang N, Ji JY, Kong B, Jiang J, Wang C. Repression of endometrial tumor growth by targeting SREBP1 and lipogenesis. Cell Cycle. 2012;11:2348–2358. doi: 10.4161/cc.20811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu N, Zhao J, Wang J, Teng H, Fu Y, Yuan H. Farnesoid X receptor ligand CDCA suppresses human prostate cancer cells growth by inhibiting lipid metabolism via targeting sterol response element binding protein 1. Am J Transl Res. 2016;8:5118–5124. [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey PR, Xing F, Sharma S, Watabe M, Pai SK, Iiizumi-Gairani M, Fukuda K, Hirota S, Mo YY, Watabe K. Elevated lipogenesis in epithelial stem-like cell confers survival advantage in ductal carcinoma in situ of breast cancer. Oncogene. 2013;32:5111–5122. doi: 10.1038/onc.2012.519. [DOI] [PubMed] [Google Scholar]
- 7.Wen YA, Xiong X, Zaytseva YY, Napier DL, Vallee E, Li AT, Wang C, Weiss HL, Evers BM, Gao TY, Gao T. Downregulation of SREBP inhibits tumor growth and initiation by altering cellular metabolism in colon cancer. Cell Death Dis. 2018;9:265. doi: 10.1038/s41419-018-0330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang PH, Ko YH, Chin HJ, Hsu C, Ding ST, Chen CY. The effect of feed restriction on expression of hepatic lipogenic genes in broiler chickens and the function of SREBP1. Comp Biochem Physiol B Biochem Mol Biol. 2009;153:327–331. doi: 10.1016/j.cbpb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Huang WC, Li X, Liu J, Lin J, Chung LW. Activation of androgen receptor, lipogenesis and oxidative stress converged by SREBP-1 is responsible for regulating growth and progression of prostate cancer cells. Mol Cancer Res. 2012;10:133. doi: 10.1158/1541-7786.MCR-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid A, Pizer ES, Moga M, Milgraum LZ, Zahurak M, Pasternack GR, Kuhajda FP, Hamilton SR. Elevated expression of fatty acid synthase and fatty acid synthetic activity in colorectal neoplasia. Am J Pathol. 1997;150:201. [PMC free article] [PubMed] [Google Scholar]
- 11.Li JN, Mahmoud MA, Han WF, Ripple M, Pizer ES. Sterol regulatory element-binding protein-1 participates in the regulation of fatty acid synthase expression in colorectal neoplasia. Exp Cell Res. 2000;261:159–165. doi: 10.1006/excr.2000.5054. [DOI] [PubMed] [Google Scholar]
- 12.Tsuruo T, Naito M, Tomida A, Fujita N, Mashima T, Sakamoto H, Haga N. Molecular targeting therapy of cancer: drug resistance, apoptosis and survival signal. Cancer Sci. 2010;94:15–21. doi: 10.1111/j.1349-7006.2003.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Zhang X, Liu F, Fan J, Wang B, Dong C. SREBP1-driven lipid desaturation supports clear cell renal cell carcinoma growth through regulation of NF-κB signaling. Biochem Biophys Res Commun. 2018;495:1383. doi: 10.1016/j.bbrc.2017.11.163. [DOI] [PubMed] [Google Scholar]
- 14.Lim W, Yang C, Bazer FW, Song G. Luteolin inhibits proliferation and induces apoptosis of human placental choriocarcinoma cells by blocking the PI3K/AKT pathway and regulating sterol regulatory element binding protein activity. Biol Reprod. 2016;95:82. doi: 10.1095/biolreprod.116.141556. [DOI] [PubMed] [Google Scholar]
- 15.Bengoechea-Alonso MT, Ericsson J. The phosphorylation-dependent regulation of nuclear SREBP1 during mitosis links lipid metabolism and cell growth. Cell Cycle. 2016;15:2753–2765. doi: 10.1080/15384101.2016.1220456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siqingaowa , Sekar S, Gopalakrishnan V, Taghibiglou C. Sterol regulatory element-binding protein 1 inhibitors decrease pancreatic cancer cell viability and proliferation. Biochem Biophys Res Commun. 2017;488:136–140. doi: 10.1016/j.bbrc.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 17.Galbraith L, Leung HY, Ahmad I. Lipid pathway deregulation in advanced prostate cancer. Pharmacol Res. 2018;131:177–184. doi: 10.1016/j.phrs.2018.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Dungan CM, Li J, Williamson DL. Caloric restriction normalizes obesity-induced alterations on regulators of skeletal muscle growth signaling. Lipids. 2016;51:905–912. doi: 10.1007/s11745-016-4168-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


