Abstract
The mutation of Kirsten rat sarcoma viral oncogene homolog (KRAS) has been reported to be prognostically important in patients with colorectal cancer (CRC). In this study, we investigated whether all KRAS mutations predict poor prognosis in patients with CRC. Our analysis of characteristics of KRAS mutations revealed the mutation rate for codon 12 was 72.7%, of which G12D was the highest (47.5%) followed by G12V (30.6%), and the mutation rate for codon 13 was 22.0%, of which all were G13D. In support of the concept that prognostic value of the KRAS codon-12 mutations is different from the codon-13 mutations, results from our Cox proportional hazard model studies showed that codon-12 mutations correlated with worse overall survival (OS; HR = 2.846, 95% CI: 1.967-4.118, P < 0.001) and progression free survival (PFS; HR = 2.011, 95% CI: 1.450-2.789, P < 0.001). No prognostic significance was revealed for codon-13 mutations. On further analysis, we found that mortality risk was significantly increased with G12D and G12V (G12D: HR = 2.802, 95% CI: 1.793-4.381, P < 0.001; G12V: HR = 2.802, 95% CI: 1.793-4.381, P < 0.001), as was the risk of disease progression (G12D: HR = 2.079, 95% CI: 1.396-3.099, P < 0.001; G12V: HR = 2.408, 95% CI: 1.517-3.822, P < 0.001). To conclude, our results support the concept that codon-12 mutations were predictive for a poor prognosis in Chinese patients with CRC. Specifically, G12D and G12V were independent prognostic factors for worse OS and PFS.
Keywords: Colorectal cancer, KRAS mutations, overall survival, prognosis, progression free survival
Introduction
KRAS is an important downstream molecular switch of cell-surface growth signal receptors such as epidermal growth factor receptor (EGFR), which is closely related to cell proliferation [1,2]. Several studies have shown that KRAS is involved in the regulation of different signaling pathways in the development of colorectal cancer (CRC), such as RAS/RAF/MAPK and RAS/PI3K/AKT [3]. The abnormal activation of KRAS is one of the primary reasons for the uncontrolled proliferation of tumor cells. The protein encoded by KRAS has GTPase activity that may affect cell proliferation, differentiation, and apoptosis by participating in the cellular signal transfer process [4]. However, the impact of KRAS mutations on the prognosis of patients with CRC remains controversial and the results from previous studies failed to reach a consensus [5-10]. Even patients with the same KRAS mutations who receive the same surgical treatment may experience different postoperative survival times. This indicates that the different codons of KRAS, and even different site mutations, may have diverse effects on tumor biological behavior [11-13].
Very few studies have focused on the characteristics of KRAS mutation subtypes in Chinese patients with CRC. In the current study, we detected seven common mutations in codons 12 and 13 of KRAS exon 2 in 1164 specimens from Chinese CRC patients and investigated the prognostic value of distinct codon-specific KRAS mutations and their association with clinicopathologic characteristics for evidence of potential clinical application in CRC.
Materials and methods
Study population
Based on the database of Fujian Provincial Hospital (Fuzhou, China), a total of 1164 patients with histologically confirmed CRC between June 2012 and September 2015 were identified, among which 26 cases had multiple mutation sites and were excluded from the analysis (Figure 1). Standard demographic and clinicopathologic data were collected on each patient, including gender, age, disease status, tumor characteristics, perioperative status, date of last follow-up, date of disease progression, and date of death. The cohort included 657 males and 481 females with an age range of 20-89 years (mean 62). Patients reported to have tumors with BRAF mutations, a history of other tumors in addition to CRC, severe heart or cerebrovascular disease, and those who received neoadjuvant therapy or anti-epidermal growth factor receptor agents in the perioperative period were excluded from the study. All patients with CRC had undergone surgery, including 1015 cases of primary radical resection and 123 cases of palliative surgery. Characteristics of the primary tumor, including tumor site, American Joint Committee on cancer T stage, nodal status, and metastasis status (8th) were recorded. The number, size, pathological type, and histological type of the tumors were determined from excised specimens. The largest lesion was used as the index lesion in the case of patients with multiple tumors. Data on KRAS mutational status, serum carcinoembryonic antigen (CEA) levels, serum carbohydrate antigen 19-9 (CA19-9) levels were also recorded. Patients with KRAS mutations were classified according to the specific KRAS mutation (G12D, G12V, G12C, G12S, G12A, G12R, and G13D). The Fujian Provincial Hospital institutional review board approved the study. Patient informed consent specific to this study was not required given its retrospective nature. Data from the clinical follow-up of the patients included progression free survival (PFS) and overall survival (OS) at last follow-up. Progression of disease was defined as the presence of a biopsy-confirmed tumor post-surgery with pathology showing colorectal adenocarcinoma cells or lesions considered suspicious in follow-up computed tomography imaging and elevated serum CEA levels.
Figure 1.

Flow chart of the current study. Cases with KRAS mutations in both codons 12 and 13 (N = 26) were excluded from the study to allow for the assessment of the effectiveness of KRAS codon 12 mutations as effective prognostic factors for patients with colorectal cancer, independent of KRAS codon 13 mutations.
DNA preparation and quantitative polymerase chain reaction (PCR)
Three 5-μm thick paraffin-embedded tissue sections were cut and placed in 1.5-mL EP tubes. After deparaffination with xylene, the genomic DNA was extracted using a DNA isolation kit (AmoyDx; Amoy Diagnostics Co., Ltd., Xiamen, China). A Nanodrop 2000 spectrophotometer was used to determine the concentration of DNA and the A260/A280 of the DNA samples were between 1.8 and 2.1. An AmoyDx® KRAS Mutation Detection Kit (Amoy Diagnostics Co., Ltd.) was used according to the manufacturer’s instructions to determine the KRAS status of each DNA sample. A Scorpions amplification refractory mutation system (Amoy Diagnostics Co., Ltd.) was used according to the kit instructions to detect the seven KRAS mutation sites in the codons 12 and 13 of exon 2 for the paraffin-embedded tissue. The real-time quantitative PCR amplification profile included one cycle of 42°C for 5 min and 95°C for 5 min; followed by 10 cycles of 95°C for 25 sec, 64°C for 20 sec, and 72°C for 20 sec; followed by 30 cycles of 93°C for 20 sec, 60°C for 35 sec and 72°C for 20 sec. The fluorescein amidite (FAM) and hexachloro-fluorescein (HEX) dye signals were measured at the 60°C step during the final 30 cycles of amplification. PCR analysis was performed in the Molecular Diagnostics Laboratory of Fujian Provincial Hospital.
Statistical analysis
Data were analyzed using the Chi-squared (χ2) test or Fisher’s exact test to compare proportions. The Kaplan-Meier method was performed for survival analysis and log rank test was used to compare the survival distributions. Cox’s proportional hazards regression model was used to identify the impact of factors on OS and PFS. Hazard ratio (HR) was calculated at 95% confidence interval (CI). P < 0.05 was considered statistically significant. All statistical analysis was performed using SPSS statistical software (version 22.0; IBM Corp., Armonk, NY, USA).
Results
Characteristics of KRAS mutations in colorectal cancer
The mutation rate of KRAS in CRC of the patient population in the current study was 42.1% (490/1164). Among the patients evaluated with KRAS mutations, codon 12 mutations were detected in 356 patients (72.7%, 356/490) and codon 13 mutations were detected in 108 patients (22.0%, 108/490). The six common mutation sites of codon 12 were G12D 47.5% (169/356), G12V 30.6% (109/356), G12C 8.2% (29/356), G12S 7.0% (25/356), G12A 5.6% (20/356), and G12R 1.1% (4/356). The codon 13 mutations were all G13D. The mutated form was dominated by single point mutations, which accounted for 94.7% (464/490) of the mutants and the composite site mutations accounted for only 5.3% (26/490). KRAS mutations in codons 12 and 13 were all missense mutations, but the amino acid base of each single-site mutation varied. We determined that G12S, G12D, and G13D were transitions (G > A) that accounted for 61.6% (302/490) of the changes; while G12C, G12R, G12V, and G12A were transversions (142 G > T, 20 G > C) that accounted for other 33.1% (162/490) changes (Table 1).
Table 1.
Characteristics of specific mutations in KRAS
| Mutation type | Mutation site | Base change | Frequency |
|---|---|---|---|
| Transitions | |||
| 1st position | G12S | GGT→AGT (Ser) | 5.1% (25/490) |
| 2nd position | G12D | GGT→GAT (Asp) | 34.5% (169/490) |
| G13D | GGC→GAC (Asp) | 22.0% (108/490) | |
| Transversions | |||
| 1st position | G12C | GGT→TGT (Cys) | 5.9% (29/490) |
| G12R | GGT→CGT (Arg) | 0.8% (4/490) | |
| 2nd position | G12V | GGT→GTT (Val) | 22.3% (109/490) |
| G12A | GGT→GCT (Ala) | 4.1% (20/490) | |
| Composite | 5.3% (26/490) |
Relationship between KRAS mutations and clinicopathological characteristics of colorectal cancer
The mutation rate of KRAS was different between genders, primary tumor sites, tumor histology types, and preoperative serum tumor marker levels (CEA and CA19-9). KRAS mutations were significantly more common in female CRC patients than that in male patients (44.5% vs 38.1%, P < 0.05). Compared with that of the left-sided colon, the right colon presented with a significantly greater number of KRAS mutations (47.2% vs 35.5%, P < 0.05). The mutation rate of KRAS in CRC patients with elevated serum CA19-9 levels preoperatively was higher than that in normal patients (48.9% vs 36.7%, P < 0.05). Additionally, KRAS mutations were also more frequently observed in patients with preoperatively elevated serum CEA levels compared to that in normal patients (46.6% vs 35.0%, P < 0.05).
The mutation rate of KRAS was different in diverse pathologic types of CRC. Mucinous adenocarcinoma demonstrated the highest rate (52.9%) followed by tubular adenocarcinoma (40.3%) with other types of CRC having the lowest rate (22.2%). The differences among the groups were statistically significant (P < 0.05). However, no other significant associations were identified for the remaining clinicopathologic characteristics (Table 2). Specifically, the mutation rate for codon 12 in rectal cancer (81.7%) was higher than that in right colon cancer (71.7%) and left colon cancer (71.9%), whereas the mutation rate of codon 13 in right colon cancer (28.3%) and left colon cancer (28.1%) was higher than that in rectal cancer (18.3%). The mutation rate for codon 12 was not significantly different between right colon cancer and left colon cancer (P > 0.05), as was codon 13 mutation (Table 2).
Table 2.
Relationship between KRAS mutation and clinicopathologic characteristics of colorectal cancer
| Characteristics | KRAS status | χ2 | P | KRAS mutant status | χ2 | P | ||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Wild-type | Mutations | Codon 12 | Codon 13 | |||||
| Total | 674 | 464 | 356 | 108 | ||||
| Gender | 4.768 | 0.029 | 0.494 | 0.482 | ||||
| Male | 407 | 250 | 195 | 55 | ||||
| Female | 267 | 214 | 161 | 53 | ||||
| Age (y) | 0.004 | 0.953 | ||||||
| ≥ 60 | 405 | 278 | ... | ... | ||||
| < 60 | 269 | 186 | ... | ... | ||||
| Size of tumor | 0.049 | 0.825 | ||||||
| ≥ 5 cm | 260 | 182 | ... | ... | ||||
| < 5 cm | 414 | 282 | ... | ... | ||||
| Tumor site | 8.082 | 0.018 | 6.425 | 0.040 | ||||
| Right colon | 134 | 120 | 86 | 34 | ||||
| Left colon | 207 | 114 | 82 | 32 | ||||
| Rectum | 333 | 230 | 188 | 42 | ||||
| Pathological type | 3.729 | 0.155 | ||||||
| Protrude | 227 | 182 | ... | ... | ||||
| Ulcerative | 422 | 265 | ... | ... | ||||
| Infiltrative | 25 | 17 | ... | ... | ||||
| Histological type | 12.806 | 0.002 | 0.468 | 0.791 | ||||
| Tubular adenocarcinoma | 590 | 399 | 304 | 95 | ||||
| Mucinous adenocarcinoma | 49 | 55 | 44 | 11 | ||||
| Others | 35 | 10 | 8 | 2 | ||||
| pT stage | 0.338 | 0.561 | ||||||
| pT1-2 | 99 | 74 | ... | ... | ||||
| pT3-4 | 575 | 390 | ... | ... | ||||
| pN stage | 0.109 | 0.741 | ||||||
| pN0 | 355 | 249 | ... | ... | ||||
| pN1-2 | 319 | 215 | ... | ... | ||||
| pM stage | 0.375 | 0.540 | ||||||
| pM0 | 598 | 417 | ... | ... | ||||
| pM1 | 76 | 47 | ... | ... | ||||
| Disease stage | 0.390 | 0.823 | ||||||
| I + II | 335 | 232 | ... | ... | ||||
| III | 263 | 185 | ... | ... | ||||
| IV | 76 | 47 | 0.585 | 0.444 | ||||
| CEA (ng/ml) | 15.674 | < 0.001 | 150 | 50 | ||||
| < 5 | 371 | 200 | 206 | 58 | ||||
| ≥ 5 | 303 | 264 | ||||||
| CA19-9 (U/ml) | 15.494 | < 0.001 | 1.175 | 0.278 | ||||
| < 27 | 482 | 280 | 210 | 70 | ||||
| ≥ 27 | 192 | 184 | 146 | 38 | ||||
CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9.
Overall survival
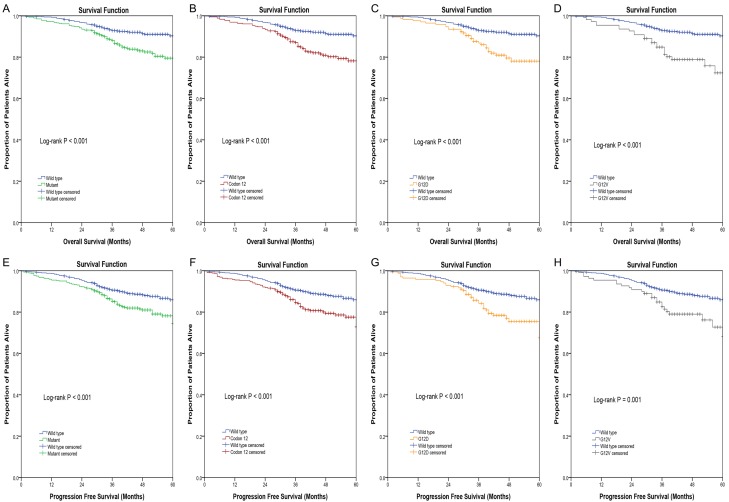
As a whole, the 5-year OS among patients with a mutated KRAS was 79.5%, compared with that of 90.3% for patients with a wild-type KRAS (P < 0.001). Patients with KRAS mutations had worse OS compared with that of patients with wild-type KRAS. Of note, the 5-year OS of patients with mutations in KRAS codon 12 and 13 were 78.2% and 83.0%, respectively, compared with 90.3% for that of patients who had a wild-type KRAS.(Figure 2A) With both log-rank Kaplan-Meier analysis (P < 0.001) and Cox regression univariate analysis (HR = 2.495, 95% CI: 1.741-3.575, P < 0.001) and multivariate analysis (HR = 2.846, 95% CI: 1.967-4.118, P < 0.001) patients with KRAS codon 12 mutations had a worse OS relative to that of patients with wild-type KRAS (Figure 2B; Table 3). In contrast, mutations in KRAS codon 13 were not associated with a worse prognosis compared with that of wild-type KRAS (P > 0.05).
Figure 2.
Kaplan-Meier curves of colorectal cancer patients according to KRAS mutation status. Overall survival in months according to (A) KRAS mutation status, (B) KRAS codon 12 mutation status, (C) KRAS G12D mutation status, (D) KRAS G12V mutation status. Progression free survival in months according to (E) KRAS mutation status, (F) KRAS codon 12 mutation status, (G) KRAS G12D mutation status, (H) KRAS G12V mutation status. Log rank test was used to compare the survival distributions. P < 0.05 was considered significant.
Table 3.
Univariate and multivariate Cox proportional hazard analysis for overall survival and progression free survival
| Prognostic Factor | Overall Survival | Progression Free Survival | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate HR (95% CI) | P | Multivariate HR (95% CI) | P | Univariate HR (95% CI) | P | Multivariate HR (95% CI) | P | |
| Gender | ||||||||
| Female | 1 (Referent) | 1 (Referent) | ||||||
| Male | 1.109 (0.785-1.566) | 0.558 | ... | 1.051 (0.770-1.434) | 0.755 | ... | ||
| Age | ||||||||
| < 60 y | 1 (Referent) | 1 (Referent) | 1 (Referent) | |||||
| ≥ 60 y | 1.636 (1.130-2.370) | 0.009 | 1.591 (1.095-2.313) | 0.015 | 1.593 (1.142-2.222) | 0.006 | 1.567 (1.122-2.188) | 0.008 |
| Tumor size | ||||||||
| < 5 cm | 1 (Referent) | 1 (Referent) | ||||||
| ≥ 5 cm | 0.892 (0.626-1.270) | 0.525 | ... | 0.848 (0.614-1.170) | 0.314 | ... | ||
| Tumor site | ||||||||
| Rectum | 1 (Referent) | 1 (Referent) | ||||||
| Right colon | 1.008 (0.641-1.586) | 0.973 | ... | 0.859 (0.564-1.308) | 0.479 | ... | ||
| Left colon | 1.378 (0.940-2.022) | 0.101 | ... | 1.275 (0.904-1.800) | 0.167 | ... | ||
| Pathological type | ||||||||
| Protrude | 1 (Referent) | 1 (Referent) | 1 (Referent) | 1 (Referent) | ||||
| Infiltrative | 3.516 (1.733-7.131) | < 0.001 | 2.704 (1.316-5.557) | 0.007 | 3.077 (1.534-6.172) | 0.002 | 2.661 (1.312-5.395) | 0.007 |
| Ulcerative | 1.747 (1.176-2.595) | 0.006 | 1.617 (1.077-2.427) | 0.020 | 1.956 (1.359-2.815) | < 0.001 | 1.804 (1.246-2.611) | 0.002 |
| Histological type | ||||||||
| Tubular adenocarcinoma | 1 (Referent) | 1 (Referent) | 1 (Referent) | |||||
| Mucinous adenocarcinoma | 0.800 (0.419-1.528) | 0.499 | 0.734 (0.382-1.413) | 0.355 | 0.773 (0.429-1.395) | 0.393 | ... | |
| Others | 2.009 (1.052-3.835) | 0.034 | 2.225 (1.141-4.342) | 0.019 | 1.787 (0.968-3.302) | 0.064 | ... | |
| T stage | ||||||||
| T1/T2 | 1 (Referent) | 1 (Referent) | ||||||
| T3/T4 | 2.387 (1.253-4.549) | 0.008 | ... | 2.251 (1.277-3.968) | 0.005 | ... | ||
| N stage | ||||||||
| N0 | 1 (Referent) | 1 (Referent) | 1 (Referent) | 1 (Referent) | ||||
| N1-2 | 2.584 (1.793-3.723) | < 0.001 | 2.089 (1.432-3.047) | < 0.001 | 2.173 (1.578-2.994) | < 0.001 | 1.822 (1.310-2.532) | < 0.001 |
| M stage | ||||||||
| M0 | 1 (Referent) | 1 (Referent) | 1 (Referent) | 1 (Referent) | ||||
| M1 | 2.565 (1.706-3.857) | < 0.001 | 2.114 (1.394-3.206) | < 0.001 | 2.283 (1.557-3.348) | < 0.001 | 1.905 (1.289-2.817) | 0.001 |
| KRAS Status | ||||||||
| Wild type | 1 (Referent) | 1 (Referent) | 1 (Referent) | 1 (Referent) | ||||
| All codon 12 mutants | 2.495 (1.741-3.575) | < 0.001 | 2.846 (1.967-4.118) | < 0.001 | 1.867 (1.349-2.584) | < 0.001 | 2.011 (1.450-2.789) | < 0.001 |
| All codon 13 mutants | 1.537 (0.839-2.817) | 0.164 | 1.776 (0.961-3.281) | 0.067 | 1.450 (0.857-2.453) | 0.166 | 1.564 (0.921-2.654) | 0.098 |
| CEA | ||||||||
| < 5 ng/ml | 1 (Referent) | 1 (Referent) | ||||||
| ≥ 5 ng/ml | 2.079 (1.452-2.976) | < 0.001 | ... | 1.795 (1.307-2.464) | < 0.001 | ... | ||
| CA19-9 | ||||||||
| < 27 U/ml | 1 (Referent) | 1 (Referent) | ||||||
| ≥ 27 U/ml | 1.341 (0.948-1.896) | 0.097 | ... | 1.247 (0.909-1.710) | 0.171 | ... | ||
CEA, carcinoembryonic antigen; CA 19-9, carbohydrate antigen 19-9; HR, hazard ratio; 95% CI, 95% confidence interval.
On further analysis of the seven most common mutations in KRAS codons 12 and 13, the G12D and G12V mutations were at the site most associated with worsened long-term prognosis. Of note, the risk of death in patients with mutations G12D (HR = 2.802, 95% CI: 1.793-4.381, P < 0.001) and G12V (HR = 3.698, 95% CI: 2.269-6.027, P < 0.001) were 2.802 and 3.698 times that of patients with a wild-type KRAS (Figure 2C and 2D; Table 3). There was no significant difference in the prognosis between patients with KRAS mutations at other sites and those with wild-type KRAS (P > 0.05).
Progression free survival
The 5-year PFS among patients with mutated KRAS was 74.5%, compared with 85.9% for that of patients with wild-type KRAS (P < 0.001). Patients with KRAS mutations had worse PFS relative to that of patients with wild-type KRAS (Figure 2E). Specifically, the 5-year PFS of patients with mutations in KRAS codons 12 and 13 were 72.8% and 79.7%, respectively, compared with 85.9% for patients who had a wild-type KRAS. According to Kaplan-Meier analysis (log-rank, P < 0.001) and both Cox regression univariate analysis (HR = 1.867, 95% CI: 1.349-2.584, P < 0.001) and multivariate analysis (HR = 2.011, 95% CI: 1.450-2.789, P < 0.001), patients with KRAS codon 12 mutations had a worse PFS compared with that of patients with wild-type KRAS (Figure 2F; Table 3). In contrast, mutations in KRAS codon 13 were not associated with progression of disease compared with that of patients with a wild-type KRAS (P > 0.05).
Further analysis of the seven most common mutations in KRAS codons 12 and 13 revealed that the G12D and G12V mutations were the site of mutations most associated with progression of disease. Specifically, the risk of disease progression in patients with mutations G12D (HR = 2.079, 95% CI: 1.396-3.099, P < 0.001) and G12V (HR = 2.408, 95% CI: 1.517-3.822, P < 0.001) were 2.079 and 2.408 times, respectively, that of patients with wild-type KRAS (Figure 2G and 2H; Table 4). There was no significant difference in the progression of disease between patients with KRAS mutations at other sites and those with wild-type KRAS (P > 0.05).
Table 4.
Univariate and multivariate Cox proportional hazard analysis for overall survival and progression free survival according to specific KRAS mutations
| KRAS | Overall Survival | Progression Free Survival | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Univariate HR (95% CI) | P | Multivariate HR (95% CI) | P | Univariate HR (95% CI) | P | Multivariate HR (95% CI) | P | |
| Wild type | 1 (Referent) | 1 (Referent) | 1 (Referent) | 1 (Referent) | ||||
| G12D | 2.566 (1.656-3.975) | < 0.001 | 2.802 (1.793-4.381) | < 0.001 | 2.039 (1.371-3.032) | < 0.001 | 2.079 (1.396-3.099) | < 0.001 |
| G12V | 3.010 (1.860-4.870) | < 0.001 | 3.698 (2.269-6.027) | < 0.001 | 2.102 (1.328-3.326) | 0.002 | 2.408 (1.517-3.822) | < 0.001 |
| G12C | 1.705 (0.617-4.710) | 0.304 | 1.780 (0.638-4.965) | 0.270 | 1.198 (0.438-3.274) | 0.725 | 1.186 (0.432-3.256) | 0.740 |
| G12S | 1.522 (0.476-4.867) | 0.479 | 1.639 (0.507-5.296) | 0.409 | 1.139 (0.359-3.610) | 0.825 | 1.371 (0.431-4.358) | 0.593 |
| G12A | 1.367 (0.333-5.609) | 0.664 | 1.834 (0.443-7.587) | 0.402 | 0.951 (0.234-3.873) | 0.944 | 1.164 (0.285-4.760) | 0.832 |
| G12R | 4.439 (0.614-32.111) | 0.140 | 6.076 (0.823-44.830) | 0.077 | 3.006 (0.418-21.629) | 0.274 | 3.985 (0.547-29.033) | 0.172 |
| G13D | 1.538 (0.839-2.818) | 0.164 | 1.774 (0.961-3.278) | 0.067 | 1.450 (0.857-2.454) | 0.166 | 1.567 (0.923-2.660) | 0.096 |
HR, hazard ratio; 95% CI, 95% confidence interval.
Discussion
The development of CRC is a complex process regulated by several genes. Abnormal signaling of pathways caused by gene mutation are involved in the process that leads to the dysregulation of intestinal epithelial cell proliferation, differentiation, and apoptosis. It has been confirmed that mutations in KRAS are key molecular events in the development of CRC with about 30%-45% of patients with CRC having a KRAS mutation [14-18]. However, there are many mutation sites in KRAS and the biological effects caused by mutations at the different sites are still controversial. To date, more than 3,000 KRAS mutation sites have been reported [19] with the most common mutations being mainly concentrated in codons 12 and 13 of exon 2. Among the 1164 Chinese patients with CRC in the current study, the total mutation rate of KRAS was 42.1%, which was consistent with most reports. All mutation forms were missense mutations with the base change mainly being G > A. Among the seven most common mutation sites in KRAS, the mutation rate of G12D was the highest (34.5%) followed by G12V (22.3%).
Different relationships between KRAS mutations, clinicopathologic characteristics, and the prognosis for CRC have been reported. Although several previous studies have demonstrated differences between the prognostic associations of KRAS mutations in codon 12 and 13, results are conflicting and none of the studies on the relationship between specific site mutations, clinicopathologic characteristics, and the prognosis of CRC have been large-scale studies with a sample size more than 300. Therefore, we evaluated specific mutations in KRAS and analyzed the relationships between the mutations and the clinicopathologic characteristics and the prognosis of CRC.
We found that the KRAS mutation rate in female Chinese CRC patients was higher than that in male patients, suggesting that the RAS signaling pathway may be affected by estrogen, which resulted in a gender-associated difference in the KRAS mutation rate. Recently, it was reported that KRAS mutations were related to the primary site of the tumor. Both the work from Li et al [20] and our research indicate that KRAS mutations are more likely to be found in the proximal colon. After further analysis, we found that the mutation rate for codon 13 in colon cancer was higher than that of rectal cancer, which was consistent with the study of Sylvester et al [21]. In addition, several previous studies have shown that mucinous adenocarcinoma has a worse prognosis than other histological types of CRC. It is also believed that the long-term survival rate of patients with elevated pre-operative serum levels of CEA and CA19-9 is lower than that of patients with normal levels. In our current study, patients with mucinous adenocarcinoma had the highest KRAS mutation rate among the different histologic types. In addition, patients with elevated pre-operative levels of serum CEA had higher KRAS mutation rates than that of patients without elevated levels. This was also the case for serum CA19-9. Both these findings suggested that KRAS may serve as a prognostic indicator.
Similar to previous studies [22-24], our current study indicated that CRC patients with KRAS mutations had a significant increased risk of postoperative tumor recurrence and death compared with that of patients with a wild-type KRAS. However, the impact of KRAS-specific codons and even specific sites within the gene on the prognosis of CRC patients is still not clear. Specifically, we found that the postoperative long-term survival rate of patients with a KRAS codon 12 mutation was significantly lower than that of patients with wild-type KRAS. In contrast, patients with a mutation in codon 13 of KRAS and patients with a wild-type KRAS showed no significant differences, which held true for recurrence as well. According to the results from cell experiments by Guerrero et al [25], codon 12 mutations increase the activation of the AKT and B-cell lymphoma 2 (bcl-2) pathways and decrease the activity of the Ras homolog gene family, member A (RhoA) pathway, thereby increasing the threshold of apoptosis. Moreover, codon 12 mutations have biological properties, such as anchorage-independent growth and cell-cell contact deregulation. These findings suggest that while they were both KRAS mutations, codon 12 and 13 mutations represent different tumor clones and cannot be generalized. KRAS codon 12 mutations conferred a more aggressive tumor phenotype compared to that of the codon 13 mutations. Thus, we may be able to determine the prognosis of CRC patients by evaluating the mutation subtype in KRAS and develop different treatment strategies according to the particular genetic status.
Notably, of the seven most common KRAS codon 12 and codon 13 mutations, mutations G12D and G12V were independent risk factors for poor prognosis and disease progression in CRC patients. This suggested that even though they were both located in codon 12, the mechanism for of the distinctive mutation sites affecting the biological behavior of tumors was different. It is well known that RAS protein expressed by KRAS has GTPase activity, but this hydrolase activity is extremely weak; thus, the stimulation of GTPase activating proteins (GAPs) is required to catalyze the process to promote the hydrolysis of GTP. Normally, the binding of RAS to GDP or GTP is in dynamic equilibrium, which allows the functional signals of RAS that regulate normal growth to be transmitted and correctly interpreted. The mutation of codon 12 may cause different degrees of damage to the balance. When the mutation is G12D, the glycine located near the catalytic site of the RAS protein is replaced with aspartic acid, which may attenuate the intrinsic GTPase activity of RAS and may also impair the stability of RAS binding with GAPs, thereby causing elevated levels of RAS-GTP in cells. Eventually, a sustained mitogenic signal for cell growth is delivered. Hunter et al [26] confirmed that the intrinsic hydrolysis rate of G12D is much smaller than that of wild-type KRAS. Even under the stimulation of GAPs, the hydrolysis rate of G12D does not obviously increase, while wild-type KRAS is able to increase by several times. Therefore, the G12D mutation not only reduces its own hydrolase activity, but also reduces its sensitivity to GAPs, thereby activating signaling pathways and promoting tumor proliferation.
The effect of the G12V mutation on the RAS signaling pathway is not the same as that of G12D. The G12V mutation is able to cause a decrease in its own GTPase activity and a decrease also in the affinity for the GTPase activating protein, thereby preventing the activation of the GTPase and causing an increase in the level of RAS-GTP in cells [27,28]. A recent study using an in vitro cell assay [29] indicated that primary tumors with the G12V mutation display significantly fewer apoptotic cells than other subtypes and the number of tumor buds was significantly higher than that of other subtypes. Furthermore, the G12V mutation is able to enhance the activity of primary tumor cells and leads to overexpression of proteins such as C-X-C chemokine receptor type 4 (CXCR4) and vascular endothelial growth factor A (VEGFA) and the abnormal activation of the AKT signaling pathway. Ultimately, the G12V mutation destroys the natural apoptosis process in tumor cells. Thus, whether it was a G12D mutation or a G12V mutation in CRC, the tumor phenotype was more aggressive than that of other subtypes. This also demonstrated that different mutations, even in a single gene, may shape distinctive biologic behaviors, which further emphasizes the importance of tumor heterogeneity in the diagnosis and treatment of cancer.
Despite these positive findings, there were several limitations to the present study. First, because it was a retrospective study, the sample population was limited, which may have introduced some selection bias. Second, although patients who received neoadjuvant therapy or anti-epidermal growth factor receptor agents were excluded from the study, the unknown remote use of those agents cannot be absolutely excluded. Finally, we did not investigate the less common mutations in KRAS codons 61 and 146; these should be evaluated in future studies.
In conclusion, among the seven most common mutations in KRAS, G12D, G12V, and G13D mutations were the most prevalent. Not all mutations of KRAS predict poor prognosis in patients with CRC. Only G12D and G12V mutations in codon 12 of KRAS were independent prognostic factors of worse OS and PFS for CRC patients. The determination of specific mutations in KRAS may help clinicians develop personalized treatment plans and follow-up strategies for patients with CRC and may even provide a reference for molecular typing of CRC.
Acknowledgements
The authors thank all study participants of the Department of Pathology for their contributions to this project. The authors also thank Amoy Diagnostics Co., Ltd. for providing KRAS Mutation Detection Kit and the corresponding primers (patent no. CN200910111501.6). The authors would also like to thank Editage [www.editage.cn] for English language editing. This work was supported by Foundation of Natural and Science of Fujian Province, China (General program No. 2015J01422), Foundation for Medical Innovation of Fujian Province (2017-CX-2), and Foundation for Youth of Fujian Province (General program No. 2018J05125).
Disclosure of conflict of interest
None.
References
- 1.Khosravi-Far R, Der CJ. The ras signal transduction pathway. Cancer Metastasis Rev. 1994;13:67–89. doi: 10.1007/BF00690419. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB, Salisbury JL, Mazzarino MC, Stivala F, Libra M. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249–279. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Downward J. Ras signalling and apoptosis. Curr Opin Genet Dev. 1998;8:49–54. doi: 10.1016/s0959-437x(98)80061-0. [DOI] [PubMed] [Google Scholar]
- 5.Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367–380. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 6.Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 7.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J. Clin. Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 8.Samowitz WS, Curtin K, Wolff RK, Tripp SR, Caan BJ, Slattery ML. Microsatellite instability and survival in rectal cancer. Cancer Causes Control. 2009;20:1763–1768. doi: 10.1007/s10552-009-9410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J, Chapusot C, Piard F. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 10.Ogino S, Meyerhardt JA, Irahara N, Niedzwiecki D, Hollis D, Saltz LB, Mayer RJ, Schaefer P, Whittom R, Hantel A, Benson AB 3rd, Goldberg RM, Bertagnolli MM, Fuchs CS Cancer and Leukemia Group B; North Central Cancer Treatment Group; Canadian Cancer Society Research Institute; Southwest Oncology Group. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Ye Y, Sun H, Shi G. Association between KRAS codon 13 mutations and clinical response to anti-EGFR treatment in patients with metastatic colorectal cancer: results from a meta-analysis. Cancer Chemother Pharmacol. 2013;71:265–272. doi: 10.1007/s00280-012-2005-9. [DOI] [PubMed] [Google Scholar]
- 12.Peeters M, Douillard JY, Van Cutsem E, Siena S, Zhang K, Williams R, Wiezorek J. Mutant KRAS codon 12 and 13 alleles in patients with metastatic colorectal cancer: assessment as prognostic and predictive biomarkers of response to panitumumab. J. Clin. Oncol. 2013;31:759–765. doi: 10.1200/JCO.2012.45.1492. [DOI] [PubMed] [Google Scholar]
- 13.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J. Clin. Oncol. 2012;30:3570–3577. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 14.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruine AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in the netherlands cohort study. Carcinogenesis. 2003;24:703–710. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 15.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 16.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 17.Sundstrom M, Edlund K, Lindell M, Glimelius B, Birgisson H, Micke P, Botling J. Kras analysis in colorectal carcinoma: analytical aspects of pyrosequencing and allele-specific PCR in clinical practice. BMC Cancer. 2010;10:660. doi: 10.1186/1471-2407-10-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao J, Li YY, Sun PN, Shen L. Comparative analysis of dideoxy sequencing, the KRAS StripAssay and pyrosequencing for detection of KRAS mutation. World J Gastroenterol. 2010;16:4858–4864. doi: 10.3748/wjg.v16.i38.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lea IA, Jackson MA, Li X, Bailey S, Peddada SD, Dunnick JK. Genetic pathways and mutation profiles of human cancers: site- and exposure-specific patterns. Carcinogenesis. 2007;28:1851–1858. doi: 10.1093/carcin/bgm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y, Shan L, Ying J, Lu N. Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer. 2015;15:340. doi: 10.1186/s12885-015-1345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sylvester BE, Huo D, Khramtsov A, Zhang J, Smalling RV, Olugbile S, Polite BN, Olopade OI. Molecular analysis of colorectal tumors within a diverse patient cohort at a single institution. Clin Cancer Res. 2012;18:350–359. doi: 10.1158/1078-0432.CCR-11-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 23.Conlin A, Smith G, Carey FA, Wolf CR, Steele RJ. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut. 2005;54:1283–1286. doi: 10.1136/gut.2005.066514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadowaki S, Kakuta M, Takahashi S, Takahashi A, Arai Y, Nishimura Y, Yatsuoka T, Ooki A, Yamaguchi K, Matsuo K, Muro K, Akagi K. Prognostic value of KRAS and BRAF mutations in curatively resected colorectal cancer. World J Gastroenterol. 2015;21:1275–1283. doi: 10.3748/wjg.v21.i4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60:6750–6756. [PubMed] [Google Scholar]
- 26.Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 27.Al-Mulla F, Milner-White EJ, Going JJ, Birnie GD. Structural differences between valine-12 and aspartate-12 ras proteins may modify carcinoma aggression. J Pathol. 1999;187:433–438. doi: 10.1002/(SICI)1096-9896(199903)187:4<433::AID-PATH273>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Bollag G, McCormick F. Intrinsic and GTpase-activating protein-stimulated ras GTPase assays. Methods Enzymol. 1995;255:161–170. doi: 10.1016/s0076-6879(95)55020-8. [DOI] [PubMed] [Google Scholar]
- 29.Alamo P, Gallardo A, Di Nicolantonio F, Pavon MA, Casanova I, Trias M, Mangues MA, Lopez-Pousa A, Villaverde A, Vazquez E, Bardelli A, Cespedes MV, Mangues R. Higher metastatic efficiency of KRas G12V than KRas G13D in a colorectal cancer model. FASEB J. 2015;29:464–476. doi: 10.1096/fj.14-262303. [DOI] [PubMed] [Google Scholar]