Abstract
Diabetic nephropathy (DN) is one of the most important microvascular diseases in diabetic patients and has been the first cause of end stage renal disease (ESRD). In this study, we are aims to investigate the genetic mechanisms of lncRNA in the regulation of DN renal fibrosis. First, we have found that the expression of lncRNA TUG1 in db/db DN mice kidney tissue and high glucose-stimulated NRK-52E cells were down-regulated and the overexpression of lncRNA TUG1 could inhibit cell fibrosis of high glucose-stimulated of NRK-52E. Second, online software program Starbase predicts that miR-21 is a target gene of lncRNA TUG1 and TIMP3 is the target gene of miR-21, which have been verified by luciferase reporter assay and RNA Binding Protein Immunoprecipitation (RIP). Last, the renal fibrosis in DN mice and cell fibrosis in high glucose-stimulated NRK-52E cells were also evaluated. We have proven that overexpression of lncRNA TUG1 can promote the expression of TIMP3 through targeting the miR-21, thereby inhibiting cell fibrosis in high glucose-stimulated NRK-52E cells and renal fibrosis in DN mice. Our results indicated that lncRNA TUG1 could indirectly regulated the expression of TIMP3 by targeting miR-21. LncRNA TUG1 inhibited high glucose-stimulated NRK-52E cell fibrosis and renal fibrosis in DN mice, which provides a theoretical basis for the treatment of DN fibrosis.
Keywords: Diabetic nephropathy, end stage renal disease, TUG1, miR-21, TIMP3
Introduction
Diabetic nephropahy (DN) is the most common and refractory microvascular complication of diabetes, and a leading cause of end-stage renal disease (ESRD) in worldwide, of which growing incidence and mortality have caused a heavy health and economic burden on society [1]. The main pathological features of diabetic nephropathy are glomerulosclerosis, deposition of extracellular matrix (ECM) and fibrosis in the tubule interstitium [2,3]. Although much effort was given explore the pathogenesis of DN, there are still many patients from DN entering ESRD, suggesting that there are still some unknown factors and mechanisms that regulate early DN events [4].
Long non-coding RNA (lncRNA) is defined as a class of non-coding RNAs over 200 nucleotides [5]. Taurine upregulated gene 1 (TUG1), a lncRNA is originally identified as contributing to the formation of photoreceptors and plays a key role in retinal development [6]. In addition, TUG1 is required for the regulation of several tumor carcinogenesis, such as osteosarcoma and melanoma [7]. In recent years, lncRNA has gained significant attention in DN, and previous studies have found that lncRNA TUG1 played an important role in DN progress [8]. Studies have revealed that lncRNA TUG1 could regulates mitochondrial bioenergetics, and alleviated ECM accumulation in DN by modulating miR-377 targeting peroxisome proliferator-activated receptor γ (PPARγ) [9,10]. However, the role of lncRNA TUG1 in renal fibrosis in DN remains unclear.
MicroRNA (miRNA) is a non-coding single-stranded RNA approximately 22 nt that binds to the 3’ untranslated region (3’UTR) of the target gene and silences the target gene to prevent its translation into protein [11]. Many studies have confirmed that miRNAs participated in the pathogenesis of DN through regulatory signaling pathways [12-14]. More than 20 miRNAs were identified involved in the molecular pathogenesis of DN [15]. Several key miRNAs, including miR-192, miR-200b, miR-200c, miR-216a, and miR-217, were found upregulated in renal tissue of DN mice [16,17]. For example, miR-192 targeted the ZEB1/2 gene and activated the TGF-β1 signaling pathway, leading to increased transcription of collagen type α 2 (Col1α2) and elevated urinary albumin levels which participated in renal fibrosis [18]. In addition, previous studies showed that miR-21 activated Akt kinase signaling pathway by targeting PTEN gene, which leaded to abnormal increase of renal fibrosis protein Col1α2, fibronectin and glomerular hypertrophy [19].
A recent study has found that high glucose or DN could significantly reduce the matrix degradation ability of membrane cells, and ECM conversion was regulated by the activity of metalloproteinases (MMPs) and tissue inhibitor of metalloproteinase (TIMPs) [20]. Among them, MMPs are the main enzymes that degrade cell matrix (ECM), and TIMPs are specific inhibitors of MMPs [21,22]. Four TIMPs (TIMP-1, TIMP-2, TIMP-3, TIMP-4) have been found to have different inhibition activity on MMPs [23]. TIMPS are non-covalently bonded with MMPs at a ratio of 1:1 to block MMPs binding with its substrates. Studies have shown that TIMP3 was the most highly expressed TIMP protein in the kidney and associated with inflammation, renal fibrosis and tubule interstitial damage in mice [24]. In DN mice, the expression of TIMP3 was greatly reduced by interaction with FoxO1/STAT1 [25]. Recent studies have shown that TIMP3 deficiency is a marker for DN in mice and humans, and the absence of TIMP3 selectively exacerbated DN [26,27]. TIMP3 expression was regulated by many miRNAs, such as miR-21, miR-181b, miR-182, miR-216 and miR-221/2 [27]. Especially studies have found that miR-21 could affect TIMP3 expression in DN mice, but the detailed mechanism was still unknown.
It was found that miR-21-5p was a target gene of lncRNA TUG1 by Starbase prediction, miR-21 promoted renal fibrosis in DN by targeting PTEN and SMAD7 [28]. It was predicted that miR-21 can target TIMP3, and the loss of TIMP3 can promote DN and renal fibrosis [27]. However, the relationship between lncRNA TUG1 and miR-21, miR-21 and TIMP3 has not been studied.
In our study, we suggested that the expression of lncRNA TUG1 was decreased in DN mice kidney tissue and high glucose-stimulated NRK-52E cells, and lncRNA TUG1 could regulate the expression of TIMP3 by targeting miR-21. Moreover, the overexpression of LncRNA TUG1 could inhibit cell fibrosis in high glucose-stimulated NRK-52E cells and renal fibrosis in DN mice, which provides a theoretical basis for the treatment of DN fibrosis.
Materials and methods
Animal model
6-8 weeks old SPF diabetic model mice (strain C57BL/KsJ-db/db mice) and control normal mice (db/m mice) purchased from Nanjing University model animals graduate School. All the animal procedures were conducted in accordance with the “Guide for the Care and Use of Laboratory Animals” published by the National Institutes of Health. During the experiment, the mice were freely eaten standard feed and drink water under constant temperature conditions, and the lights alternated for 12 hours.
Cell culture
Rat proximal tubular epithelial cells (NRK-52E) strain was purchased from the American ATCC (American Type Culture Collection) cell bank. NRK-52E was cultured in normal glucose DMEM/F12 complete medium. The DMEM medium contained 25 mm glucose and 5.5 mm glucose to simulate the high glucose status and normal physiological environment.
Protein extraction
Total cellular protein extraction: the cells were washed twice with pre-cooled PBS, then the cells were collected and putted into a 1.5 ml EP tube, centrifuged at 4°C, 800 rpm for 5 min, and the precipitate was retained. Add 100 μl of protein lysate (added protease inhibitor cocktail II), lyse on ice for 30 min, shake every 5 min, centrifuge at 4°C, 14,000 rpm for 20 min. The supernatant was collected in a new EP tube. Total protein extraction from kidney tissue: 0.1 g of kidney tissue samples were cut and washed twice with pre-cooled PBS, and tissue pellets were collected by centrifugation each time (whole process was carried out on ice). Add 500 μl of pre-cooled protein lysate, then transfer to an autoclaved tissue grinder, and fully grind the homogenate in an ice bath; collected the lysate and placed into an EP tube, placed on ice for 1 h, shaken every 10 min; centrifuged at 14,000 rpm, 4°C for 30 min, and the supernatant liquid was collected in a new EP tube.
Western blot
Protein were separated by a sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and transferred onto a nitrocellulose filter membrane (GE, USA). The membrane was blocked with 5% skimmed milk in Tris-buffered saline, and then incubated at 37°C with primary antibodies against anti-TIMP3, α-SMA, TGF-β1 (1:500, Cell Signaling, MA), anti-P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I (1:200, Abcam, USA). Subsequently, Goat anti-rabbit secondary antibody (1:3000, Biorad, USA) labeled with horseradish peroxidase (HRP) were added into the blots incubated at 37°C for 1 h. The protein signals were exposed with a storage phosphor screen. The ratio of the band of the target protein to the β-actin band of the internal reference protein were defined the quantification of protein expression. All the experiments were performed in triplicate.
Masson staining
First, dewaxed the paraffin sections into water, and placed into the solution with equal volume of hematoxylin dye solution and aqueous solution of ferric chloride, stained for 10 minutes. Rinse the excess stain on the section and place it in 1% hydrochloric acid for color separation until the chromatin and nucleus are clearly visible for about 20 sec. The sections were washed with water for 5 min, and placed in an aqueous ammonia solution for 10 sec to make the nuclei blue. The sections were washed with water for 1 min, and stained in ponceau red acid magenta staining solution for 10 min, then immersed in an aqueous acetic acid solution for 1 min, immersed in phosphomolybdic acid aqueous solution for 1 min, immersed in an aqueous acetic acid solution for 1 min. then stained with aniline blue staining solution for 1 min, and then immersed in an aqueous acetic acid solution for 1 min. Finally, sections were dehydrated with absolute ethanol, transparent with xylene, and sealed with a neutral gel.
Target prediction and luciferase assay
We successfully predict binding sites between lncRNA TUG1 and miR-21, miR-21 and 3’UTR of TIMP3 through the methods of starbase (http://starbase.sysu.edu.cn), with TargetScan (http://www.targetscan.org) website to investigate the potential targets of miR-21. The LncRNA TUG1 and 3’UTR of TIMP3 sequence as well as the mutant which interacts with miR-21 was amplified by PCR and inserted into the pRL-TK vector (Promega, USA). For dual luciferase assay, each plasmid together with miR21 mimic or a negative control mimic was co-transfected into cells. After 48 hours, the luciferase activity was detected using a dual luciferase assay system kit. Co-transfection with pRL-TK expressing Renilla luciferase was used as an internal control.
RNA extraction and qRT-PCR
We added l ml Trizol Reagent to each dish and gently shake to bring it into contact with the bottom of the bottle for 5 min to ensure adequate lysis, and transfer to a 1.5 ml Eppendorf tube. We then add 0.2 ml of chloroform, mix well and shake vigorously for 30 sec, and put at room temperature for 5 min, centrifuged 12000 rpm at 4°C for 5 min. We carefully transferred the upper aqueous phase to a new Eppendorf tube and added 0.5 ml isopropanol, mixed and put it for 10 min at room temperature, then centrifuged at 12000 rpm for 4 min at 4°C. We discarded the supernatant, added 75% alcohol to wash the RNA pellet, the centrifuged at 4°C 7500 rpm for 5 min. The supernatant was discarded and the RNA pellet was air-dried for about 10 min, dissolved in deionized water treated with DEPC. Reverse-transcribed into complementary DNA (cDNA) using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). Quantitative real-time polymerase chain reaction was performed Using an ABI PRISM 7700 System and TaqMan reagents (Applied Biosystems). Each reaction was performed in triplicate using standard Reaction conditions and U6 was used as an internal reference gene, Sequences of the Applied Biosystems primers were as follows. miR-21: 5’-GCCACCACACCAGCTAATTT-3’ (sense) 5’-CTGAAGTCGCCATGCAGATA-3’ (antisense). LncRNA TUG1 primer: 5’-GAACTACTGCGGAACCTCAA-3’ (sense), Reverse: 5’-ACTTGGTGAGCACCACTCC-3’, siTUG1: 5’-GGGAUAUAGC CAGAGAACAAUUCUA-3’ (sense), 5’-UAGAAUUGUUCUCUGGCUAUAUCCC-3’ (antisense).
RNA immunoprecipitation (RIP)
RNA immunoprecipitation assays were performed using ChIP-IT (Active Motif, Carlsbad, CA, USA), according to the Active Motif protocol. AntiAGO2 antibodies and IgG (Cell Signaling Technology, Danvers, MA, USA) were used in this study.
Statistical analysis
Results of the experimental studies are mean ± SD, as statistical analysis was performed using Student’s t test on Graph Pad Prism 5. Values of P\0.05 were considered statistically different. All experiments were performed at least three independent times.
Results
LncRNA TUG1 is down-regulated in db/db DN mice kidney tissue and high glucose-stimulated NRK-52E cells
To test the expression level of lncRNA TUG1 in kidney tissue of db/db DN mice, we constructed C57BL/KsJ-db/db mice (10 mice) and their littermate db/m mice (10 mice). First, we detected the extend of renal fibrosis between them by Masson staining, results suggested that the degree of renal fibrosis was significantly enhanced in db/db mice (Figure 1A). Subsequently, we further detected the mRNA expression level of lncRNA TUG1, the results of qRT-PCR showed that the mRNA expression level of lncRNA TUG1 was significantly down-regulated in db/db mice (Figure 1B).
Figure 1.
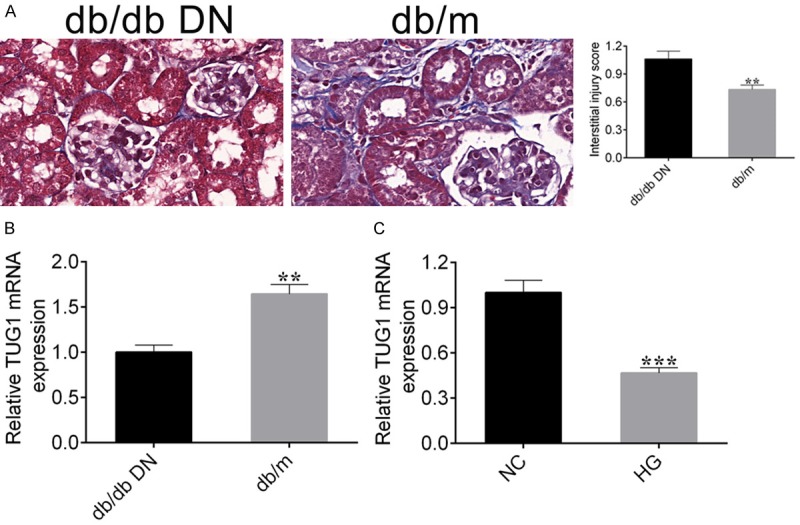
LncRNA TUG1 is down-regulated in db/db DN mice kidney tissue and high glucose-stimulated NRK-52E cells. A. Masson staining was used to detect the degree of renal fibrosis between db/db DN and db/m mice (left panel), the extent of fibrosis is quantified by Image Pro Plus, Student’s t-test: ***P < 0.001 (right panel). B. qRT-PCR detected the expression level of lncRNA TUG1 mRNA in db/db DN and db/m mice. C. qRT-PCR was also carried out to detect the expression level of lncRNA TUG1 in high glucose-stimulated NRK-52E cells. Student’s t-test: ***P < 0.001.
Next, we constructed a high glucose-induced NRK-52E cell and further detected the mRNA expression level of lncRNA TUG1 by qRT-PCR. The results showed that compared with the control mice NRK-52E-NC, the expression of lncRNA TUG1 mRNA was inhibited in NRK-52E-HG mice (Figure 1C). These results indicated that lncRNA TUG1 expression levels were down-regulated in both db/db DN mice kidney tissue and high glucose-stimulated NRK-52E cells.
Overexpression of lncRNA TUG1 inhibits high glucose-stimulated of NRK-52E cell fibrosis
To explore the effect of lncRNA TUG1 on cell fibrosis, we overexpressed lncRNA TUG1 in high glucose-suppressed NRK-52E cells and down-regulated the expression of lncRNA TUG1 by small RNA interference strategy. First, we detected the expression level of lncRNA TUG1 by qRT-PCR. Results showed that the expression level of lncRNA TUG1 was significantly up-regulated in cells overexpressing lncRNA TUG1, while the expression level of lncRNA TUG1 was down-regulated in siTUG1 cells (Figure 2A). Subsequently, we evaluated the expression level of the cell fibrosis maker gene by western blot. The results showed that in the cells overexpressing lncRNA TUG1, the fibrosis maker genes TGF-β1, α-SMA, P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I were significantly decreased, while in siTUG1 cells, these maker genes were significantly up-regulated (Figure 2B). These results indicated that lncRNA TUG1 inhibited cell fibrosis of high glucose-stimulated of NRK-52E cells.
Figure 2.
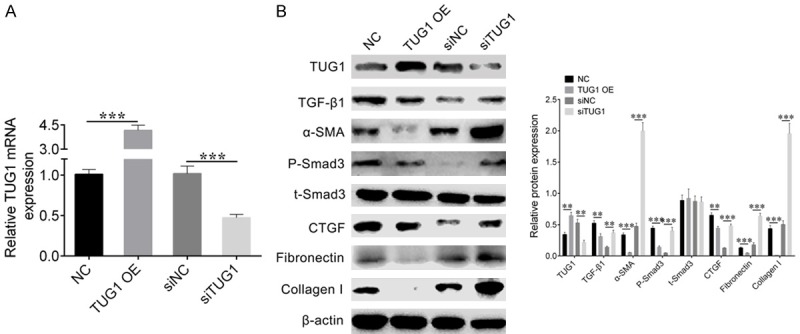
Overexpression of lncRNA TUG1 inhibits high glucose-stimulated of NRK-52E cell fibrosis. A. qRT-PCR detected the mRNA expression level of lncRNA TUG1 in high glucose-stimulated cells transfected with NC, TUG1 OE, siNC or siTUG1. Student’s t-test: ***P < 0.001. B. Western blot was performed to evaluate the expression level of TUG1 and fibrosis related maker genes TGF-β1, α-SMA, P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I. β-actin was used as loading control (left). The relative density of western blot was quantified by Image Pro Plus. Student’s t-test: **P < 0.01, ***P < 0.001.
The target gene of lncRNA TUG1 is miR-21
We performed sequence alignment of lncRNA TUG1 with miR-21, and then listed miR-214 target sites region (Figure 3A left panel). We also used a luciferase assay in NRK-52E cells to identify whether miR-21 was the target of lncRNA TUG1. We constructed a wild type (TUG1 wt) or mutated (TUG1 mut) lncRNA TUG1 vector. The NC mimics and miR-214 mimics were co-transfected with either TUG1 wt or TUG1 mut vector into NRK-52E cells, respectively. The cells were harvested and luciferase reporter activity was measured at 48 hours after co-transfection. Results showed that co-transfection with miR-214 mimics and TUG1 wt resulted in a significant reduction in luciferase reporter activity compared with control cells. After mutating the nucleotides of seeding sequence in lncRNA TUG1, the inhibitory effect of miR-214 mimics on luciferase reporter activity were largely abolished (Figure 3A right panel)). These results demonstrated miR-21 is a target gene of lncRNA TUG1. Furthermore, RIP assay showed that both TUG1 and miR-21 could bind with AGO2 protein in the RNA silencing pathway (Figure 3B).
Figure 3.
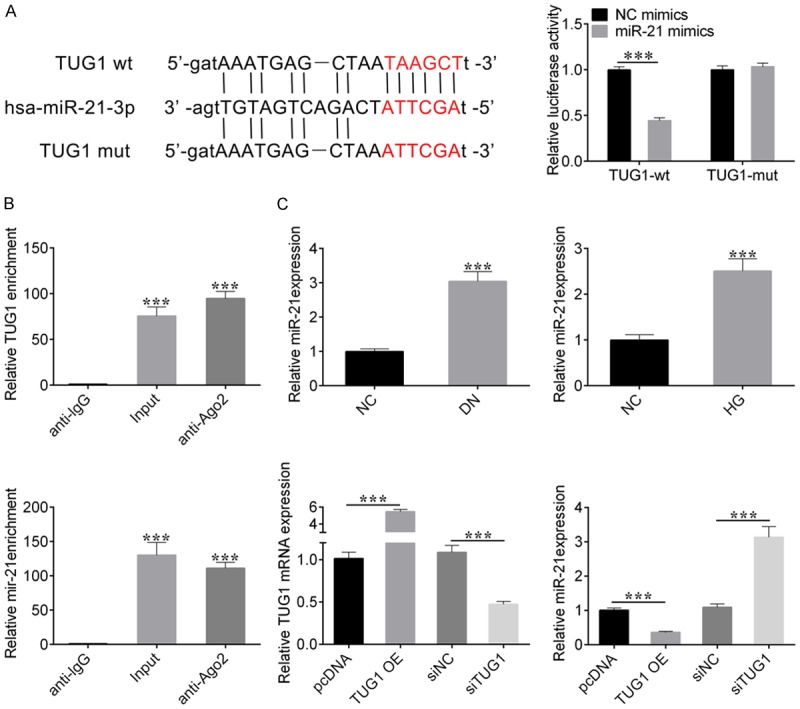
The target gene of lncRNA TUG1 is miR-21. A. Sequence alignment between lncRNA TUG1 with miR-21 (left). Dual luciferase reporter assay was used to measure the relative fluorescence activity in NRK-52E cells co-transfected with NC-mimics and TUG1-wt, miR-21 mimics and TUG1-wt, NC-mimics and TUG1-mut, or miR-21 mimics and TUG1-mut. Student’s t-test: ***P < 0.001 (right). B. RNA binding protein immunoprecipitation (RIP) assay determined the interaction between lncRNA TUG1, miR-21 with AGO2, Student’s t-test: ***P < 0.001. C. qRT-PCR detected the expression level of miR-21 in db/db DN mice and HG-stimulated NRK-52E cells (top panel). qRT-PCR was also used to evaluate the mRNA expression levels of lncRNA TUG1 or miR-21 in cells transfected with pc-DNA, pc-TUG1, si-NC, and si-TUG1 respectively (bottom panel). Student’s t-test: ***P < 0.001.
The results of qRT-PCR suggested that the expression level of miR-21 was also significantly increased in db/db DN mice and high glucose-stimulated NRK-52E cells (Figure 3C). To further verify this result, we detected the expression levels of miR-21 in cells transfected with pcDNA, TUG1 OE, si-NC, and si-TUG1, respectively. Results showed that overexpression of lncRNA TUG1 inhibited miR-21 expression. And down-regulation of lncRNA TUG1 promotes miR-21 expression (Figure 3C). Taken together, these results confirmed that miR-21 is the target gene of lncRNA TUG1.
TIMP3 is the target gene of miR-21
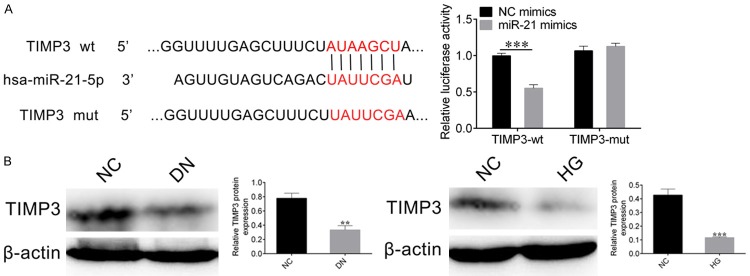
To explore the target gene of miR-21, we performed sequence alignment of miR-21 with TIMP3 3’-UTR (Figure 4A left panel), then we also carried out a luciferase assay in NRK-52E cells as above to identify whether TIMP3 was the target of miR-21. The NC mimics and miR-214 mimics were co-transfected with either TIMP3-wt or TIMP3-mut vector into NRK-52E cells, respectively. Dual luciferase reporter assay showed that miR-21 can inhibit the reporter gene when co-expressed with TIMP3-wt, but not with TIMP3-mut (Figure 4A, right panel), which demonstrated that a direct binding of miR-21 to the 3’-UTR of TIMP3.
Figure 4.
miR-21 target gene TIMP3. A. Sequence alignment between miR-21 and the 3’-UTR of TIMP3 gene (left). The wild type (wt) or mutant (mut) 3’-UTR of TIMP3 gene was fused with fluorescence reporter genes, and co-transfected with NC-mimics or miR-21 mimics into NRK-52E cells. Dual luciferase reporter assay was used to measure the relative fluorescence activity. Student’s t-test: ***P < 0.001 (right). B. Western blots detected the expression levels of TIMP3 in DN mice and high glucose-stimulated NRK-52E cells. β-actin was used as loading control. The relative intensity of bends were quantified by Image Pro Plus, Student’s t-test: ***P < 0.001.
Next, the expression levels of TIMP3 in DN mice and high glucose-stimulated NRK-52E cells were detected respectively. The western blot results showed that the expression level of TIMP3 was decreased not only in DN mice but also in high glucose-stimulated NRK-52E cells (Figure 4B). This result further proved that the differential expression of TIMP3 in DN mice and high glucose-stimulated NRK-52E cells might be regulated by lncRNA TUG1 and miR-21.
LncRNA TUG1 inhibits high glucose-stimulated NRK-52E cell fibrosis via miR-21
In order to detect whether lncRNA TUG1 regulates cell fibrosis through miR-21, we transfected NC, TUG1 OE, or co-transfected TUG1 and miR-21 into high glucose-stimulated NRK-52E cells and detected the expression levels of cell fibrosis marker genes, including α-SMA, TGF-β1, P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I. Western blot results suggested that in the lncRNA TUG1 OE cells, the expression of TIMP3 was significantly increased, while the expression of TIMP3 was decreased after co-transfected of miR-21, indicating that lncRNA TUG1 can regulate the expression of TIMP3 through miR-21. In addition, in lncRNA TUG1 OE cells, the expression level of the cell fibrosis-related marker genes was significantly decreased, but compared with the cells of the control group, the expression levels of these marker genes had no significant difference in the cells co-transfected TUG1 and miR-21, (Figure 5). These results indicate that lncRNA TUG1 can promote the expression of TIMP3 through miR-21, thereby inhibits cell fibrosis in high glucose-stimulated NRK-52E cells.
Figure 5.
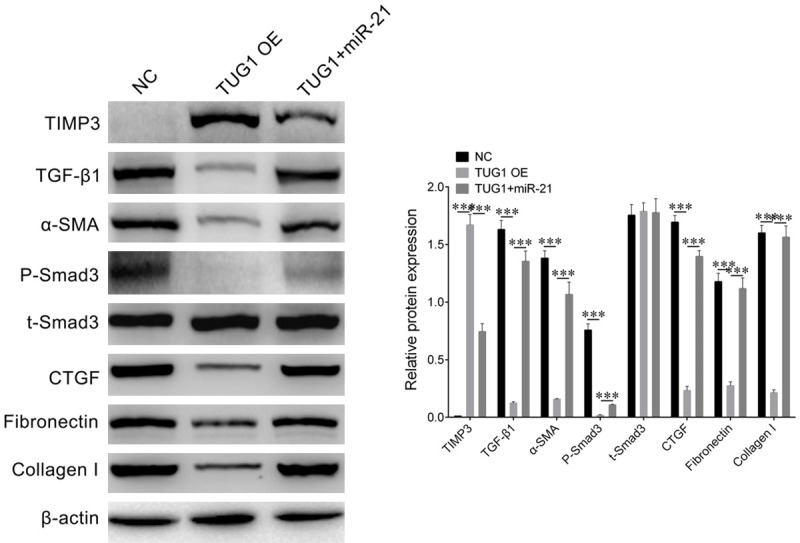
LncRNA TUG1 inhibits high glucose-stimulated NRK-52E cell fibrosis via miR-21. Western blot determined the protein accumulation of TIMP3 and cell fibrosis maker genes including TGF-β1, α-SMA, P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I in NRK-52E cells transfected with NC, TUG1 OE, or co-transfected with TUG1 and miR-21 i. β-actin was used as loading control. Relative density of western blot bends was quantified by Image Pro Plus. Student’s t-test: ***P < 0.001.
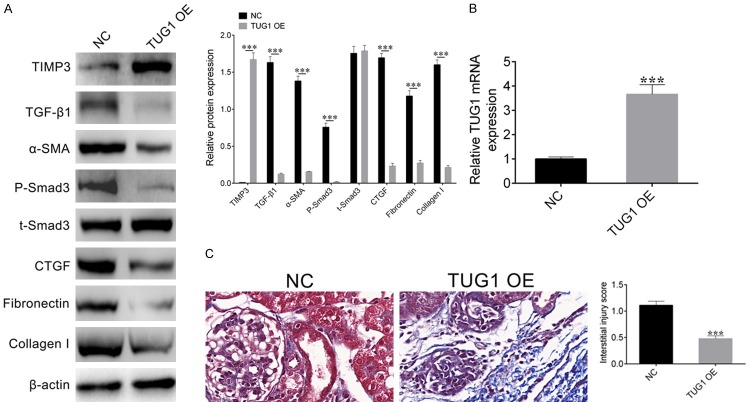
Overexpression of lncRNA TUG1 can improve renal fibrosis in DN mice
To further explore the effect of lncRNA TUG1 on renal fibrosis in DN mice, we firstly detected the expression level of TUG1 in NC mice and TUG1 OE mice by qRT-PCR. The results showed that the expression level of TUG1 was significantly increased in TUG1 OE mice (Figure 6B). Subsequently, western blot was used to detect the expression levels of the cell fibrosis-associated marker genes. Results showed that the expression of TIMP3 was significantly increased in the TUG1 OE mice, but these fibrosis marker genes α-SMA, TGF-β1, P-Smad3, t-Smad3, CTGF, Fibronectin, and Collagen I were significantly decreased (Figure 6A), which indicated that overexpression of lncRNA TUG1 could inhibit renal fibrosis in DN mice.
Figure 6.
Overexpression of lncRNA TUG1 can improve renal fibrosis in DN mice. A. Western blot was performed to detect the expression level of TIMP3 and cell fibrosis related genes in DN mice transfected with NC or TUG1 OE. β-actin was used as a loading control. Relative bends inensity was quantified by Image Pro Plus. Student’s t-test: ***P < 0.001. B. qRT-PCR detected the expression level of lncRNA TUG1 mRNA in DN mice transfected with NC or TUG1 OE. C. Masson staining and renal interstitial injury score were carried out to detect the extent of renal fibrosis in DN mice transfected with NC or TUG1 OE.
To further confirm this result, we measured the extent of cell fibrosis by Masson staining and renal interstitial lesion score. The results showed that the degree of cell fibrosis was obviously decreased in TUG1 OE mice (Figure 6C). In conclusion, these results demonstrate that overexpression of lncRNA TUG1 can reduce renal fibrosis in DN mice.
Discussion
Tubulointerstitial fibrosis is one of the main pathological features of DN. Our study demonstrated that lncRNA TUG1 can improve renal fibrosis in DN by modulating miR-21 targeting TIMP3. Our results showed that lncRNA TUG1 was down-regulated in db/db DN mice kidney tissues and high glucose-stimulated NRK-52E cells. Further experiments showed that lncRNA TUG1 could target miR-21, which could target the TIMP3 gene and regulate its expression. Finally, lncRNA TUG1 inhibited and improved fibrosis in high glucose-stimulated NRK-52E cells and DN mice by decreasing the expression of miR-21.
Recent studies have found that lncRNA plays an important role in DN [29]. Studies have shown that lncRNA-MIAT is highly expressed in diabetic mice and kidney cells cultured under high glucose conditions. Interference with lncRNA-MIAT can improve the pathologic features of diabetes and repair damaged retinal blood vessels. Knockout of the MIAT gene reduces the release of pro-inflammatory factors caused by diabetes, thereby reducing retinal vascular damage [30]. In addition, research have proven that lncRNA-HYMAI is involved in the development of transient neonatal diabetes [31]. The lncRNAs ENSMUST00000147869 and lncRNAs CYP4B1-PS1-001 protected and regulated fibrosis induced by DN [32,33]. These findings indicate that lncRNA can be involved in the development of diabetes and diabetic nephropathy, and is closely related to diabetic nephropathy, but the specific mechanism is still unclear. In our study, the expression level of lncRNA TUG1 was down-regulated in db/db DN mice and high glucose-stimulated NRK-52E cells, suggesting that TUG1 may play a role in the pathogenesis of DN. Thus, abnormal expression of TUG1 in vivo and in vitro of DN suggests that it may be involved in the occurrence of DN. At the same time, western blot analysis showed that overexpression of TUG1 in high glucose-stimulated NRK-52E cells was effective in reducing the expression levels of renal fibrosis marker proteins TGF-β1, Col-4 and FN, and inhibiting fibrosis of high glucose-stimulated NRK-52E cells.
In addition, previous studies have shown that lncRNA TUG1 could modulate ECM accumulation in DN by regulating miR-377 targeting PPARγ, and regulate mitochondrial biological energy of DN [9]. In this study, we found that TUG1 could regulate the expression of TIMP3 gene through miR-21, and ultimately affected the level of fibrosis in diabetic nephropathy, which may be a new pathway for TUG1 to participate in renal fibrosis. Many studies have shown that microRNA plays an important role in the pathogenesis of DN and renal fibrosis [14,34]. miR-21 is the most widely studied because many of its targets are related to DN [35]. Previous works have shown that miR-21 expression is up-regulated in diabetes mice and induced renal fibrosis by targeting MMP-9 or TIMPs [36], and is involved in the regulation of TGF-β1 and AKT activation [37]. In our study, lncRNA TUG1 was able to target miR-21 to regulate the expression of TIMP3, but whether it affected miR-21 targeting MMP-9 was still unknown. Moreover, overexpression of lncRNA TUG1 resulted in down-regulation of miR-21, decreased expression of fibrin and fibronectin, which was consistent with previous research.
TIMP3 was shown to be a target gene for miR-21 in this study. Previous works have suggested that TIMP3 was the most highly expressed TIMPs in kidney, and associated with inflammation, renal fibrosis and tubulointerstitial damage in mice [24]. The lack of TIMP3 has become a mark of DN in mice and humans [25]. In this study, the expression level of TIMP3 was significantly up-regulated when overexpressed lncRNA TUG1. The imbalance of MMPs/TIMPs may induce ECM accumulation. For another, MMPs (such as MMP-2, MMP-9) could degrade ECM, but TGF-β1could inhibit ECM degradation by increasing TIMPs (TIMP-1) activity which inhibited MMPs expression and activity, and finally increase the accumulation of ECM [38,39]. In our study, the expression of TGF-β1 is significantly decreased in lncRNA TUG1 overexpressed mice and high glucose-stimulated NRK-52E cells, which relieved the inhibition of MMPs and increased the degradation of ECM, and ultimately reduced the fibrosis of renal tissue.
Fibrosis caused by diabetic nephropathy can lead to loss of glomerular connection with functional tubules, abnormal renal function and persistent urinary protein, eventually leading to renal failure to form ESRD, which is one of the causes of high mortality in diabetic patients [3,40]. This study indicated that LncRNA TUG1 could regulate DN-induced fibrosis, providing a theoretical basis for a better understanding of the regulatory mechanisms of various levels of diabetic nephropathy. At the same time, LncRNA may become a new drug target, providing new ideas for the treatment of diabetic nephropathy.
Conclusion
In conclusion, we proved that lncRNA TUG1 was down-regulated in kidney tissue of db/db DN mice and high glucose-stimulated NRK-52E cells. Dual luciferase assay suggested that miR-21 was the target gene of lncRNA TUG1, and miR-21 target downstream TIMP3 gene. Increased expression of TIMP3 could attenuate the expression of renal fibrosis marker genes, and thereby inhibited cell fibrosis in high glucose-stimulated NRK-52E and renal fibrosis in DN mice. This study provides a possible theoretical basis for the future treatment of DN by editing lncRNA.
Acknowledgements
This work was funded as part of a project titled ‘PLA general hospital clinical research support fund’ (grant no. 2016FC-TSYS-2038).
Disclosure of conflict of interest
None.
References
- 1.Packham DK, Alves TP, Dwyer JP, Atkins R, de Zeeuw D, Cooper M, Shahinfar S, Lewis JB, Lambers Heerspink HJ. Relative incidence of ESRD versus cardiovascular mortality in proteinuric type 2 diabetes and nephropathy: results from the DIAMETRIC (diabetes mellitus treatment for renal insufficiency consortium) database. Am J Kidney Dis. 2012;59:75–83. doi: 10.1053/j.ajkd.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 2.Fioretto P, Mauer M. Histopathology of diabetic nephropathy. Semin Nephrol. 2007;27:195–207. doi: 10.1016/j.semnephrol.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolset SO, Reinholt FP, Jenssen T. Diabetic nephropathy and extracellular matrix. J Histochem Cytochem. 2012;60:976–986. doi: 10.1369/0022155412465073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keri KC, Samji NS, Blumenthal S. Diabetic nephropathy: newer therapeutic perspectives. J Community Hosp Intern Med Perspect. 2018;8:200–207. doi: 10.1080/20009666.2018.1500423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zampetaki A, Albrecht A, Steinhofel K. Long Non-coding RNA structure and function: is there a link? Front Physiol. 2018;9:1201. doi: 10.3389/fphys.2018.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang WY, Wang YF, Ma P, Xu TP, Shu YQ. Taurineupregulated gene 1: a vital long noncoding RNA associated with cancer in humans (review) Mol Med Rep. 2017;16:6467–6471. doi: 10.3892/mmr.2017.7472. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Shen J, Chan MT, Wu WK. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49:471–475. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li SY, Susztak K. The long noncoding RNA Tug1 connects metabolic changes with kidney disease in podocytes. J Clin Invest. 2016;126:4072–4075. doi: 10.1172/JCI90828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan LJ, Ding M, Hou LJ, Cui YT, Li CJ, Yu DM. Long noncoding RNA TUG1 alleviates extracellular matrix accumulation via mediating microRNA-377 targeting of PPARγ in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:598–604. doi: 10.1016/j.bbrc.2017.01.145. [DOI] [PubMed] [Google Scholar]
- 10.Long J, Badal SS, Ye Z, Wang Y, Ayanga BA, Galvan DL, Green NH, Chang BH, Overbeek PA, Danesh FR. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest. 2016;126:4205–4218. doi: 10.1172/JCI87927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Long JY, Wang Y, Wang WJ, Chang BHJ, Danesh FR. MicroRNA-29c Is a signature microRNA under high glucose conditions that targets sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mafi A, Aghadavod E, Mirhosseini N, Mobini M, Asemi Z. The effects of expression of different microRNAs on insulin secretion and diabetic nephropathy progression. J Cell Physiol. 2018;234:42–50. doi: 10.1002/jcp.26895. [DOI] [PubMed] [Google Scholar]
- 14.Wu H, Kong L, Zhou S, Cui W, Xu F, Luo M, Li X, Tan Y, Miao L. The role of microRNAs in diabetic nephropathy. J Diabetes Res. 2014;2014:920134. doi: 10.1155/2014/920134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewanjee S, Bhattacharjee N. MicroRNA: a new generation therapeutic target in diabetic nephropathy. Biochem Pharmacol. 2018;155:32–47. doi: 10.1016/j.bcp.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Bracken CP, Khew-Goodall Y, Goodall GJ. Network-based approaches to understand the roles of miR-200 and other microRNAs in cancer. Cancer Res. 2015;75:2594–2599. doi: 10.1158/0008-5472.CAN-15-0287. [DOI] [PubMed] [Google Scholar]
- 17.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of MicroRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol. 2010;21:438–447. doi: 10.1681/ASN.2009050530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey N, Das F, Mariappan MM, Mandal CC, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem. 2011;286:25586–25603. doi: 10.1074/jbc.M110.208066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Xiao L, Xiao P, Yang S, Chen G, Liu F, Kanwar YS, Sun L. A glimpse of matrix metalloproteinases in diabetic nephropathy. Curr Med Chem. 2014;21:3244–3260. doi: 10.2174/0929867321666140716092052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thrailkill KM, Clay Bunn R, Fowlkes JL. Matrix metalloproteinases: their potential role in the pathogenesis of diabetic nephropathy. Endocrine. 2009;35:1–10. doi: 10.1007/s12020-008-9114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valimaki J, Uusitalo H. Matrix metalloproteinases (MMP-1, MMP-2, MMP-3 and MMP-9, and TIMP-1, TIMP-2 and TIMP-3) and markers for vascularization in functioning and non-functioning bleb capsules of glaucoma drainage implants. Acta Ophthalmol. 2015;93:450–456. doi: 10.1111/aos.12654. [DOI] [PubMed] [Google Scholar]
- 23.Arpino V, Brock M, Gill SE. The role of TIMPs in regulation of extracellular matrix proteolysis. Matrix Biol. 2015;44-46:247–254. doi: 10.1016/j.matbio.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Kassiri Z, Oudit GY, Kandalam V, Awad A, Wang X, Ziou X, Maeda N, Herzenberg AM, Scholey JW. Loss of TIMP3 enhances interstitial nephritis and fibrosis. J Am Soc Nephrol. 2009;20:1223–1235. doi: 10.1681/ASN.2008050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiorentino L, Cavalera M, Menini S, Marchetti V, Mavilio M, Fabrizi M, Conserva F, Casagrande V, Menghini R, Pontrelli P, Arisi I, D’Onofrio M, Lauro D, Khokha R, Accili D, Pugliese G, Gesualdo L, Lauro R, Federici M. Loss of TIMP3 underlies diabetic nephropathy via FoxO1/STAT1 interplay. EMBO Mol Med. 2013;5:441–455. doi: 10.1002/emmm.201201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu R, Lee J, Wang Z, Patel VB, Fan D, Das SK, Liu GC, John R, Scholey JW, Oudit GY, Kassiri Z. Loss of TIMP3 selectively exacerbates diabetic nephropathy. Am J Physiol Renal Physiol. 2012;303:F1341–1352. doi: 10.1152/ajprenal.00349.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiorentino L, Cavalera M, Mavilio M, Conserva F, Menghini R, Gesualdo L, Federici M. Regulation of TIMP3 in diabetic nephropathy: a role for microRNAs. Acta Diabetol. 2013;50:965–969. doi: 10.1007/s00592-013-0492-8. [DOI] [PubMed] [Google Scholar]
- 28.McClelland AD, Herman-Edelstein M, Komers R, Jha JC, Winbanks CE, Hagiwara S, Gregorevic P, Kantharidis P, Cooper ME. miR-21 promotes renal fibrosis in diabetic nephropathy by targeting PTEN and SMAD7. Clin Sci (Lond) 2015;129:1237–1249. doi: 10.1042/CS20150427. [DOI] [PubMed] [Google Scholar]
- 29.Lu Z, Liu N, Wang F. Epigenetic regulations in diabetic nephropathy. J Diabetes Res. 2017;2017:7805058. doi: 10.1155/2017/7805058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, Tao ZF, Song YC, Chen Q, Jiang Q. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015;116:1143–1156. doi: 10.1161/CIRCRESAHA.116.305510. [DOI] [PubMed] [Google Scholar]
- 31.Ma D, Shield JP, Dean W, Leclerc I, Knauf C, Burcelin RR, Rutter GA, Kelsey G. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest. 2004;114:339–348. doi: 10.1172/JCI19876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Yao D, Wang S, Yan Q, Lu W. Long non-coding RNA ENSMUST00000147869 protects mesangial cells from proliferation and fibrosis induced by diabetic nephropathy. Endocrine. 2016;54:81–92. doi: 10.1007/s12020-016-0950-5. [DOI] [PubMed] [Google Scholar]
- 33.Wang M, Wang S, Yao D, Yan Q, Lu W. A novel long non-coding RNA CYP4B1-PS1-001 regulates proliferation and fibrosis in diabetic nephropathy. Mol Cell Endocrinol. 2016;426:136–145. doi: 10.1016/j.mce.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 34.Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2016;310:F109–118. doi: 10.1152/ajprenal.00387.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masoudi MS, Mehrabian E, Mirzaei H. MiR-21: a key player in glioblastoma pathogenesis. J Cell Biochem. 2018;119:1285–1290. doi: 10.1002/jcb.26300. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Gao Y, Ma M, Li M, Zou D, Yang J, Zhu Z, Zhao X. Effect of miR-21 on renal fibrosis by regulating MMP-9 and TIMP1 in kk-ay diabetic nephropathy mice. Cell Biochem Biophys. 2013;67:537–546. doi: 10.1007/s12013-013-9539-2. [DOI] [PubMed] [Google Scholar]
- 37.Sekar D, Venugopal B, Sekar P, Ramalingam K. Role of microRNA 21 in diabetes and associated/related diseases. Gene. 2016;582:14–18. doi: 10.1016/j.gene.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 38.Border WA, Ruoslahti E. Transforming growth factor-beta in disease: the dark side of tissue repair. J Clin Invest. 1992;90:1–7. doi: 10.1172/JCI115821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sporn MB, Roberts AB. Transforming growth factor-beta: recent progress and new challenges. J Cell Biol. 1992;119:1017–1021. doi: 10.1083/jcb.119.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shields J, Maxwell AP. Managing diabetic nephropathy. Clin Med (Lond) 2010;10:500–504. doi: 10.7861/clinmedicine.10-5-500. [DOI] [PMC free article] [PubMed] [Google Scholar]