Abstract
Background
Diabetic retinopathy, a vascular complication of diabetes mellitus, is the leading cause of visual impairment and blindness. circRNAs act as competing endogenous RNA, sponging target miRNA and thus influencing mRNA expression in vascular diseases. We investigated whether and how circDNMT3B is involved in retinal vascular dysfunction under diabetic conditions.
Methods
qRT-PCR was performed to detect expression of circDNMT3B, miR-20b-5p, and BAMBI in retinal microvascular endothelial cells under diabetic conditions. Western blot, Cell Counting Kit-8, Transwell, Matrigel tube formation, and retinal trypsin digestion assays were conducted to explore the roles of circDNMT3B/miR-20b-5p/BAMBI in retinal vascular dysfunction. Bioinformatics analysis and luciferase reporter, siRNA, and overexpression assays were used to reveal the mechanisms of the circDNMT3B/miR-20b-5p/BAMBI interaction. Electroretinograms were used to evaluate visual function.
Findings
Upregulation of miR-20b-5p under diabetic conditions promoted proliferation, migration, and tube formation of human retinal microvascular endothelial cells (HRMECs), which was mediated by downregulated BAMBI. Under diabetic conditions, circDNMT3B, which acts as a sponge of miR-20b-5p, is downregulated. circDNMT3B overexpression reduced retinal acellular capillary number and alleviated visual damage in diabetic rats. Changes in expression of circDNMT3B and miR-20b-5p were confirmed in the proliferative fibrovascular membranes of patients with diabetic retinopathy.
Interpretation
Downregulation of circDNMT3B contributes to vascular dysfunction in diabetic retinas through regulating miR-20b-5p and BAMBI, providing a potential treatment strategy for diabetic retinopathy.
Funding
National Natural Science Foundation of China, National Key Basic Research Program of China, Shanghai Municipal Science and Technology Major Project, and ZJLab.
Keywords: Diabetic retinopathy, Retinal microvascular endothelial cells, circDNMT3B, miR-20b-5p, BAMBI
Research in context
Evidence before this study
Diabetic retinopathy (DR) is one of the leading causes of visual impairment and blindness. Chronic hyperglycemia-induced microvascular dysfunction is the primary factor in the development of DR. Circular RNAs (circRNAs) are involved in regulating endothelial cells (ECs) in vascular diseases. However, the effects of circRNAs in DR and their underlying mechanisms remain unclear.
Added value of this study
We found that circDNMT3B expression was downregulated in retinal ECs both in patients with DR and diabetic rats. circDNMT3B regulated the proliferation, migration, and tube formation of human retinal microvascular endothelial cells (HRMECs) under diabetic conditions through targeting miR-20b-5p and its downstream regulator, BAMBI. circDNMT3B overexpression reduced retinal acellular capillary number and alleviated visual damage in diabetic retinas.
Implications of all the available evidence
Our results reveal the molecular mechanisms of circDNMT3B/miR-20b-5p/BAMBI function in retinal vascular dysfunction under diabetic conditions.
1. Introduction
Diabetic retinopathy (DR), a vascular complication of diabetes mellitus, is one of the major causes of visual impairment and blindness, and may affect 2•5 million people worldwide [1]. Loss of pericytes, breakdown of the blood−retina barrier (BRB) and capillary acellularity characterize the early lesions of DR. Chronic hyperglycemia has been considered to be the primary factor driving the development of DR [2]. Diabetes mellitus induces retinal microvascular dysfunction by destroying the tight junctions between the endothelial cells (ECs) comprising the BRB [3,4]. The weakening of the vascular walls, with subsequent accelerated EC proliferation and microaneurysm development, leads to vascular leakage and incompetence. Retinal vascular remodeling and capillary nonperfusion result in imbalance of normal vessels and formation of abnormal new vessels. However, the molecular mechanisms underlying this process are not yet well understood. Previous studies have shown that hyperglycemia enhanced the proliferation and migration of ECs and accelerated tube formation of ECs, which was consistent with the pathogenesis of angiogenesis in DR [5], [6], [7], [8]. Therefore, regulation of EC functions may provide a novel therapeutic strategy for the treatment of DR.
MicroRNAs (miRNAs) are small, non-coding RNAs that interact with the 3′ untranslated region (UTR) of mRNAs to regulate protein-coding genes [9]. Several miRNAs have been identified that are related to the diagnosis, prognosis, and treatment of DR [10], [11], [12]. miR-20b-5p, derived from the conserved paralogous miR-17-2∼363 cluster, is a vascular endothelial growth factor (VEGF)-inducible miRNA [13]. Previous studies have confirmed its importance in modulating cell proliferation, differentiation, apoptosis, and angiogenesis [14], [15], [16]. Circular RNAs (circRNAs), another type of non-coding RNA, are derived from exons of protein-coding genes through the backsplice mechanism [17]. The structure of circRNA is a closed loop without 5′−3′ polarity or polyadenylated tails. circRNAs are highly conserved among different species and are abundant in human cells [18]. Several studies have reported that circRNAs act as a competing endogenous RNA, sponging target miRNA and consequently influencing mRNA expression in vascular diseases [17,[19], [20], [21]]. In this study, we show that expression of circDNMT3B was downregulated in the retinal ECs both of patients with DR and of diabetic rats, and was negatively associated with miR-20b-5p. We further show that circDNMT3B regulates human retinal microvascular endothelial cell (HRMEC) function in vitro and influences the progression of DR in vivo through interaction with miR-20b-5p.
2. Materials and methods
2.1. Ethics statement
The retinal proliferative fibrovascular membranes were collected from seven patients diagnosed with proliferative DR (PDR) who underwent vitrectomy at the Eye and Ear Nose Throat (Eye & ENT) Hospital of Fudan University. Epiretinal membranes for the non-DR group were obtained from six patients diagnosed with idiopathic epiretinal membrane. All procedures were explained to patients, and informed consent was provided. Male Sprague−Dawley rats (80–100 g) were obtained from SLAC Laboratory Animal Co., Ltd (Shanghai, China), and housed under a 12-h light/dark schedule. All procedures met the ethical standards of the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and the guidelines of Fudan University. This study was approved by the Institutional Ethics Review Committee of the Eye & ENT Hospital of Fudan University.
2.2. Rat diabetic model
After fasting 12 h, each anesthetized rat was given an intraperitoneal injection of streptozotocin (STZ; Sigma-Aldrich Corp., Louis, MO, USA) (65 mg/kg). Control rats received injections of an equal volume of citrate buffer. Fasting blood glucose was measured using a glucometer (Accu-Chek Performa, Roche, Germany) at 0 days (prior to injection), 7 days, 1 month, and 3 months after the injection. Rats with blood glucose levels over 11•1 mmol/L were used in this study.
2.3. Construction of the circDNMT3B adeno-associated virus and intravitreal injection
The human circDNMT3B sequence was cloned into the pAAV2/DJ-CMV-GFP vector, which was then packaged into adeno-associated viruses (AAVs) form Vigene Sciences (Shandong, China). AAV-DJ-circDNMT3B (4 μL, 7•32 × 1013 v.g./mL) was injected into the vitreous body of rats using a Nanoject II microinjector (Drummond Scientific Company, Broomall, PA, USA) with a micropipette of 10−30 μm tip size under an OLYMPUS ZS61 microscope (Olympus, Tokyo, Japan). Control rats received injections of an equal volume of AAV-DJ-ciR-NC (empty vector). Transfection efficiency was evaluated using green fluorescent protein (GFP) signals on retinal slices, and the expression level of circDNMT3B was detected using quantitative reverse-transcription polymerase chain reaction (qRT-PCR).
2.4. Cell culture and transfection
HRMECs (Angio-Proteomie, Boston, Mass, USA) were cultured in endothelial cell medium (ECM) with 5% fetal bovine serum, 1% endothelial cell growth supplement, and 1% penicillin/streptomycin solution (ScienCell Research Laboratories, Carlsbad, CA, USA) at 37 °C under 5% CO2 atmosphere. Cells between passages three and eight were used in this study. Cells were treated with 5 mM glucose as a normal glucose (NG) control, 5 mM glucose plus 25 mM mannitol as an osmotic control, or 30 mM glucose as a high-glucose (HG) treatment. miR-20b-5p inhibitor, miR inhibitor negative control (NC), miR-20b-5p mimic, or miR mimic NC (RiboBio, Guangzhou, China) was transfected into HRMECs using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Small interfering BMP and activin membrane-bound inhibitor (siBAMBI) and siRNA NC synthesized by RiboBio were transfected using Lipofectamine RNAiMAX (Life Technologies). The human circDNMT3B sequence was inserted into the pLCDH-ciR vector, and then incorporated into a lentivirus by Geneseed Biotech (Guangzhou, China). To induce stable circDNMT3B-overexpressing HRMECs, the HRMECs at passage three were infected with lentivirus containing circDNMT3B or ciR NC (empty vector). All sequences used in this study are listed in Supplementary Table S1.
2.5. RNA extraction and qRT-PCR
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Hilden, Germany). miRNA was reverse transcribed using the miRcute Plus miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech, Beijing, China) and amplified with the miRcute Plus miRNA qPCR Detection Kit (Tiangen Biotech). RNA was reverse transcribed with the PrimeScript RT reagent Kit with gDNA Eraser (Takara, Kusatsu, Japan) and amplified using TB Green Premix Ex Taq II (Takara). circRNA was amplified using divergent primers to target the splice junction. U6 (Tiangen Biotech) and β-actin were used as internal controls for miRNA and RNA, respectively. All primers used are listed in Supplementary Table S2. qRT-PCR assays were performed on the QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, Rockford, IL, USA).
2.6. Ribonuclease R (RNase R) treatment
RNA was incubated with or without 3 U/μg RNase R (Epicentre, San Diego, CA, USA) at 37 °C for 20 min according to the manufacturer's instructions. The resulting RNA was purified with the RNeasy MinElute Cleanup Kit (Qiagen) for further analysis.
2.7. Western blotting
Western blotting was performed according to methods described in previous studies [22], [23], [24]. Briefly, proteins were extracted by using the AllPrep DNA/RNA/Protein Mini Kit (Qiagen). The primary antibodies used included monoclonal mouse anti-β-actin (A5441; 1:8000, Sigma-Aldrich), polyclonal rabbit anti-zonula occludens-1 (ZO-1) (40–2200; 1:1000; Invitrogen, Waltham, MA, USA), polyclonal rabbit anti-occludin (71–1500; 1:500; Invitrogen) and monoclonal mouse anti-claudin-5 (35–2500; 1:500; Invitrogen). The secondary antibodies used included horseradish peroxidase-conjugated anti-rabbit and anti-mouse IgG (711-035-152 and 715-035-150; 1:8000; Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Expression was visualized with the ECL enhanced chemifluorescent reagent (Thermo Scientific, Rockford, IL, USA) and the ChemiDoc XRS system (Bio-Rad, Hercules, CA, USA). Expression of β-actin was used as a control.
2.8. Cell proliferation assay
Cell Counting Kit-8 (CCK-8) (Dojindo, Tokyo, Japan) was used to detect cell proliferation. HRMECs were plated in a 96-well plate, and 10 μL CCK-8 solution was added to each well. After 4 h of incubation at 37 °C, the absorbance values were measured at 450 nm.
2.9. Cell migration assay
Cell migration was analyzed using Costar Transwell plates (Corning, Bedford, MA, USA), as described previously [25]. HRMECs were plated in the Transwell insert in a 24-well plate, and 600 μL ECM was added to the bottom of the lower chamber. After 20 h of incubation, HRMECs attached to the other side of the insert were fixed with 90% ethanol and stained with 0.1% crystal violet. Migrated cells were observed and counted under a Nikon Eclipse Ti inverted microscope (Nikon, Tokyo, Japan). In each well, at least six different fields were chosen at random, and the number of migrated cells was averaged from these fields.
2.10. Tube formation assay
Tube formation of HRMECs was tested using a previously described protocol [26]. Growth Factor Reduced Matrigel matrix (Corning) (300 μL) was laid on the bottom of a 24-well plate, onto which HRMECs were seeded. Capillary-like structures were observed under a Nikon Eclipse Ti inverted microscope (Nikon) 20 h after cell seeding. At least six different fields were randomly selected and observed in each well. Meshes, branches, and the branching length of the capillary-like structures were analyzed using ImageJ software (version1.49p; NIH, Bethesda, MD, USA).
2.11. Dual luciferase reporter assay
The 3′ UTR of the BAMBI sequence containing wild-type or mutant miR-20b-5p binding sites was inserted into the XhoI and NotI restriction sites of pmiR-RB-REPORT vector (Ribobio). The circDNMT3B sequence containing wild-type or mutant miR-20b-5p binding sites was inserted into the SgfI and NotI restriction sites of psiCHECK2 vector (Geneseed). The maps of pmiR-RB-REPORT and psiCHECK2 vectors were shown in Supplementary Fig. S1. Luciferase reporter vectors were co-transfected with miR-20b-5p mimic or miR mimic NC into 293T cells using Lipofectamine 3000 (Life Technologies). At 48 h after transfection, luciferase activities were detected with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) using the Synergy H4 Multi-Mode Reader (BioTek, Winooski, VT, USA) according to the manufacturer's instructions. The binding of miRNA on RNA sequence leads to the depressed expression of Renilla luciferase (Rluc) and affects the luciferase activity.
2.12. RNA fluorescence in situ hybridization (FISH)
HRMECs were fixed with 4% paraformaldehyde. Following pre-hybridization treatment with 0.5% Triton X-100, HRMECs were incubated with RNA probes in hybridization buffer (40% formamide, 4 × SSC, 1 mg/ml yeast tRNA, 10% Dextran sulfate, 1 × Denhardt's solution, 10 mM DDT, 1 mg/ml sheared salmon sperm DNA) for 12 h. The circDNMT3B probe was labeled with FITC and miR-20b-5p was labeled with Cy3 (GeneSeed). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA, USA). Images were captured with a Leica TCS-SP2-AOBS confocal microscope (Leica, Wetzlar, Germany). Probe sequences are listed in Supplementary Table S1.
2.13. Retinal trypsin digestion
Retinal trypsin digestion was performed following a previously described procedure [27], with some modifications. The eyes of anesthetized rats were enucleated and fixed overnight with 10% neutral buffered formalin. The retinas were dissected and gently shaken in water at room temperature overnight, then digested with 3% trypsin (1:250; Solarbio, Beijing, China) at 37 °C for 3 h. After repeated washing, the network of vessels was isolated and mounted on slides. The dried vessels were stained with the Glycogen Periodic Acid Schiff (PAS/Hematoxylin) Stain Kit (Solarbio). At least ten different fields in each retina were randomly chosen and the number of acellular capillaries was counted in each field under the microscope (Olympus DP80; Olympus, Tokyo, Japan).
2.14. Electroretinogram (ERG)
After dark adaptation overnight, the rats were anesthetized. Recording electrodes were placed in the center of the cornea. The reference electrode was placed hypodermically on the central forehead. The grounding electrode was attached to the tail. Light stimuli were provided by the ESPION ColorDome Handheld Ganzfeld stimulator (Diagnosys LLC, Littleton, MA, USA). Stimulus intensity was 0.01 or 1 cd⋅s/m2 under scotopic conditions. Full-field flash ERG was recorded with the ESPION Console (Diagnosys LLC) [28]. The amplitude of the a-wave was measured from the baseline, whereas the b-wave amplitude was determined from the trough of the a-wave to the peak of the b-wave. The responses to 10−15 successive stimuli were averaged from each recording.
2.15. Statistical analysis
Statistical analysis was performed using GraphPad Prism software (version 7.0; GraphPad Software Inc., La Jolla, CA, USA). Data were expressed as mean ± standard deviation (SD). Differences between two groups were compared using the two-tailed unpaired t-test. Multiple comparisons were calculated using one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparisons test. P values of less than 0.05 were considered statistically significant in all tests. At least 3 independent experiments were conducted for each assay.
3. Results
3.1. miR-20b-5p is upregulated in diabetic retinas and affects HRMEC function under high-glucose conditions in vitro
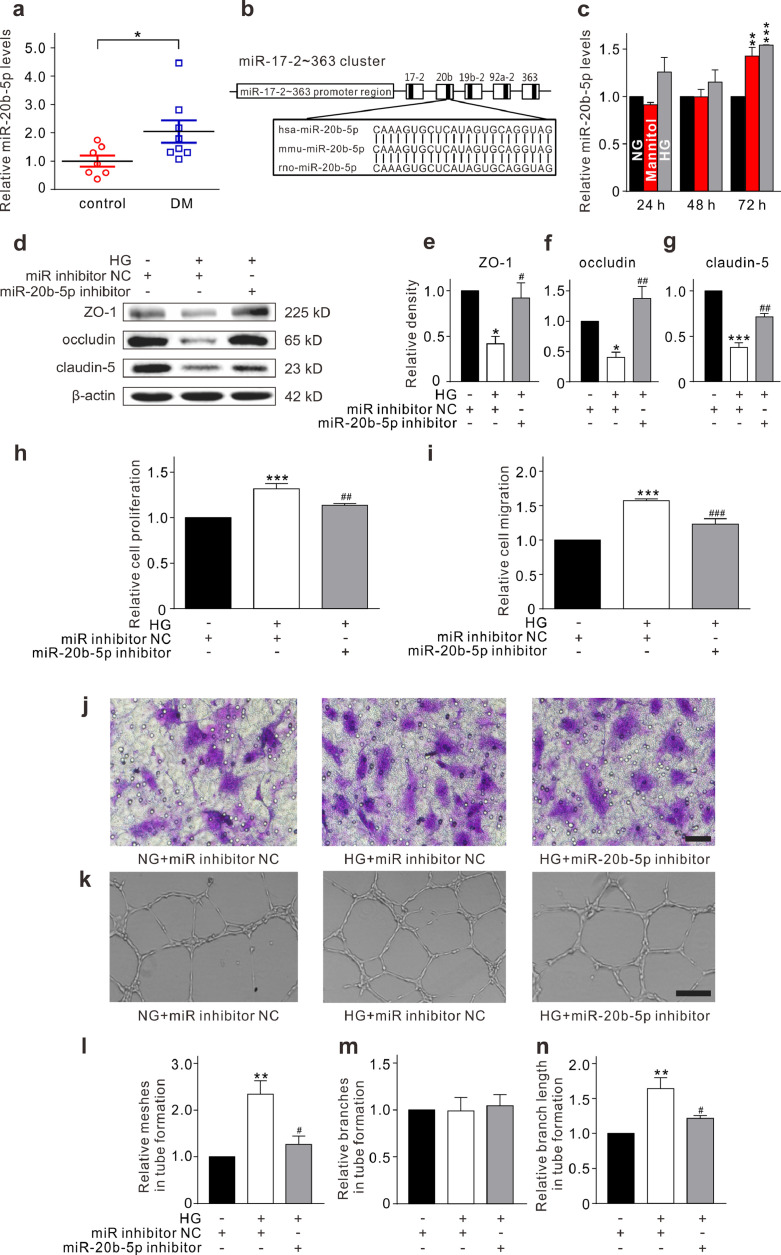
Several miRNAs are abundantly expressed in retinal ECs [13]. Because miR-20b-5p has been shown to modulate cell proliferation, differentiation, apoptosis, and angiogenesis [14], [15], [16], we examined whether miR-20b-5p expression may be altered in diabetic retinas. As shown in Fig. 1a, retinal miR-20b-5p expression was significantly upregulated in diabetic rats compared with controls based on a qRT-PCR assay. miRBase showed that the sequence of miR-20b-5p is conserved among human, mouse, and rat (Fig. 1b). To further explore the roles of miR-20b-5p in the diabetic retina, HRMECs were cultured in HG medium (30 mM) to mimic the diabetic conditions in vitro. qRT-PCR assay revealed that HG treatment significantly increased the level of miR-20b-5p (Fig. 1c). Furthermore, HG treatment markedly reduced levels of the tight junction-related proteins ZO-1, occludin, and claudin-5, as shown through Western blotting, which was alleviated by the miR-20b-5p inhibitor (Fig. 1d–g). In addition, miR-20b-5p inhibitor markedly elevated the expression of ZO-1, occludin, and claudin-5 in HRMECs under normal glucose condition (Supplementary Fig. S2). Furthermore, the CCK-8 assay showed that HG treatment enhanced the proliferation of HRMECs (Fig. 1h). Transwell and Matrigel tube formation experiments showed that HG exposure accelerated the migration and tube formation of HRMECs. The effects of HG were reversed with miR-20b-5p inhibitor (Fig. 1i–n). These results suggest that miR-20b-5p might be involved in regulating HRMECs under diabetic conditions.
Fig. 1.
miR-20b-5p is upregulated in diabetic rat retinas and affects function of HRMECs under high-glucose (HG) conditions. (a) Levels of miR-20b-5p expression, assayed by qRT-PCR, in control and diabetic rat retinas (DM). All data are normalized to control. n = 7 and 8 for control and DM groups respectively. * p < 0.05 vs. control. (b) Comparison of miR-20b-5p sequence among human, mouse, and rat. (c) Changes of miR-20b-5p levels in HRMECs cultured in 5 mM glucose (normal glucose, NG), 5 mM glucose plus 25 mM mannitol (Mannitol), or 30 mM glucose (HG) medium for 24 h, 48 h, and 72 h, respectively. Data are normalized to corresponding NG groups. n = 3 for each group. ** p < 0.01 and *** p < 0.001 vs. NG. (d–g) Representative Western blotting results show the changes in protein levels of ZO-1, occludin, and claudin-5 in HRMECs which were transfected with miR inhibitor NC (control) or miR-20b-5p inhibitor (miR-20b-5p inhibitor group) with or without the HG treatment (d). Bar chart summarizing the relative density of immunoblot bands of ZO-1 (e), occludin (f), and claudin-5 (g). All data are normalized to control. n = 3 for each group. * p < 0.05 and *** p < 0.001 vs. control; #p < 0.05 and ##p < 0.01 vs. HG. (h) Bar chart summarizing the changes of HRMEC proliferation detected by CCK-8 assay under different conditions as shown in panel d. All data are normalized to control. n = 4 for each group, *** p < 0.001 vs. control, ##p < 0.01 vs. HG. (i, j) Bar chart summarizing the migration of HRMECs determined by Transwell assay under different conditions (i). All data are normalized to control. n = 4 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. HG. Representative images are shown in panel j. Scale bar: 20 μm. (k–n) Representative images show the changes in capillary-like structure of HRMECs observed by Matrigel tube formation assay under different conditions (k). Scale bar: 100 μm. Bar chart summarizing the changes in meshes (l), branches (m) and branching length (n). All data are normalized to control. n = 3 for each group, ** p < 0.01 vs. control, and #p < 0.05 vs. HG. All in vitro experiments: n = 3 or 4 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: CCK-8 = cell counting kit-8, DM = diabetes mellitus, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, NC = negative control, NG = normal glucose, qRT-PCR = quantitative reverse-transcription polymerase chain reaction, ZO-1 = zonula occludens-1.
3.2. miR-20b-5p regulates HRMEC function through targeting BAMBI
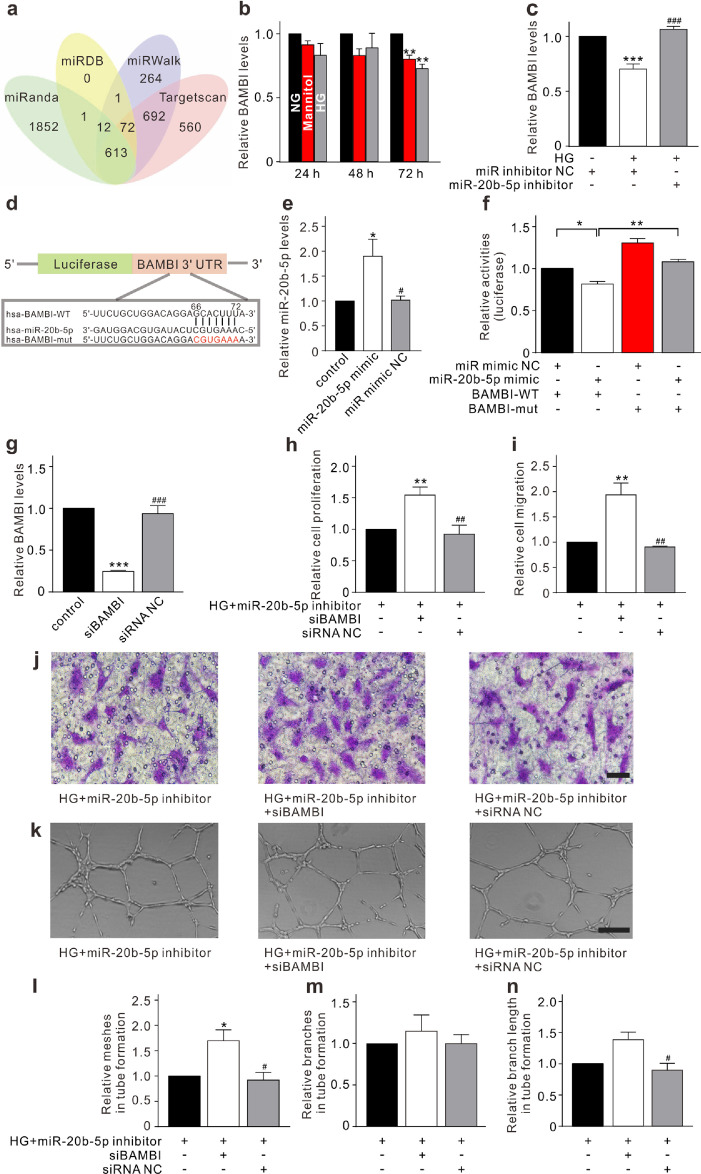
To further address how miR-20b-5p modulates HRMEC function, bioinformatics analysis was employed to predict the potential target genes of miR-20b-5p based on the miRanda, miRDB, miRWalk, and Targetscan (Fig. 2a). Genes related to EC functions were screened out. Expression levels of candidate genes, including transforming growth factor beta receptor 2 (TGFβR2), transcription factor 7 like 2 (TCF7L2), phosphatase and tensin homolog (PTEN), lkb1, cyclin-dependent kinase inhibitor 1A (CDKN1A), bone morphogenetic protein receptor type 2 (BMPR2), zinc finger and BTB domain-containing 4 (ZBTB4), and BAMBI, were examined using qRT-PCR (Supplementary Fig. S3a–g, Fig. 2b). Five genes were markedly downregulated in HRMECs under HG conditions and the downregulation of four of these genes could be reversed with miR-20b-5p inhibitor (Fig. 2c, supplementary Fig. S3h–k). Previous studies have demonstrated that BAMBI could regulate capillary growth and angiogenesis [29,30]. We further investigated whether the effects of miR-20b-5p on HRMECs were exerted through targeting BAMBI. Luciferase reporter vector containing the wild-type or mutant 3′ UTR of the BAMBI sequence was co-transfected with miR-20b-5p mimic or miR mimic NC into 293T cells (Fig. 2d and e). A significant decrease in luciferase activity was observed in the group with the wild-type BAMBI reporter (Luc-BAMBI-WT) plus miR-20b-5p mimic (Fig. 2f). Moreover, BAMBI knockdown using siRNA (Fig. 2g) enhanced the proliferation, migration, and tube formation of HRMECs and counteracted the repressive effects of miR-20b-5p inhibitor under HG conditions (Fig. 2h–n). These results indicate that miR-20b-5p regulates HRMEC function through targeting BAMBI.
Fig. 2.
miR-20b-5p regulates HRMEC function through targeting BAMBI. (a) A Veen diagram illustrates the predicted target genes of miR-20b-5p from miRanda, miRDB, miRWalk, and Targetscan. (b) Changes of BAMBI levels, assayed by qRT-PCR, in HRMECs cultured in 5 mM glucose (normal glucose, NG), 5 mM glucose plus 25 mM mannitol (Mannitol), or 30 mM glucose (HG) medium for 24 h, 48 h, and 72 h, respectively. Data are normalized to corresponding NG groups. n = 3 for each group, ** p < 0.01 vs. NG. (c) Bar chart summarizing the changes of BAMBI levels in HRMECs transfected with miR inhibitor NC (control) or miR-20b-5p inhibitor (miR-20b-5p inhibitor group) with or without HG treatment. All data are normalized to control. n = 4 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. HG. (d) Luciferase reporter vectors containing the wild-type or mutant 3′ UTR of the BAMBI were constructed according to the predicted binding sites of miR-20b-5p. (e) Bar chart summarizing the levels of miR-20b-5p in the 293T cells under different conditions. All data are normalized to control. n = 5 for each group, * p < 0.05 vs. control, and #p < 0.05 vs. miR-20b-5p mimic group. (f) Bar chart summarizing the changes of luciferase activities under different conditions. The HRMECs were co-transfected with luciferase reporter vector and miR-20b-5p mimic or miR mimic NC. n = 3 for each group, * p < 0.05, and ** p < 0.01. (g) Bar chart summarizing the changes of BAMBI levels in HRMECs transfected with siBAMBI or siRNA NC. All data are normalized to control. n = 3 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. siBAMBI group. (h) Bar chart summarizing changes of HRMEC proliferation detected by CCK-8 assay. After transfected with miR-20b-5p inhibitor under HG conditions (control), the HRMECs were transfected with siBAMBI or siRNA NC. All data are normalized to control. n = 5 for each group, ** p < 0.01 vs. control, and ##p < 0.01 vs. siBAMBI group. (i, j) Bar chart summarizing the migration of HRMECs determined by Transwell assay under different conditions as shown in panel h (i). All data are normalized to control. n = 4 for each group, ** p < 0.01 vs. control, and ##p < 0.01 vs. siBAMBI group. Representative images are shown in panel j. Scale bar: 20 μm. (k–n) Representative images show the changes of capillary-like structure of HRMECs observed by Matrigel tube formation assay under different conditions as shown in panel h (k). Scale bar: 100 μm. Bar chart summarizing the changes of meshes (l), branches (m), and branching length (n). All data are normalized to control. n = 3 for each group; * p < 0.05 vs. control, and #p < 0.05 vs. siBAMBI group. All in vitro experiments: n = 3–5 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: BAMBI = BMP and activin membrane bound inhibitor, CCK-8 = cell counting kit-8, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, miR = microRNA, mut = mutant, NC = negative control, NG = normal glucose, qRT-PCR = quantitative reverse-transcription polymerase chain reaction, siBAMBI = small interfering BMP and activin membrane bound inhibitor, siRNA = small interfering RNA, UTR = untranslated region, WT = wild-type.
3.3. circDMNT3B, a sponge of miR-20b-5p in HRMECs
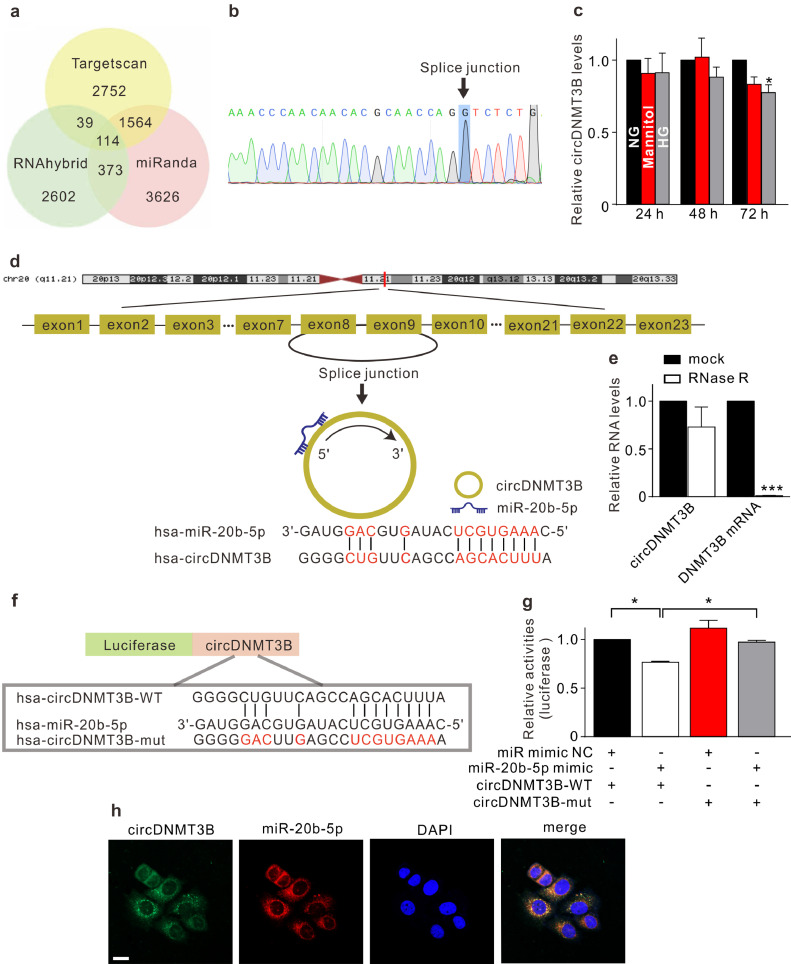
circRNAs, a novel type of non-coding RNAs, have been shown to play an important role in the pathological processes of some diseases through sponging miRNA [31], [32], [33]. We screened circRNAs that might act as a sponge of miR-20b-5p using Targetscan, RNAhybrid, and miRanda (Fig. 3a). The predictive results were overlapped with the microarray data, revealing differentially expressed circRNAs between diabetic and non-diabetic retinas [34]. Considering abundance and sequence length, the two most promising candidates, circDNMT3B (hsa_circ_0059802) and circTNFRSF21 (hsa_circ_0076699), were screened out. Divergent primers were used in qRT-PCR. Sanger sequencing confirmed that the amplified products were in accordance with sequences reported in circBase (Fig. 3b, Supplementary Fig. S4a). HG treatment led to reduced expression of circDNMT3B in HRMECs (Fig. 3c), while circTNFRSF21 expression was not affected (Supplementary Fig. S4b). circDNMT3B was derived from the eighth and ninth exons of DNA methyltransferase 3 beta (DNMT3B) (Fig. 3d). It was resistant to RNase R digestion, whereas linear DNMT3B mRNA was markedly degraded (Fig. 3e). To test whether miR-20b-5p may interact with circDNMT3B, we compared the sequences in RNAhybrid and identified one predicted binding site (Fig. 3f). The dual luciferase reporter assay demonstrated that co-transfection of the wild-type circDNMT3B reporter (Luc-circDNMT3B-WT) with miR-20b-5p mimic significantly decreased luciferase activity (Fig. 3g). RNA-FISH further showed that circDNMT3B and miR-20b-5p were co-localized in the cytoplasm of HRMECs (Fig. 3h). These results suggest that circDNMT3B acts as a sponge of miR-20b-5p in HRMECs under diabetic conditions.
Fig. 3.
circDMNT3B, a sponge of miR-20b-5p in HRMECs. (a) A Veen diagram illustrates the predicted target circRNAs of miR-20b-5p from Targetscan, RNAhybrid, and miRanda. (b) The Sanger sequencing shows that the amplified product of divergent primers was consistent with circDNMT3B sequence in circBase. (c) qRT-PCR results reveal the changes of circDNMT3B levels in HRMECs cultured in 5 mM glucose (normal glucose, NG), 5 mM glucose plus 25 mM mannitol (Mannitol), or 30 mM glucose (HG) medium for 24 h, 48 h, and 72 h, respectively. Data are normalized to corresponding NG groups. n = 3 for each group, * p < 0.05 vs. NG. (d) circDNMT3B is transcribed from the eighth and ninth exons of DNMT3B gene, and the predicted binding sites of miR-20b-5p are shown. (e) Summarized data showing the changes of circDNMT3B and DNMT3B mRNA levels in HRMECs with or without RNase R digestion. n = 3 for each group, *** p < 0.001 vs. mock group. (f) Luciferase reporter vectors containing wild-type or mutant circDNMT3B sequence were constructed. The predicted binding sites of miR-20b-5p on circDNMT3B sequence are shown. (g) Bar chart summarizing the changes of luciferase activities under different conditions. The HRMECs were co-transfected with luciferase reporter vector and miR-20b-5p mimic or miR mimic NC. n = 3 for each group, * p < 0.05. (h) RNA-FISH assay reveals that circDNMT3B (FITC-labelled) was co-localized with miR-20b-5p (Cy3-labelled) in the cytoplasm of HRMECs. Nuclei were stained with DAPI. Scale bar: 20 μm. All in vitro experiments: n = 3 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole, DNMT3B = DNA methyltransferase 3 beta, FISH = fluorescence in situ hybridization, FITC = fluorescein isothiocyanate, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, miR = microRNA, mut = mutant, NC = negative control, NG = normal glucose, qRT-PCR = quantitative reverse-transcription polymerase chain reaction, RNase R = ribonuclease R, WT = wild-type.
3.4. circDNMT3B regulates HRMEC function through targeting miR-20b-5p
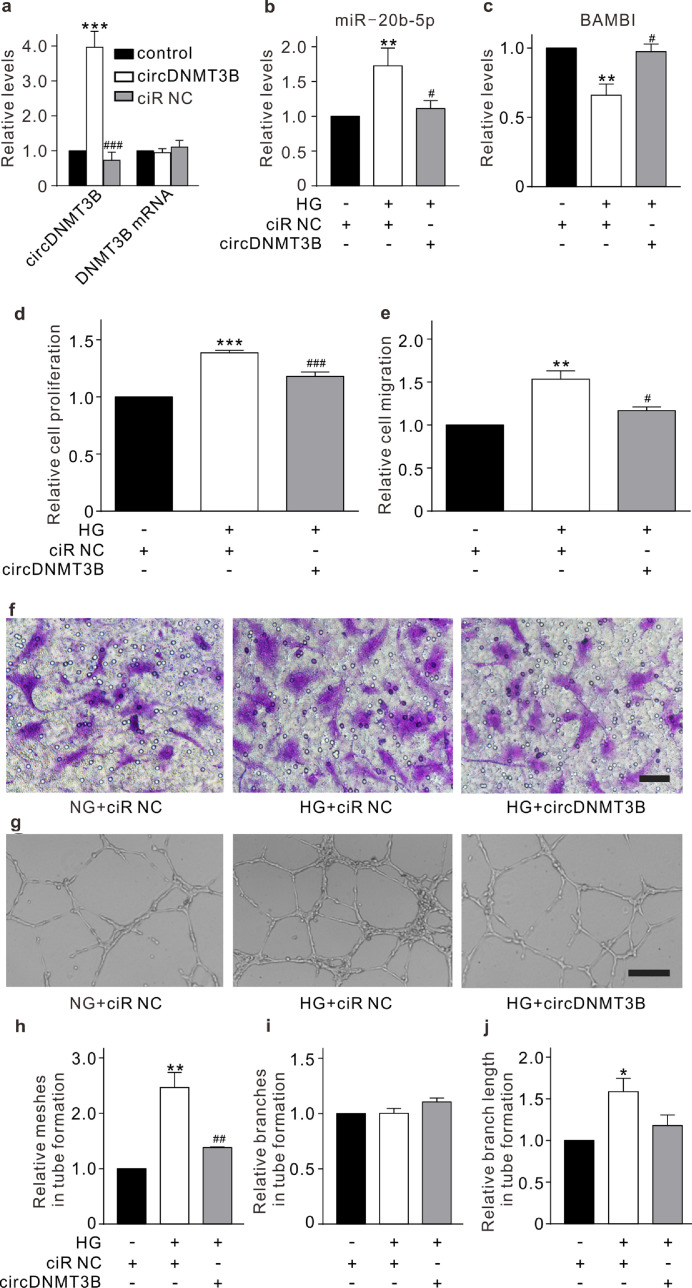
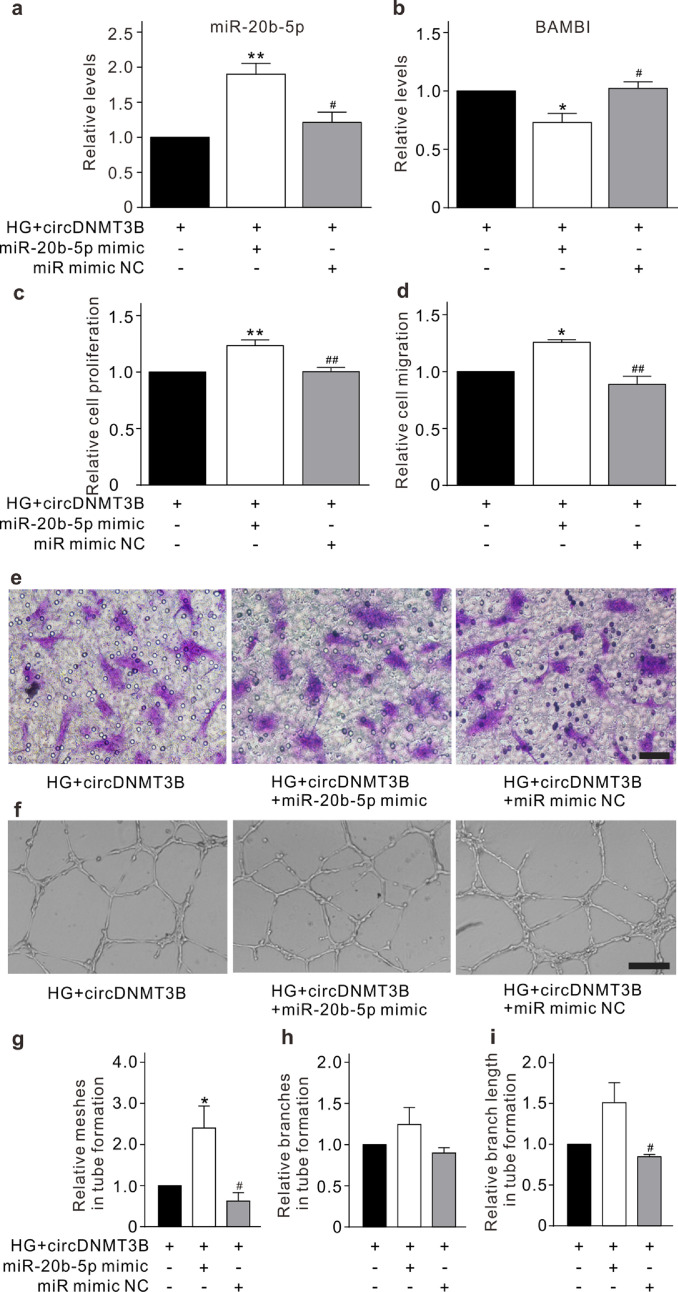
To address whether circDNMT3B regulates HRMEC function through targeting miR-20b-5p, lentivirus containing the circDNMT3B sequence was used to stably overexpress circDNMT3B in HRMECs, which did not change the level of linear DNMT3B mRNA expression (Fig. 4a). Overexpression of circDNMT3B significantly attenuated both the upregulation of miR-20b-5p and the downregulation of BAMBI in HRMECs induced by HG treatment (Fig. 4b and c). CCK-8, Transwell, and Matrigel tube formation assays were used to evaluate the roles of circDNMT3B under diabetic conditions in vitro. Overexpression of circDNMT3B alleviated the effects of HG treatment on promoting proliferation, migration, and tube formation of HRMECs (Fig. 4d–j). Furthermore, we transfected miR-20b-5p mimic into circDNMT3B-overexpressing HRMECs, which significantly increased the level of miR-20b-5p and decreased the level of BAMBI (Fig. 5a, b, and Supplementary Fig. S5). The inhibitory effects of circDNMT3B on proliferation, migration, and tube formation in HRMECs were also attenuated by miR-20b-5p mimic transfection under HG conditions (Fig. 5c–i). These results indicate that circDNMT3B regulates HRMEC function through targeting miR-20b-5p.
Fig. 4.
circDNMT3B regulates HRMEC function. (a) Summarized data showing the changes of circDNMT3B (n = 3) and DNMT3B mRNA (n = 4) levels, assayed by qRT-PCR, in HRMECs which were infected with lentivirus containing circDNMT3B or ciR NC (empty vector). All data are normalized to corresponding controls. *** p < 0.001 vs. control, and ###p < 0.001 vs. circDNMT3B group. (b, c) Summarized data showing the changes of miR-20b-5p (n = 6) (b) and BAMBI (n = 4) (c) levels in HRMECs which were infected with lentivirus containing ciR NC (control) or circDNMT3B (circDNMT3B group) with or without HG treatment. All data are normalized to control. ** p < 0.01 vs. control, and #p < 0.05 vs. HG group. (d) Bar chart summarizing the changes of HRMEC proliferation detected by CCK-8 assay under different conditions as shown in panel b. All data are normalized to control. n = 5 for each group, *** p < 0.001 vs. control, and ###p < 0.001 vs. HG. (e, f) Bar chart summarizing the migration of HRMECs determined by Transwell assay under different conditions (e). All data are normalized to control. n = 3 for each group, ** p < 0.01 vs. control, and #p < 0.05 vs. HG. Representative images are shown in panel f. Scale bar: 20 μm. (g–j) Representative images showing the changes of capillary-like structure of HRMECs observed by Matrigel tube formation assay under different conditions as shown in panel b (g). Scale bar: 100 μm. Bar chart summarizing the changes of meshes (h), branches (i) and branching length (j). All data are normalized to control. n = 3 for each group, * p < 0.05, ** p < 0.01 vs. control, and ##p < 0.01 vs. HG. All in vitro experiments: n = 3–6 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: BAMBI = BMP and activin membrane bound inhibitor, CCK-8 = cell counting kit-8, ciR = circular RNA, DNMT3B = DNA methyltransferase 3 beta, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, NC = negative control, qRT-PCR = quantitative reverse-transcription polymerase chain reaction.
Fig. 5.
circDNMT3B regulates HRMEC function through targeting miR-20b-5p. (a, b) Summarized data showing the changes of miR-20b-5p (a) and BAMBI (b) levels, assayed with qRT-PCR, in the circDNMT3B-overexpressing HRMECs. miR-20b-5p mimic (miR-20b-5p mimic group) or miR mimic NC (miR mimic NC group) was transfected into circDNMT3B-overexpressing HRMECs after HG treatment. All data are normalized to circDNMT3B group. n = 3 for each group, * p < 0.05, ** p < 0.01 vs. circDNMT3B group, and #p < 0.05 vs. miR-20b-5p mimic group. (c) Bar chart summarizing the changes in proliferation of HRMECs detected by CCK-8 assay under different conditions. All data are normalized to circDNMT3B group. n = 4 for each group, ** p < 0.01 vs. circDNMT3B group, and ##p < 0.01 vs. miR-20b-5p mimic group. (d, e) Bar chart summarizing the migration of HRMECs determined by Transwell assay under different conditions (d). All data are normalized to circDNMT3B group. n = 3 for each group, * p < 0.05 vs. circDNMT3B group, and ##p < 0.01 vs. miR-20b-5p mimic group. Representative images are shown in panel e. Scale bar: 20 μm. (f–i) Representative images showing the changes of capillary-like structure of HRMECs observed by Matrigel tube formation assay under different conditions (f). Scale bar: 100 μm. Bar chart summarizing the changes of meshes (g), branches (h), and branching length (i). All data are normalized to circDNMT3B group. n = 3 for each group, * p < 0.05 vs. circDNMT3B group, and #p < 0.05 vs. miR-20b-5p mimic group. All in vitro experiments: n = 3 or 4 biological replicates × 3 technical replicates. Data presented as means with error bars representing standard deviation (SD). Abbreviations: BAMBI = BMP and activin membrane bound inhibitor, CCK-8 = cell counting kit-8, DNMT3B = DNA methyltransferase 3 beta, HG = high glucose, HRMEC = human retinal microvascular endothelial cells, miR = micro RNA, NC = negative control, qRT-PCR = quantitative reverse-transcription polymerase chain reaction.
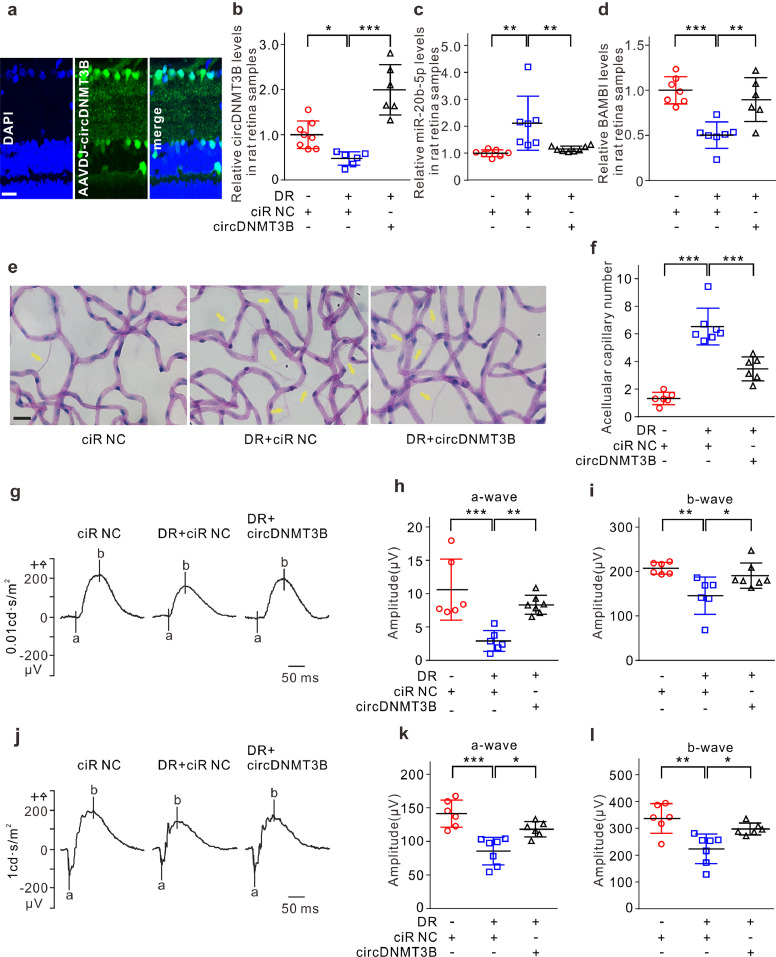
3.5. circDNMT3B alleviates retinal vascular dysfunction in a rat diabetes mellitus model
We next investigated whether circDNMT3B regulates retinal vascular dysfunction in vivo, further affecting visual function in diabetic rats. AAV-DJ-circDNMT3B was injected intravitreally into diabetic rats. Stable transfection was observed, as evidenced by GFP fluorescent signals in retinal slices (Fig. 6a). In diabetic retinas injected with AAV-DJ-ciR-NC, the level of circDNMT3B was significantly reduced. By contrast, the expression of circDNMT3B was strongly elevated in AAV-DJ-circDNMT3B-injected diabetic retinas (Fig. 6b). The elevation of circDNMT3B significantly attenuated both the upregulation of miR-20b-5p and the downregulation of BAMBI in diabetic retinas (Fig. 6c and d). DR is one of the most common microvascular complications of diabetes mellitus. Elevated circDNMT3B expression significantly reduced the effect of diabetic injury on retinal vascular function, as evidenced by the decreased number of acellular capillaries (Fig. 6e and f). ERG was employed to elucidate the effect of circDNMT3B on visual function. The amplitudes of the a-wave and b-wave in ERG were significantly smaller in diabetic retinas, and this condition was partially rescued by circDNMT3B overexpression (Fig. 6g–l), suggesting that visual damage was ameliorated by circDNMT3B.
Fig. 6.
circDNMT3B alleviates retinal vascular dysfunction in a rat DM model. (a) Fluorescence signals of GFP in retinal vertical slice taken from the AAV-DJ-circDNMT3B injected diabetic rat at 3 months after injection. Nuclei were stained with DAPI. Scale bar: 20 μm. (b) Summarized data showing the changes of circDNMT3B levels, assayed by qRT-PCR, in retinas obtained from the AAV-DJ-circDNMT3B or the AAV-DJ-ciR-NC intravitreally injected normal or diabetic rats at 3 months after injection. All data are normalized to the AAV-DJ-ciR-NC injected normal retinas (control). n = 6–8, * p < 0.05, and *** p < 0.001. (c, d) Summarized data showing the changes of miR-20b-5p (c) and BAMBI (d) levels under different conditions as shown in panel b. All data are normalized to control. n = 6–8, ** p < 0.01 and *** p < 0.001. (e, f) Retinal trypsin digestion assay showing the changes in the number of retinal acellular capillaries (yellow arrows) under different conditions (e). Scale bar: 20 μm. Bar chart summarizing the average number of retinal acellular capillaries (f). n = 6–7, *** p < 0.001. (g-i) Representative ERG recordings at stimulus intensity of 0.01 cd⋅s/m2 in diabetic rats under different conditions (g). Summarized data showing the changes of a-wave (h) and b-wave (i) amplitudes. n = 6–7, * p < 0.05, ** p < 0.01, and *** p < 0.001. (j–l) Representative ERG recordings at stimulus intensity of 1 cd⋅s/m2 in diabetic rats under different conditions (j). Summarized data showing the changes of a-wave (k) and b-wave (l) amplitudes. n = 6–7, * p < 0.05, ** p < 0.01, and *** p < 0.001. Data presented as means with error bars representing standard deviation (SD). Abbreviations: AAV = adeno-associated virus, ciR = circular RNA, DAPI = 4′,6-diamidino-2-phenylindole, DM = diabetes mellitus, DNMT3B = DNA methyltransferase 3 beta, DR = diabetic retinopathy, ERG = electroretinogram, GFP = green fluorescent protein, NC = negative control, qRT-PCR = quantitative reverse-transcription polymerase chain reaction. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
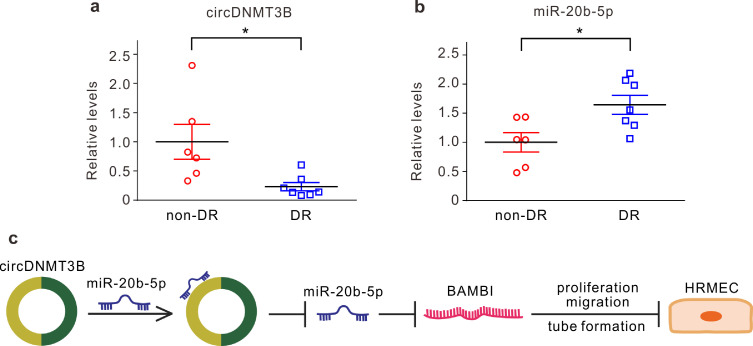
Finally, we collected the epiretinal membranes of patients without DR and the proliferative fibrovascular membranes of patients with DR. qRT-PCR showed that the level of circDNMT3B was reduced, whereas miR-20b-5p was upregulated in the DR group (Fig. 7a and b). These results further confirm that circDNMT3B and miR-20b-5p are involved in the progression of diabetic retinopathy through regulating retinal vascular dysfunction.
Fig. 7.
circDNMT3B is involved in human diabetic retinopathy (DR). (a, b) Summarized data showing the changes in circDNMT3B (a) and miR-20b-5p (b) levels, assayed by qRT-PCR, in proliferative fibrovascular membranes or epiretinal membranes collected from patients with or without DR. Data are normalized to non-DR group. n = 6–7, * p < 0.05. (c) A schematic diagram showing the hypothetical mechanism involved in the circDNMTB-mediated regulation of retinal vascular dysfunctions in DR. Data presented as means with error bars representing standard deviation (SD). Abbreviations: BAMBI = BMP and activin membrane bound inhibitor, DNMT3B = DNA methyltransferase 3 beta, DR = diabetic retinopathy, HRMEC = human retinal microvascular endothelial cells, qRT-PCR = quantitative reverse-transcription polymerase chain reaction.
4. Discussion
Retinal vascular dysfunction, characterized by microvascular remodeling, vascular hyperpermeability, capillary nonperfusion, and complementary neovascularization, is the leading cause of diabetes mellitus-induced visual damage [2], [3], [4]. Increasing evidence suggests that miRNAs play an important role in DR through targeting mRNA [11], [12], [13]. Retinal ECs are the major targets of hyperglycemic injury. Previous research has demonstrated that miRNAs regulate proliferation, activation, apoptosis, and other physiological processes of retinal ECs, and thus regulate angiogenesis [12,13]. In this study, we demonstrated that upregulation of miR-20b-5p related to retinal EC dysfunction under diabetic conditions. This relation was supported by the finding that miR-20b-5p expression was markedly elevated in retinal ECs of both diabetic rats and patients with DR, which was mimicked by HG treatment of cultured HRMECs. In addition, HG treatment significantly reduced the expression of tight junction-related proteins (ZO-1, occludin and claudin-5) in HRMECs, and this effect was reversed with miR-20b-5p inhibitor. The miR-20b-5p upregulation-induced decrease in tight junction-related protein expression could increase BRB permeability and microvascular leakage, contributing to diabetic retinal damage. circRNAs are abundant, highly conserved, and tissue-specific non-coding RNAs found in mammalian cells [35]. Although their expression patterns and functional mechanisms are not yet fully elucidated, evidence suggests that circRNAs function as miRNA sponges to regulate transcription or as RNA-binding-protein (RBP) sponges [31,36]. Previous studies have shown circRNAs engaged in the pathogenesis and intervention of several vascular diseases [19], [20], [21]. In this study, we found that circDNMT3B was downregulated in HRMECs under HG conditions, and further confirmed this finding in retinal fibrovascular membranes of patients with DR. CircDNMT3B is derived from the eighth and ninth exons of the DNMT3B gene. Consistent with the prediction from bioinformatics, the dual-luciferase reporter assay demonstrated that miR-20b-5p binds circDNMT3B at the target sites. RNA-FISH analysis showed co-localization of circDNMT3B and miR-20b-5p in the cytoplasm of HRMECs. These results demonstrate that circDNMT3B functions as a sponge of miR-20b-5p, negatively regulating miR-20b-5p expression under diabetic conditions in retinal ECs.
BAMBI, a type 1 TGFβ receptor antagonist, has been shown to play important roles in the development, tumor growth, and metastasis of certain cancers [37,38]. Recent studies have revealed that BAMBI reduces capillary growth and migration, contributing to endothelial stability and maintaining vascular homeostasis in diabetic nephropathy [29,39,40]. BAMBI deficiency results in major abnormalities of vascular systems [41]. In this study, we provide evidence that BAMBI is downregulated in retinal ECs under diabetic conditions, which is regulated by miR-20b-5p and circDNMT3B. First, the HG treatment-induced decrease in BAMBI expression was reversed with miR-20b-5p inhibitor. Second, knockdown of BAMBI reversed the inhibitory effects of miR-20b-5p inhibitor on proliferation, migration, and tube formation of HRMECs under HG conditions. Third, the dual-luciferase reporter assay showed interaction between miR-20b-5p and BAMBI, as predicted by bioinformatics. Fourth, overexpression of circDNMT3B alleviated the reduced expression of BAMBI under HG conditions.
Microvascular abnormality and capillary nonperfusion, which lead to retinal hypoxia and an inflammatory response, contribute to compensatory neovascularization in DR [3,4]. In this study, we showed that circDNMT3B/miR-20b-5p/BAMBI was involved in pathological changes of HRMECs in response to HG treatment. Excessive EC proliferation, migration, and tube formation under HG conditions was strongly inhibited by miR-20b-5p inhibitor, and this inhibition was reversed when BAMBI was silenced. Overexpression of circDNMT3B counteracted abnormal proliferation, migration, and tube formation of HRMECs induced by HG treatment. Moreover, circDNMT3B overexpression significantly reduced hyperglycemia-induced retinal acellular capillary number and partly rescued the decrease in amplitudes of a- and b-waves in ERGs in diabetic rats. These results suggest that circDNMT3B could regulate HRMEC function in vitro and vascular dysfunction in vivo under diabetic conditions. Aberrant expression of miR-20b-5p and circDNMT3B in human retinal proliferative fibrovascular membranes was detected in patients with DR, suggesting that miR-20b-5p and circDNMT3B may be involved in the clinical pathogenesis of DR. The detailed regulatory mechanism of this involvement remains to be elucidated through future research.
In conclusion, we have provided compelling evidence demonstrating that downregulation of circDNMT3B contributes to retinal vascular dysfunction in diabetic retinas through regulating miR-20b-5p and BAMBI (Fig. 7c). Interference with circDNMT3B/miR-20b-5p/BAMBI provides a potential strategy for treatment of DR.
Funding sources
This work was supported by the National Natural Science Foundation of China (No. 81790642, 31872765, 81570854, and 81770944), the National Key Basic Research Program of China (2013CB967503), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01) and ZJLab. The sponsor or funding organization had no role in the design or conduct of this research.
Author contributions
Conception and design: KZ, GZX, and ZW.
Performed the research and data collection: KZ, XH, HC, FL, NY, ALL, and KS.
Data analysis and interpretation: KZ, XH, KS, YWQ, X. Huang, QC, GZX, and ZW.
Drafted the manuscript: KZ, GZX, and ZW.
Declaration of Competing Interest
All authors declare no conflict interests.
Acknowledgments
We would like to thank Dr. Xiong-Li Yang for his helpful discussion and critical comments on the manuscript. We also thank Drs. Biao Yan, Ting-ting Jiang, Jian Yu, Juan Zhang, Yong-Heng Huang and Qing-Chen Li for the technical assistance and Yan-Qiong Zhang for the clinical sample collection.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.004.
Contributor Information
Ge-Zhi Xu, Email: drxugezhi@163.com.
Zhongfeng Wang, Email: zfwang@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Pascolini D., Mariotti S.P. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 2.Stitt A.W., Curtis T.M., Chen M. The progress in understanding and treatment of diabetic retinopathy. Prog Retin Eye Res. 2016;51:156–186. doi: 10.1016/j.preteyeres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Klaassen I., Van Noorden C.J., Schlingemann R.O. Molecular basis of the inner blood-retinal barrier and its breakdown in diabetic macular edema and other pathological conditions. Prog Retin Eye Res. 2013;34:19–48. doi: 10.1016/j.preteyeres.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Erickson K.K., Sundstrom J.M., Antonetti D.A. Vascular permeability in ocular disease and the role of tight junctions. Angiogenesis. 2007;10:103–117. doi: 10.1007/s10456-007-9067-z. [DOI] [PubMed] [Google Scholar]
- 5.Yang W.Z., Yang J., Xue L.P., Xiao L.B., Li Y. MiR-126 overexpression inhibits high glucose-induced migration and tube formation of rhesus macaque choroid-retinal endothelial cells by obstructing VEGFA and PIK3R2. J Diabetes Complications. 2017;31:653–663. doi: 10.1016/j.jdiacomp.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Mei X., Zhou L., Zhang T., Lu B., Sheng Y., Ji L. Chlorogenic acid attenuates diabetic retinopathy by reducing VEGF expression and inhibiting VEGF-mediated retinal neoangiogenesis. Vascul Pharmacol. 2018;101:29–37. doi: 10.1016/j.vph.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Huang Q., Sheibani N. High glucose promotes retinal endothelial cell migration through activation of Src, PI3K/Akt1/eNOS, and ERKs. Am J Physiol Cell Physiol. 2008;295:C1647–C1657. doi: 10.1152/ajpcell.00322.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao R., Zhu B.H., Tang S.B., Wang J.F., Ren J. Scutellarein inhibits hypoxia- and moderately-high glucose-induced proliferation and VEGF expression in human retinal endothelial cells. Acta Pharmacol Sin. 2008;29:707–712. doi: 10.1111/j.1745-7254.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Garcia de la Torre N., Fernandez-Durango R., Gomez R. Expression of Angiogenic MicroRNAs in Endothelial Progenitor Cells From Type 1 Diabetic Patients With and Without Diabetic Retinopathy. Invest Ophthalmol Vis Sci. 2015;56:4090–4098. doi: 10.1167/iovs.15-16498. [DOI] [PubMed] [Google Scholar]
- 11.Li E.H., Huang Q.Z., Li G.C., Xiong Z.Y., Zhang X. Effects of miRNA-200b on the development of diabetic retinopathy by targeting VEGFA gene. Biosci Rep. 2017;37 doi: 10.1042/BSR20160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q., Qiu F., Zhou K. Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARalpha. Diabetes. 2017;66:1671–1682. doi: 10.2337/db16-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacs B., Lumayag S., Cowan C., Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 14.Zhang D., Yi Z., Fu Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. J Cell Biochem. 2019;120:5889–5896. doi: 10.1002/jcb.27874. [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Lin B., Nie L., Li P. microRNA-20b contributes to high glucose-induced podocyte apoptosis by targeting SIRT7. Mol Med Rep. 2017;16:5667–5674. doi: 10.3892/mmr.2017.7224. [DOI] [PubMed] [Google Scholar]
- 16.Liang Z.G., Yao H., Xie R.S., Gong C.L., Tian Y. MicroRNA20b5p promotes ventricular remodeling by targeting the TGFbeta/Smad signaling pathway in a rat model of ischemiareperfusion injury. Int J Mol Med. 2018;42:975–987. doi: 10.3892/ijmm.2018.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng X., Li X., Zhang P., Wang J., Zhou Y., Chen M. Circular RNA: an emerging key player in RNA world. Brief Bioinform. 2017;18:547–557. doi: 10.1093/bib/bbw045. [DOI] [PubMed] [Google Scholar]
- 18.Jeck W.R., Sorrentino J.A., Wang K. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y., Zhang Y., Han B. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C., Yao M.D., Li C.P. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan K., Liu C., Liu B.H. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 22.Ji M., Miao Y., Dong L.D. Group I mGluR-mediated inhibition of Kir channels contributes to retinal Muller cell gliosis in a rat chronic ocular hypertension model. J Neurosci. 2012;32:12744–12755. doi: 10.1523/JNEUROSCI.1291-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong L.D., Gao F., Wang X.H. GluA2 trafficking is involved in apoptosis of retinal ganglion cells induced by activation of EphB/EphrinB reverse signaling in a rat chronic ocular hypertension model. J Neurosci. 2015;35:5409–5421. doi: 10.1523/JNEUROSCI.4376-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu K., Zhang M.L., Liu S.T. Ghrelin attenuates retinal neuronal autophagy and apoptosis in an experimental rat glaucoma model. Invest Ophthalmol Vis Sci. 2017;58:6113–6122. doi: 10.1167/iovs.17-22465. [DOI] [PubMed] [Google Scholar]
- 25.Justus C.R., Leffler N., Ruiz-Echevarria M., Yang L.V. In vitro cell migration and invasion assays. J Vis Exp. 2014:51046. doi: 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeCicco-Skinner K.L., Henry G.H., Cataisson C. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J Vis Exp. 2014:e51312. doi: 10.3791/51312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou J.C., Rollins S.D., Fawzi A.A. Trypsin digest protocol to analyze the retinal vasculature of a mouse model. J Vis Exp. 2013:e50489. doi: 10.3791/50489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang T., Chang Q., Cai J., Fan J., Zhang X., Xu G. Protective effects of melatonin on retinal inflammation and oxidative stress in experimental diabetic retinopathy. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/3528274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guillot N., Kollins D., Gilbert V. BAMBI regulates angiogenesis and endothelial homeostasis through modulation of alternative TGFbeta signaling. PLoS One. 2012;7:e39406. doi: 10.1371/journal.pone.0039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillot N., Kollins D., Badimon J.J., Schlondorff D., Hutter R. Accelerated reendothelialization, increased neovascularization and erythrocyte extravasation after arterial injury in BAMBI-/- mice. PLoS One. 2013;8:e58550. doi: 10.1371/journal.pone.0058550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 32.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K., Long B., Liu F. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 34.Zhang S.J., Chen X., Li C.P. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci. 2017;58:6500–6509. doi: 10.1167/iovs.17-22698. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Z.J., Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J., Xu W., Wang J., Wang K., Li P. The role and molecular mechanism of non-coding RNAs in pathological cardiac remodeling. Int J Mol Sci. 2017;18:608. doi: 10.3390/ijms18030608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang J., Gifford C.C., Samarakoon R., Higgins P.J. Deregulation of negative controls on TGF-beta1 signaling in tumor progression. Cancers (Basel) 2018;10:159. doi: 10.3390/cancers10060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marwitz S., Depner S., Dvornikov D. Downregulation of the TGFbeta pseudoreceptor BAMBI in non-small cell lung cancer enhances TGFbeta signaling and invasion. Cancer Res. 2016;76:3785–3801. doi: 10.1158/0008-5472.CAN-15-1326. [DOI] [PubMed] [Google Scholar]
- 39.Salles-Crawley I.I., Monkman J.H., Ahnstrom J., Lane D.A., Crawley J.T. Vessel wall BAMBI contributes to hemostasis and thrombus stability. Blood. 2014;123:2873–2881. doi: 10.1182/blood-2013-10-534024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan Y., Li X., Xiao W. BAMBI elimination enhances alternative TGF-beta signaling and glomerular dysfunction in diabetic mice. Diabetes. 2015;64:2220–2233. doi: 10.2337/db14-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ten Dijke P., Arthur H.M. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.