Abstract
The advancement in high-throughput sequencing technologies and systems biology approaches have revolutionized our understanding of biological systems and opened a new path to investigate unacknowledged biological phenomena. In parallel, the field of human microbiome research has greatly evolved and the relative contribution of the gut microbiome to health and disease have been systematically explored. This review provides an overview of the network-based and translational systems biology-based studies focusing on the function and composition of gut microbiota. We also discussed the association between the gut microbiome and the overall human physiology, as well as hepatic diseases and other metabolic disorders.
Keywords: Gut microbiome, Liver diseases, Host-microbiome interactions, Systems biology, Personalized medicine, Meta-omics, Biomarker, Metabolic models
Abbreviations
- ALD
alcoholic liver disease;
- BCAA
branched chain amino acids;
- CCL
CC chemokine ligand;
- cFBA
community Flux Balance Analysis;
- CASINO
Community And Systems-level Interactive Optimization;
- COMETS
Computation Of Microbial Ecosystems in Time and Space;
- dFBA
dynamic Flux Balance Analysis;
- DM
diabetes mellitus;
- DMMM
Dynamic Multi-species Metabolic Modeling;
- FLYCOP
FLexible sYnthetic Consortium Optimization;
- MCM
Microbial Community Modeler FXR, farnesoid X receptor; GEM, genome-scale metabolic model;
- HCC
hepatocellular carcinoma;
- IL
interleukin;
- MAPK
mitogen-activated protein kinase;
- MCM
Microbial Community Modeler; NAFLD, non-alcoholic fatty liver disease;
- NASH
non-alcoholic steatohepatitis;
- NASH
non-alcoholic steatohepatitis;
- NF-κB
nuclear factor-κB;
- SCFA
short-chain fatty acids;
- T2DM
type 2 diabetes mellitus;
- TGR5
transmembrane G protein-coupled receptor 5 (also known as GPBAR1);
- TLR
toll-like receptor.
1. Introduction
The human gastrointestinal tract is inhabited by a complex microbial community comprising more than a trillion cells of approximately 1800 genera [1]. There is increasing evidence that this diverse microbial habitat has an important contribution to the metabolism of dietary components and overall regulation of health status. This has triggered a large amount of scientific interest into the investigation of microbiota and derived products, and the recognition of metabolic links, especially along the liver and gut bidirectional relationship [2,3].
The human host benefits from the metabolism of the microorganisms in the human gut, such as degradation of dietary indigestible carbohydrates and peptides which are consequently absorbed by the host and serve as an energy source. The main end products of bacterial metabolism are short-chain fatty acids (SCFAs, namely acetate, propionate, and butyrate), branched-chain amino acids (BCAAs, namely leucine, isoleucine, and valine), tryptophan-derived metabolites (mainly indoles and tryptamine) and trimethylamine, whose association with human physiology and various diseases have been increasingly shown by recent studies [4], [5], [6]. Furthermore, the microbiota is also involved in the regulation of bile acids, which in turn, can modulate glucose and lipid metabolism through FXR and TGR5 signaling [7]. Diet, xenobiotics, the intestinal environment and microbiota composition affect metabolism of microbiome-derived metabolites; thus, identifying their exact roles in human physiology is challenging [8].
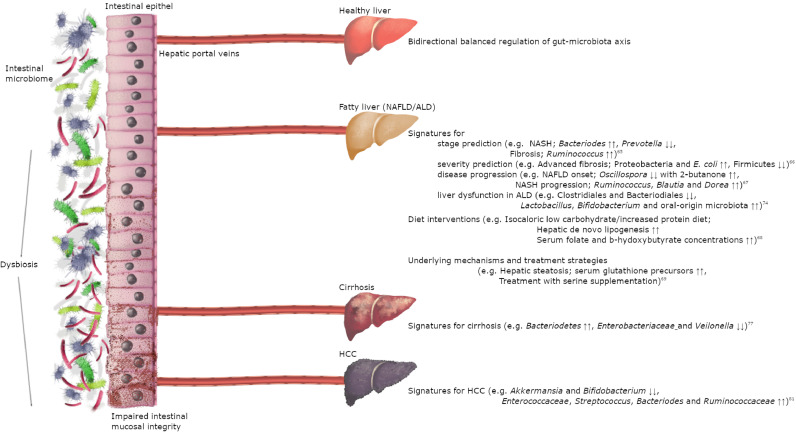
The gut microbiota also plays a crucial role in the overall health of the body through the maintenance of intestinal mucosal integrity, the synthesis of essential amino acids and vitamins, the bioconversion of dietary complex molecules, the biotransformation of oral drugs, and the production of hormones and neurotransmitters [9], [10], [11]. An imbalanced gut microbiome, with the contribution of host genetic characteristics and environmental factors (e.g. diet, drugs), may lead to the development of a range of immune-mediated diseases and conditions including diabetes mellitus (DM), obesity and various types of liver diseases (Fig. 1).
Fig. 1.
Recent systems biology studies elucidate the close connection between gut microbiota and liver. Upper part of the figure reflects the healthy state of liver-gut axis. Alterations in the microbial composition and impaired intestinal barrier elucidated as pathogenic factors in various types of liver diseases by recent systems biology studies. (NAFLD/ALD, non-alcoholic fatty liver disease/alcoholic liver disease; NASH, non-alcoholic hepatosteatosis; HCC, hepatocellular cancer).
The composition, functions, and interactions of the intestinal microbiota have been systematically studied by using novel technologies to elucidate the underlying mechanisms that might account for the pathological processes, with the purpose of prevention, diagnosis, and treatment of diseases [12]. The ultimate goal of this new era is to achieve personalized medicine, which will provide the most compatible treatment for a specific patient by increasing the efficacy of treatments, whilst reducing the adverse effects and health expenses. Therefore, data integration and knowledge discovery capabilities of novel bioinformatics methodologies have emerged as game-changing tools for the discovery of biomarkers and drug targets, as well as the development of efficient treatment strategies such as diet interventions and fecal microbial transplantation. [13,14]
From this perspective, we reviewed state-of-the-art systems biology studies in the context of how environmental changes affect microbiota function and composition, and in turn how this is associated with human physiology and liver diseases. Here, we: (1) describe the synthetic gut microbe studies that have evaluated the bacterial composition and microbiota-derived metabolites under varying conditions in human intestine; (2) summarize the systems biology studies elucidating the role of altered gut microbiota on human metabolism in healthy and obese subjects and diabetic patients, as well as the role of microbiota on drug biotransformation; and (3) outline the novel studies that provide evidence for the interplay between microbiota and hepatic diseases, including non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease (ALD), liver cirrhosis, and hepatocellular carcinoma (HCC), and its possible diagnostic and therapeutic potential.
2. Systems biology approaches in understanding gut microbial communities
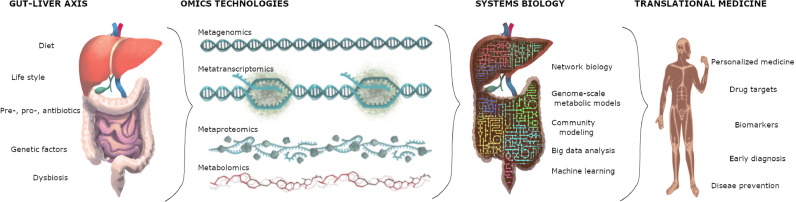
Advances in omics technologies have enabled gains in mechanistic insights of the human liver and gut microbiota in health and disease states. Omics technologies in microbiome studies are comprehensively described elsewhere [15], and are described briefly in Box 1. However, omics technologies, as a reductionist approach, have focused on describing complex biological systems in their simplest levels through concrete individual statements, but predicting the behavior of many interacting parts of systems has been highly improbable. Therefore, holistic approaches are necessary to make predictions for studying cellular and systemic functions (Fig. 2). The development and analysis of integrative biological networks can be used to make further predictions about the system-level properties through the use of genome-scale metabolic models (GEMs) [16]. GEMs are the stoichiometric reconstructions of the entire metabolism within a cell or tissue, which provide a link between genomic information and associated biochemical reactions [17]. GEMs have been presented and continuously refined to compile all gene–protein–reaction–metabolite associations with transport processes to simulate the complex relationship between the genotype and phenotype of an organism [18]. Systems biology-based studies that employ biological networks including GEMs, transcriptional regulatory, protein-protein interaction, signaling and co-expression networks in understanding the human liver physiology in healthy and diseased states have been extensively reviewed elsewhere [2].
Box 1. Omics technologies in microbiome studies.
Metagenomics
Metagenomic studies have already revealed novel insights into the diversity, population structure and dynamics of microbiota composition [83]. The linked between the functional genes of gut microorganisms and the pathogenesis of metabolic diseases, such as diabetes and obesity, are already well established [84]. Integrating omics data with biologic networks has been used in the identification of biomarkers for the development of detection and treatment strategies [85]. Although metagenomics provides useful tools for evidence-based studies, these approaches have bottlenecks in terms of high-quality annotation, assignment of functional information into uncharacterized community structures, and interpretation of comparative investigations, regardless of environmental conditions [86].
Metatranscriptomics
Actively expressed genes of the human gut microbiome enable an understanding of the potential functions of a microbial community and dynamic interactions with the host [87]. Next-generation sequencing technologies enable the identification of mRNA expression profiles, including novel non-coding RNAs such as small RNAs associated with central biologic processes [88]. The main limitation of this approach is the difficulty in the detection of bacterial mRNAs due to their short half-life on the order of minutes. In addition, assembly of non-continuous short-read sequences and repeated patterns renders the process even more problematic [89].
Metaproteomics
The determination of a complete profile of gene translation products expressed within a microbiome and their posttranslational alterations is necessary to elucidate the species involved in specific functions by assigning proteins to specific taxa [90]. Metaproteomics data integrated with computational workflows have been used to decipher active pathways in a microbial composition to explore the complex interactions of the human gut microbiome and the host [91]. As a result, metaproteomics provides new knowledge not garnered by metagenomics; however, standardized metaproteomics protocols are needed to compare studies and relate protein abundances to microbial functions [92].
Metabolomics
Analysis of the biochemical profiles of metabolism simultaneously implies the current physiology of microbial activities [93]. Measuring the compositions and concentrations of low-molecular-weight compounds in a state of flux is commonly used in the discovery of potential biomarkers and drug targets [94]. The identification of metabolite production is complicated in the context of mixed microbial communities, thus the combination of other omics technologies and stable isotope probing techniques with metabolomics has become highly useful [95].
Alt-text: Unlabelled box
Fig. 2.
Interactions between human gut and liver have been deciphered by various omics technologies. Systems biology methodologies integrate high-throughput omics data to develop high-quality translational research and personalized medicine.
Synthetic gut microbe networks decipher population interactions in multiple microbial species. Computational metabolic modeling of interspecies interactions predicts how members of the gut flora promote each other for growth and/or compete for space and nutrients [19]. Microbial relationships at different taxonomic levels have been simulated by using extended constraint-based metabolic flux analysis (e.g. OptCom, cFBA, CASINO, MMinte, SteadyCom) [20], [21], [22], [23], [24]. However, the dynamic equilibrium of the human microbiome challenges the methods that allow the prediction of interactions based on steady-state assumption. Similarly, the majority of flux prediction methods rely on a maximization of the biomass or ATP, but this is not the case for all members of the microbial community. Dynamic modeling frameworks (e.g. dFBA, DyMMM, d-optCom, COMETS, MCM, BacArena, FLYCOP) have been developed to achieve more realistic simulations of microbial ecosystems [25], [26], [27], [28], [29], [30], [31]. An overview of various community modeling frameworks is presented in Tables 1 and 2. A compilation of studies that illustrates environmental alterations on behaviors of gut microbial communities (with 4 or more species) is detailed in this review paper.
Table 1.
Summary of community modeling frameworks (steady-state) using genome-scale metabolic models.
| Name | Programming languages | Definition | Research organism | Availability | Reference |
|---|---|---|---|---|---|
| SteadyCom | MATLAB | Prediction of the flux distributions and maximum growth rate of a community (independent from number of organisms) in a time-averaged approach | 1. Bacteroides thetaiotaomicron 2. Eubacterium rectale 3. Faecalibacterium prausnitzii 4. Enterococcus faecalis 5. Lactobacillus casei 6. Streptococcus thermophilus 7. Bifidobacterium adolescentis 8. E. coli 9. Klebsiella pneumoniae |
https://github.com/maranasgroup/SteadyCom | Chan et al. [24]., |
| MMinte | Python | A compartment-based simulation of microbial interactions from an association network and assessment of 16S rDNA data | 1. Desulfovibrio piger 2. Bacteroides thetaiotaomicron 3. Bacteroides caccae 4. Bacteroides ovatus 5. Eubacterium rectale 6. Marvinbryantia formatexigens 7. Collinsella aerofaciens 8. E. coli 9. Clostridium symbiosium |
www.github.com/mendessoares/MMinte | Mendes-Soares et al. [23]., |
| CASINO | MATLAB | An optimization algorithm which incorporates the systems-level topology with iterative organism-level and multi-level optimization to predict metabolic interactions within the microbial communities | 1. Bifidobacterium adolescentis 2. Bacteroides thetaiotaomicron 3. Eubacterium rectale 4. Faecalibacterium prausnitzii 6. Lactobacillus reuteri |
– | Shoaie et al. [22]., |
| cFBA | Python | A methodology which predicts community metabolic activities at a balanced growth rate by using a simplified multi-objective optimization approach | E. coli | http://cbmpy.sourceforge.net/ | Khandewal et al. [21]., |
| optCom | UNIX/ LINUX | A pioneer multi-level and multi-objective optimization formulation to describe species- and community-level fitness analysis of microbial communities. | 1. Geobacter sulfurreducens 2. Clostridium cellulolyticum 3. Clostridium cellulolyticum 4. Methanococcus maripaludis |
– | Zomorrodi et al. [20]., |
Table 2.
Summary of community modeling frameworks (dynamic) using genome-scale metabolic models.
| Name | Programming languages | Definition | Research organism | Availability | Reference |
|---|---|---|---|---|---|
| FLYCOP | Python | A novel spatiotemporal modeling approach to explore multiple consortium configurations through stochastic local search process | 1. E. coli 2. Synechococcus elongatus 3. Pseudomonas putida |
https://github.com/beatrizgj/FLYCOP | Beatriz García-Jiménez et al. [31]., |
| BacArena | R | A rule-based spatial and temporal multi-scale modeling approach which combines FBA with individual-based modeling | 1. Anaerostipes caccae 2. Bacteroides thetaiotaomicron 3. Blautia producta 4. E. coli 5. Clostridium ramosum 6. Lactobacillus plantarum 7. Bifidobacterium longum 8. Akkermansia muciniphila 9. Pseudomonas aeruginosa |
https://github.com/euba/BacArena | Bauer et al. [30]., |
| MCM | UNIX/ LINUX | A dynamical framework for modeling microbial communities, which combines genome scale metabolic reconstructions with environmental variables and arbitrary reaction kinetics | E. coli | http://www.zoology.ubc.ca/MCM | Louca and Doebeli [29], |
| COMETS | UNIX/ LINUX | A multi-scale modeling framework that integrates spatiotemporal dynamics of microbial community with stoichiometric models | 1. E. coli 2. Salmonella enterica 3. Methylobacterium extorquens |
https://github.com/segrelab/comets | Harcombre et al. [28]., |
| d-optCom | UNIX/ LINUX | A multi-level and multi-objective simulation of microbial communities that incorporates the dynamic information of biomass concentrations and shared metabolites | 1. E. coli 2. Geobacter sulf urreducens 3. Rhodoferax ferrireducens 4. Shewanella oneidensis |
– | Zomorrodi et al. [27]., |
| DyMMM | MATLAB | A pioneer dynamic framework to integrate GEMs (add-on to the COBRA toolbox) | 1. Geobacter sulf urreducens 2. Rhodoferax ferrireducens |
https://sourceforge.net/projects/dymmm/ | Zhouang et al. [26]., |
Bacteria in the human gut microbiome inhabit the same physical location and connect instantly with each other. The sharing of a common living space with all microbial partners is regulated via metabolic cross-feeding. Predictions of metabolite production levels within the representative communities of human gut microbiota members have rendered efficient estimations of the human gut microbiota. For instance, applying a spatial and temporal multi-scale modeling approach (namely BacArena) to a sample group of seven bacteria of the human gut showed the importance of the different location-based densities of mucus glycans in terms of niche formations and ultimately, the habitat topology on the whole. The approach also demonstrated the multi-member exchange of SCFAs as a contributing factor to the concentration values of community members, which were in line with previously published experimental findings [30].
High-level methodological examination of microbial metabolic exchanges allows further discovery of microbial metabolic relationship factors relating to ecological stability and vulnerability. For instance, in silico interactions between Escherichia sp., Akkermansia muciniphila, Subdoligranulum variabile, and Intestinibacter bartlettii and their extracellular environment were evaluated with the concept of synthetic lethality analysis based on the flux balance analysis to elucidate the possible effect of metformin treatment on gut microbiota composition in patients with type 2 diabetes mellitus. Authors observed that Escherichia sp. and S. variabile were able to contribute to the production of short-chain fatty acids under aerobic and anaerobic conditions, and Escherichia sp. withstood most nutrient deficiency out of all species studied [32]. In another study, the growth interdependencies of Desulfovibrio piger between 8 other microbial species under distinct metabolic limitations (oxygen, chondroitin sulfate, and fructose) were evaluated using the MMinte framework. In this study, D. piger growth was dependent if Bacteroides ovatus, Bacteroides thetaiotaomicron, Bacteroides caccae, Clostridium symbiosium or Escherichia coli were under the absence of oxygen, but no dependence was observed if oxygen was available [23]. Similarly, a recent metabolic model-based approach of pairwise interactions of 11 gut bacteria suggested that cross-feeding behavior varied under different nutrient and atmospheric conditions of the gastrointestinal tract. For instance, the mutualistic interactions of Lactobacillus plantarum under anoxic conditions were abolished in the presence of oxygen [33]. Overall, these results may suggest that microbes rely on each other more when they are under poor nutrient and oxygen availability.
The commensal behavior of the intestinal microbes might be altered with the production and consumption of extracellular compounds under different diet conditions. The computational framework CASINO demonstrates the multifaceted anabolic and catabolic interactions amongst food intake, gut microbiota and host. The model, which accomplishes predictions in line with human values, reveals that various amino acid and SCFA levels, in addition to the microbiota composition, are impacted by microbial genome diversity and diet [22]. Likewise, metabolic flux arrangements in accordance with steady state boundaries can be estimated by using the SteadyCom optimization framework. Following an assembly of four E. coli double auxotrophic mutants, the framework was tested on a microbiota representation containing microbes from the phyla Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. With little need for constraints other than diet perturbation, SteadyCom accordingly generates predictions of abundance fluctuations, which are backed up by experimental gut microbiota representations [24].
Integration of omics data onto the metabolic models is a novel methodology for a definitive understanding of microbial functions and dynamics. A pioneer study from Shoaie et al., simulated the interactions between relevant representatives (B. thetaiotamicron, Eubacterium rectale and Methanobrevibacter smithii) of the human gut by using genome-scale metabolic models based on transcriptome data [34]. The models predicted that B. thetaiotaomicron produced more butyrate with E. rectale and more acetate with M. smithii, which resulted in more methane production of M. smithii. Predictions of the secreted SCFA profiles with in silico evaluations in different combinations of gut ecosystems were comparable with the experimental data of germ-free mice [34]. Nutrient cross-feeding between Bifidobacterium adolescentis and Faecalibacterium prausnitzii has also been explored by different groups and findings consistently indicate that SCFAs are modulated by environmental conditions, as well as microbial interactions [35,36].
3. Role of altered gut microbiota on human metabolism
To date, data from observational and comparative studies have highlighted that gut microbiota metabolism and the human liver are closely connected with each other via enterohepatic circulation, and the results of this bidirectional relation have been linked to overall health and disease [37], [38], [39]. The holistic approaches of systems biology have been gaining attention in medical management to promote health and prevent disease, which in turn renders possible the practice of predictive, preventive, personalized and participatory (P4) medicine [40]. Examples of systems biology studies highlighting the effect of environmental factors (e.g. diet, drugs) on the microbiota and the subsequent impact on host health are included in this section.
3.1. Health and microbiota
Understanding the role of the microbiota on well-being and disease necessitates the use of data mining and integration approaches as part of the current comprehensive view towards systems biology. As a seminal example, Price et al. analyzed the personal and multi-omics longitudinal data of 108 individuals and employed a correlation network to define related markers of health and disease. The authors identified several relationships between specific taxa and metabolites, as well as negative correlation of microbiome α-diversity with some immune response proteins [41]. In another study, statistical analysis of an early school-age cohort of Dutch children revealed that environmental factors (especially breastfeeding duration and dietary habits) influenced microbial composition with a significant enterotype association. High dietary fiber consumption and low plasma insulin levels were correlated with Bacteroides and Prevotella enterotypes, but not with Bifidobacterium enterotype, which has a lower microbial diversity and association with a shorter breastfeeding duration [42].
The essential role of diet in well-being or complex diseases has gained importance and landmark computational studies have evaluated the association of microbiota profiles with disease development [43,44]. Mardinoglu et al. reconstructed a generic mouse metabolic reaction GEM, together with 28 tissue-specific and 4 functional GEMs for the small intestine, colon, liver and white adipose tissues based on proteomics and transcriptomics data from conventionally raised and germ-free mice. The authors simulated the effect of the microbiota on host metabolism and highlighted that the gut microbiome leads to a depletion of host glutathione synthesis, in addition to regulating host amino acid and lipid metabolism [45].
3.2. Metabolic conditions and microbiota
The advent of omics technologies enabled the identification of key traits of microbiota associated with diet and obesity. Combined analysis of multiple omics data from a longitudinal weight perturbation study revealed that weight changes had extensive molecular signatures of chronic diseases such as hypertrophic cardiomyopathy and insulin resistance [46]. The pathophysiologic role of adipose tissue on the progression of obesity is well established; white adipose tissue can be a rational target to prevent obesity and related disorders. The generation of GEMs for adipocytes and the integration of gene expression and plasma metabolomics data onto the model enabled mechanistic explanations of the metabolic differences between lean and obese individuals. Model predictions (e.g. decreased glutaminolysis and alterations in the glutamate, pyruvate, and α-ketoglutarate metabolism) were consistent with the results from human subjects [47]. Moreover, a recent study revealed that energy-dense diets altered the diversity of the microbial composition and increased mucosa permeability of the ileum and colon in obese mice with NAFLD. The authors also found that plasma SCFA levels were correlated with specific groups of bacteria, and specific bacterial taxa associated with disease-associated factors were also positively correlated with isoacid SCFAs. In addition, dietary fermentable fibers were found to alter microbiota-derived signals and regulate the gene expression and metabolic pathways of the liver by reducing host nitrogen and amino acid homeostasis [48].
Diabetes mellitus (DM) is a major public health concern, affecting over 425 million people worldwide [49]. Increasing evidence indicates a contributing role of gut microbiome in the pathophysiology of DM. An integrative taxonomic and functional analysis of multiple meta-omics data revealed that intra- and inter-individual variation of microbiota composition in the context of type 1 DM were strongly affected by family membership [50]. Similarly, two large cohorts of healthy individuals showed that gut microbiome composition was more affected by environmental influences (e.g. household sharing, diet, lifestyle) instead of host genetics [51]. Several predictive metagenomic tools described an association with obesity, insulin resistance and T2DM. Gut microbiome 16S rRNA sequencing and metagenomics profiling of T2DM zebrafish model revealed similar features with human T2DM, such as lower taxa richness, higher faecal and plasma fructose and BCAA concentrations, reduced fecal butyrate levels, and altered pathways of amino acid and sugar metabolism [52].
3.3. Drug metabolism and microbiota
The bidirectional interaction of gut microbiota and drug metabolism have been reported for more than 50 pharmaceuticals [53]. Gut microbiome composition has recently been associated with the efficacy and toxicity profiles of commonly used drugs, but the extent of this relationship remains largely unknown. Drug–microbe networks serve as a novel step towards revealing the role of the microbiome in drug metabolism. For instance, Zimmerman et al. identified 30 human gut microbiome-encoded enzymes responsible for the biotransformation of 20 drugs to 59 candidate metabolites, which suggests that drug-metabolizing activities of human gut microbiota may differ on the basis of interpersonal variation of microbial genomic contents [54]. In another comprehensive study, screening of more than 1000 marketed non-antibiotic drugs against the growth of gut bacterial strains revealed that 24% of the drugs showed antibiotic-like effects and the inhibition of bacterial growth was strongly correlated between non-antibiotic and antibiotic drugs, which implies antibiotic resistance on the basis of gut microbiome composition, after regular consumption of non-antibiotic drugs [55]. Similarly, a recent study proposed an interplay between decreased Bacteroides fragilis increased glycoursodeoxycholic acid – inhibition of intestinal FXR signaling; which might explain how metformin improves hyperglycemia. Hence, this study identified glycoursodeoxycholic acid as a potential target for the treatment of T2D [56]. The findings of this study are concordant with accumulating evidence that associates the antihyperglycemic effect of metformin being modulated by gut microbiota, which was recently proven in double-blind metagenomics and targeted metabolomics research [57].
4. Role of microbiota in hepatic diseases
In the context that the enterohepatic circulation links the gut microbiota and liver through the transport of the intermediate end-products of microbial metabolism, efforts to uncover the potential physiological impact of the gut microbiome on liver damage have lately been increased. In recent years, several lines of human and animal research indicated that the gut microbiome represented a significant environmental factor that contributed to the development of several liver diseases and its progression into end-stage cirrhosis, as well as cancer [58,59]. In addition to prompting the use of microbiome-based approaches for the diagnosis of liver diseases, evidence also shows that this may be a primary target for the treatment of diseases. Taking advantage of the individual gut microbiota makeup, which can be manipulated via a range of methods, selection of suitable fecal mass transplant donors and designation of potential effective pro- and/or prebiotics can be achieved so as to alleviate the pathologies as well as conceivably devise personalized therapies for various liver diseases.
4.1. Non-alcoholic fatty liver disease
NAFLD has been defined as global burden because it is the leading cause of chronic liver disease and affects around 25% of the global population [60]. The pathogenesis of NAFLD begins with the simple accumulation of lipid in hepatocytes and progresses to hepatocellular damage and inflammation (non-alcoholic steatohepatitis, NASH), which can progress to cirrhosis [61]. The pathophysiology of NAFLD has not yet been elucidated, but several human and animal studies have confirmed the contribution of intestinal microbiota as a driver through the compositional changes, altered microbiota-derived metabolites, and impaired gut-barrier integrity [62], [63], [64]. A definite understanding of the molecular basis of the gut and liver bidirectional relation is a prerequisite for developing non-invasive approaches with a robust discriminative ability for determining the presence and severity of NAFLD, as well as treating NAFLD and NASH by the use of precision pharmacotherapies.
Despite the common use of advanced fibrosis as the primary determinant in predicting liver destruction, its rigorous detection by way of using markers associated with gut microbiota lacks extensive and concordant data. For this aim, a study in a biopsy-proven population of adult patients with NAFLD evaluated the association between gut dysbiosis and the severity of NAFLD lesions by using 16S rRNA gene sequencing of stool samples. Multivariate analysis of the results indicated that Bacteroides abundance and Prevotella depletion were observed in people with NASH and Ruminococcus abundance was associated with fibrosis [65]. Likewise, accurate stage prediction of liver disease (from mild/moderate NAFLD to advanced fibrosis) was characterized using a panel of intestinal microbiota-derived signatures. Metagenomics of stool microbiome and serum metabolome analysis data were used to build a random forest classifier model with a set of 40 features, with which advanced fibrosis was distinguished by an increased abundance of Proteobacteria and E. coli and a decreased abundance in Firmicutes [66]. Another multiomics study that claims specific gut microbiota states signal to pathologic conditions in differing stages is supported with a metagenomics and metabolomics study of pediatric patients with NAFLD. The findings suggested that a decrease in Oscillospira coupled with 2-butanone enrichment was found to be a microbiome signature for NAFLD onset and Ruminococcus, Blautia, and Dorea were significantly increased with NASH progression [67].
Integration of high-quality genome-scale metabolic models with multi-omics data of patients has been of considerable interest in exploring diet–microbiota interactions to understand the pathogenesis and prevention of NAFLD. In a recent study, authors showed the dramatic benefits of an isocaloric low-carbohydrate/increased protein diet in obese subjects with NAFLD. The researchers performed in-depth multi-omics profiling (included plasma metabolomics and liver transcriptomics) after a short-term dietary intervention and combined the data with a genome-scale metabolic model of hepatocytes. They observed rapid and marked reductions in hepatic de novo lipogenesis, augmented serum b-hydroxybutyrate concentrations compatible with mitochondrial beta-oxidation, and significant microbial changes toward folate-producing Streptococcus resulting in increased serum folate concentrations. Overall, the results indicated that carbohydrate-restricted diets shaped the gut microbiome composition and held potential for the treatment of NAFLD [68]. Another comprehensive study investigated the underlying metabolic differences in NAFLD using systems-level approaches coupled with mice experiments and a proof-of-concept human study. The investigators integrated the metabolomic measurements of each subject with a liver GEM to simulate individual liver metabolism. Their analysis revealed that plasma levels of glycine, serine, betaine, and N-acetyl-glycine (precursors for glutathione and NAD+ biosynthesis) were negatively correlated in subjects with high degrees of hepatic steatosis, and dietary serine supplementation was likely an effective treatment strategy [69].
4.2. Alcoholic liver disease
ALD is damage to the liver caused by excess alcohol intake. The spectrum of disease ranges from fatty liver, to hepatitis and cirrhosis. Misuse of alcohol affects the composition and function of gut microbiota, which could initiate or potentiate liver disease [70]. Several animal studies indicated that composition of gut microbiota might be responsible for the consequences of ALD including raised liver inflammation, weakened immune system, and alterations in microbial metabolism products [71], [72], [73]. Correspondingly, shotgun metagenomes of patients with alcohol dependence and liver dysfunction were associated with depletion of many commensal gut taxa and community shifts, including the reduction of multiple Clostridiales and Bacteroidales members, but enrichment in Bifidobacterium and Lactobacillus and oral-origin microbiota. Molecular changes in the course of ALD encompass bacterial overgrowth due to sparsity of the geni Blautia, Lachnospiraceae, Faecalibacterium and Roseburia, and raised serum concentrations of endotoxins that leak from the gut wall in the presence of acetaldehyde [74].
Computational modeling and manipulation of the ALD network provides the means to form sound estimations regarding the chief inflammatory cascades and other molecular interactions active in this disease. For instance, interactome and transcriptome data of ALD were used to reconstruct static and dynamic networks that were consistent with the important role of key signaling pathways (TLR4, NF-κB, MAPK and apoptosis) in ALD. The findings of this study allow for the emergence of varied classes of data, enabling the representation of molecular networks and signaling cascades, and thus paving the way for new drugs targeting ALD [75].
4.3. Liver cirrhosis
Cirrhosis is scarring of the hepatic tissue, mainly seen at the late stages of chronic liver disease. Alterations of gut microbiota composition and function is significant in liver cirrhosis, but the underlying mechanisms are still unresolved. Disruption of the intestinal barrier by the altered gut microbiome and systemic inflammation with bacterial products are the proposed key players in the advancement and related complications of liver cirrhosis [76].
Combined metagenomic and metabolic assays could provide significant advantages to decipher the physiology of gut microbiome in cirrhosis states. For instance, a comparison of fecal microbiota of patients with cirrhosis and controls revealed a decrease in Bacteroidetes and enrichment in Enterobacteriaceae and Veillonella. This functional diversity resulted in an enrichment in the metabolism of toxins depredation and nutrient absorption, but also a depletion of bile acid and cell cycle-related metabolism [77]. Likewise, a recent meta-omics–based study directly linked microbial dynamics to liver cirrhosis through metabolites by reporting a significant decrease of bacteria involved in the digestion of non-starch polysaccharides (e.g. Alistipes sp.HG5, Clostridium thermocellum) and butyrate-producing bacteria, and a marked increase of opportunistic pathogens during disease progression. This impaired homeostasis of the gut may be responsible for the disorganized intestinal barrier, which allows endotoxins and pathogens into the hepatic circulation, resulting in systemic inflammation [78].
Systems biology approaches of human gut microbiota are promising tools for the estimation of disease progression. For instance, Bajaj et al. described a quantitative index of dysbiosis accompanying cirrhosis severity in a comprehensive, well-characterized population within a range of individuals from healthy controls to those with end-stage cirrhosis. The cirrhosis dysbiosis ratio reflects changes of autochthonous to non-autochthonous taxa such as the negative correlation with disease progression as well as endotoxemia; however, the ratio was stable if disease remained unchanged, which may be useful in clinical practice to evaluate changes in microbiome accompanying cirrhosis progression [79].
4.4. Hepatocellular carcinoma
HCC is the most common primary malignancy of the liver. Accumulating evidence suggests that the gut microbiota has been involved in the pathogenesis of HCC. Advancement of chronic liver disease to HCC has been linked to inflammatory pathways, which are activated by the disruption of intestinal mucosa and translocation of endotoxins to the portal veins as a result of imbalanced gut microbiota [80]. As novel evidence, a correlation model of the features (such as microbiota profile, intestinal permeability, inflammatory status, and circulating mononuclear cells), which possibly link microbiota with HCC, demonstrated that Enterococaceae, Streptococcus, Bacteroides, and Ruminococcaceae were increased in patients with HCC, whereas Akkermansia and Bifidobacterium were reduced. Plasma levels of inflammatory markers (IL8, IL13, CCL3, CCL4, and CCL5) and circulating monocytes were higher in the HCC group. These findings propose that replacement of mucosa protective bacteria with LPS-producing bacteria is associated with intestinal and systemic inflammation and may promote the development of HCC [81].
By reason of symptoms being nonspecific in initial stages, there is an urgent need for novel diagnostic and therapeutic targets for HCC. Alterations of gut microbiota will likely provide a potential biomarker for the prediction of early stage HCC. A systematic investigation of microbiota in early HCC across a large clinical cohort from three different regions of China demonstrated a depletion of Akkermansia and butyrate-producing bacteria (e.g. Ruminococcus, Oscillibacter, Faecalibacterium, Clostridium IV, and Coprococcus), and enrichment of gram-negative species (e.g. Klebsiella and Haemophilus). The authors proposed 30 biomarkers identified by random forest models that might be used as non-invasive diagnostic tools of HCC through further validation of these results in different cohorts from different countries and ethnicities [82].
5. Conclusion and outstanding questions
The interactions between the gut microbiome and human host metabolism has received increasing attention in the context of understanding the potential role of the gut microbiome in health and various complex conditions including obesity, DM and liver diseases. Regardless of the fact that increasing evidence from numerous studies suggest that gut microbiota contributes to overall health and disease, one should be careful whilst interpreting the results; (1) the strict description of a healthy microbiota is still obscure due to the taxonomic composition and the definitive interactions between gut microorganisms shared among healthy individuals are not yet well-characterized; (2) considering the complex ecosystem of the microbiome is ever-changing, determination of the relevant measures of functional capacity and stability are affected by multiple factors including environmental and host-related influences; (3) the genes in the microbiome were mapped to their functions; however, the genetic information may not always be reflected in the phenotype or may not perform the expected function; (4) although the taxonomic profile of the human gut microbiome is not affected in the long term, the dynamic structure of the microbiota makes it difficult to predict the outcome of dietary interventions, and the long-term results of these interventions are unknown; (5) the species-level findings in the gut microbiota research are not consistent, which may be a consequence of the study-related discrepancies, such as patient cohorts, comparison groups, definition of disease; (6) the reproducibility of results is low and may result in bias in the interpretation of results because sample collection, sequencing, and analysis of high-throughput methodologies are not standardized; (7) systems biology methodologies have proven to be a valuable tool in individual cells and tissues, but the computational models for bacteria and the algorithms developed for understanding microbial communities are still in their infancy. Nevertheless, through the advancement in high throughput technologies and methods that are used in system biology-based studies, the effect of the gut microbiome on human health may be better understood, which could lead to the development of preventive and personalized treatment strategies.
Search strategy and selection criteria
Data for this review were identified by searches of PubMed, Google Scholar, and references from relevant articles using the search terms “microbiome”, “microbiota”, “liver”, “omics”, “biomarker”, “personalized medicine”, and “genome-scale metabolic model”. Only articles published in English between 2010 and 2019 were included, with a preference for those published after 2016.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We would like to thank Stephen Doran and Simon Lam of King's College London for editorial comments. This work was supported by Knut and Alice Wallenberg Foundation.
Contributor Information
Jens Nielsen, Email: nielsenj@chalmers.se.
Mathias Uhlen, Email: mathias.uhlen@scilifelab.se.
Jan Boren, Email: Jan.Boren@wlab.gu.se.
Adil Mardinoglu, Email: adilm@scilifelab.se.
References
- 1.Stilling R.M., Bordenstein S.R., Dinan T.G., Cryan J.F. Friends with social benefits: host-microbe interactions as a driver of brain evolution and development? Front Cell Infect Microbiol. 2014;29(4):147. doi: 10.3389/fcimb.2014.00147. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardinoglu A., Boren J., Smith U., Uhlen M., Nielsen J. Systems biology in hepatology: approaches and applications. Nat Rev Gastroenterol Hepatol. 2018;15(6):365–377. doi: 10.1038/s41575-018-0007-8. [DOI] [PubMed] [Google Scholar]
- 3.Human Microbiome Project C. structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canfora E.E., Meex R.C.R., Venema K., Blaak E.E. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019May;15(5):261–73. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed]
- 5.Lynch C.J., Adams S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol. 2014;10(12):723–736. doi: 10.1038/nrendo.2014.171. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Zhao Y. Gut microbiota derived metabolites in cardiovascular health and disease. Protein Cell. 2018;9(5):416–431. doi: 10.1007/s13238-018-0549-0. MayEpub 2018 May 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia W., Xie G., Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15(2):111–128. doi: 10.1038/nrgastro.2017.119. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. SepEpub 2013 Jul 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macpherson A.J., Heikenwalder M., Ganal-Vonarburg S.C. The liver at the nexus of host-microbial interactions. Cell Host Microbe. 2016;20(5):561–571. doi: 10.1016/j.chom.2016.10.016. Nov 9. [DOI] [PubMed] [Google Scholar]
- 10.De Vadder F., Grasset E., Mannerås Holm L., Karsenty G., Macpherson A.J., Olofsson L.E., Bäckhed F. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci U S A. 2018;115(25):6458–6463. doi: 10.1073/pnas.1720017115. Jun 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crommen S., Simon M.C. Microbial regulation of glucose metabolism and insulin resistance. Genes (Basel) 2017;9(1) doi: 10.3390/genes9010010. Dec 29pii: E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashyap P.C., Chia N., Nelson H., Segal E., Elinav E. Microbiome at the frontier of personalized medicine. Mayo Clin Proc. 2017;92(12):1855–1864. doi: 10.1016/j.mayocp.2017.10.004. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaffer M., Armstrong A.J.S., Phelan V.V., Reisdorph N., Lozupone C.A. Microbiome and metabolome data integration provides insight into health and disease. Transl Res. 2017;189:51–64. doi: 10.1016/j.trsl.2017.07.001. NovEpub 2017 Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunkwall L., Orho-Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia. 2017;60(6):943–951. doi: 10.1007/s00125-017-4278-3. JunEpub 2017 Apr 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight R., Vrbanac A., Taylor B.C. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. Jul. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen J. Systems biology of metabolism: a driver for developing personalized and precision medicine. Cell Metab. 2017;25(3):572–579. doi: 10.1016/j.cmet.2017.02.002. Mar 7. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson A., Mardinoglu A., Nielsen J. Predicting growth of the healthy infant using a genome scale metabolic model. NPJ Syst Biol Appl. 2017;3:3. doi: 10.1038/s41540-017-0004-5. Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C., Hua Q. Applications of genome-scale metabolic models in biotechnology and systems medicine. Front Physiol. 2016;6:413. doi: 10.3389/fphys.2015.00413. Jan 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zomorrodi A.R., Segrè D. Synthetic ecology of microbes: mathematical models and applications. J Mol Biol. 2016;428(5 Pt B):837–861. doi: 10.1016/j.jmb.2015.10.019. Feb 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zomorrodi A.R., Maranas CD. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS Comput Biol. 2012 Feb;8(2) doi: 10.1371/journal.pcbi.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khandelwal R.A., Olivier B.G., Röling W.F., Teusink B., Bruggeman F.J. Community flux balance analysis for microbial consortia at balanced growth. PLoS ONE. 2013;8(5):e64567. doi: 10.1371/journal.pone.0064567. May 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoaie S., Ghaffari P., Kovatcheva-Datchary P., Mardinoglu A., Sen P., Pujos-Guillot E. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell Metab. 2015;22(2):320–331. doi: 10.1016/j.cmet.2015.07.001. Aug 4. [DOI] [PubMed] [Google Scholar]
- 23.Mendes-Soares H., Mundy M., Soares L.M., Chia N. MMinte: an application for predicting metabolic interactions among the microbial species in a community. BMC Bioinformatics. 2016;17(1):343. doi: 10.1186/s12859-016-1230-3. Sep 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan S.H.J., Simons M.N., Maranas C.D. SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput Biol. 2017;13(5) doi: 10.1371/journal.pcbi.1005539. May 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanly T.J., Henson M.A. Dynamic flux balance modeling of microbial co-cultures for efficient batch fermentation of glucose and xylose mixtures. Biotechnol Bioeng. 2011;108(2):376–385. doi: 10.1002/bit.22954. Feb. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang K., Izallalen M., Mouser P., Richter H., Risso C., Mahadevan R., Lovley D.R. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 2011;5(2):305–316. doi: 10.1038/ismej.2010.117. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zomorrodi A.R., Islam M.M., Maranas C.D. d-OptCom: dynamic multi-level and multi-objective metabolic modeling of microbial communities. ACS Synth Biol. 2014;3(4):247–257. doi: 10.1021/sb4001307. Apr 18. [DOI] [PubMed] [Google Scholar]
- 28.Harcombe W.R., Riehl W.J., Dukovski I., Granger B.R., Betts A., Lang A.H. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7(4):1104–1115. doi: 10.1016/j.celrep.2014.03.070. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louca S., Doebeli M. Calibration and analysis of genome-based models for microbial ecology. Elife. 2015;4:e08208. doi: 10.7554/eLife.08208. Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer E., Zimmermann J., Baldini F., Thiele I., Kaleta C. BacArena: individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS Comput Biol. 2017;13(5) doi: 10.1371/journal.pcbi.1005544. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Jiménez B., García J.L., Nogales J. FLYCOP: metabolic modeling-based analysis and engineering microbial communities. Bioinformatics. 2018;34(17):i954–i963. doi: 10.1093/bioinformatics/bty561. Sep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosario D., Benfeitas R., Bidkhori G., Zhang C., Uhlen M., Shoaie S., Mardinoglu A. Understanding the representative gut microbiota dysbiosis in metformin-treated type 2 diabetes patients using genome-scale metabolic modeling. Front Physiol. 2018;9:775. doi: 10.3389/fphys.2018.00775. Jun 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinken A., Thiele I. Anoxic conditions promote species-specific mutualism between gut microbes in silico. Appl Environ Microbiol. 2015;81(12):4049–4061. doi: 10.1128/AEM.00101-15. Jun 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoaie S., Karlsson F., Mardinoglu A., Nookaew I., Bordel S., Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci Rep. 2013;3:2532. doi: 10.1038/srep02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rios-Covián D., Gueimonde M., Duncan S.H., Flint H.J., De Los Reyes-Gavilán C.G. Enhanced butyrate formation by cross-feeding between faecalibacterium prausnitzii and bifidobacterium adolescentis. FEMS Microbiol. Lett. 2015;362 doi: 10.1093/femsle/fnv176. fnv176. [DOI] [PubMed] [Google Scholar]
- 36.El-Semman I.E., Karlsson F.H., Shoaie S., Nookaew I., Soliman T.H., Nielsen J. Genome-scale metabolic reconstructions of bifidobacterium adolescentis L2-32 and faecalibacterium prausnitzii A2-165 and their interaction. BMC Syst Biol. 2014;8:41. doi: 10.1186/1752-0509-8-41. Apr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng X., Li S., Li Y., Gan R.Y., Li H.B. Gut microbiota's relationship with liver disease and role in hepatoprotection by dietary natural products and probiotics. Nutrients. 2018;10(10):E1457. doi: 10.3390/nu10101457. Oct 8pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adolph T.E., Grander C., Moschen A.R., Tilg H. Liver-microbiome axis in health and disease. Trends Immunol. 2018;39(9):712–723. doi: 10.1016/j.it.2018.05.002. Sep. [DOI] [PubMed] [Google Scholar]
- 39.Boulangé C.L., Neves A.L., Chilloux J., Nicholson J.K., Dumas M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8(1):42. doi: 10.1186/s13073-016-0303-2. Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hood L., Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol. 2012;29(6):613–624. doi: 10.1016/j.nbt.2012.03.004. Sep 15Epub 2012 Mar 18. [DOI] [PubMed] [Google Scholar]
- 41.Price N.D., Magis A.T., Earls J.C., Glusman G., Levy R., Lausted C. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat Biotechnol. 2017;35(8):747–756. doi: 10.1038/nbt.3870. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong H., Penders J., Shi Z., Ren H., Cai K., Fang C. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome. 2019;7(1):2. doi: 10.1186/s40168-018-0608-z. Jan 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonnenburg J.L., Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535(7610):56–64. doi: 10.1038/nature18846. Jul 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh R.K., Chang H.W., Yan D., Lee K.M., Ucmak D., Wong K. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15(1):73. doi: 10.1186/s12967-017-1175-y. Apr 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mardinoglu A., Shoaie S., Bergentall M., Ghaffari P., Zhang C., Larsson E. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol. 2015;11(10):834. doi: 10.15252/msb.20156487. Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piening B.D., Zhou W., Contrepois K., Röst H., Gu Urban G.J., Mishra T. Integrative personal omics profiles during periods of weight gain and loss. Cell Syst. 2018;6(2):157–170. doi: 10.1016/j.cels.2017.12.013. Feb 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mardinoglu A., Kampf C., Asplund A., Fagerberg L., Hallström B.M., Edlund K. Defining the human adipose tissue proteome to reveal metabolic alterations in obesity. J Proteome Res. 2014;13(11):5106–5119. doi: 10.1021/pr500586e. Nov 7. [DOI] [PubMed] [Google Scholar]
- 48.Kieffer D.A., Piccolo B.D., Marco M.L., Kim E.B., Goodson M.L., Keenan M.J. Mice fed a high-fat diet supplemented with resistant starch display marked shifts in the liver metabolome concurrent with altered gut bacteria. J Nutr. 2016;146(12):2476–2490. doi: 10.3945/jn.116.238931. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toniolo A., Cassani G., Puggioni A., Rossi A., Colombo A., Onodera T. The diabetes pandemic and associated infections: suggestions for clinical microbiology. Rev Med Microbiol. 2019;30(1):1–17. doi: 10.1097/MRM.0000000000000155. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heintz-Buschart A., May P., Laczny C.C., Lebrun L.A., Bellora C., Krishna A. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat Microbiol. 2016;2:16180. doi: 10.1038/nmicrobiol.2016.180. Oct 10. [DOI] [PubMed] [Google Scholar]
- 51.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. Mar 8. [DOI] [PubMed] [Google Scholar]
- 52.Okazaki F., Zang L., Nakayama H., Chen Z., Gao Z.J., Chiba H. Microbiome alteration in type 2 diabetes mellitus model of zebrafish. Sci Rep. 2019;9(1):867. doi: 10.1038/s41598-018-37242-x. Jan 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spanogiannopoulos P., Bess E.N., Carmody R.N., Turnbaugh P.J. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol. 2016;14(5):273–287. doi: 10.1038/nrmicro.2016.17. AprEpub 2016 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A.L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570(7762):462–467. doi: 10.1038/s41586-019-1291-3. JunEpub 2019 Jun 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maier L., Pruteanu M., Kuhn M. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555(7698):623–628. doi: 10.1038/nature25979. Mar 29Epub 2018 Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L., Xie C., Wang G. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat Med. 2018;24(12):1919–1929. doi: 10.1038/s41591-018-0222-4. DecEpub 2018 Nov 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu H., Esteve E., Tremaroli V. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. doi: 10.1038/nm.4345. JulEpub 2017 May 22. [DOI] [PubMed] [Google Scholar]
- 58.Tripathi A., Debelius J., Brenner D.A. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. doi: 10.1038/s41575-018-0011-z. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Safari Z., Gérard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell Mol Life Sci. 2019;76(8):1541–1558. doi: 10.1007/s00018-019-03011-w. AprEpub 2019 Jan 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease- Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. Jul. [DOI] [PubMed] [Google Scholar]
- 61.Lovric A., Granér M., Bjornson E., Arif M., Benfeitas R., Nyman K. Characterization of different fat depots in NAFLD using inflammation-associated proteome, lipidome and metabolome. Sci Rep. 2018;8(1):14200. doi: 10.1038/s41598-018-31865-w. Sep 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Roy T., Llopis M., Lepage P., Bruneau A., Rabot S., Bevilacqua C. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- 63.Lambert J.E., Parnell J.A., Eksteen B., Raman M., Bomhof M.R., Rioux K.P. Gut microbiota manipulation with prebiotics in patients with non-alcoholic fatty liver disease: a randomized controlled trial protocol. Gut. 2013;62(12):1787–1794. doi: 10.1186/s12876-015-0400-5. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engstler A.J., Aumiller T., Degen C., Dürr M., Weiss E., Maier I.B. Insulin resistance alters hepatic ethanol metabolism: studies in mice and children with non-alcoholic fatty liver disease. Gut. 2016;65(9):1564–1571. doi: 10.1136/gutjnl-2014-308379. Sep. [DOI] [PubMed] [Google Scholar]
- 65.Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63(3):764–775. doi: 10.1002/hep.28356. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loomba R., Seguritan V., Li W., Long T., Klitgord N., Bhatt A. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25(5) doi: 10.1016/j.cmet.2017.04.001. May 21054-1062.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Del Chierico F., Nobili V., Vernocchi P., Russo A., Stefanis C., Gnani D. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65(2):451–464. doi: 10.1002/hep.28572. Feb. [DOI] [PubMed] [Google Scholar]
- 68.Mardinoglu A., Wu H., Bjornson E., Zhang C., Hakkarainen A., Räsänen S.M. An integrated understanding of the rapid metabolic benefits of a carbohydrate-restricted diet on hepatic steatosis in humans. Cell Metab. 2018;27(3) doi: 10.1016/j.cmet.2018.01.005. Mar 6559-571.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mardinoglu A., Bjornson E., Zhang C., Klevstig M., Söderlund S., Ståhlman M. Personal model-assisted identification of NAD+ and glutathione metabolism as intervention target in NAFLD. Mol Syst Biol. 2017;13(3):916. doi: 10.15252/msb.20167422. Mar 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol. 2019;16(4):235–246. doi: 10.1038/s41575-018-0099-1. Apr. [DOI] [PubMed] [Google Scholar]
- 71.Sarin S.K., Pande A., Schnabl B. Microbiome as a therapeutic target in alcohol-related liver disease. J Hepatol. 2019;70(2):260–272. doi: 10.1016/j.jhep.2018.10.019. Feb. [DOI] [PubMed] [Google Scholar]
- 72.Wang H., Yan Y., Yi X. Histopathological features and composition of gut microbiota in rhesus monkey of alcoholic liver disease. Front Microbiol. 2019;10:165. doi: 10.3389/fmicb.2019.00165. Feb 8eCollection 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciocan D Rebours V., Voican C.S. Characterization of intestinal microbiota in alcoholic patients with and without alcoholic hepatitis or chronic alcoholic pancreatitis. Sci Rep. 2018;8(1):4822. doi: 10.1038/s41598-018-23146-3. Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dubinkina V.B., Tyakht A.V., Odintsova V.Y., Yarygin K.S., Kovarsky B.A., Pavlenko A.V. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. 2017;5(1):141. doi: 10.1186/s40168-017-0359-2. Oct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shafaghati L., Razaghi-Moghadam Z., Mohammadnejad J. A systems biology approach to understanding alcoholic liver disease molecular mechanism: the development of static and dynamic models. Bull Math Biol. 2017;79(11):2450–2473. doi: 10.1007/s11538-017-0336-8. Nov. [DOI] [PubMed] [Google Scholar]
- 76.Acharya C., Sahingur S.E., Bajaj J.S. Microbiota, cirrhosis, and the emerging oral-gut-liver axis. JCI Insight. 2017;2(19):94416. doi: 10.1172/jci.insight.94416. Oct 5pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei X., Yan X., Zou D., Yang Z., Wang X., Liu W. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013;13:175. doi: 10.1186/1471-230X-13-175. Dec 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bajaj J.S., Heuman D.M., Hylemon P.B., Sanyal A., White M., Monteith P. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940–947. doi: 10.1016/j.jhep.2013.12.019. MayHepatol. 2013;60(5):940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao L., Ling Z., Chen D., Liu Y., Yang F., Li L. Disorganized gut microbiome contributed to liver cirrhosis progression: a meta-omics-based study. Front Microbiol. 2018;9:3166. doi: 10.3389/fmicb.2018.03166. Dec 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu L.X., Schwabe R.F. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14(9):527–539. doi: 10.1038/nrgastro.2017.72. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ponziani F.R., Bhoori S., Castelli C., Putignani L., Rivoltini L., Del Chierico F. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology. 2019;69(1):107–120. doi: 10.1002/hep.30036. Jan. [DOI] [PubMed] [Google Scholar]
- 82.Ren Z., Li A., Jiang J., Zhou L., Yu Z., Lu H. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2018 doi: 10.1136/gutjnl-2017-315084. Jul 25pii: gutjnl-2017-315084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garud N.R., Good B.H., Hallatschek O., Pollard K.S. Evolutionary dynamics of bacteria in the gut microbiome within and across hosts. PLoS Biol. 2019;17(1) doi: 10.1371/journal.pbio.3000102. Jan 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korem T., Zeevi D., Suez J., Weinberger A., Avnit-Sagi T., Pompan-Lotan M. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349(6252):1101–1106. doi: 10.1126/science.aac4812. Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang L., Brunell D., Stephan C., Mancuso J., He B., Thompson T.C. Driver network as a biomarker: systematic integration and network modeling of multi-omics data to derive driver signaling pathways for drug combination prediction. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz109. Feb 15. pii: btz109[Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hiraoka S., Yang C.C., Iwasaki W. Metagenomics and bioinformatics in microbial ecology: current status and beyond. Microbes Environ. 2016;31(3):204–212. doi: 10.1264/jsme2.ME16024. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bashiardes S., Zilberman-Schapira G., Elinav E. Use of metatranscriptomics in microbiome research. Bioinform Biol Insights. 2016;10:19–25. doi: 10.4137/BBI.S34610. Apr 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leimena M.M., Ramiro-Garcia J., Davids M., van den Bogert B., Smidt H., Smid E.J. A comprehensive metatranscriptome analysis pipeline and its validation using human small intestine microbiota datasets. BMC Genomics. 2013;14:530. doi: 10.1186/1471-2164-14-530. Aug 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gosalbes M.J., Durbán A., Pignatelli M., Abellan J.J., Jiménez-Hernández N., Pérez-Cobas A.E. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 2011;6(3):e17447. doi: 10.1371/journal.pone.0017447. Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kolmeder C.A., Salojärvi J., Ritari J., de Been M., Raes J., Falony G. Faecal metaproteomic analysis reveals a personalized and stable functional microbiome and limited effects of a probiotic intervention in adults. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0153294. Apr 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiong W., Abraham P.E., Li Z., Pan C., Hettich R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics. 2015;15(20):3424–3438. doi: 10.1002/pmic.201400571. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zuñiga C., Zaramela L., Zengler K. Elucidation of complexity and prediction of interactions in microbial communities. Microb Biotechnol. 2017;10(6):1500–1522. doi: 10.1111/1751-7915.12855. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Koppel N., Balskus E.P. Exploring and understanding the biochemical diversity of the human microbiota. Cell Chem Biol. 2016;23(1):18–30. doi: 10.1016/j.chembiol.2015.12.008. Jan 21. [DOI] [PubMed] [Google Scholar]
- 94.Peng B., Li H., Peng X.X. Functional metabolomics: from biomarker discovery to metabolome reprogramming. Protein Cell. 2015;6(9):628–637. doi: 10.1007/s13238-015-0185-x. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol. 2016;17(7):451–459. doi: 10.1038/nrm.2016.25. Jul. [DOI] [PMC free article] [PubMed] [Google Scholar]