Abstract
Background
Sunitinib is one of the multi-targeted tyrosine kinase inhibitors for the treatment of renal cell carcinoma (RCC) at present. However, its clinical efficacy is limited by chemoresistance of RCC. Our previous study found that eukaryotic translation initiation factor 3 subunit D (EIF3D) was an oncogene in the development and progression of RCC but little is known about whether EIF3D participated in sunitinib resistance of RCC.
Methods
The expression of EIF3D in the tumor tissue specimen was detected by immunohistochemistry. The effect of EIF3D on sunitinib-resistance of RCC cells was evaluated by colony formation, IC50 proliferation and in vivo tumor growth assays. The interaction between EIF3D and glucose regulated protein 78 (GRP78) was assessed by Co-IP and Western blot assays.
Finding
EIF3D expression was found higher in 786-OR and ACHN-R cells with acquired sunitinib resistance than that in 786-O and ACHN cells sunitinib to sensitive. The EIF3D level was also up-regulated in sunitinib-chemoresistant tumor tissues compared with chemosensitive tumor tissues. Functional study showed that EIF3D knockdown decreased cell viability with sunitinib treatment. Mechanistical study demonstrated that EIF3D interacted with GRP78 and enhanced protein stability through blocking the ubiquitin-mediated-proteasome degradation of GRP78. GRP78 overexpression induced sunitinib resistance of RCC cells by triggering the unfolded protein response, whereas GRP78 silencing inhibited cell viability. Forced expression of GRP78 eliminated the inhibitory effect of EIF3D silencing on cell growth in vitro and in vivo.
Interpretation
our results indicate that EIF3D played an important role in sunitinib resistance of RCC cells, suggesting that it may prove to be a potential therapeutic target for RCC.
Keywords: Renal cell carcinoma, EIF3D, GRP78, Sunitinib, Chemoresistance
Research in context
Evidence before this study
Sunitinib, tyrosine kinase inhibitor (TKI), is the mainstay of treatment for renal cell cancer (RCC), but chemoresistance of RCC cells limits its clinical efficacy. Eukaryotic initiation factor 3 subunit d (EIF3D) functions as an oncogene in the RCC occurrence and progression reported in our previous study, while little is known about whether EIF3D regulates sunitinib resistance of RCC.
Added value of this study
Our results indicate the important role of EIF3D facilitates sunitinib resistance of RCC by interacting with GRP78, suggesting its candidacy as a potential therapeutic target for RCC.
Implications of all the available evidence
EIF3D expression was found higher in RCC cell lines with acquired sunitinib resistance than in its sunitinib sensitive parent cells, and EIF3D level was also regulated in sunitinib-chemoresistant tumor tissues compared with chemosensitive tumor tissues. Functional studies showed that knockdown of EIF3D resulted in decreased cell viability with sunitinib treatment. Mechanistical studies demonstrated that EIF3D interacted with glucose regulated protein 78 (GRP78) and enhanced the protein stability through blocking the ubiquitin-mediated-proteasome degradation of GRP78. GRP78 overexpression induced sunitinib resistance of RCC cells by triggering the unfolded protein response (UPR), whereas GRP78 silencing inhibited cell viability. Forced expression of GRP78 eliminated the inhibition effect of EIF3D silencing on cell growth in vitro and in vivo.
1. Introduction
Kidney cancer is among the 10 most common cancers in both men and women, representing 3.7% of all new cancer cases. It is reported that 63,990 individuals were diagnosed as having kidney cancer in the United States in 2017. Renal cell carcinoma (RCC) is the most common form of kidney cancer, accounting for about 85% of all kidney cancers [1]. According to the histomorphology, immunophenotype and genetic characteristics, RCC is classified into three major histological subtypes: clear cell RCC (ccRCC), comprising about 75–80%; papillary RCC (pRCC), comprising about 10–15%; and chromophobe RCC (cRCC), comprising about 4–5% [2,3]. Currently, surgical remains the mainstay of treatment for localized or early-stage RCC [3]. Unfortunately, some patients with localized early-stage RCC will develop metastatic or advanced RCC after radical nephrectomy. Given the poor prognosis for patients who present with high‐stage disease and the complexity of outcome prediction, it is urgent to develop new biomarkers for early diagnosis and identification of underlying mechanisms causing cancer metastasis.
Sunitinib is a molecular targeted therapy drug used as the first-line chemotherapy agent for metastatic RCC [4]. Sunitinib works as a multi-targeted tyrosine-kinase inhibitor whose targets include vascular endothelial growth factor (VEGF) receptors, stem cell growth factor receptor (KIT), Platelet-derived growth factor receptor (PDGFR) and macrophage colony-stimulating factor receptor (MCSFR) [4,5]. Although the initial response rate to sunitinib was reported to be high as 47% [6,7], drug resistance and tumor progression often occur after 9–12 months of sunitinib therapy [8,9], suggesting the presence of acquired sunitinib resistance. However, the biological mechanism underlying sunitinib resistance remains to be elucidated.
Eukaryotic translation initiation factor 3 (EIF3) is one of the largest and most complex eukaryotic translation initiation factors containing 13 subunits (eIF3a-eIF3m), which plays an important role in the regulation of eukaryotic translation [10]. EIF3 promotes mRNA binding to 40S subunits and prevents premature binding of large ribosomal subunits to regulate eukaryotic protein synthesis [3]. Emerging studies have demonstrated that eIF3 plays a role in tumorgenesis and tumor progression [11]. Zang et al. reported that eIF3b was a potential prognostic indicator and therapeutic target for ccRCC [11], that EIF3b knockdown inhibited the Akt signaling pathway, thus suppressing cell proliferation by triggering apoptosis. In addition, EIF3b inhibition also impaired epithelial-to-mesenchymal transition (EMT), thus suppressing migration and invasion in vitro and in vivo [11]. It was found in our previous study that the expression level of EIF3D in the RCC tissue was higher than that in the paired adjacent normal tissues, and that the expression level of EIF3D was positively correlated with TNM stage and tumor size [3]. Functionally, EIF3D knockdown significantly inhibited cell proliferation and colony formation. Our previous study also demonstrated that EIF3D functions as a proto-oncogene in the development and progression of RCC [3]. However, the role of EIF3D in sunitinib resistance of RCC remained unknown.
Based on the above analyses, the present study was intended to determine whether aberrant expression of EIF3D in RCC was correlated with sunitinib resistance. The result showed that EIF3D expression is upregulated in sunitinib-resistant RCC cell lines and chemoresistant tumor tissues after sunitinib treatment. Knockdown of EIF3D inhibited cell viability with sunitinib treatment. EIF3D contributed to sunitinib resistance by enhancing the protein stability of GRP78, thus triggering the unfolded protein response.
2. Materials and methods
2.1. Tissue samples and cell lines
The present study followed the reporting recommendations for tumor marker prognostic studies (REMARK). RCC tissues were obtained upon the approval of the Institutional Clinical Ethics Review Board of Ruijin Hospital Affiliated to Shanghai Jiaotong University School of Medicine (Shanghai, China). Seventy-one patients provided informed consent and agreed to participate in the study. The patient characteristics (including 59 chemosensitive and 12 chemoresistant tumor tissues) are showed in Table 1. The pathologic specimens were evaluated by a surgical pathologist (Y.X.Q.), with the stage and grade determined according to the 2010 American Joint Committee on Cancer guidelines and Fuhrman grading system, respectively. The clinical data of these RCC patients at the time of diagnosis included age, gender, Fuhrman nuclear grade, and TNM stage.
Table 1.
Correlation between EIF3D expression and patient characteristics.
| Characteristics | EIF3D expression | Total | P value | |
|---|---|---|---|---|
| Low | High | |||
| Total cases | 50 | 21 | 71 | |
| Gender | 35 | 14 | 49 | 0.115 |
| Male | 15 | 7 | 22 | 0.777 |
| Female | 14 | 7 | 21 | |
| Age | 36 | 14 | 50 | |
| <60 | ||||
| ≥60 | ||||
| Tumor size⁎⁎⁎ | <0.001 | |||
| <7cm | 38 | 6 | 44 | |
| ≥7cm | 12 | 15 | 27 | |
| T stage⁎⁎⁎ | <0.001 | |||
| I/II | 37 | 3 | 40 | <0.001 |
| III/IV | 13 | 18 | 31 | |
| Fuhrman⁎⁎⁎ | 29 | 6 | 35 | <0.001 |
| 1–2 | 21 | 15 | 36 | |
| 3–4 | ||||
| Sunitinib | <0.001 | |||
| resistance⁎⁎⁎ | 3 | 9 | 12 | |
| Yes | 47 | 12 | 59 | |
| No | ||||
| Metastases⁎⁎ | 0.002 | |||
| Yes | 9 | 12 | 21 | |
| No | 41 | 9 | 50 | |
Abbreviations: Group performance status T-test for continuous variables and χ2 test or Fisher's exact test for categorical variables. * indicate P < 0.05,.
indicate P < 0.01,
indicate P < 0.00, P < 0.05 is considered statistically significant.
Human RCC cell lines 786-O and ACHN were obtained from ATCC (Manassas, VA, USA) and maintained in DMEM medium (Gibco, Carlsbad, CA, USA) supplemented with 10% FBS (Gibco). The sunitinib-resistant 786-OR and ACHN-R cells were established through continuously exposing cells to sunitinib for three months as previously described [12]. Cells used in the study were kept in a humidified atmosphere with 5% CO2 at 37 °C.
2.2. Overexpression and RNA interference (RNAi)
Recombinant plasmids pcDNA3-EIF3D and pcDNA3-GRP78 were constructed to overexpress EIF3D and GRP78 in our laboratory, respectively. Recombinant lentiviruses targeting EIF3D (Lv-shEIF3D), GRP78 (Lv-shGRP78) or the scrambled nontargeting vector (Lv-shNC) was obtained from GeneChem Co., Ltd (Shanghai, China). Transfection of cells was carried out by exposing them to dilutions of the viral supernatant in the presence of polybrene (1 μg/mL) for 48 h, and the silencing efficiency of EIF3D or GRP78 was detected by Western blotting. The sequence of shEIF3D was: 5′ -GCGTCATTGACATCTGCATGACTCGAGTCATGCAGATGTCAATGACGCTTTTTT-3′. The shGRP78 sequence was CCGGCTTGTTGGTGGCTCGACTCGACTCGAGTCGAGTCGAGCCACCAACAAGTTTTT. The sequence of the control shRNA (shNC) was 5′-TTCTCCGAA CGTGTCACGTCTCGAGACGTGA-3′.
2.3. Real-time quantitative PCR (qPCR)
Total RNA was isolated from the RCC tissues treated with or without sunitinib and 786-O, 786-OR, ACHN and ACHN-R cells by Trizol reagent (Invitrogen, Carlsbad, CA). Total RNA samples were reversely transcribed to cDNAs with the reverse transcription kit (Takara, Japan) as the manufacturer's description. qPCR was conducted using SYBR Green PCR kit (Takara) on CFX Connect™ Real-Time PCR Detection System. The relative EIF3D levels were normalized to β-actin levels by using 2−ΔΔCT method. Primers used to detect EIF3D expression were 5′ -CTGGAGGAGGGCAAATACCT-3′ (forward) and 5′ -CTCGGTGGAAGGACAAACTC-3′ (reverse). β-actin primers were 5′ -TGGACATCCGCAAAGAC-3′ (forward) and 5′ -AAAGGGTGTAACGCAACTA-3′ (reverse).
3. Western blot
Total protein was extracted from the tumor tissues or whole cells using lysis buffer. Protein quantification was determined using Bicinchoninic acid (BCA) assay kit (Pierce, Rockford, IL, USA). An equal amount of proteins (80 µg) was electrophoresed through a 10% SDS-PAGE gel, and then electro-transferred onto nitrocellulose Hybond membranes (Millipore, Billerica, MA, USA). The blots were incubated overnight at 4 °C with the indicated antibodies (EIF3D: ab85951, 1:5000, Abcam, CA, USA; GRP78: PA5-34941, 1:2000, Invitrogen, CA, USA; ATF6: H00022926-D01, 1:1000, Novus Biologicals, CO,USA; PERK: ab65142, 1:500, Abcam, CA, USA; CHOP: ab10444, 1:200, Abcam, CA, USA; IRE-1: ab37073, 1 μg/ml, Abcam, CA, USA; GAPDH: 60004-1-Ig, 1:5000, PeproTech, NJ, USA), and then incubated with corresponding HRP-linked secondary antibody (1:5000, Sigma-Aldrich, CA, USA). Protein bands were visualized with an enhanced chemiluminescence substrate (Milipore, Schwalbach, Germany). The intensity of Western blot bands was quantified using Image J.
3.1. Immunohistochemistry (IHC)
IHC assay was carried out to detect EIF3D expression as described previously [13] Briefly, paraffin‐embedded samples were sliced into 5 μm sections. After incubation in EIF3D primary antibodies (10219-1-AP, 1:2000, proteintech) overnight at 4 °C, the slides were then labeled with biotinylated goat anti-rabbit serum streptavidin-peroxidase conjugate in quarter at room temperature, and reacted with diaminobenzidine (DAB, Invitrogen) for 10 min. IHC images were captured using a high-resolution camera. Cases were scored semiquantitatively as negative, weakly positive, moderately positive, or strongly positive along with the percentage of positive cells, after which an H-score was generated as a score of 0–3 intensity multiplied by the 0–100 percentage of positive cells (range 0–300). H score ≤ 150 refers to low expression, while H score > 150 refers to high expression. And the H score of each patient was calculated independently by two experienced pathologists in a double blind way.
3.2. Half maximal inhibitory concentration (IC50)
The cells were seeded into 96-well plates at a density of 3 × 103 cells/well and treated with or without pcDNA3-GRP78, pcDNA3-EIF3D, Lv-shNC or Lv-shEIF3D for 48 h. 10 µl CCK-8 was added to each well and incubated for additional 2 h. The data were then recorded with a Bio-Rad microplate reader. IC50 was obtained by probit analysis and calculated using GraphPad Prism 5.0 software.
4. Colony formation
RCC cells were seeded in 6-well plate and cultured for a period of time until the density reached 1 × 103 cells per well. Cells were exposed to pcDNA3-GRP78, Lv-shNC, Lv-shEIF3D or Lv-shGRP78 and then colony formation was detected after 10-day culture. Colonies were fixed with 4% paraformaldehyde (Sangon Biotech, Shanghai, China) for 15 min, and then stained with 1% crystal violet (Sangon Biotech) for 15 min. After washing with PBS, images were taken for comparison and analysis. The experiments were repeated at least three times in each group.
4.1. In vivo experiments
All animal procedures were performed in accordance with the Animal Care and Use Committee policies of Shanghai Jiaotong University School of Medicine. The athymic BALB/C mice (5 weeks old) were (Chinese Academy of Sciences, Shanghai, China) were maintained in a specific pathogen-free facility. Twelve nude mice were equally randomized into four groups: (1) 786-OR cells (5 × 105 cells) with stable expression of control were subcutaneously (s.c.) injected into the flanks of nude mice and then treated with saline by oral gavage daily (n = 3); (2) 786-OR cells (5 × 105 cells) with stable expression of control were injected s.c. into the flanks of the nude mice and then treated with 20 mg/kg/day sunitinib by oral gavage daily (n = 3); (3) 786-OR cells (5 × 105 cells) with Lv-shEIF3D were injected s.c. into the flanks of the nude mice and then treated with 20 mg/kg/day sunitinib by oral gavage daily (n = 3); and(4) 786-OR cells (5 × 105 cells) with Lv-shEIF3D and GRP78 were injected s.c. into the flanks of the nude mice and then treated with 20 mg/kg/day sunitinib by oral gavage daily (n = 3). Tumor growth was monitored by tumor volume, which was calculated at the indicated time using the formula: (length × width2)/2.
4.2. Statistical analyses
All data are presented as mean ± standard deviation (SD). Each experiment was carried out at least in triplicates. Two-tailed student's t-test was performed to analyze the difference between two groups using the SPSS software version 16 (SPSS, Chicago, IL, USA). A value of p < 0.05 was considered statistically significant.
5. Results
5.1. EIF3D expression is upregulated in the sunitinib-resistant RCC tissue
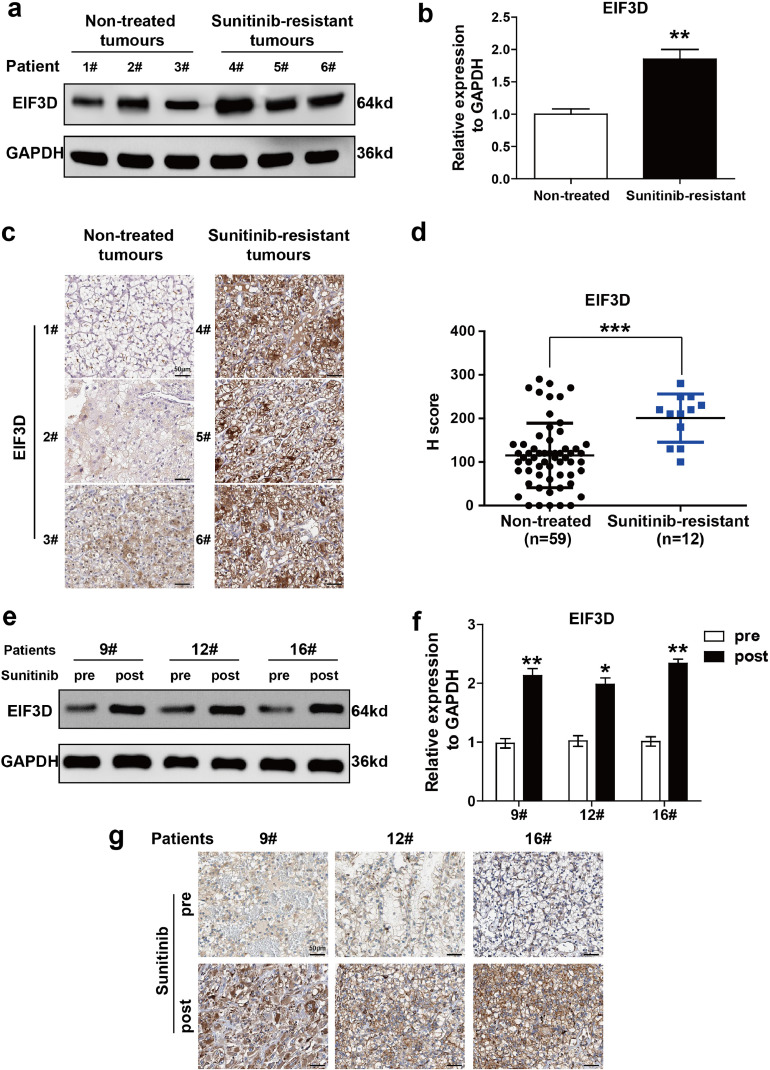
Sunitinib resistance is one of the main obstacles in the treatment of advanced RCC. Revealing the underlying mechanisms of sunitinib resistance is crucial for developing novel therapeutic agents. Our previous study demonstrated that EIF3D functioned as an oncogene in the progression of RCC [3]. To determine whether EIF3D dysregulation was associated with sunitinib resistance in RCC, the present study first analyzed the correlation between EIF3D expression and sunitinib treatment in RCC. Fresh RCC tissues resistant to sunitinib (n = 3) and non-treated RCC samples (n = 3) were analyzed by Western blotting. As shown in Fig. 1a and b, the expression level of EIF3D in sunitinib-resistant tumor was significantly higher than that in the non-treated tumor tissue. IHC staining was used to assess the correlation between EIF3D expression and sunitinib treatment in the specimens obtained from 71 patients. As shown in Fig. 1c–d, the EIF3D expression in sunitinib-resistant tumors (n = 12) was higher than that in the non-treated RCC tissue (n = 59). In addition, we quantified the expression of EIF3D and compared the pathologic features in the dichotomized EIF3D status in this sample group. As summarized in Table 1, EIF3D expression was positively correlated with tumor size (P < 0.001), T stage (P < 0.001), Fuhrman grade (P < 0.001), sunitinib resistance (P < 0.001) and the metastatic status (P = 0.002). Sequentially, EIF3D expression before and after sunitinib treatment in the same patient (n = 3) was assessed by Western blot and IHC. The results indicated that EIF3D was more prevalent in the acquired resistant tissues which were shown in Fig. 1e–g, suggesting that EIF3D upregulation may contribute to sunitinib resistance of RCC.
Fig. 1.
EIF3D expression is upregulated in sunitinib-resistant RCC tissues. (a–b) Western blot analysis was performed in six cases of fresh RCC tissues with or without sunitinib treatment. (P value: t-test), values are represented as the mean ± SD (c) Representative IHC staining in sunitinib-resistant RCC tissues and non-treated RCC samples (scale bar= 50 µm). (d) The EIF3D expression was determined by immunohistochemistry in 59 chemosensitive and 12 chemoresistant tumour tissue specimens. The expression of EIF3D was evaluated by H-score, which has been listed in Materials and Methods (P value: Wilcoxon test), values represented as the mean ± SD. (e–g) Western blot analysis (e–f) and IHC assay (g) were performed in three cases of fresh RCC tissues before or after sunitinib treatment (scale bar = 50 µm) (P value: t-test), values represented as the mean ± SD.(*: P < 0.05, **: P < 0.01; ***: P < 0.001).
5.2. EIF3D is positively correlated with sunitinib resistance of RCC
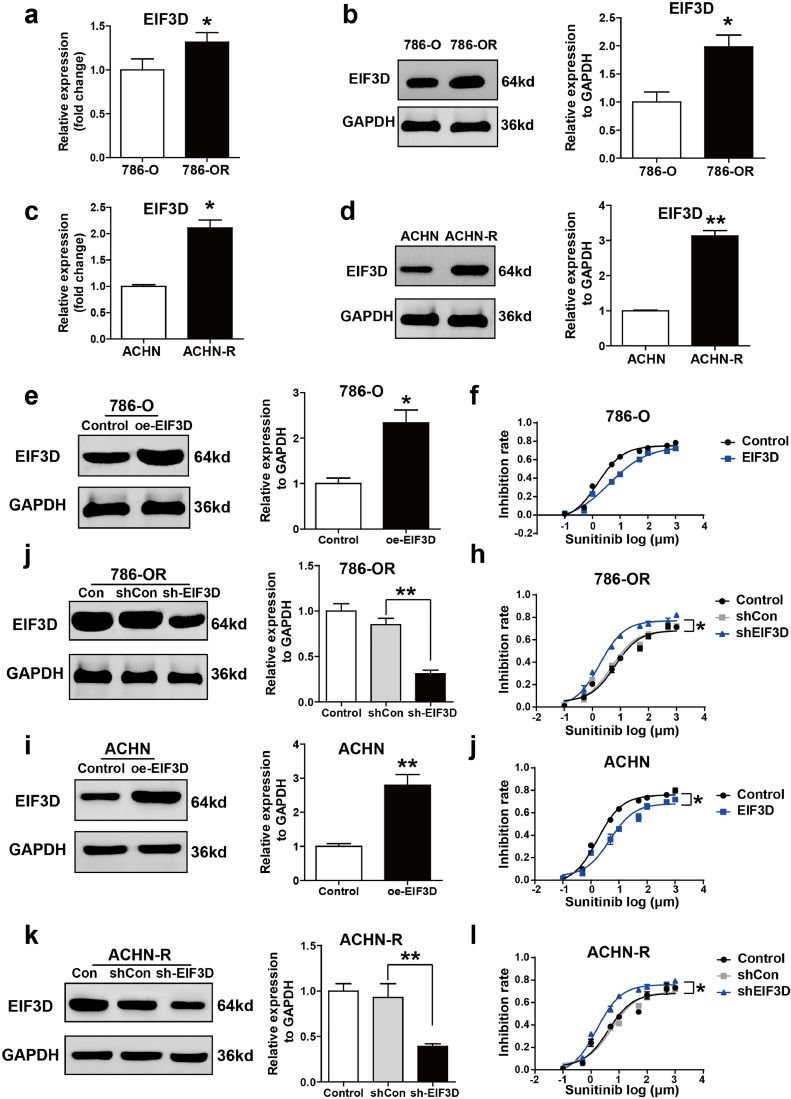
We further investigated the cellular functions of EIF3D established sunitinib-resistant 786-O cell line (786-OR) and sunitinib-resistant ACHN cell line (ACHN-R) through continuously exposing them to sunitinib for three months. The IC50 of 786-O, 786-OR, ACHN and AHCN-R cells is shown in Supplementary Figure 1 a and b. The expression level of EIF3D was detected in 786-O, 786-OR, ACHN and ACHN-R cells by qPCR and Western-blot. It was found that EIF3D mRNA and protein levels were enhanced in sunitinib-resistant cell lines 786-OR and ACHN-R as compared with those in sunitinib-sensitive cell (Fig. 2a–d).
Fig. 2.
EIF3D is correlated with sunitinib resistance of RCC. To establish sunitinib-resistant cell lines 786-OR and ACHN-R, 786-O and ACHN cell lines were treated with 10 μM sunitinib for three months. (a-d) Western blot and RT-PCR was performed to analyze the expression of EIF3D in two pairs of cell lines. (e and i) Ectopic expression of EIF3D was confirmed by Western blot assay in 786-O and ACHN cell lines. (f and j) Ectopic expression of EIF3D passivated 786-O and ACHN cell to the cytotoxic effect of sunitinib as shown in inhibition rate assay. (g and k) Lentivirus-mediated EIF3D knockdown was confirmed by Western blot assay in 786-OR and ACHN-R cells. (h and l) Knocking down the expression of EIF3D sensitized 786-OR and ACHN-R cells to the cytotoxic effect of sunitinib as shown in inhibition rate assay. (P value: t-test), values are represented as the mean ± SD, (*: P < 0.05, **: P < 0.01; ***: P < 0.001).
To elucidate the effect of EIF3D in sunitinib resistance, two sunitinib-sensitive RCC cell lines were applied to gain-of-function study. pcDNA-EIF3D was constructed and used to overexpress EIF3D in 786-O and ACHN cells. As shown in Fig. 2e and i showed that EIF3D protein expression was observably increased in the two cell lines after pcDNA-EIF3D treatment. We then assessed the inhibition rate in these two paired cell lines and found that EIF3D overexpression significantly increased sunitinib resistance of the tumor cells (Fig. 2f and j). On the other wise, 786-OR and ACHN-R cells were subjected to loss-of-function study. Lentivirus-mediated shRNA was used to silence EIF3D expression in 786-OR and ACHN-R cells. As shown in Fig. 2g and k showed that the expression level of EIF3D was observably suppressed in 786-OR and ACHN-R cells following Lv-shEIF3D treatment. We also assessed the inhibitory rate in these two cell lines, and found that EIF3D knockdown significantly inhibited sunitinib resistance of tumor cells (Fig. 2h and l). These data demonstrate that dysregulated expression of EIF3D was closely related to sunitinib resistance of RCC.
5.3. EIF3D exerts sunitinib resistance through regulating GRP78 and endoplasmic reticulum stress
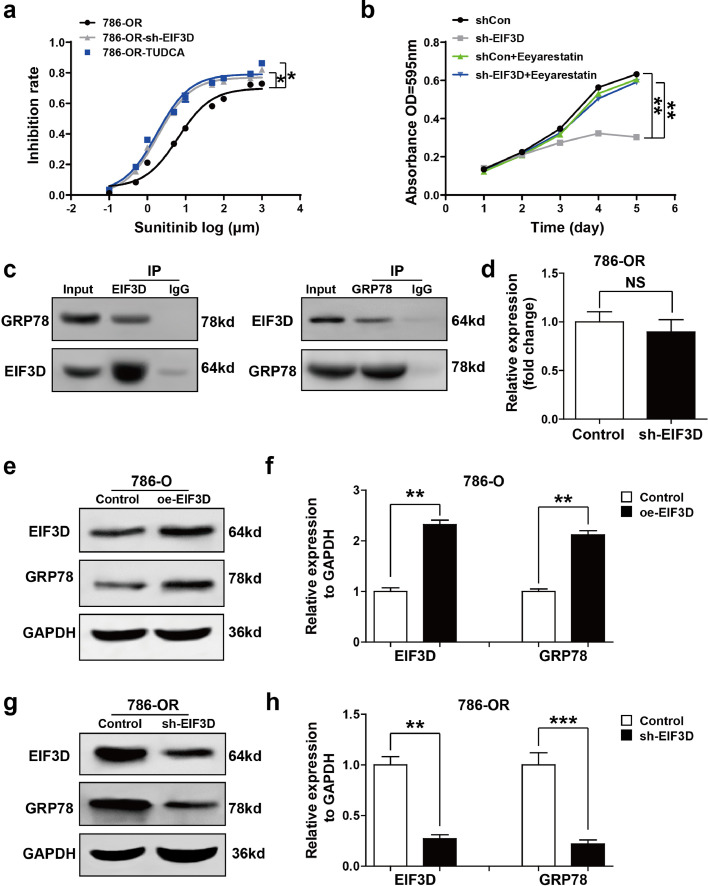
Endoplasmic reticulum stress (ERS) and subsequent unfolded protein response (UPR) are known to be correlated with chemoresistance of a variety of tumor cells [14]. To see whether ERS was associated with sunitinib resistance in RCC, 786-OR cell was selected for inhibition rate assay. The ERS was inhibited by tauroursodeoxycholate (TUDCA) as previously described [15]. As shown in Fig. 3a, knockdown of EIF3D or treatment with TUDCA (100 μM) at 37 °C for 30 min overcame sunitinib resistance in 786-OR cells. To clarify whether ERS mediated EIF3D-induced sunitinib resistance, 786-OR cells transfected with Lv-shEIF3D were treated with or without Eeyarestatin, which is a small molecule shown to cause ERS via inhibiting the endoplasmic reticulum-associated degradation (ERAD). As shown in Fig. 3b, additional treatment with Eeyarestatin markedly destroyed the effect of EIF3D knockdown on reduced cell viability. All these results suggest that EIF3D induced sunitinib resistance of RCC, at least in part by activating ERS.
Fig. 3.
EIF3D exerts sunitinib resistance through regulating GRP78 and ERS. (a) ERS inhibitor TUDCA (100 μM) has the same effect as shEIF3D sensitized 786-OR at 37 °C for 30 min to the cytotoxic effect of sunitinib as shown in inhibition rate assay. Values represented as the mean ± SD. (b) 786-OR cells, transfected with Lv-shEIF3D in the presence or absence of Eeyarestatin, were exposed to sunitinib for 5 days, and the cell viabilities were assessed by the absorbance OD 590. Values are represented as the mean ± SD. (c) EIF3D physically interacts with GRP78. The endogenous proteins from 786-OR cells were IP with IgG or antibodies against EIF3D and GRP78, followed by western-blot analysis and cell lysis for input. (d) Real-time PCR detected the GRP78 mRNA expression in the EIF3D knockdown and control 786-OR cell. Values are represented as the mean ± SD. (n = 3). (e-h) Western-blot assay was performed to detect the GRP78 protein levels in the 786-O cell lines expressing EIF3D or in the 786-OR cell lines knocking-down EIF3D. Values are represented as the mean ± SD (n = 3). (P value: t-test) (*: P < 0.05, **: P < 0.01; ***: P < 0.001., NS = no significate).
Knowing that glucose regulated protein 78 (GRP78) is a typical ERS marker and central regulator of the UPR [16], we investigated whether EIF3D regulated ERS/UPR and subsequent sunitinib resistance by regulating GRP78 and explored whether GRP78 and EIF3D had a functional interaction by reciprocal co-immunoprecipitation assay. As shown in Fig. 3c, positive GRP78 signal was observed in co-immunoprecipitation complex pulled-down by anti-EIF3D-specific antibody. Meanwhile, EIF3D was also detected in the protein pool pulled-down by anti-GRP78 antibody (Fig. 3c). Interestingly, EIF3D did not change GRP78 mRNA level (Fig. 3d), indicating that the effect of EIF3D on the expression of GRP78 may be due to autophagy or ubiquitin-proteasome-mediated degradation. These results suggest that there was a physical interaction between EIF3D and GRP78 in RCC cell line. We then tested whether EIF3D directly regulated GRP78 expression by analyzing the expression level of GRP78 in the EIF3D-overexpressed cell line, and found that EIF3D overexpression increased GRP78 protein level in 786-O cell (Fig. 3e and f). On the other hand, knockdown of EIF3D in 786-OR cell decreased the GRP78 protein expression level (Fig. 3g and h).
5.4. EIF3D enhances the protein stability of GRP78 by blocking the ubiquitin mediated-proteasome degradation of GRP78
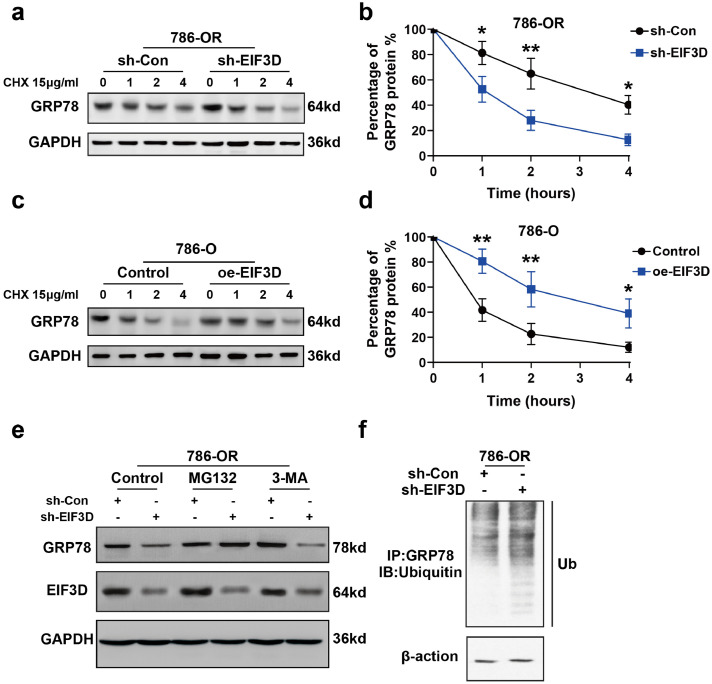
To clarify the mechanism underlying the regulatory effect of EIF3Don GRP78 expression, we used a cycloheximide chase assay to analyze how GRP78 expression underwent changes over time in 786-OR cells transfected with Lv-shEIF3D. It was found that the half-life of GRP78 was markedly reduced in the EIF3D knockdown cells (Fig. 4a and b). Conversely, overexpression of EIF3D in 786-O cells delayed the degradation of GRP78 protein (Fig. 4c and d). The ubiquitin-proteasome and autophagic-lysosome pathways are the main pathways of protein degradation in eukaryotic cells [17,18]. To explore the mechanism of the regulatory effect of EIF3D on GRP78 degradation, we treated EIF3D-knockdown 786-OR cells (4 × 105 cells/well) with proteasome inhibitors MG132 (20 μM) or autophagy-lysosome inhibitor 3-MA (10 mmol/L). MG132, but not 3-MA, restored GRP78 levels in EIF3D-knockdown 786-OR cells (Fig. 4e), implying that the ubiquitin-proteasome pathway participated in GRP78 degradation. To validate this, we detected polyubiquitination of GRP78 with or without EIF3D expression and found that knockdown of EIF3D decreased polyubiquitination of GRP78 (Fig. 4f). These data suggest that EIF3D may inhibit degradation of GRP78 by interacting with it.
Fig. 4.
EIF3D enhances the protein stability of GRP78 by blocking the ubiquitin mediated-proteasome degradation of GRP78. (a–b) EIF3D regulated stability of GRP78 protein. Knockdown of EIF3D was performed in 786-OR cells, followed by the treatment with cycloheximide (CHX, 15 µg/ml) for various times as indicated. Cell lysates were then analyzed by immunoblotting with the GRP78 antibodies. The relative GRP78 protein level was determined using densitometry scanning (ImageJ software). (P value: t-test) (*P < 0.05, **P < 0.01). (c–d) Ectopic expression of EIF3D increased GRP78 stability. EIF3D expression vector or empty vector was transfected into in 786-O cells. Treatment of cycloheximide and western-blot were performed as described in A. Values are represented as the mean ± SD. of three independent experiments. (P value: t-test) (*P < 0.05, **P < 0.01). (e) The regulation of GRP78 stability by EIF3D was through proteasome-mediated ubiquitination and degradation pathway. The 786-OR cells were transfected with EIF3D shRNA or scrambled shRNA. At 48 h post transfection, the cells were treated with either 3-MA (10 mmol/L) or MG132 (20 μM) for additional 4 h. The whole-cell lysates were subjected to the immunoblotting with the indicated antibodies. (f) Depletion of EIF3D increased ubiquitination of GRP78. The scramble and EIF3D shRNA infected cell lysates were immunoprecipitated with anti-GRP78 antibody. The IP was then analyzed by immunoblotting with anti-ubiquitin antibody.
5.5. GRP78 is involved in sunitinib resistance of RCC cells
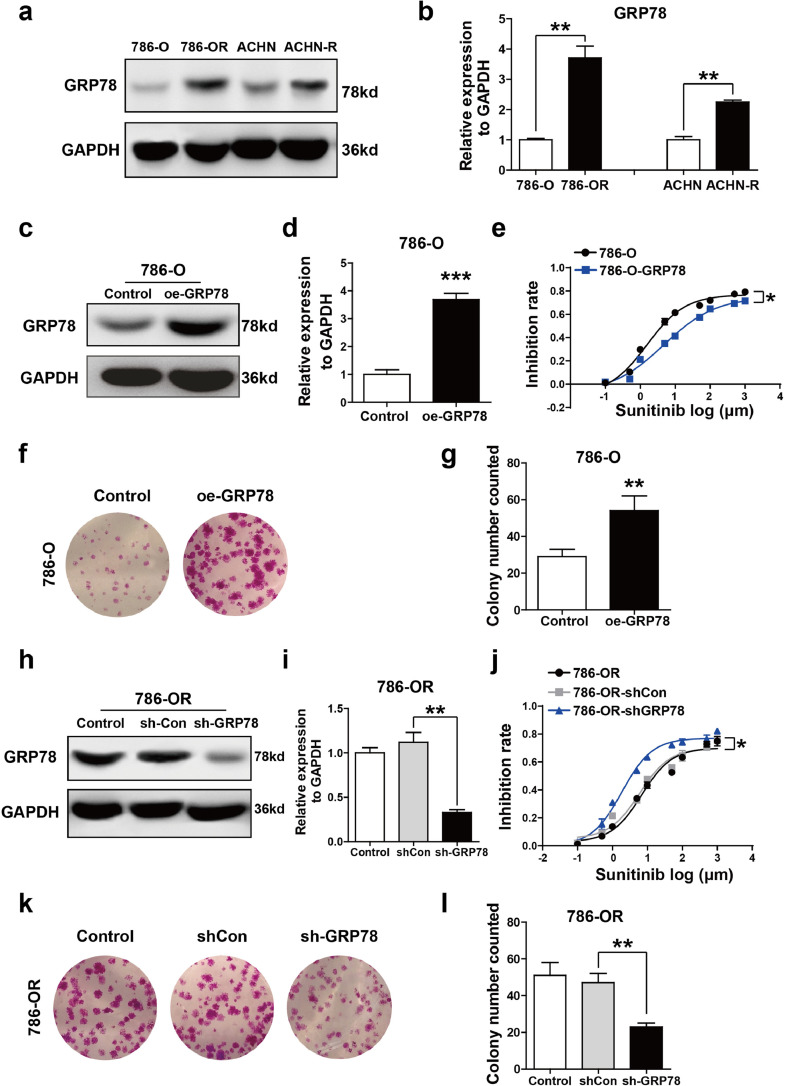
We then analyzed the expression of GRP78 in 786-O, 786-OR, ACHN and ACHN-R cells by Western blot. As shown in Fig. 5a and b, GRP78 levels were increased in sunitinib-resistant cell lines 786-OR and ACHN-R as compared with that in sunitinib-sensitive cell lines (786-O and ACHN cells). The effect of GRP78 on sunitinib resistance of RCC cells was then investigated. 786-O was selected for gain-of-function study. pcDNA-GRP78 plasmid was used to overexpress GRP78 in 786-O cell lines. As shown in Fig. 5c and d, GRP78 protein expression was significantly upregulated in 786-O cells after pcDNA-GRP78 treatment. We then assessed the inhibition rate of sunitinib in 786-O cells, data showed that overexpression of GRP78 obviously increased sunitinib resistance of tumor cell (Fig. 5e). The proliferation of 786-O cell was analyzed by colony formation assay. It was found that GRP78 upregulation remarkably improved cell proliferative capacity (Fig. 5f and g). Conversely, 786-OR cell lines were selected for loss-of-function study. Lentivirus-mediated shRNA was used to silence GRP78 expression in 786-OR cell line. Fig. 5h and i showed that the expression level of GRP78 was significantly suppressed in 786-OR cell following Lv-shGRP78 treatment. We also assessed the inhibition rate of sunitinib in the 786-OR cell lines, and found that knockdown of GRP78 significantly inhibited sunitinib resistance or 786-OR cells (Fig. 5j). In addition, the colony formation assay was performed to analyze the cell proliferation. As shown in Fig. 5k and l, knockdown of GRP78 significantly reduced 786-OR cell proliferation. These results indicate that GRP78 played a crucial role in promoting sunitinib resistance in RCC.
Fig. 5.
GRP78 is involved in sunitinib resistance of RCC cells. (a and b) The levels of GRP78 were detected in 786-O, 786-OR, ACHN, ACHN-R cells by Western blot analysis. GAPDH was used as the loading control, (P value: t-test). (c and d) Ectopic expression of GRP78 in 786-O cells was detected by Western blot analysis, (P value: t-test). (e) Ectopic expression of GRP78 passivated 786-O cell to the cytotoxic effect of sunitinib as shown in inhibition rate assay, (P value: t-test). (f and g) Ectopic expression of GRP78 increased the colony formation capacity of 786-O cells. The number of Colonies is shown in the bar graph, (P value: t-test). (h and i) Lentivirus-mediated GRP78 knockdown in 786-OR cells, (P value: t-test). (j) Knocking down the expression of GRP78 sensitized 786-OR cell to the cytotoxic effect of sunitinib, (P value: t-test). (k and l) GRP78 knockdown decreased the colony formation capacity of 786-OR. Colony numbers were shown in the bar graph, (P value: t-test). Values are represented as the mean ± SD. (n = 3) (*P < 0.05, **P < 0.01).
5.6. EIF3D exerts sunitinib resistance through GRP78-mediated activation of UPR pathway
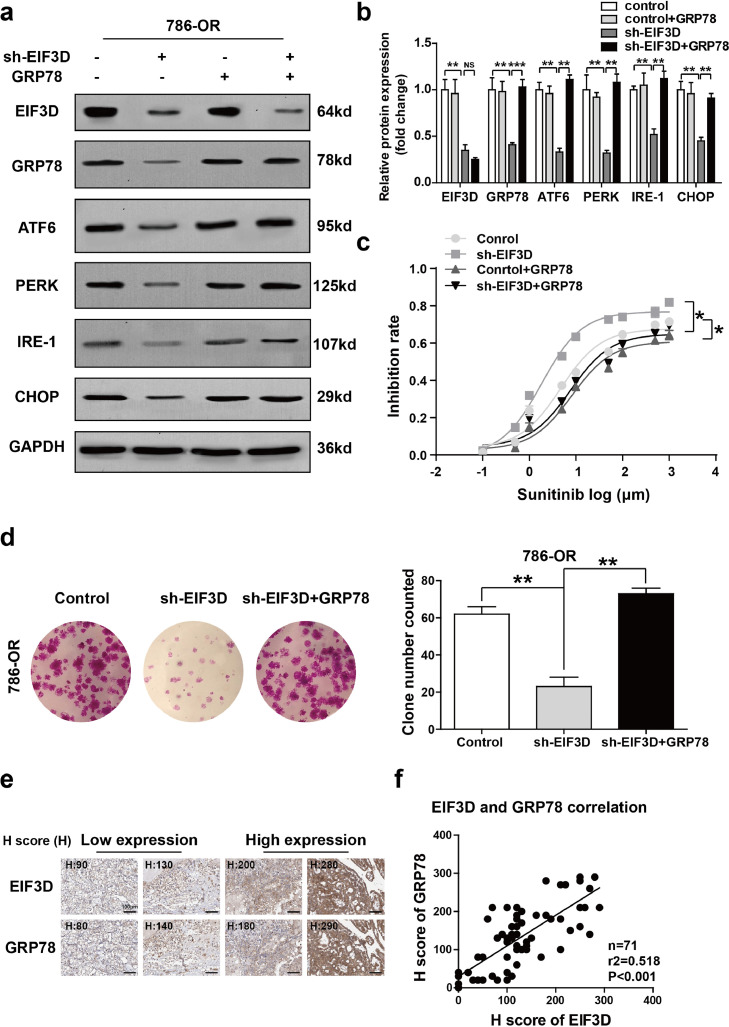
To see whether the sunitinib-resistant effect of EIF3D was dependent on GRP78 upregulation, we next conducted a rescue experiment and found that EIF3D was inhibited in the presence or absence of GRP78 overexpression. knowing that GRP78 is a central regulator of the UPR via sequestration of PERK-eIF2a-ATF4, IRE1a-XBP1s, and ATF6 pathways [19], we first detected the activation of the three UPR signaling pathways by Western blot analysis. It was found that the expression of PERK, IRE1a, and ATF6 was decreased in EIF3D-knockdown 786-OR cell line (Fig. 6a and b). In addition, downregulation of CHOP, a key mediator of UPR, mediated the apoptosis pathway in EIF3D-knockdown 786-OR cell line (Fig. 6a and b). Reconstitution of GRP78 expression restored PERK, IRE1a, ATF6 and CHOP expression in EIF3D knockdown 786-OR cells (Fig. 6a and b), implying that EIF3D regulated UPR by mediating GRP78. In addition, sunitinib inhibition rate and colony formation assays showed that the GRP78 overexpression restored the sunitinib resistance and colony formation capacity of EIF3D-konckdown cells to the baseline level in 786-OR cells (Fig. 6c and d). Additionally, the IHC assay of GRP78 expression in RCC tissue was performed and statistical analysis about the relationship between the expression level of GRP78 and EIF3D was done (Fig. 6e and f). Interestingly, a positive correlation was observed between EIF3D expression and GRP78, the spearman r2 being 0.518 with P < 0.001. In addition, we quantified the expression of GRP78 and compared the pathologic features in the dichotomized GRP78 status in our sample group. As summarized in supplementary Table 1, GRP78 expression was positively correlated with tumor size (P = 0.001), T stage (P < 0.001), and Sunitinib resistance (P = 0.007). These results showed that EIF3D facilitated the resistance of RCC cell to sunitinib through increasing GRP78 expression and inducing subsequent ERS and UPR.
Fig. 6.
EIF3D exerts sunitinib resistance through GRP78-mediated activation of UPR pathway. (a and b) LV-shEIF3D virus and pcDNA3-GRP78 expression vector were used to kno7ck down EIF3D and re-constitutively express GRP78 respectively, in 786-OR cells. The protein level of EIF3D, GRP78, ATF6, PERK, IRE-1, and CHOP was detected by Western blot analysis. GAPDH was used as the loading control, (P value: t-test). (c) The inhibition rate of sunitinib in 786-OR cell transfected with Lv-shEIF3D in the presence or absence of GRP78 was determined by CCK-8 assay (P value: t-test). (d) Colony formation assay showed that ectopic GRP78 expression restored the number of cell colonies in EIF3D knockdown 786-OR cells. The number of colonies numbers is shown in the bar graph (P value: t-test). (e–f) The IHC assay of EIF3D and GRP78 expression in RCC tissues was performed and statistical analysis about the relationship between the expression level of GRP78 and EIF3D was shown, (scale bar= 100 µm), (P value: spearman correlation coefficient) (*: P < 0.05, **: P < 0.01; ***: P < 0.001., NS = no significate).
5.7. EIF3D inhibits sunitinib resistance of RCC cells is mediated by GRP78 in vivo
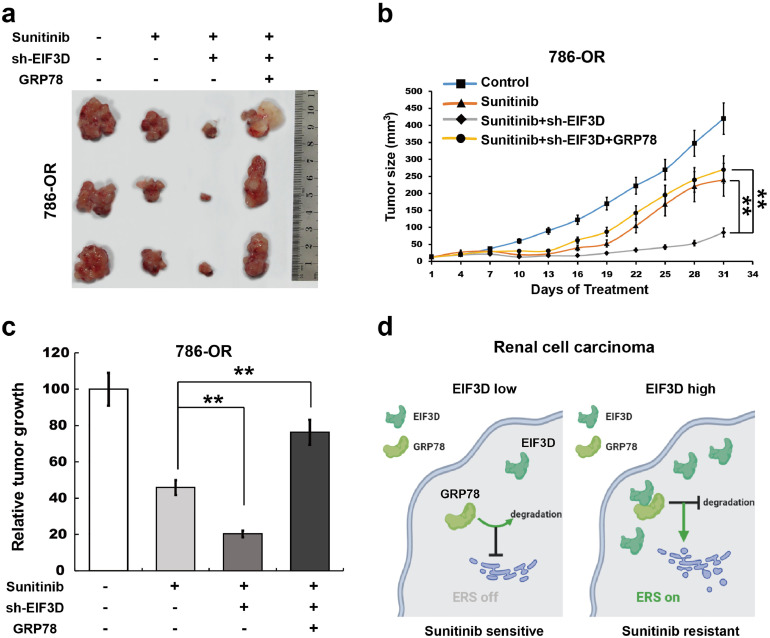
To further verify the effect of EIF3D and GRP78 on sunitinib resistance of RCC in vivo, we explored the impact of EIF3D and GRP78 expression in the nude mice tumorigenicity assay. 786-OR cell lines with stable expression of control, Lv-shEIF3D or Lv-shEIF3D with GRP78 were injected into nude mice and then sunitinib was used to treat these mice. It was found that tumor growth was markedly inhibited in mice injected with 786-OR-shEIF3D cells compared with 786-OR-empty vector cells after sunitinib treatment, and restoration of GRP78 expression significantly increased the tumor growth (Fig. 7a–c). All these data suggest that increased level of EIF3D promoted the sunitinib resistance of RCC at least partially through interacting with GRP78 and inhibiting its degradation, which further activated UPR signaling pathways, leading to the activation of ERS (Fig. 7d).
Fig. 7.
EIF3D inhibits sunitinib resistance of RCC cells is mediated by GRP78 in vivo. (a) Representative images of nude mice tumourigenicity assay with 786-OR cell line stably transfected with empty vector or shEIF3D with or without GRP78. (b and c) Tumour growth curve was measured every 3 days; **P < 0.01 versus shRNA of EIF3D group. Relative tumour growth indicated an overall decrease in EIF3D knockdown group, and re-constitutive expression of GRP78 restored the tumour growth. (P value: t-test) Values represented as the mean ± SD. (** indicates P < 0.01, *** indicates P < 0.001). (d) Proposed mechanistic scheme of EIF3D stabilizes GRP78 to activate the UPR signaling pathway in RCC. Overexpression of EIF3D in RCC physically interacts with the GRP78 and, eventually abrogated ubiquitin–proteasome degradation of GRP78, thus activating the UPR signaling pathway and ERS, and finally promoting the resistance of RCC cells to sunitinib.
6. Discussion
In the present study, we investigated the role of EIF3D in regulating sunitinib resistance of RCC cells and its potential molecular mechanism. The current data demonstrate that (i) EIF3D expression was increased in sunitinib-resistant RCC tissues and cells, (ii) EIF3D upregulation promoted sunitinib resistance of RCC, (iii) EIF3D interacted with GRP78 and increased protein stability of GRP78, (iv) EIF3D induced ERS and UPR by regulating GRP78, and (v) EIF3D exerted sunitinib resistance of RCC cells by regulating GRP78/ERS/UPR. These results demonstrated the important role and underlying mechanism of EIF3D in regulating sunitinib resistance of RCC and may provide a new therapeutic opportunity to chemoresistant RCC.
The role of EIF3D in regulating cancer progression has been well established [20,21]. Our previous study also demonstrated that EIF3D functioned as a potential proto-oncogene in the carcinogenesis and development of RCC [3]. EIF3D expression was observed to be upregulated in RCC tissue as compared with that in the normal tissue, and knockdown of EIF3D inhibited cell proliferation and colony formation, caused G2/M arrest [3]. The present study was designed to further explore whether EIF3D is correlated with chemoresistance of RCC. The result showed that EIF3D expression was upregulated in the sunitinib-resistant RCC tissues. Functional studies verified that EIF3D knockdown observably inhibited tumor cell resistance to sunitinib.
The endoplasmic reticulum (ER) is a dynamic intracellular structure that serves multiple functions in protein synthesis, lipid metabolism, cellular homeostasis and stress responsiveness [22,23]. Cellular adaptation to ERS is mediated by the UPR, which aims at repairing the homeostasis of ER via facilitating the degradation of misfolded proteins [24]. The biological role of the UPR in oncogenesis, cancer development is well demonstrated in many cancers including breast, renal, prostate, and colon cancer [25,26]. Emerging evidence indicates thatUPR is a mechanism of cancer cells to ensure survival after exposure to chemotherapy drugs [25]. The inhibition of UPR results in the resensitization of chemoresistant cells [27,28]. It is acknowledged that UPR is mediated by three highly specific signaling protein molecules: activating transcription factor 6 (ATF6), double-stranded RNA-activated protein kinase (PKR)-like ER kinase (PERK) and inositol requiring enzyme 1 (IRE1) and regulated by master regulator protein, Glucose-regulated protein GRP78 or Binding immunoglobulin Protein (BiP/GRP78) and GRP94. Under normal conditions, the ER luminal domain of these transmembrane molecules including ATF6, PERK, and IRE1 is bound to the chaperone protein BiP/GRP78, a regulator of the UPR system When cells are exposed to some aberrant stress, this system can rescue cell apoptosis in different ways by either exhibiting its chaperone activity or preventing UPR sensor activation and preserving ER calcium homeostasis [29]. In the present study we tested the role of EIF3D in regulating ERS and UPR and found EIF3D inhibition decreased the expression of ATF6, PERK, IRE-1 and CHOP, indicating that EIF3D positively regulates ERS and UPR. Functionally, knockdown of EIF3D overcame sunitinib resistance in 786-OR cells. To further verify whether ERS mediated EIF3D-induced sunitinib resistance, EIF3D was inhibited in 786-OR cells in the presence or absence of Eeyarestatin, and data showed that EIF3D knockdown decreased cell growth, whereas additional treatment of Eeyarestatin restored cell viability during sunitinib treatment. The current results suggest that ERS activation is important for EIF3D-mediated sunitinib resistance of RCC. However, the underlying mechanisms EIF3D-induced ERS in regulating RCC resistance merit further evaluation.
GRP78 is a typical ERS marker and a central regulator of the UPR. Given the important of EIF3D in regulating ERS and sunitinib resistance, we then investigated whether EIF3D activated ERS through GRP78 mediation. The results obtained from co-immunoprecipitation showed that EIF3D interacts directly with GRP78 in RCC cell line. Functionally, we found that EIF3D overexpression enhanced the expression level of GRP78, whereas knockdown of EIF3D decreased GRP78 protein level. Interestingly, EIF3D did not change GRP78 mRNA level, suggesting that EIF3D increased GRP78 expression through autophagy or ubiquitin-proteasome-mediated degradation, which are known as two major pathways of protein degradation in eukaryotic cells [17,18]. It was the proteasome inhibitor MG132 rather the autophagy inhibitor 3-MA () that repaired GRP78 levels in EIF3D-knockdown 786-OR cells, indicating that the ubiquitin-proteasome pathway participated in GRP78 degradation. Moreover, increased GRP78 expression was reported to be associated with a higher pathologic grade, an increased probability of recurrence and poor patient survival in breast, liver, prostate, and colon cancers [30]. Our study also demonstrated that GRP78 expression was increased in sunitinib resistant tissue, which was positively correlated with the malignant features of RCC. GRP78 was also found to be involved in tumor progression, spread and drug resistance, while dormant tumor cells and quiescent tumor cells could rely on GRP78 to escape chemotherapy as well [30]. In addition to its effect of activating ERS, other potential mechanisms of GRP78 need to be further explored.
Finally, we demonstrated that EIF3D promoted the sunitinib resistance of RCC at least partially through interacting with GRP78 and inhibiting its degradation, which further activated UPR signaling pathways and maintaining the homeostasis of ERS.
Funding
The National Natural Science Foundation of China (No. 81572525 and 81602238) and the Medical Guidance Science and Technology Support Project(No. 18411960100). Authors declare that the funders were not involved in the study design, data collection, data analysis, interpretation, writing of the report or decision to publish.
CRediT authorship contribution statement
Hai Huang: Conceptualization, Data curation, Formal analysis, Writing - original draft, Writing - review & editing. Yi Gao: Formal analysis. Ao Liu: Formal analysis. Xiaoqun Yang: Supervision. Fang Huang: Formal analysis. Le Xu: Formal analysis. Xu Danfeng: Writing - review & editing. Lu Chen: Conceptualization, Data curation, Formal analysis, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest in this study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.030.
Contributor Information
Xu Danfeng, Email: xdf12036@rjh.com.
Lu Chen, Email: cl12063@rjh.com.cn.
Appendix. Supplementary materials
References
- 1.Barata P.C., Rini B.I. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. Nov 2017;67(6):507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 2.Garje R., An J.J., Sanchez K. Current landscape and the potential role of hypoxia-inducible factors and selenium in clear cell renal cell carcinoma treatment. Int J Mol Sci. 2018;19(12) doi: 10.3390/ijms19123834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan X.W., Chen L., Hong Y. EIF3D silencing suppresses renal cell carcinoma tumourigenesis via inducing G2/M arrest through downregulation of cyclin B1/CDK1 signaling. Int J Oncol. 2016;48(6):2580–2590. doi: 10.3892/ijo.2016.3459. [DOI] [PubMed] [Google Scholar]
- 4.Schmid T.A., Gore M.E. Sunitinib in the treatment of metastatic renal cell carcinoma. Ther Adv Urol. 2016;8(6):348–371. doi: 10.1177/1756287216663979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito T., Kutikov A. Kidney cancer in 2014: key advances promise progress for kidney cancer patients. Nat Rev Urol. Feb 2015;12(2):69–70. doi: 10.1038/nrurol.2014.356. [DOI] [PubMed] [Google Scholar]
- 6.Dornbusch J., Zacharis A., Meinhardt M. Analyses of potential predictive markers and survival data for a response to sunitinib in patients with metastatic renal cell carcinoma. PLoS One. 2013;8(9):e76386. doi: 10.1371/journal.pone.0076386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motzer R.J., Rini B.I., Bukowski R.M. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006;295(21):2516–2524. doi: 10.1001/jama.295.21.2516. [DOI] [PubMed] [Google Scholar]
- 8.Eisen T., Loembe A.B., Shparyk Y. A randomised, phase II study of nintedanib or sunitinib in previously untreated patients with advanced renal cell cancer: 3-year results. Br J Cancer. Oct 20 2015;113(8):1140–1147. doi: 10.1038/bjc.2015.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joosten S.C., Hamming L., Soetekouw P.M. Resistance to sunitinib in renal cell carcinoma: from molecular mechanisms to predictive markers and future perspectives. Biochim Biophys Acta. 2015;1855(1):1–16. doi: 10.1016/j.bbcan.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y., Long J., Sun Y. The function and clinical significance of eIF3 in cancer. Gene. 2018;673:130–133. doi: 10.1016/j.gene.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 11.Zang Y., Zhang X., Yan L. Eukaryotic translation initiation factor 3b is both a promising prognostic biomarker and a potential therapeutic target for patients with clear cell renal cell carcinoma. J Cancer. 2017;8(15):3049–3061. doi: 10.7150/jca.19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai I., Miyake H., Fujisawa M. Acquired resistance to sunitinib in human renal cell carcinoma cells is mediated by constitutive activation of signal transduction pathways associated with tumour cell proliferation. BJU Int. 2013;112(2):E211–E220. doi: 10.1111/j.1464-410X.2012.11655.x. [DOI] [PubMed] [Google Scholar]
- 13.Gao J., Zhao C., Liu Q. Cyclin G2 suppresses wnt/beta-catenin signaling and inhibits gastric cancer cell growth and migration through Dapper1. J Exp Clin Cancer Res. 2018;37(1):317. doi: 10.1186/s13046-018-0973-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thakur P.C., Miller-Ocuin J.L., Nguyen K. Inhibition of endoplasmic-reticulum-stress-mediated autophagy enhances the effectiveness of chemotherapeutics on pancreatic cancer. J Transl Med. 2018;16(1):190. doi: 10.1186/s12967-018-1562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keestra-Gounder A.M., Byndloss M.X., Seyffert N. NOD1 and NOD2 signalling links ER stress with inflammation. Nature. 2016;532(7599):394–397. doi: 10.1038/nature17631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han K.S., Li N., Raven P.A. Inhibition of endoplasmic reticulum chaperone protein glucose-regulated protein 78 potentiates anti-angiogenic therapy in renal cell carcinoma through inactivation of the PERK/eIF2alpha pathway. Oncotarget. 2015;6(33):34818–34830. doi: 10.18632/oncotarget.5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korolchuk V.I., Menzies F.M., Rubinsztein D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010;584(7):1393–1398. doi: 10.1016/j.febslet.2009.12.047. [DOI] [PubMed] [Google Scholar]
- 18.Ugun-Klusek A., Tatham M.H., Elkharaz J. Continued 26S proteasome dysfunction in mouse brain cortical neurons impairs autophagy and the Keap1-Nrf2 oxidative defence pathway. Cell Death Dis. Jan 5 2017;8(1):e2531. doi: 10.1038/cddis.2016.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian Y., Wong C.C., Xu J. Sodium channel subunit scnn1b suppresses gastric cancer growth and metastasis via GRP78 degradation. Cancer Res. 2017;77(8):1968–1982. doi: 10.1158/0008-5472.CAN-16-1595. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F., Xiang S., Cao Y. EIF3D promotes gallbladder cancer development by stabilizing GRK2 kinase and activating PI3K-AKT signaling pathway. Cell Death Dis. 2017;8(6):e2868. doi: 10.1038/cddis.2017.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu G.Z., Liu J.Z., Li X.Q. Knockdown of eukaryotic translation initiation factor 3 subunit D (eIF3D) inhibits proliferation of acute myeloid leukemia cells. Mol Cell Biochem. 2018;438(1–2):191–198. doi: 10.1007/s11010-017-3127-5. [DOI] [PubMed] [Google Scholar]
- 22.Bravo R., Parra V., Gatica D. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol. 2013;301:215–290. doi: 10.1016/B978-0-12-407704-1.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz D.S., Blower M.D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cellular Mole Life Sci. 2016;73(1):79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevet E., Hetz C., Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov. Jun 2015;5(6):586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- 25.Madden E., Logue S.E., Healy S.J., Manie S., Samali A. The role of the unfolded protein response in cancer progression: from oncogenesis to chemoresistance. Biol Cell. 2019;111(1):1–17. doi: 10.1111/boc.201800050. [DOI] [PubMed] [Google Scholar]
- 26.Ojha R., Amaravadi R.K. Targeting the unfolded protein response in cancer. Pharmacol Res. 2017;120:258–266. doi: 10.1016/j.phrs.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J., Yin Y., Hua H. Blockade of GRP78 sensitizes breast cancer cells to microtubules-interfering agents that induce the unfolded protein response. J Cell Mol Med. 2009;13(9B):3888–3897. doi: 10.1111/j.1582-4934.2009.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J.K., Kang K.A., Piao M.J. Endoplasmic reticulum stress induces 5-fluorouracil resistance in human colon cancer cells. Environ Toxicol Pharmacol. 2016;44:128–133. doi: 10.1016/j.etap.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 29.Lee A.S. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67(8):3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- 30.Dong D., Ni M., Li J. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68(2):498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.