Abstract
Background: Hepatitis B surface antigen (HBsAg) is one of the important clinical indexes for hepatitis B virus (HBV) infection diagnosis and sustained seroconversion of HBsAg is an indicator for functional cure. However, the level of HBsAg could not be reduced by interferons and nucleoside analogs effectively. Therefore, identification of a new drug targeting HBsAg is urgently needed.
Methods: In this study, 6-AN was screened out from 1500 compounds due to its low cytotoxicity and high antiviral activity. The effect of 6-AN on HBV was examined in HepAD38, HepG2-NTCP and PHHs cells. In addition, the antivirus effect of 6-AN was also identified in mouse model.
Findings: 6-AN treatment resulted in a significant decrease of HBsAg and other viral markers both in vitro and in vivo. Furthermore, we found that 6-AN inhibited the activities of HBV SpI, SpII and core promoter by decreasing transcription factor PPARα, subsequently reduced HBV RNAs transcription and HBsAg production.
Interpretation: We have identified a novel small molecule to inhibit HBV core DNA, HBV RNAs, HBsAg production, as well as cccDNA to a minor degree both in vitro and in vivo. This study may shed light on the development of a novel class of anti-HBV agent.
Keywords: Hepatitis B virus, Hepatitis B surface antigen (HBsAg), 6-Aminonicotinamide (6-AN), Anti-HBV drugs
Abbreviation
- 6-AN
6-Aminonicotinamide
- HBsAg
Hepatitis B surface antigen
- HBV
hepatitis B virus
- ETV
Entecavir
- rcDNA
relaxed circular DNA
- cccDNA
covalently closed circular DNA
- HBeAg
hepatitis B e antigen
- IFN-α
interferon
- NAs
nucleoside analogs
- CC50
half maximal cytotoxicity concentration
- EC50
concentration for 50% of maximal effect
- SI
sensitive index
- LD50
lethal dose 50%
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- G6PD
glucose 6-phosphatedehydrogenase
- PPP
pentose phosphate pathway
- DMSO
dimethyl sulfoxide
1. Introduction
Hepatitis B virus (HBV) infection remains a global public health problem. According to current estimates, more than 250 million people worldwide have infected with chronic HBV [1]. As a member of the Hepadnaviridae family [2], HBV harbors a partially double-stranded relaxed circular DNA (rcDNA) genomes. Once internalized by sodium taurocholate co-transporting polypeptide (NTCP), the rcDNA is delivered into the nucleus and converted into covalently closed circular DNA (cccDNA) which serves as the template for viral RNA synthesis and progeny virions generation [3]. In clinical definition, complete cure of HBV infection is defined as the clearance of HBsAg and HBV DNA. Compared with the reduction of HBV DNA load, the level of HBsAg works as a more accurate indicator to evaluate the drug efficacy and disease prognosis [4], [5], [6], [7]. HBsAg seroconversion represents achievement of a strong suppression of viral replication and a low risk of off-treatment virological relapse, and data from the REVEAL study suggest that higher HBsAg in low viremic patients is associated with an increased risk for HCC [8], [9], [10]. Notably, HBsAg comprises the primary antigenic determinant in HBV infection which mainly determines the host antiviral response [11]. A recent study showed that the serological level of HBsAg is positively correlated with intrahepatic HBV replication and transcription in HBeAg negative chronic hepatitis B (CHB) carriers [12]. Despite strong inhibition of anti-HBV drugs interferon (IFN) and nucleoside analogs (NAs) on HBV DNA, they have limited effect on HBsAg [13,14]. The NAs such as Lamivudine, Adefovir dipivoxil, Entecavir, Telbivudi can rarely cut down HBsAg, and only a small part of treated patients can achieve HBsAg loss by IFN-based therapy [15]. Therefore, identification of a new drug targeting HBsAg is urgently needed.
6-Aminonicotinamide, an analogue of niacin which commonly refers to as 6-AN, has been applied in preclinical trials as an adjuvant of anticancer drugs in cancer therapy because of its ability to potentiate the effects of radiation and enhance the efficacy of anticancer agents [16,17]. 6-AN has been used in combination with other chemicals to increase the effectiveness of treatment in many cancers, including breast, ovarian, leukemia [18], [19], [20] and liver cancer [21,22]. Additionally, it was well-known that 6-AN functions as an inhibitor of G6PD (glucose-6-phosphate dehydrogenase), key enzyme of the pentose phosphate pathway (PPP) [18,23]. 6-AN was shown to specifically inhibit PPP activity in mouse oocyte–cumulus complexes [24] and in bovine pronuclear oocytes [25]. The inhibition of meiosis resumption by 6-AN following sperm penetration into zona-free oocytes was also associated with the PPP [26]. Furthermore, 6-AN directly modulates the PPP signal pathway to trigger metabolic disorders, especially the drug metabolism, in HepG2 and other cancer cells [27], indicating that 6-AN generated metabolic disorders of liver cancer. However, there are no reports of 6-AN on HBV replication [28]. In our study, we explored the function of 6-AN on HBV replication, aiming to find a new therapeutic approach targeting HBV.
By screening 1500 compounds from a small molecular compound library, we found that 6-AN significantly inhibited HBsAg, total HBV RNAs, 3.5-kb RNA and HBV DNA expression in vitro and in vivo. Meanwhile, 6-AN suppressed the cccDNA transcription activity to inhibit HBV RNAs synthesis and HBsAg expression to achieve the anti-HBV function, which was a profound insight about pharmacodynamics of 6-AN. These findings identified that 6-AN might work as a valuable candidate for the current HBV treatments.
2. Materials and methods
2.1. Drug and plasmid
6-AN was purchased from Bide Pharmatech Ltd (Catalog no. 329-89-5). 6-AN was dissolved in water and stored at −80 °C with the concentration of 20 mM and then diluted in medium at concentrations ranging from 0 μM to 500 μM.
PPARα was constructed by in-frame insertion of full-length PPARα into pcDNA3.1. pGEM-HBV1.3 was a gift from U. Protzer (University of Heidelberg, Heidelberg, Germany). pCH9/3091 was obtained from Lin Lan (The Third Military Medical University, China).
The plasmid with dimers of HBV genotypes A, B, C and D were transfected into HepG2 cells, 12 h after transfected, the cells were cultured with different concentrations of 6-AN and continued to incubate 72 h. Then the HBsAg, HBeAg, HBV DNA in supernatant and cells, total RNAs and 3.5-kb RNA were measured.
The short-interferon RNA (siRNA) targeting G6PD (si-G6PD1, si-G6PD2, si-G6PD3) or nontargeting siRNA (si-control) were purchased from GenePharma and transfected into HBV infected HepG2-NTCP cells, 72 h after transfected, harvested the cells to detect the viral markers. The primer sequences of the experimental primers are listed in Supplementary Table.
2.2. Cell culture
PHHs were purchased from Sciencell (San Diego, USA) and maintained in hepatocyte medium (Catalog no. 5210; Sciencell).
HepAD38 were purchased from the Shanghai Second Military University. The Dulbecco's Modified Eagle Medium (DMEM) was purchased from the Corning Incorporated. The fetal Bovine Serum (FBS) were purchased from Gibco BRL. The HBV producing cell lines HepG2-NTCP and HepAD38 were selected by 400 ug/ml G418 (Merck) to maintain the replication of HBV.
HepG2 were purchased from ATCC (American Type Culture Collection) and Huh-7 were purchased from Health Science Research Resource Blank. All the hepatocellular carcinoma cell lines HepG2, Huh-7 and PLC/PRF/5 were cultured in DMEM with 10% FBS and 100 U/ml penicillin and 100 ug/ml streptomycin (Thermo). All cells were maintained in an incubator containing 5% CO2 at 37 °C. Transfection was carried out using a DNA transfection reagent (Roche) according to the manufacturer's instructions.
2.3. Viruses and infection
HBV infection in the HepG2-NTCP/PHH cell lines was carried out as previously reported [29]. HepG2-NTCP cells or PHHs were infected with 2 × 103 genome equivalents/cell of HBV particles in the presence of 4% PEG8000 for 24 h and were then rinsed three times with PBS, and maintained in the normal medium or in the medium with different concentrations of 6-AN. Medium was changed every 2 days.
2.4. MTT
The cytotoxic effects of 6-AN on various cells were assessed by (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sangon Biotech). Cells were seeded into 96-well plates and incubated with different concentrations (multiple proportion dilution from 500 μM to 0.98 μM) of 6-AN for 72 h. Next, 40 ug/ml MTT were added into cell culture media and incubated for 4 h under the condition of protection from light. After that, dimethyl sulfoxide (DMSO) (Solarbio) was added and incubated overnight in the dark to lyses the cells and solubilize formazan dissolving in media. Fifty-percent cytotoxicity concentrations (CC50) and Fifty-percent effective concentrations (EC50) were determined with a microplate reader and calculated via non-linear regression using GraphPad Prism 5.0.
2.5. Alamar blue assay
The alamar blue (Invitrogen) assay is designed to measure quantitatively the cellular health and viability. The alamar blue (AB) assay was carried out according to manufacturer's instructions. Briefly, the medium of HepAD38 cells treated with 6-AN and ETV for 9 days and HepG2-NTCP cells treated for 10days in 12 wells were removed; the cells were rinsed with PBS and 1 ml of alamar blue solution (10% [v/v] solution of AB dye) prepared in fresh medium were added to each well. Following 3 h incubation in dark, transferred 100 μl medium to a new 96 wells. AB fluorescence was quantified at the respective excitation and emission wavelength of 560 and 590 nm using a microplate reader.
2.6. Enzyme-linked immunosorbent assay (ELISA)
Cells were treated with the different concentrations of 6-AN, and HBsAg in cell culture supernatant and mouse serum were collected and detected using commercial enzyme-linked immunosorbent assay kits (KHB) according to the manufacturer's instructions. Each experiment included 3 negative controls, 1 positive control, 1 blank hole and the sample holes. We compared absorbance of 6-AN treated group with vehicle. The average absorbance of negative control and blank hole were always 0.05 ± 0.003, the absorbance of positive control was always 3.9 ± 0.005. Supernatant from HepAD38 would be diluted with 20 folds, serum from HBV transgenic mouse would be diluted with 5000 folds, and others use primary sample. All OD values were between 0.5–3.5. Alanine aminotransferase (“ALT”, Nanjing Jiancheng Bioengineering Institute) and Aspartate aminotransferase (“AST”, Nanjing Jiancheng Bioengineering Institute) concentrations in mouse serum were measured by ELISA assay according to the manufacturer's protocol.
2.7. Luciferase reporter assay
The plasmid of Renilla were transfected into HepG2 cells, the luciferase report plasmids (pGL3-Cp, pGL3-Xp, pGL3-Sp1, and pGL3-Sp2) were transfected into Huh-7 cells, pRL-TK was co-transfected with reporter plasmid to normalize the transfection efficiency, 12 h after transfected, the cells were cultured with different concentrations of 6-AN and continued to incubate 48 h. Luciferase activity was assayed by dual-luciferase reporter assay system (Promega, USA) and measured by GloMax microplate luminometer (Promega, USA).
2.8. HBsAg dot blot assay and western blot
The HepAD38 cell culture medium was harvested after 6 days drug treating. centrifuged at 1000 rpm for 10 min, four microliters of the supernatant were spotted onto a nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK), air dried, soaked in 2.5% formaldehyde-PBS for 30 min, rinsed with water at room temperature for 5 min, and then soaked in 50% methanol for 30 min. After three 5-min washes with water, the membrane was blocked with 5% skim milk, incubated with a designated primary antibody, anti-HBsAg antibody (Novus, Colorado, USA), and further incubated with the corresponding HRP-conjugated secondary antibody. Blots were visualized with ECL Western blot reagents (Millipore, Massachusetts, USA).
The HBV-infected PHH cells were collected using a RIPA lysis buffer with protease inhibitors (Roche, Mannheim, Germany) after 10 days drug treated. The protein concentration was determined using protein assay reagent (Bio-Rad, California, USA). 30 μgof protein each sample was separated on SDS–polyacrylamide gels and transferred to polyvinylidene fluoride membranes (GE Healthcare, Buckinghamshire, UK). Subsequent experiments are same as above.
2.9. Nascent RNA synthesis assay and 7-aminoactinomycin D assay
PHH cells were seeded in 12-well plates, 5-ethynyl Uridine (5-EU) and 7-aminoactinomycin D (7-AAD) (Sangon Biotech, A606804-0001) were add into medium after 6 Days drug treated. Total RNA was collected after 5-EU sustain 24 h or 7-AAD treated 24 h and 48 h. Nascent RNA was captured and subjected to real-time PCR analyzed according to instruction of Nascent RNA Capture Kit (Thermo).
2.10. Apolipoprotein B and albumin assay
After 9 days treated with 6-AN in HepAD38 cells, the supernatant was collected to examine the secretion Albumin and Apolipoprotein B according to Albumin test kit (Nanjing Jiancheng Bioengineering Institute) and ApoB ELISA Kit (Shanghai Ze Ye Biotech)
2.11. Real-time PCR
HBV core DNA were obtained as described [30]. HBV genome DNA in mouse serum and liver were extracted by using Biospin Virus DNA Extraction Kit and Biospin Tissue Genomic DNA Extraction Kit, respectively. Absolute real-time PCR were quantified by FastStart Universal SYBR Green Master Mix (Roche) (Bio-Rad, CFX Connect Real-time System), the efficiency was 95–105%, r2 > 99%. (the LOD = 40, LOQ = 1.0 × 102)
Total RNA both cells and tissue were extracted using TRNzol Reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized from 1μg of RNA using the iScriptTM cDNA Synthesis kit (Bio-Rad). Relative RNA expression levels were quantified by FastStart Universal SYBR Green Master Mix (Roche) and β-actin mRNA was used as an internal control. Values were analyzed using the 2-△△Ct method. (QuantStudio 6 Flex, appliedbiosystems).
The primer sequences of the experimental primers are listed in Supplementary Table.
2.12. Hirt extraction of cccDNA and analysis
HBV cccDNA were extracted as previously described [29]. Briefly, the equal number of cells were lysed by SDS lysis buffer and mixed with 2.5 M KCl at 4 °C overnight with gentle rotation. cccDNA were extracted by using phenol chloroform and washed with ethanol and finally resuspended in Elution Buffer. The Hirt DNA sample was then treated with 1000 IU/ml T5 exonuclease (New England Biolabs, USA) for 1 h at 37 °C to eliminate DNAs except for double-stranded closed circular DNA, and then incubated at 99 °C for 5 min to deactivate T5 exonuclease. The treated Hirt DNA was subjected to Taq-man probe qRT-PCR to detect HBV cccDNA levels, the specific primers and probe are listed in Supplementary Table.
Mice were sacrificed to collected liver tissue after 20 days treatment. The tissue block of similar size and location were collected and fixed in formalin for immunohistochemistry, the remaining tissue samples were ground to fine powder in a mortar and pestle under liquid nitrogen. We weighed same mass of the powdered tissue each group for subsequent cccDNA, HBV DNA and HBV RNA detection. For cccDNA, tissue was continuedly grinded and lysed by SDS lysis buffer. The subsequent experiments were same with the cccDNA extracting in cells.
2.13. Southern blot
HBV core DNA was obtained as described [30]. HBV core DNA were separated by 0.9% agarose gels, and then the DNA was transferred onto nylon membranes (Roche), which was hybridized with Dig-labeled full-length HBV genome probe at 42 °C all night. The next day, the membrane was washed in different concentration of SSC/SDS, washing buffer, detection buffer. After blocked and incubated with anti-Dig antibody, the signal was detected by X-ray film.
2.14. Northern blot
Total RNA was detected by DIG Northern Starter Kit (Roche) manufacturer's protocol. The extracted RNA was separated by 1.4% formaldehyde-agarose gel and was stained with ethidium bromide to evaluate the quality of the target RNA under UV light. The RNA was transferred onto nylon membrane by capillary siphon method. The membrane was hybridized with DIG-labeled HBV RNA probe, and washed in different concentration of SSC/SDS, washing buffer, detection buffer. After blocked and incubated with secondary antibody, the membrane was exposed by using X-ray film.
2.15. Mouse efficacy study
HBV-transgenic mice (HBV-Tg C57 BL/6), encoding a 1.2-overlength copy of the HBV genome (serotype awy), were kindly provided by Prof. Xia Ningshao (Xia Men University, China). These mice were fed and maintained under specific conditions isolating from pathogen. According to the Chinese Council on Animal Care In all experiments, all the mice studies were carried out and approved by Chongqing Medical University. Mice were selected as age (6 to 8 weeks), weight (21±1 g), and possess of basically the same serum HBsAg and HBV DNA levels. Next, the mice were assigned to four groups of 10 individuals per group at random: negative control (0.9% saline), positive control (0.02 mg/kg Entecavir (ETV)), low concentration test group (6-AN 2.5 mg/kg), high concentration test group (6-AN 5 mg/kg). Animals received 0.9% saline or 6-AN via intraperitoneal injection and animals received ETV via oral gavage every two days. The serum samples were collected via orbital blood every four days after injection. All blood was collected for analysis of serum HBsAg and HBV markers. 6 mice from each group were sacrificed by cervical dislocation at day 20 and the liver samples were used for intrahepatic HBV DNA and RNA determination. The remaining animals were monitored without treatment up to day 32.
2.16. Mouse model of HBV infection involving HBV recombinant (r) cccDNA construction
Wide type (wt) male mice (C57 BL/6) aged 4–5 weeks were hydrodynamically injected with 4 ug Precursor plasmid rcccDNA (prcccDNA) and 4 ug plasmid pCMV-KRAB-Cre (which encodes a KRAB domain-fused Cre recombinase under control of a cytomegalovirus (CMV) promoter) through tail veins. Those plasmids were dissolved in a volume of phosphate-buffered saline (PBS) equivalent to 8% of the mouse body weight [31], [32], [33]. After a week, serum HBV DNA copies were detected by q-PCR to determine whether the model was built successfully. Next, the successfully infected mice were assigned to four groups of 6 individuals per group at random, administrated with the target drug, positive control (ETV) and negative control (0.9% saline). All animal studies were approved by the Animal Ethics Committee of Chongqing Medical University.
2.17. Immunohistochemistry
The deparaffinization of formalin-fixed paraffin-embedded tissue sections were performed in xylene before the dehydration through a graded alcohol series. then microwaved-heated in sodium citrate buffer (10 mmol/L, pH 6.0) for antigen retrieval. the sections were incubated with primary antibody (anti-HBs, working solution, ZM-0122, Zhongshan Jinqiao Biological Technology) overnight at 4 °C followed by incubation with secondary antibody. Diaminobenzidine (DAB) staining was used for detecting immunoreactivity. Counterstaining was performed using hematoxylin.
2.18. Statistical analysis
Statistics were performed with the nonparametric Mann–Whitney U test. A value of P < 0.05 was considered significant (*P < 0.05; ⁎⁎P < 0.01; n.s., not significant). All statistical analyses were performed using SPSS 19.0 software. Results are expressed as the average of four independent experiments. The mean value ± standard error is indicated.
3. Results
3.1. Identification of 6-AN as an inhibitor of HBV surface antigen secretion
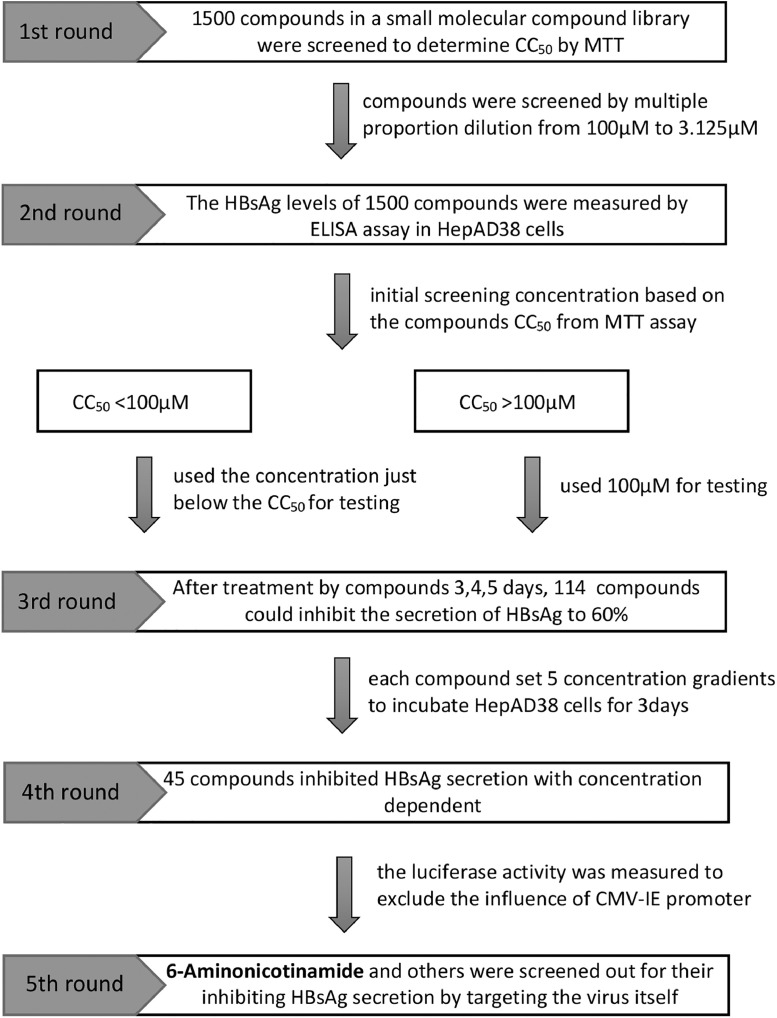
To identify the effective inhibitor on the secretion of HBV surface antigen, 1500 small molecules of a compound library purchased from Topscience (Target Molecule Corp, USA), were screened as shown in the schematic procedure in Fig. 1. Cytotoxicity of the 1500 compounds were first evaluated by MTT assay in HepAD38 cells which HBV replication can be regulated by tetracycline. Based on the MTT results, all tested compounds were divided into 2 groups (CC50 > 100 μM or CC50 ≤ 100 μM). For compounds with CC50 ≤ 100 μM, a working concentration lower than CC50 was used for further experiments, while 100 μM was used as the maximum concentration of compounds with CC50 > 100 μM. The inhibitory effect of those compounds on HBsAg secretion were determined by ELISA assay. 114 compounds were able to decrease the level of HBsAg by 40% or above. These selected 114 compounds were then further validated over a specified concentration range. 45 compounds were found to inhibit the secretion of HBsAg in a dose-dependent manner. It is noteworthy that HepAD38 cell lines direct HBV expression under the control of the tetracycline-repressible CMV-IE promoter instead of the native viral core promoter [34,35], therefore luciferase activity was measured to exclude the possibilities of interaction of drugs with the CMV-IE promoter. Finally, 6-AN was selected as the candidate for further investigation in this study due to its low cytotoxicity, high antiviral activity and negative effect to CMV-IE promoter.
Fig. 1.
Identification of 6-AN as an inhibitor of HBV surface antigen secretion. A flow chart illustrated the process of screening drug targeting HBsAg. The box on the left represented the five rounds of screening. The box on the right detailed the contents of each round of screening. The annotation on the right side of the arrow indicated the concentration used for each screening.
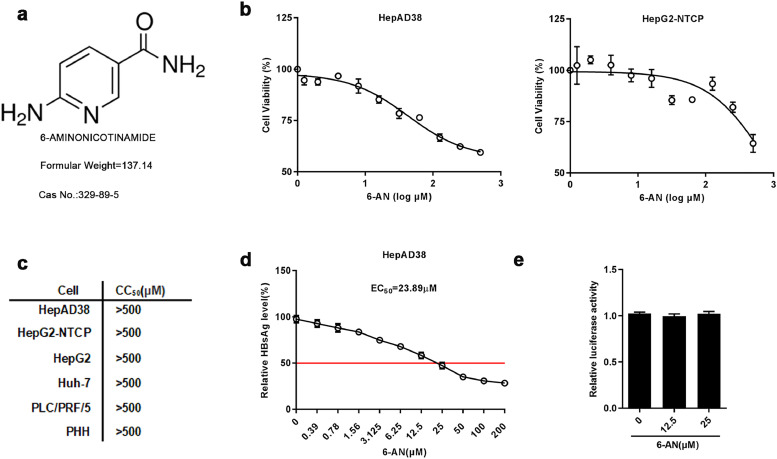
3.2. Characterization of 6-AN
The chemical structure of 6-AN was shown in Fig. 2a. The cytotoxicity of 6-AN on HepAD38 and HepG2-NTCP cells were examined by MTT assay. 6-AN exhibited no significant cytotoxicity in both cell lines after 72 h of 6-AN treatment from 0 μM to 500 μM (Fig. 2b). The cytotoxicity was further examined in additional HCC cell lines including Huh-7, HepG2, PLC/PRF/5 and primary human hepatocytes (PHH) (Fig. S1a) and tabulated in Fig. 2c. Moreover, the EC50 of 6-AN on the inhibition of surface antigen (HBsAg) is approximately 23.89 μM in HepAD38 cells with a sensitive index (SI) > 20 (Fig. 2d). Furthermore, we excluded that 6-AN acted as an anti-HBV agent by affecting the CMV-IE promotors’ activities (Fig. 2e) in HepAD38 cells. During the experiment, 6-AN were treated in HepAD38 cells and HepG2-NTCP cells up to 9 days and 10 days respectively. In order to assess whether the long-term treatment of 6-AN would affect the cellular activity, alamar blue assay was used to measure the number of metabolic active cells and found long-term treatment with 6-AN did not resulted in significant change on cellular viability (Fig.S1b).
Fig. 2.
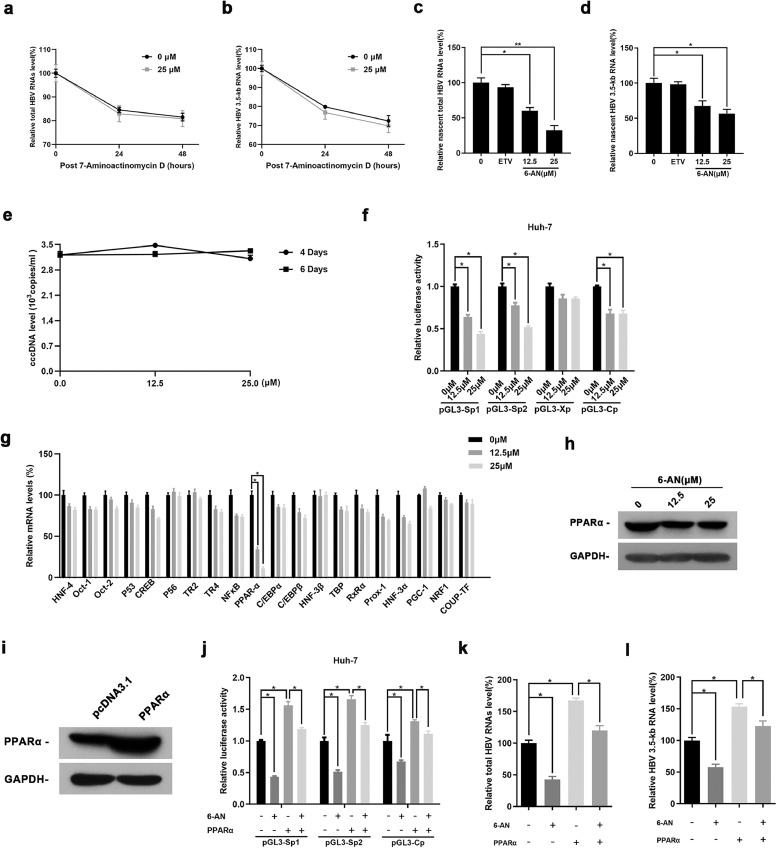
Characterization of 6-AN. (a) Chemical Structural formula and basic information. (b) HepAD38 and HepG2-NTCP cells were treated with the indicated concentrations of 6-AN for 72 h. Effects of 6-AN on the viability of HepAD38 and HepG2-NTCP cells were determined by the MTT assay. (c) The CC50 determined by the MTT assay in the indicated cell lines. (d) EC50 determined by the HBsAg ELISA. Working concentration of 6-AN were set from 200 μM to 0.39 μM in a descending order and 0 μM was set as control. HepAD38 cells were incubated with the medium containing different concentrations of 6-AN for 3days. (e) Effect of 6-AN on CMV promoter. Renilla luciferase reporter vector was transfected into HepG2 cells, which were treated with 12.5 μM and 25 μM 6-AN at 12 h post-transfection. The luciferase activity was measured at 48 h after 6-AN treatment. Results are expressed as the average of three independent experiments (n = 3 per group). The mean value ± standard error is indicated. (*P < 0.05; ⁎⁎P < 0.01).
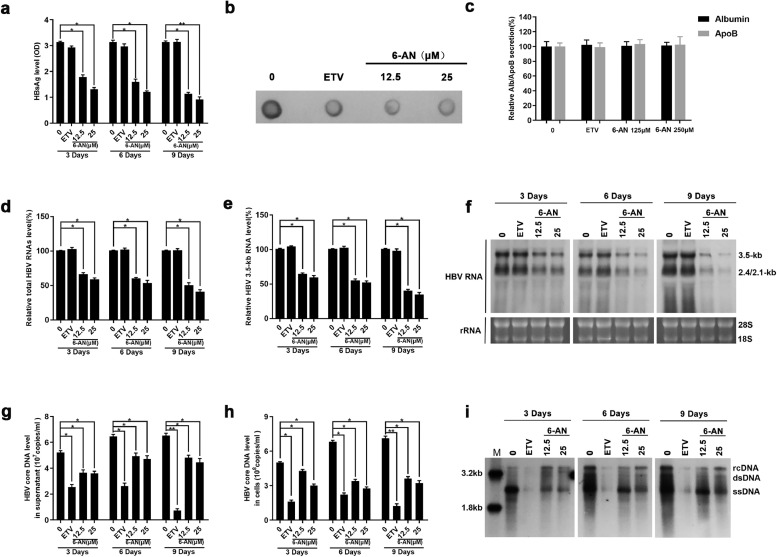
3.3. 6-AN inhibits HBV replication and HBsAg production
To further investigate the anti-HBV activity of 6-AN, HBV markers including HBV RNA, core DNA, cccDNA as well as HBsAg in HepAD38 cells were assessed after 3, 6, 9 days of drug treatment, respectively. ELISA assay showed that 6-AN significantly reduced the level of HBsAg in supernatant. To be specific, at 3, 6 and 9 days, 12.5 μM of 6-AN reduced the level of HBsAg to 57%, 51% and 36% and 25 μM of 6-AN reduced that to 42%, 39% and 29% (Fig. 3a). Dot blot enzyme immunoassay got similar results on total L, M and S secretion (Fig. 3b). To exclude the possibility that HBsAg inhibition was consequent of common secretion pathways inhibiting, we examined the secretion level of albumin and apolipoprotein B in HepAD38 cells and found that 6-AN didn't affect the secretion of albumin and apolipoprotein B, even at the concentration of 250 μM which was 12 times higher than the EC50 (Fig.3c). These data suggest that 6-AN decreases HBsAg production but not blocks its secretion.
Fig. 3.
6-AN inhibited HBV antigen secretion and blocked virus transcription and replication. HepAD38 cells were treated with 0 μM, 12.5 μM, 25 μM 6-AN and 25 nM ETV, after 3, 6 and 9 days of treatment, the level of HBsAg secreted in medium, HBV core DNA in supernatant and cells, total RNAs were extracted (n = 5 per group). (a-b) The relative HBsAg secretion levels were examined by HBsAg ELISA and Dot blot assay. (c) HepAD38 cells were treated with 250 μM 6-AN for 6 days. Culture media were collected and tested for the secretion of human albumin and apolipoprotein B with ELISA. Medium from cells treated with entecavir 25 nM each were included as controls. (d-e) 6-AN inhibited HBV transcription dose-dependently in HepAD38 cells. Total RNAs was extracted after treatment. Relative real-time PCR was subjected to detect the total HBV RNAs (d) and 3.5-kb RNA levels (e), The mRNA level of β-actin was used as internal control. (f) Northern blot was applied to determine the total HBV RNAs and 3.5-kb RNA levels. The rRNA level of 28 s/18 s were used as an internal control. (g-h) 6-AN treatment decreased the level of HBV core DNA in supernatant (g) and in cells (h). The absolute quantification PCR and Southern blot (i) were performed to determine the level of HBV core DNA after 3, 6 and 9days treatment. M, marker; rcDNA, relaxed circular DNA; dsDNA, double-strand DNA; ssDNA, single-strand DNA. Results are expressed as the average of five independent experiments (n = 5 per group). The mean value ± standard error is indicated. (*P < 0.05; ⁎⁎P < 0.01).
HBV cccDNA serves as the template for transcription of all four viral mRNAs (3.5, 2.4, 2.1 and 0.7 kb). HBsAg is composed of large, middle and small surface proteins and is translated from the 2.4/2.1 kb mRNA. To determine if the reduction of HBsAg was due to decreased 2.4/2.1 kb mRNA levels, HBV RNAs were analyzed by real-time PCR and Northern blotting analysis. As expected, real-time PCR showed that 6-AN decreased total HBV RNAs and 3.5-kb RNA levels in a dose-dependent manner, whereas ETV had no effect (Fig. 3d and e). Northern blotting analysis confirmed that 6-AN markedly decreased the level of 3.5-kb, 2.4-kb and 2.1-kb HBV RNAs (Fig.3f). Consequently, a significant reduction of HBV core DNA in supernatant and cell was observed in 6-AN treated HepAD38 cells (Fig. 3g–i), which may be caused by the decline of up-stream 3.5-kb RNA.
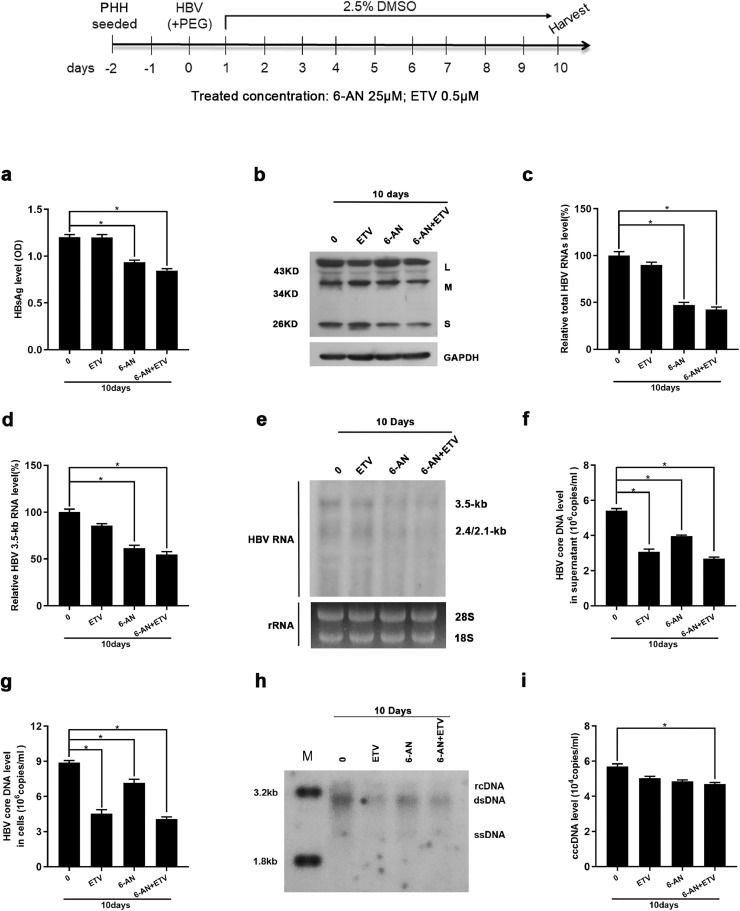
To determine if 6-AN was also effective against HBsAg produced from cccDNA, we infected HepG2-NTCP cells and primary human hepatocytes (PHH) with HBV virions. Consistently, 6-AN alone significantly reduced HBsAg expression in supernatant and cells as evidenced by ELISA and West blotting analysis (Fig 4a and b and Fig.S2a). Moreover, reduced HBV RNAs and core DNA were also observed in 6-AN-treated cells (Fig 4c–h and Fig.S2b–e). Importantly, Taq-man probe qRT-PCR found that 6-AN reduced the level of transcription template cccDNA moderately both in PHHs and HepG2-NTCP cells (Fig. 4i and Fig.S2f). However, ETV alone only reduced HBV core DNA without affecting HBsAg levels and HBV RNAs. Combination with entecavir and 6-AN improved the antiviral activity of 6-AN (Fig. 4a–i). Collectively, these data suggest that 6-AN-reduced HBV replication and HBsAg is resulted from inhibition of 3.5-kb RNA and 2.4/2.1 kb mRNA levels, partially via reducing cccDNA level.
Fig. 4.
6-AN displayed anti-HBV activity in an in vitro HBV infection model. PHH cells were infected with 2 × 103 genome equivalents/cell of HBV particles in the presence of 4% PEG8000 and then co-cultured with 6-AN (0 μM, 25 μM), ETV (0.5 μM) and 6-AN combined ETV for 10 days. (a) Cell culture supernatant were collected for HBsAg analysis via ELISA. 6-AN caused an obvious reduction of HBsAg level of secretion. (b) Western blot proved 6-AN significantly reduced HBsAg in cells. (c-d) 6-AN inhibited the level of total HBV RNAs (c) and 3.5-kb RNA (d) dose-dependently in PHH cells. (e) Northern blot proved that 6-AN not only reduced the 3.5-kb RNA, but also 2.4/2.1-kb RNA. (f-g) 6-AN treatment decreased the level of HBV core DNA in supernatant (f) and in cells (g). (h) Southern blot got a consistent decline. (i) 6-AN treatment showed a little degree reduction of HBV cccDNA level. Results are expressed as the average of four independent experiments (n = 4 per group). The mean value ± standard error is indicated. (*P < 0.05).
3.4. Effect of 6-AN on the promoter activity of HBV genes
To investigate the mechanism by which 6-AN acts on HBV RNAs, we performed RNA dacay assay and a nascent RNA capture assay to examine the effect of 6-AN on HBV RNAs transcription or stability. HBV-infected PHHs were treated with 7-aminoactinomycin D which inhibits DNA-primed RNA polymerase to block RNA synthesis. The data showed that 6-AN did not alter half-life of total HBV RNAs and 3.5-kb RNA, suggesting 6-AN had no effect on HBV RNAs stability (Fig. 5a and b). Meanwhile, a nascent RNA capture assay association with qRT-PCR confirmed that 6-AN decreased the level of nascent HBV RNAs (Fig. 5c and d), suggesting 6-AN inhibited new HBV RNA synthesis without affecting RNAs stability. Moreover, we also excluded that 6-AN partially reduced the level of cccDNA by affecting cccDNA degradation process (Fig. 5e), which implied the reduce may be a result of decrease of nucleic acid synthesis.
Fig. 5.
Effect of 6-AN on the promoter activity of HBV genes. (a-b) HBV-infected PHH cells were treated with 7-aminoactinomycin D (5μg/ml). Total HBV RNAs and 3.5-kb RNA were quantified by RT-qPCR and the amount of RNA at time zero was set at 100%. (c-d) HBV-infected PHH cells incubated with 0.2 mM EU for another 24 h after 6-AN treated 6 days. The newly synthesized EU-labeled RNA was purified from the total RNA, and EU-labeled HBV RNAs were quantified by RT-qPCR. (e) HepAD38 cells were cultured with Tet (2 μg/ml) and ETV (25 nM) during 12 days to eliminate rcDNA, the source of cccDNA. And then, 6-AN was added into the medium, changed every two days. After 6-AN treated 4 days and 6 days, cells were collected to detect the level of cccDNA by Taq man probe real-time PCR. (f) The activities of four HBV promotors were detected by dual-luciferase reporter assay system after 6-AN treated 36 h in Huh-7 cells. (g)Twenty transcription factors related to HBV SpI, SpII and core promotors were screened by RT-qPCR in HepG2-NTCP cells after treated 6-AN 6 days, PPARα were screened out from it. (h) Western blot assay further proved 6-AN reduced the expression of PPARα. (i) Western blotting of PPARα proteins in cells transfected with pcDNA3.1 vector expressing PPARα. GAPDH was used as a loading control. (j-l) The activities of HBV promoters (j) and the level of HBV RNAs (i-m) were detected in Huh-7 cells and HBV infected HepG2-NTCP cells after overexpressed PPARα or 6-AN treated. PPARα overexpression rescued the inhibiting effect caused by 6-AN treated. Results are expressed as the average of three independent experiments (n = 3 per group). The mean value ± standard error is indicated. (*P < 0.05; ** P < 0.01).
6-AN is a well-known inhibitor of G6PD, to exclude the possibility that its anti-HBV function is due to inhibiting G6PD. We transfected HepG2-NTCP with three independent siRNAs targeting G6PD (siG6PD-1, siG6PD-2, siG6PD-3). Real-time PCR results showed efficient knockdown of 6-AN (>70%) compared to RNAi negative control (sicont) (Fig. S3a), and western blot assay showed similar trends (Fig. S3b). However, the effect of G6PD knockdown on HBV transcription and replication was not obvious. ELISA assay and real-time PCR showed no significance influence happened to HBsAg (Fig. S3c), HBV RNAs (Fig. S3d–e), cccDNA (Fig. S3f) and HBV core DNA (Fig. S3g–h). These data suggest 6-AN possess the anti-HBV activity of 6-AN was not dependent on G6PD inhibition. Next, we examined the effect of 6-AN on four HBV promoters that closely association with cccDNA transcription. Consistently, the data showed that 6-AN significantly reduced SpI, SpII and core promoter activities (Fig. 5f). To further examine the mechanism of transcriptional inhibition of 6-AN on HBV promoters, a panel of transcription factors, including HNF-4, OCT-1, OCT-2, CREB, TBP, NF-κB, PPARα, HNF-3β, HNF-3α et.al., was screened by real-time PCR. Therein, peroxisome proliferators-activated receptors-α (PPARα), control of all the three promoters [36], were markedly reduced by 6-AN (Fig. 5g–h). Collectively, 6-AN inhibited the activities of HBV SpI, SpII and core promoter by decreasing transcription factor PPARα, subsequently reduced HBV RNAs transcription and HBsAg production. Meanwhile, we demonstrated that overexpression of PPARα significantly accelerated the activities of three promoters and the production of HBV RNAs, and rescued the decline of HBV promoters and HBV RNAs caused by 6-AN treatment (Fig. 5i–l).
3.5. Anti-HBV activity of 6-AN in transient transfection cell model
HBV is classified into eight genotypes (A to H) [37,38], and A, B, C and D are the four most prevalent genotypes among them. However, the HBV genome in HepAD38 cells was of genotype D and PHHs/HepG2-NTCP cells were infected with HBV virions harvested from HepAD38 cells, these data only showed the activity of 6-AN on HBV genotype D. To examine if 6-AN possess a broad genotypic activity, plasmids of HBV genotypes A, B, C and D were transfected into HepG2 cells, respectively. After treatment of 6-AN for 3 days, the amount of HBsAg, HBeAg and HBV core DNA secreted into the culture medium, as well as the level of intracellular HBV core DNA, total HBV RNAs and 3.5-kb RNA were determined. As shown in Table 1 and Fig. S4, the viral makers were markedly decreased after treated with 6-AN, suggesting that all 4 tested genotypes of HBV were sensitive to 6-AN.
Table. 1.
6-AN activity across HBV genotypes.
| HBV Genotype | 6-AN (μM) | HBsAg (%) | HBeAg (%) | HBV core DNA level (%) | Total HBV RNAs (%) | HBV 3.5-kb RNA (%) | |
|---|---|---|---|---|---|---|---|
| Supernatant | Intracellular | ||||||
| Genotype A | 12.5 | 62.81* | 80.37* | 100.82n.s. | 101.33n.s. | 80.06* | 75.39* |
| 25 | 49.02* | 68.92* | 91.60n.s. | 97.42n.s. | 67.55* | 71.96* | |
| Genotype B | 12.5 | 76.74* | 86.00* | 81.26* | 71.67* | 67.95* | 73.45* |
| 25 | 72.16* | 83.44* | 78.63* | 66.16* | 47.03* | 54.00* | |
| Genotype C | 12.5 | 81.47* | 31.59* | 81.05* | 78.90* | 71.52* | 72.40* |
| 25 | 77.96* | 23.86* | 70.16* | 74.82* | 69.00* | 59.33* | |
| Genotype D | 12.5 | 41.61* | 33.18* | 69.76* | 59.97* | 65.90* | 58.38* |
| 25 | 36.18* | 31.02* | 60.54* | 30.61* | 58.33* | 51.71* | |
The plasmid with dimers of HBV genotypes A, B, C and D were transfected into HepG2 cells, 12 h after transfected, the cells were incubated with 12.5 μM, 25 μM 6-AN and continued to incubate 72 h. Then viral markers were detected (n = 5 per group). (* P < 0.05, n.s., not significant, compared with the normal group).
6-AN, 6-Aminonicotinamide; HBsAg, Hepatitis B surface antigen; HBeAg, Hepatitis B e antigen.
3.6. 6-AN exhibited antiviral activity in vivo
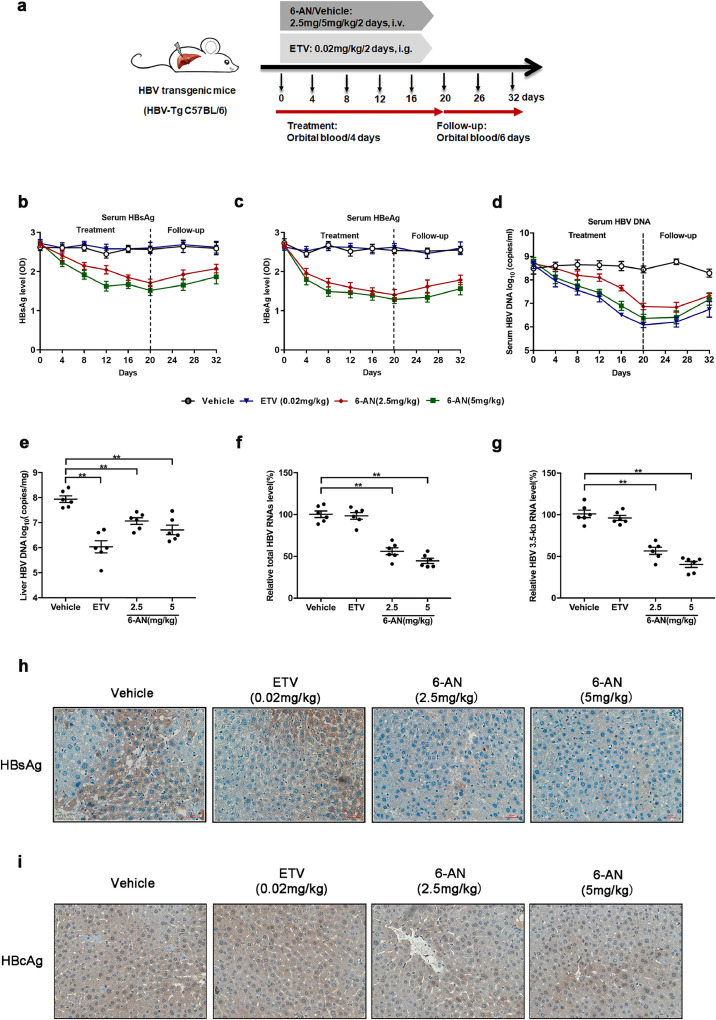
To detect the biological anti-HBV activity of 6-AN in vivo, HBV-transgenic mice were adopted for studying the antiviral effect of 6-AN. Each group of mice with an average body weight of 21 ± 1 g were then treated with 2.5 mg/kg, 5 mg/kg, 10 mg/kg and 20 mg/kg of 6-AN, respectively, by intravenous injection every 2 days for the evaluation of the lethal dose 50 (LD50) of 6-AN. All the mice were in a good condition except that a phenomenon of muscle weakness was appeared in the mice treated with 20 mg/kg of 6-AN for 2 weeks. Therefore, 2.5 mg/kg and 5 mg/kg of 6-AN were selected for the subsequent experiments (Fig. 6a). The level of serum ALT/AST indicated no significant hepatotoxicity (Fig. S5 a-b). Moreover, no significant difference in body and liver weight were observed in 6-AN treated group when compared to the vehicle group (Fig. S5c and d). Moreover, the level of serum HBsAg secretion in mice treated with 2.5 mg/kg or 5 mg/kg of 6-AN was decreased by 35% and 42% respectively (Fig. 6b). In addition, the level of HBeAg was decreased by 47% and 51% at the end of treatment period at day 20 (Fig. 6c). And both 6-AN and ETV markedly reduced serum HBV DNA after or during treatment (Fig. 6d). The level of HBV viral markers had an uptrend after cessation of treatment, but level of serum HBV DNA restored more slowly in 6-AN group when compared to ETV group. The level of HBsAg and HBV DNA showed no significant changes in vehicle group during the experiment (Table 2). Consistently, 6-AN treatment decreased intrahepatic HBV DNA, total HBV RNAs and 3.5-kb RNA in HBV-transgenic mice model (Fig. 6e–g). In addition, immunohistochemistry results demonstrated that 6-AN lead to a significant reduction of cytoplasmic HBsAg level when compared to mice receiving vehicle or ETV treatment (Fig. 6h), and HBcAg both in cytoplasmic and nuclear were decreased in 6-AN treated group (Fig. 6i). These data indicated the antiviral effects of 6-AN in vivo.
Fig. 6.
6-AN exhibited antiviral activity in vivo. (a) Flow chart explaining the way and concentration of 6-AN and ETV administration as well as the intervals of orbital blood collection. The mice were randomly assigned to 4 groups (n = 10 per group). 6-AN was injected at a dose of 0 mg/kg, 2.5 mg/kg and 5 mg/kg once in two days. ETV by administering oral doses at 0.2 mg/kg/2day. Blood samples were taken every 4 days until the 20th day. At day 20, 6 animals from each group were sacrificed to harvest the liver samples. The remaining animals (n = 4 per group) were monitored for the follow-up period and blood samples were taken every 6 days until to the 32th day. (b-c) HBsAg, HBeAg were determined in serum from HBV-transgenic mice with 6-AN during the treatment and follow-up period up to 32 days by ELISA. Shown are mean ratio to baseline values ± standard error. (d)The serum level of HBV DNA was extracted and analyzed by absolute quantification PCR. Shown are mean values ± standard error. At day 20, 6 animals from each group were sacrificed and the livers were harvested for hepatic HBV DNA, total HBV RNAs and 3.5-kb RNA determination and immunohistochemistry examination of HBsAg in HBV-transgenic mice tissues were determined after 6-AN treatment. (e)Liver HBV DNA was analyzed by absolute quantification PCR. (f-g) Relative real-time PCR was subjected to detect the total HBV RNAs (f) and 3.5-kb RNA (g) levels, the mRNA level of β-actin was used as an internal control. (h-i) Representative images of immunohistochemistry of HBsAg (h) and HBcAg (i) in liver tissue. (*P < 0.05; n.s., not significant).
Table. 2.
Serum HBV Markers Analysis from HBV-Transgenic Mice.
| Treatmenta | HBV DNA (log10 ± SD) | HBsAg (%) | HBeAg (%) | |
|---|---|---|---|---|
| Day 20b | Day 20 – Baselinec | Day 20/Baselined | Day 20/Baselinee | |
| Vehicle | 8.44 ± 0.46 | −0.04 ± 0.70n.s. | 97.24 ± 9.75n.s. | 96.17 ± 9.58n.s. |
| ETV(0.02 mg/kg) | 6.09 ± 0.37 | −2.55 ± 0.65* | 98.62 ± 17.24n.s. | 99.33 ± 12.03n.s. |
| 6-AN(2.5 mg/kg) | 6.87 ± 0.44 | −1.83 ± 0.75* | 64.82 ± 11.57⁎⁎ | 53.35 ± 16.25⁎⁎ |
| 6-AN(5 mg/kg) | 6.37 ± 0.54 | −2.43 ± 0.86* | 57.50 ± 15.28⁎⁎ | 48.81 ± 12.49⁎⁎ |
At day 20, the levels of serum HBV DNA and the ratio of HBsAg from HBV-transgenic mouse was shown in Table 3 (n = 10 per group). (* P < 0.05, ⁎⁎p < 0.01, n.s., not significant, compared with the vehicle group).
6-AN, 6-Aminonicotinamide; HBsAg, Hepatitis B surface antigen; HBeAg, Hepatitis B e antigen; ETV, Entecavir.
Treatment doses and schedules as described in the Methods section.
Mean log10 values ± standard error (copies/ml).
Mean log10 change values ± standard error (copies/ml) from baseline.
Mean ratio values ± standard error (%) of HBsAg to baseline.
Mean ratio values ± standard error (%) of HBeAg to baseline.
3.7. Combination treatment with 6-AN and ETV induced a profound reduction of both viremia and HBsAg levels in (r) cccDNA recombination mice
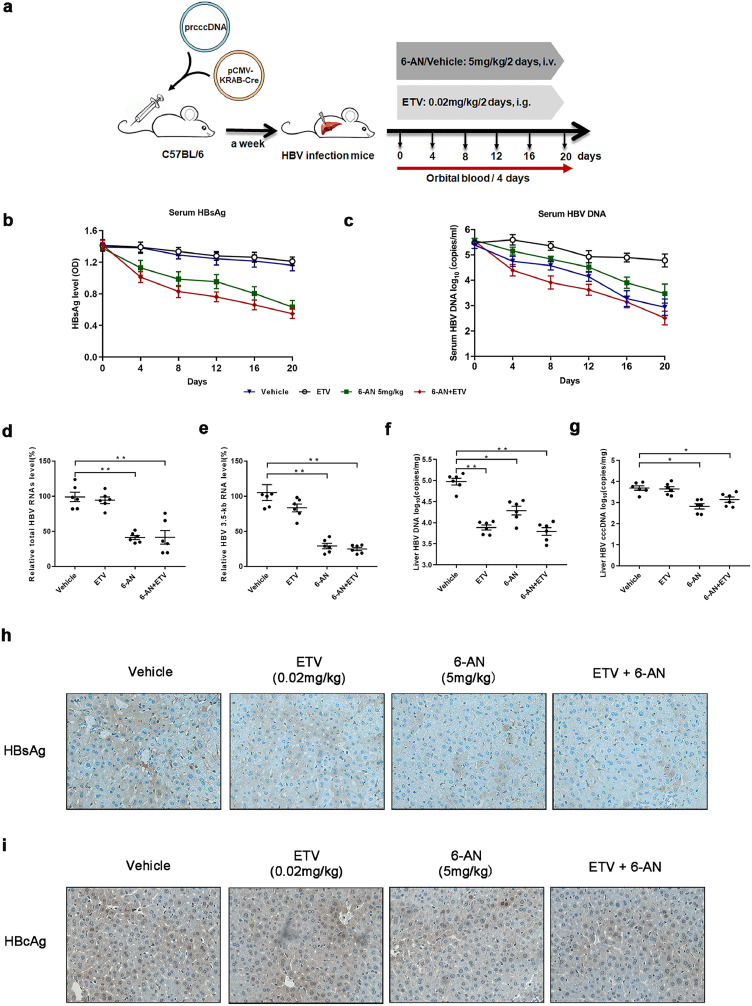
Next, the antiviral efficacy of 6-AN in combination with ETV was evaluated in a mouse model of HBV infection involving HBV recombinant (r) cccDNA using site-specific DNA recombination as described previously [39]. The infected mice were treated with vehicle, with 6-AN at 5 mg/kg once two days, with ETV at 0.02 mg/kg once two days (Fig. 7a). Similarly, the level of serum ALT/AST indicated no significant hepatotoxicity (Fig. S6a and b), body and liver weight did not show a significant change in all the group (Fig. S6c and d). Moreover, caspase3 and ki67 staining in cytoplasmic were almost undetectable, suggested that there was no obvious apoptosis and malignant proliferation in the mice administrated with 6-AN (Fig. S6e–f). As before, 6-AN alone significantly reduced both circulating HBsAg (by 48%) and HBV DNA (2.10 log10) (Fig. 7b and c). ETV alone reduced HBV DNA (2.46 log10) with no markedly effect on HBsAg levels compared to baseline. Combining 6-AN and ETV showed a pronounced reduction of both HBsAg (by 55%) and HBV DNA (3.03 log10) (Fig. 7c and Table3). To further evaluate the levels of HBV RNAs, cccDNA, and HBV DNA in mouse liver, all animals were sacrificed to collected liver tissue after 20 days of treatment. Similar decrease of intrahepatic HBV RNAs, DNA, cccDNA, HBcAg as well as HBsAg were observed in 6-AN treated group (Fig. 7d–i). Combination therapy of entecavir and 6-AN also improved the antiviral activity of 6-AN (Fig. 7d–i). These data suggest the combination therapy resulted in a robust reduction of all viral markers and intracellular antigen levels tested in our study.
Fig. 7.
Combination treatment with 6-AN and ETV induced a profound reduction of both viremia and HBsAg levels in (r) cccDNA recombination mice. (a) Flow chart explaining the way and concentration of 6-AN and ETV administration as well as the intervals of orbital blood collection. The mouse model established by injecting prcccDNA and pCMV-KRAB-Cre from tail vein. The mouse model of HBV infection involving HBV recombinant (r) cccDNA constructed successfully were randomly assigned to 4 groups (n = 6 per group). 6-AN was dissolved and diluted in saline and tail intravenous injected at a dose of 0 mg/kg and 5 mg/kg once in two days. ETV was dissolved in water by administering oral doses at 0.2 mg/kg/2day. Blood samples were taken every 4 days until the 20th day. (b) HBsAg were determined in serum during the treatment by ELISA. Shown are mean ratio to baseline values ± standard error from six mice. (c)The serum level of HBV DNA was extracted and analyzed by absolute quantification PCR. Shown are mean values ± standard error from six mice. At day 20, All animals from each group were sacrificed to harvest the liver samples. Hepatic HBV DNA, HBV cccDNA, total HBV RNAs and 3.5-kb RNA and immunohistochemistry examination of HBsAg were determined in those mice tissues after 6-AN treatment. (d-e) Relative real-time PCR was subjected to detect the total HBV RNAs (d) and 3.5-kb RNA (e) levels, the mRNA level of β-actin was used as an internal control. (f) Liver HBV DNA was analyzed by absolute quantification PCR. (g) Liver HBV cccDNA was analyzed by absolute quantification PCR, using cccDNA-specific primers. (h-i) Representative images of immunohistochemistry of HBsAg (h) and HBcAg (i) in liver tissue. (*P < 0.05; ⁎⁎P < 0.01; n.s., not significant).
Table. 3.
Serum HBV Markers Analysis from Mouse Model of HBV Infection Involving HBV Recombinant (r) cccDNA.
| Treatmenta | HBV DNA (log10 ± SD) | HBsAg (%) | |||
|---|---|---|---|---|---|
| Day 20b | Day 20 – Baselinec | Change from vehicled | Day 20/Baselinee | Ratio to vehiclef | |
| Vehicle | 4.78 ± 0.57 | −0.70 ± 0.54 | n.s. | 86.41 ± 8.68 | n.s. |
| ETV(0.02 mg/kg) | 2.93 ± 0.73 | −2.46 ± 0.90 | −1.85 ± 0.73** | 82.78 ± 11.07 | 95.80 ± 12.82 |
| 6-AN(5 mg/kg) | 3.47 ± 0.84 | −2.10 ± 0.91 | −1.31 ± 0.84* | 45.12 ± 13.26 | 52.22 ± 15.35** |
| 6-AN+ETV | 2.51 ± 0.61 | −3.03 ± 0.63 | −2.27 ± 0.61** | 39.11 ± 9.71 | 45.26 ± 11.23** |
At day 20, the levels of serum HBV DNA and the ratio of HBsAg from mouse model of HBV infection involving HBV recombinant (r) cccDNA were shown in Table 3 (n = 6 per group). (* P < 0.05, ⁎⁎p < 0.01, n.s., not significant, compared with the vehicle group).
6-AN, 6-Aminonicotinamide; HBsAg, Hepatitis B surface antigen; ETV, Entecavir.
Treatment doses and schedules as described in the Methods section.
Mean log10 values ± standard error (copies/ml).
Mean log10 change values ± standard error (copies/ml) from baseline.
Mean log10 change values ± standard error (copies/ml) from vehicle.
Mean ratio values ± standard error of HBsAg to baseline.
Mean ratio values ± standard error of HBsAg to vehicle.
4. Discussion
Despite new advances in therapeutic strategies over recent decades, chronic hepatitis B remains a major global health burden, and patients are still at high risk of developing some serious diseases including liver cirrhosis and hepatocellular carcinoma [1]. Current therapies available to HBV-infected patients, such as interferon-α (IFN-α) and nucleoside analogues, seldom achieve functional cure, and many of them cause different extents of liver damage with serious adverse effects [40,41]. Therefore, new therapeutic approaches for chronic HBV infection is desirable. Although many new HBsAg inhibitors have been reported in recent years, these inhibitors seldom possess comprehensive anti-HBV efficacy. For example, HBF-0259 could inhibit HBsAg secretion but possess no effect on other viral markers [42]. A benzimidazole derivative, BM601, inhibited HBsAg secretion, but without effects on HBV DNA replication and HBeAg secretion [43]. Cyclophilin A inhibitors are well-known to have antiviral properties [44], which significantly decreased the level of HBsAg secretion in a dose- and time-dependent manner, but with no effect on HBV DNA. Moreover, the in vivo anti-HBV efficiency of all the candidates mentioned were not evaluated. On the contrary, our study showed a novel class of small molecule which significantly inhibited the transcription and replication of HBV in several HBV cell models, and with the in vivo anti-HBV efficiency validated successfully on mouse model of HBV-transgenic mouse and HBV infection involving HBV recombinant (r) cccDNA, respectively.
By screening 1500 compounds from a small molecule compound library, we identified 5 compounds that exhibited potent inhibition of HBsAg secretion in a dose-dependent manner without obvious cytotoxicity in the HepAD38 cell model. Therein, 6-Aminonicotinamide (6-AN), an analogue of niacin, showed the most effective anti-HBV activity. It inhibited significantly the expression levels of HBsAg both in vitro and in vivo. Previous studies have demonstrated that 6-AN has been clinically used to treat some advanced cancers as well as serving as a topical treatment for skin diseases [45]. In addition, some microorganisms and parasites, such as Leishmania and Plasmodium falciparum [46], exhibited sensitive and strong response to 6-AN, highlighting the possible functional roles of 6-AN in clinical application. Here, we first reported that 6-AN exhibited potent anti-HBV activity in vitro. Our results showed that 6-AN exhibited no obvious cytotoxicity in a panel of liver cell lines, and reduced significantly the HBsAg secretion with relative low working concentrations at 12.5 μM. Furthermore, we found that 6-AN significantly decreased the levels of total HBV RNAs and 3.5-kb RNA with a dose- and time-dependent manner in both HepAD38 and HBV-infected HepG2-NTCP/PHHs cell lines, suggesting its effective inhibitory role in HBV transcription. Moreover, we confirmed that the HBV RNAs reduction was not due to the acceleration of RNA decay but 6-AN suppressed nascent HBV RNAs synthesis. We further investigated the role of 6-AN on HBV cccDNA replication and transcription. Interestingly, we first found that 6-AN only decreased the level of cccDNA in a minor degree, but significantly reduced the ratios of total HBV RNAs to cccDNA or 3.5-kb RNA to cccDNA which was on behalf of cccDNA transcription activity. Moreover, we also found that the level of HBV DNA was reduced in a dose- and time-dependent manner both in vitro and in vivo, and we guess that may be due to the decline of 3.5-kb RNA.
Besides, the cccDNA minichromosome, as a transcription template, directs the viral RNAs synthesis by using polymerase II [47]. Meanwhile, it is well known that HBV transcription is modulated by four promoters, including the core, SpI, SpII and X promoters. The core promoter is responsible for the transcription of 3.5-kb RNA, including pregenomic RNA (pgRNA) and precore/core RNA which encodes for polymerase protein, core protein, and the precore protein. The SpI promoter controls the transcription of the 2.4-kb mRNA, which encodes the large surface protein. The SpII promoter supports the transcription of the 2.1-kb mRNA, which encodes the middle and small surface protein. The X promoter controls the transcription of the 0.7-kb mRNA encoding the X protein, a regulatory protein of HBV [48,49]. We next confirmed that 6-AN significantly reduced the activities of HBV SpI, SpII and core promoter which are closely association with cccDNA transcription activity. Moreover, we screened out a transcription factor, PPARα, control of all the three promoters, was reduced significantly when treated with 6-AN. On the Contrary, the decline of HBV promoters and HBV RNAs were reversed when PPARα overexpression. These data demonstrated that 6-AN inhibited the activities of HBV promoters by decreasing expression of PPARα to interfere cccDNA transcription activity, then inhibited RNA synthesis and HBsAg expression to act as an anti-HBV agent.
Additionally, HBV genotype can also affect the response of HBV infected patients to nucleos(t)ide analogues treatment [50,51]. To evaluate the antiviral effect of 6-AN on various HBV genotypes, plasmids expressing HBV genotypes A, B, C and D, respectively, were transfected into HepG2 cells. Our data confirmed that similar inhibitory effect of 6-AN were observed on all tested genotypes, suggesting 6-AN possesses a broad cross-genotype activity against wild-type HBV A to D.
Notably, we are the first group to perform in vivo investigation on the efficacy of 6-AN against HBV by using the mouse model of HBV-transgenic mouse and HBV infection involving HBV recombinant (r) cccDNA. ALT and AST activities showed no obvious hepatotoxicity during and after the treatment of 6-AN. Consistently, no significant changes in body weights of animals were observed, immunohistochemical analysis also did not detect the expression of caspase3 and ki67 in cytoplasmic, suggested that 6-AN did not induce the cell to malignantly proliferate or accelerate the cell apoptosis. More importantly, viral markers in serum and tissue were both significantly reduced after administrating with 6-AN alone rather than ETV alone. Therein, HBsAg levels were reduced more profoundly than other markers, which is highly consistent with the results in vitro. Furthermore, the rebound of serum HBsAg and HBV DNA level in HBV-transgenic mice was found after cessation of therapy, which suggesting that curative effect is dependent on the continued presence of 6-AN or ETV.
In HBV curative, combination therapy is more efficient than monotherapy. In our study, 6-AN is the main driver of HBsAg, while ETV is more efficient on HBV DNA reduction. we combined 6-AN with ETV to achieve a complementary plate, and result in a balanced antiviral situation that effectively reduced HBV DNA and HBsAg both in vitro and in vivo.
Taken together, the findings in this study has revealed that 6-AN exhibits a potent anti-HBV effect both in vitro and in vivo, via affecting HBsAg production as well as HBV transcription and replication, thus may provide a valuable alternative or complementary therapy for the current and future antiviral treatments.
CRediT authorship contribution statement
Fang Ren: Writing - original draft. Xiao Yang: Writing - original draft. Zhong-Wen Hu: Writing - original draft. Vincent Kam Wai Wong: Methodology. Hong-Yan Xu: Methodology. Ji-Hua Ren: Methodology. Shan Zhong: Methodology. Xiao-Jiong Jia: . Hui Jiang: Methodology. Jie-Li Hu: Methodology. Xue-Fei Cai: Data curation. Wen-Lu Zhang: Data curation. Fang-Long Yao: Data curation. Hai-Bo Yu: Data curation. Sheng-Tao Cheng: Formal analysis. Hong-Zhong Zhou: Formal analysis. Ai-Long Huang: Formal analysis. Betty Yuen Kwan Law: Conceptualization. Juan Chen: Conceptualization.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81861168035, 81871656 and 81922011, JC), Chongqing Natural Science Foundation (cstc2018jcyjAX0114, JC) and Creative Research Group of CQ University (CXQT19016, JC), National Science and Technology Major Project (Grant no. 2017ZX10202203 to AL H), and scientific research project jointly funded by National Natural Science Foundation of China and the Macao Science and Technology Development Fund) (0036/2018/AFJ to YK L) .
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.022.
Contributor Information
Betty Yuen Kwan Law, Email: yklaw@must.edu.mo.
Juan Chen, Email: chenjuan2014@cqmu.edu.cn.
Appendix. Supplementary materials
References
- 1.Schweitzer A., Horn J., Mikolajczyk R.T. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. 2015/08/02. [DOI] [PubMed] [Google Scholar]
- 2.Chen D.S. From hepatitis to hepatoma: lessons from type b viral hepatitis. Science. 1993;262:369–370. doi: 10.1126/science.8211155. 1993/10/15. [DOI] [PubMed] [Google Scholar]
- 3.Yan H., Zhong G., Xu G. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. 2012/11/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCaffrey A.P., Nakai H., Pandey K. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–644. doi: 10.1038/nbt824. 2003/05/13. [DOI] [PubMed] [Google Scholar]
- 5.Lindh M., Rydell G.E., Larsson S.B. Impact of integrated viral DNA on the goal to clear hepatitis B surface antigen with different therapeutic strategies. Curr Opin Virol. 2018;30:24–31. doi: 10.1016/j.coviro.2018.01.011. 2018/02/18. [DOI] [PubMed] [Google Scholar]
- 6.European association for the study of the liver. electronic address eee and european association for the study of the L. Easl 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. 2017/04/22. [DOI] [PubMed] [Google Scholar]
- 7.Revill P.A., Chisari F.V., Block J.M. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. 2019;4:545–558. doi: 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuen M.F., Wong D.K., Fung J. HBsAg Seroclearance in chronic hepatitis B in Asian patients: replicative level and risk of hepatocellular carcinoma. Gastroenterology. 2008;135:1192–1199. doi: 10.1053/j.gastro.2008.07.008. 2008/08/30. [DOI] [PubMed] [Google Scholar]
- 9.Kim G.A., Lee H.C., Kim M.J. Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: a need for surveillance. J Hepatol. 2015;62:1092–1099. doi: 10.1016/j.jhep.2014.11.031. 2014/12/03. [DOI] [PubMed] [Google Scholar]
- 10.Lin C.L., Kao J.H. Risk stratification for hepatitis B virus related hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:10–17. doi: 10.1111/jgh.12010. 2012/10/26. [DOI] [PubMed] [Google Scholar]
- 11.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. 2000/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J., Sun X., Fang J. Analysis of intrahepatic total HBV DNA, cccDNA and serum HBsAg level in Chronic Hepatitis B patients with undetectable serum HBV DNA during oral antiviral therapy. Clin Res Hepatol Gastroenterol. 2017;41:635–643. doi: 10.1016/j.clinre.2017.03.004. 2017/04/26. [DOI] [PubMed] [Google Scholar]
- 13.Lok A.S., McMahon B.J., Brown R.S., Jr. Antiviral therapy for chronic hepatitis B viral infection in adults: a systematic review and meta-analysis. Hepatology. 2016;63:284–306. doi: 10.1002/hep.28280. 2015/11/14. [DOI] [PubMed] [Google Scholar]
- 14.EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. 2017/04/22. [DOI] [PubMed] [Google Scholar]
- 15.Marcellin P., Sang H.A., Ma X. Combination of tenofovir disoproxil fumarate and peginterferon alfa-2 a increases loss of hepatitis B surface antigen in patients with chronic hepatitis B. Gastroenterology. 2016;150:134–144. doi: 10.1053/j.gastro.2015.09.043. e110. [DOI] [PubMed] [Google Scholar]
- 16.Perlia C.P., Kofman S., Sky-Peck H. Clinical use of 6-aminonicotinamide in patients with disseminated neoplastic disease. Cancer. 1961;14:644–648. doi: 10.1002/1097-0142(199005/06)14:3<644::aid-cncr2820140328>3.0.co;2-n. 1961/05/01. [DOI] [PubMed] [Google Scholar]
- 17.Koutcher J.A., Alfieri A.A., Stolfi R.L. Potentiation of a three drug chemotherapy regimen by radiation. Cancer Res. 1993;53:3518–3523. 1993/08/01. [PubMed] [Google Scholar]
- 18.Stolfi R.L., Colofiore J.R., Nord L.D. Biochemical modulation of tumor cell energy: regression of advanced spontaneous murine breast tumors with a 5-fluorouracil-containing drug combination. Cancer Res. 1992;52:4074–4081. 1992/08/01. [PubMed] [Google Scholar]
- 19.Catanzaro D., Nicolosi S., Cocetta V. Cisplatin liposome and 6-amino nicotinamide combination to overcome drug resistance in ovarian cancer cells. Oncotarget. 2018;9:16847–16860. doi: 10.18632/oncotarget.24708. 2018/04/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims J.L., Sikorski G.W., Catino D.M. Poly(adenosinediphosphoribose) polymerase inhibitors stimulate unscheduled deoxyribonucleic acid synthesis in normal human lymphocytes. Biochemistry. 1982;21:1813–1821. doi: 10.1021/bi00537a017. 1982/04/13. [DOI] [PubMed] [Google Scholar]
- 21.Dewey D.L., Hawes C. 6-Aminonicotinamide and the radiosensitivity of human liver cells in culturE. Nature. 1963;200:1176–1178. doi: 10.1038/2001176a0. 1963/12/21. [DOI] [PubMed] [Google Scholar]
- 22.Dietrich L.S., Muniz O., Farinas B. 6-aminonicotinamide-14 C utilization by the 755 tumor and host liver tissue. Cancer Res. 1968;28:1652–1654. 1968/08/01. [PubMed] [Google Scholar]
- 23.Yin X., Tang B., Li J.H. ID1 promotes hepatocellular carcinoma proliferation and confers chemoresistance to oxaliplatin by activating pentose phosphate pathway. J Exp Clin Cancer Res. 2017;36:166. doi: 10.1186/s13046-017-0637-7. 2017/11/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Downs S.M., Humpherson P.G., Leese H.J. Meiotic induction in cumulus cell-enclosed mouse oocytes: involvement of the pentose phosphate pathway. Biol Reprod. 1998;58:1084–1094. doi: 10.1095/biolreprod58.4.1084. 1998/04/18. [DOI] [PubMed] [Google Scholar]
- 25.Comizzoli P., Urner F., Sakkas D. Up-regulation of glucose metabolism during male pronucleus formation determines the early onset of the s phase in bovine zygotes. Biol Reprod. 2003;68:1934–1940. doi: 10.1095/biolreprod.102.011452. 2003/02/28. [DOI] [PubMed] [Google Scholar]
- 26.Urner F., Sakkas D. Involvement of the pentose phosphate pathway and redox regulation in fertilization in the mouse. Mol Reprod Dev. 2005;70:494–503. doi: 10.1002/mrd.20222. 2005/02/03. [DOI] [PubMed] [Google Scholar]
- 27.Xiao W.J., Ma T., Ge C. Modulation of the pentose phosphate pathway alters phase I metabolism of testosterone and dextromethorphan in HepG2 cells. Acta Pharmacol Sin. 2015;36:259–267. doi: 10.1038/aps.2014.137. 2015/01/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Zhang Y., Zhu X. DEAD box protein 5 inhibits liver tumorigenesis by stimulating autophagy via interaction with p62/SQSTM1. Hepatology. 2018 doi: 10.1002/hep.30300. 2018/10/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J.H., Hu J.L., Cheng S.T., et al. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3-9 homolog 1 and set domain containing 1 A histone methyltransferases. Hepatology2018. 10.1002/hep.29912. [DOI] [PubMed]
- 30.Ren J.H., Tao Y., Zhang Z.Z. Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J Virol. 2014;88:2442–2451. doi: 10.1128/JVI.02861-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang P.L., Althage A., Chung J. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. 2002/10/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang P.L., Althage A., Chung J. Immune effectors required for hepatitis B virus clearance. Proc Natl Acad Sci USA. 2010;107:798–802. doi: 10.1073/pnas.0913498107. 2010/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang L.R., Wu H.L., Chen P.J. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci USA. 2006;103:17862–17867. doi: 10.1073/pnas.0608578103. 2006/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cullen B.R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986;46:973–982. doi: 10.1016/0092-8674(86)90696-3. 1986/09/26. [DOI] [PubMed] [Google Scholar]
- 35.Ladner S.K., Otto M.J., Barker C.S. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. 1997/08/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quasdorff M., Protzer U. Control of hepatitis B virus at the level of transcription. J Viral Hepat. 2010;17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. 2010/06/16. [DOI] [PubMed] [Google Scholar]
- 37.Norder H., Courouce A.M., Magnius L.O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. 1994/02/01. [DOI] [PubMed] [Google Scholar]
- 38.Stuyver L., De Gendt S., Van Geyt C. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. 2000/01/21. [DOI] [PubMed] [Google Scholar]
- 39.Qi Z., Li G., Hu H. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88:8045–8056. doi: 10.1128/JVI.01024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brahmania M., Janssen H.L.A. Future therapy for HBV: role of cell cycle inhibitors. Curr Hepatol Rep. 2016;15:245–251. [Google Scholar]
- 41.Zoulim F., Durantel D. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021501. 2015/04/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dougherty A.M., Guo H., Westby G. A substituted tetrahydro-tetrazolo-pyrimidine is a specific and novel inhibitor of hepatitis B virus surface antigen secretion. Antimicrob Agents Chemother. 2007;51:4427–4437. doi: 10.1128/AAC.00541-07. 2007/09/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y.B., Yang L., Wang G.F. Benzimidazole derivative, BM601, a novel inhibitor of hepatitis B virus and HBsAg secretion. Antiviral Res. 2014;107:6–15. doi: 10.1016/j.antiviral.2014.04.002. 2014/04/22. [DOI] [PubMed] [Google Scholar]
- 44.Dawar F.U., Tu J., Khattak M.N. Cyclophilin A: a key factor in virus replication and potential target for anti-viral therapy. Curr Issues Mol Biol. 2017;21:1–20. doi: 10.21775/cimb.021.001. 2016/04/02. [DOI] [PubMed] [Google Scholar]
- 45.Zackheim H.S. Topical 6-aminonicotinamide plus oral niacinamide therapy for psoriasis. Arch Dermatol. 1978;114:1632–1638. 1978/11/01. [PubMed] [Google Scholar]
- 46.Almugadam S.H., Trentini A., Maritati M. Influence of 6-aminonicotinamide (6AN) on Leishmania promastigotes evaluated by metabolomics: beyond the pentose phosphate pathway. Chem Biol Interact. 2018;294:167–177. doi: 10.1016/j.cbi.2018.08.014. 2018/09/01. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W., Chen J., Wu M. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology. 2017;66:398–415. doi: 10.1002/hep.29133. 2017/02/27. [DOI] [PubMed] [Google Scholar]
- 48.Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4–s16. doi: 10.1016/j.jhep.2016.01.027. 2016/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang H., Oishi N., Kaneko S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006;97:977–983. doi: 10.1111/j.1349-7006.2006.00299.x. 2006/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcellin P., Heathcote E.J., Buti M. Tenofovir disoproxil fumarate versus adefovir dipivoxil for chronic hepatitis B. N Engl J Med. 2008;359:2442–2455. doi: 10.1056/NEJMoa0802878. 2008/12/05. [DOI] [PubMed] [Google Scholar]
- 51.Zeuzem S., Gane E., Liaw Y.F. Baseline characteristics and early on-treatment response predict the outcomes of 2 years of telbivudine treatment of chronic hepatitis B. J Hepatol. 2009;51:11–20. doi: 10.1016/j.jhep.2008.12.019. 2009/04/07. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.