Abstract
Background
Colon cancer stem cells (CSCs), considered responsible for tumor initiation and cancer relapse, are constantly exposed to regulatory cues emanating from neighboring cells present in the tumor microenvironment. Among these cells are enteric glial cells (EGCs) that are potent regulators of the epithelium functions in a healthy intestine. However, whether EGCs impact CSC-driven tumorigenesis remains unknown.
Methods
Impact of human EGC primary cultures or a non-transformed EGC line on CSCs isolated from human primary colon adenocarcinomas or colon cancer cell lines with different p53, MMR system and stemness status was determined using murine xenograft models and 3D co-culture systems. Supernatants of patient-matched human primary colon adenocarcinomas and non-adjacent healthy mucosa were used to mimic tumor versus healthy mucosa secretomes and compare their effects on EGCs.
Findings
Our data show that EGCs stimulate CSC expansion and ability to give rise to tumors via paracrine signaling. Importantly, only EGCs that were pre-activated by tumor epithelial cell-derived soluble factors increased CSC tumorigenicity. Pharmacological inhibition of PGE2 biosynthesis in EGCs or IL-1 knockdown in tumor epithelial cells prevented EGC acquisition of a pro-tumorigenic phenotype. Inhibition of PGE2 receptor EP4 and EGFR in CSCs inhibited the effects of tumor-activated EGCs.
Interpretation
Altogether, our results show that EGCs, once activated by the tumor, acquire a pro-tumorigenic phenotype and stimulate CSC-driven tumorigenesis via a PGE2/EP4/EGFR-dependent pathway.
Funding
This work was supported by grants from the French National Cancer Institute, La Ligue contre le Cancer, the ‘Région des Pays de la Loire’ and the UNC Lineberger Comprehensive Cancer Center.
Keywords: Enteric glial cells, Colon cancer stem cells, Colorectal cancer: Tumor microenvironment, PGE2
Abbreviations: CM, Conditioned medium; CSC, Cancer stem cell; EGC, Enteric glial cell; TEC, Tumor epithelial cell
Research in context.
Evidence before this study
Enteric glial cells are part of the enteric nervous system, which is the local nervous system of the gastrointestinal tract. Research conducted over the past two decades has demonstrated that enteric glial cells are essential to all gastrointestinal functions in healthy conditions and are implicated in several digestive and extradigestive diseases. However, whether enteric glial cells impact colon cancer development has never been studied.
Added value of this study
In this study, we demonstrate that enteric glial cells are part of the tumor microenvironment and that, in response to tumor-derived ligands, they release a soluble factor, which promotes tumorigenesis via the activation of the subset of cancer cells with increased tumor-initiating abilities, called cancer stem cells. Specifically, we show that tumor epithelial cell-derived IL-1 activates enteric glial cells to produce and release increased quantities of PGE2 leading to increased cancer stem cell tumor-initiating capabilities and tumor growth.
Implications of all the available evidence
This study demonstrates that enteric glial cells are active players of colon carcinogenesis and indicates that a better understanding of the molecular pathways involved in the bi-directional crosstalk between enteric glial cells and colon cancer (stem) cells may lead to the identification of new therapeutic targets for anti-cancer therapies.
Alt-text: Unlabelled Box
1. Introduction
Compelling evidence has well-established that within a colonic tumor, only a relatively small fraction of cells are able to give rise to a de novo tumor identical to the original one [1,2]. These cells, termed tumor-initiating cells or cancer stem cells (CSCs), have been extensively studied over the last decade and are considered as a highly valuable therapeutic target. Indeed not only do these cells initiate tumor development, but they also have been associated with a high metastatic potential [3] and increased chemoresistance [4]. Although their origin is still controversial, CSCs are thought to be derived from genetically and/or epigenetically-damaged colonic epithelial stem cells [5]. Regardless of their origin, and similar to normal intestinal stem cells, CSCs are tightly regulated by their neighboring cells that compose the so-called ‘tumor microenvironment’ [6]. For instance, elegant studies by the Medema group have demonstrated that tumor-activated fibroblasts activate ‘differentiated’ tumor cells to re-acquire stemness via paracrine pathways [7] and this was associated with increased chemoresistance [8]. In the same vein, recent work suggests that adipose tissue adjacent to the tumor provides adipose-derived stem cells that enhance tumor initiation and growth via the liberation of IL-6 [9]. Thus, while fibroblasts, adipocytes, as well as immune cells of the tumor microenvironment are being closely investigated, to the best of our knowledge, the impact of enteric glial cells (EGCs) on CSCs and associated tumorigenesis remains completely unknown.
EGCs are the most abundant cell type of the enteric nervous system and form a dense network that runs all along the gastrointestinal tract and extends into all layers of the intestinal wall [10]. Work from our group and others has demonstrated that EGCs are essential for the maintenance of intestinal homeostasis and functions [11,12]. Indeed complete loss of EGCs leads to a massive breakdown of the intestinal epithelium followed by a fulminant and fatal jejunoileitis in transgenic mice [13]. Furthermore, EGCs regulate all the major functions of the epithelium via the release of specific paracrine factors. For instance, they promote barrier function and mucosal healing via the secretion of S-Nitrosoglutathione (GSNO) and pro-EGF, respectively [14,15]. While the impact of EGCs on the epithelium has been well-studied in the healthy intestine, there is very little information about EGCs in colorectal cancer. Descriptive studies from our group and others have reported architectural alterations of the glial network in colorectal pre-cancerous lesions and adenocarcinomas [16,17]. Nevertheless, while a hypothetic role for glial cells in colon carcinogenesis has been proposed elsewhere [18], there is no direct evidence available that EGCs affect colon tumorigenesis and more importantly, whether EGCs impact colon CSCs has not been addressed as yet.
In the present study we used xenograft and 3D co-culture systems of CSCs isolated from human primary colon adenocarcinomas and several human colon cancer cell lines, grown in the presence of human primary cultures of EGCs or a non-transformed EGC line to test our hypothesis that EGCs affect CSC-derived tumorigenesis. We also used the supernatant of human primary colon adenocarcinoma to mimic the tumor microenvironment and compare its impact on EGCs versus the supernatant of patient-matched healthy mucosa to demonstrate that tumor-derived ligands activate prostaglandin E2 (PGE2) synthesis and release in EGCs, but not in fibroblasts. Overall, combining pharmacological and genetic approaches, we demonstrate that once activated by tumor cells, EGCs acquire pro-tumorigenic effects and activate CSCs and associated tumorigenesis via a PGE2/EP4/EGFR-dependent pathway.
2. Methods
2.1. Reagents
The selective mPGES-1 inhibitor CAY10526, the EP1 receptor antagonist SC-19220 and the EP4 receptor antagonist L-161,982 were purchased from Cayman Chemical. PGE2, Human IL-1α, β and Ra were purchased from Sigma-Aldrich. Antibodies phycoerythrin (PE) anti-human CD44 (clone BJ18), Brilliant Violet 421 (BV421) anti-human CD24 (clone ML5), Alexa Fluor 488 anti-human CD326 (EpCAM) and APC anti-human CD44 (clone BJ18) antibodies were purchased from Biolegend. APC anti-human CD133/2 antibody was purchased from Miltenyi Biotec. The EGFR Kinase inhibitor AG 1478 was purchased from Merck Millipore.
2.2. Tissue collection
All collected tissues originated from patients having undergone surgery for colorectal cancer with a histologic diagnosis of colon adenocarcinoma confirmed by pathologists at Nantes University Hospital and at University of North Carolina (UNC) Hospital according to international criteria. Two types of tissues were collected: 1) tumor specimens to establish primary cultures of tumor epithelial cells and 2) macroscopically healthy colon specimens located at >10 cm from the tumor area to isolate primary EGCs [19]. Patients gave their informed consent to take part in the study and all procedures were performed according to the guidelines of the French Ethics Committee for Research on Human and the Office of Human Research Ethics at UNC (UNC, Chapel Hill, NC). All experimental protocols were approved by local Committee on Ethics and Human Research and Inserm (Institut national de la santé et de la recherche médicale) and the Institutional Review Board at UNC, and were registered respectively as #DC-2008-402 and IRB # 90-0573.
2.3. Preparation of human tissue supernatants
Supernatants were obtained from human primary colon adenocarcinomas or from macroscopically healthy human colon mucosa. Briefly, tissues were incubated for 1 h at 37 °C in 1 mL Krebs-Hepes solution per 30 mg tissue and frozen in liquid nitrogen. Supernatants were centrifuged using Spin-X Centrifuge Tube Filter 0.22 µm (Corning) and diluted 1:8 in culture medium prior testing.
2.4. Enteric glial cell cultures
JUG-EGC line was obtained as described in Van Landeghem et al. [14]. JUG-EGCs were cultured in DMEM (4.5 g/l glucose; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 50 U/ml penicillin, and 50 µg/ml streptomycin (Gibco). Cells were routinely confirmed to be mycoplasma-free using PCR testing [20].
Human primary HOG-EGC cultures were isolated and grown following procedures previously established in Soret et al. [19]. Briefly, the mucosa, the submucosa and the circular muscle were removed under a dissection microscope, and the remaining longitudinal muscle/myenteric plexus (LMMP) layer was cut into small pieces (∼0.5 × 0.5 cm) and placed into GentleMACS tubes C (MiltenyiBiotec) containing 5 mL of DMEM/F12 complete medium (Dulbecco's modified Eagle's medium/F12; Gibco) supplemented with 100 IU/mL penicillin, 100 µg/mL streptomycin, 1.1 µg/mL amphotericin B, 20 µg/mL gentamicin, 6 mM glutamine, and 2.1 g/L NaHCO3 (all from Invitrogen); 250 µL protease (type I from bovine pancreas; stock solution: 5 g/L), 250 µL collagenase (Clostridium histolyticum; stock solution: 20 g/L), and 400 µL bovine serum albumin (BSA; stock solution: 50 g/L) (all from Sigma‐Aldrich). The specimens were then mechanically dissociated using a GentelMACS dissociator (MiltenyiBiotec) and incubated under gentle agitation on a rocker at 37 °C in a dry incubator for 45 min. Digestion was stopped using 10 mL of DMEM/F12 supplemented with 10% heat‐inactivated fetal calf serum (FCS, Gibco). Cell preparations were washed with Krebs solution and resuspended in 10 mL of DMEM/F12 complete medium. The suspensions were then transferred in Petri dishes (Ø 10 cm) and individual ganglia or interganglionic fiber strands were picked up using a pipette fitted with a 200 µL sterile round gel‐loading tip under a binocular microscope. Ganglionic structures numbering 10 to 20 were then seeded on gelatin-(gelatin type A from Porcine skin 90–110; 0.5% in phosphate buffered saline (PBS), Sigma‐Aldrich) coated 24‐well plates and cultured in 500 µL of DMEM/F12 complete medium for 48 h at 37 °C in a 5% CO2 incubator. After a 48 h culture period, half of the medium was replaced with DMEM 4.5 g/L glucose supplemented with 10% FCS, 2 mM glutamine, 50 IU/mL penicillin, 50 µg/mL streptomycin. Only wells with clearly identified ganglia were maintained and expanded. Only cultures with more than 80% cells positive for EGC markers were used. HOG-EGCs were maintained in DMEM (4.5 g/l glucose; Gibco) supplemented with 10% heat-inactivated FCS, 2 mM L-glutamine (Gibco), 50 U/ml penicillin, and 50 µg/ml streptomycin.
2.5. Colon cancer cell cultures
Human colon adenocarcinoma HT29, HCT116 and HCT15 cell lines were purchased from ATCC. Human colon adenocarcinoma SW1222 cell line was from Sigma-Aldrich. HT29, HCT116, HCT15 and SW1222 cell lines were cultured in DMEM (4.5 g/l glucose; Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 50 U/ml penicillin, and 50 µg/ml streptomycin (Gibco). All cell lines were routinely confirmed to be mycoplasma-free using PCR testing [20].
To establish primary cultures of tumor epithelial cells, tumors harvested at the time of surgical resection were immediately placed into Nunclon Sphera 90 mm culture dishes (Thermo Fisher Scientific) after dissociation using the gentleMACs Dissociator (Miltenyi) according to the manufacturer's instructions. Cells were then cultured in defined medium (Advanced DMEM/F12 containing 2 mM Glutamax, 100 U/mL penicillin, 100 µg/mL streptomycin, HEPES 10 mM, 1X B27 supplement, 1X N2 supplement, 2 µg/ml heparin, 40 ng/ml bFGF, 40 ng/ml EGF, gastrin 10 nM and 1 mM N-acetylcysteine). Medium was changed every 2 to 3 days, and yielded floating spheres containing primary tumor epithelial cells (Supplementary Fig. 5) were maintained until CSCs were FACS-isolated.
2.6. Generation of IL-1α/β knock-down HT29 cells
Stable HT29 cell lines expressing short hairpin RNA (shRNA) sequences targeting IL-1α (sc-39613-V) or IL-1β (sc-39615-V) were established using lentiviral particles purchased from Santa Cruz Biotechnology (MOI 5). Non-Target control HT29 cells were infected with control shRNA lentiviral particles (sc-108080). Stable cell lines were selected with 5 µg/mL puromycin dihydrochloride (Sigma-Aldrich) for 3 consecutive weeks prior to testing and were maintained constantly in selective medium.
2.7. Fibroblast cultures
The human colon fibroblast CCD18-Co cell line was purchased from ATCC. CCD18-Co cells were grown in MEM medium (Gibco) with 10% heat-inactivated FBS, 2 mM l-glutamine, 0.1 mM non-essential amino acid (Gibco), 50 U/ml penicillin, and 50 µg/ml streptomycin. Cells were routinely confirmed to be mycoplasma-free using PCR testing [20].
2.8. Fluorescence-activated cell sorting (FACS)-based isolation of CSCs
HT29, HCT116, HCT15 and SW1222 CSCs were sorted on a BD FACS Aria based on the levels of expression of the cell surface markers CD44 and CD24 [21], CD44 and CD133 [22], or CD44, CD24 and CD133. Briefly, cells were trypsinized, resuspended in culture medium, and incubated for 30 min on ice with CD44 antibody (1:40), CD24 antibody (1:20) and/or CD133 antibody (1:100). Sorted CSCs corresponded to the highest 1–2% CD24/CD44, CD44/CD133, or CD24/CD44/CD133-expressing cells, and sorted non-CSCs corresponded to the lowest 1–2% CD24/CD44, CD44/CD133, or CD24/CD44/CD133-expressing cells.
Primary CSCs were sorted on a BD FACS Aria based on the level of expression of the cell surface markers EpCAM and CD44 as previously described [23]. Briefly, dissociation of the spheres into single cells was performed in 1X PBS containing 0.5% BSA and 2 mM EDTA and using a 1 ml insulin syringe. Dissociated cell preparations were then incubated for 30 min on ice with EpCAM antibody (1:20) and CD44 antibody (1:40). Sorted CSCs corresponded to EpCAM positive cells and to the highest 1–2% CD44-expressing cells. Sorted non-CSCs corresponded to EpCAM positive cells and CD44 negative cells.
2.9. Limiting dilution assay
HT29-derived CSCs (highest 1–2% CD24/CD44) and non-CSCs (lowest 1–2% CD24/CD44) were automatically sorted into an ultra-low attachment 96 well plate (Corning Costar) at increasing densities comprised between 1 and 256 cells/well using a SONY SH800 cell sorter (SONY Biotechnology). Floating tumorsphere formation was evaluated after 2 days in serum-free DMEM/F12 (Gibco) supplemented with 20 ng/ml epidermal growth factor (EGF) (Peprotech), 5 µg/ml insulin (Sigma-Aldrich), 0.4% bovine serum albumin (Sigma-Aldrich) and 2% B27 (Gibco).
2.10. Aldefluor assay
Whether CD44High-CD24High CSCs exhibited higher ALDH1 enzymatic activity levels than CD44Low-CD24Low cells was assessed by flow cytometry using the Aldefluor kit from Stemcell Technologies according to the manufacturer's instructions. Briefly, 106 cells were suspended in 1 ml of Aldefluor assay buffer containing the ALDH1 substrate (Bodipy-Aminoacetaldehyde). To set up the ALDH negative gate, half of the cells were immediately transferred to a control tube containing the specific ALDH enzyme inhibitor diethylaminobenzaldehyde (DEAB; 15 µM). After a 45-minute incubation at 37 °C, cells were then stored on ice and stained with conjugated antibodies against CD24 (CD24-BV421) and CD44 (CD44-APC). Cells were analyzed using a SONY SH800 cell sorter in analyzer mode. Cells with high or low levels of ALDH1 enzymatic activity were gated, and levels of expression of CD24 and CD44 were analyzed in ALDHHi vs. ALDHLo populations.
2.11. Xenografts
Scid Beige mice (6 week old; 6 males and 6 females) were purchased from Charles River and housed in specific pathogen-free conditions at the Experimental Therapy Unit animal facility of Nantes (Nantes, France) in accordance with the institutional guidelines of the French Ethical Committee (CEEA, authorization # 2010.49). Appropriate experimental sample size was estimated from previous pilot studies aiming to determine which number of CSCs and which immunodeficient mouse model allowed for the most robust and reproducible tumor growth. Mice were anaesthetized by inhalation of an isoflurane/air mixture (2%, 1 L/min) before injection of 200 CSCs isolated from luciferase-expressing HT29 cells (Bioware HT29-luc-D6 PerkinElmer) alone or with JUG-EGCs (ratio 1:1) in opposite flanks subcutaneously. Mice were divided into two groups: (Group 1) 3 males and 3 females were injected with CSCs in the left flank and with CSCs + EGCs in the right flank, and (Group 2) 3 males and 3 females were injected with CSCs in the right flank and with CSCs + EGCs in the left flank. Furthermore CSCs and EGCs from different passages/sorts were used in the same group. At 4 weeks post-injection, bioluminescent imaging was performed after luciferin injection using a PhotonIMAGER (Biospace Lab). At 5 weeks post-injection, tumor burden was evaluated by volume calculation using caliper measurement by an observer blinded to treatment groups.
2.12. CSC tumorsphere formation assay
HT29, HCT116, HCT15 and SW1222 CSCs were plated at a density of 400 cells per drop (50 µL) of Growth Factor Reduced Matrigel Matrix (Corning) in 24-well plates, and cultured alone or in the presence of JUG-EGCs, HOG-EGCs, CCD18-Co cells, conditioned medium (CM) of JUG-EGCs pre-activated (TA EGC-CM) or not by HT29 cell-CM (TEC-CM). Medium was changed every day. Tumorsphere number was determined at day 8 post-plating by two observers blinded to treatment groups, and tumorsphere size was determined at day 9 using an inverted system microscope Olympus IX50 (objective 10X FN 22) and ImageJ software.
In co-culture experiments, JUG-EGCs, HOG-EGCs and CCD18-Co cells were seeded onto porous filters at 45,000 cells per filter (24-well Transwell Clear, 0.4 µm porosity; Corning) one day prior to testing. In conditioned medium (CM)-based experiments, HT29 cells, IL-1α/β knock-down HT29 cells and JUG-EGCs were seeded in 24-well plates at a density of 15,000 cells per well at least one day prior to testing. CM were collected after a 24-hour incubation, and centrifuged for 5 min at 3000 rpm to eliminate the presence of potential cells/debris. HT29 cell-CM was incubated for 24 h with JUG-EGCs to obtain CM of tumor-activated EGCs (TA EGC-CM), or placed in an empty well for 24 h (TEC-CM). To evaluate PGE2 impact on tumorsphere numbers, PGE2 (10 µM) was added to the culture medium daily. To assess CAY10526 effects, JUG-EGCs were pre-incubated for 30 min with CAY10526 (10 µM), and then cultured with TEC-CM supplemented with CAY10526 (10 µM). To test the involvement of EP receptors and EGFR respectively, CSCs were pre-incubated for 30 min with EP1 or EP4 antagonists (50 µM), or EGFR tyrosine kinase inhibitor AG 1478 (10 µM), and then grown in the presence of EGC-CM or TA EGC-CM supplemented with EP1 or EP4 antagonists (50 µM), or AG 1478 (10 µM).
Primary CSCs were plated at a density of 5,000 cells per drop (50 µL) Matrigel Matrix hESC-qualified (Corning) in 24-well plates, and cultured in defined medium alone or in the presence of HOG-EGCs. Tumorsphere number and size were determined at days 8, 10, 12, 16 and 20 post-plating using an inverted system microscope Olympus IX50 (objective 10X FN 22) and ImageJ software.
2.13. CSC expansion assay
HT29 CSCs were cultured in 3D-Matrigel as described above for 8 days alone or in the presence of JUG-EGCs. Yielded tumorspheres were then extracted from Matrigel using the Cell Recovery Solution (Corning) according to the manufacturer's protocol. Tumorspheres were trypsinized, and dissociated cells were then replated in a 50 µL drop of Matrigel and grown without JUG-EGCs for another 8 days before determining the number of tumorspheres that were de novo formed.
2.14. Evaluation of glial PGE2 production
To study mPGES-1 expression and PGE2 release, HOG-EGCs were seeded onto porous filters (24-well Transwell Clear, 0.4 µm porosity; Corning) and cultured until they reached confluence. HOG-EGCs were then cultured without FBS for 24 h in control medium, HT29 cell-CM, or supernatants of human primary colon adenocarcinomas or macroscopically healthy human colon mucosa. To determine the impact of IL-1α/β on glial mPGES-1 expression, HOG-EGCs were incubated for 24 h without FBS in the presence of 1 nM or 10 nM human IL-1α or IL-1β, or with tumor vs. healthy supernatants supplemented or not with IL-1Ra (10 µM, after a pre-incubation of 30 min with IL-1Ra alone). After 24 h, supernatants of HOG-EGCs treated or not were collected and processed for PGE2 EIA, and HOG-EGCs were lysed in RA1 buffer (Macherey-Nagel) to assess mPGES-1 gene expression by Real-Time qPCR.
2.15. EIA/ELISAs
Supernatants were collected and centrifuged for 5 min at 3,000 rpm to eliminate the presence of potential cells/debris. PGE2 quantity was measured using the PGE2 EIA Kit (Cayman Chemical) according to the manufacturer's protocol. IL-1α/β concentrations were assessed using human IL-1α/IL-1F1 and IL-1β/IL-1F2 Quantikine ELISA Kits (R&D).
2.16. Real-time quantitative polymerase chain reaction analysis
Extraction of total RNA was performed using the Nucleospin RNA kit (Macherey-Nagel) following the manufacturer's recommendations. Reverse transcription was performed using the Superscript III reverse transcriptase (Invitrogen). Real-Time qPCR studies were carried out using TaqMan gene expression assays (Applied Biosystems), and run on StepOnePlus system (Life Technologies). All qRT-PCR data were normalized to the invariant control gene RPS6. The following TaqMan primer/probe sets were used: RPS6: Hs041950248g1/Rn00820815_g1 (human/rat), CD24: Hs03044178_g1, CD44: Hs01075862_m1, PTK7: Hs00897151_m1, CEACAM6 (CD66c): Hs03645554_m1, PTGER1: Hs00168752_m1, PTGER2: Hs04183523, PTGER3: Hs00168755_m1, PTGER4: Hs00168761_m1, IL1A: Hs00174092_m1, IL1B: Hs01555410_m1, PTGES: Hs01115610_m1, IL1R1: Hs00991002_m1/Rn00565482_m1 (HOG-EGC/JUG-EGC).
2.17. BioMark dynamic array real-time quantitative polymerase chain reaction analysis
Extraction of total RNA was performed using the RNAqueous Total RNA Isolation Kit (Ambion) following the manufacturer's recommendations. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). High throughput multiplex real-time qPCR studies were carried out using BioMark dynamic arrays (Fluidigm) and run on the BioMark Real Time qPCR system (Fluidigm). RT-qPCR data were normalized to the invariant control genes RPS6 or GAPDH, and analyzed using the 2–∆∆Ct method. Heatmaps have been generated using the BioMark Real Time PCR Analysis software (Fluidigm). The following TaqMan primer/probe sets were used: (human, HOG-EGC) GAPDH Hs02786624_g1, PTGES: Hs01115610_m1, PTGES2: Hs00228159_m1, PTGES3: Hs04187819_g1, PTGS1: Hs00377726_m1, PTGS2: Hs00153133_m1, TGFB1: Hs00998133_m1, EGF: Hs01099999_m1, JAG1: Hs01070032_m1, RSPO1: Hs00543475_m1, NOG: Hs00271352_s1, FGF2: Hs00266645_m1, EPCAM: Hs00158980_m1, S100B: Hs00902901_m1, SOX10: Hs00366918_m1; (rat, JUG-EGC) Rps6: Rn00820815_g1, S100b Rn00566139_m1, Sox10: Rn00569909_m1, Gfap: Rn00566603_m1, Plp1: Rn00456892_m1.
2.18. Western blotting
HT29 CSCs were 3D-cultured in Matrigel as described above for 4 days alone or in the presence of JUG-EGCs. Tumorspheres were then extracted from Matrigel using the Cell Recovery Solution (Corning) according to the manufacturer's protocol. JUG-EGCs were incubated for 3 days with or without TEC-CM (HT29 cell conditioned medium) in the presence or absence of CAY10526 (10 µM). Cells were lysed with 2X Laemmli buffer with 5% β-Mercaptoethanol heated to 60 °C. Samples were processed for electrophoresis using NuPAGE MOPS SDS buffer (Life Technologies) and separated on 4–12% or 12% Bis-Tris gels (Life Technologies). Proteins were transferred onto nitrocellulose membranes using the iBlot System (Life Technologies). After blocking, blots were incubated overnight at 4 °C with primary antibodies diluted in 1X TBS with 5% BSA and 0.1% Tween for ERK (1:500, Cell Signaling), pERK (1:500, Cell Signaling), EGFR (1:500, Cell Signaling), pEGFR (1:4000, Cell Signaling), mPGES-1 (1:200, Santa cruz), Cox2 (1:1000, Invitrogen) and GAPDH (1:1000, Cell Signaling). Immunoblots were probed with appropriate horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature (Thermo Scientific) and visualized by chemiluminescence (Clarity Western ECL Substrate; Bio-Rad) using a Gel-Doc imager and the Image Lab Software (Bio-Rad).
2.19. 2D. imaging of human specimens
Tissues were fixed in 4% paraformaldehyde overnight at 4 °C. Following paraffin-embedding and sectioning, slides were baked at 60 °C for 2 h, deparaffinized with successive incubations in xylene, absolute ethanol, 95% ethanol and 70% ethanol, and incubated in 1X Target Retrieval Solution (Dako) at 110 °C for 90 s. Sections were then successively incubated in blocking solution (Dako) for 1 h, primary and secondary antibodies diluted in antibody diluent solution (Dako) overnight at 4 °C and 1 h at room temperature, respectively. The following primary and secondary antibodies were used: rabbit polyclonal anti-S-100β (1:1000, DAKO), rabbit polyclonal anti-GFAP (1:500, DAKO), mouse polyclonal anti-EpCAM (1:200, Biolegend), anti-rabbit-Cy3 (1:500, Jackson) and anti-mouse-Alexa Fluor 488 (1:500, Invitrogen). Stained sections were imaged using an OLYMPUS BX51 microscope (objectives 20X FN 26.5, 40X FN 26.5) and with an OLYMPUS IX83 inverted microscope (objectives 20X FN 22, 40X FN 22).
2.20. 3D imaging of human specimens
Tissues were fixed in 4% paraformaldehyde overnight at 4 °C. After extensive 1X PBS washes, samples were dehydrated, bleached, immunostained and clarified according to the iDISCO protocol described by Renier and colleagues [24]. Briefly, samples were dehydrated for 1 h at room temperature (RT) in ascending concentrations of methanol (20%, 40%, 60%, 80%, 100%) and incubated overnight at RT in 66% dichloromethane-33% methanol. After methanol washes, tissues were bleached in a 5% hydrogen peroxide solution-100% methanol overnight at 4 °C and re‐hydrated on the following day in descending concentrations of methanol (80%, 60%, 40%, 20%) followed by 1X PBS washes. Samples were permeabilized in 1X PBS containing 23 g/L glycine (Sigma‐Aldrich), 0.2% Triton X‐100 (Sigma‐Aldrich) and 20% DMSO (Sigma‐Aldrich) for 2 days at 37 °C and blocked in 1X PBS containing 0.2% Triton X‐100, 10% DMSO and 6% donkey serum (Jackson Immunoresearch) for 2 days at 37 °C. Immunostaining for EpCAM (Biolegend, 324202, 1:1250), S-100β (DAKO, Z0311, 1:500), GFAP (DAKO, Z0334, 1:500), Tuj (βIII‐tubulin) (Biolegend, 801201, 1:200) and TH (tyrosine hydroxylase) (Millipore, AB9702, 1:500) was performed in 1X PBS containing 0.2% Tween 20 (Sigma‐Aldrich), 10 µg/ml Heparin (Sigma‐Aldrich), 5% DMSO and 3% donkey serum at 37 °C for 4 days, followed by 5 washes of 1 h in 1X PBS with 0.2% Tween 20 and 10 µg/ml Heparin at RT. Tissues were then incubated for 4 days at 37 °C with secondary antibodies (donkey anti-mouse AF647, donkey anti-rabbit AF488, donkey anti-chicken AF790, Invitrogen, 1:1000) in 1X PBS with 0.2% Tween 20, 10 µg/ml Heparin and 3% donkey serum. After washes in 1X PBS with 0.2% Tween 20 and 10 µg/ml Heparin at RT, tissues were mounted in 1% agarose (Invitrogen) diluted in 1X TAE (Tris-Acetate-EDTA buffer) and left in 0.2% Tween 20 and 10 µg/ml Heparin at RT overnight. The next day, samples were dehydrated in ascending concentrations of methanol (20%, 40%, 60%, 80%, 100%) for 1 h at RT and left overnight in 100% methanol. Samples were then cleared in 66% dichloromethane-33% methanol for 3 h at RT and then washed in dichloromethane. Samples were subsequently transferred and stored in 100% dibenzyl ether solution (DBE, Sigma‐Aldrich). Samples were imaged using a LaVision BioTec UltraMicroscope II light sheet system equipped with zoom body optics, an Olympus MV PLAPO 2XC 2X/0.5 objective, and a LaVision BioTec corrected DBE dipping cap with a 5.7 mm working distance. The zoom setting was always 1.6X, for a combined magnification of 3.2X, corresponding to a 1.91 µm pixel size. Samples were imaged sequentially from both sides, with sheet NA set to 0.026 (estimated Z thickness at waist = 28 µm), and sheet waist centered on the midpoint of each side of the sample. LaVision's InspectorPro software automatically combined the two images into one by merging the left side of the image illuminated with the left light sheet, and the right side of the right image illuminated with the right light sheet, with a small interpolation region in the middle. Multi-channel images were acquired by sequentially taking Z stacks in each channel, from shortest to longest wavelength, with 5 µm Z spacing. Channels for each antigen were as follows (excitation laser wavelength, emission filter): EpCAM (647 nm, Chroma ET690/50 m), S-100β (488 nm, Chroma ET525/50 m), PGP9.5 (785 nm, Chroma ET900LP), GFAP (488 nm, Chroma ET525/50 m), TH (785 nm, Chroma ET900LP). Images were generated with Imaris 9.2 (Bitplane) software by cropping in 3D to trim away rough borders of the sample and adjusting each channel's display settings.
2.21. Statistical analysis
Data were expressed as means ± SEM. Statistical analyses were performed on data representing independent experiments (biological replicates) using SigmaPlot software. Non-parametric tests were used to compare different groups as indicated in the figure legends if Shapiro-Wilk Normality Test or Equal Variance Test failed. Differences were considered as significant for a p-value of less than 0.05.
2.22. . Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
3. Results
3.1. Enteric glial cells are part of the tumor microenvironment
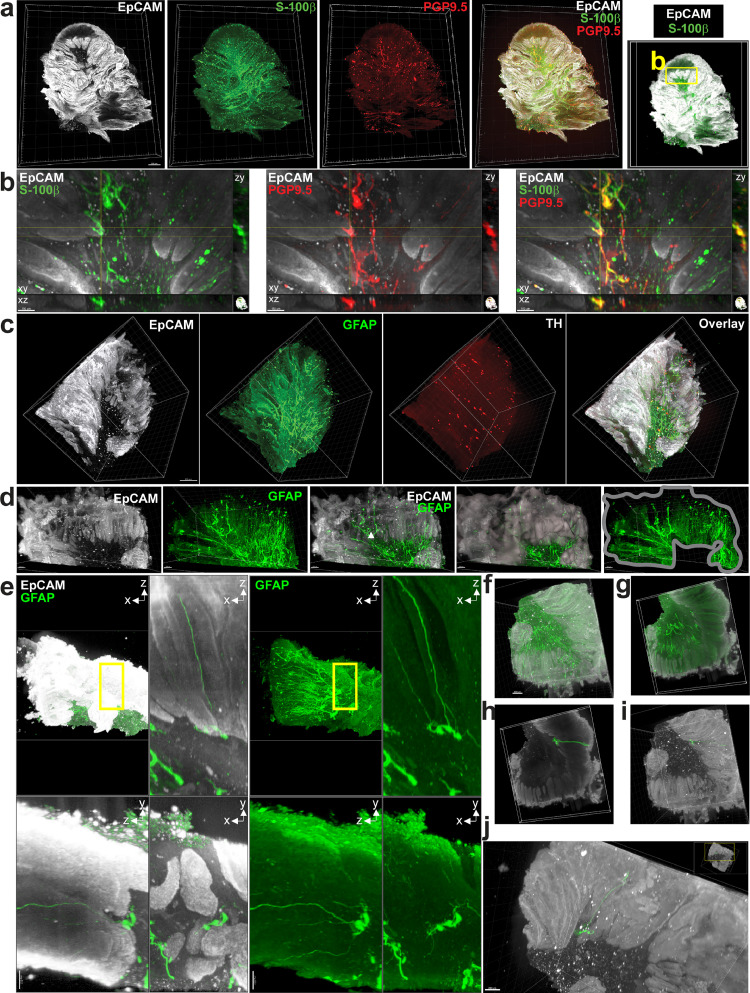
Three-dimensional imaging of full thickness human colon adenocarcinomas (Fig. 1) and macroscopically healthy colonic walls taken at distance from the tumor (Supplementary Fig. 1a, b) cleared and stained using the iDISCO method [24] and acquired using light-sheet microscopy was first used to visualize the organization and the density of the EGC network. Data shown in supplementary figures 1a and 1b illustrate that in macroscopically healthy colons, S-100β- and GFAP-expressing EGCs form a dense network readily observable in the two major plexi of the enteric nervous system, the submucosal and the myenteric plexi, where it co-localizes with the neuronal network observed using PGP9.5, a general neuronal marker, and tyrosine hydroxylase (TH), which labels serotonin-producing neurons. The EGC network further extends beyond the two plexi and is also present in the muscle layers, as well as in the mucosa, where it is especially dense at the base of the crypts identified using the epithelial marker EpCAM (Supplementary Fig. 1a, b). In adenocarcinomas, while we did not detect any TH immunoreactivity (Fig. 1c), we observed PGP9.5 staining, predominantly at the tumor front (Fig. 1a-b). Strikingly, S-100β and GFAP staining revealed a dense arborization of the EGC network massively infiltrating all parts of the tumor (Fig. 1a, c). This is further demonstrated in Fig. 1d showing a dense GFAP positive network found in a region positive for EpCAM. We further observed regions with S-100β (glial) and PGP9.5 (neuronal) positive cell bodies and extensions closely intertwined (Fig. 1b). Classical 2D immunofluorescence studies on human primary colon adenocarcinomas confirmed that S-100β-expressing EGCs are located close to tumor epithelial cells, some being immediately adjacent (Supplementary Fig. 1c, d). Using 3D imaging, we were also able to observe clusters of EGC bodies and processes immediately adjacent to tumor cells (Fig. 1e) with some processes fully spanning the tumor from the stroma to the top of the tumor (Fig 1f–j). These results demonstrate not only that EGCs are densely present in the tumor microenvironment, but also that paracrine communication can occur between EGCs and tumor epithelial cells (TECs).
Fig. 1.
EGCs are part of the tumor microenvironment. a. 3D imaging of human colon adenocarcinoma using light sheet microscopy after clearing and staining for EpCAM (epithelial cell marker), S-100β (enteric glial cell marker) and PGP9.5 (pan-neuronal marker). Scale bar: 500 µm. b. Zoom-in of 1a (xy) and orthogonal projections centered on a cluster of neuronal and glial cell bodies indicated by quadrant (xz and yz). c. 3D imaging of human colon adenocarcinoma using light sheet microscopy after clearing and staining for EpCAM (epithelial cell marker), GFAP (enteric glial cell marker) and TH (marker of serotonin-producing neurons). Scale Bar: 500 µm. d. Zoom-in of 1c. A mask (grey) was created to isolate GFAP signal only where EpCAM signal is present (2 images on right side). e. Upper left panel shows xz maximal projection of the entire specimen. Upper right, lower right and lower left panels show respectively xz, xy and yz maximal projections of a 250 µm section inside the specimen (yellow rectangle in upper left panel). (EpCAM: white, GFAP: green). f. Other view of specimen shown in 1c (EpCAM: white, GFAP: green). g. 250 µm section of specimen shown in 1f (EpCAM: white, GFAP: green). h. Isolation (green mask) of GFAP-positive structure containing cell bodies and processes (green) overlaid with EpCAM staining (white) within the 250 µm section shown in 1 g. i/j. Views of isolated GFAP-positive structure containing cell bodies and processes (in green) overlaid with EpCAM staining (white) shown in full thickness specimen. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Enteric glial cells activate cancer stem cells to promote tumorigenesis
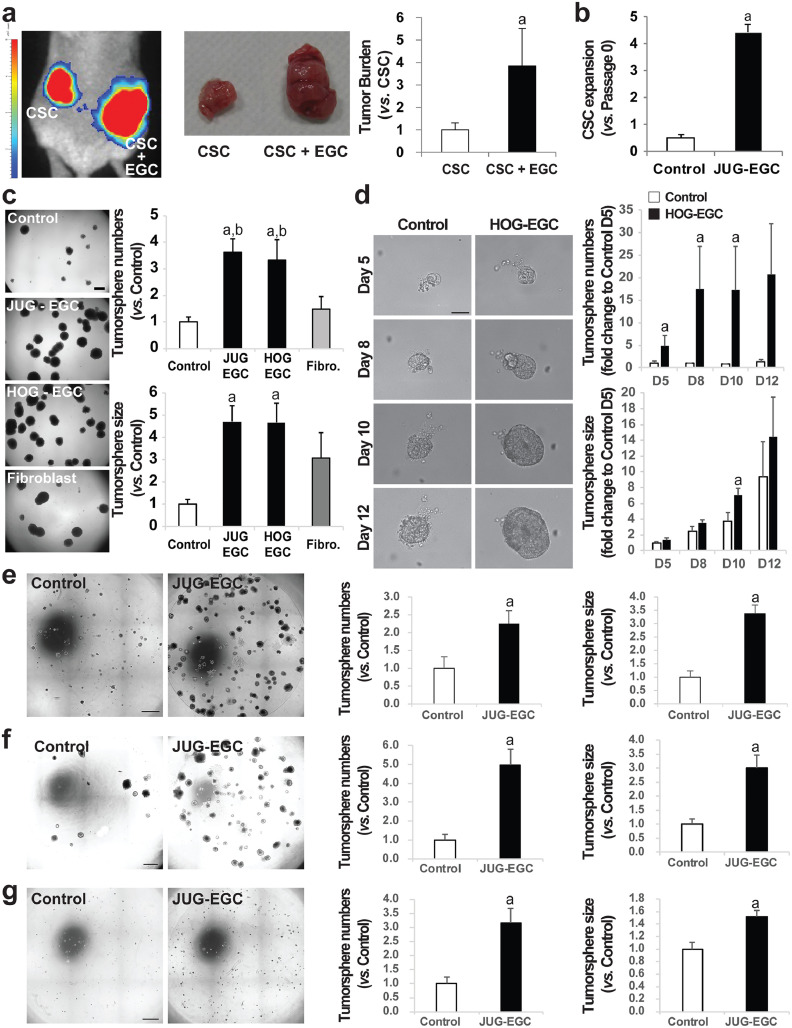
We next sought to evaluate whether EGCs could impact CSC-driven tumorigenesis. To do this, 200 CD44High-CD133High CSCs isolated from the human colon cancer HT29 cell line (Supplementary Fig. 2) stably expressing luciferase were injected alone or concomitantly with EGCs subcutaneously in immunodeficient mice (ratio 1:1). Our data demonstrate that EGCs increased tumor load and this was correlated with enhanced bioluminescence intensity (Fig. 2a).
Fig. 2.
EGCs stimulate CSC-driven tumorigenesis. (a–c.CSCs were isolated from the human HT29 cell line.)a.(left panel) Bioluminescence imaging of a representative mouse at 4 weeks after injection of luciferase-expressing CSCs alone (CSC) and a mix of CSCs + EGCs (CSC + EGC) at a 1:1 ratio in opposite flanks. (middle panel) Photographs of representative tumors at 6 weeks after injection of CSCs alone and a mix of CSCs + EGCs in opposite flanks. (right panel) Tumor burden corresponds to the tumor volume and is expressed as fold change relative to control (CSC) (mean ± SEM). n = 12; Mann and Whitney test, a: p < 0.01. b. Quantification of CSC expansion corresponds to [number of tumorspheres (Passage 1, P1) yielded from single cell suspension obtained from dissociation of P0 tumorspheres and cultured alone] / [initial number of tumorspheres (Passage 0, P0) yielded from CSCs cultured alone or in the presence of EGCs]. n = 5; Mann and Whitney test, a: p < 0.01. c.(left panel) Representative photographs of CSCs cultured alone (Control), in the presence of JUG-EGCs (non-transformed EGC line from rat embryo), HOG-EGCs (primary cultures of human EGC) or normal human fibroblasts (CCD18-Co). Scale Bar: 500 µm. Quantification of tumorsphere number (right, upper panel) and size (right, lower panel) is expressed as fold change relative to Control (mean ± SEM). n ≥ 3; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. fibroblasts. d. Representative photographs (left panel) of tumorspheres derived from CSCs isolated from primary adenocarcinomas from colorectal cancer patients cultured alone (Control) or in the presence of primary cultures of human EGCs (HOG-EGC) from day 5 to day 12 post-plating illustrating that HOG-EGCs increased tumorsphere number (right, upper panel) and size (right, lower panel). n = 5; Mann and Whitney test, a: p < 0.05. Scale Bar: 20 µm. e-g. Representative photographs (left panel) of HCT116 (e), HCT15 (f) and SW1222 (g)-derived CSCs cultured alone (Control) or in the presence of EGCs (JUG-EGC) illustrating that EGCs increased tumorsphere number (middle panel) and size (right panel). n ≥ 4; Mann and Whitney test, a: p < 0.05. Scale Bar: 1 mm.
To dissect the molecular crosstalk between EGCs and CSCs, we developed a co-culture system allowing bi-directional paracrine communication between EGCs and CSCs, in which HT29-derived CSCs were 3D-grown in Matrigel in the presence or absence of EGCs seeded on Transwell filters. Since there is no human EGC line available, we used JUG-EGCs, which correspond to a non-transformed rat embryonic EGC line immortalized by serial passages [14,19] that expresses Sox10, S-100β, GFAP and PLP-1, all well-characterized EGC markers (Supplementary Fig. 3a). In the presence of JUG-EGCs, the number and the size of the tumorspheres formed by CSCs were dramatically increased as compared to CSCs grown alone (Fig. 2c). JUG-EGCs increased tumorsphere formation to the same extent in CD44High-CD133High, CD44High-CD24High and CD44High-CD133High-CD24High HT29-derived CSCs [21,22] (Supplementary Fig. 4). Using procedures developed by our group [19], we established primary cultures of human EGCs (HOG-EGCs) that express Sox10, S-100β and GFAP (PLP-1 not tested) (Supplementary Fig. 3b-d). HOG-EGCs had similar impact to JUG-EGCs on tumorsphere formation and growth (Fig. 2c). In contrast, normal human colonic fibroblasts (CCD18-Co) had no significant impact on tumorsphere yield or size (Fig. 2c). Since EGCs or fibroblasts were seeded on Transwell filters which porosity only allowed for paracrine crosstalk between CSCs and EGCs or fibroblasts, we inferred that EGC impact on CSC ability to form tumorspheres was mediated by soluble factors released by EGCs, but not (at least not enough) by fibroblasts.
To assess whether EGCs impact CSC expansion, tumorspheres derived from CSCs grown in the presence or absence of JUG-EGCs were retrieved, dissociated, replated in Matrigel and cultured alone for 8 additional days. CSC expansion was evaluated by dividing the number of tumorspheres obtained after replating by the initial number of tumorspheres yielded in the presence or absence of EGCs. Quantification showed that the tumorspheres grown in the presence of EGCs were composed of a significant increased proportion of cells that had the ability to form tumorspheres (Fig. 2b). Thus this strongly suggests that EGCs stimulated CSC expansion.
Together these results demonstrate that EGC-derived paracrine factor(s) stimulate CSC expansion and ability to give rise to tumors. These findings were further validated by additional data demonstrating that JUG-EGCs had a similar stimulatory impact on number and size of tumorspheres yielded from CSCs isolated from several human colon cancer cell lines associated with different p53, MMR system and stemness status [21,25] that are HCT116 (Fig. 2e), HCT15 (Fig. f) and SW1222 (Fig. 2g). Furthermore, we also confirmed EGC pro-tumorigenic effects on CSCs using exclusively primary cultures derived from human specimens. Indeed, primary cultures of human EGCs (HOG-EGCs) significantly increased the number and size of tumorspheres formed from primary CSCs isolated from adenocarcinomas from colon cancer patients (Fig. 2d; Supplementary Fig. 5).
3.3. Tumor epithelial cells activate EGCs to acquire pro-tumorigenic abilities by stimulating glial PGE2 release
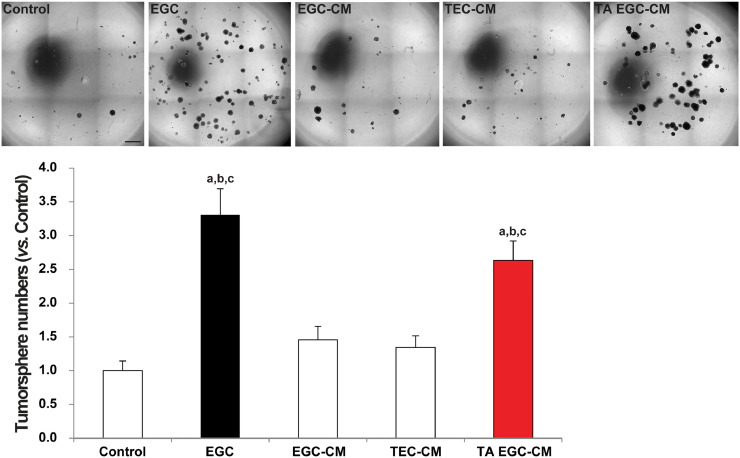
Considering that the in vitro co-culture system only permits paracrine communication, we sought to test whether EGC-conditioned medium (EGC-CM) could reproduce EGC effects on CSCs. Our data show that EGC-conditioned medium (CM) had no significant impact on tumorsphere formation (Fig. 3), indicating that at basal state EGCs do not release pro-tumorigenic factor(s) or not in sufficient quantities to activate CSCs. Since Transwell filters allow bi-directional exchanges between EGCs and CSCs/tumor epithelial cells (TECs) composing the tumorspheres, we reasoned that EGCs may be activated to acquire a pro-tumorigenic phenotype in response to TEC-derived ligands. To test this, we evaluated whether conditioned medium (CM) of EGCs that had been pre-incubated with HT29 cell-conditioned medium (TEC-CM) could activate CSCs. Our results demonstrate that conditioned medium (CM) of EGCs that had been activated by tumor epithelial cells (TA EGC-CM) significantly increased tumorsphere numbers (Fig. 3). Of note conditioned medium of HT29 cells (TEC-CM) had no significant effect on CSC ability to give rise to tumorspheres (Fig. 3).
Fig. 3.
Tumor epithelial cells activate EGCs to acquire pro-tumorigenic effects. Representative photographs and bar graph demonstrating that in contrast to the presence of EGCs on Transwell filters (EGC), EGC-conditioned medium (EGC-CM) did not impact CSC-derived tumorsphere yield. However, CM of tumor-activated EGCs (TA EGC-CM) induced a significant increase in tumorsphere numbers. CM of tumor epithelial cells (TEC-CM) had no effect. Quantification of tumorsphere number is expressed as fold change relative to Control (mean ± SEM). n ≥ 9; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. EGC-CM, c: p < 0.05 vs. TEC-CM. Scale Bar: 1 mm.
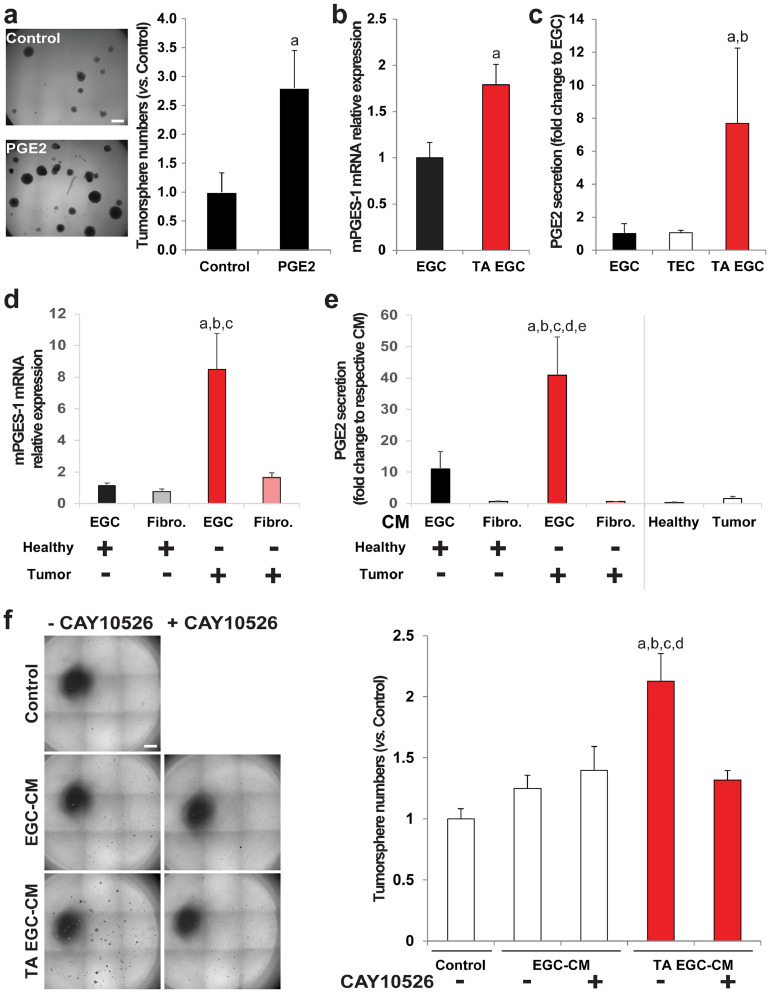
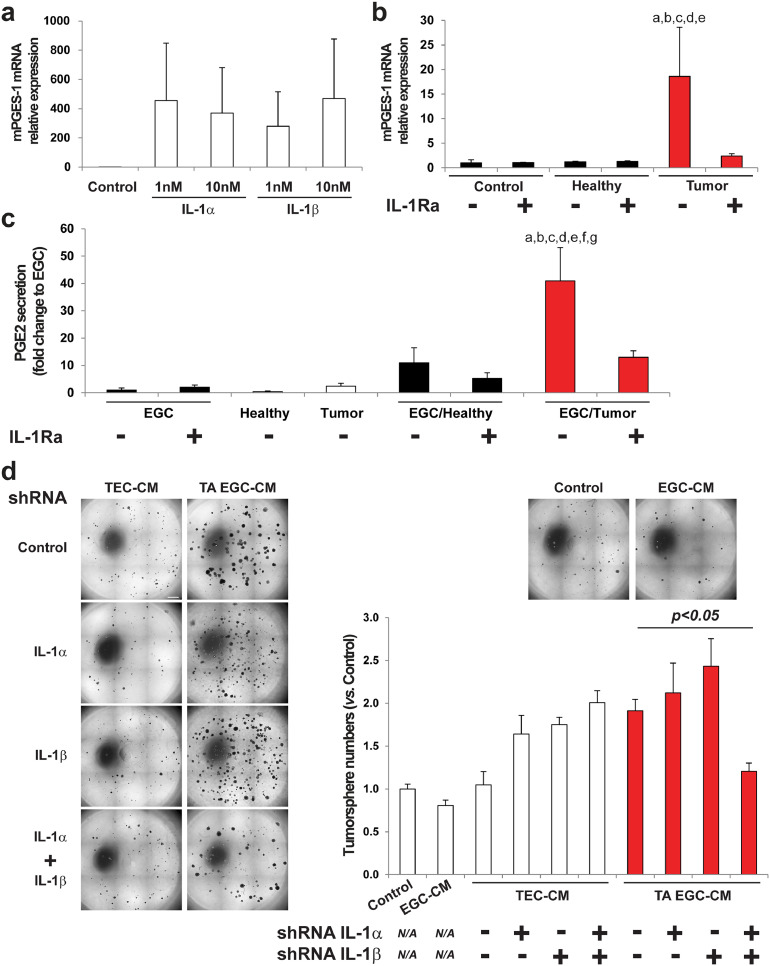
To identify the EGC-derived soluble ligand(s) responsible for CSC activation, we tested several glial derived factors (Supplementary Fig. 6a) and only PGE2 [26] reproduced EGC effects (Fig. 4a). RT-qPCR on EGCs incubated with control medium or with TEC-CM showed that TEC-CM increased gene expression of microsomal prostaglandin E synthase 1 (mPGES-1), the inducible terminal synthase in the PGE2 biosynthesis pathway (Fig. 4b). EIA (Enzyme Immunoassay) demonstrated that this was accompanied by increased glial release of PGE2 (Fig. 4c). Next, primary adenocarcinomas from colon cancer patients and patient-matched macroscopically healthy mucosa taken at a distance from the tumor were incubated for one hour, and the supernatants, which thus contained soluble factors released by the tumor or by a normal colonic mucosa, were collected and incubated with primary cultures of human EGCs (HOG-EGCs) or colonic fibroblasts (CCD18-Co). Supernatant of human colon adenocarcinomas led to an increase in mPGES-1 gene expression and PGE2 release in EGCs as compared to supernatant of healthy colon mucosa (Fig. 4d, e). In contrast, supernatant of colon adenocarcinomas and healthy mucosa did not differentially impact mPGES-1 gene expression or PGE2 release in colonic fibroblasts (Fig. 4d, e), suggesting that TEC activate PGE2 production in EGCs in a selective manner. Supporting our initial screening, mRNA levels of other glial derived factors (TGFB1, JAG1, NOG, FGF2 and EGF) show no significant changes in EGCs incubated with supernatant of colon adenocarcinomas versus healthy mucosa (Supplementary Fig. 6b). Additional data indicate that gene expression of Cox2, the rate-limiting enzyme upstream of mPGES-1, showed a trend towards an increase in EGCs incubated with supernatant of colon adenocarcinomas as compared to EGCs incubated with supernatant of healthy mucosa, although this was not significant (Supplementary Fig. 7a). Of note, no significant changes in mRNA levels were observed for COX1 and PTGES2, which encodes mPGES-2 (Supplementary Fig. 7a). Transcripts of PTGES3, which encodes cPGES, were hardly detectable. Western blot analyses confirmed that, while TEC-CM induced a significant increase in mPGES-1 protein level in EGCs, it only led to a non-significant minor increase in Cox2 protein levels (Supplementary Fig. 7b). Finally, to functionally assess whether mPGES-1 inhibition in EGCs could block their pro-tumorigenic effects, we used CAY10526, a selective inhibitor of mPGES-1 expression. Western blot analyses confirmed that CAY10526 abolished TEC-CM-induced increase in mPGES-1 expression in EGCs (Supplementary Fig. 7b). Importantly, TEC-CM supplemented with CAY10526 failed to activate EGCs to stimulate CSC-derived tumorsphere formation (Fig. 4f). Altogether these data indicate that TECs stimulate mPGES-1 expression and PGE2 release in EGCs, which, in turn, increase CSC-driven tumorigenesis.
Fig. 4.
Tumor-activated EGCs stimulate CSC ability to give rise to tumorspheres via increased glial PGE2. a. (left panel) Photographs illustrating that PGE2 treatment (10 µM) increased the number of tumorspheres yielded from CSCs. Scale Bar: 500 µm. (right panel) Quantification is expressed as fold change to Control (mean ± SEM). n = 3; two tailed t-test, a: p < 0.05. b. RT-qPCR data showing that mPGES-1 gene expression was increased in tumor-activated EGCs (TA EGC) vs. control EGCs. n = 5; Mann and Whitney test, a: p < 0.01. c. PGE2 EIA analysis showed a significantly higher PGE2 concentration in TA EGC-CM vs.EGC-CM or TEC-CM. n ≥ 3; Kruskal-Wallis ANOVA, Dunn's multiple comparison test, a: p < 0.05 vs. EGC, b: p < 0.05 vs. TEC. d. Supernatant of human primary colon adenocarcinomas (Tumor) stimulated mPGES-1 gene expression in EGCs, but not in fibroblasts (CCD18Co), compared to supernatant of patient-matched healthy colon mucosa. n ≥ 4; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. EGC/Healthy, b: p < 0.05 vs. Fibro./Healthy, c: p < 0.05 vs. Fibro./Tumor. e. PGE2 EIA validated that supernatant of human primary colon adenocarcinomas (Tumor) stimulated PGE2 release in EGCs, but not in fibroblasts, compared to supernatant of healthy mucosa (CM = conditioned medium). n ≥ 4; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. EGC/Healthy, b: p < 0.05 vs. Fibro./Healthy, c: p < 0.05 vs. Fibro./Tumor, d: p < 0.05 vs. Healthy, e: p < 0.05 vs. Tumor. f. Representative photographs and quantification to illustrate that the addition of a specific inhibitor of mPGES-1 expression (CAY10526; 10 µM) to tumor epithelial cell-CM prevented TA-EGCs from acquiring pro-tumorigenic effects. n = 5; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. EGC CM, c: p < 0.05 vs. EGC-CM+CAY10526, d: p < 0.05 vs. TA EGC-CM+CAY10526. Scale Bar: 1 mm.
3.4. Tumor-activated EGCs stimulate CSCs via PGE2 receptor EP4
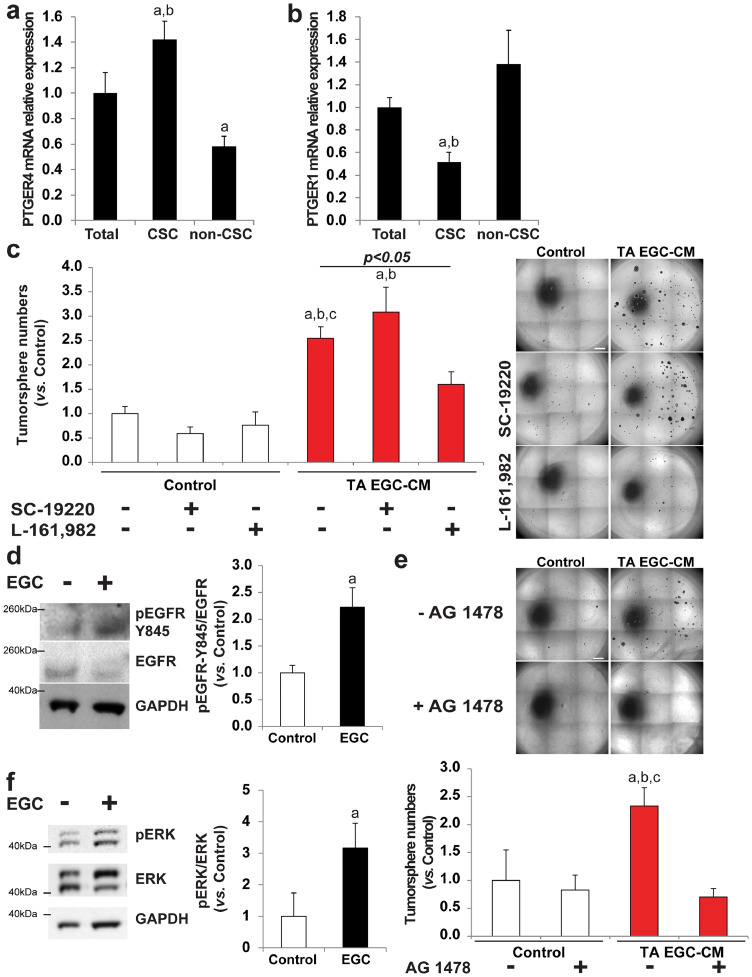
PGE2 signals through four distinct G protein–coupled receptors termed EP 1 to 4. We sought to define which EP subtype(s) was expressed in HT29-derived CSCs. Out of the 4 receptors, only PTGER1 and PTGER4 mRNAs, encoding EP1 and EP4 respectively, were expressed at detectable levels in the (unsorted) HT29 cell line. We next compared the levels of expression of PTGER1 and PTGER4 in HT29 CD44High-CD24High CSCs versus HT29 CD44Low-CD24Low cells (Supplementary Fig. 8a-d). We termed CD44Low-CD24Low cells “non-CSCs” since, when compared with CD44High-CD24High CSCs, (1) they show decreased/limited tumor-initiating abilities in vitro in 3D Matrigel (Supplementary Fig. 8e) and in free-floating cultures as assessed by limiting dilution assay (Supplementary Fig. 8f), (2) they express lower levels of colon (cancer) stem cell markers PTK7 [27] and CEACAM6/CD66c [28] (Supplementary Fig. 8 g, h) and (3) they exhibit lower levels of enzymatic activity of aldehyde dehydrogenase 1 (ALDH1), which is a well-characterized marker of CSCs in multiple cancers including colon cancer [29] (Supplementary Fig. 8i). RT-qPCR data demonstrated that CSCs were significantly enriched for PTGER4 mRNA as compared to non-CSCs (Fig. 5a). In contrast, PTGER1 showed a profile of expression opposite to PTGER4, i.e. PTGER1 mRNA was enriched in non-CSCs while being repressed in CSCs (Fig. 5b). These data suggest that EP4 has a prominent role in CSCs as compared with other EP receptors and notably EP1. To investigate whether EP4 was implicated in EGC pro-tumorigenic effects on CSCs, we added a selective antagonist of EP4 (L-161,982) or of EP1 (SC-19220) to tumor-activated (TA) EGC-CM. Our data show that while EP1 antagonist had no significant impact, EP4 antagonist abolished EGC pro-tumorigenic impact on CSCs (Fig. 5c). These results demonstrate that glial-derived PGE2 stimulates CSC clonogenicity via EP4 activation.
Fig. 5.
TA EGC stimulate CSC clonogenicity via EP4 activation as well as EGFR- and ERK-dependent pathways. a/b. RT-qPCR data showing that PTGER4 (EP4) mRNA was significantly enriched in CSCs compared to unsorted HT29 cells (Total) and compared to cells expressing lower levels of CSC markers (non-CSC) (a). In contrast, PTGER1 (EP1) was expressed at significantly lower levels in CSCs than in unsorted HT29 cells and non-CSCs (b). Data are expressed as fold change to Total (mean ± SEM). n = 5; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Total, b: p < 0.05 vs. non-CSC. c. Representative photographs (right panel) and quantification (left panel) demonstrated that the addition of a selective antagonist of EP4 (L-161,982, 50 µM) but not EP1 (SC-19220, 50 µM) to TA EGC—CM abolished its pro-tumorigenic effects. n ≥ 4; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. SC-19220, c: p < 0.05 vs. L-161,982. Scale Bar: 1 mm. d. Representative western blots and quantification demonstrating that EGCs increased pEGFR-Y845/EGFR in CSCs. Quantification is expressed as fold change to Control (mean ± SEM). n = 4, Mann and Whitney test, a: p < 0.05. e. Representative photographs and quantification of tumorspheres yielded from CSCs cultured alone (Control) or with tumor-activated EGC-CM (TA EGC-CM) supplemented or not with an inhibitor of EGFR tyrosine kinase activity (AG 1478; 10 µM). Data are expressed as fold change relative to Control (mean ± SEM). n = 4; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. AG 1478, b: p < 0.05 vs. TA EGC-CM + AG 1478. Scale bar: 1 mm. f. Representative immunoblots illustrating that pERK/ERK was increased in CSCs cultured in the presence of EGCs compared to CSCs cultured alone. n = 4; Mann and Whitney test, a: p < 0.05.
3.5. CSC activation by tumor-activated EGCs involves EGFR and ERK pathways
Previous studies indicated that PGE2 could promote colon cancer growth by transactivating EGFR and consequently activating downstream ERK pathway [30,31]. Therefore we first sought to assess whether tumor-activated (TA) EGCs stimulate CSCs in an EGFR-dependent manner. Western blot data showed that EGFR phosphorylation on tyrosine 845 (Y845), which is a site required for EGFR transactivation, was increased in CSC-derived tumorspheres grown in the presence of EGCs as compared to controls (Fig. 5d). Importantly, addition of a selective inhibitor of EGFR tyrosine kinase activity (AG 1478) to TA EGC-CM abolished EGC pro-tumorigenic effects (Fig. 5e). Activation of the downstream ERK pathway was also increased in CSC-derived tumorspheres grown in the presence of EGCs, as assessed by immunoblotting showing a significant increase in ERK1/2 phosphorylation (Fig. 5f). These results indicate that EGC-induced CSC activation involves EGFR and ERK signaling pathways.
3.6. Tumor cells activate EGCs to acquire a pro-tumorigenic phenotype via IL-1 secretion
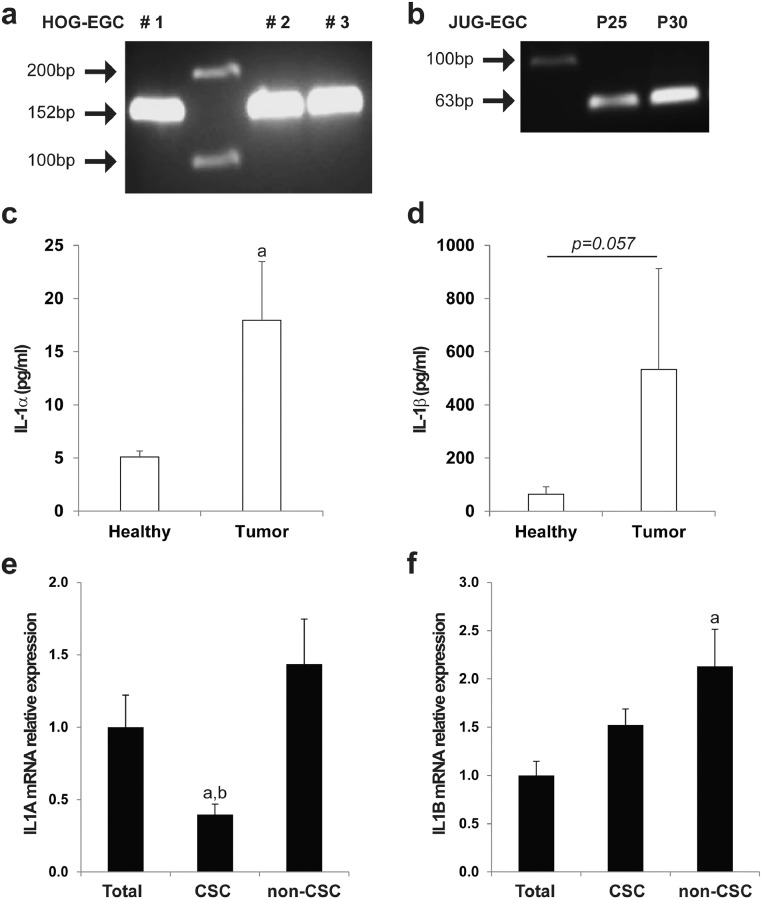
Since IL-1 signaling has been reported to modulate the production of various prostaglandins including PGE2 in different organs [32], [33], [34], [35], we tested whether tumor epithelial cells (TECs) activate EGCs to acquire a pro-tumorigenic phenotype via the release of IL-1. Consistent with previous findings [36], IL-1 receptor (IL-1R) was expressed in primary cultures of human EGCs (HOG-EGCs) (Fig. 6a) and in rat EGCs (JUG-EGCs) (Fig. 6b). We next sought to determine the effect of IL-1α and IL-1β on mPGES-1 gene expression in EGCs. RT-qPCR data showed that both IL-1α and IL-1β markedly induced mPGES-1 gene expression in primary cultures of human EGCs (HOG-EGCs) (>270 fold increase vs. control) (Fig. 7a). Supernatants of human primary colon adenocarcinomas were significantly enriched in IL-1α and IL-1β as compared with supernatants of patient-matched healthy mucosa as assessed by ELISA (Fig. 6c, d). Importantly, the addition of a naturally occurring IL-1R antagonist (IL-1Ra) abolished the stimulatory impact of the supernatant of human primary colon adenocarcinomas on glial mPGES1 expression and PGE2 release (Fig. 7b, c). Consistent with these results, CM of EGCs that had been pre-activated with CM from IL-1α and IL-1β double knock-down TECs failed to stimulate tumorsphere formation as compared to CM of EGCs pre-activated with CM of TECs control or single knock-down for IL-1α or IL-1β (Fig. 7d; Supplementary Fig. 9). Interestingly, IL-1α and IL-1β mRNAs were significantly enriched in CD44Low-CD24Low HT29 cells (non-CSCs) as compared to unsorted HT29 cells, suggesting that the tumor cells responsible for EGC activation towards a pro-tumorigenic phenotype correspond to non-CSCs/a more differentiated cellular fraction of the tumor (Fig. 6e, f). Altogether these results demonstrate that TEC-derived IL-1α/β activates the synthesis and release of glial PGE2 that, in return, stimulates CSCs.
Fig. 6.
EGCs express IL-1R, and IL-1α/β are highly enriched in the tumor microenvironment. a/b. Agarose gel electrophoresis of PCR products corresponding to IL-1R cDNA amplicons in human primary EGC cultures (a) (HOG-EGC, amplicon size 152 bp) and in rat non-transformed EGC line (b) (JUG-EGC, amplicon size 63 bp). Lanes #1, #2 and #3 (a) correspond to 3 cultures derived from 3 different patients. Lanes P25 and P30 (b) correspond to 2 different passages. c/d. ELISA data showed that IL-1α (c) and IL-1β (d) were highly enriched in supernatants of human colon adenocarcinomas (Tumor) compared with supernatants of patient-matched healthy colonic mucosa (Healthy). n = 4; Wilcoxon rank-sum test, a: p < 0.05 vs. Healthy. e/f. Real-Time qPCR data demonstrated that IL-1α gene expression (e) was downregulated in CD44High-CD24HighCSCs as compared to unsorted HT29 (Total) and CD44Low-CD24Lownon-CSCs, and IL-1β mRNA (f) was enriched in non-CSCs. Data are expressed as fold change to unsorted HT29 cells (Total) (mean ± SEM). n = 4, ANOVA, a: p < 0.05 vs. Total, b: p < 0.05 vs. non-CSC.
Fig. 7.
Tumor epithelial cell-derived IL1α/β activates a pro-tumorigenic phenotype in EGCs. a. RT-qPCR data demonstrating that addition of IL-1α or IL-1β strongly induced mPGES-1 gene expression in EGCs. n = 3. b. RT-qPCR data showing that addition of an antagonist of IL-1R (IL-1Ra, 10 µM) to supernatants of human primary colon adenocarcinomas (Tumor) abolished the tumor-induced up-regulation of mPGES-1 expression in EGCs. Healthy represents supernatants of patient-matched healthy colonic mucosa. n = 8; ANOVA, Holm-Sidak multiple comparison test, a: p < 0.05 vs. Control, b: p < 0.05 vs. IL-1Ra, c: p < 0.05 vs. Healthy, d: p < 0.05 vs. Healthy+IL-1Ra, e: p < 0.05 vs. Tumor+IL-1Ra. c. PGE2 EIA confirmed that addition of IL-1Ra blocked the increase in PGE2 release in EGCs activated by supernatants of human colon adenocarcinomas (EGC/Tumor). EGC, Healthy, Tumor and EGC/Healthy represent EGC-CM, supernatant of patient-matched healthy mucosa, supernatant of tumors, and CM of EGCs stimulated with supernatant of healthy mucosa, respectively. n ≥ 4; ANOVA, a: p < 0.05 vs. EGC, b: p < 0.05 vs. EGC+IL-1Ra, c: p < 0.05 vs. Healthy, d: p < 0.05 vs. Tumor, e: p < 0.05 vs. EGC/Healthy, f: p < 0.05 vs. EGC/Healthy+IL-1Ra, g: p < 0.05 vs. EGC/Tumor+IL-1RA. d. Representative photographs (left panel) and quantification (right panel) to show that double IL-1α and β knock-down tumor epithelial cells (TEC) had lost their abilities to activate a pro-tumorigenic phenotype in EGCs, while single IL-1α or β knock-down TECs had impact similar to that of Control TECs. n = 4, ANOVA. Scale Bar: 1 mm.
4. Discussion
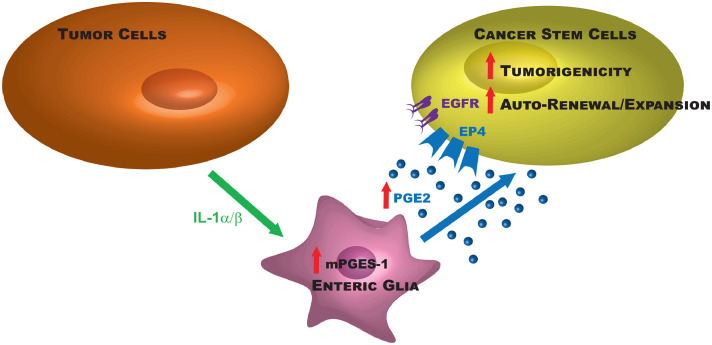
Consistent with the ‘seed and soil’ theory from Fuchs and Paget at the end of the 19th century, current views in the field suggest that tumor initiation driven by a subset of cancer cells depends on cues emanating from resident or recruited cells present at the tumor site that have been altered to favor tumor formation and growth. Altogether our data give strong evidence in vivo and in vitro that EGCs are critical cells of the tumor microenvironment that are remodeled by tumor epithelial cells to acquire a pro-tumorigenic phenotype and activate CSC-driven colon tumorigenesis. This work further identifies the causal bidirectional molecular crosstalk between tumor epithelial cells and EGCs (Fig. 8).
Fig. 8.
Molecular crosstalk between EGCs and colon cancer (stem) cells. The schematic summarizes the molecular pathways involved in the pro-tumorigenic effects of tumor-activated EGCs on colon CSC-driven tumorigenesis.
One major finding of this study is that EGCs stimulate CSC tumor-initiating abilities and CSC-derived tumor growth as demonstrated by our xenograft and co-culture results. This might be surprising considering previous work from our group demonstrating that EGCs inhibit intestinal epithelial cell proliferation partly via the release of TGF-β1 [16]. However, the former study tested the impact of EGCs on epithelial cells grown in 2D and more importantly on unsorted cells, i.e. it did not focus on the cell fraction with higher stemness potential. Thus, it is likely that EGCs differentially impact CSCs and ‘more differentiated’ cells via distinct glial-derived paracrine factors and also via the expression of distinct receptor panels in CSCs vs. non-CSCs. This is consistent with our data indicating that EP4 mRNA is enriched, while EP1 is repressed, in CSCs vs. non-CSCs. It would be interesting to assess the impact of EGC-induced activation of EP1 in non-CSCs.
Another major finding of this work is that, at basal state, EGCs do not produce (or not enough) pro-tumorigenic factors, and thus do not impact CSC-derived tumorsphere growth. However, once activated by tumor epithelial cell (TEC)-derived IL-1, EGCs acquire a pro-tumorigenic impact and stimulate CSC tumor-initiating abilities and tumorsphere growth (Fig. 8). This is consistent with a growing body of evidence demonstrating that tumor initiation and progression is dependent on the activation of different cell types present in the stroma such as fibroblasts, macrophages or adipocytes in response to stimuli emanating from the tumor such as cytokines or reactive oxygen species [37], [38], [39]. While to our knowledge no study has ever reported a positive correlation between IL-1 and mPGES expressions in colon cancer, previous work from others had reported a positive correlation between IL-1α, IL-1β and Cox2 in breast and colon carcinomas [32], with Cox2 being the inducible enzyme upstream mPGES-1 in the PGE2 synthesis cascade. Here using the supernatant of human primary colon adenocarcinomas to mimic tumor-derived ligands, we show that the addition of IL-1Ra to the supernatant of human colon cancer completely blocked EGC activation as assessed by glial PGE2 release. In the same vein, in our co-culture system, knocking down IL-1α and IL-1β in TECs inhibited EGC pro-tumorigenic effect. Altogether these data are in favor of a model where IL-1 released by TECs activates EGCs. Our results even tend to indicate that ‘more differentiated’ tumor cells (as opposed to CSCs) predominantly produce IL-1α/β. However, we cannot rule out that other cell types of the microenvironment also release IL-1, thereby further activating EGCs and glial PGE2 production. In our study, the activation of EGCs by the tumor was associated with increased mPGES-1 expression and PGE2 production. However it is very likely that the tumor also impacts other cellular mechanisms and molecular pathways in EGCs as is the case for other stromal cell types of the tumor microenvironment [40]. It is also interesting to note that the supernatant of human primary colon adenocarcinomas did not impact mPGES-1 gene expression or PGE2 release in colonic fibroblasts, suggesting that increased PGE2 production in response to tumor-derived factors may be specific to EGCs.
According to our results, PGE2 derived from tumor-activated EGCs was responsible for CSC activation and induced an increase in tumorsphere formation and growth. This strengthens the concept arising from a growing body of studies that EGCs are a major source of prostaglandins of the stroma that regulate all the main functions of the intestine [41], [42], [43], [44]. Our data using the CAY10526 inhibitor demonstrate a complete blockade of EGC pro-tumorigenic effects. Combined to our western blot data showing little to no change in Cox2 protein levels in tumor-activated EGCs in the presence or absence of the CAY10526 compound, this indicates that the tumor-induced increase in PGE2 release was dependent on the up-regulation of mPGES-1 gene expression, but not Cox2 [45]. Other studies have suggested that IL-1 activates mPGES-1 expression via Egr-1, NAB-1, NFκB or DNA methylation-related pathways [46], [47], [48], and it would be interesting to investigate in further studies the molecular mechanism involved in our system.
Altogether the results of this study propose a model where tumor-activated EGCs release increased levels of PGE2 that targets CSCs and activates their ability to form tumorspheres via the activation of a pathway that involves EP4, EGFR and ERK1/2 (Fig. 8). This is consistent with a recent study by the group of DuBois demonstrating that PGE2 induces CSC expansion via an EP4/ERK1 pathway [49]. The transactivation of EGFR by PGE2/EP4 has already been proposed by other groups [30,50]. Here our data suggest that PGE2 activation of the pathway EP4/EGFR/ERK may be specific to CSCs as indicated by our results demonstrating that EP4 is enriched in CSCs as compared to ‘more differentiated’ tumor cells that predominantly express EP1.
This study is, to the best of our knowledge, the first to define EGCs as active players of the tumor microenvironment. Indeed, from a general perspective, the role of the enteric nervous system in the development and progression of colon cancer remains poorly understood [18,51]. Nevertheless, recent work from our group has shown that colon cancer cells adhere and migrate along the enteric neuronal network that thus represents a privileged route for local dissemination of cancer cells [52]. Here we show that the EGC network densely innervates human colon adenocarcinomas and engages in crosstalk with tumor cells that ultimately leads to increased CSC tumor-initiating abilities. Notably our data give strong and direct evidence that EGCs of the tumor microenvironment undergo a switch towards a pro-tumorigenic phenotype induced by the cytokine IL-1. One can therefore speculate that chronic inflammatory stress associated with enhanced local levels of cytokines and increased risk of colorectal cancer [53] may activate a similar switch of phenotype in EGCs that might thus accompany or even precede tumor initiation. Supporting this hypothesis, previous descriptive studies have reported that alterations in the glial network architecture occur as early as in adenomas [17], implying that EGC remodeling is an early event during colon carcinogenesis. Thus, chronic inflammatory stress may alter the bi-directional molecular crosstalk between EGCs and tumor epithelial cells, which would favor CSC-derived tumor formation. This strengthens the tissue organization field theory (TOFT) of carcinogenesis, which proposes that it is the disruption of normal signaling between cells due to carcinogenic insults that compromises genomic integrity leading to tumor development [54]. It will be very interesting to evaluate whether EGC acquisition of a ‘pro-tumorigenic phenotype’ occurs before or concomitantly with tumor initiation in future studies.
Finally, from a therapeutic perspective, a better understanding of the crosstalk between EGCs and tumor cells may lead to the development of new therapies aiming to block CSC-driven tumor initiation and cancer relapse. Here we show that the tumor-induced remodeling of EGCs activating stemness in colon tumors is driven by an IL-1/PGE2 paracrine signaling. Cooperative effects between cancer cells and their niche via an IL-1/PGE2 paracrine signaling have been previously suggested in other systems [55,56], and interestingly, recent studies in head and neck squamous cell carcinoma suggest that targeting IL-1 may hinder carcinogenesis by reprogramming the tumor microenvironment [57]. Together with our study, this may suggest that the IL-1/PGE2 paracrine signaling might be a common target to limit stemness in various cancers, and it urges for studies aiming to identify specific molecular actors downstream IL-1/PGE2 that may represent safe therapeutic targets, along with in vivo validation studies.
Funding sources
This work was supported by grants from the French National Cancer Institute (INCA_7904 to L. Van Landeghem), La Ligue contre le Cancer(to M. Neunlist from the Loire-Atlantique, Morbihan, Mayenne, Loiret and Sarthe Committees, a post-doctoral fellowship to L. Van Landeghem from the Loire-Atlantique and Sarthe Committees, and a PhD fellowship to S. Valès from La Ligue Nationale contre le Cancer), the ‘Région des Pays de la Loire’ and the UNC Lineberger Comprehensive Cancer Center (Developmental grant to L. Van Landeghem).
Author contributions
SV, GB and LVL designed and performed the experiments. SV, GB and LVL analyzed and interpreted the data. MB, MT, FG, KAD, SL and LO participated in the in vitro experiments. AB, SML and TO participated in the in vivo experiments. GB and MT performed histology procedures and analyses. SV, GB, FG, CB, MF, ED, LO and LVL performed the collection of human colon specimens. SV, LO, MN and FMV participated in the study concept and the manuscript writing. LVL supervised the study and wrote the manuscript.
Declaration of Competing Interest
All the authors declare no potential conflict of interest.
Acknowledgments
The authors wish to thank Tony Durand, Philippe Aubert, Julien Chevalier, Alice Prigent, Julie Jaulin, Elise Beneteau, Elaine Glenny, Bradley Wieland, Carlton Anderson, Gabrielle Cannon and Nathalie Vaillant for their invaluable technical assistance. Discussions with Geneviève Aillet, Pablo Ariel, Scott Magness and Christophe Guilluy are gratefully acknowledged. The work was assisted by services of the Cytometry and cell sorting Cytocell core facility of Nantes (France), the Experimental Therapy Unit animal facility of Nantes (France), the Histopathology Institute of Nantes (IHP, France), the UNC Microscopy Services Laboratory (Department of Pathology and Laboratory Medicine, UNC-CH, Chapel Hill, USA), which is supported in part by a Cancer Center Core Support Grant (P30 CA016086 to the UNC Lineberger Comprehensive Cancer Center) and a North Carolina Biotech Center InstitutionalSupport Grant (2016-IDG-1016), the UNC LCCC Tissue Procurement Facility(IRB# 90–0573 “LCCC 9001 Procurement of Biospecimens”), and the UNC Center for Gastrointestinal Biology and Disease Advanced Analytics Core, which is supported by a National Institutes of Health grant (P30 DK34987).
References
- 1.Ricci-Vitiani L., Lombardi D.G., Pilozzi E., Biffoni M., Todaro M., Peschle C. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007 4;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 2.O'Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007 4;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 3.Gao W., Chen L., Ma Z., Du Z., Zhao Z., Hu Z. Isolation and phenotypic characterization of colorectal cancer stem cells with organ-specific metastatic potential. Gastroenterology. 2013;145(3):636–646. doi: 10.1053/j.gastro.2013.05.049. [DOI] [PubMed] [Google Scholar]
- 4.Colak S., Zimberlin C.D., Fessler E., Hogdal L., Prasetyanti P.R., Grandela C.M. Decreased mitochondrial priming determines chemoresistance of colon cancer stem cells. Cell Death Differ. 2014;21(7):1170–1177. doi: 10.1038/cdd.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huels D.J., Sansom O.J. Stem vs non-stem cell origin of colorectal cancer. Br J Cancer. 2015;113(1):1–5. doi: 10.1038/bjc.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medema J.P., Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature. 2011;474(7351):318–326. doi: 10.1038/nature10212. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen L., De Sousa E., Melo F., van der Heijden M., Cameron K., de Jong J.H., Borovski T. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 8.Colak S., Medema J.P. Human colonic fibroblasts regulate stemness and chemotherapy resistance of colon cancer stem cells. Cell Cycle Georget Tex. 2014 doi: 10.4161/15384101.2014.973321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei H.-J., Zeng R., Lu J.-H., Lai W.-F.T., Chen W.-H., Liu H.-Y. Adipose-derived stem cells promote tumor initiation and accelerate tumor growth by interleukin-6 production. Oncotarget. 2015;6(10):7713–7726. doi: 10.18632/oncotarget.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulbransen B.D., Sharkey K.A. Novel functional roles for enteric glia in the gastrointestinal tract. Nat Rev Gastroenterol Hepatol. 2012;9(11):625–632. doi: 10.1038/nrgastro.2012.138. [DOI] [PubMed] [Google Scholar]
- 11.Sharkey K.A. Emerging roles for enteric glia in gastrointestinal disorders. J Clin Invest. 2015 2;125(3):918–925. doi: 10.1172/JCI76303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neunlist M., Van Landeghem L., Mahé M.M., Derkinderen P., des Varannes S.B., Rolli-Derkinderen M. The digestive neuronal-glial-epithelial unit: a new actor in gut health and disease. Nat Rev Gastroenterol Hepatol. 2013;10(2):90–100. doi: 10.1038/nrgastro.2012.221. [DOI] [PubMed] [Google Scholar]
- 13.Bush T.G., Savidge T.C., Freeman T.C., Cox H.J., Campbell E.A., Mucke L. Fulminant jejuno-ileitis following ablation of enteric glia in adult transgenic mice. Cell. 1998;93(2):189–201. doi: 10.1016/s0092-8674(00)81571-8. [DOI] [PubMed] [Google Scholar]
- 14.Van Landeghem L., Chevalier J., Mahé M.M., Wedel T., Urvil P., Derkinderen P. Enteric glia promote intestinal mucosal healing via activation of focal adhesion kinase and release of proEGF. Am J Physiol Gastrointest Liver Physiol. 2011;300(6):G976–G987. doi: 10.1152/ajpgi.00427.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savidge T.C., Newman P., Pothoulakis C., Ruhl A., Neunlist M., Bourreille A. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology. 2007;132(4):1344–1358. doi: 10.1053/j.gastro.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 16.Neunlist M., Aubert P., Bonnaud S., Van Landeghem L., Coron E., Wedel T. Enteric glia inhibit intestinal epithelial cell proliferation partly through a TGF-beta1-dependent pathway. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G231–G241. doi: 10.1152/ajpgi.00276.2005. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y.A., Chung Y.C., Pan S.T., Shen M.Y., Hou Y.C., Peng S.J. 3-D imaging, illustration, and quantitation of enteric glial network in transparent human colon mucosa. Neurogastroenterol Motil Off J Eur Gastrointest Motil Soc. 2013;25(5):e324–e338. doi: 10.1111/nmo.12115. [DOI] [PubMed] [Google Scholar]
- 18.Garcia S.B., Stopper H., Kannen V. The contribution of neuronal–glial–endothelial–epithelial interactions to colon carcinogenesis. Cell Mol Life Sci. 2014;71(17):3191–3197. doi: 10.1007/s00018-014-1642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soret R., Coquenlorge S., Cossais F., Meurette G., Rolli-Derkinderen M., Neunlist M. Characterization of human, mouse, and rat cultures of enteric glial cells and their effect on intestinal epithelial cells. Neurogastroenterol Motil Off J Eur Gastrointest Motil Soc. 2013;25(11):e755–e764. doi: 10.1111/nmo.12200. [DOI] [PubMed] [Google Scholar]
- 20.Wong-Lee J.G., Lovett M. Rapid and sensitive PCR method for identification of Mycoplasma species in tissue culture. Diagn Mol Microbiol Princ Appl Am Soc Microbiol Wash DC USA. 1993 [Google Scholar]
- 21.Yeung T.M., Gandhi S.C., Wilding J.L., Muschel R., Bodmer W.F. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci. 2010 23;107(8):3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botchkina G.I., Zuniga E.S., Das M., Wang Y., Wang H., Zhu S. New-generation taxoid SB-T-1214 inhibits stem cell-related gene expression in 3D cancer spheroids induced by purified colon tumor-initiating cells. Mol Cancer. 2010;9:192. doi: 10.1186/1476-4598-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalerba P., Dylla S.J., Park I.-K., Liu R., Wang X., Cho R.W. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007 12;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renier N., Wu Z., Simon D.J., Yang J., Ariel P., Tessier-Lavigne M. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159(4):896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Adamsen B.L., Kravik K.L., De Angelis P.M. DNA damage signaling in response to 5-fluorouracil in three colorectal cancer cell lines with different mismatch repair and TP53 status. Int J Oncol. 2011;39(3):673–682. doi: 10.3892/ijo.2011.1080. [DOI] [PubMed] [Google Scholar]
- 26.Murakami M., Ohta T., Otsuguro K.-I., Ito S. Involvement of prostaglandin E2 derived from enteric glial cells in the action of bradykinin in cultured rat myenteric neurons. Neuroscience. 2007;145(2):642–653. doi: 10.1016/j.neuroscience.2006.12.052. [DOI] [PubMed] [Google Scholar]
- 27.Jung P., Sommer C., Barriga F.M., Buczacki S.J., Hernando-Momblona X., Sevillano M. Isolation of human colon stem cells using surface expression of PTK7. Stem Cell Rep. 2015;5(6):979–987. doi: 10.1016/j.stemcr.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gemei M., Mirabelli P., Di Noto R., Corbo C., Iaccarino A., Zamboli A. CD66c is a novel marker for colorectal cancer stem cell isolation, and its silencing halts tumor growth in vivo. Cancer. 2013;119(4):729–738. doi: 10.1002/cncr.27794. [DOI] [PubMed] [Google Scholar]
- 29.Huang E.H., Hynes M.J., Zhang T., Ginestier C., Dontu G., Appelman H. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pai R., Soreghan B., Szabo I.L., Pavelka M., Baatar D., Tarnawski A.S. Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med. 2002;8(3):289–293. doi: 10.1038/nm0302-289. [DOI] [PubMed] [Google Scholar]
- 31.Buchanan F.G., Gorden D.L., Matta P., Shi Q., Matrisian L.M., DuBois R.N. Role of beta-arrestin 1 in the metastatic progression of colorectal cancer. Proc Natl Acad Sci U S A. 2006;103(5):1492–1497. doi: 10.1073/pnas.0510562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H.-J., Reinhardt F., Herschman H.R., Weinberg R.A. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2012;2(9):840–855. doi: 10.1158/2159-8290.CD-12-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinarello C.A., Wolff S.M. The role of interleukin-1 in disease. N Engl J Med. 1993;328(2):106–113. doi: 10.1056/NEJM199301143280207. [DOI] [PubMed] [Google Scholar]
- 34.Postlethwaite A.E., Raghow R., Stricklin G.P., Poppleton H., Seyer J.M., Kang A.H. Modulation of fibroblast functions by interleukin 1: increased steady-state accumulation of type I procollagen messenger RNAs and stimulation of other functions but not chemotaxis by human recombinant interleukin 1 alpha and beta. J Cell Biol. 1988;106(2):311–318. doi: 10.1083/jcb.106.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer-Barber K.D., Andrade B.B., Oland S.D., Amaral E.P., Barber D.L., Gonzales J. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511(7507):99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoffels B., Hupa K.J., Snoek S.A., van Bree S., Stein K., Schwandt T. Postoperative ileus involves interleukin-1 receptor signaling in enteric glia. Gastroenterology. 2014;146(1):176–187. doi: 10.1053/j.gastro.2013.09.030. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz B., Yehuda-Shnaidman E. Putative role of adipose tissue in growth and metabolism of colon cancer cells. Front Oncol. 2014;4:164. doi: 10.3389/fonc.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catalano V., Turdo A., Di Franco S., Dieli F., Todaro M., Stassi G. Tumor and its microenvironment: a synergistic interplay. Semin Cancer Biol. 2013;23(6 Pt B):522–532. doi: 10.1016/j.semcancer.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Mariani F., Sena P., Roncucci L. Inflammatory pathways in the early steps of colorectal cancer development. World J Gastroenterol. 2014;20(29):9716–9731. doi: 10.3748/wjg.v20.i29.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taddei M.L., Giannoni E., Comito G., Chiarugi P. Microenvironment and tumor cell plasticity: an easy way out. Cancer Lett. 2013;341(1):80–96. doi: 10.1016/j.canlet.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 41.Le Loupp A.-G., Bach-Ngohou K., Bourreille A., Boudin H., Rolli-Derkinderen M., Denis M.G. Activation of the prostaglandin D2 metabolic pathway in Crohn's disease: involvement of the enteric nervous system. BMC Gastroenterol. 2015;15:112. doi: 10.1186/s12876-015-0338-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pochard C., Coquenlorge S., Jaulin J., Cenac N., Vergnolle N., Meurette G. Defects in 15-HETE production and control of epithelial permeability by human enteric glial cells from patients with crohn's disease. Gastroenterology. 2016;150(1):168–180. doi: 10.1053/j.gastro.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Coquenlorge S., Van Landeghem L., Jaulin J., Cenac N., Vergnolle N., Duchalais E. The arachidonic acid metabolite 11β-ProstaglandinF2α controls intestinal epithelial healing: deficiency in patients with Crohn's disease. Sci Rep. 2016;6:25203. doi: 10.1038/srep25203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bach-Ngohou K., Mahé M.M., Aubert P., Abdo H., Boni S., Bourreille A. Enteric glia modulate epithelial cell proliferation and differentiation through 15-deoxy-12,14-prostaglandin J2. J Physiol. 2010;588(Pt 14):2533–2544. doi: 10.1113/jphysiol.2010.188409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerrero M.D., Aquino M., Bruno I., Terencio M.C., Paya M., Riccio R. Synthesis and pharmacological evaluation of a selected library of new potential anti-inflammatory agents bearing the gamma-hydroxybutenolide scaffold: a new class of inhibitors of prostanoid production through the selective modulation of microsomal prostaglandin E synthase-1 expression. J Med Chem. 2007;50(9):2176–2184. doi: 10.1021/jm0700823. [DOI] [PubMed] [Google Scholar]
- 46.El Mansouri F.E., Nebbaki S.-S., Kapoor M., Afif H., Martel-Pelletier J., Pelletier J.-P. Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1β-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes. Arthritis Res Ther. 2014;16(3):R113. doi: 10.1186/ar4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zayed N., El Mansouri F.E., Chabane N., Kapoor M., Martel-Pelletier J., Benderdour M. Valproic acid suppresses interleukin-1ß-induced microsomal prostaglandin E2 synthase-1 expression in chondrocytes through upregulation of NAB1. J Rheumatol. 2011;38(3):492–502. doi: 10.3899/jrheum.100907. [DOI] [PubMed] [Google Scholar]
- 48.Deckmann K., Rörsch F., Steri R., Schubert-Zsilavecz M., Geisslinger G., Grösch S. Dimethylcelecoxib inhibits mPGES-1 promoter activity by influencing EGR1 and NF-κB. Biochem Pharmacol. 2010;80(9):1365–1372. doi: 10.1016/j.bcp.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 49.Wang D., Fu L., Sun H., Guo L., DuBois R.N. Prostaglandin E2 promotes colorectal cancer stem cell expansion and metastasis in mice. Gastroenterology. 2015;149(7):1884–1895. doi: 10.1053/j.gastro.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doherty G.A., Byrne S.M., Molloy E.S., Malhotra V., Austin S.C., Kay E.W. Proneoplastic effects of PGE2 mediated by EP4 receptor in colorectal cancer. BMC Cancer. 2009;9:207. doi: 10.1186/1471-2407-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rademakers G., Vaes N., Schonkeren S., Koch A., Sharkey K.A., Melotte V. The role of enteric neurons in the development and progression of colorectal cancer. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbcan.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 52.Duchalais E., Guilluy C., Nedellec S., Touvron M., Bessard A., Touchefeu Y. Colorectal cancer cells adhere to and migrate along the neurons of the enteric nervous system. Cell Mol Gastroenterol Hepatol. 2018;5(1):31–49. doi: 10.1016/j.jcmgh.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pesic M., Greten F.R. Inflammation and cancer: tissue regeneration gone awry. Curr Opin Cell Biol. 2016;43:55–61. doi: 10.1016/j.ceb.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Soto A.M., Sonnenschein C. The tissue organization field theory of cancer: a testable replacement for the somatic mutation theory. BioEssays News Rev Mol Cell Dev Biol. 2011;33(5):332–340. doi: 10.1002/bies.201100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou Z., Falcone D.J., Subbaramaiah K., Dannenberg A.J. Macrophages induce COX-2 expression in breast cancer cells: role of IL-1β autoamplification. Carcinogenesis. 2011;32(5):695–702. doi: 10.1093/carcin/bgr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dudás J., Fullár A., Bitsche M., Schartinger V., Kovalszky I., Sprinzl G.M. Tumor-produced, active interleukin-1 β regulates gene expression in carcinoma-associated fibroblasts. Exp Cell Res. 2011;317(15):2222–2229. doi: 10.1016/j.yexcr.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu T., Hong Y., Jia L., Wu J., Xia J., Wang J. Modulation of IL-1β reprogrammes the tumor microenvironment to interrupt oral carcinogenesis. Sci Rep. 2016;6:20208. doi: 10.1038/srep20208. [DOI] [PMC free article] [PubMed] [Google Scholar]