Abstract
Background
Mothers are the primary source of bacteria for newborns, but it is unclear whether mother-to-newborn transmission occurs prior to, during or after birth. Similarly, the effect of the delivery mode on neonatal microorganisms has been the focus of controversy.
Methods
Healthy maternal and neonatal pairs that underwent vaginal birth and caesarean section were enrolled in this study. Meconium, placenta, membrane and amniotic fluid samples for newborns and vaginal, rectal and oral samples for mothers were collected. All samples were amplified and sequenced by a 16S rRNA gene primer set targeting bacteria and archaea.
Findings
A total of 550 samples from 36 mother-neonate pairs with vaginal births and 42 mother-neonate pairs with caesarean sections were included in this study. The negative controls showed that the data analysis in this study was not affected by contamination. There was a high diversity of microbial communities in the pregnancy environment of the foetus. Meconium samples could be divided into three distinct types that were not influenced by the delivery method.
Interpretation
The distribution patterns of bacterial communities in the meconium, placenta, and foetal membranes were highly similar and had nothing to do with the mode of delivery. For approximately half of the placental microorganisms, the same sequence could be found in the vaginal, rectal, and oral samples of the mother.
Keywords: Meconium, Microbiota, Delivery mode, Archaea
Research in context
Evidence before this study
Database: Web of Science. Date: In the past 10 years.
De Goffau's publication in August 2019 was contrary to multiple investigators prior publications, and suggested that microorganisms in the human placenta could not be reliably distinguished from possible contamination. However, bacterial translocation from the intestine to the maternal blood stream and from the blood stream to other organ systems is increased during pregnancy. Multiple recent studies employing meconium as a proxy for in utero bacterial communities suggested that bacterial transmission from the mother to the foetus is a regular occurrence during human pregnancies. Seferovic et al. suggested that the taxonomic makeup of more than 80% of placental microbes was distinct from that of contamination controls by both 16S rRNA in situ hybridization and Illumina sequencing of the V4 region of the rRNA gene. This study also used strict negative controls, including “kit-negative” and sampling negative controls. In addition to bacteria and archaea, fungi also play a complex role in the health of newborns. Fungal colonization of the primordial gut was initially found by sequencing and culture-based techniques with negative controls, and perinatal exposures known to shape the postnatal microbiome did not significantly alter the community composition. Cultivatable bacteria in the foetal intestine were found during mid-gestation but not late gestation, as confirmed by a recent study.
Another puzzle was the effect of delivery mode on the microbial composition in the meconium. Several studies have reported significant differences in the composition of vaginally delivered newborns versus C-section-delivered newborns, with vaginally born neonates harbouring an early microbiome that resembles that of the vagina and C-section-delivered neonates harbouring an early microbiome that resembles that of the human skin. However, at the same time, several studies suggested that the bacterial community of the meconium was not affected by the delivery mode. Chu and colleagues found that in neonatal stool, the microbiota community was similar in neonates with different delivery modes. Our other small-sample study also showed no significant difference in the meconium between neonates delivered by C-section and neonates delivered vaginally. Hu et al.’s study of 23 neonates (10 of whom were delivered by C-section) reported no significant differences in the meconium microbiota of full-term vaginally delivered and C-section-delivered neonates. Similarly, Mshvildadze et al. reported no significant differences in the meconium microbiota of pre-term (23–32 weeks of gestational age) vaginally delivered and C-section-delivered neonates.
Added value of this study
Our results were mainly divided into two parts: 1. With strict negative controls, samples from the foetal environment still showed high microbial diversity and a consistent microbial community composition. 2. Unlike most studies, the microbial composition in the meconium in this study could be divided into three types and was not affected by the mode of delivery. In addition, although archaea comprised a small part of the community, this is an interesting topic of study.
Implications of all the available evidence
Understanding the composition and source of intestinal microorganisms in neonates is of great significance to the health of infants, children and adults. Our findings herein demonstrate that most of the microbes in the meconium originate from the foetal environment. Therefore, we indirectly refute the findings of de Goffau, and suggest that it is necessary to further study the role of microorganisms in the process of normal fetal development.
1. Introduction
In recent years, the impact of the mode of delivery (i.e., vaginal vs. caesarean) on healthy infants has been subjected to scrutiny due to the increased rate of caesarean deliveries worldwide and their potential association with allergic and autoimmune diseases [1,2]. Interestingly, various authors who have performed studies on the composition of bacterial communities in the meconium have drawn completely inconsistent conclusions. Several studies have reported significant differences in the composition of bacterial communities in vaginally delivered versus C-section-delivered newborns, with vaginally born neonates harbouring an early microbiome that resembles that of the vagina and neonates born by C-section harbouring an early microbiome that resembles that of the human skin [3], [4], [5]. However, at the same time, several studies suggested that the bacterial community of the meconium was not affected by the delivery mode. Chu and colleagues found that in neonatal stool, the microbiota community was similar in neonates with different delivery modes [6]. Our other small-sample study also showed no significant difference in meconium between neonates delivered by C-section and neonates delivered vaginally [7]. Hu et al.’s study of 23 neonates (10 of whom were delivered by C-section) reported no significant differences in the meconium microbiota of full-term vaginally delivered and C-section-delivered neonates [8]. Similarly, Mshvildadze et al. reported no significant differences in the meconium microbiota of pre-term (23–32 weeks of gestational age) vaginally delivered and C-section-delivered neonates [9].
Another myth about the gestational environment is that although many studies have shown that the meconium or placenta contains microbes, there are some studies that contradict this conclusion [10], [11], [12]. Stout and colleagues [13] suggested instead that the endometrial epithelium of the nonpregnant uterus might harbour occult microbes that become incorporated into the basal plate at the time of placental implantation. Aagaard and colleagues [14] determined the microbiome of pre-term and full-term placentas and identified a unique bacterial community consisting of a large number of species with low abundance. These studies propose that the foetus, the placenta, and the amniotic fluid are not sterile and that microbial acquisition and colonization of the human gastrointestinal tract begins in utero [11,14,15]. However, until recently, the existence of microorganisms in the foetal environment has been controversial, and some studies have shown that the microorganisms detected in the placenta by conventional methods originate from environmental pollution [16,17]. Whether the presence of a placental microbiome is advantageous to the growing embryo remains to be established — at least it is now clear that such bacteria are not always harmful. If this “bacteria in uterus” hypothesis proves correct, there would be major repercussions on our understanding of the establishment of the human microbiome and clinical practices — for example, the idea that C-sections do not affect the establishment of the foetal intestinal flora [11].
Therefore, an in-depth study on the microorganisms that are contained in the foetal growth environment and whether the meconium is related to the mode of delivery is essential to improve our understanding of the establishment of intestinal microorganisms in neonates and the subsequent potential impact on health. In this study, a total of 38 vaginally delivered and 42 C-section-delivered mother-neonate pairs were sampled. Rectal, vaginal, and oral mucosal samples for the mothers and placental, foetal membrane, amniotic fluid and meconium samples for the neonates were collected, and their taxonomic compositions were assessed to determine the impact of the caesarean mode of delivery and its potential confounders or modifiers on the neonatal microbiota structure.
2. Materials and methods
2.1. Subject recruitment and clinical information
To reduce the impact of climate and the hospital environment at different times, all samples were collected within two weeks at the Obstetrics Department of Yan'an Affiliated Hospital of Kunming Medical University in Yunnan Province, China. During this period, healthy maternal and neonate pairs were recruited until the number of samples for each delivery mode reached nearly 40 (Supplementary Table S1). The mothers did not receive probiotic supplementation during pregnancy. This project was approved by the Ethics Committee of Yan'an Affiliated Hospital of Kunming Medical University (2019-077-01). The mothers were made aware of the nature of the study, specifically consented to give their personal information, and gave written informed consent for their and their child's participation.
2.2. Sample collection
A total of seven samples were collected for each mother-neonate pair. During the delivery process, swabs were taken from the rectal, vaginal, and oral mucosa of the mother at approximately 1 h before delivery. After delivery, approximately 5 g of placenta and 10 cm2 of foetal membrane samples were removed with a sterile scalpel and placed into sterile single-use collection tubes. Approximately 4 mL of amniotic fluid was collected using a sterile primary syringe as soon as possible after delivery. Samples from prematurely ruptured membranes were not included. Internal portions of the first-pass meconium stools before breastfed meconium were collected from sterile single-use diapers into sterile single-use collection tubes. All samples were stored at −20 °C prior to DNA extraction.
To detect contamination in the sampling process and the experimental process, strict negative controls were carried out. When each swab was sampled, two blank swabs were placed and mock sampled at the same time. For samples placed in centrifuge tubes, approximately the same amount of sterile water was placed in the same batch of sterile single-use collection tubes. All negative control samples were also used in the subsequent experimental process.
2.3. DNA preparation, PCR amplification, and sequencing
A frozen aliquot (1 g) of each placenta, foetal membrane and meconium sample was ground with liquid nitrogen four to five times to generate a uniform fine powder for DNA extraction. To avoid contamination, each mortar used in the grinding process was cleaned and dried at approximately 200 °C. All 4 mL of the amniotic fluid sample was centrifuged for 5 min at 12,000 r/min, and the pellet was used for DNA extraction. The cotton sections of the swabs were cut for DNA extraction. Total DNA was extracted as previously described [18]. In the DNA extraction process, all reagents were separated immediately after purchase in a newly purchased (no microbial or molecular biology-related experiments) super-clean table, and each sample was treated by an independent reagent group. were DNA yield was assessed using the Nanodrop 2000 (Thermo Fisher, USA).
For each sample, the bacterial and archaeal 16S rRNA gene was amplified with the barcoded primer set 515F [19] and 909R [20] containing the Illumina MiSeq adapter sequences. For all PCR amplifications, reactions were performed with pfu DNA MasterMix (Tiangen, China) in a total volume of 50 μl with 15 ng of DNA added as template, and the annealing temperature was 51 °C. During each DNA extraction and PCR batch, two negative controls with the same reagents and consumables were operated as the same procedure. Amplicons were cleaned using the UltraClean PCR Clean-up kit (MO BIO, USA), and an equivalent amount of PCR product was mixed for sequencing using the Illumina MISEQ™ system (Illumina, USA).
2.4. Sequence analysis and statistics
All sequences were demultiplexed using the barcodes of each sample. Sequence processing was performed by combining features of Mothur v1.42.0 [21] according to MiSeq SOP. The SSU rRNA database sequences and taxonomic information from SILVA (v132) [22] were downloaded directly from the Mothur website. Chimera checking was performed after the sequences were aligned. Similar sequences were clustered into OTUs with a minimum identity of 97% or 100%. Phylogenetic trees were constructed via MEGA version 6.0 [23] with the neighbour joining method. The distance matrix was analysed by thetaYC [21] methods, and the larger principal components were selected for principal component analysis (PCA). The shared OTU index was calculated with sorabund, which returned the abundance-based Sorenson dissimilarity index [24]. A singleton was an OTU with only one sequence, and a doubleton was an OTU with two sequences. The analysis of similarities (ANOSIM) function in the program Mothur v1.42.0 was used to test for differences in community composition among various sample groups. Population levels between different groups of samples were analysed by Metastats and LEfSe. The community types were defined based on Dirichlet multinomial mixtures, as described by Holmes et al., and this approach was used because it allows for clustering from unevenly sampled populations [25].
2.5. Nucleotide sequence availability
The PCR product sequencing data in this study were deposited in the Short Read Archive of NCBI under the accession number PRJNA559967.
3. Results
3.1. Distinct microbiota abundance and composition in different mother-neonate pairs
A total of 78 mother-neonate pairs (36 vaginally delivered and 42 C-section-delivered) were enrolled, and the details are provided in Tables 1 and S1. There was no significant difference in age and birth weight between the two modes of delivery. There were significant differences in the gestational times and gestational ages between the two modes of delivery.
Table 1.
General characteristics of the mother-neonate pairs enrolled in this study, including birth mode, gender, sampling time, birth weight, height, and antibiotic exposure.
| Characteristic | All | Vaginally born | Caesarean-born | p |
|---|---|---|---|---|
| Sample numbers | 78 | 42 | 36 | – |
| Maternal age (year) | 28.9 ± 4.5 | 28.7 ± 4.5 | 29.1 ± 4.5 | 0.663 |
| Gestational times | 2.4 ± 1.2 | 2.0 ± 1.0 | 2.7 ± 1.2 | 0.012 |
| Gestational age (day) | 275 ± 9 | 277 ± 10 | 273 ± 7 | 0.007 |
| Birth weight (g) | 3278 ± 400 | 3334 ± 373 | 3230 ± 419 | 0.252 |
| Gender | ||||
| Male (%) | 54% | 56% | 53% | – |
| Female (%) | 46% | 44% | 47% | – |
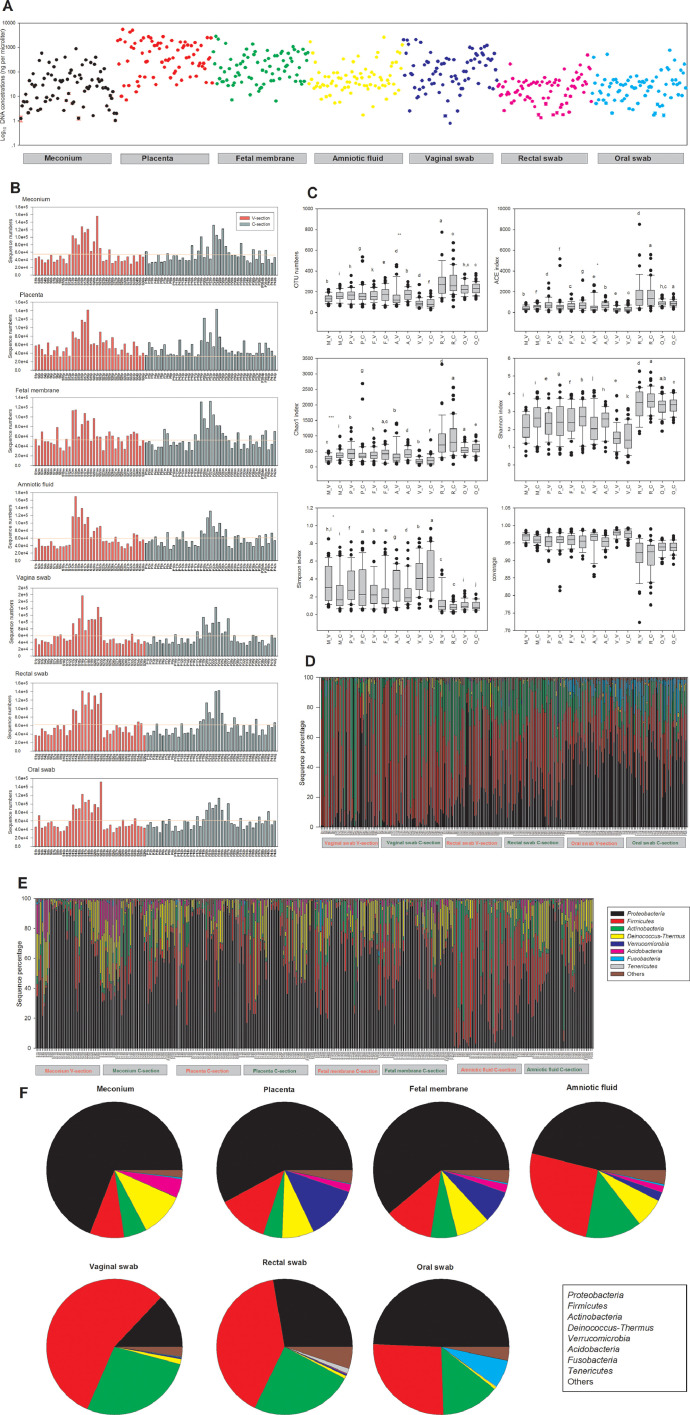
All the negative control samples contained less DNA than the lower detection limit of the Nanodrop 2000, and when the “DNA” extracted from the negative control samples was used as template, no corresponding bands were obtained under the same PCR conditions. The DNA contents of all samples are shown in Fig. 1A. Considering the different amounts of DNA extracted from the different types of samples, most of the samples contained approximately the same range of DNA. With respect to different sequencing runs, the sequence numbers of each sample were quite different, but all samples contained more than 30,000 sequences (Fig. 1B). Considering the different α-diversity indices (OTU numbers and ACE, Chao1, Shannon and Simpson indices), there were significant differences among most different types of samples (different sampling sites for both delivery modes) (Fig. 1C). Generally, the α-diversity indices of the meconium, placenta, foetal membrane, and amniotic fluid samples of neonates were most similar to those of the other samples. Compared to the samples from the mothers, the vaginal samples had the lowest diversity, and the rectal samples from the mothers had the highest diversity. The bacterial composition of each sample at the phylum level is shown in Fig. 1D (mother-related samples) and 1E (neonate-related samples), and the overall community composition of each site is shown in Fig. 1F. Firmicutes was the most prevalent phylum in the rectal and vaginal swab samples, while Proteobacteria was the most predominant phylum in the oral swab and neonate-related samples.
Fig. 1.
Microbiota abundance and composition in different mother-neonate pairs. (A) Concentration of total DNA extracted from each sample. (B) Number of sequences obtained by 16S rRNA gene sequencing. The numbers refer to individual mother-neonate pairs. The horizontal line reflects the average number of sequences for all samples. (C) Alpha-diversity box-whisker plots of OTU number, taxon richness (ACE and Chao1 indices) and diversity (Shannon–Wiener and Simpson indices) in samples analysed by 16S rRNA gene sequencing. The letters above the bars indicate the results of the Tukey HSD test following a significant 1-way ANOVA. Mean values that do not share the same letters were significantly different from each other (p < 0.05). (D) Relative abundances of bacterial taxa identified at the phylum level for each mother-related sample. (E) Relative abundances of bacterial taxa identified at the phylum level for each neonate-related sample. (F) Diversity of the most abundant bacterial taxa identified at the phylum level in different sample types (meconium, placenta, foetal membrane, amniotic fluid, vaginal swab, rectal swab and oral swab). Taxonomic assignment of 16S rRNA sequences was carried out with the SILVA SSU database v132 with a cutoff of 80% homology.
3.2. OTU-based analysis of different sampling sites
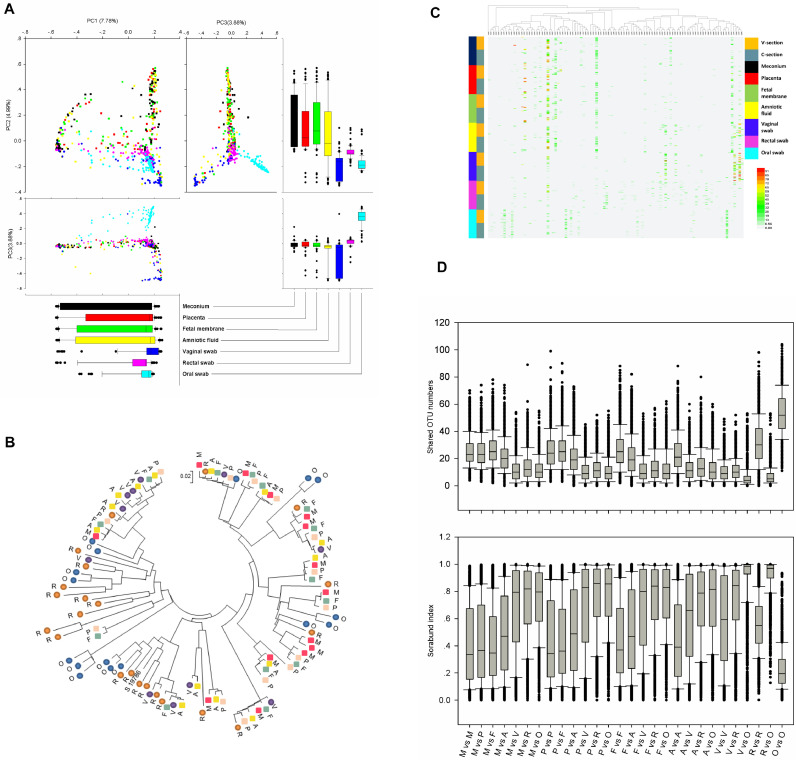
The seven sampling sites selected in this study were very useful for studying the relationship between the microorganisms in the mother and neonate. Thus, PCA was performed using OTU data. Fig. 2A revealed that the mother and neonate were the most significant contributors. For PC1 (7.78%), the meconium samples were the most widely distributed, and the other three types of neonatal samples had little difference from PC1. The samples from the mothers were concentrated. PC2 contributed 4.99% of the principal components, and the maternal samples were significantly separated from the neonatal samples. The representative OTU of each type of sample was used to construct a phylogenetic tree for overall comparison (Fig. 2B). The clustering of major OTUs on the phylogenetic trees clearly showed that the neonatal samples were clustered together. Detailed information and specific distributions for the top 100 OTUs in all samples are shown in Fig. 2C. The distribution of major OTUs in mothers and newborns was significantly different. Among the top 10 OTUs named by OTU abundance, the OTUs mainly distributed in the neonatal samples were OTU0001, OTU0003, OTU0004, OTU0005, OTU0006, and OTU0010, while the OTUs that mainly belonged to the maternal samples were OTU0002, OTU0007, OTU0008, and OTU0009. The comparison among different types of samples is shown by shared OTU and sorabund parameters (Fig. 2D). Meconium had more shared OTU numbers with placenta, foetal membrane, and amniotic fluid than with the mother-related samples. The fewest shared OTUs were found between the vaginal or rectal swabs and the oral samples. The sorabund parameters showed similar community similarity.
Fig. 2.
Comparison of the microbiomes of meconium, placenta, foetal membrane, amniotic fluid, vaginal swab, rectal swab and oral swab samples. (A) PCA plot based on the relative taxon abundance. Samples are marked by the group type. (B) Phylogenetic tree based on 16S rRNA gene sequences of major OTUs (more than 1% with a 0.03 cutoff) for each sample type. Meconium: red square, M; placenta: pink square, P; foetal membrane: grey-green square, F; amniotic fluid: yellow square, A; vaginal swab: purple circle, V; rectal swab: brown circle, R; oral swab: blue circle, O. (C) Heat map and phylogenetic analysis of the 100 most abundant OTUs (0.03 cutoff) in all samples. Vaginally delivered, yellow; C-section-delivered, grey blue; meconium, dark; placenta, red; foetal membrane, green; amniotic fluid, yellow; vaginal swab, blue; rectal swab, pink; oral swab, sky-blue. (D) Shared OTU numbers (0.03 cutoff) and sorabund index reflecting the similarities between different samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. The effect of delivery mode on the foetus-related microbiome
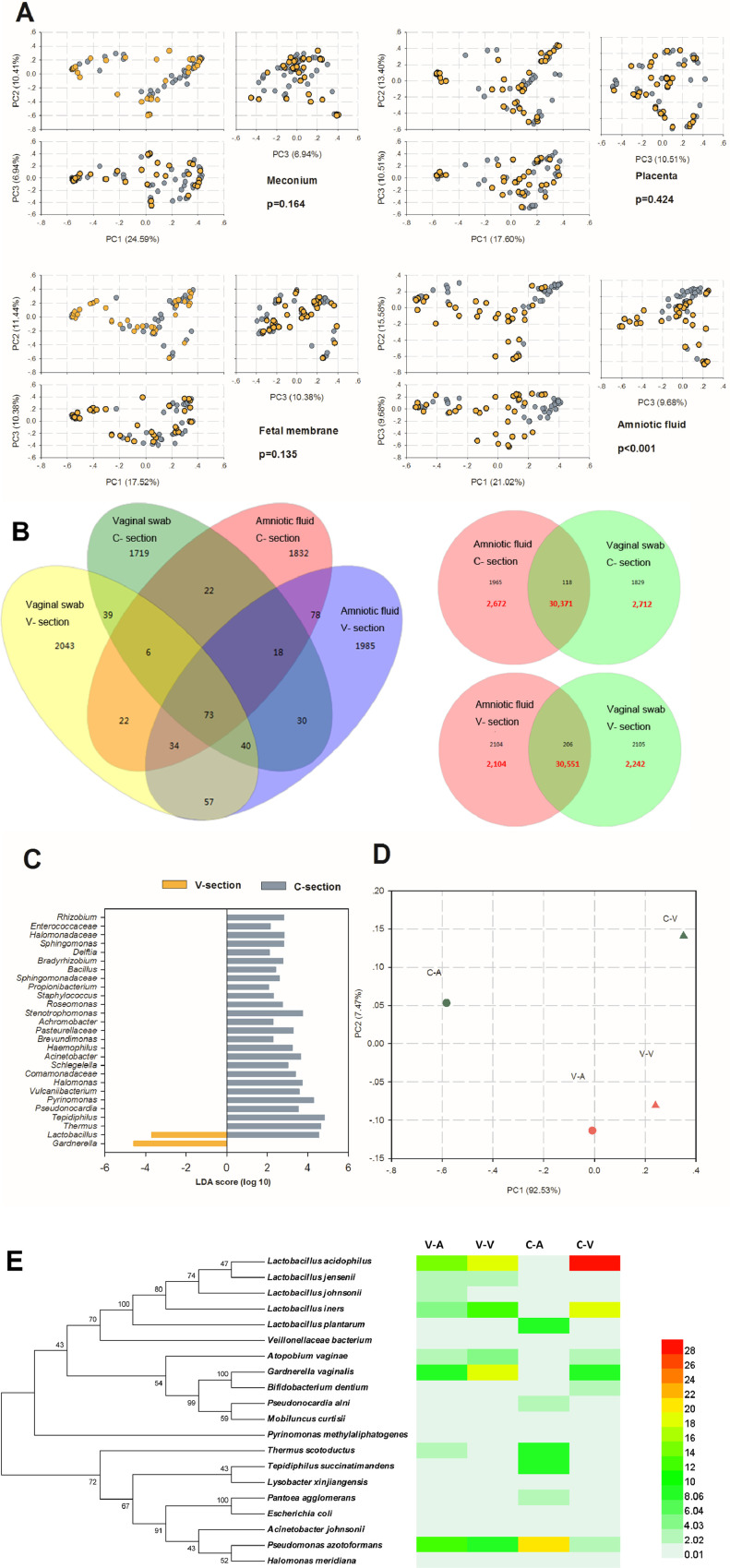
To illustrate the relationship between the samples from the vaginally delivered and C-section-delivered neonates, PCA was performed using species data at all sample levels (Fig. 3A). Only amniotic fluid samples showed significant differences (p < 0.001), while meconium, placenta, and foetal membrane samples showed no significant difference between the vaginally delivered and C-section-delivered neonates.
Fig. 3.
Comparison of the microbiomes of meconium, placenta, foetal membrane and amniotic fluid from vaginally delivered and C-section-delivered neonates. (A) PCA plot based on the relative taxon abundance in meconium, placenta, foetal membrane, and amniotic fluid samples from vaginally delivered and C-section-delivered neonates. The analysis of ANOSIM similarities for differences in community composition among various sample groups and expressed by the p-value. (B) Venn diagram of shared OTUs and sequences among different sample types at a 0.03 cutoff. A total of 20,000 randomly selected raw sequences of one sample type were used. (C) LEfSe comparison of microbiomes from amniotic fluid samples from vaginally delivered and C-section-delivered neonates. Enriched taxa in samples from vaginally delivered neonates with a negative linear discriminant analysis (LDA) score are shown in yellow; C-section samples with a positive LDA score are shown in grey. (D) PCA plot of amniotic fluid and vaginal swab samples based on the relative taxon abundance. (E) Heat map analysis generated by major OTUs (more than 1% with a 0.03 cutoff) in amniotic fluid and vaginal swab samples and their phylogenetic tree with taxonomic information. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Therefore, the difference in the bacterial community in the amniotic fluid samples between the vaginally delivered and C-section-delivered neonates deserves further study. First, the shared sequences between the amniotic fluid and vaginal swab samples from the vaginally delivered neonates contributed 88% of the total sequences used in these analyses (20,000 raw sequences in each sample type), and the percentage from the C-section neonates was 85% (Fig. 3B). Second, a LEfSe analysis was performed to investigate differences in the community composition between groups (Fig. 3C). The genera Lactobacillus and Gardnerella were significantly enriched in the vaginally delivered amniotic fluid samples, whereas Thermus, Tepidiphilus, and the other 24 genera were higher in abundance in the C-section delivered group. Third, a more accurate PCA analysis is shown in Fig. 3D. Amniotic fluid and vaginal swab samples showed more similar bacterial community structures for the vaginally delivered neonates than for the C-section-delivered neonates. Finally, a heat map of major OTUs in the four groups mentioned above and a phylogenetic tree with taxonomic information were constructed and are shown in Fig. 3E. The most obvious difference was an OTU belonging to L. acidophilus, which was similar in the amniotic fluid and vaginal swab samples from the vaginally delivered neonates.
3.4. Influencing factors and sources of the meconium microbiome
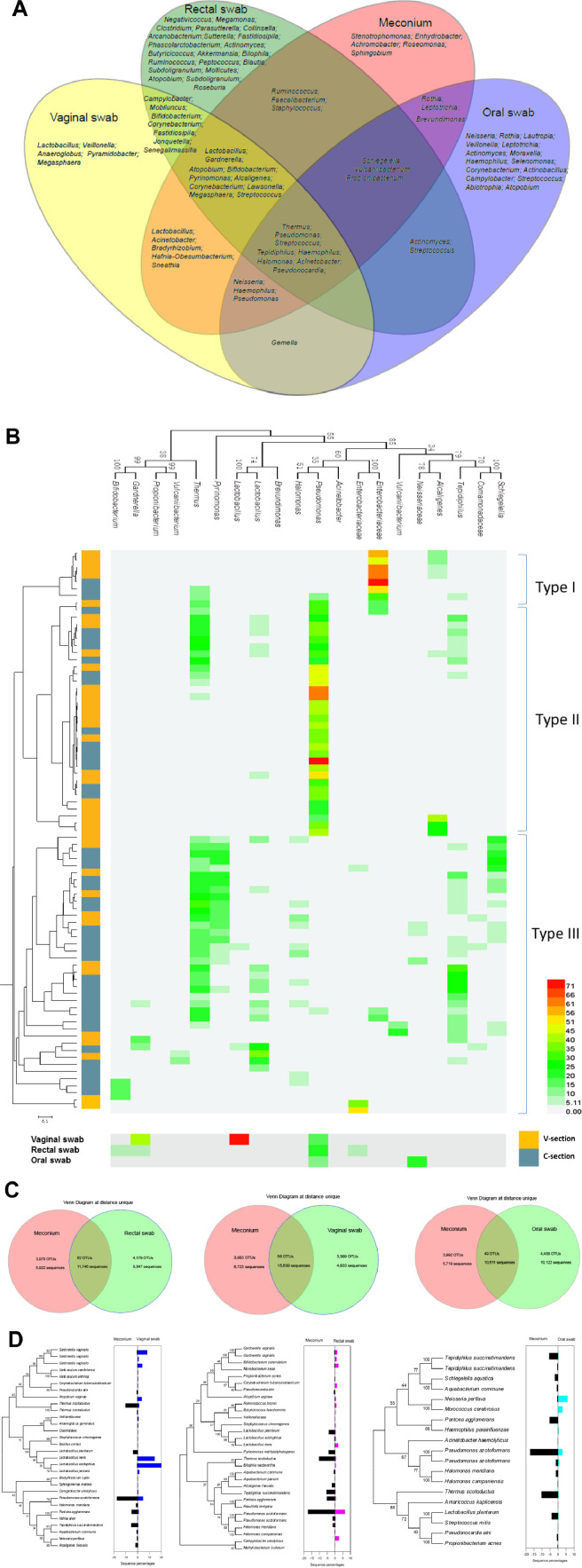
Samples from different maternal body sites were considered to be potential sources of bacteria for each neonatal sample. All bacterial genera shared between the meconium and mother-related samples are shown by a Venn diagram in Fig. 4A. The genera Thermus, Pseudomonas, Streptococcus, Tepidiphilus, Haemophilus, Halomonas, Acinetobacter and Pseudonocardia were shared by the meconium and vaginal, rectal, and oral swab samples. The rectal and oral samples had more unique genera. Next, we performed OTU analysis on all meconium samples (Fig. 4B). At a cutoff of 0.03, the meconium samples could be divided into three community types. The community types were not irrelevant to the delivery mode, maternal age, gestational times, gestational age, birth weight, or neonatal gender.
Fig. 4.
The correlation between meconium and maternal microorganisms. (A) Venn diagram of bacterial genera hypothesized to contribute to the meconium microbiome. (B) Heat map and phylogenetic tree of the major OTUs (more than 1% with a 0.03 cutoff) in each meconium sample and all mother-related sample types. All OTUs could be divided into three types; vaginally delivered, yellow; C-section-delivered, grey blue. (C) Venn diagrams of shared OTUs and sequence numbers between meconium and mother-related samples at a unique cutoff (the exact same sequence was treated as an OTU). A total of 20,000 randomly selected raw sequences of one sample type were used. (D) Phylogenetic tree of shared OTUs at a unique cutoff (all OTUs excluding singletons and doubletons) between meconium and mother-related samples. The histogram on the right side of the phylogenetic tree shows the percentage of each OTU. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To more accurately determine the origin of meconium microorganisms, in subsequent analysis, the OTUs were determined more strictly, and the same sequences were classified as OTUs. Under such an algorithm, the most common sequence (58%) was shared between the meconium and vaginal samples, with a percentage of 45% between the meconium and rectal swabs. Only 40% of the oral sample sequences were similar to the meconium sample sequences (Fig. 4C). In detail, all unique OTUs except singletons and doubletons were used to construct the phylogenetic tree comparing the meconium, vaginal, rectal and oral samples (Fig. 4D). The OTUs affiliated with Thermus scotoductus, Pseudomonas azotoformans, Pantoea agglomerans, and Tepidiphilus succinatimandens were more abundant in the meconium samples than in the vaginal samples, while the OTUs belonging to Gardnerella vaginalis, L. iners, and L. acidophilus showed the opposite trend. Between the meconium and rectal samples, the percentages of almost all shared OTUs were low in the faecal samples.
3.5. Archaea: friends or enemies?
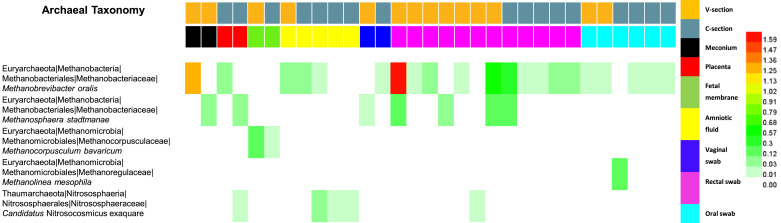
The primer sets 515F and 909R used in this study were designed to amplify both bacteria and archaea. A total of 45 samples contained 1210 sequences belonging to archaea (Fig. 5). The most abundant OTU was affiliated with Methanobrevibacter oralis, which was detected in all sample types except the foetal membrane samples. The second most abundant OTU belonged to Methanosphaera stadtmanae, which was detected in the meconium, placenta, vaginal, and rectal samples. The third and fourth most abundant OTUs were Methanocorpusculum bavaricum and Methanolinea mesophila, which were detected only in the foetal membrane and oral samples, respectively. Delivery methods had little effect on the detection of archaea.
Fig. 5.
Heat map and taxonomic information for the five archaeal OTUs (all OTUs excluding singletons and doubletons with a 0.03 cutoff) in different samples. Vaginally delivered, yellow; C-section-delivered, grey blue; meconium, dark; placenta, red; foetal membrane, green; amniotic fluid, yellow; vaginal swab, blue; rectal swab, pink; oral swab, sky-blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Whether the microbiota is present at birth or develops after birth is a controversial issue, and the same applies to the production of microorganisms in newborns. It has been suggested that microbial contact may start before birth, although this hypothesis is still controversial. The first 1000 days after conception (including the pregnancy period and the first two years of life), which are considered a “window of opportunity”, are crucial for the development and health of the future adult and key to the establishment of the intestinal microbiota and the maturation of the immune system [26]. To answer this question, we systematically studied the effects of delivery mode on the microorganisms of newborns and the sources of microorganisms of newborns.
In this study, the enrolled mother-neonate pairs for two different delivery modes were randomly selected over a two-week sampling period. The weight of the newborn and the age of the mother had no significant effect on delivery mode. In contrast, the number of pregnancies could affect the choice of C-section on the mother's own initiative. Similarly, as China allows second births, scar pregnancy and other factors have also led mothers to select C-sections without having other unhealthy factors; however, the probability of uterine rupture is very small.
With de Goffau's research showing that there are no microorganisms in the human placenta [17], whether there are microbial groups in the environment of foetal gestation has aroused great controversy. Before our research began, it was certain that even if microorganisms were present in the human gestation environment, the microbial content would be much lower than reported in the intestinal and oral environments. Therefore, at each sampling point, we set up more than two negative controls, such as centrifugal tubes and cotton swabs that were opened in the delivery room and operating room, to simulate the sampling process. Such strict negative control settings can effectively avoid the existence of false positives in the results, although none of these negative control samples suggested that the DNA content reached the detection limit of the Nanodrop 2000. This result at least indicated that samples such as placenta, foetal membranes and amniotic fluid contained more microorganisms than contaminated samples. The placenta and foetal membranes from vaginal delivery were sampled in the delivery room (on the 8th floor of the hospital), samples from C-section were sampled in the operation room (on the 14th floor), and both types of meconium were sampled in the ward (on the other part of the 8th floor). These three types of samples were sampled in different places but had highly similar microbial groups, indicating that the main microorganisms were not contaminated. These conclusions challenge the recent conclusion that the placenta is sterile [17]. The transfer of bacteria from the pregnant mother to the foetus is universal in the animal kingdom. Bacterial translocation from the intestine to the maternal blood stream and from there to other organ systems is increased during pregnancy [27]. Multiple recent studies employing meconium as a proxy for in utero bacterial communities suggest that bacterial transmission from the mother to the foetus is a regular occurrence during human pregnancies [15,28]. Seferovic et al. suggested that the taxonomic makeup of more than 80% of placental microbes was distinct from that of contamination controls by both 16S rRNA in situ hybridization and Illumina sequencing of the V4 region in the rRNA gene. This study also used strict negative controls, including “kit-negative” and sampling negative controls [29]. In addition to bacteria and archaea, fungi also play a complex role in the health of newborns. The initial fungal colonization of the primordial gut was found by sequencing and culture-based techniques under negative controls, and perinatal exposures known to shape the postnatal microbiome did not significantly alter community composition [30]. Cultivatable bacteria in the foetal intestine were found during mid-gestation but not late gestation, as confirmed using surgical deliveries of pregnant mice under highly controlled, sterile conditions in the laboratory by a recent study [31]. These studies also proved that although there must be contamination during sampling and the nucleic acid extraction period, this contamination did not affect the subsequent microbial analysis to a large extent.
The meconium microbiota appeared to have a distinct maternal origin from several body sites. Having extensively sampled multiple maternal body sites in parallel with equivalent neonatal samples, we sought to predict the most likely maternal origin of the neonatal microbiota. Our results demonstrated that meconium microbiota had more bacterial similarity with the vaginal samples from the mother regardless of the delivery mode. Meconium also shared more than 40% of unique sequences with the maternal gut and oral microbiota. Probiotic bacteria such as Lactobacilli detected in the meconium as well as the maternal samples have been detected repeatedly in the human placenta [32].
The important impact of intestinal microorganisms on human health has been recognized. The meconium, as the embryonic form of the intestinal microbiome, is undoubtedly of great significance to the long-term health of newborns. In a previous study, there were many different conclusions about the effect of vaginal or C-section deliveries on neonates. However, the effect of the delivery mode on the meconium is still a mystery, and different studies have drawn opposite conclusions [3,4,7,14]. This interesting phenomenon might be caused by small and unrepresentative sample sizes, population differences, experimental contamination and other factors.
Thus, in this study, relatively large sample sizes (36 vaginal deliveries and 42 C-sections) and various sampling sites (meconium, placenta, foetal membrane, amniotic fluid, vaginal swabs, rectal swabs, and oral swabs) were used to assess the effects of delivery methods on the microorganisms of the neonatal meconium. In our study, several results showed that the delivery mode had no effect on the microbiota of the meconium, placenta, and foetal membrane samples. Amniotic fluid was the only sampled site that was significantly affected by the delivery mode. The significant difference observed appeared to be related to the amniotic fluid sampling method. The amniotic fluid samples from vaginal delivery were bound to be contaminated by the vagina with Lactobacillus spp., but not the C-section amniotic fluid samples; therefore, our analysis confirms that the amniotic fluid samples from vaginal deliveries were indeed closer to the microbial community of the vaginal swabs. Since there were microorganisms in the environment of foetal gestation and since the microbial community in the meconium was very similar to the microorganisms in the placenta and foetal membranes, this observation indicated that the formation of the microbiome in the meconium might occur earlier than delivery; therefore, the mode of delivery may not affect the meconium microbiome. On the other hand, this result confirmed that there was not enough contamination from our experimental process to affect the experimental results.
The transfer of bacteria from the pregnant mother to the foetus is universal in the animal kingdom. At least in mammals, the maternal faecal microbiome appears to shape the microbiota in the foetal intestine, and therefore, the maternal gastrointestinal tract is the most likely source [8,33]. Bacterial translocation from the intestine to the maternal blood stream and from there to other organ systems is increased during pregnancy [34]. Jiménez et al. [15] orally inoculated pregnant mice with genetically labelled Enterococcus that had been previously isolated from the breast milk of a healthy woman. The labelled bacteria were retrieved from the internal meconium of pups after C-section delivery. Multiple recent studies employing meconium as a proxy for in utero bacterial communities suggest that bacterial transmission from the mother to the foetus is a regular occurrence during human pregnancies [15,28]. The meconium microbiota appeared to have a distinct maternal origin from several body sites. Having extensively sampled multiple maternal body sites in parallel with equivalent neonatal samples, we thus sought to predict the most likely maternal origin of the neonatal microbiota. Our results demonstrated that the meconium microbiota had more bacterial similarity with the vaginal samples from the mother regardless of the delivery mode. Meconium also shared more than 40% of unique sequences with the maternal gut and oral microbiota. Probiotic bacteria such as Lactobacilli detected in the meconium as well as the maternal samples have been detected repeatedly in the human placenta [32].
Methane has been associated with gastrointestinal disorders, mainly chronic constipation and constipation-predominant irritable bowel syndrome [35], as well as metabolic diseases such as obesity [36]. Most archaea detected in this study were from the rectal and oral samples of mothers; however, at the beginning of life, it seems that there were also archaea present. For neonatal samples, although only a few samples contained archaeal sequences, this was important evidence of the existence of archaea. It is worth mentioning that in the amniotic fluid samples, most of the samples containing archaeal sequences were from C-section samples, which were likely not contaminated by vaginal environments, which confirms that archaea exist in the environment of foetal gestation, although the proportion was not high.
In summary, by comparing a large number of samples and sequences and by using a rigorous experimental design, we draw the following important conclusions from this study. First, among the neonatal samples, including the meconium, placenta and foetal membranes, microbial communities existed, and the communities in samples from the same neonate were highly similar. Second, the distribution of bacterial communities in the meconium, placenta, and foetal membranes had nothing to do with the mode of delivery. Amniotic fluid samples were indeed related to the delivery mode due to the limitations of the sampling methods. The bacterial communities in the amniotic fluid from V-section were closer to those in the vaginal samples. Third, for approximately half of the placental microorganisms, the same sequence could be found in the vaginal, rectal, and oral samples of the mother. Fourth, although very rare, methanogens do exist in the foetal gestation system.
Funding
The National Natural Science Foundation of China (81860437), the Yunnan Province Innovation Team of Intestinal Microecology-Related Disease Research and Technological Transformation (China)and the Fundamental Application Research Foundation of Yunnan Province(2017FE468-092).
CRediT authorship contribution statement
Chen-Jian Liu: Data curation, Writing - original draft. Xiao Liang: Data curation, Writing - original draft. Zhao-Yi Niu: Investigation, Resources. Qing Jin: Investigation, Resources. Xue-Qin Zeng: Methodology, Formal analysis, Validation. Wen-Xue Wang: Methodology, Formal analysis, Validation. Meng-Yue Li: Methodology, Formal analysis, Validation. Xue-Rong Chen: Investigation, Resources. Hai-Yun Meng: Investigation, Resources. Ran Shen: Investigation, Resources. Shi-Yi Sun: Methodology, Formal analysis, Validation. Yi-Yong Luo: Software, Visualization. En Yang: Software, Visualization. Jia-Wei Geng: Funding acquisition, Writing - review & editing, Project administration. Xiao-Ran Li: Funding acquisition, Writing - review & editing, Project administration.
Declaration of Competing Interest
We declare no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.045.
Contributor Information
Jia-Wei Geng, Email: jia_wei_geng@163.com.
Xiao-Ran Li, Email: starkeyran@163.com.
Appendix. Supplementary materials
References
- 1.Chen H., Tan D. Cesarean section or natural childbirth? cesarean birth may damage your health. Front Psychol. 2019;10:351. doi: 10.3389/fpsyg.2019.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osterman M.J.K., Martin J.A. Trends in low-risk cesarean delivery in the United States, 1990-2013. national vital statistics reports: from the centers for disease control and prevention, national center for health statistics. Natl Vital Statistics Syst. 2014;63(6):1–16. [PubMed] [Google Scholar]
- 3.Dominguez-Bello M.G. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backhed F. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Nagpal R. Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu D.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23(3):314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong X.-D. Bacterial communities in neonatal feces are similar to mothers' placentae. Can J Infect Dis Med Microbiol. 2015;26(2):90–94. doi: 10.1155/2015/737294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu J. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0078257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mshvildadze M. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010;156(1):20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tribe R.M. Parturition and the perinatal period: can mode of delivery impact on the future health of the neonate? J Physiol-Lond. 2018;596(23):5709–5722. doi: 10.1113/JP275429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaser M.J., Dominguez-Bello M.G. The human microbiome before birth. Cell Host Microbe. 2016;20(5):558–560. doi: 10.1016/j.chom.2016.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Haahr T. Vaginal seeding or vaginal microbial transfer from the mother to the caesarean-born neonate: a commentary regarding clinical management. Bjog-an Int J Obstet Gynaecol. 2018;125(5):533–536. doi: 10.1111/1471-0528.14792. [DOI] [PubMed] [Google Scholar]
- 13.Stout M.J. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol. 2013;208(3) doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aagaard K. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237) doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez E. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008;159(3):187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Leiby J.S. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018;6 doi: 10.1186/s40168-018-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Goffau M.C. Human placenta has no microbiome but can contain potential pathogens. Nature. 2019;572(7769) doi: 10.1038/s41586-019-1451-5. 329-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt T.M., DeLong E.F., Pace N.R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reysenbach A.L. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58(10):3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunk C.F., Eis N. Quantitative measure of small-subunit rRNA gene sequences of the kingdom Korarchaeota. Appl Environ Microbiol. 1998;64(12):5064–5066. doi: 10.1128/aem.64.12.5064-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss P.D. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruesse E. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao A. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett. 2005;8(2):148–159. [Google Scholar]
- 25.Holmes I., Harris K., Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE. 2012;7(2) doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dzidic M. Gut microbiota and mucosal immunity in the neonate. Med Sci (Basel) 2018;6(3) doi: 10.3390/medsci6030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shokry E. Investigation of the impact of birth by cesarean section on fetal and maternal metabolism. Arch Gynecol Obstet. 2019;300(3):589–600. doi: 10.1007/s00404-019-05213-w. [DOI] [PubMed] [Google Scholar]
- 28.Moles L. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seferovic M.D. Visualization of microbes by 16S in situ hybridization in term and preterm placentas without intraamniotic infection. Am J Obstet Gynecol. 2019;221(2) doi: 10.1016/j.ajog.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willis K.A. Fungi form interkingdom microbial communities in the primordial human gut that develop with gestational age. FASEB J. 2019 doi: 10.1096/fj.201901436RR. fj201901436RR-fj201901436RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Younge N. Fetal exposure to the maternal microbiota in humans and mice. JCI Insight. 2019;4(19) doi: 10.1172/jci.insight.127806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satokari R. Bifidobacterium and lactobacillus DNA in the human placenta. Lett Appl Microbiol. 2009;48(1):8–12. doi: 10.1111/j.1472-765X.2008.02475.x. [DOI] [PubMed] [Google Scholar]
- 33.Thum C. Can nutritional modulation of maternal intestinal microbiota influence the development of the infant gastrointestinal tract? J Nutr. 2012;142(11):1921–1928. doi: 10.3945/jn.112.166231. [DOI] [PubMed] [Google Scholar]
- 34.Perez P.F. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119(3):E724–E732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 35.Sahakian A.B., Jee S.-.R., Pimentel M. Methane and the gastrointestinal tract. Dig Dis Sci. 2010;55(8):2135–2143. doi: 10.1007/s10620-009-1012-0. [DOI] [PubMed] [Google Scholar]
- 36.Mathur R. Methane and hydrogen positivity on breath test is associated with greater body mass index and body fat. J Clin Endocrinol Metab. 2013;98(4):E698–E702. doi: 10.1210/jc.2012-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.