Abstract
Background
Numerous studies have shown that cell-free DNA (cfDNA) levels may serve as a non-invasive biomarker of a broad spectrum of acute and chronic pathologies. However, in order to make clinical decisions based on cfDNA measurements, it is essential to understand the magnitude of biological variation so this variation is not confused with a variation that actually represent a clinically relevant change. The present study was designed to evaluate the biological variation of cfDNA in healthy subjects and lung cancer patients.
Methods
Plasma samples were collected from 33 healthy subjects and ten lung cancer patients over three days, as well as during the same day. CfDNA was quantified using droplet digital PCR. Biological variation data was estimated using mixed models.
Findings
The within-subject variation was 25% and the between-subject variation was 30%. The reference change value for the healthy subjects was 70%. There was no systematic difference in cfDNA levels from day-to-day (p = 0⋅61), but there was a significant decline during the day (p<0⋅01). The within-subject variation in cancer patients was comparable to healthy subjects, whereas the between-subject variation was much larger (139%). No systematic differences from day-to-day were observed for the cancer patients (p>0⋅3).
Interpretation
Our findings show that cfDNA levels fluctuate significantly during the day and exhibit considerable within-subject variation. Thus, the data presented offer a substantial contribution to the interpretation of the clinical significance of cfDNA.
Funding
Læge Sofus Carl Emil Friis og hustru Olga Doris Friis' Legat, Harboefonden, and Dagmar Marshalls Fond.
Keywords: CfDNA, Biological variation, Biomarker, Monitoring, Droplet digital PCR, Cancer
Introduction
Circulating cell-free DNA (cfDNA) are fragments of DNA found in the circulation. Exactly how cfDNA is released into the blood remains unknown, but apoptosis, necrosis, and active secretion all represent possible mechanisms.1 Elevated levels of cfDNA can be found in various pathological conditions such as cancer2, acute and chronic inflammatory diseases3,4, trauma5,6, and cardiovascular disease7,8, as well as following physical exercise.9,10 Furthermore, high sensitivity techniques allow detection of low-abundant DNA fractions from the total pool of cfDNA, e.g. tumour-derived DNA fragments in cancer patients11, foetal DNA in pregnant women12 and donor-derived DNA in transplant recipients.13 As cfDNA levels have been found to significantly correlate with several clinical parameters, it has been extensively studied as a potential diagnostic, prognostic, and predictive biomarker for numerous diseases. Longitudinal monitoring has similarly attracted much attention, owing to the minimal invasiveness of sample collection and short half-life of cfDNA.
Research in context.
Evidence before this study
Circulating cell-free DNA (cfDNA) is a promising biomarker for numerous clinical conditions such as cancer, infections, and inflammatory diseases. Several studies have proposed a role for cfDNA in the clinic, showing that cfDNA levels are associated with clinical indications. Yet, despite intensive research into the standardization of preanalytical and analytical approaches in recent years, the biological aspects of cfDNA are still largely unknown. One important issue to consider is the inherent fluctuations of cfDNA. Without knowledge of the magnitude of these, the risk of wrongly interpreting cfDNA level changes is great. Thus, before clinical decisions can be based on cfDNA measurements, it is of great significance to understand the magnitude of between- and within-subject variation of cfDNA levels.
Added value of this study
In the present paper, we examine the biological variation of circulating cell-free DNA in both healthy subjects (N = 33) and lung cancer patients (N = 10). We demonstrate that the within-subject biological variation is considerable, which warrants caution when interpreting results based on cfDNA measurements. Interestingly, we further found that the that the cfDNA level systematically decreased during the day and that a change in cfDNA level between two samples from the same individual had to exceed 70% to be significant.
Implications of all the available evidence
The estimated biological variation of cfDNA can guide clinicians to determine if a found cfDNA level change can be attributed to biological variation or actually represent a clinically relevant status. This is an important step towards a more comprehensive understanding of cfDNA fluctuations, which will lay the groundwork for the adaption of cfDNA as a reliable and unambiguous clinical biomarker.
Alt-text: Unlabelled box
Despite the great potential of cfDNA as a biomarker, analysis of cfDNA has yet to be implemented into clinical practice. There have been reported many conflicting and ambiguous findings and, in particular in cancer, it is widely debated whether cfDNA levels actually are correlated to disease status.14 These conflicting findings could be attributed to inadequate preanalytical standardization and the fundamental lack of knowledge concerning both cfDNAs origin and function. Thus, it is paramount to understand all the analytical and biological aspects of cfDNA prior to using it in clinical practice.
Although considerable research has been devoted to the need for standardization of preanalytical approaches in recent years[15], [16], [17], [18], less attention has been paid to the natural dynamics of cfDNA. In order to base clinical decisions on cfDNA measurements, it is essential to understand the magnitude of between- and within-subject variation and whether cfDNA level changes can be attributed to biological variation or actually represent a clinically relevant status. This is of utmost importance for both longitudinal monitoring of disease, as well as for distinguishing between pathological conditions. Nevertheless, studies have yet to investigate the fluctuations of cfDNA levels resulting from inherent biological variation in healthy subjects.19
The present study was designed to evaluate the magnitude of biological variation of cfDNA in a cohort of healthy individuals, as well as in a cohort of lung cancer patients. This will help clarify how to interpret cfDNA analyses and thus assist the translation of these analyses into clinical practice.
Materials and methods
Subjects
Healthy individuals were recruited from June 2018 until October 2018 at Aarhus University Hospital, Denmark. The subjects were eligible for enrolment if the following criteria were fulfilled: age ≥ 18 years, non-smoker, non-pregnant, no medication, and no chronic infection or inflammation. Subjects were excluded if they had crossed more than one time zone, had worked nightshifts, or had acute infections one week prior to the study. The study was conducted according to the Helsinki Declaration and approved by the Central Denmark Region Committees on Biomedical Research Ethics (1–10–72–452–17). Subjects provided written informed consent before inclusion. Lung cancer patients were recruited from May 2016 until June 2018 at the Department of Oncology, Aarhus University Hospital, Denmark as described in.20 The following inclusion criteria were met: advanced-stage lung cancer, age ≥ 18 years, response or stable disease by the response evaluation criteria in solid tumours (RESIST) criteria21 on the latest computed tomography scan, and no current anti-cancer treatment. Patients were excluded if they had active cancer other than lung cancer or an infection. The study was conducted according to the Helsinki Declaration and approved by the Central Denmark Region Committees on Biomedical Research Ethics (1–10–72–55–15). Patients provided written informed consent before inclusion.
Study design
Healthy subjects were submitted to blood draws at 9 AM for three days in a row. On the second day, blood samples were drawn every third hour for 12 h (12 noon, 3 PM, 6 PM, and 9 PM). All blood draws were performed by the same four technicians. During the study period, subjects were not allowed to drink alcohol or engage in high-impact exercise defined as exercise harder than walking or low intensity bicycling. Furthermore, subjects had to refrain from physical activity and food an hour prior to each blood draw. The study was designed with the checklists for biological variation studies in mind.22,23 For the lung cancer cohort, each patient had five blood samples drawn; two samples were collected one hour apart on the first two days and one sample was collected at the first time point on the third day. Samples were taken at the same time of the day for the individual patient, but the time points varied between patients. The patients were instructed to minimize physical activity before blood draws.
Blood sample collection and processing
Whole blood was collected in 10 ml EDTA tubes (BD Vacutainer) and processed immediately. After 30 min of incubation, the samples were centrifuged for 10 min at 1800 g. The supernatant plasma fractions were carefully transferred to new tubes and particular attention was devoted to not disturb the buffy coat layer. The plasma was subjected to a second centrifugation for 10 min at 13,000 g, aliquoted into new tubes, and stored at −80 °C within two hours of blood draw. All samples were processed by the same four analysts.
DNA extraction
CfDNA was extracted from 4 ml plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer's protocol. The isolated DNA was eluted in 100 µL elution buffer and stored at −80 °C until analysis. To minimize analytical variance, DNA extraction was performed on all samples from each individual at the same time and within two weeks of blood draw by a single analyst. We have previously estimated the cfDNA extraction to have a CV% of 3⋅9.
Droplet digital PCR
cfDNA was quantified using droplet digital PCR (ddPCR) targeting four distinct genes (B2M, EIF2C1, RNaseP, TERT). The ddPCR reaction volume was 22 µL consisting of 13 µL master mix (900 nM primers, 250 nM probe, 2x Supermix for probes (no UTP) (Bio-Rad laboratories)) and 9 µL cfDNA. The cycling conditions for the ddPCR were as follows: 95 °C for 10 min, 40 cycles of 94 °C for 30 s, and 60 °C for 1 min, and 98 °C for 10 min. Blood samples collected from ten anonymous blood donors at the Blood Bank at Aarhus University Hospital were subjected to cfDNA extraction as described above and cfDNA was pooled. This cfDNA pool was used as a positive control for each analysis and was used to estimate the intra-run variation (Supplemental Figure 1). Furthermore, a non-template control was also included in each analysis. To test for linearity, a pool of cfDNA from ten donors was concentrated using a ScanSpeed 40 (Labogene), and subjected to seven two-fold dilutions (Supplemental Figure 1). Samples were analysed on a QX200™ AutoDG™ Droplet Digital™ PCR System (Bio-Rad). All assays were purchased from Life Technologies (Supplemental Table 1). Assays were run as multiplex reactions, and tests were performed to ensure the same efficacy of reactions as for singleplex analyses. To minimize analytical variance, all samples from each individual were analysed within a single run, and all analyses were performed by a single analyst. Each sample was analysed in triplicates. Quantification of cfDNA levels of the four selected reference genes was performed on the first 21 healthy subjects. CfDNA levels of the remaining 12 subjects and the ten lung cancer patients were only quantified using the EIF2C1 and TERT assays.
Statistical analysis
Outlier analyses were performed on three levels; analytical, within-subject, and between-subject. Cochran's C test was used for determining analytical and within-subject outliers, and the Dixon-Reed criterion was used for between-subject outliers.24,25 The analytical variation (CVA) was estimated from triplicate results of every specimen according to24. The within-subject variation (CVI) and between-subject variation (CVG) were calculated using linear mixed effects models with day and sample as fixed effects and subject as a random effect. Normality of the residuals was confirmed visually and using Shapiro-Wilks test. Data from the lung cancer patients followed a log-normal distribution and was thus transformed accordingly. Data is presented as median and range. Pair-wise comparisons of day-to-day and semidiurnal mean values were adjusted for multiple comparisons with the Bonferroni correction. Reference change values (RCVs), index of individuality (II), and number of samples that are required to estimate a subject's homeostatic set point (n) within ± 15% with 80% confidence were calculated using the following equations: RCV=√2 • Z • √(CVI2 +CVA2); II=√(CVI2 +CVA2)/CVG; n=(z • √(CVI2 +CVA2)/D)2, where z is the z-score and D is the desired percentage closeness to the homeostatic set point according to24. For ln-transformed data, RCV was calculated as RCV = exp(± z • √2 • σ) – 1, where σ = √ln (CVI+A2+1).26 CfDNA levels in healthy subjects and cancer patients were compared using Welch's t-test on ln-transformed data. P<0⋅05 was considered statistically significant. All statistical calculations were performed in STATA 14 (StataCorp).
Results
Description of the healthy subjects
A total of 186 blood samples were collected from 33 healthy individuals during the study. Of these, 23 were women and the median age of all individuals was 45 years (range 22–66). Fifteen participants were subjected to blood draws on all seven time points, thirteen participants were subjected to all but the 6 PM and 9 PM blood draws, while five participants only participated in the three 9 AM blood draws. Outliers were identified on three levels for all four genes quantified (Supplemental Table 2). Identification of outliers on the within-subject level disqualified all samples from one participant for B2M quantification, while another participant was excluded for all four genes.
Components of variation in healthy subjects
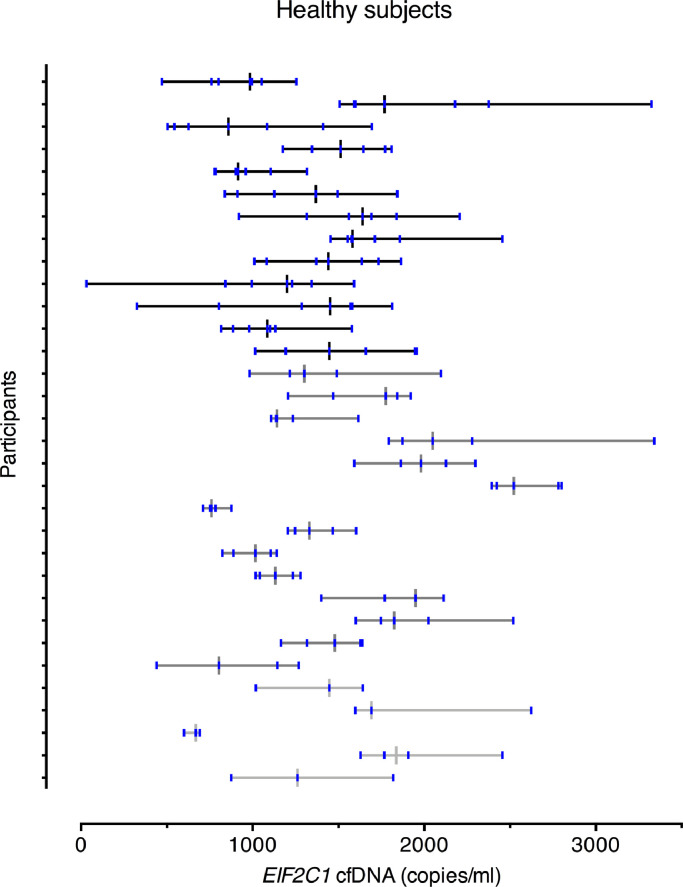
All four assays demonstrated very similar cfDNA levels and CVs, so the data presented is estimated with the EIF2C1 assay, which had the lowest CVA. Median values, ranges, and variance components for all assays are reported in Table 1. The median value of cfDNA was 1399⋅62 copies/ml plasma. Fig. 1 shows the median and range of cfDNA levels for all subjects individually. The CVA was generally low for all assays (6⋅4% for EIF2C1) and well below the recommended analytical goal of CVA ≤ 0⋅5 CVI.24 For all assays, both the CVI and the CVG (23⋅9% and 29⋅6% for EIF2C1) were of similar magnitude, and the CVI was lower than the CVG. The latter was reflected in the II, which was below 1 for all four assays. The RCV was approximately 70% for all assays, reflecting that a change in two serial measurements must exceed 70% to be classified as a significant change at the 95% confidence level. For all assays, approximately 4 samples would be needed to provide an estimate of the homeostatic setting point within ± 15% with 80% confidence.
Table 1.
Components of biological variation in healthy subjects.
| B2M | EIF2C1 | RNaseP | TERT | |
|---|---|---|---|---|
| Number of samplesa | 114 | 177 | 121 | 177 |
| Median (copies/ml) | 1434⋅11 | 1399⋅62 | 1056⋅03 | 1532⋅84 |
| Range (copies/ml) | 42⋅57–2509⋅30 | 30⋅60–3340⋅73 | 22⋅34–2194⋅19 | 34⋅23–3481⋅28 |
| CVI (%) | 23⋅6 | 23⋅9 | 25⋅4 | 23⋅3 |
| CVG (%) | 25⋅5 | 29⋅6 | 27⋅1 | 30⋅0 |
| CVA (%) | 6⋅6 | 6⋅4 | 8⋅4 | 6⋅6 |
| IIb | 0⋅96 | 0⋅84 | 0⋅99 | 0⋅81 |
| RCVc (%) | 67⋅9 | 68⋅5 | 74⋅1 | 67⋅1 |
| nd | 4 | 4 | 5 | 4 |
Number of samples analysed after exclusion of outliers;.
II, Index of individuality;.
RCV, reference change value at 95% significance;.
n, number of samples required to estimate homeostatic set point. Abbreviations: CVI, within-subject coefficient of variation; CVG, between-subject coefficient of variation; CVA, analytical coefficient of variation.
Fig. 1.
EIF2C1 cfDNA levels in healthy subjects after exclusion of outliers. Subjects are color-coded according to the number of samples they contributed with: black, 7 samples; dark grey, 5 samples; light grey, 3 samples and the black/grey vertical line illustrates the median value. The vertical blue lines indicate each cfDNA level measured in the patient. The horizontal line illustrates the range from minimum to maximum. 1000 copies/ml correspond to 3⋅3 ng/ml.
Day-to-day and semidiurnal variance in healthy subjects
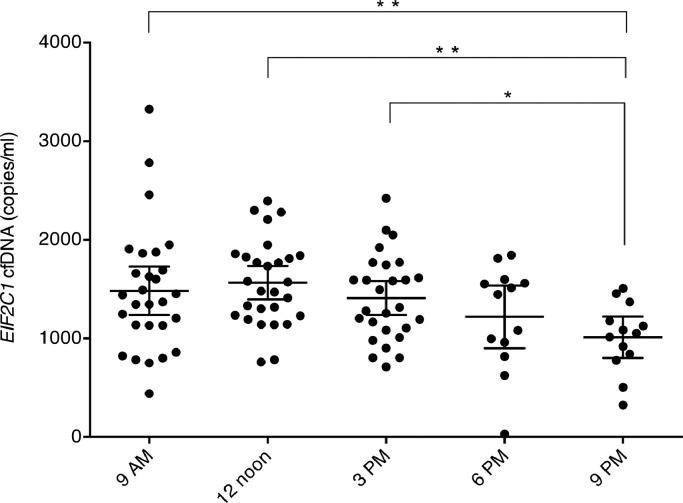
Day-to-day variance components were obtained for the 9 AM measurements on each of the three days. There was no significant difference in cfDNA levels between the three days (p = 0⋅61; Wald test). Semidiurnal variance components were obtained for all the measurements on day 2 (9 AM, 12 noon, 3 PM, 6 PM, 9 PM). There was a significant difference in cfDNA levels during the day (p<0⋅01; Wald test) with the level decreasing throughout the day (Fig. 2). The day-to-day and semidiurnal variance components were comparable to the total and are shown in Table 2.
Fig. 2.
Within-day EIF2C1 cfDNA levels on day 2. Individual results are shown with mean (longest horizontal line) and 95% confidence intervals (the smallest horizontal lines). Significant differences from a pairwise comparison are marked with *p<0⋅05 and **p<0⋅005.
Table 2.
Day-to-day and within-day components of biological variation in healthy subjects.
| B2M | EIF2C1 | RNaseP | TERT | |
|---|---|---|---|---|
| Day-to-day | ||||
| Day 1 (copies/ml) | 1574⋅86 | 1589⋅82 | 1113⋅81 | 1746⋅23 |
| Day 2 (copies/ml) | 1528⋅13 | 1404⋅82 | 1038⋅50 | 1586⋅47 |
| Day 3 (copies/ml) | 1407⋅81 | 1433⋅51 | 1134⋅37 | 1581⋅10 |
| CVI (%) | 20⋅9 | 23⋅0 | 23⋅5 | 21⋅8 |
| CVG (%) | 29⋅6 | 33⋅7 | 32⋅2 | 33⋅5 |
| Within-day | ||||
| 12 noon (copies/ml) | 1865⋅89 | 1524⋅45 | 1344⋅41 | 1712⋅91 |
| 03 PM (copies/ml) | 1440⋅16 | 1314⋅36 | 1059⋅58 | 1621⋅22 |
| 06 PM (copies/ml) | 1371⋅18 | 1446⋅10 | 1018⋅73 | 1532⋅84 |
| 09 PM (copies/ml) | 1159⋅03 | 1052⋅96 | 799⋅70 | 1100⋅44 |
| CVI (%) | 22⋅9 | 24⋅0 | 24⋅3 | 27⋅3 |
| CVG (%) | 23⋅1 | 26⋅5 | 25⋅0 | 27⋅6 |
Day-to-day and within-day cfDNA levels are presented as medians. Abbreviations: CVI, Within-subject coefficient of variation; CVG, Between-subject coefficient of variation.
Components of variation in lung cancer patients
EIF2C1 and TERT cfDNA levels were also measured in a total of 50 blood samples from ten lung cancer patients with stable disease. The median age was 66⋅5 years (range 58–77) and six patients were women. One patient was identified as an outlier on the within-subject level, and all measurements for this patient were thus excluded (Supplemental Table 2).
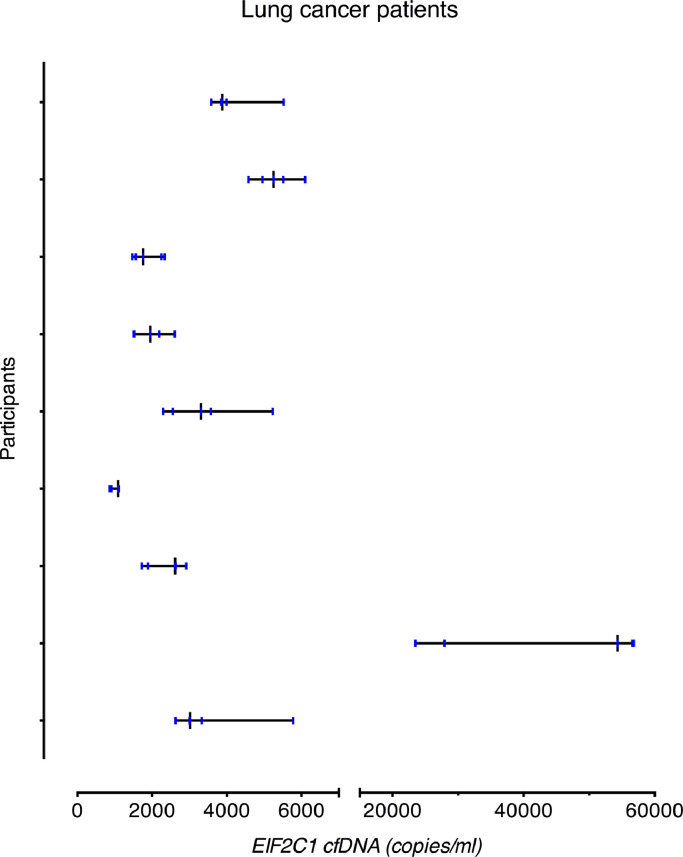
The cfDNA level was significantly higher in the lung cancer cohort compared to the healthy cohort (p = 0⋅026; Welch's t-test). Fig. 3 shows the median and range of cfDNA levels for all patients individually. The CVA was comparable to that for the healthy subjects (4⋅7% for EIF2C1). For both assays, the CVI was in the same range as for the healthy subjects, whereas the CVG was much larger (23⋅8% and 139⋅3%, respectively, for EIF2C1). The II values were below 0⋅2, meaning that the cfDNA showed very high individuality. Moreover, cfDNA levels had to increase approximately 95% between two measurements to be considered significant. Variance components for both assays are reported in Table 3.
Fig. 3.
EIF2C1 cfDNA levels in lung cancer patients after exclusion of outliers. All subjects contributed with 5 samples. The black vertical line illustrates the median value and the vertical blue lines indicate each cfDNA level measured in the patient. The horizontal line illustrates the range from minimum to maximum. 1000 copies/ml correspond to 3⋅3 ng/ml.
Table 3.
Components of biological variation in lung cancer patients.
| EIF2C1 | TERT | |
|---|---|---|
| Number of samplesa | 45 | 45 |
| Median (copies/ml) | 2918⋅78 | 3226⋅91 |
| Range (copies/ml) | 863⋅19–56,810⋅07 | 950⋅84–64,512⋅68 |
| CVI (%) | 23⋅8 | 22⋅7 |
| CVG (%) | 139⋅3 | 143⋅8 |
| CVA (%) | 4⋅7 | 5⋅6 |
| IIb | 0⋅17 | 0⋅16 |
| RCVc (%) | −48⋅4–93⋅9 | −47⋅3–89⋅6 |
| nd | 4 | 4 |
Number of samples analysed after exclusion of outliers.
II, Index of individuality.
RCV, outer limits for reference change values at 95% significance.
n, number of samples required to estimate homeostatic set point. Abbreviations: CVI, within-subject coefficient of variation; CVG, between-subject coefficient of variation; CVA, analytical coefficient of variation.
Day-to-day variance components were obtained for the first measurement on each of the three days (Table 4). There was no significant difference in cfDNA levels between the three days (p = 0⋅90; Wald test). Hour-to-hour variance components were obtained for the two time points on day 1 and 2 (Table 4). There was no significant difference in the cfDNA levels from hour-to-hour (p = 0⋅35; Wald test).
Table 4.
Day-to-day and hour-to-hour components of biological variation in lung cancer patients.
| EIF2C1 | TERT | |
|---|---|---|
| Day-to-day | ||
| Day 1 (copies/ml) | 3883⋅50 | 4301⋅14 |
| Day 2 (copies/ml) | 3310⋅75 | 3615⋅40 |
| Day 3 (copies/ml) | 2629⋅59 | 3103⋅89 |
| CVI (%) | 26⋅4 | 25⋅9 |
| CVG (%) | 141⋅2 | 143⋅8 |
| Hour-to-hour | ||
| Baseline (copies/ml) | 3320⋅79 | 3616⋅34 |
| After 1 h (copies/ml) | 2590⋅16 | 2903⋅50 |
| CVI (%) | 24⋅6 | 24⋅6 |
| CVG (%) | 137⋅1 | 141⋅3 |
Day-to-day and hour-to-hour cfDNA levels are presented as medians. Abbreviations: CVI, Within-subject coefficient of variation; CVG, Between-subject coefficient of variation.
Sensitivity analysis
The outlier analyses dictated removal of all measurements from a single lung cancer patient (ID2). However, since cfDNA levels may be influenced by the disease, it can be difficult to assess whether this patient represents a true statistical outlier or whether the naturally occurring variation is elevated due to possible aberrant cfDNA dynamics in cancer patients. If the latter is the case, excluding this patient will potentially underestimate the magnitude of biological variation in the cancer cohort. To determine the effect of removing the outlier, a secondary sensitivity analysis was conducted that included all ten cancer patients (Supplemental Table 3). The CVI was twice as large when the outlier was included (23⋅8% in the original analysis vs. 46⋅2%), meaning cfDNA levels had to increase approximately 250% between two measurements to be considered significant. Furthermore, there was still no significant difference in cfDNA levels between days (p = 0⋅75; Wald test), but a significant difference was found from hour-to-hour (p = 0⋅02; Wald test) with the level decreasing 18% from the first to the second measurement.
Discussion
Here, we report the magnitude of biological variation of cfDNA levels in a cohort of healthy subjects and lung cancer patients. We show that the variation was considerable on both the within-subject and between-subject levels. There were no systematic changes in the level of cfDNA from day-to-day; however, cfDNA levels significantly declined during the day. In addition, we found that the between-subject variation was notably higher in lung cancer patients than in healthy subjects. To our knowledge, no other reported studies have been designed specifically to estimate the biological variation of cfDNA.
CfDNA is a putative biomarker for diagnosis and monitoring of disease severity in numerous pathologies. However, caution must be taken when interpreting cfDNA analyses without knowledge of the inherent biological variation of cfDNA levels and it is thus of great clinical interest to estimate. We found a CVG of up to 30% and a CVI of up to 25%. The II reflected that cfDNA showed marked individuality, meaning that comparison of measurements to a conventional reference interval will be of limited use. A RCV of approximately 70% suggested that the difference between two serial cfDNA measurements must vary more than 70% to represent a significant change that exceeds the natural variation. This is of utmost importance to consider when interpreting serial results. Many studies have investigated the use of cfDNA levels for longitudinal monitoring. One study reported that the changes in cfDNA levels over five days were significantly different for acute myocardial infarction patients with complications than for those without (maximum change from baseline of 1⋅49 vs. 1.1).8 Another study found that a cfDNA increase over the first 48 h in patients with shock could predict a fatal outcome within 28 days.27 The best cut-off value for a cfDNA change was an increase of 16% (sensitivity 69%, specificity 90%). This cut-off does not exceed the RCV of 70% found in our study. These are a few of many studies that do not take the biological variation into consideration when interpreting cfDNA level changes.
We did not find the cfDNA levels from day-to-day to vary systematically. This is in accordance with Xie et al., who measured cfDNA levels in 30 healthy subjects over five days and five months and reported that the mean levels varied insignificantly.8 However, they did not estimate the magnitude of biological variation. Interestingly, we found that the level of cfDNA declined significantly during the day. The reason for this phenomenon is unknown as knowledge about the biology of cfDNA is sparse. However, it could be speculated that this is a trend coupled with other biological processes. Sennels et al28 have shown that hematological parameters show significant diurnal rythms with some parameters declining throughout the day and others increasing. Since cfDNA mostly originates from blood cells in healthy subjects, some connection to the amount of blood cells is to be expected. Although most blood samples are collected during the day, the decline in cfDNA in the evening suggests that the exact time of blood draw affects the cfDNA level and that comparing results from samples collected at different time points may be biased in either direction. These results warrant that cfDNA samples that are to be longitudinally compared have to be taken at approximately similar time points during the day. Additionally, we found that up to five samples should be analysed to estimate a subject's homeostatic setting point within 15% with 80% confidence, indicating that multiple samples should be obtained before basing any clinical decision on the results of cfDNA analyses.
In some areas of research, the cfDNA of interest, such as tumour-derived cfDNA in cancer patients and donor-derived cfDNA in transplantation patients, is normalized to total cfDNA, yielding a ratio or frequency. We have previously shown that tumor-derived and total cfDNA levels in cancer patients do not necessarily display comparable dynamics20, a phenomenon also seen with foetal and maternal cfDNA levels.29,30 In contrast, Moreira et al. found that donor-derived and total cfDNA levels had similar dynamics in kidney-transplant patients that experienced acute rejection.31 In the present study, we find that the day-to-day biological variation of total cfDNA levels is not only random, but also of a considerable magnitude. This will significantly affect ratios and frequencies and extreme caution must therefore be taken when interpreting these in longitudinal monitoring. The fact that the fluctuations of the cfDNA of interest and total cfDNA may not be synchronized further stresses that ratios will be highly impacted by the total cfDNA variation.
The cfDNA level was higher in the cancer patients compared to healthy subjects, as has been demonstrated previously.2 Nonetheless, we demonstrated that the CVI for the cancer patients was approximately of the same magnitude as for the healthy subjects. However, it should be noted that including an individual, which was identified as an outlier on the within-subject level led to a two-fold increase in the CVI. This was caused by the fact that the cfDNA levels in this patient decreased 5-fold between the two measurements on day 1, while it increased 13-fold on day 2. Considering that these changes occur within the span of one hour, it may be suspected that the particular patient had disregarded the instruction to rest between sampling. CfDNA levels are known to increase during exercise and up to 8-fold rises have been reported following moderate aerobic running.10 In agreement with our primary analysis, a vast number of analytes have been demonstrated to display comparable biological variation in healthy and diseased subjects.19 This supports the applicability of cfDNA CVs and RCVs estimated from healthy subjects to cancer patients and possibly patients with other chronic illnesses as well. However, as we cannot conclude with absolute confidence that the outlying subject has violated the study instructions, we report a CVI range in the cancer cohort of 24–46%.
Since standardization of which reference genes to use for quantification of cfDNA is still non-existing, we measured four different genes. We found that the variation components were comparable for the four genes, even though the mean values differed. These differences in mean level are most likely owing to the chromosomal position of the genes, as the most telomeric genes (TERT, EIF2C1) had the highest mean values.15
Despite its strengths, there are some limitations to our study, which should be considered. Our study has an overrepresentation of female subjects; yet, we saw no significant difference in cfDNA levels between the sexes (data not shown), and we expect the variation of cfDNA to be the same in male and female subjects. Furthermore, various studies have been unable to demonstrate a correlation between cfDNA level and gender.8,[31], [32], [33] Another limitation is the short duration of the study period, which might underestimate the within-subject variation. Rotterdam et al. showed that serial samples for total and HDL-cholesterol taken less than four days apart resulted in a smaller CVI than samples taken farther apart.34 However, our estimations of the CVI and the RCV are comparable to values estimated for donor-derived cfDNA levels in stable renal transplant recipients measured monthly for up to one year.35 Lastly, the observed decline in cfDNA levels during the day could be linked to higher physical activity of the participants during the day compared with the evening. However, all participants performed only sedentary tasks during the day, and thus activity was the same throughout the day and evening. We therefore believe that our finding reflect the biological variation of cfDNA.
Based on the results of the current study, our recommendation is that blood samples for the assessment of cfDNA levels in a monitoring setting should be taken at least a day apart and, if possible, collected at approximately the same time of day. CfDNA might not be suitable as a biomarker in pathologies where short-term, e.g. hour-to-hour, measurements are needed or at least clinicians need to pay very close attention to the declining trend in cfDNA levels in the evening. Moreover, caution must be exhibited when drawing conclusions based on a single cfDNA measurement. A study investigating the variation over weeks or even months would be of great relevance to assess if the magnitude of long-term biological variation is similar and whether cfDNA is a suitable biomarker for long-term monitoring of chronic pathologies. This study is the first step towards a more comprehensive understanding of cfDNA fluctuations, which will lay the groundwork for the adaption of cfDNA as a reliable and unambiguous clinical biomarker.
Funding sources
The work was supported by Læge Sofus Carl Emil Friis og hustru Olga Doris Friis' Legat, Harboefonden, and Dagmar Marshalls Fond. The funders did not have any role in the study design, data collection, data analysis, interpretation, or writing of the report.
Author contributions
ATM, JAH, and AWL collected data. ATM and AWL conceived study, performed statistical data analysis, and data interpretation. ATM performed experiments, analysed data, and drafted the manuscript. BSS and AWL supervised experiments. All authors reviewed and revised the manuscript.
Declaration of Competing Interest
The authors have no conflicts of interest to declare
Acknowledgements
The authors wish to thank the biomedical laboratory technicians at the research department of the Department of Clinical Biochemistry, Aarhus University Hospital. Additionally, they thank the Biostatistical Advisory Service at Aarhus University for providing statistical help.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.008.
Appendix. Supplementary materials
References
- 1.Aucamp J., Bronkhorst A.J., Badenhorst C.P S, Pretorius P.J. The diverse origins of circulating cell-free dna in the human body: a critical re-evaluation of the literature. Biol Rev. 2018;93:1649–1683. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 2.Sozzi G., Conte D., Leon M.E. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol. 2003;21:3902–3908. doi: 10.1200/JCO.2003.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto T., Yoshida K., Hashimoto N. Circulating cell free DNA: a marker to predict the therapeutic response for biological DMARDs in rheumatoid arthritis. Int J Rheum Dis. 2017;20:722–730. doi: 10.1111/1756-185X.12959. [DOI] [PubMed] [Google Scholar]
- 4.Hou Y.Q., Liang D.Y., Lou X.L., Zhang M., Zhang Z.H., Zhang L.R. Branched DNA-based Alu quantitative assay for cell-free plasma DNA levels in patients with sepsis or systemic inflammatory response syndrome. J Crit Care. 2016;31:90–95. doi: 10.1016/j.jcrc.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Lam N.Y.L, Rainer T.H., Chan L.Y.S, Joynt G.M., Lo Y.M.D. Time course of early and late changes in plasma DNA in trauma patients. Clin Chem. 2003;49:1286–1291. doi: 10.1373/49.8.1286. [DOI] [PubMed] [Google Scholar]
- 6.Brodbeck K., Kern S., Schick S. Quantitative analysis of individual cell-free DNA concentration before and after penetrating trauma. Int J Legal Med. 2019;133:385–393. doi: 10.1007/s00414-018-1945-y. [DOI] [PubMed] [Google Scholar]
- 7.Lou X., Hou Y., Liang D. A novel Alu-based real-time PCR method for the quantitative detection of plasma circulating cell-free DNA: sensitivity and specificity for the diagnosis of myocardial infarction. Int J Mol Med. 2015;35:72–80. doi: 10.3892/ijmm.2014.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie J., Yang J., Hu P. Correlations of circulating cell-free DNA with clinical manifestations in acute myocardial infarction. Am J Med Sci. 2018;356:121–129. doi: 10.1016/j.amjms.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Atamaniuk J., Vidotto C., Tschan H., Bachl N., Stuhlmeier K.M., Müller M.M. Increased concentrations of cell-free plasma DNA after exhaustive exercise. Clin Chem. 2004;50:1668–1670. doi: 10.1373/clinchem.2004.034553. [DOI] [PubMed] [Google Scholar]
- 10.Haller N., Tug S., Breitbach S., Jörgensen A., Simon P. Increases in circulating cell-free DNA during aerobic running depend on intensity and duration. Int J Sports Physiol Perform. 2017;12:455–462. doi: 10.1123/ijspp.2015-0540. [DOI] [PubMed] [Google Scholar]
- 11.Bettegowda C., Sausen M., Leary R.J. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo Y.M.D, Tein M.S.C, Lau T.K. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo Y.M.D, Tein M.S.C, Pang C.C.P. Presence of donor-specific DNA in plasma of kidney and liver-transplant recipients. Lancet. 1998;351:1329–1330. doi: 10.1016/s0140-6736(05)79055-3. [DOI] [PubMed] [Google Scholar]
- 14.van der Vaart M., Pretorius P.J. Is the role of circulating DNA as a biomarker of cancer being prematurely overrated. Clin Biochem. 2010;43:26–36. doi: 10.1016/j.clinbiochem.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Devonshire A.S., Whale A.S., Gutteridge A. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406:6499–6512. doi: 10.1007/s00216-014-7835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Ginkel J.H., van den Broek D.A., van Kuik J. Preanalytical blood sample workup for cell-free DNA analysis using Droplet Digital PCR for future molecular cancer diagnostics. Cancer Med. 2017;6:2297–2307. doi: 10.1002/cam4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Messaoudi S., Rolet F., Mouliere F., Thierry A.R. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta. 2013;424:222–230. doi: 10.1016/j.cca.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Meddeb R., Pisareva E., Thierry A.R. Guidelines for the preanalytical conditions for analyzing circulating cell-free DNA. Clin Chem. 2019;65:623–633. doi: 10.1373/clinchem.2018.298323. [DOI] [PubMed] [Google Scholar]
- 19.Ricós C., Iglesias N., García-Lario J.-.V. Within-subject biological variation in disease: collated data and clinical consequences. Ann Clin Biochem. 2007;44:343–352. doi: 10.1258/000456307780945633. [DOI] [PubMed] [Google Scholar]
- 20.Hojbjerg J.A., Madsen A.T., Schmidt H.H. Intra‐individual variation of circulating tumour DNA in lung cancer patients. Mol Oncol. 2019:1–9. doi: 10.1002/1878-0261.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer E.A., Therasse P., Bogaerts J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett W.A., Braga F., Carobene A. A checklist for critical appraisal of studies of biological variation. Clin Chem Lab Med. 2015;53:879–885. doi: 10.1515/cclm-2014-1127. [DOI] [PubMed] [Google Scholar]
- 23.Aarsand A.K., Røraas T., Fernandez-Calle P. The biological variation data critical appraisal checklist: a standard for evaluating studies on biological variation. Clin Chem. 2018;64:501–514. doi: 10.1373/clinchem.2017.281808. [DOI] [PubMed] [Google Scholar]
- 24.Fraser G.G., Harris E.K. Generation and application of data on biological variation in clinical chemistry. Crit Rev Clin Lab Sci. 1989;27:409–437. doi: 10.3109/10408368909106595. [DOI] [PubMed] [Google Scholar]
- 25.Braga F., Panteghini M. Generation of data on within-subject biological variation in laboratory medicine: an update. Crit Rev Clin Lab Sci. 2016;53:313–325. doi: 10.3109/10408363.2016.1150252. [DOI] [PubMed] [Google Scholar]
- 26.Fokkema M.R., Herrmann Z., Muskiet F.A.J, Moecks J. Reference change values for brain natrluretic peptides revisited. Clin Chem. 2006;52:1602–1603. doi: 10.1373/clinchem.2006.069369. [DOI] [PubMed] [Google Scholar]
- 27.Xia D.L., Zhang H., Luo Q.L., Zhang A.F., Zhu L.X. Cell-free DNA increase over first 48 hours in emergency intensive care unit predicts fatal outcome in patients with shock. J Int Med Res. 2016;44:1002–1012. doi: 10.1177/0300060516650785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sennels H.P., Jørgensen H.L., Hansen A.L.S, Goetze J.P., Fahrenkrug J. Diurnal variation of hematology parameters in healthy young males: the Bispebjerg study of diurnal variations. Scand J Clin Lab Invest. 2011;71:532–541. doi: 10.3109/00365513.2011.602422. [DOI] [PubMed] [Google Scholar]
- 29.Zhong X.Y., Bürk M.R., Troeger C., Kang A., Holzgreve W., Hahn S. Fluctuation of maternal and fetal free extracellular circulatory DNA in maternal plasma. Obstet Gynecol. 2000;96:991–996. doi: 10.1016/s0029-7844(00)01065-6. [DOI] [PubMed] [Google Scholar]
- 30.Chiu R.W.K, Poon L.L.M, Lau T.K., Leung T.N., Wong E.M.C, Lo Y.M.D. Effects of blood-processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem. 2001;47:1607–1613. [PubMed] [Google Scholar]
- 31.Moreira V.G., García B.P., Martín J.M.B, Suárez F.O., Alvarez F V. Cell-free DNA as a noninvasive acute rejection marker in renal transplantation. Clin Chem. 2009;55:1958–1966. doi: 10.1373/clinchem.2009.129072. [DOI] [PubMed] [Google Scholar]
- 32.Tamkovich S.N., Bryzgunova O.E., Rykova E.Y., Permyakova V.I., Vlassov V V., Laktionov P.P. Circulating nucleic acids in blood of healthy male and female donors. Clin Chem. 2005;51:1317–1319. doi: 10.1373/clinchem.2004.045062. [DOI] [PubMed] [Google Scholar]
- 33.Beiter T., Fragasso A., Hudemann J., Nieß A.M., Simon P. Short-term treadmill running as a model for studying cell-free DNA kinetics in vivo. Clin Chem. 2011;57:633–636. doi: 10.1373/clinchem.2010.158030. [DOI] [PubMed] [Google Scholar]
- 34.Rotterdam E.P., Katan M.B., Knuiman J.T. Importance of time interval between repeated measurements of total or high-density lipoprotein cholesterol when estimating an individual's baseline concentrations. Clin Chem. 1987;33:1913–1915. [PubMed] [Google Scholar]
- 35.Bromberg J.S., Brennan D.C., Poggio E. Biological variation of donor-derived cell-free DNA in renal transplant recipients: clinical implications. J Appl Lab Med. 2017;2:309–321. doi: 10.1373/jalm.2016.022731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.