Abstract
Background
Understanding how cells respond to mitotic poisons is of great biomedical and clinical significance. However, it remains unknown how cell-death or survival is determined during exposure to anti-mitotic drugs.
Methods
The biological effects of SLC39A6 (LIV-1) and GrpE-like 1 (GRPEL1) on mitotic exit and apoptosis were evaluated both in vitro and in vivo using flow cytometry, western blotting, xenografts and time-lapse imaging. The interactions between proteins and the ubiquitination of GRPEL1 were assessed by GST pull down, immunoprecipitation and mass spectrometry analysis. The expression of LIV-1 in cancers was assessed by immunohistochemistry.
Findings
Overexpression of LIV-1 led to direct apoptosis. Depleted for LIV-1 evade anti-mitotic agent-induced killing through a rapid exit from arrested mitosis. LIV-1 interacts with GRPEL1 and Stabilizes GRPEL1 Protein by Preventing Ubiquitylation of GRPEL1. LIV-1-GRPEL1 axis depletion works to reduce the mitotic arrest by inducing PP2A-B55α phosphates activity, while inhibit apoptosis by banding AIF and preventing the latter's release into the nucleus. Loss of function in this axis was frequent in multiple types of human epithelial cancer.
Interpretation
These data demonstrate that LIV-1-GRPEL1 axis dually regulates mitotic exit as well as apoptosis by interacting with PP2A B55α and AIF. Its discovery constitutes a conceptual advance for the decisive mechanism of cell fate during damaged mitosis.
Fund
National Clinical Research Center for Obstetric and Gynecologic Diseases, the National Natural Science Foundation of China.
Keywords: LIV-1-GRPEL1 axis, Mitotic exit, Apoptosis, Anti-mitotic agents, Human cancer
Research in context section.
Evidence before this study
Understanding how cells respond to mitotic poisons is of great biomedical and clinical significance. Previous studies proposed a model in which cell fate is dictated by two competing but independent networks: one activates cell death and the other is related to the degradation of cyclin B1. However, little is known about how cell death or survival is determined during exposure to anti-mitotic drugs. The key molecules/pathways that are capable of dually wiring these two opposite signaling networks during MC remains unclear and necessitates further exploration.
Added value of this study
We identified that LIV-1 and its downstream target GRPEL1 promote cell death by simultaneously regulating cell-death signaling and mitotic arrest interacting with PP2A B55α and AIF. Loss of the LIV-1-GRPEL1 axis enabled tumour cells to resist mitotic poisons by blocking apoptosis, facilitating slippage, and recapitulating the cell cycle. Loss of function in this axis was frequent in multiple types of human cancer. Our discovery of the LIV-1-GRPEL1 axis provides a new starting point for elucidating how mitotic exit is controlled after exposure to mitotic poisons.
Implications of all the available evidence
In this study, we provide in vitro and in vivo evidence that LIV-1 and its downstream mediator GRPEL1 act as a critical death signal that accumulates during mitotic arrest and is indispensable for anti-mitotic agent-induced cell death. As such, identification of the LIV-1-GRPEL1 axis constitutes a conceptual framework for understanding tumorigenesis and developing a new generation of mitosis-targeting therapies.
Alt-text: Unlabelled box
1. Introduction
To date, one of the most successful anti-cancer strategies has been the use of anti-mitotic drugs to disrupt normal mitotic progression [1]. Drugs such as the taxanes and the vinca alkaloids, which target microtubule dynamics, have successfully been used for the treatment of various human malignancies and have demonstrated outstanding therapeutic efficacy [2], [3]. Moreover, novel anti-mitotic agents that target mitotic kinases and components other than microtubules have been developed [4], [5], [6]; their benefits are currently under investigation in clinical trials [7], [8], [9], [10], [11], [12], [13].
During anti-mitotic drug-induced mitotic checkpoint (MC), some cancer cells can survive and enter a second round of mitosis [14]. Several mechanisms have been proposed to guarantee cancer-cell survival during damaged mitosis [15]. First, these cells may fail to execute apoptosis efficiently due to defects in apoptosis pathways. For example, failure to degrade an anti-apoptosis protein MCL1 during exposure to anti-tubulin chemotherapeutics confers resistance to these agents in some primary tumours [16]. Second, cancer cells may slip out of mitotic arrest before they die, a phenomenon that is commonly termed “slippage” or “adaptation” [17,18]. Gascoigne and Taylor proposed a model in which cell fate is dictated by two competing but independent networks: one activates cell death and the other is related to the degradation of cyclin B1 [14], [19], [20]. During prolonged mitotic arrest, these two networks work in opposite directions. Consistent with this model, premature exit from mitotic arrest due to a weakened MC is known to decrease sensitivity to anti-mitotic agents; blocking of mitotic exit is a more-effective anti-mitotic strategy than perturbing spindle assembly [21].
LIV-1 (SLC39A6) is a member of the Zrt/Irt-like protein family of zinc transporters [22]. In the zebrafish gastrula organiser, LIV-1 regulates the epithelial-mesenchymal transition as a downstream target of STAT3 [23]. Clinically, elevated LIV-1 transcriptional expression is associated with tumour progression in certain tumour types [24]. A recent genome-wide association study on esophageal carcinoma identified common variants in LIV-1 that were associated with survival [25]. These results have provided important clues that point to a potentially important role of LIV-1 in the progression of human cancer.
In this study, we provide in vitro and in vivo evidence that LIV-1 acts as a critical death signal that accumulates during mitotic arrest and is indispensable for anti-mitotic agent-induced cell death. LIV-1 and its downstream mediator GrpE-like 1 (GRPEL1) forms the LIV-1-GRPEL1 axis to adjust cell fates on response to mitotic poisons. As such, identification of the LIV-1-GRPEL1 axis constitutes a conceptual framework for understanding tumorigenesis and developing a novel generation of mitosis-targeting therapies.
2. Materials and methods
2.1. Plasmids and constructs
Pcep4 vector carrying anti-sense LIV-1 cDNA was named AS-LIV-1 as previously described [26]. EGFP/LIV-1 plasmid was generated by ligating the amplified coding region of human LIV-1 cDNA to EGFP in the EGFP-C1 plasmid (Clontech). To generate the inducible expression lentivirus LV/LIV-1, a LIV-1/V5 fusion plasmid was generated by cloning LIV-1 into pcDNA3.1/V5-his-TOPOR, which was subcloned into a lentivirus tetracycline-inducible expression vector. LIV-1 deletion constructs were generated by fusing base pairs 1–141, 1–282, 283–1149, or 283–1299 of the LIV-1 cDNA to EGFP, respectively named aa1-47, aa1-94, aa95-383, aa95-433. Each construct was confirmed by DNA sequencing. The N-terminal 141 bp of the LIV-1 gene was synthesized and cloned into the pGEX-4T1 vector to generate the GST aa1-47 plasmid. The coding region of GRPEL1 was cloned into the EGFP-C1 plasmid to generate the EGFP/GRPEL1. Each construct was confirmed by DNA sequencing. Mito-DsRed and pDsRed-ER were purchased from Addgene. Lentivirus carrying anti-sense LIV-1 shRNA was named LV-sh/LIV-1. Lentivirus carrying GFP-α-tubulin was named GFP-tubulin.
2.2. Antibodies
Rabbit anti-human LIV-1 was obtained from Abcam (ab61307). Rabbit anti-AIF (ab32516), rabbit anti-β-actin (ab8226), rabbit anti-Phospho-histone H3 (H3P) (Ser10, ab5176), rabbit antibody against GFP (ab290), rabbit antibody against ubiquitin (ab137025), rabbit anti-V5 tag (ab9116) and rabbit anti-Ki67 (ab92742) were obtained from Abcam. Mouse anti-cyclin B1 (4135), rabbit anti-Phospho-(Ser) CDKs substrate (9477), and rabbit anti-cleaved PARP (5625) were purchased from Cell Signaling Technology. Mouse anti-B55α (sc-81606) was purchased from Santa. Rabbit anti-GRPEL1 (12720–1-AP), rabbit anti-GST (10000–0-AP), rabbit anti-MCL1 (16225–1-AP), rabbit anti-COX2 (12375–1-AP) and rabbit, rabbit anti-α-tubulin (11224–1-AP), and anti-Hsp90 (13171–1-AP) were purchased from Proteintech. Rabbit anti-cleaved caspase 3 (AC033), and rabbit anti-histone 3 (AH433) were obtained from Beyotime Biotechnolog.
2.3. Cell culture
The OV2008 ovarian cancer cell line was kind gifts from Dr. Rakesh Goel (Ottawa Regional Cancer Center, Ottawa, Canada). HeLa, 293 (HEK-293), A549, and HepG2 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). MCF-7 cells were cultured in DMEM containing 0.01 mg/ml bovine insulin.
The T-REx™-293 cell line (293T) was purchased from Invitrogen. To generate a 293T/LIV-1/V5 stable cell line, 293T cells were transfected with the inducible expression lentivirus LV/LIV-1/V5 and selected in medium containing blasticidin (5 µg/ml) and zeocin (200 µg/ml) for 14 days. The expression of inserted gene could be induced with tetracycline (1 µg/ml). To generate a HeLa/GFP-tubulin stable cell line, HeLa cells were transfected with LV-GFP-tubulin and cultured in medium containing puromycin (600 ng/ml). To generate HeLa/LV-sh/LIV-1 or HeLa/LV-sh/mock stable cell lines, HeLa cells were stably transfected with LV-sh/LIV-1 or LV-sh/mock and cultured in medium containing puromycin (600 ng/ml).
2.4. siRNAs
The siRNAs against LIV-1 are siRNA1 (AUA AAG GAC AGC CUG CUU AAC GGU C) and siRNA2 (AUU AUG GUC AGA GUG AUG CUC GUG G). siRNA against MAD2 (AAG AGU CGG GAC CAC AGU UUA), siRNA against GRPEL1 (UUU CGU GGC UGU GCA CAA CAA CCG G), siRNA against AIF (AUA GCA UUG GGC AUC ACC UUA ACC C), siRNA against B55α (GCA GAU GAU UUG CGG AUU ATT) and a negative universal siRNA duplex were all obtained from Invitrogen. In general, siRNA transfection was performed at a final concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. The sequence of anti-sense LIV-1 shRNA was used as following: ACA GGA AGT CTA CAA TGA A.
2.5. Real-time PCR
Real-time PCR was performed using the SYBR Green real-time PCR method. Each sample was tested in triplicate, and relative gene expression was determined using the 2−∆∆CT method. Results are expressed as fold induction compared with the untreated group. Real-time PCR was performed with the CFX96™ Real-Time PCR Detection System (Bio-Rad). The following primer sequences were used: LIV-1 primer, forward 5′- CAC TTA CTG GCT TAT-3′, reverse 5′- GTT GGT CAA TAA TGT AAT AAG AC-3′; CyclinB1 primer, forward 5′-TCG AGC AAC ATA CTT TGG CCA-3′, reverse 5′-GCA AAA AGC TCC TGC TGC AA-3′; GAPDH primer, forward 5′- TGC ACC ACC AAC TGC TTA GC-3′,reverse 5′- GGC ATG GAC TGT GGT CAT GAG-3′.
2.6. Apoptosis
For apoptosis detection, cells were assayed with an Annexin V-FITC/PI apoptosis kit (MultiSciences Biotech Co., Ltd. Nanjing, China) and analyzed by flow cytometry. The GFP transfected cells were assayed with an Annexin PI/7AAD apoptosis kit (MultiSciences Biotech Co., Ltd. Nanjing, China) and analyzed by flow cytometry.
2.7. Tumour xenografts in Nod/SCID mice
HeLa cells were labelled with luciferase (Shanghai Genechem Co.) using a lentiviral transduction method. Luciferase-labelled HeLa cells were stably transfected with lentivirus carrying LIV-1 shRNA (HeLa /LIV-1 shRNA cells) or mock shRNA (HeLa/Mock cells) (Shanghai Genechem Co.). 1 × 106 tumour cells were implanted subcutaneously into the right flanks of Nod/SCID mice. The mice were injected i.p once a week (for four weeks) with Taxol (12 mg/kg) [27,28] or DMSO when tumour sizes reached 200 mm3. The bioluminescence signal intensity (BLI) and tumour volumes were measured using the Xenogen IVIS spectrum in vivo imaging system (Caliper Life Sciences, Inc.). All animals were obtained from BEIJING HFK BIOSCIENCE Co., Ltd. (Beijing, China), and experiments were approved by the Committee on the Ethics of Animal Experiments of Tongji Medical College. Mice were maintained in the accredited animal facility of Tongji Medical College.
2.8. Cell synchronization and cell cycle assays
Cells were synchronized via double-thymidine (2 mM) blockade, then released and harvested at the indicated time points. For mitotic synchronization, cells were treated with double-thymidine, then released to nocodazole-containing medium (0.1 µM) for 12 h before mitotic cells shaken off. For thymidine-taxol, cells were synchronized via double-thymidine blockade and released into taxol-containing medium (0.1 µM). Cell-cycle distributions were analyzed via flow cytometry as previously described [29].
2.9. H3P staning
The cells were trypsizined, pelleted and fixed in ice cold Ethanol. Then, the cells were incubated in rabbit anti-histone H3 (phosphoS10) antibody (1/1000; ab5176; Abcam) in 1% bovine serum albumin (BSA)/phosphate-buffered saline (PBS), washed, incubated in sheep anti-rabbit-FITC antibody in 1% BSA/PBS and counterstained with PI. Debris was eliminated and Mitotic cells as defined by H3P immunofluorescence, were counted via flow cytometry using FL2-A/FL2-W. At least 15000 cells were counted.
2.10. Metaphase spreads
Cells were hypotonically swollen in 40% full medium/60% tap water for 5.5 min. Hipotonic treatment is stopped by adding an equal volume of Carnoy's solution (75% pure methanol, 25% glacial acetic acid), cells are then spun down and resuspended and fixed with Carnoy's solution for 10 min. After fixation cells are dropped from a 5 cm height onto glass slides. Slides are stained with 5% Giemsa in PBS for 7 min. To evaluate premature separation of sister chromatids, pre-treated cells were harvested for metaphase chromosome spreads according to standard protocols. Cells with separated sister chromatids were observed via Olympus FV1000 confocal laser scanning microscopy. Chromatids were counted in 400 mitotic cells.
2.11. Time-lapse imaging
For time-lapse imaging, phase-contrast and fluorescence images of live cells were captured every 2 min with a 100 × objective and processed using an Olympus FV1000 confocal laser scanning microscope Viewer. The temperature of the imaging medium was maintained at 37 °C and 5% CO2. Cellular behaviour was evaluated manually. To evaluate cell fate in response to anti-mitotic agents, pre-transfected HeLa/GFP-tubulin cells were released into taxol-containing medium. Image acquisition was performed for a further 45 h.
2.12. Immunofluorescence in paraffin sections or cultured cells
Paraffin sections were removed the paraffin and dehydrated with standard protocol. Cultured cells were fixed with cold methanol during 15 min at 20 °C, washed in PBS containing 0.1% BSA (Sigma). Sections or cells were incubated with 0.1% Triton X for permeabilization. Sections or cells were then blocked with 3% BSA and incubated with primary antibodies for 2–4 hr at 37 °C. The matching secondary antibodies (Alexa 488 or Alexa 594) are from Molecular Probes (Invitrogen). Images were obtained using an Olympus FV1000 confocal laser scanning microscopy or Leica DFC490.
2.13. Co-immunoprecipitation (Co-IP)
Co-immunoprecipitation analysis was performed with Pierce Crosslink Immunoprecipitation Kit (cat. no. 26147; Thermo Scientific) according to the manufacturer's instructions. The eluted fractions were subjected to liquid chromatography and tandem mass spectrometry (LC-MS/MS; The Beijing Genomics Institute, China).
2.14. GST pull-down
GST pull-down assays were performed using a GST protein interaction pull-down kit (cat. no. 21516; Thermo Scientifific). GST or GST aa1-47 fusion protein was produced in Rosetta-gami B (Novagen) E. coli strains for 4 h at 37 °C after the addition of 1 mM IPTG (Sigma). The protein complexes were further processed for immunoblot or mass spectrometry analysis.
2.15. LIV-1 knockout mouse study
LIV-1 knockout mice (B6; 129S5-Slc39a6tm1Lex/Mmucd, #032587-UCD) were purchased from the Mutant Mouse Regional Resource Center. Genomic DNA was isolated from tails and PCR was used to screen mouse genotypes using a common primer outside of target exon 2 and a primer in the neo gene. The following primer sequences were used: Primers target LIV-1 exon 2, forward 5′-ATC TGT AAT CAT GAT CTT GAC C-3′, reverse 5′-AAA GCG AGT TCC TTC AGA GA-3′; Primers target Neo gene, forward 5′-GAA GAG GCG TAC ACA T-3′, reverse 5′-GCA GCG CAT CGC CTT CTA TC-3′. The RNA was isolated from tails and RT-PCR was used to detect LIV1 transcript isform. The following primer sequences were used: forward 5′-AAT CAA AAG AAA CCT GAA AAT-3′; reverse 5′-GAA ATT TAT ACG AAA CAC AAT-3′.
LIV-1+/− females were crossed to LIV-1+/− males, DMSO or taxol (Sigma; 30 mg/kg, every other day) was injected (i.p.) into pregnant females from E10.5 to E12.5. After the last administration of drug, mice were sacrificed 3 days later and embryos were isolated for the subsequent experiments. Six weeks old mice (LIV-1+/+or LIV-1−/−) were injected (i.p.) with DMSO or taxol (30 mg/kg, once a week for four weeks). After the last administration of drug, mice were sacrificed 3 days later and tissues were isolated for the subsequent experiments.
MEFs were isolated from E13.5 embryos and cultured in Dulbecco's modified Eagle medium with 10% fetal bovine serum containing 100 nM glutamine medium (CM) [30]. The fibroblasts (P0, passage 0) are the only cells that have the ability to attach to the gelatine-coated flasks. Ideally, cells are 80–90% confluent after 24 h and at this stage a major part of P0 cells is frozen for further usage. MEFs of the second to fifth passages (P1-P3) were used for experiments.
2.16. Subcellular fractionation
Mitochondria isolation was performed using the Mitochondria Isolation Kit (cat no.89874; Thermo Scientific) according to the manufacturer's instructions. Factions enriched in muclear and mitochondrial proteins were obtained from 1.5 × 107 cells.
3. Phosphatase activity assay
For in vivo phosphatase assays, pre-treated cells (5∼8 × 106 cells, ∼5000 µg protein) were suspended in lysis buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, 5% glycerol, protease inhibitors; pH 7.4), cleared from debris by centrifugation, incubated with anti-rabbit B55α beads (40µl) or with control anti-rabbit IgG beads (40µl). The beads were washed with lysis buffer, resuspended in phosphatase reaction buffer and determined for PP2A-B55α activity using Serine/Threonine Phosphatase Assay kit (Millipore) according to Malachite Green Phosphatase Assay Protocol. Colorimetric assay was measured using microplate reader at 620 nM (Hyperion).
For in vitro phosphatase assays, 293 cells (5∼8 × 106 cells, ∼5000 µg protein) were suspended in lysis buffer, cleared from debris by centrifugation, incubated with anti-rabbit B55α beads (40 µl) or with control anti-rabbit IgG beads (40 µl) at 4 °C for 8 h. Next, the beads was washed with lysis buffer for three times and then incubated with different concentration of recombinant GST or GST/GRPEL1 proteins at 4 °C for 8 h. Finally, the beads were washed with lysis buffer, resuspended in phosphatase reaction buffer and analyzed for PP2A-B55α activity.
3.1. Ethical approval
The tissues were obtained from Tongji Hospital (Wuhan, China). The tissue microarray was obtained from Xi'an Alena Biotechnology Ltd., Co. The study was approved by the Ethical Committee. The specimens were completely anonymous and had no direct identifiers and no codes or indirect identifiers that link back to subjects.
3.2. Immunohistochemical statistics
For semi-quantitative evaluation, an immunoreactivity-scoring (IRS) system was applied as reported previously [31]. In briefly, the scoring method was based on the fact that the specimens clearly showed a varying degree of staining intensity and percentage of cells staining. Therefore, a combined assessment of staining intensity and the percentage of positively stained cells was used. The staining intensity was scored as follows: 0 = no expression, 1 = weak expression, 2 = moderate expression, 3 = strong expression. For each intensity score, the percentage of cells with that score was estimated visually. A combined weighted score, consisting of the sum of the percentage of cells with each score, was calculated for each sample. For example, a sample with 65% strong staining, 15% moderate staining, and 20% weak staining would be scored as follows: 65 × 3 + 15 × 2 + 20 × 1 = 245. The maximum score should be 300. The slides were evaluated by two independent pathologists.
3.3. Statistical analysis
Statistical comparisons between two groups were conducted with Unpaired 2-tailed t test. One-way analysis of variance was used for multiple-group comparisons. The Pearson correlation coefficient was used to measures the linear association between the two scale variables. In general, statistical analysis was performed using SPSS version 16.0. All data are shown as mean ± SEM; Probabilities of P < 0.05 were considered significant.
4. Results
4.1. LIV-1 is activated and indispensable for the killing of tumour cells by mechanistically distinct anti-mitotic agents
Our recent genome-wide search for mediators of histone deacetylase inhibitor (HDACi) action revealed that LIV-1 is potentially responsible for HDACi-induced apoptosis [32]. Surprisingly, LIV-1 was also indispensable for the killing of tumour cells by mechanistically distinct anti-mitotic agents, but not by other commonly used chemotherapeutics (Fig. S1A-D). Knockdown of LIV-1 was required for HeLa cells to resist killing by anti-mitotic agents (Fig. S1E-F and Fig. 1A). To assess the effect of LIV-1 on resisting killing by anti-mitotic agents in vivo, HeLa /LIV-1 shRNA cells or HeLa/Mock cells were subcutaneously injected into Nod/SCID mice, followed by injecting i.p once a week with Taxol. We found that the luciferase activity and tumour volume of HeLa/mock group were significantly lower than that of the HeLa/LIV-1 shRNA group after Taxol treatment, suggesting that tumour cells in the LIV-1 shRNA group are more resistant to Taxol (Fig. 1B–D).
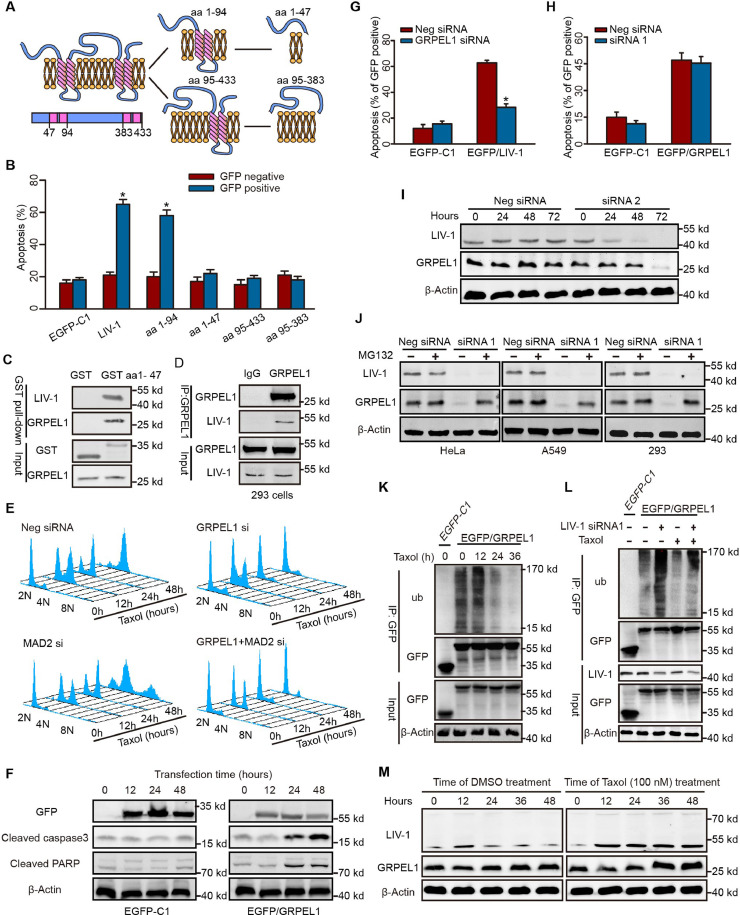
Fig. 1.
LIV-1 is Required for Anti-Mitotic Agent-Induced Killing. (A) HeLa cells were transfected with various siRNAs for 72 h and then exposed to anti-mitotic agents. At 48 h, cells were assayed for apoptosis. (Unpaired 2-tailed t-test, *P < 0.01 versus neg siRNA). (B-D) HeLa/Mock cells or HeLa /LIV-1 shRNA cells were implanted subcutaneously into the right flanks of Nod/SCID mice. The mice were injected i.p. with Taxol. (Day 10; arrow) (n = 7 each group). (B) Bioluminescence images of mice with tumor at day 50. (C) Bioluminescence intensity of tumor at indicated time points (One-way ANOVA, *P < 0.01). (D) Weights (mg) of tumors from the experiment shown in (B) (One-way ANOVA, *P < 0.05, **P < 0.01). (E) HeLa cells were synchronized via double-thymidine blockade, released, and sampled at 2-h intervals for immunoblotting. (F) HeLa cells transfected with the indicated siRNAs for 72 h were synchronized and released into DMSO- or taxol-containing culture medium. (G-H) HeLa cells were transfected with EGFP/LIV-1 or EGFP-C1 and evaluated for apoptosis. (G) The fates of transfected cells were tracked via video confocal microscopy. Scale bar, 5 µm. Numbers represented the recording time. (H) The percentage of apoptosis was determined in GFP-positive and -negative HeLa cells via flow cytometry 72 h after transfection (Unpaired 2-tailed t-test, *P < 0.01 versus EGFP-C1). (I) GFP-positive cells were sorted and immunoblotted 72 h after transfection. Date are represented as Mean ± SEM in triplicate from a representative experiment of three independent experiments.
To determine the expression of LIV-1 during cell-cycle progression, LIV-1 displayed a cell cycle-specific expression pattern that was identical to that of cyclin B1 (Fig. S1G and Fig. 1E). More strikingly, treatment with taxol induced a dramatic and persistent elevation in LIV-1 protein levels that was coordinated with gradual activation of caspase-3 and cleavage of PARP (Fig. 1F, left and middle panels). However, the apoptotic cascade was attenuated by knockdown of LIV-1 (Fig. 1F, right panel). Thus, LIV-1 is activated and indispensable for the killing of tumour cells by mechanistically distinct anti-mitotic agents.
To address the functional consequences of LIV-1 overexpression, HeLa cells were transfected with EGFP/LIV-1. While cells transfected with control vector underwent normal mitosis, ectopic expression of LIV-1 directly induced apoptosis (Fig. 1G). Apoptosis in EGFP-positive cells were therefore analyzed by flow cytometry and western blot (Fig. 1H,I). Ectopic expression of LIV-1 led to direct apoptosis.
Taken together, these results demonstrate that LIV-1 was indispensable for the killing of tumour cells by mechanistically distinct anti-mitotic agents, while knockdown of LIV-1 was required for HeLa cells to resist killing by anti-mitotic agents and overexpression of LIV-1 led to direct apoptosis.
4.2. Tumour cells depleted for LIV-1 evade anti-mitotic agent-induced killing through a rapid mitosis exit
Since anti-mitotic agents kill tumour cells by inducing mitotic arrest, we investigated how depletion of LIV-1 impacts mitotic arrest. Simultaneous knockdown of LIV-1 T1 and T2 overrode taxol-induced mitotic arrest in HeLa cells (Fig. 2A,B). In many other tumour cell lines, LIV-1 inhibition reproducibly attenuated cell-cycle arrest induced by various anti-mitotic agents (Fig. S2A). Surprisingly, LIV-1 inhibition enabled tumour cells to slip out of mitotic arrest with cytokinesis and return to the G1 phase in a diploid state (Fig. 2C). The tetraploidization conferred by MAD2 siRNA was completely rectified by knockdown of LIV-1 (Fig. 2C).
Fig. 2.
Tumor Cells Depleted for LIV-1 Evade Anti-Mitotic Agent-Induced Killing. (A-B) HeLa cells were transfected with the indicated siRNAs for 72 h and then treated with taxol, sampled, and analyzed for mitotic arrest. (A) The cell cycle was analyzed by flow cytometry at the indicated time points. (B) Mitotic cells (black circles, the percentage of H3P-positive cells), as defined by H3P immunofluorescence, were counted via flow cytometry 24 h after taxol exposure. (C) HeLa cells transfected with the indicated siRNAs were analyzed for cell cycle and DNA ploidy by flow cytometry. (D) HeLa cells were transfected with siRNAs for 72 h and synchronized with nocodazole for an additional 12 h. Mitotic cells were shaken off, replated, and subjected to mitotic-exit analysis. (E) HeLa cells were transfected with various siRNAs, exposed to taxol for 24 h, and harvested for metaphase chromosome spreads. Scale bar, 2 µm. (F) GFP-Tubulin HeLa cells were transfected with neg siRNA or siRNA1 and then treated with Taxol. Dynamic changes in cell fate were tracked over 45 h from Taxol beginning with time-lapse confocal imaging. Each row indicates the life cycle of an individual cell. Division, continuation of cell-cycle progression; Exit, exited mitosis without division; Dormancy, persistent cell-cycle arrest in interphase; Slippage, slipping out of mitosis in a diploid state.
To examine the direct impact of LIV-1 inhibition on mitosis, synchronized HeLa cells were released and the kinetics of mitosis was monitored. Upon release, knockdown of LIV-1 dramatically accelerated the kinetics of mitosis (Fig. 2D). In asynchronous HeLa cells, > 60% of LIV-1 siRNA-transfected mitotic cells showed premature separation of sister chromatids, in contrast to < 10% of mitotic cells transfected with control siRNA (Fig. 2E).
Then, time-lapse confocal imaging was employed to track cell fates following exit from anti-mitotic agent-induced mitotic arrest with LIV-1 knockdown. After exposure to taxol, HeLa cells typically underwent several fates (Fig. S2B). Some cells died in mitosis or exited mitosis and died in the next interphase. The other cells underwent persistent cell-cycle arrest in the interphase (dormancy). In contrast, LIV-1 depleted cells successfully entered mitosis and underwent cytokinesis in the presence of taxol (Fig. 2F and Video S1-5). Nevertheless, LIV-1 depleted cells took much shorter time to complete cell division than that of the negative control (Fig. 2D, Fig. S2C and Video S1-2), and LIV-1 depleted cells in the presence of taxol took much longer time to divide their DNA and execute cytokinesis than those of HeLa cells in the absence of taxol (Fig. 2F, Video S1-4). Depletion of LIV-1 did not affect the intracellular concentration of taxol at all the time points examined by high-performance liquid chromatography and tandem mass spectrometry (LC-MS/MS), indicating effects of LIV-1 knockdown on mitosis is specific (Fig. S2D-F). The results suggest that LIV-1 inhibition enabled tumour cells to slip out of mitotic arrest and return to the G1 phase in a diploid state, which did not affect the intracellular concentration of taxol.
4.3. GRPEL1 interacts with LIV-1 and functions downstream of LIV-1
To unravel the mechanisms by which LIV-1 regulates apoptosis and mitosis, we investigated the proteins that interact with LIV-1. LIV-1 localizes prominently to the endoplasmic reticulum (ER) rather than to mitochondria (Fig. S3A). During mitosis, LIV-1 displayed a diffuse intracellular distribution (Fig. S3B). To identify the proteins that interact with LIV-1, we initially sought to increase intracellular LIV-1 levels by transfecting HeLa cells with a LIV-1 expression plasmid. However, this transfection yielded low numbers of EGFP/LIV-1-positive cells because LIV-1 overexpression is toxic and induces instant apoptosis (Fig. 1G–I). LIV-1 contains 433 amino acids (aa) and four putative ER transmembrane domains predicted with the SOSUI algorithm (Fig. 3A). Four LIV-1 deletion mutants (aa1-94, 1–47, 95–433, and 95–383) were constructed based on the putative topological structure. Subcellular localization analysis of the deletion mutants justified the predicted topological structure of the deletion constructs (Fig. S3C). A functional transfection assay showed that mutant aa1-94 displayed potent pro-apoptotic activity similar to that of LIV-1 (Fig. 3B). Since aa1-94 contains the cytoplasmic region of LIV-1 and exhibits key pro-apoptotic activity, mutant aa1-47, the cytoplasmic portion of mutant aa1-94, was chosen for subsequent co-ip experiments to identify proteins that interact with LIV-1.
Fig. 3.
GRPEL1 Functions Downstream of LIV-1. (A) Topological structures of LIV-1 and truncated mutants in relation to the ER membrane were predicted by the SOSUI algorithm. (B) HeLa cells were transfected with various LIV-1 mutant plasmids, cultured for 72 h, and assayed for apoptosis (*P < 0.01 versus GFP-negative, Unpaired 2-tailed t-test). (C) Endogenous GRPEL1 was pulled down by GST aa1-47 in 293 cells. (D) Lysates of 293 cells were immunoprecipitated with GRPEL1 antibody or rabbit IgG and immunoblotted with LIV-1 antibody. (E) HeLa cells were transfected with various siRNAs, treated with taxol, and sampled for analyses of the cell cycle and DNA ploidy. (F) HeLa cells were transfected with EGFP-C1 or EGFP/GRPEL1. Expression of apoptosis-related proteins was determined. (G) HeLa cells were transfected various siRNAs and cultured for 48 h. Then the cells were transfected with EGFP-C1 or EGFP/LIV-1. Apoptosis of GFP-positive cells was evaluated via flow cytometry (*P < 0.01 versus neg siRNA, Unpaired 2-tailed t-test). (H) HeLa cells were transfected various siRNAs and cultured for 48 h. Then the cells were transfected with EGFP-C1 or EGFP/GRPEL1 and cultured for 72 h. Apoptosis of GFP-positive cells was evaluated via flow cytometry (*P < 0.01 versus neg siRNA, Unpaired 2-tailed t-test). (I) HeLa cells were transfected with the indicated siRNAs, sampled, and the levels of LIV-1 and GRPEL1 were determined. (J) Cells were transfected with indicated siRNAs, and cultured for 72 h. Cells were cultured in medium with or without 10 µM MG132 for an additional 6 h and immunoblotted for GRPEL1. (K) HeLa cells were transfected with EGFP-C1 or EGFP/GRPEL1 for 12 h and cultured in medium with 100 nM taxol. Cells were sampled and lysed for immunoprecipitation with GFP antibody. The immunoprecipitate was analyzed for polyubiquitylation. (L) HeLa cells were transfected with indicated siRNAs for 72 h. Cells were then transfected with EGFP-C1 or EGFP/GRPEL1 for 12 h before treatment with taxol or DMSO for another 24 h before polyubiquitylation was assayed. (M) HeLa cells were synchronized and released into DMSO- or taxol-containing medium. Protein levels of LIV-1 and GRPEL1 were determined at various time points. Date are represented as Mean ± SEM in triplicate from a representative experiment of three independent experiments.
Recombinant GST aa1-47 was incubated with a lysate from 293 cells, and the polypeptides that co-ip with it were subjected to LC/MS/MS (Fig. S3D). Notably, GRPEL1 was the only protein bound to GST aa1-47 that was identified reproducibly in all three independent experiments (Fig. 3C; Table S1). LIV-1/V5 precipitated endogenous GRPEL1 in 293 cells that inducibly expressed LIV-1/V5 (Fig. S3E). In 293 cells, endogenous GRPEL1 precipitated LIV-1 protein (Fig. 3D).
Knockdown of GRPEL1 facilitated rapid slippage of HeLa cells from taxol-arrested mitosis into a 2 N G1 phase (Fig. 3E). In many other tumour cell lines exposed to various anti-mitotic agents, knockdown of GRPEL1 reproducibly overrode mitotic arrest (Fig. S3F). Overexpression of GRPEL1 was sufficient to induce apoptosis in HeLa cells (Fig. S3G). In EGFP/GRPEL1-transfected cells, ectopic expression of GRPEL1 led to direct activation of caspase-3 and cleavage of PARP (Fig. 3F). These data demonstrate that at least some of the functions of GRPEL1 are identical to those of LIV-1. Notably, overexpression of LIV-1 failed to induce apoptosis when GRPEL1 was knocked down (Fig. 3G). On the other hand, the pro-apoptotic activity due to GRPEL1 overexpression was not affected by the status of LIV-1 expression (Fig. 3H). These data addressed LIV-1 interacts with GRPEL1, and acts through GRPEL1
4.4. LIV-1 stabilizes GRPEL1 protein by preventing ubiquitylation of GRPEL1
Currently, the function of human GRPEL1 is not known. GRPEL1 is a mitochondria-localized protein in E. coli and has been proposed to act as a co-chaperone [33], [34], [35]. In this study, however, GRPEL1 protein displayed a diffuse intracellular distribution, which display in cytoplasm and nucleus localization and didn't display any mitochondrial locazation (Fig. S3H-I).
In HeLa cells, knockdown of LIV-1 directly reduced GRPEL1 levels, with a lag of 24 h behind the reduction in LIV-1 levels (Fig. 3I). Depletion of LIV-1 reproducibly led to a dramatic decline in GRPEL1 levels; this decrease was abolished by the addition of proteasome inhibitors (Fig. 3J). While GRPEL1 was effectively polyubiquitylated in HeLa cells, taxol exposure attenuated GRPEL1 ubiquitylation (Fig. 3K). On the other hand, knockdown of LIV-1 substantially enhanced GRPEL1 ubiquitylation both at baseline and after taxol exposure (Fig. 3L). GRPEL1 protein expression did not display an obvious cell cycle-specific pattern in HeLa cells released from a synchronized state (Fig. 3M, left panels). HeLa cells treated with taxol increased their GRPEL1 levels with a lag of 24 h behind the increase in LIV-1 levels (Fig. 3M, right panels). These data indicate that LIV-1 function is a potent upstream signal that prevents GRPEL1 ubiquitylation and subsequent proteasomal degradation.
4.5. Genetic ablation of LIV-1 results in lower levels of GRPEL1 and overrides taxol-induced mitotic arrest in vivo
We next tested whether depletion of LIV-1 in vivo reproduced the functional phenotype observed in vitro. LIV-1 knockout mice were generated via homologous recombination in embryonic stem cells (Fig. 4A). LIV-1−/− mice were viable and fertile and did not develop obvious illness. Genomic DNA and RNA were isolated from tails and amplified by PCR to confirm the deletion of LIV-1 (Fig. 4B,C). In line with our findings in vitro, mouse embryonic fibroblasts (MEFs) derived from LIV-1−/− mice harbored substantially less endogenous GRPEL1 (Fig. 4D) as well as dramatically enhanced ubiquitylation of GRPEL1 versus cells from LIV-1+/+ or LIV-1+/− mice (Fig. 4E). MEFs derived from LIV-1−/− embryos overrode taxol-induced mitotic arrest (Fig. 4F). In contrast, LIV-1+/+ and LIV-1+/− MEFs displayed striking taxol-induced mitotic arrest.
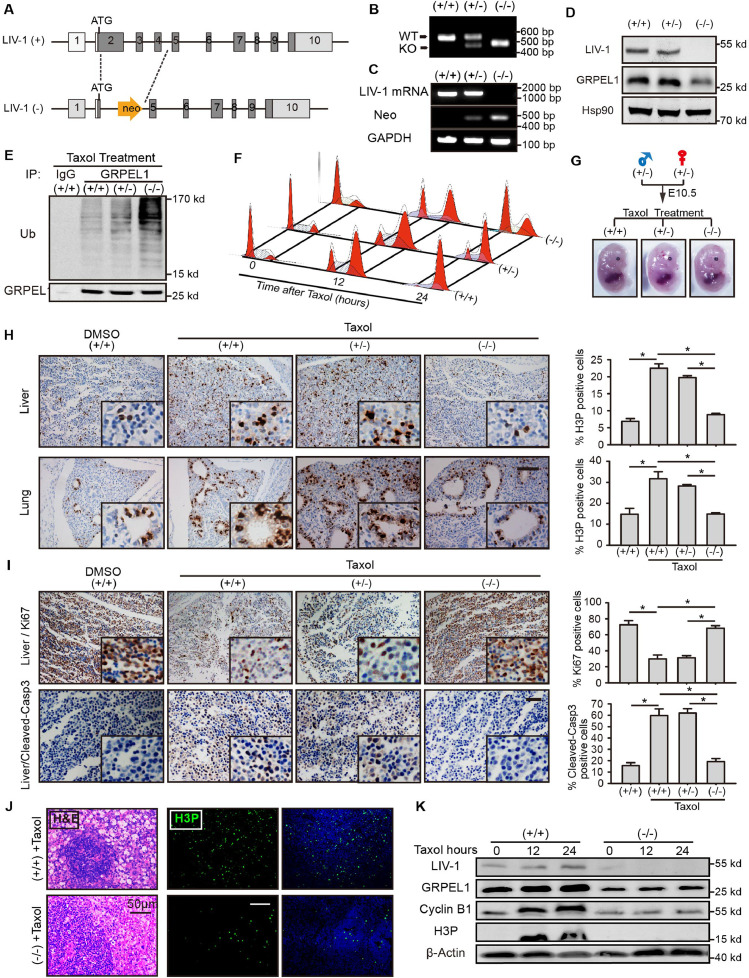
Fig. 4.
Genetic Ablation of LIV-1 Results in a Weakened Anti-Mitotic Agent-Induced MC in vivo. (A) LIV-1−/− mice were generated via targeting disruption of exons 2–4. (B) Genomic DNA was isolated from tails and assayed for deletion of the targeted exons. WT, wildtype; KO, knockout. (C) mRNA was isolated from tails and assayed for LIV-1 transcriptional expression. (D) MEFs derived from the indicated genotypes were immunoblotted. (E) GRPEL1 from MEFs of the indicated genotypes was immunoprecipitated and polyubiquitylation was evaluated. (F) MEFs derived from the indicated genotypes were treated with taxol-containing culture medium and sampled for cell-cycle analysis. (G-I) LIV-1+/- females were crossed with LIV-1+/- males. Females were exposed to DMSO or 30 mg/kg taxol. (G) Representative appearances of embryos of the indicated genotypes. (H) Representative immunohistochemical staining for mitotic marker H3P (brown) in liver (left upper panels) and lung (left lower panels) from indicated embryos. Right panels, percentage of H3P-positive cells. Scale bar: 100 µm. (I) Representative immunohistochemical staining for Ki-67 and apoptotic marker cleaved caspase-3 in liver from indicated embryos. Right panels, percentages of Ki-67-positive and cleaved caspase3-positive cells (* P < 0.01, One-way ANOVA). Scale bar, 100 µm. (J) Mice (6 weeks old) were injected with DMSO or taxol. After the last administration of drug, mice were sacrificed 3 days later and tissues were isolated. Left panel, hematoxylin and eosin staining in spleen tissues. Right panel, representative immunofluorescence images of H3P (green) and nucleus (blue) were shown in spleen from taxol-exposed mice. Scale bar, 50 µm in HE; 100 µm in H3P. (K) MEFs derived from the indicated genotypes were treated with taxol-containing culture medium and sampled for immunoblotting. Date are represented as Mean ± SEM in triplicate from a representative experiment of three independent experiments.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To evaluate the functional status of taxol-induced mitotic arrest in LIV-1−/− mice, LIV-1+/− female mice were crossed with LIV-1+/− males. Females were exposed to DMSO or 30 mg/kg taxol. There were no evident abnormalities in the appearance of LIV-1+/+, LIV-1+/− and LIV-1−/- embryos (Fig. 4G). Immunohistochemistry revealed that metaphase arrest was significantly relieved in LIV-1−/− embryos after taxol treatment, indicating that impaired mitotic arrest was caused by deletion of LIV-1 (Fig. 4H). Strikingly, taxol-exposed LIV-1−/- embryonic tissues retained high proliferative capacity and exhibited lower frequencies of apoptosis than tissues from LIV-1+/+ or LIV-1+/− embryos (Fig. 4I). Moreover, adult LIV-1−/- mice also exhibited impaired mitotic arrest after taxol treatment. Four week-treatment of LIV-1−/− and LIV-1+/+ mice with taxol did not significantly affect the viability of these mice. Spleen tissue from LIV-1+/+ mice displayed hydropic degeneration of splenic sinus cells, increased hemogonia in the splenic sinus, and dramatic metaphase arrest; no similar histological abnormalities were evident in taxol-treated LIV-1−/− mice (Fig. 4J).
MEFs derived from LIV-1−/− embryos overrode taxol-induced mitotic arrest and immunoblotting revealed that LIV-1−/− cells failed to increase the levels of cyclin B1 and H3P after treatment with taxol (Fig. 4K). Notably, loss of LIV-1 led to prominently lower GRPEL1 levels that did not increase after taxol exposure (Fig. 4K). Collectively, these data demonstrate that genetic ablation of LIV-1 results in reduced endogenous GRPEL1 levels and impaired mitotic arrest induced by taxol in vivo.
4.6. GRPEL1 dually regulates mitotic exit as well as apoptosis by interacting with PP2A B55α and AIF
To address how GRPEL1 regulates apoptosis and mitosis, we sought to identify the proteins that interact with GRPEL1. HeLa cells were transfected with EGFP-C1 or EGFP/GRPEL1 and lysed for co-ip with GFP antibody. Polypeptides that co-ip with EGFP-C1 or EGFP/GRPEL1 were analyzed with LC/MS/MS (Fig. S4A-C). More than 100 bound polypeptides co-ip with EGFP/GRPEL1; these polypeptides cover a wide range of cellular functions, such as the regulation of cell cycle and apoptosis (Fig. S4C). Notably, several well-known critical regulators of apoptosis and the MC, such as apoptosis inducing factor (AIF), Bcl-2-associated transcription factor (BTF), and regulatory subunits of PP2A phosphatase complexes (B55α) were identified (Fig. S4A-C).
We were intrigued by our identification of B55α and AIF because these proteins are known to play critical roles in the control of mitotic exit and apoptosis [36], [37], [38]. In 293 cells, endogenous GRPEL1 precipitated AIF and B55α (Fig. S4D), and both endogenous AIF and B55α precipitated GRPEL1 (Fig. S4E-F). In HeLa cells transfected with EGFP-C1 or EGFP/GRPEL1, endogenous AIF and B55α were pulled down by EGFP/GRPEL1 in a time-dependent fashion (Fig. 5A). Knockdown of AIF partially rescued EGFP/GRPEL1-induced apoptosis (Fig. 5B). In response to pro-apoptotic stimuli, AIF induces apoptosis by detaching from the mitochondria in a truncated form and translocating to the nucleus [38], [39]. Interestingly, elevated GRPEL1 expression directly induced the release and translocation of AIF in a time-dependent fashion (Fig. 5C–E), suggesting that GRPEL1 promotes apoptosis partially through the action of AIF. On the other hand, knockdown of B55α substantially rescued GRPEL1 siRNA-impaired mitotic arrest (Fig. 5F and Fig. S4G,H), indicating that B55α is the downstream target of GRPEL1 to regulate mitotic exit.
Fig. 5.
GRPEL1 Is a Key Dual Regulator of Mitotic Exit and Apoptosis Through Its Interactions with AIF and PP2A B55α. (A) HeLa cells transfected with EGFP-C1 or EGFP/GRPEL1 were lysed for co-immunoprecipitation with anti-GFP antibody. AIF, full-length AIF; tAIF, truncated AIF. (B) HeLa cells were transfected with AIF siRNA for 48 h and transfected with EGFP/GRPEL1 or EGFP-C1. Apoptosis in GFP-positive cells was examined via flow cytometry (*P < 0.01 versus neg siRNA, Unpaired 2-tailed t-test). (C-E) HeLa cells were transfected with EGFP-C1 or EGFP/GRPEL1. (C) Representative images of the release and translocation of AIF (red; anti-AIF antibody) into the nucleus (blue; Hoechst 33342) induced by GRPEL1. Scale bar: 20 µm in EGFP-C1; 10 µm in EGFP/GRPEL1. (D) Percentage of HeLa cells displaying AIF translocation induced by GRPEL1 (n = 100 cells per treatment). (E) Transfected cells were fractionated (Nucle, nuclear; Mito, mitochondrial) and immunoblotted for AIF. (F) HeLa cells were transfected with various siRNAs for 72 h, treated or not treated with 100 nM taxol for 24 h, and analyzed for mitotic arrest. (G) B55α protein immunoprecipitated with anti-B55α agarose from 293 cell extracts was incubated with indicated concentration of recombinant GST or recombinant GST/GRPEL1 for 8 h. Left panel, Immunoblots of eluates with GST (top) or B55α (bottom) antibody. Right panel, B55α agarose was subjected to the PP2A phosphatase assay (*P < 0.01 versus GST, Unpaired 2-tailed t-test). (H-I) The PP2A protein was immunoprecipitated from the indicated treated cells by anti-B55α antibody and analyzed for PP2A phosphatase activity. (H) HeLa cells were transfected with the indicated siRNAs for 72 h and treated with or without taxol. (I) LIV-1+/+ and LIV-1−/- MEFs cells were treated with or without taxol (*P < 0.01, One-way ANOVA). Date are represented as Mean ± SEM in triplicate from a represeBntative experiment of three independent experiments.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To examine how GRPEL1 affects the PP2A phosphatase activity of B55α, B55α was immunoprecipitated from 293 cells with anti-B55α agarose and then incubated with recombinant protein GST or GST/GRPEL1 before phosphatase activity assay. B55α directly pulled down GST/GRPEL1 (Fig. 5G, left panel), and GRPEL1 inhibited the PP2A phosphatase activity in a dose-dependent fashion (Fig. 5G, right panel). In HeLa cells, taxol treatment significantly reduced the phosphates activity; this inhibition was completely abolished when GRPEL1 was knocked down (Fig. 5H). Further, taxol treatment dramatically decreased the PP2A activity in LIV-1+/+ but not in LIV-1−/− MEFs (Fig. 5I). These data indicate that GRPEL1 is required for the down-regulation of PP2A-B55α activity after exposure to taxol.
Collectively, these data demonstrate that the LIV-1-GRPEL1 axis depletion works to reduce the mitotic arrest by inducing PP2A-B55α phosphates activity, while inhibit apoptosis by banding AIF and preventing the latter's release into the nucleus.
4.7. Inactivation of the LIV-1-GRPEL1 axis occurs frequently in multiple types of human cancer
To address the clinical relevance of the LIV-1-GRPEL1 axis in human cancer, we examined LIV-1 status in a wide range of human malignancies. Immunohistochemical analysis of LIV-1 protein from a tissue array including multiple types of human cancers demonstrated that LIV-1 preferentially expressed in most of normal epithelial tissues. Strikingly, expression of LIV-1 protein was dramatically lower in most tumours of epithelial origin than in corresponding non-neoplastic tissues (Fig. S5A–C).
We further examined LIV-1 protein expression in a large pool of primary samples from gastric and ovarian cancers to confirm these observations. Neoplastic gastric and ovarian samples displayed remarkably lower levels of LIV-1 protein than that of normal tissues (Fig. 6A,B, left panels). Notably, lower LIV-1 levels in gastric and ovarian cancer samples were consistently related to tumour grade (Fig. 6A,B, middle panels). In contrast, LIV-1 levels in gastric cancer samples were not related to tumour stage (Fig. 6A, right panel). Interestingly, samples from primary ovarian cancer patients who were clinically sensitive to taxol-based chemotherapy displayed higher levels of LIV-1 than samples from patients who were resistant to chemotherapy (Fig. 6B, right panel).
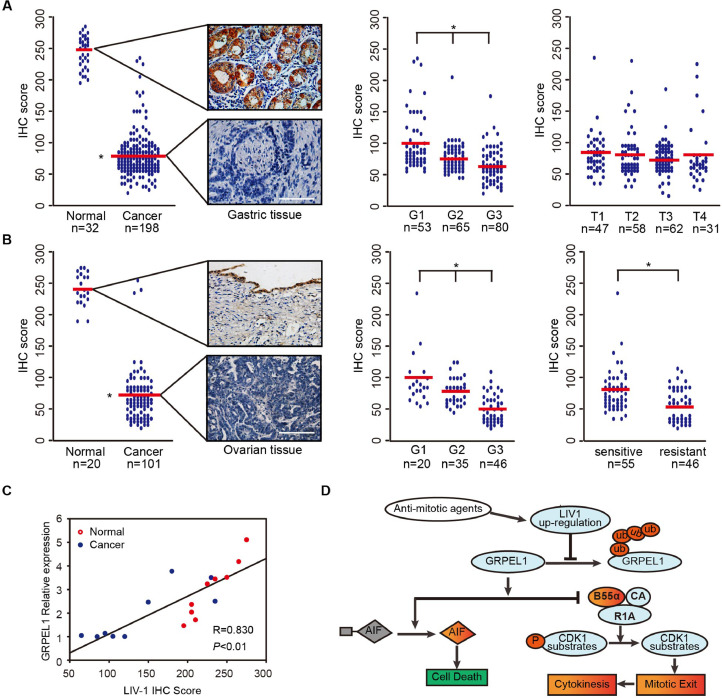
Fig. 6.
Significantly Decreased Expression of LIV-1 and GRPEL1 Occurs Frequently in Human Cancer. (A-B) Assessment of LIV-1 expression via semi-quantitative immunoreactivity H-scoring in primary gastric (A) and ovarian (B) tumor samples. Red line, mean expression score in each group. Left panels of (A-B), typical images representing the mean LIV-1 expression in gastric (A) and ovarian (B) samples. Middle panels of (A-B), comparison of samples with different tumor grades. Right panel of (A), comparison of tumor samples with different tumor-nodes-metastasis staging. Right panel of (B) comparison of tumor samples from patients who were sensitive or resistant to taxol-based chemotherapy. Scale bar, 200 µm. (*P < 0.01, One-way ANOVA). (C) Correlation between LIV-1 expression and GRPEL1 expression in gastric samples. Relative GRPEL1 expression was detected by immunoblotting and analyzed with ImageJ. LIV-1 expression was determined with immunohistochemistry scores. Normal, non-neoplastic tissues (red circles). Cancer, tumor tissues (blue circles) (*P < 0.01, Pearson correlation coefficient). (D) Working model of the control of mitotic exit by the LIV-1-GRPEL1 axis during anti-mitotic agent-induced mitotic arrest.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In primary gastric cancer samples, LIV-1 levels significantly correlated with GRPEL1 levels (r = 0.446, P < 0.01; Fig. 6C), which is in good consistence with our previous findings. Collectively, these data demonstrate that frequent inactivation of the LIV-1-GRPEL1 axis occurs in multiple types of human cancer through a post-transcriptional mechanism.
5. Discussion
In this study, we present in vitro and in vivo data supporting a model in which cell fate during anti-mitotic agent-damaged mitosis is regulated by a previously unrecognized LIV-1-GRPEL1 axis (Fig. 6D). LIV-1 expression was elevated after treatment with Anti-mitotic agents, which in turn led to the accumulation of endogenous GRPEL1 by preventing its ubiquitylation and proteasomal degradation. One of the LIV-1-GRPEL1 axis elevation works to maintain the mitotic condition by inhibiting PP2A-B55α phosphates activity, while the other works to induce apoptosis by banding AIF and promoting the latter's release into the nucleus. On the other hand, LIV-1-GRPEL1 axis inhibition enabled tumor cells to slip out of anti-mitotic agent caused mitotic arrest with cytokinesis and return to the G1 phase in a diploid state.
The current investigation adds critical knowledge to our understanding of cancer biology. First, we have shown that the LIV-1-GRPEL1 axis is indispensable for the killing of tumour cells by a wide spectrum of mechanistically distinct anti-mitotic agents. Second, it is the previously unrecognized pathway identified to date whose functional status adjusts cell fate during damaged mitosis by dually regulating mitotic exit and apoptosis. Third, we observed that mice lacking the gene that encodes LIV-1 reproduce the functional features of the LIV-1-GRPEL1 axis. Fourth, inactivation of the LIV-1-GRPEL1 axis apparently leads to a distinct slippage phenotype that facilitates the survival of anti-mitotic agent-damaged cells. The consequences of a defective checkpoint reduce the duration of mitotic arrest in taxol and other anti-mitotic agents. However, virtually all slipped cells were reported to exit mitosis in a tetraploid state without undergoing chromosome segregation and cytokinesis before endocycling. Surprisingly, here, loss of the LIV-1-GRPEL1 axis resulted in a previously unrecognized slippage phenotype in which cells exited damaged mitosis with chromosome segregation and cytokinesis in the presence of microtubule stabilizing/destabilizing drugs. At present, we do not have a clear molecular explanation for this slippage phenotype. Given that GRPEL1 regulates PP2A-B55α phosphates activity and binds importinβ1, the LIV-1-GRPEL1 axis may also play a critical role in the regulation of post-mitotic assembly events such as chromosome segregation and cytokinesis. Finally, our findings strongly argue that the LIV-1-GRPEL1 axis is clinically relevant. Dramatically reduced or absent LIV-1 expression was common across a large variety of primary tumours and was closely correlated with chemotherapeutic sensitivity and tumour grade. As such, our findings highlight a regulatory mechanism that adjusts cell fate during damaged mitosis.
Several studies have proposed mechanistic explanations of how cancer cells slip out of mitotic arrest and survive damaged mitosis [40]. Mitotic slippage involves several factors, including the accelerated destruction of cyclinB1 [41] and gain or loss of the function of mitotic kinases, all of which lead to premature inactivation of cyclin-dependent kinase-1 (CDK1) during the damaged mitotic arrest [42], [43]. The roles of mitotic-exit regulators that act downstream of the CDC20-CDK1 axis in anti-mitotic agent-associated slippage have never been addressed. Here we observed that taxol treatment inhibited PP2A-B55α activity in a LIV-1-GRPEL1 axis-dependent fashion. Inactivation of this axis failed to reduce PP2A-B55α activity and was therefore unable to prevent mitotic exit during an activated MC. Therefore, the LIV-1-GRPEL1 axis appears to be the previously unappreciated key regulator to act downstream of the CDC20-CDK1 axis and to operate independent of MC activity.
Despite a long history of research, the mechanism that regulates PP2A-B55α activity during cell-cycle progression is still poorly understood. The only key regulator identified to date is Greatwall kinase (also known as MASTL) [44], [45], [46]. In the current study, GRPEL1 appears to constitute a novel mechanism for inhibiting PP2A-B55α activity during damaged mitosis through direct protein-protein interactions. Our discovery of the LIV-1-GRPEL1 axis provides a new starting point for elucidating how mitotic exit is controlled after exposure to mitotic poisons.
Our findings have important clinical implications. Our identification of the LIV-1-GRPEL1 axis provides a conceptual framework for developing revolutionary anti-mitotic agents. Given our observation of differential LIV-1 expression in neoplastic and corresponding non-neoplastic tissues, tumour-selective killing is likely to be achieved with this strategy; here, reactivating the LIV-1-GRPEL1 axis by overexpressing GRPEL1 effectively killed tumour cells. Finally, inactivation of the LIV-1-GRPEL1 axis led to the production of genetically unstable progeny. Accumulating evidence has shown that haploinsufficiency of spindle assembly checkpoint components leads to a defective checkpoint as well as a high rate of tumour production. Further elucidation of the contributions of an inactivated LIV-1-GRPEL1 axis to tumorigenesis is a matter of great clinical significance.
Declaration of Competing Interest
The authors declare that there are no conflicts of interest with this work.
Acknowledgments
Funding sources
This work was supported by National Clinical Research Center for Obstetric and Gynecologic Diseases (grant no. 2015BAI13B05), the National Natural Science Foundation of China (grant nos. 81702572, 81630060, 81472783, 81272859, 81372801, 81572570, 81772787, 81702574).
Author Contributions
GC, QLG and JFZ supervised the project, interpreted the data, and wrote the article. PBC, BBW and QQM designed the research, and analyzed all the data. PW, YF and YT pervided the technical support; XJ, YW and YG performed the experiments. YC and YZ designed and analysis of LC/MS/MS experiments. LX, SXW, JBH and DM supervised the study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.09.054.
Contributor Information
Jianfeng Zhou, Email: jfzhou@tjh.tjmu.edu.cn.
Qinglei Gao, Email: qlgao@tjh.tjmu.edu.cn.
Gang Chen, Email: tjchengang@hust.edu.cn.
Appendix. Supplementary materials
References
- 1.Jordan M.A., Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 2.McGuire W.P., Rowinsky E.K., Rosenshein N.B., Grumbine F.C., Ettinger D.S., Armstrong D.K. Taxol: a unique antineoplastic agent with significant activity in advanced ovarian epithelial neoplasms. Ann Intern Med. 1989;111(4):273–279. doi: 10.7326/0003-4819-111-4-273. [DOI] [PubMed] [Google Scholar]
- 3.Kaur R., Kaur G., Gill R.K., Soni R., Bariwal J. Recent developments in tubulin polymerization inhibitors: an overview. Eur J Med Chem. 2014;87:89–124. doi: 10.1016/j.ejmech.2014.09.051. [DOI] [PubMed] [Google Scholar]
- 4.Keen N., Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4(12):927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 5.Strebhardt K., Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6(4):321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 6.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17(2):93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedberg J.W., Mahadevan D., Cebula E., Persky D., Lossos I., Agarwal A.B. Phase II study of alisertib, a selective Aurora A kinase inhibitor, in relapsed and refractory aggressive B- and T-cell non-Hodgkin lymphomas. J Clinic Oncol. 2014;32(1):44–50. doi: 10.1200/JCO.2012.46.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr P.M., Li H., Spier C., Mahadevan D., LeBlanc M., Ul Haq M. Phase II intergroup trial of alisertib in relapsed and refractory peripheral T-cell lymphoma and transformed mycosis fungoides: SWOG 1108. J Clinic Oncol. 2015;33(21):2399–2404. doi: 10.1200/JCO.2014.60.6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nokihara H., Yamada Y., Fujiwara Y., Yamamoto N., Wakui H., Nakamichi S. Phase I trial of volasertib, a polo-like kinase inhibitor, in Japanese patients with advanced solid tumors. Invest New Drugs. 2016;34(1):66–74. doi: 10.1007/s10637-015-0300-0. [DOI] [PubMed] [Google Scholar]
- 10.DuBois S.G., Mosse Y.P., Fox E., Kudgus R.A., Reid J.M., McGovern R. Phase II trial of alisertib in combination with irinotecan and temozolomide for patients with relapsed or refractory neuroblastoma. Clinic Cancer Res. 2018;24(24):6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penna L.S., Henriques J.A.P., Bonatto D. Anti-mitotic agents: are they emerging molecules for cancer treatment? Pharmacol Ther. 2017;173:67–82. doi: 10.1016/j.pharmthera.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Dickson M.A., Mahoney M.R., Tap W.D., D'Angelo S.P., Keohan M.L., Van Tine B.A. Phase II study of MLN8237 (Alisertib) in advanced/metastatic sarcoma. Ann Oncol. 2016;27(10):1855–1860. doi: 10.1093/annonc/mdw281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly K.R., Friedberg J.W., Park S.I., McDonagh K., Hayslip J., Persky D. Phase I study of the investigational aurora a kinase inhibitor alisertib plus rituximab or rituximab/vincristine in relapsed/refractory aggressive B-cell lymphoma. Clinic Cancer Res. 2018;24(24):6150–6159. doi: 10.1158/1078-0432.CCR-18-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gascoigne K.E., Taylor S.S. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14(2):111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Shi J., Mitchison T.J. Cell death response to anti-mitotic drug treatment in cell culture, mouse tumor model and the clinic. Endocr Relat Cancer. 2017;24(9):T83–T96. doi: 10.1530/ERC-17-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wertz I.E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D.J. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471(7336):110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 17.Mantel C., Guo Y., Lee M.R., Han M.K., Rhorabough S., Kim K.S. Cells enter a unique intermediate 4 N stage, not 4N-G1, after aborted mitosis. Cell Cycle. 2008;7(4):484–492. doi: 10.4161/cc.7.4.5316. [DOI] [PubMed] [Google Scholar]
- 18.Sinha D., Duijf P.H.G., Khanna K.K. Mitotic slippage: an old tale with a new twist. Cell Cycle. 2019;18(1):7–15. doi: 10.1080/15384101.2018.1559557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gascoigne K.E., Taylor S.S. How do anti-mitotic drugs kill cancer cells? J Cell Sci. 2009;122(Pt 15):2579–2585. doi: 10.1242/jcs.039719. [DOI] [PubMed] [Google Scholar]
- 20.Topham C., Tighe A., Ly P., Bennett A., Sloss O., Nelson L. MYC is a major determinant of mitotic cell fate. Cancer Cell. 2015;28(1):129–140. doi: 10.1016/j.ccell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manchado E., Guillamot M., de Carcer G., Eguren M., Trickey M., Garcia-Higuera I. Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55alpha,delta phosphatase. Cancer Cell. 2010;18(6):641–654. doi: 10.1016/j.ccr.2010.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Taylor K.M., Nicholson R.I. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta. 2003;1611(1–2):16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 23.Hogstrand C., Kille P., Ackland M.L., Hiscox S., Taylor K.M. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3) Biochem J. 2013;455(2):229–237. doi: 10.1042/BJ20130483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasper G., Weiser A.A., Rump A., Sparbier K., Dahl E., Hartmann A. Expression levels of the putative zinc transporter LIV-1 are associated with a better outcome of breast cancer patients. Int J Cancer. 2005;117(6):961–973. doi: 10.1002/ijc.21235. [DOI] [PubMed] [Google Scholar]
- 25.Wu C., Li D., Jia W., Hu Z., Zhou Y., Yu D. Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat Genet. 2013;45(6):632–638. doi: 10.1038/ng.2638. [DOI] [PubMed] [Google Scholar]
- 26.Ma X., Ma Q., Liu J., Tian Y., Wang B., Taylor K.M. Identification of LIV1, a putative zinc transporter gene responsible for HDACi-induced apoptosis, using a functional gene screen approach. Mol Cancer Ther. 2009;8(11):3108–3116. doi: 10.1158/1535-7163.MCT-08-0772. [DOI] [PubMed] [Google Scholar]
- 27.Au Yeung C.L., Co N.N., Tsuruga T., Yeung T.L., Kwan S.Y., Leung C.S. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel N., Chatterjee S.K., Vrbanac V., Chung I., Mu C.J., Olsen R.R. Rescue of paclitaxel sensitivity by repression of Prohibitin1 in drug-resistant cancer cells. Proc Natl Acad Sci U S A. 2010;107(6):2503–2508. doi: 10.1073/pnas.0910649107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin X., Mo Q., Zhang Y., Gao Y., Wu Y., Li J. The p38 MAPK inhibitor BIRB796 enhances the antitumor effects of VX680 in cervical cancer. Cancer Biol Ther. 2016;17(5):566–576. doi: 10.1080/15384047.2016.1177676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Higuera I., Manchado E., Dubus P., Canamero M., Mendez J., Moreno S. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10(7):802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 31.Luo J., Zha S., Gage W.R., Dunn T.A., Hicks J.L., Bennett C.J. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62(8):2220–2226. [PubMed] [Google Scholar]
- 32.Wu P., Tian Y., Chen G., Wang B., Gui L., Xi L. Ubiquitin B: an essential mediator of trichostatin A-induced tumor-selective killing in human cancer cells. Cell Death Differ. 2010;17(1):109–118. doi: 10.1038/cdd.2009.142. [DOI] [PubMed] [Google Scholar]
- 33.Naylor D.J., Stines A.P., Hoogenraad N.J., Hoj P.B. Evidence for the existence of distinct mammalian cytosolic, microsomal, and two mitochondrial GrpE-like proteins, the Co-chaperones of specific Hsp70 members. J Biol Chem. 1998;273(33):21169–21177. doi: 10.1074/jbc.273.33.21169. [DOI] [PubMed] [Google Scholar]
- 34.Choglay A.A., Chapple J.P., Blatch G.L., Cheetham M.E. Identification and characterization of a human mitochondrial homologue of the bacterial co-chaperone GrpE. Gene. 2001;267(1):125–134. doi: 10.1016/s0378-1119(01)00396-1. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava S., Savanur M.A., Sinha D., Birje A., R V., Saha P.P. Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J Biol Chem. 2017;292(44):18075–18090. doi: 10.1074/jbc.M117.788463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz M.H., Held M., Janssens V., Hutchins J.R., Hudecz O., Ivanova E. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat Cell Biol. 2010;12(9):886–893. doi: 10.1038/ncb2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Susin S.A., Lorenzo H.K., Zamzami N., Marzo I., Snow B.E., Brothers G.M. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 38.Joza N., Susin S.A., Daugas E., Stanford W.L., Cho S.K., Li C.Y. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 39.Cheung C.H., Chen H.H., Kuo C.C., Chang C.Y., Coumar M.S., Hsieh H.P. Survivin counteracts the therapeutic effect of microtubule de-stabilizers by stabilizing tubulin polymers. Mol Cancer. 2009;8:43. doi: 10.1186/1476-4598-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieder C.L., Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7(5):637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Brito D.A., Rieder C.L. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16(12):1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malumbres M. Keeping order in anaphase. Dev Cell. 2015;35(4):403–404. doi: 10.1016/j.devcel.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 43.Grallert A., Boke E., Hagting A., Hodgson B., Connolly Y., Griffiths J.R. A PP1-PP2A phosphatase relay controls mitotic progression. Nature. 2015;517(7532):94–98. doi: 10.1038/nature14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mochida S., Maslen S.L., Skehel M., Hunt T. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 2010;330(6011):1670–1673. doi: 10.1126/science.1195689. [DOI] [PubMed] [Google Scholar]
- 45.Alvarez-Fernandez M., Sanz-Flores M., Sanz-Castillo B., Salazar-Roa M., Partida D., Zapatero-Solana E. Therapeutic relevance of the PP2A-B55 inhibitory kinase MASTL/Greatwall in breast cancer. Cell Death Differ. 2018;25(5):828–840. doi: 10.1038/s41418-017-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang P., Malumbres M., Archambault V. The Greatwall-PP2A axis in cell cycle control. Method Molecul Biol. 2014;1170:99–111. doi: 10.1007/978-1-4939-0888-2_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.