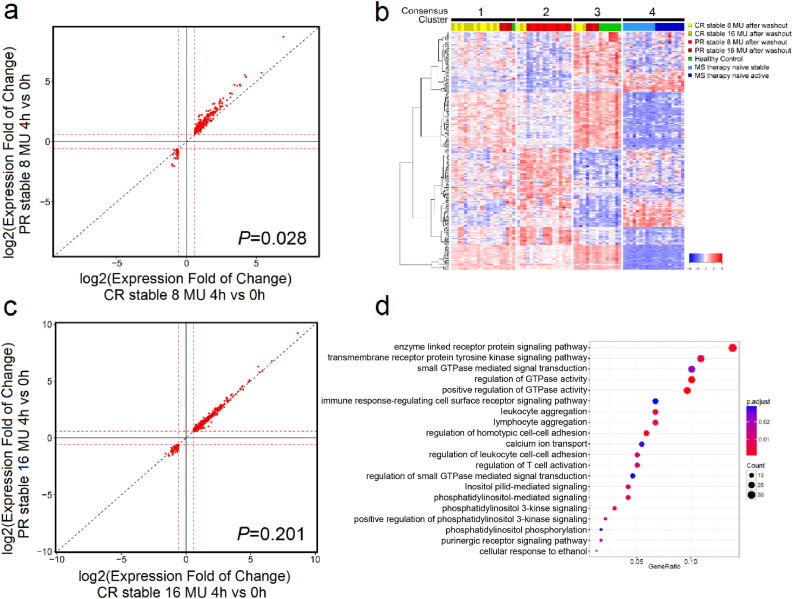
Fig. 2.
Short-term IFN-β-induced gene expression distinguishes clinically stable PR from CR patients. (a) 250 ug (8 MU) or (b) 500 ug (16 MU) log2-transformed fold change (FC) of gene expression at time 4 vs. 0 h is shown for CR stable (x-axis) and PR stable (y-axis). A statistically significant stronger response is observed in PR compared to CR patients after 250 ug IFN-β injection, indicated by upward deviation of data points from the diagonal line (p-value = 0•028, Welch's t-test, two-sided), which is not observed after 500 ug (p-value = 0•201, Welch's t-test, two-sided). Only significant DEGs are shown. The red dashed line represents the 1•5 FC threshold. (c) 277 PR-specific genes in long-term IFN-β-induced gene expression patterns with FDR-corrected p-value<0•05 (gene list provided in Table S5). 50 samples from stable IFN-β-treated NAb-negative CR and PR after a 4-day washout, healthy controls, and therapy-naïve stable and active MS were grouped into 4 consensus clusters (cluster 1 to 4). (d) Gene Ontology (GO) terms significantly enriched in the 277 protein-coding genes from (c). The size of the dots reflects the number of genes; the colour indicates the enrichment p-value after multi-testing correction. The ratio of (number of genes from the 277-gene set) divided by (total number of genes annotated with this GO term) is shown on the x-axis; GO terms are shown on the y-axis.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)