Abstract
Rationale: The oropharyngeal microbiome is a primary source of lung microbiota, contributes to lower respiratory infection, and is also a driver of oral health.
Objectives: We sought to understand oropharyngeal microbial communities in advanced lung disease, community dynamics after lung transplantation, and ecological features of dysbiosis.
Methods: Oropharyngeal wash samples were obtained from individuals with end-stage disease awaiting transplantation (n = 22) and longitudinally from individuals at 6 weeks, 3 months, and 6 months after transplantation (n = 33), along with healthy control subjects (n = 14). Bacterial 16S and fungal internal transcribed spacer rRNA regions were deep-sequenced, and bacterial community respiratory patterns were imputed from taxonomic composition.
Results: Healthy subjects’ oropharyngeal microbiomes showed a gradient of community types reflecting relative enrichment of strictly anaerobic, aerobic, or facultative anaerobic bacteria. Patients with end-stage lung disease showed severe dysbiosis by both taxonomic composition and respiration phenotypes, with reduced richness and diversity, increased facultative and decreased aerobic bacteria, and absence of communities characterized by obligate aerobes. In patients at 6 weeks and 3 months post-transplant, richness and diversity were intermediate between healthy and pretransplant subjects, with near-normal distribution of community types. However, by 6 months post-transplant, oropharyngeal wash resembled the low-diversity facultative-dominated profile of pretransplant subjects. Community ecotype correlated with Candida abundance.
Conclusions: End-stage lung disease is associated with marked upper respiratory tract dysbiosis involving both community structure and respiratory metabolism profiles of constituent bacteria. Dynamic changes occur after lung transplantation, with partial normalization early but later appearance of severe dysbiosis similar to pretransplant patients. Aberrant oropharyngeal communities may predispose to abnormal lung microbiota and infection risk both in advanced lung disease and after transplantation.
Keywords: microbiome, dysbiosis, lung transplantation, bacteria, fungi
People with advanced pulmonary disease are at increased risk of lung infection, which may be due to structural abnormalities, mechanical and immune defense defects, and consequences of treatment. Lung transplantation is often the only option for many end-stage pulmonary diseases but carries a high risk of infection due to immunosuppression required to avoid allograft rejection, defective cough reflex, and damaged mucociliary clearance. In healthy people, the lung contains only low levels of bacterial DNA that closely match sequences found in the oropharynx, suggesting that the microbiome of the healthy lung is largely derived passively from the upper respiratory tract through physiological microaspiration (1, 2). In disease states, the upper respiratory tract is typically the origin for pathogens that infect the lung (3), and disruption of oropharyngeal communities (dysbiosis) is believed to increase susceptibility to pathogen colonization (4, 5).
We previously conducted a cross-sectional analysis of lung transplant recipients that revealed decreased diversity of the oropharyngeal microbiome compared with healthy subjects (6). This finding therefore raised questions about the upper respiratory tract microbiome in subjects with end-stage lung diseases pretransplant, how it evolves post-transplant, and the relationship between individual taxa and broader community ecotypes. However, the upper respiratory tract microbiome in people with advanced lung disease has not previously been investigated.
Here we investigated the upper respiratory bacterial and fungal microbiome in people with end-stage lung disease awaiting transplantation and individuals who underwent serial sampling during the first 6 months after lung transplantation. Because a principal feature distinguishing oropharyngeal bacteria (and also contributing to oral health) is respiratory metabolism, we investigated both taxonomic composition and respiration phenotypes of bacterial communities. We show that 1) healthy oropharyngeal communities exhibit a gradient of bacterial respiration phenotypes characterized by anaerobic, aerobic, and facultative profiles; 2) individuals with end-stage lung disease have oropharyngeal communities that are markedly dysbiotic both in taxonomic composition and in disrupted balance among bacterial respiration phenotypes; and 3) early post-transplantation subjects show more modest dysbiotic patterns, but by 6 months post-transplant they approach the severe dysbiosis seen pretransplantation.
Methods
Study Subjects and Samples
Lung transplant subjects (Post-Tx; n = 33) were prospectively enrolled into a longitudinal study that sampled the oropharyngeal microbiome at approximately 6 weeks, 3 months, and 6 months after transplantation (Table 1; see Table E1 in the online supplement). An additional group of subjects with end-stage lung disease awaiting transplant (Pre-Tx; n = 22), all of whom were ambulatory outpatients, were recruited and sampled at a single time point. Healthy volunteers (n = 14) were sampled as previously described (1). Subjects provided written informed consent under protocols approved by the University of Pennsylvania Institutional Review Board.
Table 1.
Samples and subjects
| Healthy (n = 14) | Pre-Tx (n = 22) | Post-Tx (n = 33) | |||
|---|---|---|---|---|---|
| Age, yr | 37.8 (±11.5) | 59.6 (±8.4) | 57.9 (±10.3) | ||
| Lung disease, n | |||||
| COPD | — | 8 | 13 | ||
| ILD | — | 14 | 20 | ||
| Transplant type, S/B | — | 17/16 | |||
| Lung Allocation Score | — | 41.66 (±12.8) | 47.3 (±18.9) | ||
| Longitudinal | 6 wk (n = 32) | 3 mo (n = 33) | 6 mo (n = 32) | ||
| Days post-transplant | — | — | 56 (±16.2) | 99.6 (±21.5) | 190.3 (±31.1) |
| Rejection (grade >A2), n | — | — | 6 | 4 | 0 |
| Medications at sample time | |||||
| Immunosuppressive regimen | |||||
| Pred, Tac, MMF/AZA | — | 0 (0) | 29 (90.6) | 32 (97) | 29 (90.6) |
| Pred, Tac | — | 0 (0) | 3 (9.3) | 1 (3) | 3 (9.4) |
| Pred | — | 7 (31.8) | 0 (0) | 0 (0) | 0 (0) |
| Other | — | 4 (18.2) | 0 (0) | 0 (0) | 0 (0) |
| None | 14 (100) | 11 (50) | 0 (0) | 0 (0) | 0 (0) |
| Antibiotics | |||||
| TMP-S | — | 4 (18.2) | 24 (75) | 24 (72.7) | 17 (53.1) |
| TMP-S and/or other | — | 3 (13.6) | 4 (12.5) | 7 (21.2) | 12 (37.5) |
| None | 14 (100) | 15 (68.2) | 4 (12.5) | 2 (6.1) | 3 (9.4) |
| Antifungal | |||||
| Nystatin | — | 0 (0) | 19 (59.4) | 18 (56.2) | 16 (50) |
| Nystatin and/or other | — | 0 (0) | 9 (28.1) | 9 (27.2) | 8 (25) |
| None | 14 (100) | 22 (100) | 4 (12.5) | 6 (18.2) | 8 (25) |
Definition of abbreviations: AZA = azathioprine; B = bilateral; COPD = chronic obstructive pulmonary disease; ILD = interstitial lung disease; MMF = mycophenolate; Pred = prednisone; S = single; Tac = tacrolimus; TMP-S = trimethoprim/sulfamethoxazole; Tx = transplant.
Data shown as mean (±SD) or n (%) unless otherwise noted. Rejection indicates acute cellular rejection grade > A2.
Clinical data were collected for age, sex, lung disease, transplant type, and lung allocation score (LAS) at the time of listing (Pre-Tx) or transplantation (Post-Tx) (Tables 1 and E1). Medication information was collected for immunosuppression, antibacterial, antifungal, and antiviral drugs at the time of sample collection. Formal oral health examinations were not available, but all patients were screened for active dental issues, which were corrected before listing for transplant.
Oropharyngeal wash (OW) was collected as previously described (1) and stored at −80°C. OW pH was measured using a microelectrode pH meter (Hanna-HI4222).
DNA Extraction and Polymerase Chain Reaction Amplification
OW (1.8 ml) was pelleted (10,000 g × 1 min) and DNA extracted (PowerSoil DNA Kit; MoBio Laboratories) following manufacturer’s instructions, with an additional 10-minute incubation at 95°C to improve fungal DNA recovery (7). As an environmental control for microbial DNA contamination arising through the entire pipeline, sterile OW saline was processed in parallel. Extraction and polymerase chain reaction amplification of the bacterial 16S rRNA gene variable 1-2 (V1-V2) region was performed with Golay-barcoded universal primers 27F and 338R as described (8). Amplicons were purified and adjusted to 5-ng/μl concentration before pooling. For samples that did not reach the target DNA concentration (such as environmental controls), 20 μl of amplicon were added to the final pool. The pooled library was sequenced by 2 × 250 bp paired-end MiSeq Illumina technology. Amplification of the fungal internal transcribed spacer 1 (ITS1) region was done using ITS-1/ITS-2 primers as described (9) and sequenced using the same platform.
Bacterial 16S rRNA and ITS Gene Analysis and Imputed Bacterial Respiratory Patterns
Bacterial analysis was performed as described using the QIIME v1.91 pipeline (6, 10). Briefly, 16S rRNA gene sequences were clustered into operational taxonomic units (OTUs) at 97% similarity with UCLUST (11) and taxonomic assignments obtained using the GreenGenes database v.13_8 (12). Richness and diversity were analyzed after rarefaction to 3,000 reads per sample. Bacterial respiration phenotypes were assigned at the genus level, obtained from the literature (Table E2), as strictly aerobic, facultative anaerobic, or obligate anaerobic. To identify fungal taxa, sequences were processed with the PIPITS pipeline (13) followed by additional annotation of representative sequences with BROCC (14) and manual curation with BLASTn. Figures and statistical tests were conducted in R. Sequence data are available in the National Center for Biotechnology Information Short Read Archive (accession #PRJNA518155).
Statistical Analysis
Principal coordinate analysis (PCoA) was performed using the R package “ape” (15). The R package “vegan” was used for analyzing UniFrac distances between communities (16, 17). The Kruskal-Wallis test was used to assess for significant differences in relative abundances of dominant taxa. The Dunn’s test with Benjamini-Hochberg correction was performed for multiple comparisons if the Kruskal-Wallis test was significant. The same procedure was used for respiration phenotypes. Identification of discriminant taxa used linear discriminant analysis effect size analysis (18). Clustering of bacterial communities into ecotypes was done by partitioning around medoids (PAM), with the number of clusters estimated by maximum average silhouette width (19).
Results
Oropharyngeal Microbiota in Lung Transplant Recipients
OW samples were obtained from patients awaiting transplantation (Pre-Tx) and from individuals at approximately 6 weeks, 3 months, and 6 months post-transplantation (Post-Tx) (Tables 1 and E2). To exclude disease that directly impacts upper respiratory tract microbiota, individuals with cystic fibrosis were not included in this study. We also analyzed OW from healthy volunteers. A total of 202 specimens were analyzed for bacterial 16S rRNA gene content, including 151 OW samples and 51 environmental controls. OW yielded a median of 51,450 sequences per sample. Sequences were grouped into OTUs as an indication of bacterial taxa present. Genera with >1% abundance across samples are shown as a heat map in Figure E1.
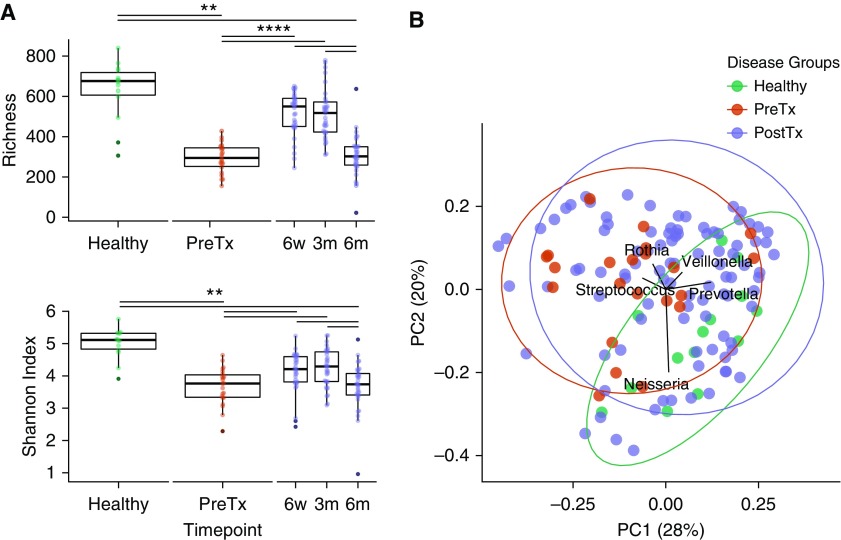
We found significant differences between groups in bacterial richness (Figure 1A). Median numbers of OTUs were 676 for healthy subjects but only 294 for Pre-Tx subjects and 550, 517, and 302 for 6 weeks, 3 months, and 6 months after transplantation, respectively (healthy vs. Pre-Tx P = 0.002, and Post-Tx P < 0.0001, Wilcoxon test). A similar pattern was seen for diversity, in that healthy subjects had the highest diversity measure, Pre-Tx and 6-month Post-Tx samples had the lowest, and 6-week and 3-month post-transplant samples were intermediate (Figure 1A). Thus, end-stage lung disease exhibits a marked decrease in oropharyngeal microbiome richness and diversity, which is also seen post-transplant, although less severely at early than later time points.
Figure 1.
Oropharyngeal microbiome richness and diversity in health and lung disease. (A) Richness is represented by Chao1 index (upper panel) and diversity by Shannon index (lower panel). Samples are grouped by subject category and, for post-transplant, by time of sampling. Dots in each category show the distribution within group. Asterisks indicate statistical significance (Wilcoxon rank sum test; ** P < 0.01 and **** P < 0.001). (B) Principal coordinate analysis (PCoA) of weighted UniFrac distances among oropharyngeal wash samples. Colors correspond to disease status: Healthy (green), pretransplant (PreTx) (red), and post-transplant (PostTx) (purple). Vectors show the taxa present at >5% abundance that are responsible for ordination on the PCoA plot. UniFrac distances were significantly different between healthy and disease groups: Healthy versus PreTx (P = 0.002) and PostTx (P < 0.0001); there were no significant differences between PreTx and PostTx (permutational multivariate analysis of variance). PC1 = principal coordinate axis 1; PC2 = principal coordinate axis 2.
We then analyzed the OW communities using the UniFrac distance metric, which uses phylogenetic relatedness to compare bacterial populations (Figure 1B). The weighted UniFrac principal coordinate analysis (PCoA) showed that healthy individuals were significantly different from both Pre-Tx and Post-Tx samples (P = 0.002 and P < 0.0001 for Pre-Tx and Post-Tx, respectively; permutational multivariate analysis of variance), whereas no difference was seen between Pre-Tx and Post-Tx samples. Within subjects with lung disease, there was no difference between chronic obstructive pulmonary disease and interstitial lung disease groups at any time point (Figure E2).
Identifying taxa present at >5% abundance that were responsible for the PCoA ordination (Figure 1B) showed that the separation in the three main directions was driven by Rothia and Streptococcus (both of which are facultative) in one direction, Veillonella and Prevotella (both anaerobic) in another direction, and Neisseria (a strict aerobe). All of these lineages are characteristic of normal commensal oral microbiota (20).
Bacterial Dominance of Oropharyngeal Microbiota and Taxa that Differ between Groups
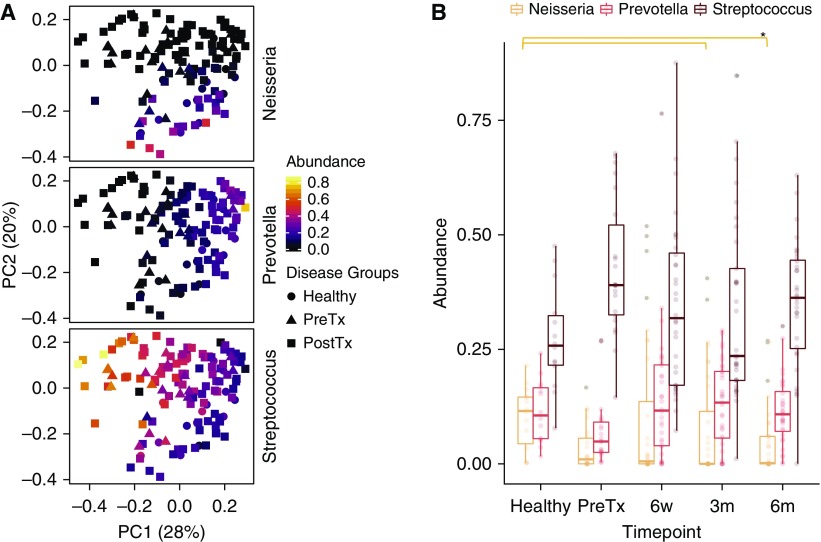
The most abundant three genera across subjects were Streptococcus, Prevotella, and Neisseria, reflected the three poles on the PCoA (Figures 1B and 2A). We therefore compared the relative abundances of these taxa between specimen groups (Figure 2B). Neisseria was significantly different between groups (P = 0.048; Kruskal-Wallis). Individually, Neisseria was lower in post-transplant samples at 3- and 6-month samples than healthy samples (P = 0.028 and P = 0.05, respectively; Dunn’s test). In contrast, Streptococcus and Prevotella only showed nonsignificant trends across sample groups (P = 0.08 and P = 0.09, respectively).
Figure 2.
Disease groups differ in relative dominant oral bacteria. (A) Principal coordinate analysis on the basis of weighted UniFrac distances colored to show relative abundances of Neisseria, Prevotella, and Streptococcus. The shapes correspond to disease groups. (B) Box plots showing relative abundances of the three dominant genera. There were significant differences for Neisseria, but not for Prevotella and Streptococcus, between time points (P = 0.048, P = 0.059, and P = 0.08, respectively; Kruskal-Wallis). Individual differences between sample groups are indicated by *P < 0.05 (Dunn’s multiple comparison test). PC1 = principal coordinate axis 1; PC2 = principal coordinate axis 2; PostTx = post-transplant; PreTx = pretransplant.
We applied linear discriminant analysis effect size analysis to identify taxa that best discriminate among sample groups (Figure E3). End-stage lung disease communities demonstrated loss of multiple taxa typical of the healthy oropharynx in addition to loss of Neisseria and Prevotella, whereas a small number of taxa were overrepresented along with Streptococcus (Figure E3A). A generally similar pattern was seen for Post-Tx subjects (Figure E3B). In contrast, when Pre-Tx and Post-Tx communities were compared, only a few taxa showed significant enrichment in one group or the other (Figure E3C). Thus, at the individual taxonomic level, both Pre-Tx and Post-Tx communities differ extensively from healthy communities and only modestly from each other.
Oropharyngeal Microbiome Bacterial Respiration Patterns
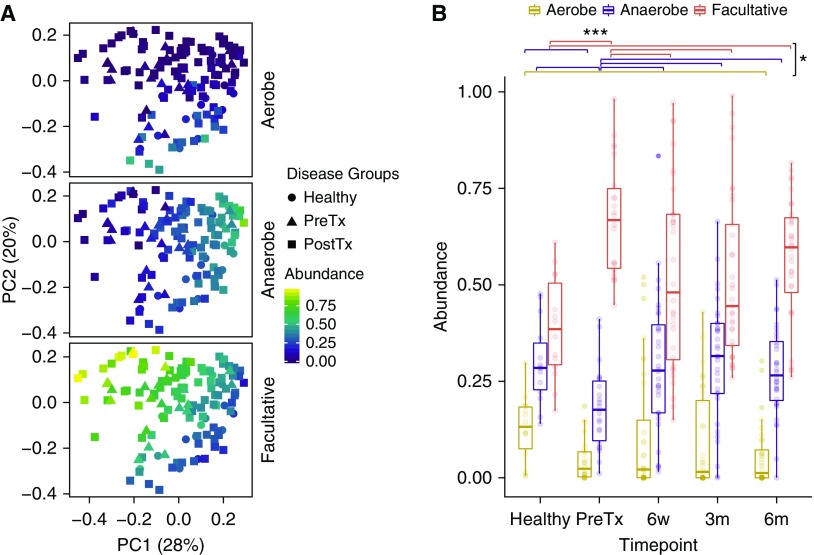
To investigate community profiles on the basis of functional criteria, we queried whether groups might differ in bacterial respiration phenotypes and whether such phenotypic features might better capture group differences than taxonomy alone. We classified each bacterial genus with an abundance >1% across samples into aerobe, anaerobe, or facultative anaerobe categories (Table E2), which was used to impute respiration phenotypic profiles for full bacterial communities. Relative abundance of bacteria in aerobe, anaerobe, and facultative categories for each sample was then compared (Figure 3).
Figure 3.
Bacterial respiration phenotype identifies differences between groups. (A) Principal coordinate analysis on the basis of weighted UniFrac distances colored based on relative abundances of strictly aerobic, obligate anaerobic, and facultative anaerobic bacteria respiration phenotypes. The shapes correspond to disease groups. (B) Box plots show relative abundances of bacterial respiration phenotypes by disease groups. There were significant differences for facultative and anaerobic bacteria between sample groups and a trend for aerobic bacteria (P = 0.001, P = 0.016, and P = 0.057, respectively; Kruskal-Wallis). Individual differences between sample groups are indicated by *P < 0.05 or ***P < 0.001 (Dunn’s multiple comparison test). PC1 = principal coordinate axis 1; PC2 = principal coordinate axis 2; PostTx = post-transplant; PreTx = pretransplant.
Across sample groups, there were marked differences in the abundances of facultative bacteria (P = 0.001; Kruskal-Wallis test). Individually, both pretransplant and 6-month post-transplant samples had significantly greater facultative bacteria than healthy (P < 0.001 and P = 0.015, respectively; Dunn’s test). Interestingly, both the 6-week and 3-month samples had lower abundances of facultative bacteria than pretransplant time point (P = 0.02 and P = 0.02, respectively; Dunn’s test), whereas 6-month samples were not significantly different from pretransplant. There was also a significant difference among sample groups for anaerobes (P = 0.016; Kruskal-Wallis); pretransplant samples were significantly lower than healthy samples (P = 0.04; Dunn’s test), whereas the post-transplant samples at 6 weeks, 3 months, and 6 months were all higher than pretransplant (P = 0.029, P = 0.013, and P = 0.03, respectively; Dunn’s test) and not different from healthy. Finally, aerobic bacteria showed a trend toward differences across samples (P = 0.057; Kruskal-Wallis), with pretransplant and post-transplant samples all appearing lower than healthy, although this reached statistical significance only for the 6-month time point (P = 0.04; Dunn’s test).
Thus, oropharyngeal microbial communities of subjects with end-stage lung disease pretransplant show a marked increase in facultative anaerobes and a decrease in strict anaerobic bacteria compared with healthy individuals. This pattern is present but significantly less pronounced in the early post-transplant samples but devolves toward pretransplant levels by 6 months, also accompanied by loss of aerobes. Figure E4 shows community type dominance in each sample on an individual subject basis.
Community Clustering of Upper Respiratory Tract Microbiomes
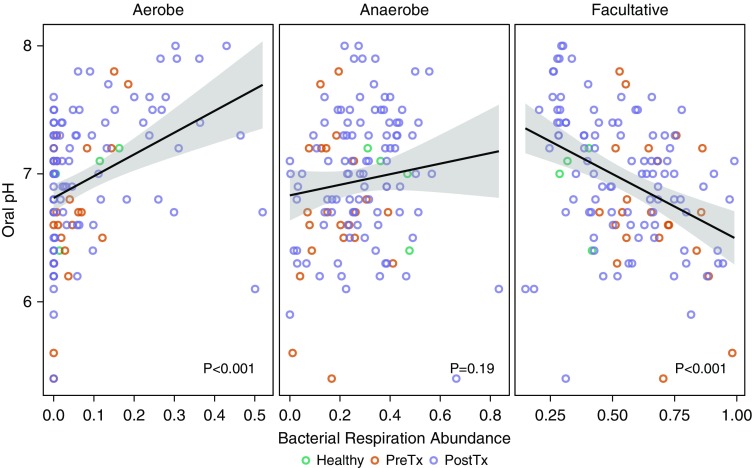
Because oral pH is associated with oral microbiome structure and function (21), we measured oropharyngeal wash pH. As expected, OW pH was significantly correlated with bacterial respiration phenotypes (Figure 4). We assessed the relationship among phenotypic variables including relative abundance of dominant bacteria, respiratory phenotype, and pH (Table E3), which revealed that respiration phenotypes were the primary independent variables among those measured. We then performed clustering analysis by PAM, with three optimum clusters estimated by maximum average silhouette width. The three clusters corresponded to anaerobe-enriched (cluster 1), facultative-enriched (cluster 2), and aerobe-enriched (cluster 3) groups.
Figure 4.
Oral pH is correlated with bacterial respiration phenotype. Relative abundances of bacterial taxa belonging to three respiration phenotypes (strictly aerobe, obligate anaerobe, and facultative anaerobe bacteria) are shown relative to oral pH. Aerobic bacteria and pH are positively correlated (first panel), anaerobe abundance is not significantly correlated with oral pH (middle panel), and facultative abundance and pH are inversely correlated (third panel). Lines indicate the tendency, and the gray area corresponds to standard error. P values were obtained from a linear model (Wilcoxon rank sum test). PostTx = post-transplant; PreTx = pretransplant.
We categorized each sample by PAM community type and analyzed the proportion of each in the different groups (Figure E5). All oral community types were represented in the healthy group. In contrast, Pre-Tx OW samples were dominated by facultative-enriched communities (86%), and aerobic-enriched oropharyngeal communities were absent. Post–lung transplantation samples at earlier time points had similar proportions of the three respiration phenotype groups, but by 6 months aerobic-type communities were almost completely lost.
Fungal Taxa in Oropharyngeal Communities
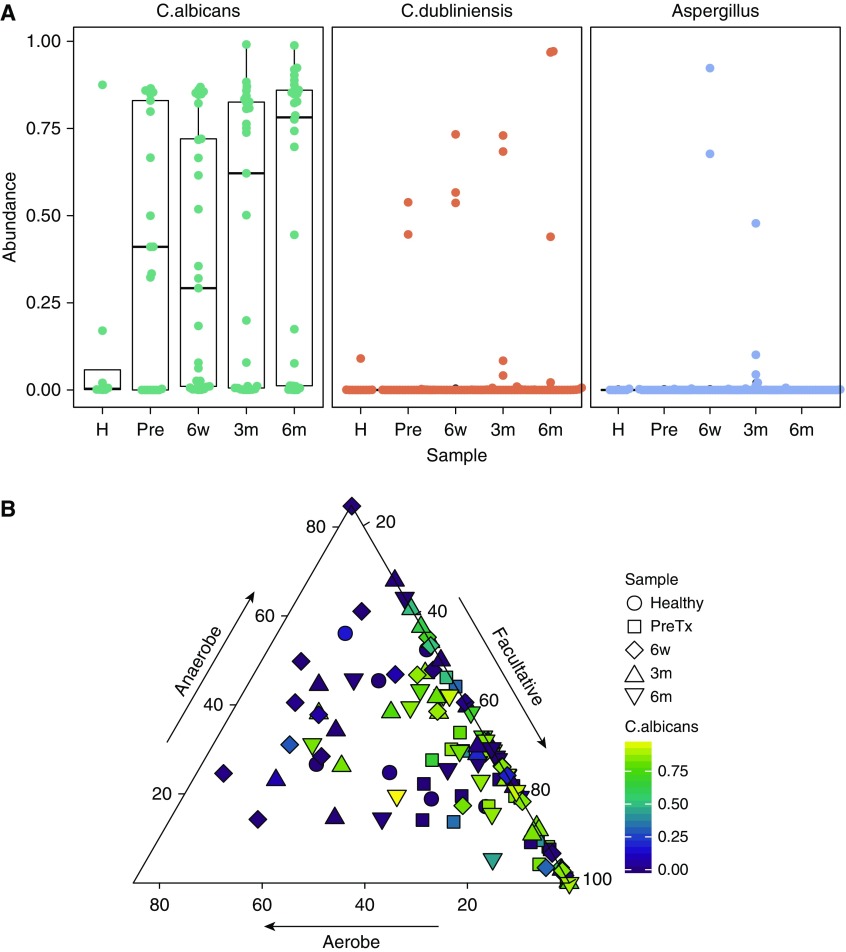
The fungal mycobiome was investigated using ITS sequences (Figure E6). Candida albicans was the most prevalent fungus across all subject groups, followed by Candida dubliniensis. Aspergillus and several other taxa were also present. Overall, subjects with lung disease had higher abundance of C. albicans than healthy people (Figure 5A), although the difference was nonsignificant. Aspergillus was found in several post-transplant subjects’ upper respiratory tract but was absent in healthy and Pre-Tx oropharyngeal subjects.
Figure 5.
Oropharyngeal fungi and relation to bacterial communities. Fungal communities were analyzed using internal transcribed spacer (ITS) marker gene sequencing. (A) Box plot shows relative abundance of Candida albicans, Candida dubliniensis, and Aspergillus among sample sets (H = healthy; Pre = pretransplant; 6w = 6 weeks post-transplant; 3m = 3 months post-transplant; 6m = 6 month post-transplant). (B) Relationship between Candida albicans abundance and abundance of respiratory bacterial phenotypes. Each point corresponds to a sample, and shapes represent time points. Gradient color indicates Candida albicans abundance per sample. Location of the point into the ternary plot depicts the relative abundance of facultative, anaerobic, and aerobe bacteria in each sample. Samples with relative abundance of facultative near to 100% (right corner of the triangle) have higher Candida albicans population (P = 0.0049, analysis of variance). Samples dominated by aerobic bacterial populations (left area of the plot) have the lowest fungi abundances (P = 0.0038, ANOVA).
We then asked if C. albicans, the most abundant fungus, correlated with bacterial respiration groups (Figures 5B and E7). The relative abundance of C. albicans was positively correlated with facultative communities (P = 0.0049, analysis of variance) and negatively correlated with aerobic communities (P = 0.0038, analysis of variance). Thus, the fungal microbiome is also linked to broader microbial community composition reflected in bacterial respiration phenotype.
Relationship of the Oropharyngeal Microbiome and Clinical Status
We queried whether OW richness, diversity, PAM group, or other metrics were associated with disease severity in pretransplant subjects on the basis of LAS and found no significant relationship. Neither was LAS at time of transplant correlated with community measures in post-transplant subjects.
More than 90% of Post-Tx subjects were taking a combination of three immunosuppressants, and nearly all were also on antimicrobials, including prophylactic trimethoprim-sulfamethoxazole and/or other agents (Table 1), limiting our ability to analyze antibiotic and immunosuppressive drug effects within this group. However, less than one-third of pre-Tx subjects were on antimicrobial therapy at time of sampling, and only half were on any immunosuppressive therapy (generally low-dose prednisone). These data suggest that the dysbiosis shared by both Pre-Tx and Post-Tx subjects was not mainly driven by current antimicrobial or immunosuppressive therapy.
We also queried whether dysbiosis within the Pre-Tx group was linked to these factors (Table E4). There was no difference between Pre-Tx subjects taking versus not taking antibiotics in diversity (mean Shannon index, 3.88 vs. 3.56, respectively; P = 0.45) or richness (mean species, 311 vs. 285, respectively; P = 0.45), nor in community ecotype on the basis of PAM group distribution. Similarly, there was no difference in diversity, richness, or ecotype on the basis of whether Pre-Tx subjects were on immunosuppression (Table E4). We thus conclude that oropharyngeal dysbiosis in these Pre-Tx and Post-Tx patients is not primarily due to antibiotic or immunosuppressive use at the time of sampling, although an impact of prior use cannot be excluded.
Discussion
We investigated the upper respiratory tract microbiome in patients with end-stage lung disease awaiting transplantation, individuals after lung transplantation, and healthy people. We made three principal observations. First, incorporating the ecological feature of bacterial respiratory metabolism provides information beyond simple taxonomic composition for this niche, is linked to other features such as pH and fungal taxa, and better captures dysbiosis. Second, marked dysbiosis is present both in subjects with end-stage lung disease awaiting transplantation and in individuals post lung transplantation. Third, dysbiosis is dynamic, with early post-transplant time points less severe but later on reaching the degree seen in patients with end-stage lung disease. The finding of dysbiotic communities has implications for oropharyngeal and lung complications in both advanced lung disease and after transplantation.
The extreme upper respiratory tract microbiome dysbiosis in patients with end-stage lung disease awaiting transplantation was unexpected. Compared with healthy people, their communities had decreased richness and diversity, loss of strict aerobes, overrepresentation of facultative anaerobes, and a preponderance of the facultative/acidic oropharyngeal bacterial communities. We did not include subjects with cystic fibrosis who have intrinsic abnormalities of the mucosa that affect bacterial colonization. Thus, the abnormalities before transplantation may be a consequence of advanced lung disease itself, possibly due to abnormal lower airway microbiota and increased burden that could then impact the upper respiratory tract in a retrograde manner via cough. Alternatively, dysbiosis could be related to treatment of these lung diseases, including prior use of antibiotics, prior hospitalizations, and/or debilitation that may alter oropharyngeal flora. We previously described modest dysbiosis of the upper respiratory tract associated with smoking (22), but none of the subjects here were current smokers, and the extent of dysbiosis found here far exceeds that seen in healthy smokers. Previous studies examining the oropharyngeal microbiome in lung disease reported modest differences in diversity and composition compared with healthy subjects (23–25). However, ours is the first to our knowledge to study subjects with severe end-stage disease. The relationship between advanced lung disease and oropharyngeal dysbiosis merits further investigation.
Upper respiratory tract dysbiosis after lung transplantation had a similar pattern to that seen in subjects with end-stage lung disease, although less severe at early time points (6 wk and 3 mo) but equivalent to that of pretransplant by 6 months. It is possible that this pattern reflects partial but transient normalization associated with transplantation, although the potential mechanisms are not clear. This notion that transplantation might improve (even if temporarily) the oropharyngeal microbiome rather than induce dysbiosis is counter to what we expected. We previously reported upper respiratory tract dysbiosis in a cross-sectional study of people after lung transplantation (median, 4 mo post-transplant; range, 1–114 mo) (6). The temporal data shown here suggest that dysbiosis actually begins pretransplant and that the microbiome is dynamic and less aberrant early after transplantation as compared with later on.
The findings here are strikingly different from those reported for renal and cardiac transplantation, which found no differences in diversity or community structure compared with healthy individuals for the oral (salivary) microbiome (26). On the other hand, enrichment of several potential pathogens was reported, which we did not find (data not shown). Although different specimen types were examined in the two studies, the distinct results nevertheless suggest that the oropharyngeal dysbiosis in our subjects is specific to the lung transplantation population rather than a general consequence of transplant-related immunosuppression. Taken together, these data suggest that solid organ transplantation itself may have only modest effects on the upper respiratory tract microbiome and that preexisting dysbiosis associated with advanced lung disease is key to the upper respiratory tract dysbiosis of our post–lung transplantation subjects.
The relationship between the aberrant oropharyngeal microbiome and advanced lung disease may be bidirectional. Our findings suggest that severe lung disease (or its treatment) is a driver of aberrant oropharyngeal microbiota in the pretransplant subjects. Conversely, colonization of the upper respiratory tract is believed to be the origin for most pathogens that cause lower respiratory tract infections, and a normal oropharyngeal microbiome is believed to restrict colonization by virulent or opportunistic pathogens (4, 5, 26). Both patients with advanced lung disease and post-lung transplant subjects are at high risk for lung infections. There are multiple reasons for increased infection risk, including structural airway abnormalities and damaged mucociliary clearance in severe lung disease, plus anastomotic barriers, defective cough reflex, and immunosuppression in post-transplant subjects. Our data suggest that dysbiotic upper respiratory microbiomes may also contribute to elevated risk of lower respiratory tract infection in both patients with advanced lung disease and post-transplant patients.
We sought to define characteristic community and ecological profiles that might provide broader information and better classification than purely taxonomic data. By incorporating the imputed respiratory metabolism of the total bacterial community as well as oral pH, we could classify communities into three general community types, characterized by facultative-enriched, anaerobe-enriched, and aerobe-enriched, respectively, that provided improved discriminatory power among healthy subjects and those with disease. These community types were not discrete clusters; rather, they reflected poles on a gradation among oropharyngeal samples. Although all types of OW communities were found in healthy people, subjects with lung disease were overrepresented for facultative-enriched and underrepresented for aerobic-enriched communities. In addition, beyond the potential for regulating opportunistic or pathogenic infections, the impact of microbial communities is linked in large part to metabolic activity and products. The oral microbiome also regulates local oral health and may contribute to systemic inflammation (27–29). Further studies will be needed to determine the local and systemic consequences of the skewed metabolic profile in lung disease microbiomes.
A limitation of our study is that pretransplant specimens were not from the same subjects followed longitudinally post-transplantation. However, the similarities in pre- and post-transplantation communities suggest that the end-stage lung disease specimens studied here would accurately reflect those of our post-transplant subjects. Despite a large number of OW samples (n = 151), the number of subjects in any specific clinical outcome group was small, limiting the power to identify associations with clinical events. Thus, future studies will be needed to determine whether oral microbiome ecotypes are associated with clinical outcomes. We did not have detailed oral examination data on our subjects, so we cannot correlate findings to specific oral health factors. However, all subjects receive comprehensive oral medicine correction of any active processes before listing for transplant and are followed closely after transplantation, so it is unlikely that these findings are substantially confounded by concurrent oral diseases unrelated to lung disease or transplantation.
In summary, community ecotypes incorporating bacterial respiratory patterns provided insight into dysbiosis beyond taxonomic composition alone. Patients with end-stage lung diseases have abnormal upper respiratory microbial communities characterized by loss of richness and diversity, loss of obligate aerobic communities, and overrepresentation of bacteria with facultative anaerobic metabolism. Communities are partially but incompletely normalized at 6 weeks and 3 months post-transplantation but by 6 months are similar to the profiles seen in advanced lung disease. Upper respiratory tract microbiome dysbiosis may contribute to increased susceptibility to infection in advanced lung disease and post-transplantation, as well as to local complications after transplantation. Future studies might investigate whether this could offer a potential target for therapeutic intervention.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the pre- and post-transplantation subjects and healthy volunteers, the clinical staff who provide care for the patients and assisted with the study, and members of the Collman and Bushman labs for helpful suggestions. They also acknowledge assistance and support from the PennCHOP Microbiome Program and the Penn Center for AIDS Research (P30-AI045008).
Footnotes
Supported by National Institutes of Health grants R01-HL113252 and R61-HL137063.
Author Contributions: Conceptualized studies: A.S.-S., J.D.C., F.D.B., and R.G.C.; enrolled subjects and collected samples and clinical data: M.C.B., E.C., and J.M.D.; performed wet bench studies: A.S.-S., M.C.B., I.I., and V.R.K.; analyzed data: A.S.-S., M.B.S., J.E.M., A.B., E.L.C., E.C., H.L., and K.B.; interpreted the data: A.S.-S., M.B.S., J.E.M., H.L., K.B., J.M.D., J.D.C., F.D.B., and R.G.C.; wrote manuscript: A.S.-S., F.D.B., and R.G.C.; reviewed revised and or approved manuscript: all authors.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson RP, Huffnagle GB. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog. 2015;11:e1004923. doi: 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heo SM, Haase EM, Lesse AJ, Gill SR, Scannapieco FA. Genetic relationships between respiratory pathogens isolated from dental plaque and bronchoalveolar lavage fluid from patients in the intensive care unit undergoing mechanical ventilation. Clin Infect Dis. 2008;47:1562–1570. doi: 10.1086/593193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ewan VC, Reid WDK, Shirley M, Simpson AJ, Rushton SP, Wade WG. Oropharyngeal microbiota in frail older patients unaffected by time in hospital. Front Cell Infect Microbiol. 2018;8:42. doi: 10.3389/fcimb.2018.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Steenhuijsen Piters WA, Huijskens EG, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 2016;10:97–108. doi: 10.1038/ismej.2015.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, et al. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bittinger K, Charlson ES, Loy E, Shirley DJ, Haas AR, Laughlin A, et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 2014;15:487. doi: 10.1186/s13059-014-0487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, et al. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing MBio 20167e01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 12.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gweon HS, Oliver A, Taylor J, Booth T, Gibbs M, Read DS, et al. PIPITS: an automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol Evol. 2015;6:973–980. doi: 10.1111/2041-210X.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dollive S, Peterfreund GL, Sherrill-Mix S, Bittinger K, Sinha R, Hoffmann C, et al. A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 2012;13:R60. doi: 10.1186/gb-2012-13-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 16.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oksanen J Blanchet FG Kindt R Legendre P Minchin P, O’Hara RB, et al. Vegan: community ecology package. R Package. Version 2.2-1. 2015 [accessed 2019 Apr 9]. Available: https://CRAN.R-project.org/package=vegan.
- 18.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufman L, Rousseeuw PJ. Finding groups in data: an introduction to cluster analysis. Hoboken, NJ: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 20.Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:259. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleinberg I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Bio Med. 2002;13:108–125. doi: 10.1177/154411130201300202. [DOI] [PubMed] [Google Scholar]
- 22.Charlson ES, Chen J, Custers-Allen R, Bittinger K, Li H, Sinha R, et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5:e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garzoni C, Brugger SD, Qi W, Wasmer S, Cusini A, Dumont P, et al. Microbial communities in the respiratory tract of patients with interstitial lung disease. Thorax. 2013;68:1150–1156. doi: 10.1136/thoraxjnl-2012-202917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park H, Shin JW, Park SG, Kim W. Microbial communities in the upper respiratory tract of patients with asthma and chronic obstructive pulmonary disease. PLoS One. 2014;9:e109710. doi: 10.1371/journal.pone.0109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diao W, Shen N, Du Y, Qian K, He B. Characterization of throat microbial flora in smokers with or without COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:1933–1946. doi: 10.2147/COPD.S140243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz PI, Hong BY, Frias-Lopez J, Dupuy AK, Angeloni M, Abusleme L, et al. Transplantation-associated long-term immunosuppression promotes oral colonization by potentially opportunistic pathogens without impacting other members of the salivary bacteriome. Clin Vaccine Immunol. 2013;20:920–930. doi: 10.1128/CVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita Y, Takeshita T. The oral microbiome and human health. J Oral Sci. 2017;59:201–206. doi: 10.2334/josnusd.16-0856. [DOI] [PubMed] [Google Scholar]
- 28.Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. 2014;4:4828. doi: 10.1038/srep04828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demmer RT, Breskin A, Rosenbaum M, Zuk A, LeDuc C, Leibel R, et al. The subgingival microbiome, systemic inflammation and insulin resistance: The Oral Infections, Glucose Intolerance and Insulin Resistance Study. J Clin Periodontol. 2017;44:255–265. doi: 10.1111/jcpe.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.