Abstract
Engineered proteins provide an interesting template for designing fluorine-19 (19F) magnetic resonance imaging (MRI) contrast agents, yet progress has been hindered by the unpredictable relaxation properties of fluorine. Herein, we present the biosynthesis of a protein block copolymer, termed “fluorinated thermoresponsive assembled protein” (F-TRAP), which assembles into a monodisperse nanoscale micelle with interesting 19F NMR properties and the ability to encapsulate and release small therapeutic molecules, imparting potential as a diagnostic and therapeutic (theranostic) agent. The assembly of the F-TRAP micelle, composed of a coiled-coil pentamer corona and a hydrophobic, thermoresponsive elastin-like polypeptide core, results in a drastic depression in spin-spin relaxation (T2) times and unaffected spin-lattice relaxation (T1) times. The nearly unchanging T1 relaxation rates and linearly dependent T2 relaxation rates have allowed for detection via zero echo time 19F MRI, and the in vivo MR potential has been preliminarily explored using 19F magnetic resonance spectroscopy (MRS). This fluorinated micelle has also demonstrated the ability to encapsulate the small-molecule chemotherapeutic doxorubicin and release its cargo in a thermoresponsive manner owing to its inherent stimuli-responsive properties, presenting an interesting avenue for the development of thermoresponsive 19F MRI/MRS-traceable theranostic agents.
Keywords: protein engineering, micelle, 19F MRI, self-assembly, theranostic, drug delivery, thermoresponsiveness
Graphical Abstract
Successful production of a protein-based magnetic resonance imaging (MRI) contrast agent is a challenging yet worthy endeavor,1–3 as proteins possess malleable architectures and functionalities4–13 not yet achievable with synthetic materials. Prior attempts of imaging proteins have focused on exploiting the relatively inert properties of serum proteins such as albumin3 or metal-binding proteins1,2 that are modified by appending paramagnetic metals (Gd3+, Mn2+) or manipulating superparamagnetic metals (iron oxide) in order to reduce the proton nuclei (1H) spin-lattice (T1) or spin-spin (T2) relaxation times, respectively. Reduction in T1 relaxation times enables high-intensity image acquisition, while a decrease in T2 relaxation times produces negative contrast images against a high-intensity background.1,2 Another method for imaging proteins via MRI hinges upon lysine-rich and/or arginine-rich proteins and peptides that contribute exchangeable protons with surrounding water molecules for altering 1H signal in a technique called chemical exchange saturation transfer (CEST).14 To date, protein and peptide imaging via MRI has largely required either chemical modification post-biosynthesis or indirect environmental imaging methods.15–20
One alternative technique for imaging proteins is the introduction of fluorine nuclei (19F). The 19F nucleus remains a standard NMR probe because it serves as a steric replacement for hydrogen. In recent years, however, 19F nanoprobes have been exploited for 19F MRI and magnetic resonance spectroscopy (MRS) as an alternative to 1H MRI. This application of 19F is made possible as it exists in 100% natural abundance and elicits a sensitivity of 83% compared to hydrogen,21–24 yet it is nearly absent in organisms, with the exception of trace amounts found in bone and teeth.21,25 Therefore, 19F is often pursued as an MRI contrast agent, providing agent-specific signal that can be easily co-registered with a 1H anatomical background.26–29 While the signal-to-noise (SNR) ratio of 19F MR is not as strong as that of 1H studies, this drawback is mitigated by the lack of background signal in 19F MRI/MRS such that the signal is directly proportional to the concentration of the introduced 19F agent.30,31
19F MRI and MRS have been used for a variety of biomedical applications including tumor imaging32–36 and measuring tumor cell proliferation,37 cell tracking,30,38,39 assessing physiological oxygen tension40 and inflammation,31 detecting venous thrombosis,41 and monitoring enzyme activity.42–45 Recently developed 19F agents take the form of perfluorocarbon nanoemulsions,36,38,40,41 perfluoropo-lyethers,30,33,39,46 polymer-35,47 and peptide-based nanop-robes,44 small molecular weight ligands31 and substrates,37,43 lipid nanoemulsions,34 liposomes,48 metal organic frame-works,32 nanogels,49 nanocrystals,50 and dendrimers.45 19F-incorporated peptides and proteins have also been explored, but to date have only been studied using NMR and phantom MRI.42,51–55 Solid-phase synthesis of peptides has been used to incorporate fluorinated amino acids;51 however, this technique can be limited by low yield with increasing peptide chain length.56 Structural biologists and protein chemists have also biosynthetically incorporated 19F nuclei into proteins, either site-specifically52 or residue-specifically,42,53,57 for use as NMR probes to understand structure-function relationships involved in ligand binding53 and protein folding.58
Direct MR visualization of 19F nuclei in proteins is often hindered by unfavorable relaxation properties, which necessitate the inclusion of high concentrations of magnetically equivalent 19F nuclei, long imaging times, and/or chemical conjugation of lanthanide chelates to shorten relaxation times.25 As the incorporation of fluorinated amino acids can impart benefits to the native protein function, direct imaging of 19F nuclei alone, unadulterated by synthetic linkers or metals, would be an advantageous development for multifunctional imaging agents.
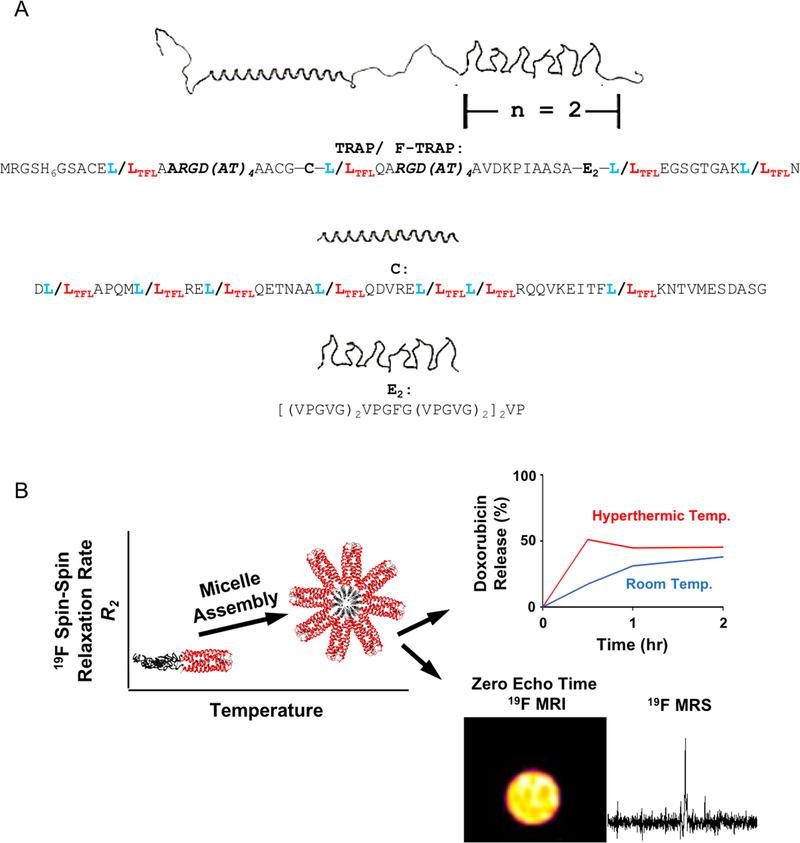
Here we describe the biosynthesis and characterization of a protein block copolymer, or fluorinated thermoresponsive assembled protein (F-TRAP) (Figure 1A), composed of a coiled-coil domain and two repeats of elastin-like polypeptide domains. When subjected to concentration and temperature increases, this construct assembles into nanoscale micelles characterized by a drastic decrease in 19F T2 relaxation times and nearly constant 19F T1 relaxation times. The precipitous decrease in 19F T2 relaxation times results from micelle assembly where NMR-active nuclei in the corona become more ordered and structurally constricted. Here, we overcome the signal loss associated with the dynamic T2 relaxation properties using a zero echo time (ZTE) 19F MRI pulse sequence, demonstrating the direct imaging of fluorinated amino acids within a protein (Figure 1B) without the aid of lanthanide chelates.59,60 Furthermore, as a proof-of-concept, this agent was introduced intratumorally into a mouse xenograft model of human breast cancer, enabling the acquisition of an agent-specific 19F MRS signal (Figure 1B) and demonstrating that the agent is detectable in vivo.
Figure 1.
(A) TRAP monomer and sequence showing the coiled-coil (C) domain and elastin-like polypeptide (E) domain. F-TRAP results in replacement of leucine (L, blue) with trifluoroleucine (TFL, red). (B) Schematic representation of F-TRAP assembly, thermoresponsive drug release, and detection by 19F MRS and by zero-echo time (ZTE) 19F MRI when sufficient fluorine is present.
In addition to 19F MRI/MRS detection, the inclusion of therapeutic capability with diagnostic visualization (Figure 1B) has led to the generation of a dual-purpose theranostic agent.61 Previous work has shown that the coiled-coil domain of our protein copolymer can bind an array of small molecules within its hydrophobic pore.5–7,62–66 This pore, along with the hydrophobic core of the assembled micelle, plays a role in binding doxorubicin (Dox), an anthracycline-class chemotherapeutic agent that has broad therapeutic efficacy against an array of tumor types.67 However, Dox also maintains profound off-target effects,68 spurring the need for research into Dox-carrying vehicles that provide controlled release.69–73 We anticipate that this work will encourage additional use of rational design principles in protein engineering to further exploit self-assembly for the controlled release of small-molecule therapeutics and the modification of relaxation properties in 19F MRI/MRS.
RESULTS AND DISCUSSION
Rationale and Protein Synthesis.
We have designed TRAP and F-TRAP to contain an N-terminal hexahistidine tag for purification purposes followed by an RGD motif present within an ARGD(AT)6 linker, a modified cartilage oligomeric matrix protein coiled-coil (C) domain,63 another ARGD(AT)6 linker, and an elastin-like polypeptide (E) region.5–7 While TRAP is a nonfluorinated protein, F-TRAP bears 5,5,5-DL-trifluoroleucines (TFLs), provided during protein expression as a racemic mixture of the two diastereoisomers (2S,4S)-5,5,5-trifluoroleucine and (2S,4R)-5,5,5-trifluoroleucine.4,8–12 The C domain is known to self-assemble into a parallel coiled-coil pentamer,11 while the E domain, composed of a [(VPGVG)2VPGFG(VPGVG)2]2 repeat,5–7 exhibits an inverse transition temperature (Tt) characterized by a random coil-to-β-spiral conformational change and subsequent chain association dependent on sequence composition.5–7,74–76
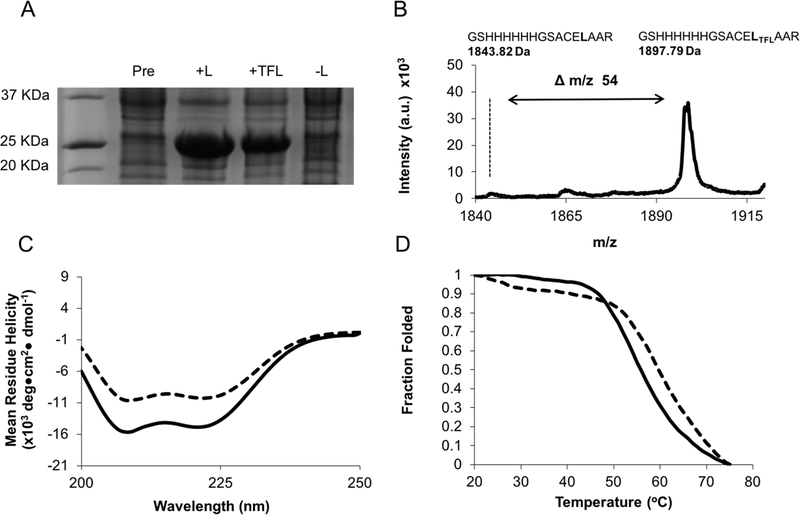
The TRAP plasmid construct was transformed into the leucine auxotrophic E. coli strain LAM1000 for recombinant expression and integration of TFL4,8–12 to generate F-TRAP (Figures 2A, S1, Table S1). Purified F-TRAP (Figure S1) was assessed for TFL incorporation using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) and amino acid analysis (AAA) (Figures 2B, S2, and S3, Table S2). MALDI-TOF MS and AAA confirmed TFL incorporation of 80.90 ± 1.02% and 81.36 ± 5.22%, respectively (Table S2).
Figure 2.
(A) SDS-PAGE of TRAP and F-TRAP overexpression in the presence and absence of L (leucine) or TFL (trifluoroleucine). (B) MALDI-TOF spectrum of tryptic fragment GSHHHHHHGSACELTFLAAR showing a 54 Da mass shift compared to wild-type TRAP. (C) Circular dichroism spectra of TRAP (solid line) and F-TRAP (dashed line) at 0.2 mg/mL and 20 °C. (D) Melt curves for TRAP (solid line) and F-TRAP (dashed line).
Secondary Structure and Thermostability.
Circular dichroism (CD) spectra acquired at 20 °C revealed a 30% loss in negative signal in F-TRAP by comparing the mean residue ellipticity values at 222 and 208 nm to TRAP (Figure 2C, Table S3). Moreover, F-TRAP (Tm = 59.3 ± 2.3 °C) possessed a 5.3 °C higher melting temperature than TRAP (Tm = 54.0 ± 2.0 °C), presumably due to the increased hydrophobicity of TFL9 (Figure 2D, Table S4). The K2D3 CD deconvolution software77 corroborated the differences in α-helicity at 20 °C and demonstrated an inversely proportional relationship between α-helical content and β-strand content as a function of temperature. This structural change was an expected result based on the Tt properties of the E domain6 (Figure S4).
Micellar Assembly and Stability.
A highly specific probe, 5-dodecanoyl amino fluorescein (5-daf), was used in fluorescence anisotropy experiments to accurately assess the critical micelle concentration (CMC).78,79 The 5-daf probe has two functional moieties: (i) a solvatochromic fluorescent component activated in the nonpolar micelle core and (ii) a long saturated aliphatic hydrocarbon chain that inserts slightly into the micelle corona.78 CMC experiments using 5-daf revealed CMC values of 0.59 ± 0.02 μM and 0.75 ± 0.11 μM for F-TRAP and TRAP, respectively (Table 1). The lower CMC value for F-TRAP could be indicative of either a more cooperative assembly of fluorinated protein-based micelles, a result shown in synthetic fluorinated micelles,80 or simply a probe-specific effect where the long aliphatic hydrocarbon tail of 5-daf fit tightly into the more hydrophobic fluorinated pore of F-TRAP.
Table 1.
Fluorescence Anisotropy and Static Light Scattering Data for F-TRAP and TRAP
| protein | CMCa (μM) | mol wtb (KDa) | Naggc (monomer) | Naggc (pentamer) |
|---|---|---|---|---|
| F-TRAP | 0.59 ± 0.02 | 717.09 ± 22.77 | 42.43 ± 1.34 | 8.48 ± 0.26 |
| TRAP | 0.75 ± 0.11 | 682.42 ± 4.77 | 42.12 ± 0.55 | 8.42 ± 0.11 |
Critical micelle concentration.
Data determined from Debye plot fit.
Aggregation number.
We further studied the assembly and stability of F-TRAP and TRAP as a function of temperature (Figure 3A,B) using the small molecular weight solvatochromic probe Nile red.81 Nile red demonstrates negligible fluorescence in aqueous solution, but shows increased fluorescence when bound within an assembling micellar core.81–83 An increase in fluorescence intensity was observed at 20 °C with the maximum fluorescence intensity at approximately 35 °C, followed by an emission decrease as the Tt was approached (Figure 3A,B). The increased fluorescent signal up to 35 °C was indicative of increased micelle assembly and, therefore, optimal Nile red uptake, while the subsequent signal decrease suggested micelle disassembly at higher temperatures (Figure 3A,B). Debye plots generated from static light scattering experiments revealed macromolecular F-TRAP assemblies measured at 717.09 ± 22.77 kDa, corresponding to 42.43 ± 1.34 monomeric units or 8.48 ± 0.26 pentameric units (Table 1). TRAP possessed a molecular weight of 682.42 ± 4.77 kDa and showed a similar monomer aggregation number of 42.12 ± 0.55 units and 8.42 ± 0.11 pentamer units (Table 1).
Figure 3.
Micelle stability of (A) F-TRAP and (B) TRAP plotted as a function of temperature and Nile red probe fluorescence. Particle diameters of (C) F-TRAP and (D) TRAP by dynamic light scattering at different concentrations and increasing temperature. (E) Inverse transition temperatures (Tt) of F-TRAP (dashed line) and TRAP (solid line) at different concentrations. Error bars represent the standard deviation of three trials.
The supramolecular assembly of the proteins was further assessed by dynamic light scattering (DLS) and UV-vis turbidometry analyses with particle morphologies confirmed by transmission electron microscopy (TEM) (Figures 3, S5, S6, S7, Tables S5, S6, S7, S8). As expected,7 at 20 °C, 0.5 mg/ mL F-TRAP assembled into nanoparticles 30.30 ± 0.60 nm in diameter as assessed by DLS, and spherical particles 33.78 ± 6.06 nm in diameter were observed via TEM (Figures 3C, S5, S6A, Tables S5, S7). At the same temperature, 0.5 mg/mL TRAP demonstrated slightly larger nanoparticles by DLS (32.14 ± 1.20 nm) with TEM results showing spherical particles of similar size to F-TRAP (34.96 ± 7.30 nm) (Figures 3D, S5, S6C, Tables S6, S8). Increasing F-TRAP and TRAP concentrations, to 0.75 and 1.0 mg/mL, showed a negligible difference in size via DLS at 20 °C (Tables S5, S6). As the temperature was increased to 50 °C, a dramatic change in size was observed by DLS with near-micrometer size aggregates of F-TRAP (952.06 ± 300.17 nm) and TRAP (1715.96 ± 912.70 nm) at concentrations ranging from 0.5 to 1.0 mg/mL (Figures 3C,D, Tables S5, S6). Proteins prepared at 0.5 mg/mL and 50 °C revealed aggregates on TEM similar in size to those observed by DLS, but with larger polydispersity (724.67–3057.73 nm) (Figure S6B,D, Tables S7, S8).84,85 The large aggregate size range was reflective of the stochastic nature of the coacervation. The Tt was known to decrease with increasing concentration,74,76 and here we observed this trend in both F-TRAP and TRAP, with fluorination imparting resistance to coacervation as the Tt was raised by 4.10 ± 0.43 °C (Figure 3E).
DLS was also employed to assess the z-average diameters of F-TRAP and TRAP over 24 h at both 35 °C, the temperature at which Nile red fluorescence studies revealed peak micelle formation (Figure S7A,B), and the hyperthermic temperature 42 °C86 (Figure S7C,D). Upon reaching 35 °C, F-TRAP and TRAP demonstrated similar z-average diameters among all three concentrations studied from 0.5 to 1.0 mg/mL with 28.64 ± 2.46 nm for F-TRAP (Figure S7A) and 24.19 ± 4.49 nm for TRAP (Figure S7B). At 35 °C held over 24 h, both F-TRAP and TRAP showed no statistically significant change in z-average diameter according to a paired two-tailed Student’s t-test between 0 and 24 h. These results suggest micelle stability of at least 24 h at 35 °C for both protein assemblies. By contrast, at 42 °C, both F-TRAP (Figure S7C) and TRAP (Figure S7D) exhibited an increase in size. The size of detectable protein aggregates peaked within the first 4 h at 42 °C for 0.75 and 1.0 mg/mL F-TRAP and then drastically reduced as aggregates precipitated out of solution. F-TRAP at 0.5 mg/mL, however, showed peak aggregation size after approximately 8 h at 42 °C before precipitating. TRAP samples showed peak aggregation at 8 h for all concentrations, with 0.75 and 1.0 mg/mL samples subsequently precipitating out of solution, while 0.5 mg/mL remained as large detectable aggregates. These time course studies suggested that F-TRAP and TRAP micelle assemblies were stable for at least 24 h under physiological conditions and again confirmed their capacity for temperature-stimulated coacervation at 42 °C.
Doxorubicin Encapsulation and Thermoresponsive Release.
On the basis of previous work, we hypothesized that the C domain of F-TRAP and TRAP would serve as a depot for binding and delivering small molecules.62,63,66,87 Here, we explored the binding capacity of F-TRAP and TRAP assemblies for Dox, aimed at controlling its release through thermosensitive coacervation of the protein. A 2:1 ratio of Dox:protein was used, and free Dox was removed by size exclusion chromatography (SEC). F-TRAP encapsulated 175% more Dox than TRAP (Table S9). In addition, comparison of the weighted Dox loading revealed 55% greater loading by F-TRAP compared to TRAP (Table S9), likely due to the increased hydrophobicity of the coiled-coil pore of F-TRAP upon incorporation of TFL.
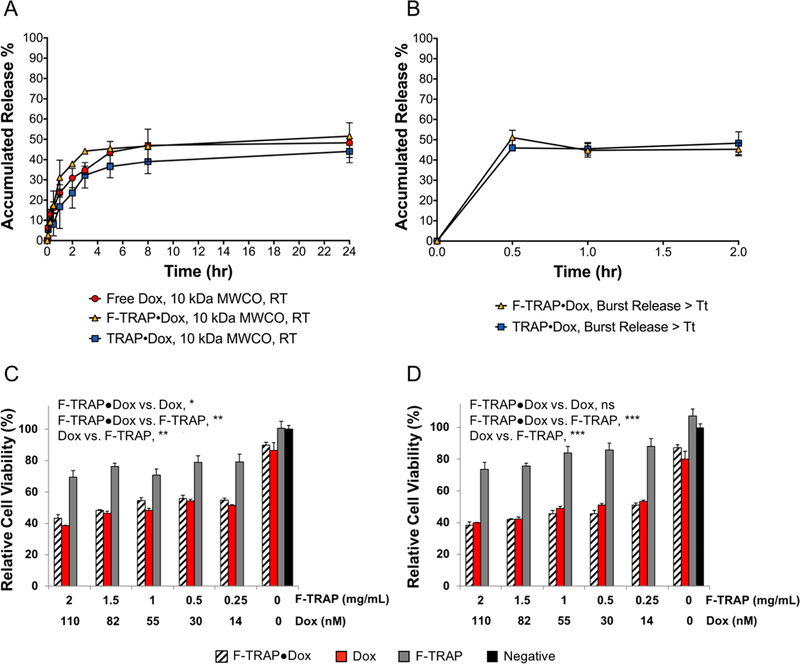
As both F-TRAP and TRAP were capable of binding Dox, we assessed the drug-carrying capabilities of these constructs. Initial experiments quantified the passive release of free Dox or Dox encapsulated by F-TRAP or TRAP at their corresponding loading efficiencies (Table S9) through a 10 kDa MWCO dialysis membrane at room temperature over 24 h (Figure 4A). The dialysis buffer was replaced every 1 h for up to 8 h to prevent the Dox concentration from reaching an equilibrium state. A slow release of Dox through the membrane was exhibited by free Dox, F-TRAP·Dox, and TRAP·Dox with 46.99 ± 7.99%, 46.68 ± 1.67%, and 38.99 ± 5.99% of Dox being collected at 8 h, respectively (Figure 4A). At 24 h, free Dox, F-TRAP·Dox, and TRAP·Dox had released 48.30 ± 9.83%, 51.58 ± 1.39%, and 44.06 ± 3.04% of the initial Dox concentration, respectively (Figure 4A).
Figure 4.
(A) Passive release of free base doxorubicin (Dox) at room temperature (RT) compared to F-TRAP- or TRAP-encapsulated Dox through 10 kDa MWCO dialysis membranes over 24 h. (B) Burst release for Dox from F-TRAP and TRAP at 45 °C (>Tt) over 2 h. MTT cell viability assay for F-TRAP·Dox, Dox alone, or F-TRAP alone at (C) 37 °C or (D) subjected to a 42 °C hyperthermic incubation prior to 37 °C. Symbols in panels C and D indicate the results of Tukey’s HSD test for multiple comparisons, applied to assess the significance between treatments on cell viability at each temperature over the range of concentrations studied, where ns = no significance, *p < 0.05, **p < 0.01, and ***p < 0.001. The test also assessed the significance of temperature on Dox and F-TRAP·Dox at 37 °C vs 42 °C, where Dox 37 °C vs Dox 42 °C = ns and F-TRAP·Dox 37 °C vs F-TRAP·Dox 42 °C = *.
To determine whether an increase in temperature could impact Dox release through thermally induced coacervation of the proteins in vitro, F-TRAP·Dox and TRAP·Dox were heated at 45 °C for 30 min, 1 h, or 2 h, and the supernatant was assessed for free Dox. By 30 min, Dox release by F-TRAP and TRAP peaked at 51.07% ± 3.60% and 45.96% ± 0.78%, respectively, without further release after 1–2 h of heating (Figure 4B). Therefore, both proteins demonstrated improved drug release at elevated temperature.
In vitro Therapeutic Delivery of Doxorubicin to MCF-7 Breast Adenocarcinoma Cells.
The release of Dox by F-TRAP and its therapeutic efficacy was assessed in mammalian tumor cells. MCF-7 human breast adenocarcinoma cells were subjected to treatment with free and F-TRAP-encapsulated Dox in addition to F-TRAP and buffer-only controls (Figure 4C,D). Treatment effects on cell viability were compared via Tukey’s honestly significant difference (HSD) test for multiple comparisons. MCF-7 cell viability at 37 °C was significantly reduced by both Dox and F-TRAP·Dox, ranging from 0 to 110 nM Dox, compared to F-TRAP alone (**, p < 0.01), but the overall effect of Dox on cell viability was significantly stronger than that of F-TRAP·Dox (*, p < 0.05). However, while a thermal onus of 42 °C for 1 h and subsequent incubation at 37 °C for 47 h did not affect cell viability under Dox treatment (p = 0.72), the hyperthermic state resulted in significantly reduced cell viability with F-TRAP·Dox treatment compared to F-TRAP·Dox at 37 °C (*, p < 0.05). This difference suggested that the therapeutic effect of F-TRAP·Dox could be improved under hyperthermic conditions. Again, both Dox and F-TRAP·Dox were significantly more effective at reducing cell viability than F-TRAP alone (***, p < 0.001).
19F NMR Detection and Relaxation Analysis.
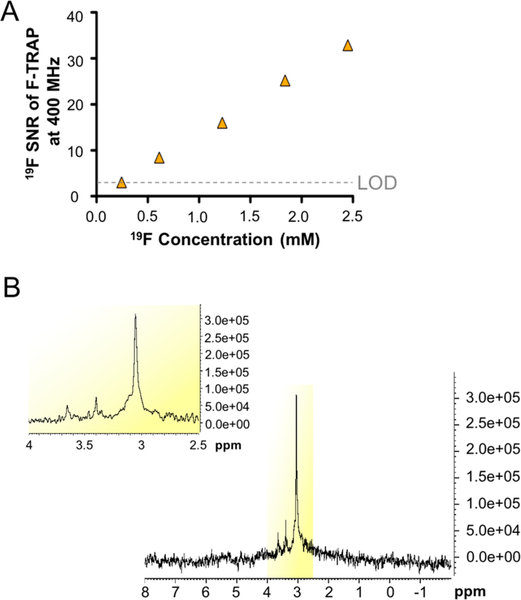
F-TRAP was further investigated as a potential dynamic probe for noninvasive 19F NMR, MRI, and MRS using radiofrequency (RF) probes tuned to a 19F Larmor frequency. The limit of detection (LOD)88–91 for 19F within F-TRAP was assessed using a 400 MHz (9.4 T) NMR spectrometer, with a spectral resolution of 1.36 Hz/pt (0.004 ppm/pt) (Figure 5A); an example 19F NMR spectrum was displayed at 1.125 mg/mL protein concentration (1.84 mM 19F) (Figure 5B). As demonstrated in the example spectrum (Figure 5B), all F-TRAP NMR spectra revealed three peaks spread over approximately 0.6 ppm. The main resonance peak, detected at 3.05 ppm, made up 82.19% of the signal contributed by the three peaks. A second peak, observed at 3.40 ppm, contributed 12.07% of the overall signal, and a third peak, at 3.65 ppm, contributed 5.74% of the signal. The SNRs of the main resonance peak at 3.05 ppm were calculated using the Bruker SINO command at 19F concentrations of 2.5 mM and lower until an SNR = 3 was reached, indicative of the LOD88 (Figure 5A). The LOD of F-TRAP at 400 MHz, acquired over 264 scans in 3 min 13 s, was reached at 0.25 mM 19F concentration (corresponding to 0.15 mg/mL protein), while the LOD of a commercially available perfluorocarbon nanoemulsion, V-Sense (Celsense), measured using its main resonance peak detected at −15.04 ppm, was found to be 100-fold lower than that of F-TRAP (Figure S8) at a concentration of 0.0025 mM 19F. Furthermore, the full width of the main resonance peak at half-maximum (fwhm) for F-TRAP and V-Sense were compared at 0.75 mM 19F concentration and found to be 21.14 Hz (0.056 ppm) and 31.60 Hz (0.084 ppm), respectively.
Figure 5.
(A) Signal-to-noise ratios (SNR) from 400 MHz one-dimensional 19F NMR spectra acquired over 264 scans in 3 min 13 s for F-TRAP diluted from 2.5 mM 19F to its limit of detection (LOD), at which SNR = 3,88 of 0.25 mM 19F. (B) Representative 400 MHz one-dimensional 19F NMR spectra of F-TRAP at 1.125 mg/mL protein (1.84 mM 19F), with the inset focused on the three peaks of the F-TRAP spectrum.
19F NMR spectra of F-TRAP at 1 mg/mL protein concentration (1.63 mM 19F) were then further explored at temperatures of 26.85, 31.85, and 37.0 °C and magnified within a 2 ppm range to compare the F-TRAP peaks (Figure S9). The main resonance peaks demonstrated a minor chemical shift of 0.03 ppm with increasing temperatures, resonating at 3.87, 3.88, and 3.90 ppm for 26.85, 31.85, and 37.0 °C, respectively.
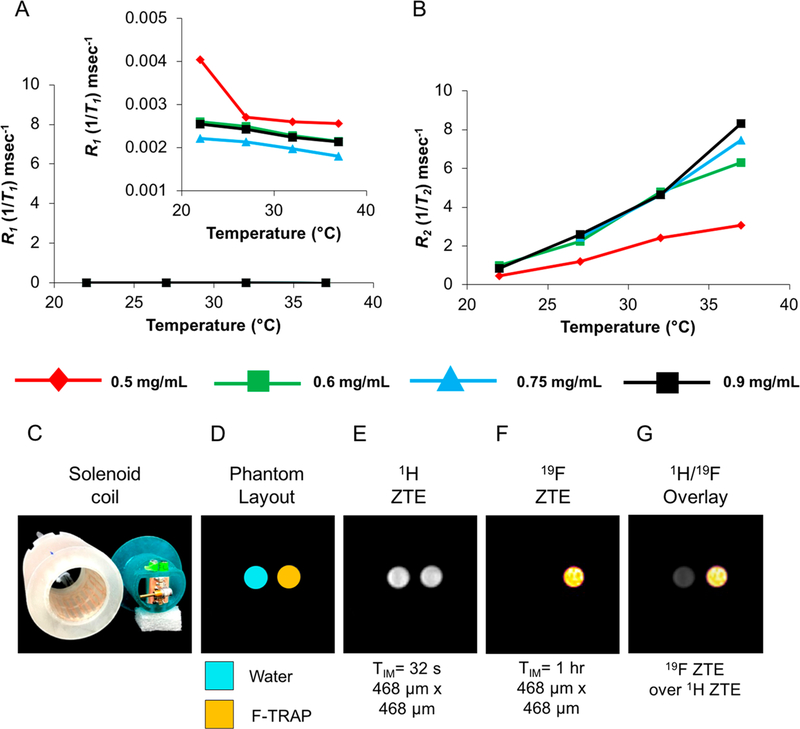
The longitudinal (R1 = 1/T1) and transverse (R2 = 1/T2) relaxation times of F-TRAP were assessed via inversion recovery and Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence experiments, respectively, via 19F NMR (Figure 6A,B). The relaxation experiments were performed to not only attain a deeper understanding of the macromolecular assembly of F-TRAP, but to also serve as an aid in selecting pulse sequence experiments for acquiring reliable MRI signals. Inversion recovery experiments, performed in predominantly water-based solvent systems, revealed a near-uniform resistance of the longitudinal relaxation rates (R1 = 1/T1) to both temperature and concentration increases, yielding values in the range of approximately 0.002–0.004−1 ms (Figure 6A, Table S10). The 19F spin-spin relaxation times, however, presented an interesting depiction of protein assembly as R2 (=1/T2) relaxation times increased with restricted motion of nuclei,92,93 a characteristic observed in synthetic micelles.92 This assembly-driven restriction of internal 19F atom mobility was further corroborated by the observed increase in R2/R1 relaxation rate (Tables S10, S11).52,92,93 The increase in R2 relaxation rates illustrated linearity while maintaining the same enhanced sensitivity when reaching the F-TRAP CMC (Figure 6B, Table S11). A thorough quantitative analysis revealed a 17000-fold increase in sensitivity when comparing the r2/r1 relaxivity values within the 22–37 °C temperature range (F-TRAP exhibited the following R1 and R2 relaxivity ranges [−0.09: −0.03] s−1/°C and [2009.16:17396.80] s−1/°C, respectively). Compared to the consistent temperature-dependent slope, the observed concentration effect is minor within the range of 0.6–0.9 mg/mL F-TRAP concentration (0.98–1.47 mM 19F, respectively) (Figure 6B). This temperature-dependent R2 increase could be exploited to monitor the accumulation of F-TRAP at a site of interest (e.g., tumor) and its response to external heating.
Figure 6.
(A) 19F R1 and (B) 19F R2 relaxometry plots. Inset in A shows R1 plot on a smaller scale. (C) Homemade whole body linear birdcage coil (left, in white) inductively coupled with a slotted resonator (right, in blue) capable of holding two glass tubes (o.d. 6 mm) and broadband tuning via a variable capacitor. (D) MRI phantom layout, (E) zero echo time (ZTE) imaging of 1H nuclei, (F) ZTE imaging of 19F nuclei, and (G) overlay of 1H and 19F ZTE. TIM = total acquisition time.
Phantom 19F MRI Detection.
Since R2 demonstrated a much greater linear dependence on temperature than R1, F-TRAP was deemed a likely candidate as a T2-MRI temperature sensor that would require strategies for spatial localization reliant on rapid acquisition of echo-trains and short repetition times in order to generate an appropriate SNR.94 Given that shortened T2 times restrict high signal intensity attainment and that imaging is further hampered by low 19F content, we acquired zero-echo time pulse sequence MRI on F-TRAP. ZTE MRI overcomes signal loss as a result of very short T2 relaxation times95,96 and is often used for the detection of tendons, cortical bone, and ligaments.97,98 A homemade whole body birdcage coil was inductively coupled to a slotted resonator for broadband tuning via a variable capacitor capable of housing two glass tubes (Figure 6C). Two samples, water as a control for 1H signal and F-TRAP (13.40 mg/mL or 0.82 mM protein, equivalent to 21.90 mM 19F) for the experimental 19F MRI signal, were scanned. The phantom samples for the water and F-TRAP (Figure 6D) both generated a positive signal for 1H nuclei (Figure 6E). The 19F signal from F-TRAP was examined using a ZTE MRI sequence while ensuring that the total imaging time was realistically achievable in vivo (in the present case: 1 h) (Figure 6F). In 1 h, ZTE imaging with a spatial resolution of (468 μm)2 achieved an SNR of 43 (Figure 6F,G). The corresponding limit of detection,89 at which the SNR = 3, of the ZTE sequence at 7 T under current experimental conditions was measured to be 0.91 mg/mL F-TRAP (1.49 mM 19F). 19F MRS, with a spectral resolution of 97.66 Hz/pt (0.35 ppm/pt), was also performed on the 13.40 mg/mL F-TRAP phantom sample in order to assess the difference in spectrum compared to that achieved via 19F NMR (Figure S10). Notably, only one peak was obtained by 19F MRS, in contrast to the three peaks observed on NMR. This F-TRAP peak resonated at 3.11 ppm (Figure S10), showing little shift from the 3.05 ppm main resonance peak observed on NMR (Figure 5B). However, the fwhm of the 19F MRS peak was 540.37 Hz (1.91 ppm), likely broadened by the lower static magnetic field homogeneity inherent to MRI scanners99 and resulting in a lower spectral resolution requirement. The width of this single peak encompassed the entire 0.6 ppm spread of the three F-TRAP peaks detected via 19F NMR.
In vivo Detection in an MCF-7 Xenograft Mouse Model.
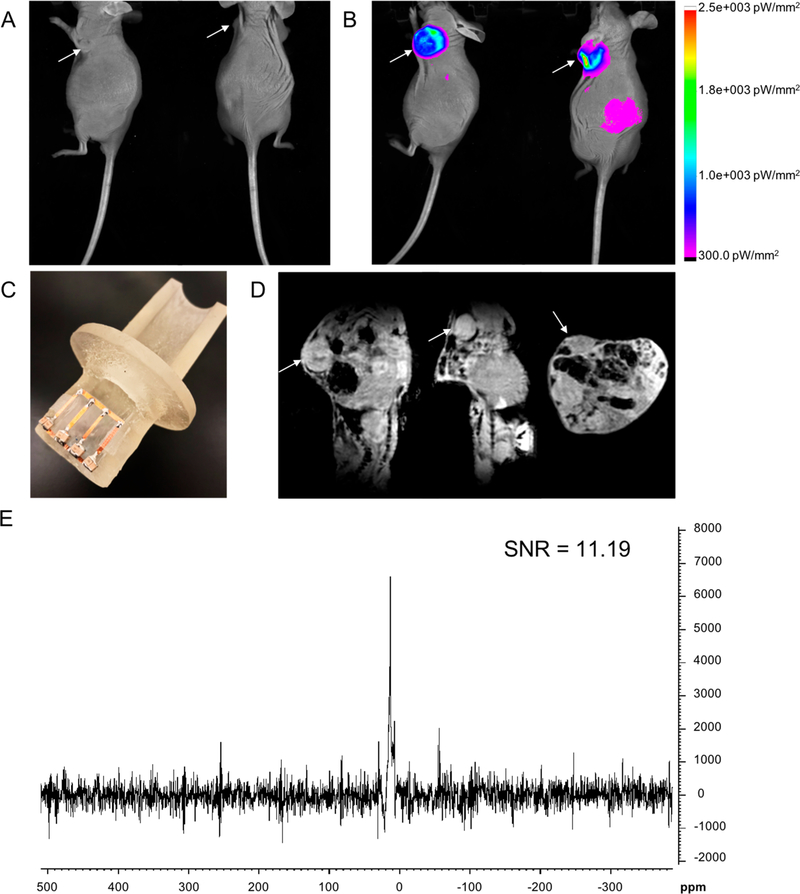
Following ultrasound imaging to confirm a tumor size of at least 100 mm3 in MCF-7 xenograft CrTac:NCr-Foxn1nu mice (Figure S11), in vivo detection of intratumorally injected near-infrared (NIR) fluorochrome-conjugated F-TRAP (NIR-F-TRAP) was assessed using fluorescence imaging and 19F MRS (Figure 7). The route of intratumoral injection was chosen as a preliminary assessment of the in vivo detection of F-TRAP by 19F MRS because it has previously proven successful for other elastin-like polypeptide-containing constructs, providing retention of the chemotherapeutic at the tumor site with minimal exposure to healthy tissues and is most often used in cases where surgery is not advised or when the tumor size must be reduced prior to surgery.100–105 Pre-injection fluorescence imaging overlaid onto an anatomical reflectance image revealed baseline fluorescence values that did not exceed 10 picowatts per mm2 (pW/mm2) (Figure 7A). Following the injection of 13.40 mg/mL NIR-F-TRAP (equivalent to 0.82 mM F-TRAP and 21.90 mM 19F), tumor fluorescence exhibited an intensity of up to 2.5 × 10003 pW/mm2 (Figure 7B), confirming successful intratumoral injection of NIR-F-TRAP. A 24 h post-injection dual fluorescence-reflectance image of the same subject showed the fluorescence signal had cleared from the tumor, with only a low level (∼300 pW/mm2) of fluorescence visible at the periphery of the tumor (Figure S12). Injected mice were followed for up to 1 week post-injection and showed no signs of acute toxicity.
Figure 7.
In vivo fluorescence-reflectance imaging of MCF-7 xenograft mouse model (A) pre-injection of NIR-F-TRAP and (B) immediately following intratumoral injection of NIR-F-TRAP. (C) Homemade animal holder attached to a 19F-tunable low-pass ladder surface coil for inductive coupling and fitting with the whole body coil described in Figure 6C. (D) Sagittal, coronal, and axial views from a 3D in vivo 1H T1-weighted FLASH MRI of a NIR-F-TRAP-injected mouse with FOV focused around the MCF-7 tumor. (E) In vivo 19F MRS, and corresponding SNR, of a NIR-F-TRAP-injected mouse acquired in 6 min 40 s.
Immediately following the initial post-injection fluorescence imaging, in vivo MR studies were conducted using a house-made setup composed of a 19F-tunable low-pass ladder surface coil inductively coupled to the same homemade whole body coil used for phantom imaging (Figure 7C), which was placed directly over the tumor. A low-resolution (234.375 μm)3 1H T1-weighted 3D fast low-angle shot (FLASH) MRI was used to identify the location of the tumor within the MRI field of view (FOV) and confirm the appropriate time window for 19F MRS acquisition (Figure 7D). The superficial tumor was identified in all three dimensions within the FOV (Figure 7D). In vivo 19F MRS, with a spectral resolution of 97.66 Hz/pt (0.35 ppm/ pt), was obtained from the entire tissue covered by the surface coil. Similar to the 19F MRS spectrum of the F-TRAP phantom (Figure S10), in vivo MRS of NIR-tagged and intratumorally injected NIR-F-TRAP revealed a single 19F peak (Figure 7E). The single peak resonated at 13.50 ppm with an SNR of 11.19 acquired over 200 scans in 6 min 40 s. The fwhm of the peak was 332.00 Hz (1.17 ppm), again encompassing the 0.6 ppm spread of the three F-TRAP peaks detected via 19F NMR.
CONCLUSIONS
We present the biosynthesis and characterization of protein-engineered block copolymers that assemble into micelles for 19F MRS and MRI. We employ ZTE MRI, an approach with stringent requirements, but one that enables near spin density images that align with the nanometer-sized micellar assembly properties of F-TRAP. 19F ZTE MRI was utilized as a proof-of-concept imaging sequence to demonstrate the ability to visualize F-TRAP within just 1 h acquisition time at 13.40 mg/mL F-TRAP (0.82 mM F-TRAP, 21.90 mM 19F). 19F MRS was able to detect intratumorally injected F-TRAP signal in a mouse model in under 7 min, allowing for rapid detection and monitoring of the agent in vivo. While the F-TRAP monomer bears a measured 26.77 19F atoms (1136 19F atoms per micelle), it demonstrates significant signal intensity via ZTE MRI despite the R2 alterations. Improvements in coil design and pulse calibration,106 or the use of more effective pulse sequence schemes,95,107 will improve our ability to detect F-TRAP in vivo using 19F ZTE MRI, allowing for further exploitation of F-TRAP’s temperature-dependent R2 sensitivity. By measuring the R2 values, after at least 0.6 mg/mL F-TRAP has accumulated, or the 19F MRS intensity after in vivo administration and imaging, we could determine the F-TRAP localization and concentration at specific organ sites. Furthermore, localized heating could increase this R2 effect, creating an on/off MRI switch. This T2-nanothermometer application is currently being explored, and initial experiments (Figures 6A,B, S13) suggest promise in monitoring F-TRAP’s theranostic drug thermorelease function. The theranostic aspect of F-TRAP is apparent when examining its ability to bind Dox, due to both the hydrophobic coiled-coil pore and micellar core, and its effective drug release to MCF-7 breast adenocarcinoma cells particularly under hyperthermic conditions. Promising in vivo 19F MRS studies and in vitro Dox delivery encourage future in vivo dual therapeutic-diagnostic studies under normal physiological and hyperthermic conditions to assess the theranostic potential of F-TRAP, including targeting through its RGD sequences or other tumor-targeting domains, by monitoring its in vivo drug release with simultaneous 19F MRI/MRS detection.
In conclusion, this work describes a biosynthesized fluorinated protein for MR detection that does not require post-translational modification. In terms of protein-based MRI contrast agent development, these results provide a template for rationally designing self-assembling biomaterials. The ability to design MR molecular probes at the genetic level allows for construct modification tailored to specific needs, such as targeting sequences unique to a tissue or cell type of interest, sequence mutations that alter the drug-loading capacity of the construct and stimuli-responsive cargo delivery, or an alteration in the number of potential sites for fluorinated residue incorporation.4–12 Protein engineering, therefore, has great potential in the development of fluorinated theranostic agents.
EXPERIMENTAL SECTION
Materials.
PfuUltra HotStart DNA polymerase and 10× PfuUltra HotStart reaction buffer were purchased from Agilent Technologies. DpnI enzyme was purchased from New England BioLabs. The miniprep plasmid purification kit was purchased from Zymogen. Deoxynucleotide triphosphates mix, sodium phosphate dibasic, sodium chloride, urea, imidazole, isopropyl β-D-1-thiogalactopyrano-side (IPTG), glucose monohydrate, tryptone, tryptic soy agar, yeast extract, ampicillin, kanamycin, magnesium chloride, calcium chloride, thiamine hydrochloride, HisPur Ni-NTA agarose, tricine, Pierce bicinchoninic acid (BCA) assay kit, Pierce snakeskin dialysis tubing 3.5 kDa molecular weight cut off (MWCO), Pierce high-capacity endotoxin removal spin columns, 5-dodecanoyl amino fluorescein, Vybrant MTT cell viability assay, methanol, and sodium dodecyl sulfate were acquired from Thermo Fisher Scientific. Racemic 5,5,5-trifluoroleucine was purchased from Oakwood Chemical. Nile red, acetonitrile, trifluoroacetic acid, all 20 amino acids, α-cyano-4-hydrocinnamic acid, sequencing grade modified trypsin, Proteomass peptide and protein MALDI-MS calibration kit, and β-estradiol were purchased from Sigma-Aldrich. Formvar square mesh copper TEM grids were purchased from Electron Microscopy Sciences. Macrosep and Microsep advance centrifugal 3 kDa MWCO devices and acrodisc 0.2 μm syringe filters were purchased from Pall. C18 packed zip-tips and Durapore 0.2 μm membrane filters were purchased from EMD Millipore. Acrylamide/bis solution (30%) 29:1, Precision Plus protein dual-color standards, and Gene Pulser electroporation cuvettes were purchased from Bio-Rad. Free base doxorubicin was purchased from MedKoo Sciences. VivoTag-S 750 was purchased from PerkinElmer. Corning Matrigel matrix was purchased from Corning. MCF-7 cells were purchased from ATCC, and CrTac:NCr-Foxn1nu nude mice were acquired from Taconic Biosciences.
TRAP and F-TRAP Construction and Production.
TRAP is derived from the CE2 parent construct that was previously generated and thoroughly characterized.5,6 Two RGD mutations located at sites flanking the C domain were introduced by site-directed mutagenesis in the pQE30/CE2 plasmid. Mutant constructs were generated by a polymerase chain reaction (PCR) using high-fidelity PfuUltra HotStart Taq polymerase (2.5 units/μL), 10× PfuUltra reaction buffer, deoxynucleotide triphosphates mix (10 mM), and the following primers (125 ng/μL) from MWG Operon: RGDlink1fwd 5′-GAGCTCGCTGCTCGTGGCGACGCCACTGCTACG-3′, RGDlink1rev 5′-CGTAGCAGTGGCGTCGCCACG-AGCAGCGAGCTC-3′, RGDlink2fwd 5′-CTGCAGGCTGCC-CGTGGCGACGCTACTGCAACC-3′, and RGDlink2rev 5′-GGTTGCAGTAGCGTCGCCACGGGCAGCCTGCAG-3′. After PCR, the methylated parent strand was digested for 3 h at 37 °C with DpnI enzyme. The resultant mutant constructs were transformed into chemically competent XL1-Blue E. coli cells and permitted to grow on tryptic soy agar plates for approximately 16 h at 37 °C under ampicillin (200 μg/mL) antibiotic selection control. DNA was extracted, and the resulting purified plasmid pQE30/TRAP was confirmed via sequencing (Eurofins Operon).
pQE30/TRAP plasmid was transformed into electrocompetent leucine auxotrophic LAM1000 E. coli cells,4,8–10,12 a gift from David Tirrell (California Institute of Technology) and permitted to grow on tryptic soy agar plates for approximately 12 h at 37 °C under both kanamycin (35 μg/mL) and ampicillin (200 μg/mL) antibiotic selection control. Single colonies were selected for growth in starter cultures (5 mL) of minimal media M9 (0.5 M Na2HPO4, 0.22 M KH2PO4, 0.08 M NaCl, 0.18 M NH4Cl) in the presence of kanamycin (35 μg/mL), ampicillin (200 μg/mL), vitamin B1 (35 μg/mL), MgSO4 (100 mM), CaCl2 (0.1 mM), and 20 amino acids (40 μg/mL) for approximately 16 h at 37 °C and 350 rpm. The starter cultures were then transferred to a large-scale expression flask (200 mL for TRAP, 500 mL for F-TRAP) at volumetric percentages of 2% containing the aforementioned M9 media and associated chemicals used for starter culture preparation. The large-scale cultures were grown at 37 °C and 300 rpm until the optical density at 600 nm reached 0.8–1.0. At this step, TRAP expression was induced with the addition of IPTG (200 μg/mL). Following 3 h of protein expression, at 37 °C and 300 rpm, the cells were pelleted by centrifugation at 4000 rpm for 15 min at 4 °C. The cell pellets were stored at −80 °C until purification. For F-TRAP expression, when the optical density at 600 nm reached 0.8–1.0, the cells were pelleted by centrifugation at 4000 rpm for 15 min at 4 °C and washed three times with ice cold 0.9% NaCl. After the final centrifugation, the pellet was resuspended in the aforementioned M9 media with 19 amino acids, minus leucine, and all other associated chemicals. The resuspended cell mixture was permitted to grow for 15 min at 37 °C and 300 rpm to exhaust any additional leucine. Following this growth period, TFL (555 μg/mL) and IPTG (200 μg/mL) were added to induce expression and residue-specific incorporation of TFL.9,10,108 After 3 h of protein expression at 37 °C and 300 rpm, the cells were pelleted by centrifugation at 4000 rpm for 15 min at 4 °C. The cell pellets were stored at −80 °C until purification.
Cell pellets were permitted to thaw at 4 °C for 30 min and then resuspended in lysis buffer (6 M urea, 50 mM Na2HPO4, 0.5 M NaCl, pH 8.0). Lysis was achieved by two freeze-thaw cycles between −80 °C and room temperature followed by ultrasonic probe sonication (Q500 sonicator, amplitude 50%, pulse 5 s on and 30 s off, in an ice water bath for a total sonication time of 1 min and 30 s) until cell lysate was visually transparent. The cell lysate was centrifuged at 13500 rpm for 1 h at 4 °C. The supernatant was then combined with HisPur Ni-NTA agarose slurry, pre-equilibrated with lysis buffer according to the manufacturer’s instructions, and left to incubate with end-over-end mixing (Labquake Rotisserie, Barnstead Thermolyne) overnight at 4 °C. The protein of interest was eluted by increasing elution buffer (6 M urea, 50 mM Na2HPO4, 1 M imidazole, pH 8.0) with effective imidazole concentrations ranging from 25 to 1000 mM. All fractions were subjected to 12% SDS-PAGE to confirm purity. Pure protein, typically in the elutions spanning 100–1000 mM imidazole, was then dialyzed in snakeskin tubing, 3.5 kDa MWCO, across four buckets (3.5 L each) of 50 mM Na2HPO4 and 0.5 M NaCl, pH 8.0 at 4 °C. Following dialysis, the protein was concentrated with a Macrosep Advance centrifugal device (3 kDa MWCO) at 3500 rpm at 4 °C. After 0.2 μm syringe filtration, the concentration of each protein was determined by BCA assay. Proteins were stored at 4 °C.
Confirmation of TRAP and F-TRAP Composition.
TRAP and F-TRAP samples at effective concentrations of 0.2 mg/mL were subjected to targeted enzymatic digestion with sequencing-grade modified trypsin (0.2 mg/mL, 2% v/v), pre-warmed to 30 °C for 15 min prior to addition, in 60 mM ammonium bicarbonate buffer for 6 h at 37 °C with 300 rpm mixing. Trypsin digestion was quenched with trifluoroacetic acid (TFA) (10% v/v in diH2O) until the pH reached approximately 4.0. Digested TRAP and F-TRAP were then subjected to zip-tip preparation using C18 packed tips. Tips were wetted in 50% v/v acetonitrile in deionized water (diH2O) and equilibrated in 0.1% v/v TFA in diH2O prior to protein binding and subsequent elution from the C18 column with 0.1% v/v TFA in 75% acetonitrile into the saturated α-cyano-4-hydrocinnamic acid matrix at 1:1 protein:matrix volumetric ratio. TRAP and F-TRAP samples were spotted in 1 μL volumes on a MALDI-TOF MS steel target plate and permitted to dry under vacuum for 1 h. A theoretical trypsin digestion of TRAP was performed with the online tool Expasy PeptideMass (http://web.expasy.org/peptidemass/ ) with adjustments made after theoretical digestion to account for size increase upon TFL substitution at all leucine sites for F-TRAP. The UltrafleXtreme MALDI-TOF (Bruker) was calibrated with the following standards: bradykinin fragment 1–7, angiotensin II, P14R (synthetic peptide), adrenocorticotropic hormone fragment 18–39, insulin chain B oxidized (bovine origin), and insulin (bovine origin), all at working solutions of 10 pmol/μL. Equal volumes of standards were collected in a single tube and combined in a 1:1 volumetric ratio with the α-cyano-4-hydrocinnamic acid matrix. The mass spectra were analyzed for TFL incorporation according to peak intensities. Amino acid analysis was also performed by John Schulze and the team at Molecular Structure Facility (via Science Exchange) at University of California, Davis, on the same proteins and analyzed on an L-8800 Hitachi analyzer, utilizing a sodium citrate buffer system.
Secondary Structure and Thermostability Analysis.
Circular dichroism spectroscopy was used to assess the secondary structure and thermostability of TRAP and F-TRAP. This was accomplished using a Jasco J-815 CD spectrometer with a PTC-423S single-position Peltier temperature control system. TRAP and F-TRAP proteins were diluted to 10 μM concentrations in 50 mM Na2HPO4 and 0.5 M NaCl, pH 8.0, and dispensed into quartz cuvettes. The wavelength spectrum was measured over the range of 200–250 nm with a bandwidth (step size) of 1 nm and path length of 1 cm.5–7,9 The scanning mode was continuous at 20 nm/min. All wavelength CD runs were conducted in triplicate at 20 °C. Data represent an average of three independent protein preparations. Temperature scans were performed at 222 nm from 20 to 75 °C, at a heating rate of 1 °C/min. The melting temperature (Tm) was determined by taking the first derivative of the normalized mean residue ellipticity as a function of temperature.5–7 The Tm was confirmed with additional temperature scans using protein concentrations of 5 and 15 μM so as to separate the constant Tm from the dynamic inverse transition temperature (Tt). Using spectral analysis software provided by the instrument manufacturer (Jasco), all spectra underwent a 50 mM Na2HPO4/0.5 M NaCl, pH 8.0, buffer spectrum subtraction followed by a smoothing operation according to the Savitzky-Golay method at a convolution width of 21.109 In terms of calculations for CD analysis, mean residue ellipticity (θMRE) was calculated from ellipticity (θ) using the following equation:63
Deconvolution of spectra at each temperature, accounting for buffer subtraction, smoothing, and mean residue ellipticity calculations, was carried out using K2D3 software (http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d3//index.html ) to determine the α-helix and β-strand composition.77
Turbidometry and Inverse Transition Temperature Analysis.
TRAP and F-TRAP at varying concentrations were loaded into a type 21 quartz cuvette with a 10 mm path length (BuckScience). Proteins were characterized for their inverse transition temperature (Tt) behavior at 320 nm using a UV-vis Cary-50 (Varian Inc.) equipped with a TC125 temperature regulator (Quantum North-west). Samples were heated at a rate of 1 °C/min from 20 to 80 °C. The Tt was determined by taking the first derivative of the normalized absorbance at 320 nm as a function of temperature. At least three separate protein preparations were subjected to UV-vis spectroscopy across all concentrations.
Self-Assembly and Analysis.
Fluorescence anisotropy experiments were carried out on a Molecular Devices Flexstation 3 plate reader equipped with the necessary hardware for fluorescence polarization measurements. 5-Dodecanoyl amino fluorescein (10 nM) dissolved in methanol was added to clear glass threaded vials and dried to a thin film under vacuum. Protein was then added to each tube at increasing concentrations, and the tubes were sealed tightly and gently agitated (80 rpm orbital mixing) at 25 °C for 24 h before measuring the fluorescence polarization (excitation 485 nm, emission 528 nm, cutoff 515 nm, G factor 1.0). The critical micelle concentration was determined to be the concentration at which fluorescence polarization increased dramatically.78 These experiments were conducted at least three times with two different protein preparations with the average CMC taken for all trials.
Fluorescence experiments were carried out to measure micelle stability as a function of temperature using a Biotek Synergy HT plate reader. Nile red (1 μM) dissolved in methanol was added to PCR tubes and dried to a thin film under vacuum. Protein was added to each tube at a static concentration of 0.5 mg/mL at 4 °C. Tubes were quickly transferred to a Bio-Rad thermocycler and then slowly heated (0.1 °C/s) with 10 min incubation periods at each measured temperature. At the conclusion of each incubation period, the contents of each tube were subjected to fluorescence measurements (excitation 550 nm, emission 635 nm). These experiments were conducted at least three times with two different protein preparations.
Static light scattering experiments were carried out using a Zetasizer Nano Series model Nano ZS90 (Malvern Instruments). Proteins ranging in concentration from 0.25 to 2.0 mg/mL were incubated at 35 °C for 24 h with mild agitation (80 rpm orbital mixing). After 24 h, the tubes were visually inspected for aggregation, and after confirming that coacervation had not occurred, the protein molecular weights were measured using Debye plot fitting of KC/Rθ (where K is the Debye constant, C is the concentration, and Rθ is the Rayleigh ratio) plotted against the concentration. The y intercept was determined to be the molecular weight.110 These experiments were conducted at least three times with two different protein preparations with the average molecular weights taken for all trials.
Dynamic light scattering measurements were performed on a Zetasizer ZS90 in a low-volume disposable cuvette with the applied settings: material protein (refractive index 1.450), absorption 0.001, dispersant phosphate buffer with a viscosity of 1.0200 cP, and refractive index of 1.335. Approximately 1.0 mL of 0.5, 0.75, or 1.0 mg/mL TRAP or F-TRAP in 50 mM Na2HPO4/0.5 M NaCl, pH 8.0, was double filtered (0.2 μm) to minimize contamination and then added to the cuvette for analysis. Three separate protein preparations were subjected to DLS measurements, and the average z-average diameters among the trials were plotted. To assess the effect of temperature on protein assembly, three independent measurements were taken, conducting 10 runs for each measurement (5 s per run), with a delay time of 2 s from 20 to 50 °C. Stability of the protein assembly was also investigated via DLS using the above parameters to measure the z-average diameters from 20 °C to either 35 or 42 °C, at which point measurements were taken every 30 min for 12 h and, subsequently, every 1 h through 24 h.
TEM experiments were undertaken to visualize and confirm particle assembly and sizes. The TRAP and F-TRAP proteins (0.5 mg/mL) were heated to 20 or 50 °C for 1.5 h followed by a 2 μL application of the protein onto a 400 square mesh copper TEM grid for 30 s. Excess protein was blotted from the grid with filter paper and then subjected to two washes with diH2O. Uranyl acetate (1% w/v) in a 1 μL volume was applied to the grid, quickly blotted, and followed by an additional 1 μL application of uranyl acetate. After 20 s, the excess uranyl acetate was blotted and permitted to air-dry until imaging. The TEM grids were imaged using a FEI Titan Halo 300 kV electron microscope at the Applied Science and Research Center (ASRC) at City University of New York (CUNY). These experiments were repeated using three grids per sample to confirm morphological characteristics.
Doxorubicin Binding and Release.
To investigate the potential of F-TRAP as a drug delivery vehicle, TRAP and F-TRAP were incubated with free base Dox for assessment of loading efficiency and release. To determine the encapsulation efficiency of TRAP and F-TRAP, Dox was dissolved in DMSO and added to TRAP or F-TRAP at a 2:1 molar ratio of drug to protein (100 μM Dox:50 μM protein) in an amber tube and incubated at 4 °C for 12 h. Free Dox was removed via SEC using a G-25 medium Sephadex resin. The resin was hydrated in diH2O overnight at room temperature before being added to a column (0.5 cm internal diameter, 15 cm length). The resin was equilibrated in degassed mobile phase buffer (50 mM Na2HPO4/0.5 M NaCl, pH 8.0) followed by application of the protein-drug complex. The mobile phase buffer was constantly added to maintain continuous flow and elution from the column. Elutions (1 mL each) were collected for absorbance readings of Dox at 485 nm.111 Elution absorbances were compared to those of a standard curve of known Dox concentrations prepared in 50 mM Na2HPO4/0.5 M NaCl, pH 8.0, to determine Dox concentrations. Elution fractions with the highest absorbance values were also subjected to an enhanced BCA assay for assessment of protein concentration. The loading efficiency of Dox by TRAP and F-TRAP was calculated from the amount of Dox quantified in SEC elutions using the following equation:112
The weighted loading was calculated as μg Dox per mg protein.113
Passive, room-temperature, release of Dox by TRAP and F-TRAP was assessed following passive loading of Dox at the corresponding loading efficiencies, determined by SEC studies, overnight. The protein-drug complex, or free base Dox alone as a control, was added to a 10 kDa MWCO dialysis membrane and suspended in 50 mM Na2HPO4/0.5 M NaCl, pH 8.0, at room temperature, with mixing at 80 rpm. Over a period of 24 h, samples were subjected to absorbance readings at 485 nm to measure the concentration of Dox remaining compared to a standard curve of known Dox concentrations also prepared in 50 mM Na2HPO4/0.5 M NaCl, pH 8.0. Released Dox was plotted as an accumulated percentage of the original amount of Dox loaded. The dialysis buffer outside of the membrane was replaced hourly for the first 8 h to spur drug release.
Dox release was further investigated under hyperthermic conditions in which TRAP or F-TRAP-bound Dox complexes were added to amber tubes in triplicate and suspended in a 45 °C water bath.114 At 30 min, 1 h, and 2 h, one tube was removed from the water bath and centrifuged for 5 min at 13200 rpm to pellet the coacervate protein. The supernatant was subjected to absorbance readings at 485 nm.114 The concentration of released Dox was compared to a standard curve of known Dox concentrations, and the amount of Dox released was plotted as an accumulated percentage of the original amount of Dox loaded.
Cell Viability Studies.
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell viability assays were conducted to assess the efficacy of free and F-TRAP-delivered Dox on the MCF-7 breast adenocarcinoma cell line.115 MCF-7 cells were seeded into a 96-well plate at a density of 10000 cells/well in minimum essential media (MEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 0.01 mg mL−1 human recombinant insulin and incubated overnight at 37 °C with 5% CO2. MEM was subsequently removed, and 90 μL of fresh MEM was added per well. Cells were treated with 10 μL of Dox, F-TRAP, or Dox-loaded F-TRAP (F-TRAP·Dox) in quadruplicates at increasing concentrations with a static final DMSO concentration of 0.55% v/v. Treated cells were incubated either for 48 h at 37 °C with 5% CO2 or for 1 h in a 42 °C water bath followed by 37 °C with 5% CO2 for 47 h. After treatment, the media was removed; the cells were rinsed once with 1× PBS, and 100 μL of fresh MEM was added per well. To assess the cytotoxicity of Dox and the Dox-loaded protein complexes, 10 μL of 5 mg/mL MTT stock in 1× PBS was added per well and incubated for 4 h at 37 °C with 5% CO2. Following incubation, all but 25 μL of the medium was removed per well and 50 μL of DMSO was added per the manufacturer’s protocol. Cells were incubated for 10 min at 37 °C with 5% CO2. The absorbance of each well was assessed using a Biotek Synergy HT microplate reader at 540 nm. The cell viability of treated cells was calculated as a percentage normalized to untreated cell controls. Data represent the average of four replicates per sample per trial, and error bars represent the standard deviations.
Nuclear Magnetic Resonance Measurements.
Limit of 19F detection studies were conducted on a Bruker AVIII-400 (9.4 T) instrument equipped with a BB(F)O probehead. F-TRAP sensitivity was compared to the commercially available 19F nanoemulsion V-Sense (Celsense) using a 19F one-pulse sequence with the following parameters: echo time (TE) = 300 ms, flip angle (FA) = 90°, sweep width (SW) = 89.285 kHz, acquisition time = 0.73 s, with number of scans (NS) =264, resulting in an experiment time TIM = 3 min 13 s. The 1D 19F NMR spectra of F-TRAP and V-sense at 19F concentrations ranging from 0.25 to 2.5 mM and 0.0025 to 1 mM, respectively, were acquired. A standard composed of 10% TFA/10% D2O/80% H2O, acquired with the same sequence, was used to calibrate all spectra in Topspin 3.2 software. Topspin 3.2 was further used to quantify SNRs, calculated via the Bruker SINO command to identify the LOD, the concentration at which the SNR is equal to 3.88–91 The Bruker PEAKW command was employed to determine the fwhm for F-TRAP and V-Sense. The intensities of the F-TRAP peaks were quantified using Bruker’s integration function. 1D 19F NMR spectra of F-TRAP samples at 1 mg/mL protein concentration were also obtained as a function of temperature at 26.85 °C (300 K), 31.85 °C (305 K), and 37.0 °C (310.15 K), each calibrated to a 10% trifluoroethanol/10% D2O/80% H2O standard acquired at the corresponding temperature using the identical sequence. Both the F-TRAP sample and calibration standard were acquired using the above-described 19F one-pulse sequence. Topspin 3.2 software was used to overlay the spectra acquired at variable temperatures.
The relaxation properties of F-TRAP were obtained as a function of temperature on a 500 MHz (11.7 T) Bruker Avance II instrument equipped with a 5 mm room-temperature BB(F)O SMART probe. Both the inversion recovery and CPMG 19F NMR experiments58 were performed on F-TRAP at 0.5, 0.6, 0.75, and 0.9 mg/mL (corresponding to 0.82 mM 19F, 0.98 mM 19F, 1.23 mM 19F, and 1.47 mM 19F) at varying temperatures (21.85, 26.85, 31.85, 37 °C). Inversion recovery experiments were used to determine the T1 relaxation times with an SW of 38 311 Hz (corresponding to 76.6219 ppm) and delay time of 2.0 s with 1024 scans at each of the following variable delay times: 0.001, 0.05, 0.1, 0.2, 0.3, 0.5, 0.8, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 4.5, 5.0 s. CPMG experiments were used to determine the T2 relaxation times with an SW of 38 311 Hz (corresponding to 76.6219 ppm) and delay time of 2.0 and 1024 scans at each of the following variable counter times: 2, 20, 50, 100, 200, 400, 600, 800, 1000, 1400, 1600, 1800, 2500 μs. The relaxation times (T1 and T2) were calculated by using curve fitting analysis in Topspin 3.2 software.
Phantom Magnetic Resonance Experiments.
Magnetic resonance imaging was conducted using 1H nuclei to identify all samples containing water and using 19F nuclei to locate and quantify the sample containing 19F-TRAP. All MRI experiments were performed on a Biospec 7030 micro-MRI system (Bruker) composed of an Avance-3 HD console and a zero-boil-off 7 T (300 MHz) 300 mm horizontal bore magnet equipped with an actively shielded gradient coil insert (Bruker BGA-12S-HP; i.d. 114 mm, 660 mT/m gradient strength, 130 μs rise time). The MRI experiments were performed with a house-made whole body birdcage coil inductively coupled to a slotted resonator, capable of holding two glass tubes (o.d. 6 mm) and broadband tuning via a variable capacitor to cover both Larmor frequencies of 1H proton (300.16 MHz) and 19F (282.4 MHz) at 7 T (Figure 6C). The coil was first tuned and matched for 1H for rapid scout scans due to the greater proton signal sensitivity, which was used as a spatial reference for 19F scans.
ZTE imaging was employed for 1H and 19F imaging (repetition time (TR) = 2.5 ms, TE = 20 μs, FA = 4°, SW = 100 kHz, NS = 128, resulting in 781 Hz/pixel) with the number of repetitions (NR) = 1 and TIM = 32 s for 1H imaging and NR = 110 and TIM = 1 h for 19F imaging. The ZTE imaging sequence is based on a 3D radial acquisition method for which center-out readouts are performed in the presence of a constant gradient in a steady state of the magnetization. A nonselective excitation with very short RF pulse was employed in order to capture signals with extremely short T2 values (TE = 20 μs), while a very small flip angle (FA = 4°) in combination with a very short repetition time (TR = 2.5 ms) was used to achieve efficient SNR scanning. Despite the apparent high duty cycle, the very short RF excitation pulse, in combination with the low flip angle used, prevented any RF-induced heating effect as assessed by the continuous monitoring of the temperature of the sample imaged.
For imaging experiments, a set of solutions with concentrations ranging from 5 to 13.40 mg/mL F-TRAP (equivalent to 8.17 mM 19F and 21.90 mM 19F, respectively), at a total volume of ∼500 μL, and a negative control volume of water were placed in separate 5 mm NMR tubes. The temperature of the 5 mm glass tubes containing the F-TRAP solutions was systematically and continuously monitored during the MRI scans via a thermocouple probe implanted at the tip of the NMR glass tube across the rubber cap to ensure proper sealing. No temperature changes that may be induced by RF heating were observed during the MRI scans.
The 13.40 mg/mL F-TRAP phantom sample was also acquired using a 19F MRS sequence on the same 7 T (300 MHz) system (TR = 2000 ms, NS = 200, FA = 90°, TIM = 6 min 40 s). The MRS spectra was calibrated in Topspin 3.2 software to a standard composed of 10% TFA/90% H2O acquired with the same sequence.
Endotoxin Removal.
Pierce high-capacity endotoxin removal spin columns were used to remove endotoxins from the F-TRAP sample to be studied in vivo. After regeneration with 0.2 N NaOH, endotoxin removal resin was washed using 2 M NaCl solution, followed by endotoxin-free, ultrapure water. The resin was then equilibrated with endotoxin-free 1× PBS, pH 7.4, for a total of three times. The protein sample at a concentration of 2.7 mg/mL was added to the resin and incubated for 1 h at 4 °C. The sample was eluted in a sterile 15 mL tube via centrifugation at 500g for 1 min.
Near-Infrared Fluorochrome Conjugation.
Endotoxin-free F-TRAP was subsequently diluted in endotoxin-free 50 mM carbonate/bicarbonate buffer, pH 8.5, to 1 mg/mL and conjugated to the NIR fluorochrome VivoTag-S 750 (PerkinElmer) at room temperature for 1 h using end-over-end mixing (Labquake Rotisserie, Barnstead Thermolyne). Nonconjugated fluorochrome was removed using an Amicon Ultra-15 centrifugal filter 3 kDa MWCO, while buffer exchanging fluorochrome-conjugated F-TRAP (NIR-F-TRAP) into endotoxin-free 1× PBS, pH 7.4, and concentrating to 13.40 mg/mL.
In vivo Experiments.
The use of F-TRAP for in vivo tumor imaging was assessed in an MCF-7 xenograft mouse model using optical imaging and magnetic resonance imaging. Animal work was performed in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and using procedures approved by the Institutional Animal Care and Use Committee at the New York University School of Medicine. At 4–6 weeks old, female CrTac:NCr-Foxn1nu nude mice acquired from Taconic Biosciences were injected with 3 × 106 MCF-7 cells embedded in 150 μL Corning Matrigel matrix. One week after MCF-7 cell injection, mice were injected over the right scapula with Matrigel embedded with 100 μg of β-estradiol twice a week. Tumors were permitted to grow until a 5 mm diameter, or 100 mm3 volume, was reached as measured via high-frequency ultrasound (Fujifilm Vevo 3100 VisualSonic Ultrasound) prior to ultrasound-guided intratumoral injection of endotoxin-free NIR-F-TRAP. Mice were anesthetized with a cocktail of ketamine (100 mg/mL)/xylazine (20 mg/mL) at 0.1 mL/g mouse weight. Prior to injecting NIR-F-TRAP,mice were subjected to baseline in vivo multispectral vis-NIR fluorescence imaging overlaid onto an anatomical reflectance image on a Bruker In-Vivo Xtreme, with an excitation of 730 nm and emission acquisition at 790 nm over 30 s exposure. Mice were subsequently injected with 50 μL of 13.40 mg/mL NIR-F-TRAP (equivalent to 0.82 mM F-TRAP and 21.90 mM 19F) intratumorally under high-frequency ultrasound using a microfine 26 gauge 1/2 in. needle and immediately reimaged using dual fluorescence-reflectance imaging. A 24 h post-injection fluorescence-reflectance image was also acquired to confirm clearance of the construct. All fluorescence images were presented on the same intensity scale provided in picowatts per mm2.
In vivo 1H MR imaging and 19F MRS of NIR-F-TRAP were conducted immediately following fluorescence-reflectance imaging using a house-made setup composed of a 19F-tunable house-made low-pass ladder surface coil inductively coupled to the same homemade birdcage coil used for phantom imaging, which was placed directly over the tumor. The tumor was visualized using a standard 1H T1-weighted 3D FLASH imaging sequence (TR = 610.94 ms, TE = 2 ms, FA = 30°, matrix = (256 mm)3, resolution = (234.375 μm)3, SW = 67 kHz, TIM = 1 min, 18 s), and NIR-F-TRAP injected into the tumor was detected using a 19F MRS sequence (TR = 2000 ms, NS = 200, FA = 90°, TIM = 6 min 40 s), which maintained a higher sensitivity than was obtainable with 19F MR imaging. The MRS spectra were calibrated in Topspin 3.2 software to a standard composed of 10% TFA/90% H2O acquired with the same sequence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NSF-MRSEC Program under Award Number DMR 1420073, NSF-DMREF under Award Number DMR 1728858, the NYU Shiffrin-Myers Breast Cancer Discovery Fund, and the NYU CTSA grant UL1 TR000038 from the National Center for Advancing Transla-tional Sciences, National Institutes of Health. Some of the experiments were performed at the NYU Langone Health Preclinical Imaging Laboratory, a shared resource partially supported by the NIH/SIG 1S10OD018337-01, the Laura and Isaac Perlmutter Cancer Center Support Grant NIH/NCI 5P30CA016087, and the NIBIB Biomedical Technology Resource Center Grant NIH P41 EB017183. The authors thank J. Schulze at the Molecular Structure Facility at UC Davis for assistance in AAA, C. Lin for his guidance with 19F NMR and subsequent analysis, J. Baquero Buitrago for supplying us with the MCF-7 xenograft CrTac:NCr-Foxn1nu mice, and O. Aristizábal for his assistance with ultrasound-guided intratumoral injection and training on the Bruker In vivo Xtreme II.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.8b07481.
Supporting figures and tables related to MALDI-TOF MS, protein expression yields, protein purification, turbidity, CD deconvolution, TEM, DLS, NMR, MRS, MRI, ultrasound, vis-NIR fluorescence and reflectance imaging, and doxorubicin loading efficiency (PDF)
The authors declare no competing financial interest.
NOTE ADDED AFTER ASAP PUBLICATION
This paper published ASAP on 2/19/2019. Due to a production error, “molar residue”‘ was corrected to “mean residue”, and the revised version was reposted on 2/20/2019.
REFERENCES
- (1).Deans AE; Wadghiri YZ; Bernas LM; Yu X; Rutt BK; Turnbull DH Cellular MRI Contrast via Coexpression of Transferrin Receptor and Ferritin. Magn. Reson. Med. 2006, 56, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Korkusuz H; Ulbrich K; Welzel K; Koeberle V; Watcharin W; Bahr U; Chernikov V; Knobloch T; Petersen S; Huebner F; Ackermann H; Gelperina S; Kromen W; Hammerstingl R; Haupenthal J; Gruenwald F; Fiehler J; Zeuzem S; Kreuter J; Vogl TJ; Piiper A Transferrin-Coated Gadolinium Nanoparticles as MRI Contrast Agent. Mol. Imaging Biol. 2013, 15, 148–154. [DOI] [PubMed] [Google Scholar]
- (3).Kremer S; Lamy J; Magnus A; Oesterle H; Jeantroux J; Trunet S; Armspach J-P; Dietemann J-L; de Sèze J Evaluation of an Albumin-Binding Gadolinium Contrast Agent in Multiple Sclerosis. Neurology 2013, 81, 206–210. [DOI] [PubMed] [Google Scholar]
- (4).Montclare JK; Tirrell DA Evolving Proteins of Novel Composition. Angew. Chem., Int. Ed. 2006, 45, 4518–4521. [DOI] [PubMed] [Google Scholar]
- (5).Haghpanah JS; Yuvienco C; Civay DE; Barra H; Baker PJ; Khapli S; Voloshchuk N; Gunasekar SK; Muthukumar M; Montclare JK Artificial Protein Block Copolymers Blocks Comprising Two Distinct Self-Assembling Domains. ChemBioChem 2009, 10, 2733–2735. [DOI] [PubMed] [Google Scholar]
- (6).Dai M; Haghpanah J; Singh N; Roth EW; Liang A; Tu RS; Montclare JK Artificial Protein Block Polymer Libraries Bearing Two SADs: Effects of Elastin Domain Repeats. Biomacromolecules 2011, 12, 4240–4246. [DOI] [PubMed] [Google Scholar]
- (7).Yuvienco C; More HT; Haghpanah JS; Tu RS; Montclare JK Modulating Supramolecular Assemblies and Mechanical Properties of Engineered Protein Materials by Fluorinated Amino Acids. Biomacromolecules 2012, 13, 2273–2278. [DOI] [PubMed] [Google Scholar]
- (8).Montclare JK; Son S; Clark GA; Kumar K; Tirrell DA Biosynthesis and Stability of Coiled-Coil Peptides Containing (2S,4R)-5,5,5-Trifluoroleucine and (2S,4S)-5,5,5-Trifluoroleucine. ChemBioChem 2009, 10, 84–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).More HT; Zhang KS; Srivastava N; Frezzo JA; Montclare JK Influence of Fluorination on Protein-Engineered Coiled-Coil Fibers. Biomacromolecules 2015, 16, 1210–1217. [DOI] [PubMed] [Google Scholar]
- (10).Panchenko T; Zhu WW; Montclare JK Influence of Global Fluorination on Chloramphenicol Acetyltransferase Activity and Stability. Biotechnol. Bioeng. 2006, 94, 921–930. [DOI] [PubMed] [Google Scholar]
- (11).Voloshchuk N; Montclare JK Incorporation of Unnatural Amino Acids for Synthetic Biology. Mol. BioSyst. 2010, 6, 65–80. [DOI] [PubMed] [Google Scholar]
- (12).Voloshchuk N; Lee MX; Zhu WW; Tanrikulu IC; Montclare JK Fluorinated Chloramphenicol Acetyltransferase Thermostability and Activity Profile: Improved Thermostability by a Single-Isoleucine Mutant. Bioorg. Med. Chem. Lett. 2007, 17, 5907–5911. [DOI] [PubMed] [Google Scholar]
- (13).Yin L; Yuvienco C; Montclare JK Protein Based Therapeutic Delivery Agents: Contemporary Developments and Challenges. Biomaterials 2017, 134, 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).van Zijl PCM; Yadav NN Chemical Exchange Saturation Transfer (CEST): What Is in a Name and What Isn’t? Magn. Reson. Med. 2011, 65, 927–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Schmiedl U; Ogan M; Paajanen H; Marotti M; Crooks LE; Brito AC; Brasch RC Albumin Labeled with Gd-DTPA as an Intravascular, Blood Pool-Enhancing Agent for MR Imaging: Biodistribution and Imaging Studies. Radiology 1987, 162, 205–210. [DOI] [PubMed] [Google Scholar]
- (16).Bogdanov AA; Weissleder R; Frank HW; Bogdanova AV; Nossif N; Schaffer BK; Tsai E; Papisov MI; Brady TJ A New Macromolecule as a Contrast Agent for MR Angiography: Preparation, Properties, and Animal Studies. Radiology 1993, 187, 701–706. [DOI] [PubMed] [Google Scholar]
- (17).Wadghiri YZ; Sigurdsson EM; Sadowski M; Elliott JI; Li Y; Scholtzova H; Tang CY; Aguinaldo G; Pappolla M; Duff K; Wisniewski T; Turnbull DH Detection of Alzheimer’s Amyloid in Transgenic Mice Using Magnetic Resonance Microimaging. Magn. Reson. Med. 2003, 50, 293–302. [DOI] [PubMed] [Google Scholar]
- (18).Artemov D; Mori N; Ravi R; Bhujwalla ZM Magnetic Resonance Molecular Imaging of the HER-2/Neu Receptor. Cancer Res. 2003, 63, 2723–2727. [PubMed] [Google Scholar]
- (19).Mulder WJM; Strijkers GJ; Habets JW; Bleeker EJW; van der Schaft DWJ; Storm G; Koning GA; Griffioen AW; Nicolay K MR Molecular Imaging and Fluorescence Microscopy for Identification of Activated Tumor Endothelium Using a Bimodal Lipidic Nanoparticle. FASEB J. 2005, 19, 2008–2010. [DOI] [PubMed] [Google Scholar]
- (20).Wadghiri YZ; Li J; Wang J; Hoang DM; Sun Y; Xu H; Tsui W; Li Y; Boutajangout A; Wang A; de Leon M; Wisniewski T Detection of Amyloid Plaques Targeted by Bifunc-tional USPIO in Alzheimer’s Disease Transgenic Mice Using Magnetic Resonance Microimaging. PLoS One 2013, 8, No. e57097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Marsh ENG; Suzuki Y Using (19)F NMR to Probe Biological Interactions of Proteins and Peptides. ACS Chem. Biol. 2014, 9, 1242–1250. [DOI] [PubMed] [Google Scholar]
- (22).Yu J-X; Hallac RR; Chiguru S; Mason RP New Frontiers and Developing Applications in 19F NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 70, 25–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Srinivas M; Boehm-Sturm P; Figdor CG; de Vries IJ; Hoehn M Labeling Cells for In vivo Tracking Using (19)F MRI. Biomaterials 2012, 33, 8830–8840. [DOI] [PubMed] [Google Scholar]
- (24).Ahrens ET; Bulte JWM Tracking Immune Cells In vivo Using Magnetic Resonance Imaging. Nat. Rev. Immunol. 2013, 13, 755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ratner AV; Quay S; Muller HH; Simpson BB 19F Relaxation Rate Enhancement and Frequency Shift with Gd-DTPA. Invest. Radiol. 1989, 24, 224–227. [DOI] [PubMed] [Google Scholar]
- (26).Fluorine Magnetic Resonance Imaging; Flögel U; Ahrens E, Eds.; Pan Stanford: New York, 2016. [Google Scholar]
- (27).Flögel U; Ding Z; Hardung H; Jander S; Reichmann G; Jacoby C; Schubert R; Schrader J In vivo Monitoring of Inflammation after Cardiac and Cerebral Ischemia by Fluorine Magnetic Resonance Imaging. Circulation 2008, 118, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Janjic JM; Srinivas M; Kadayakkara DKK; Ahrens ET Self-Delivering Nanoemulsions for Dual Fluorine-19 MRI and Fluorescence Detection. J. Am. Chem. Soc. 2008, 130, 2832–2841. [DOI] [PubMed] [Google Scholar]
- (29).Ruiz-Cabello J; Walczak P; Kedziorek DA; Chacko VP; Schmieder AH; Wickline SA; Lanza GM; Bulte JWM In vivo “Hot Spot” MR Imaging of Neural Stem Cells Using Fluorinated Nanoparticles. Magn. Reson. Med. 2008, 60, 1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Moonshi SS; Zhang C; Peng H; Puttick S; Rose S; Fisk NM; Bhakoo K; Stringer BW; Qiao GG; Gurr PA; Whittaker AK A Unique 19F MRI Agent for the Tracking of Non Phagocytic Cells In Vivo. Nanoscale 2018, 10, 8226–8239. [DOI] [PubMed] [Google Scholar]
- (31).Yu M; Bouley BS; Xie D; Enriquez JS; Que EL 19F PARASHIFT Probes for Magnetic Resonance Detection of H2O2 and Peroxidase Activity. J. Am. Chem. Soc. 2018, 140, 10546–10552. [DOI] [PubMed] [Google Scholar]
- (32).Guo C; Xu S; Arshad A; Wang L A PH-Responsive Nanoprobe for Turn-on 19F-Magnetic Resonance Imaging. Chem. Commun. 2018, 54, 9853–9856. [DOI] [PubMed] [Google Scholar]
- (33).Zhang C; Moonshi SS; Wang W; Ta HT; Han Y; Han FY; Peng H; Král P; Rolfe BE; Gooding JJ; Gaus K; Whittaker AK High F-Content Perfluoropolyether-Based Nanoparticles for Targeted Detection of Breast Cancer by 19F Magnetic Resonance and Optical Imaging. ACS Nano 2018, 12, 9162–9176. [DOI] [PubMed] [Google Scholar]
- (34).Iima R; Takegami S; Konishi A; Tajima S; Minematsu N; Kitade T Thermal Behavior of 19F Nuclear Magnetic Resonance Signal of 19F-Containing Compound in Lipid NanoEmulsion for Potential Tumor Diagnosis. AAPS PharmSciTech 2018, 19, 2679–2686. [DOI] [PubMed] [Google Scholar]
- (35).Huang P; Guo W; Yang G; Song H; Wang Y; Wang C; Kong D; Wang W Fluorine Meets Amine: Reducing Microenviron-ment-Induced Amino-Activatable Nanoprobes for 19F-Magnetic Resonance Imaging of Biothiols. ACS Appl. Mater. Interfaces 2018, 10, 18532–18542. [DOI] [PubMed] [Google Scholar]
- (36).Shin SH; Park E-J; Min C; Choi SI; Jeon S; Kim Y-H; Kim D Tracking Perfluorocarbon Nanoemulsion Delivery by 19F MRI for Precise High Intensity Focused Ultrasound Tumor Ablation. Theranostics 2017, 7, 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Ko IO; Jung K-H; Kim MH; Kang KJ; Lee KC; Kim KM; Noh I; Lee YJ; Lim SM; Kim JY; Park J-A Preliminary 19F-MRS Study of Tumor Cell Proliferation with 3′-Deoxy-3′-Fluorothymidine and Its Metabolite (FLT-MP). Contrast Media Mol. Imaging 2017, 2017, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Makela AV; Foster PJ Imaging Macrophage Distribution and Density in Mammary Tumors and Lung Metastases Using Fluorine-19 MRI Cell Tracking. Magn. Reson. Med. 2018, 80, 1138–1147. [DOI] [PubMed] [Google Scholar]
- (39).Constantinides C; Basnett P; Lukasiewicz B; Carnicer R; Swider E; Majid QA; Srinivas M; Carr CA; Roy I In vivo Tracking and 1H/19F Magnetic Resonance Imaging of Biodegradable Polyhydroxyalkanoate/Polycaprolactone Blend Scaffolds Seeded with Labeled Cardiac Stem Cells. ACS Appl. Mater. Interfaces 2018, 10, 25056–25068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shi Y; Oeh J; Hitz A; Hedehus M; Eastham-Anderson J; Peale FV; Hamilton P; O’Brien T; Sampath D; Carano RAD Monitoring and Targeting Anti-VEGF Induced Hypoxia within the Viable Tumor by 19F-MRI and Multispectral Analysis. Neoplasia 2017, 19, 950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Temme S; Grapentin C; Quast C; Jacoby C; Grandoch M; Ding Z; Owenier C; Mayenfels F; Fischer JW; Schubert R; Schrader J; Flogel,¨ U. Noninvasive Imaging of Early Venous Thrombosis by 19F Magnetic Resonance Imaging with Targeted Perfluorocarbon Nanoemulsions. Circulation 2015, 131, 1405–1414. [DOI] [PubMed] [Google Scholar]
- (42).Zhang F; Zhou Q; Yang G; An L; Li F; Wang J A Genetically Encoded 19F NMR Probe for Lysine Acetylation. Chem. Commun. 2018, 54, 3879–3882. [DOI] [PubMed] [Google Scholar]
- (43).Hu J; Wu Q; Cheng K; Xie Y; Li C; Li Z A19 F NMR Probe for the Detection of β-Galactosidase: Simple Structure with Low Molecular Weight of 274.2, “Turn-on” Signal without the Background, and Good Performance Applicable in Cancer Cell Line. J. Mater. Chem. B 2017, 5, 4673–4678. [DOI] [PubMed] [Google Scholar]
- (44).Yuan Y; Ge S; Sun H; Dong X; Zhao H; An L; Zhang J; Wang J; Hu B; Liang G Intracellular Self-Assembly and Disassembly of (19)F Nanoparticles Confer Respective “Off” and “On” (19)F NMR/MRI Signals for Legumain Activity Detection in Zebrafish. ACS Nano 2015, 9, 5117–5124. [DOI] [PubMed] [Google Scholar]
- (45).Wang H; Raghupathi KR; Zhuang J; Thayumanavan S Activatable Dendritic (19)F Probes for Enzyme Detection. ACS Macro Lett. 2015, 4, 422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Vernikouskaya I; Pochert A; Lindén M; Rasche V Quantitative 19F MRI of Perfluoro-15-Crown-5-Ether Using Uniformity Correction of the Spin Excitation and Signal Reception. MAGMA 2018, 1–12, DOI: 10.1007/s10334-018-0696-6. [DOI] [PubMed] [Google Scholar]
- (47).Xu M; Guo C; Hu G; Xu S; Wang L Organic Nanoprobes for Fluorescence and19 F Magnetic Resonance Dual-Modality Imaging. Chin. J. Chem. 2018, 36, 25–30. [Google Scholar]
- (48).Bo S; Yuan Y; Chen Y; Yang Z; Chen S; Zhou X; Jiang Z-X In vivo Drug Tracking with 19F MRI at Therapeutic Dose. Chem. Commun. 2018, 54, 3875–3878. [DOI] [PubMed] [Google Scholar]
- (49).Kolouchova K; Sedlacek O; Jirak D; Babuka D; Blahut J; Kotek J; Vit M; Trousil J; Konefał R; Janouskova O; Podhorska B; Slouf M; Hruby M Self-Assembled Thermoresponsive Polymeric Nanogels for 19F MR Imaging. Biomacromolecules 2018, 19, 3515–3524. [DOI] [PubMed] [Google Scholar]
- (50).Ashur I; Allouche-Arnon H; Bar-Shir A Calcium Fluoride Nanocrystals: Tracers for In vivo 19 F Magnetic Resonance Imaging. Angew. Chem. 2018, 130, 7600–7604. [DOI] [PubMed] [Google Scholar]
- (51).Kirberger SE; Maltseva SD; Manulik JC; Einstein SA; Weegman BP; Garwood M; Pomerantz WCK Synthesis of Intrinsically Disordered Fluorinated Peptides for Modular Design of High-Signal 19 F MRI Agents. Angew. Chem., Int. Ed. 2017, 56, 6440–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jackson JC; Hammill JT; Mehl RA Site-Specific Incorporation of a (19)F-Amino Acid into Proteins as an NMR Probe for Characterizing Protein Structure and Reactivity. J. Am. Chem. Soc. 2007, 129, 1160–1166. [DOI] [PubMed] [Google Scholar]
- (53).Gee CT; Arntson KE; Urick AK; Mishra NK; Hawk LML; Wisniewski AJ; Pomerantz WCK Protein-Observed (19)F-NMR for Fragment Screening, Affinity Quantification and Druggability Assessment. Nat. Protoc. 2016, 11, 1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Preslar AT; Tantakitti F; Park K; Zhang S; Stupp SI; Meade TJ (19)F Magnetic Resonance Imaging Signals from Peptide Amphiphile Nanostructures Are Strongly Affected by Their Shape. ACS Nano 2016, 10, 7376–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Hawk LML; Gee CT; Urick AK; Hu H; Pomerantz WCK Paramagnetic Relaxation Enhancement for Protein-Observed 19F NMR as an Enabling Approach for Efficient Fragment Screening. RSC Adv. 2016, 6, 95715–95721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hansen PR; Oddo A Fmoc Solid-Phase Peptide Synthesis. Methods Mol. Biol. 2015, 1348, 33–50. [DOI] [PubMed] [Google Scholar]
- (57).Tang Y; Tirrell DA Biosynthesis of a Highly Stable Coiled-Coil Protein Containing Hexafluoroleucine in an Engineered Bacterial Host. J. Am. Chem. Soc. 2001, 123, 11089–11090. [DOI] [PubMed] [Google Scholar]
- (58).Aramini JM; Hamilton K; Ma L-C; Swapna GVT; Leonard PG; Ladbury JE; Krug RM; Montelione GT (19)F NMR Reveals Multiple Conformations at the Dimer Interface of the Nonstructural Protein 1 Effector Domain from Influenza A Virus. Structure 2014, 22, 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chalmers KH; De Luca E; Hogg NHM; Kenwright AM; Kuprov I; Parker D; Botta M; Wilson JI; Blamire AM Design Principles and Theory of Paramagnetic Fluorine-Labelled Lanthanide Complexes as Probes for (19)F Magnetic Resonance: A Proof-of-Concept Study. Chem. - Eur. J 2010, 16, 134–148. [DOI] [PubMed] [Google Scholar]
- (60).Srivastava K; Weitz EA; Peterson KL; Marjańska M; Pierre VC Fe- and Ln-DOTAm-F12 Are Effective Paramagnetic Fluorine Contrast Agents for MRI in Water and Blood. Inorg. Chem. 2017, 56, 1546–1557. [DOI] [PubMed] [Google Scholar]
- (61).Kelkar SS; Reineke TM Theranostics: Combining Imaging and Therapy. Bioconjugate Chem. 2011, 22, 1879–1903. [DOI] [PubMed] [Google Scholar]
- (62).Gunasekar SK; Anjia L; Matsui H; Montclare JK Effects of Divalent Metals on Nanoscopic Fiber Formation and Small Molecule Recognition of Helical Proteins. Adv. Funct. Mater. 2012, 22, 2154–2159. [Google Scholar]
- (63).Gunasekar SK; Asnani M; Limbad C; Haghpanah JS; Hom W; Barra H; Nanda S; Lu M; Montclare JK N-Terminal Aliphatic Residues Dictate the Structure, Stability, Assembly, and Small Molecule Binding of the Coiled-Coil Region of Cartilage Oligomeric Matrix Protein. Biochemistry 2009, 48, 8559–8567. [DOI] [PubMed] [Google Scholar]
- (64).Haghpanah JS; Yuvienco C; Roth EW; Liang A; Tu RS; Montclare JK Supramolecular Assembly and Small Molecule Recognition by Genetically Engineered Protein Block Polymers Composed of Two SADs. Mol. BioSyst. 2010, 6, 1662–1667. [DOI] [PubMed] [Google Scholar]
- (65).Yin L; Agustinus AS; Yuvienco C; Minashima T; Schnabel NL; Kirsch T; Montclare JK Engineered Coiled-Coil Protein for Delivery of Inverse Agonist for Osteoarthritis. Biomacromolecules 2018, 19, 1614–1624. [DOI] [PubMed] [Google Scholar]
- (66).Liu CF; Chen R; Frezzo JA; Katyal P; Hill LK; Yin L; Srivastava N; More HT; Renfrew PD; Bonneau R; Montclare JK Efficient Dual SiRNA and Drug Delivery Using Engineered Lipoproteoplexes. Biomacromolecules 2017, 18, 2688–2698. [DOI] [PubMed] [Google Scholar]
- (67).Speth PA; van Hoesel QG; Haanen C Clinical Pharmacokinetics of Doxorubicin. Clin. Pharmacokinet. 1988, 15, 15–31. [DOI] [PubMed] [Google Scholar]
- (68).Tsai CJ; Li J; Gonzalez-Angulo AM; Allen PK; Woodward WA; Ueno NT; Lucci A; Krishnamurthy S; Gong Y; Yang W; Cristofanilli M; Valero V; Buchholz TA Outcomes After Multidisciplinary Treatment of Inflammatory Breast Cancer in the Era of Neoadjuvant HER2-Directed Therapy. Am. J. Clin. Oncol. 2015, 38, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kim MK; Kang YK Positional Preference of Proline in Alpha-Helices. Protein Sci. 1999, 8, 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gaucher G; Dufresne M-H; Sant VP; Kang N; Maysinger D; Leroux J-C Block Copolymer Micelles: Preparation, Characterization and Application in Drug Delivery. J. Controlled Release 2005, 109, 169–188. [DOI] [PubMed] [Google Scholar]
- (71).Fang J; Sawa T; Maeda H Factors and Mechanism of “EPR” Effect and the Enhanced Antitumor Effects of Macromolecular Drugs Including SMANCS. Polymer Drugs in the Clinical Stage 2004, 519, 29–49. [DOI] [PubMed] [Google Scholar]
- (72).Mohan P; Rapoport N Doxorubicin as a Molecular Nanotheranostic Agent: Effect of Doxorubicin Encapsulation in Micelles or Nanoemulsions on the Ultrasound-Mediated Intracellular Delivery and Nuclear Trafficking. Mol. Pharmaceutics 2010, 7, 1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Bandyopadhyay A; Wang L; Agyin J; Tang Y; Lin S; Yeh I-T; De K; Sun L-Z Doxorubicin in Combination with a Small TGFbeta Inhibitor: A Potential Novel Therapy for Metastatic Breast Cancer in Mouse Models. PLoS One 2010, 5, No. e10365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Trabbic-Carlson K; Meyer DE; Liu L; Piervincenzi R; Nath N; LaBean T; Chilkoti A Effect of Protein Fusion on the Transition Temperature of an Environmentally Responsive Elastin-like Polypeptide: A Role for Surface Hydrophobicity? Protein Eng., Des. Sel. 2004, 17, 57–66. [DOI] [PubMed] [Google Scholar]
- (75).Garanger E; MacEwan SR; Sandre O; Brûlet A; Bataille L; Chilkoti A; Lecommandoux S Structural Evolution of a Stimulus-Responsive Diblock Polypeptide Micelle by Temperature Tunable Compaction of Its Core. Macromolecules 2015, 48, 6617–6627. [Google Scholar]
- (76).Meyer DE; Chilkoti A Quantification of the Effects of Chain Length and Concentration on the Thermal Behavior of Elastin-like Polypeptides. Biomacromolecules 2004, 5, 846–851. [DOI] [PubMed] [Google Scholar]
- (77).Louis-Jeune C; Andrade-Navarro MA; Perez-Iratxeta C Prediction of Protein Secondary Structure from Circular Dichroism Using Theoretically Derived Spectra. Proteins: Struct., Funct., Genet. 2012, 80, 374–381. [DOI] [PubMed] [Google Scholar]
- (78).Thorsteinsson MV; Richter J; Lee AL; DePhillips P 5-Dodecanoylaminofluorescein as a Probe for the Determination of Critical Micelle Concentration of Detergents Using Fluorescence Anisotropy. Anal. Biochem. 2005, 340, 220–225. [DOI] [PubMed] [Google Scholar]
- (79).Olsen AJ; Katyal P; Haghpanah JS; Kubilius MB; Li R; Schnabel NL; O’Neill SC; Wang Y; Dai M; Singh N; Tu RS; Montclare JK Protein Engineered Triblock Polymers Composed of Two Sads: Enhanced Mechanical Properties and Binding Abilities. Biomacromolecules 2018, 19, 1552–1561. [DOI] [PubMed] [Google Scholar]
- (80).Mukerjee P Fluorocarbon hydrocarbon Interactions in Micelles and Other Lipid Assemblies, at Interfaces, and in Solutions. Colloids Surf., A 1994, 84, 1–10. [Google Scholar]
- (81).Kim W; Thévenot J; Ibarboure E; Lecommandoux S; Chaikof EL Self-Assembly of Thermally Responsive Amphiphilic Diblock Copolypeptides into Spherical Micellar Nanoparticles. Angew. Chem., Int. Ed. 2010, 49, 4257–4260. [DOI] [PubMed] [Google Scholar]
- (82).Gillies ER; Fréchet JMJ A New Approach towards Acid Sensitive Copolymer Micelles for Drug Delivery. Chem. Commun. 2003, 1640–1641. [DOI] [PubMed] [Google Scholar]
- (83).Kurniasih IN; Liang H; Mohr PC; Khot G; Rabe JP; Mohr A Nile Red Dye in Aqueous Surfactant and Micellar Solution. Langmuir 2015, 31, 2639–2648. [DOI] [PubMed] [Google Scholar]
- (84).Cho Y; Zhang Y; Christensen T; Sagle LB; Chilkoti A; Cremer PS Effects of Hofmeister Anions on the Phase Transition Temperature of Elastin-like Polypeptides. J. Phys. Chem. B 2008, 112, 13765–13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Abramoff MD; Magalhães PJ; Ram SJ Image Processing with ImageJ. Biophotonics International 2004, 11, 36–42. [Google Scholar]
- (86).Khan VR; Brown IR The Effect of Hyperthermia on the Induction of Cell Death in Brain, Testis, and Thymus of the Adult and Developing Rat. Cell Stress Chaperones 2002, 7, 73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Hume J; Sun J; Jacquet R; Renfrew PD; Martin JA; Bonneau R; Gilchrist ML; Montclare JK Engineered Coiled-Coil Protein Microfibers. Biomacromolecules 2014, 15, 3503–3510. [DOI] [PubMed] [Google Scholar]
- (88).Shrivastava A; Gupta V Methods for the Determination of Limit of Detection and Limit of Quantitation of the Analytical Methods. Chron. Young Sci. 2011, 2, 21–25. [Google Scholar]
- (89).Lacey ME; Subramanian R; Olson DL; Webb AG; Sweedler JV High-Resolution NMR Spectroscopy of Sample Volumes from 1 NL to 10 ΜL. Chem. Rev. 1999, 99, 3133–3152. [DOI] [PubMed] [Google Scholar]
- (90).Currie LA Detection and Quantification Limits: Origins and Historical Overview. Anal. Chim. Acta 1999, 391, 127–134. [Google Scholar]
- (91).Nic M; Jirat J; Kosata B IUPAC Compendium of Chemical Terminology. International Union of Pure and Applied Chemistry 2014, 1–1622. [Google Scholar]
- (92).Wong TC Micellar Systems: Nuclear Magnetic Resonance Spectroscopy. In Encyclopedia of Surface and Colloid Science, third ed.; Taylor & Francis, 2015; pp 4391–4409. [Google Scholar]
- (93).Cheng HN; Asakura T; English AD NMR Spectroscopy of Polymers: Innovative Strategies for Complex Macromolecules; ACS Symposium Series; American Chemical Society: Washington, DC, 2011; Vol. 1077, pp 3–16. [Google Scholar]
- (94).Bin Na H; Hyeon T MRI Contrast Agents Based on Inorganic Nanoparticles In Nanoplatform-Based Molecular Imaging; Chen X, Ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011; pp 279–308. [Google Scholar]
- (95).Schmid F; Höltke C; Parker D; Faber C Boosting (19) F MRI-SNR Efficient Detection of Paramagnetic Contrast Agents Using Ultrafast Sequences. Magn. Reson. Med. 2013, 69, 1056–1062. [DOI] [PubMed] [Google Scholar]
- (96).Robson MD; Gatehouse PD; Bydder M; Bydder GM Magnetic Resonance: An Introduction to Ultrashort TE (UTE) Imaging. J. Comput. Assist Tomogr 2003, 27, 825–846. [DOI] [PubMed] [Google Scholar]
- (97).Larson PEZ; Han M; Krug R; Jakary A; Nelson SJ; Vigneron DB; Henry RG; McKinnon G; Kelley DAC. Ultrashort Echo Time and Zero Echo Time MRI at 7T. MAGMA 2016, 29, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Weiger M; Pruessmann KP MRI with Zero Echo Time In Encyclopedia of Magnetic Resonance; Harris RK, Ed.; John Wiley & Sons, Ltd: Chichester, UK, 2007. [Google Scholar]
- (99).Juchem C; de Graaf RA B0Magnetic Field Homogeneity and Shimming for In vivo Magnetic Resonance Spectroscopy. Anal. Biochem. 2017, 529, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).McDaniel JR; Callahan DJ; Chilkoti A Drug Delivery to Solid Tumors by Elastin-like Polypeptides. Adv. Drug Delivery Rev. 2010, 62, 1456–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Liu W; Dreher MR; Furgeson DY; Peixoto KV; Yuan H; Zalutsky MR; Chilkoti A Tumor Accumulation, Degradation and Pharmacokinetics of Elastin-like Polypeptides in Nude Mice. J. Controlled Release 2006, 116, 170–178. [DOI] [PubMed] [Google Scholar]
- (102).Liu W; MacKay JA; Dreher MR; Chen M; McDaniel JR; Simnick AJ; Callahan DJ; Zalutsky MR; Chilkoti A Injectable Intratumoral Depot of Thermally Responsive Polypeptide-Radionuclide Conjugates Delays Tumor Progression in a Mouse Model. J. Controlled Release 2010, 144, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Bier J; Benders P; Wenzel M; Bitter K Kinetics of 57Co-Bleomycin in Mice after Intravenous, Subcutaneous and Intratumoral Injection. Cancer 1979, 44, 1194–1200. [DOI] [PubMed] [Google Scholar]
- (104).Tomita T Interstitial Chemotherapy for Brain Tumors: Review. J. Neuro-Oncol. 1991, 10, 57–74. [DOI] [PubMed] [Google Scholar]
- (105).Wang Y; Hu JK; Krol A; Li YP; Li CY; Yuan F Systemic Dissemination of Viral Vectors during Intratumoral Injection. Mol. Cancer Ther. 2003, 2, 1233–1242. [PubMed] [Google Scholar]
- (106).Goette MJ; Lanza GM; Caruthers SD; Wickline SA Improved Quantitative (19) F MR Molecular Imaging with Flip Angle Calibration and B1-Mapping Compensation. J. Magn. Reson. Imaging 2015, 42, 488–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Goette MJ; Keupp J; Rahmer J; Lanza GM; Wickline SA; Caruthers SD Balanced UTE-SSFP for19 F MR Imaging of Complex Spectra. Magn. Reson. Med. 2015, 74, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Gao Z-G; Tian L; Hu J; Park I-S; Bae YH Prevention of Metastasis in a 4T1Murine Breast Cancer Model by Doxorubicin Carried by Folate Conjugated PH Sensitive Polymeric Micelles. J. Controlled Release 2011, 152, 84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Savitzky A; Golay MJE Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar]
- (110).Pisárčik M; Devínsky F; Lacko I Aggregation Number of Alkanediyl-α, ω-Bis (Dimethylalkylammonium Bromide) Surfactants Determined by Static Light Scattering. Colloids Surf., A 2000, 172, 139–144. [Google Scholar]
- (111).Zhang X; Jackson JK; Burt HM Determination of Surfactant Critical Micelle Concentration by a Novel Fluorescence Depolarization Technique. J. Biochem. Biophys. Methods 1996, 31, 145–150. [DOI] [PubMed] [Google Scholar]
- (112).Laginha KM; Verwoert S; Charrois GJR; Allen TM Determination of Doxorubicin Levels in Whole Tumor and Tumor Nuclei in Murine Breast Cancer Tumors. Clin. Cancer Res. 2005, 11, 6944–6949. [DOI] [PubMed] [Google Scholar]
- (113).Thao LQ; Byeon HJ; Lee C; Lee S; Lee ES; Choi YW; Choi H-G; Park E-S; Lee KC; Youn YS Doxorubicin-Bound Albumin Nanoparticles Containing a TRAIL Protein for Targeted Treatment of Colon Cancer. Pharm. Res. 2016, 33, 615–626. [DOI] [PubMed] [Google Scholar]
- (114).Bae S; Ma K; Kim TH; Lee ES; Oh KT; Park E-S; Lee KC; Youn YS Doxorubicin-Loaded Human Serum Albumin Nanoparticles Surface-Modified with TNF-Related Apoptosis-Inducing Ligand and Transferrin for Targeting Multiple Tumor Types. Biomaterials 2012, 33, 1536–1546. [DOI] [PubMed] [Google Scholar]
- (115).Soule HD; Vazquez J; Long A; Albert S; Brennan M A Human Cell Line from a Pleural Effusion Derived from a Breast Carcinoma 2. JNCI: Journal of the National Cancer Institute 1973, 51, 1409–1416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.