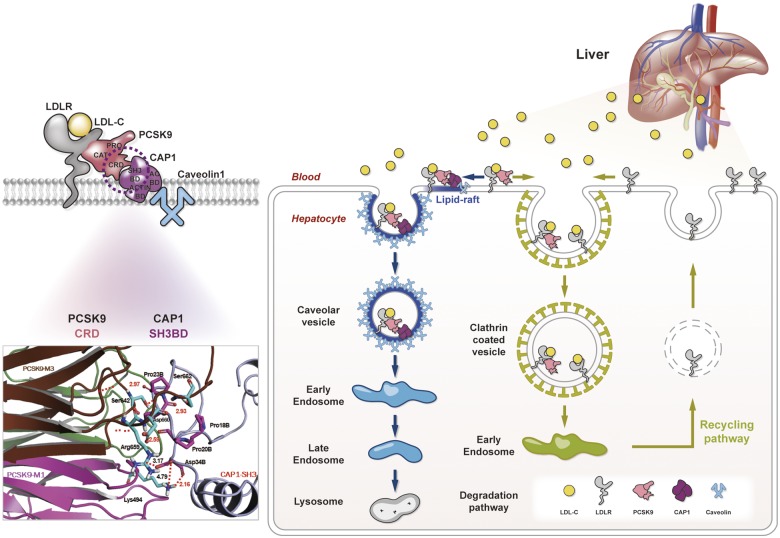
Abstract
Aims
Proprotein convertase subtilisin/kexin type-9 (PCSK9), a molecular determinant of low-density lipoprotein (LDL) receptor (LDLR) fate, has emerged as a promising therapeutic target for atherosclerotic cardiovascular diseases. However, the precise mechanism by which PCSK9 regulates the internalization and lysosomal degradation of LDLR is unknown. Recently, we identified adenylyl cyclase-associated protein 1 (CAP1) as a receptor for human resistin whose globular C-terminus is structurally similar to the C-terminal cysteine-rich domain (CRD) of PCSK9. Herein, we investigated the role of CAP1 in PCSK9-mediated lysosomal degradation of LDLR and plasma LDL cholesterol (LDL-C) levels.
Methods and results
The direct binding between PCSK9 and CAP1 was confirmed by immunoprecipitation assay, far-western blot, biomolecular fluorescence complementation, and surface plasmon resonance assay. Fine mapping revealed that the CRD of PCSK9 binds with the Src homology 3 binding domain (SH3BD) of CAP1. Two loss-of-function polymorphisms found in human PCSK9 (S668R and G670E in CRD) were attributed to a defective interaction with CAP1. siRNA against CAP1 reduced the PCSK9-mediated degradation of LDLR in vitro. We generated CAP1 knock-out mice and found that the viable heterozygous CAP1 knock-out mice had higher protein levels of LDLR and lower LDL-C levels in the liver and plasma, respectively, than the control mice. Mechanistic analysis revealed that PCSK9-induced endocytosis and lysosomal degradation of LDLR were mediated by caveolin but not by clathrin, and they were dependent on binding between CAP1 and caveolin-1.
Conclusion
We identified CAP1 as a new binding partner of PCSK9 and a key mediator of caveolae-dependent endocytosis and lysosomal degradation of LDLR.
Keywords: Proprotein convertase subtilisin/kexin type-9, Low-density lipoprotein receptor, Cyclase-associated protein 1, LDL cholesterol, Endocytosis, Caveolae
Translational perspective
Although monoclonal antibodies to proprotein convertase subtilisin/kexin type-9 (PCSK9) have been approved to lower plasma low-density lipoprotein (LDL) cholesterol levels and improve clinical outcomes in patients with atherosclerotic cardiovascular diseases, the precise mechanism of how PCSK9 directs LDL receptor (LDLR) to lysosomal degradation is unclear. We have disputed the previous notion that LDLR degradation is mediated by clathrin. We identified adenylyl cyclase-associated protein 1 as a liaison between the PCSK9-LDLR complex and caveolin, which directs the complex to caveolae-mediated endocytosis and lysosomal degradation. Our findings will change LDLR degradation biology from a PCSK9-centric view to a broader and more balanced view, contributing to the emergence of another new therapeutic strategy for dyslipidaemia and atherosclerotic cardiovascular diseases.
Introduction
Proprotein convertase subtilisin/kexin type-9 (PCSK9) determines plasma low-density lipoprotein (LDL) cholesterol (LDL-C) levels by regulating internalization and lysosomal degradation of the LDL receptor (LDLR)1 and has emerged as a promising therapeutic target.2 Inhibitors of PCSK9 have reduced plasma LDL-C levels and improved cardiovascular outcomes.3,4 However, the precise mechanism by which PCSK9 determines the fate of LDLR has not been elucidated. The LDLR enters cells while bound to LDL-C, dissociates from LDL-C in endosomes, and is recycled to the cell surface, whereas LDL-C is directed to lysosomes for degradation. In contrast, if bound to PCSK9, LDLR is internalized and escorted to lysosomes for degradation through an unknown mechanism.1 Although PCSK9 is a proteinase K-like serine protease, after autocatalytic cleavage, the terminal portion of the pro-domain (amino acids 32-152) covers the catalytic triad, preventing further proteolytic activity.1,2 The catalytic domain of PCSK9 binds LDLR, and another part of PCSK9, i.e. the C-terminal cysteine-rich domain (CRD), is suspected to interact with a putative membrane-bound protein that escorts the protein complex towards lysosomal degradation.1 Recently, PCSK9-CRD has been reported to have structural homology with the human resistin C-terminal domain homotrimer,5 which is a pro-inflammatory cytokine inducing atherosclerosis.6 In our previous paper, we firstly reported that adenylyl cyclase-associated protein 1 (CAP1), which is a conserved protein known as a component of Saccharomyces cerevisiae adenylyl cyclase complex,7 is a surface receptor that binds the C-terminal domain of homo-trimer human resistin.8 Thus, we think that CAP1 binds to resistin as well as CRD of PCSK9. Furthermore, CAP1 is known to regulate actin filament dynamics important for cell morphology, migration, and endocytosis.9 Therefore, we hypothesized that CAP1 is the unknown protein that interacts with PCSK9-CRD to escort the LDLR-PCSK9 complex towards lysosomal degradation.
Materials and methods
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the Clinical Research Institute in Seoul National University Hospital, Korea. For a detailed description of all methods, see the Supplementary material online, Experimental Procedures.
Results
CRD of PCSK9 directly binds CAP1
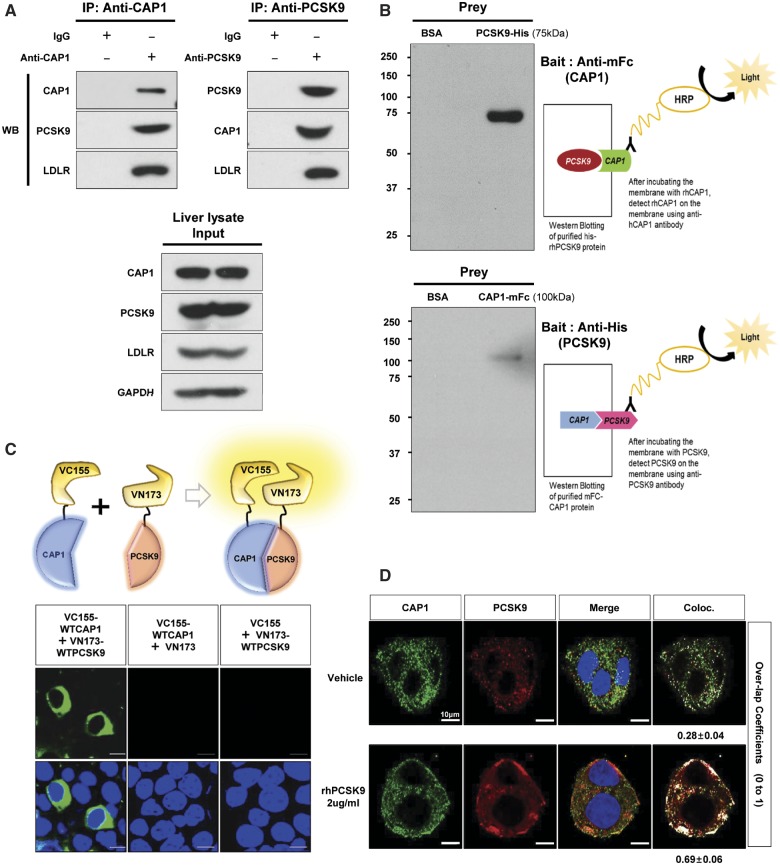
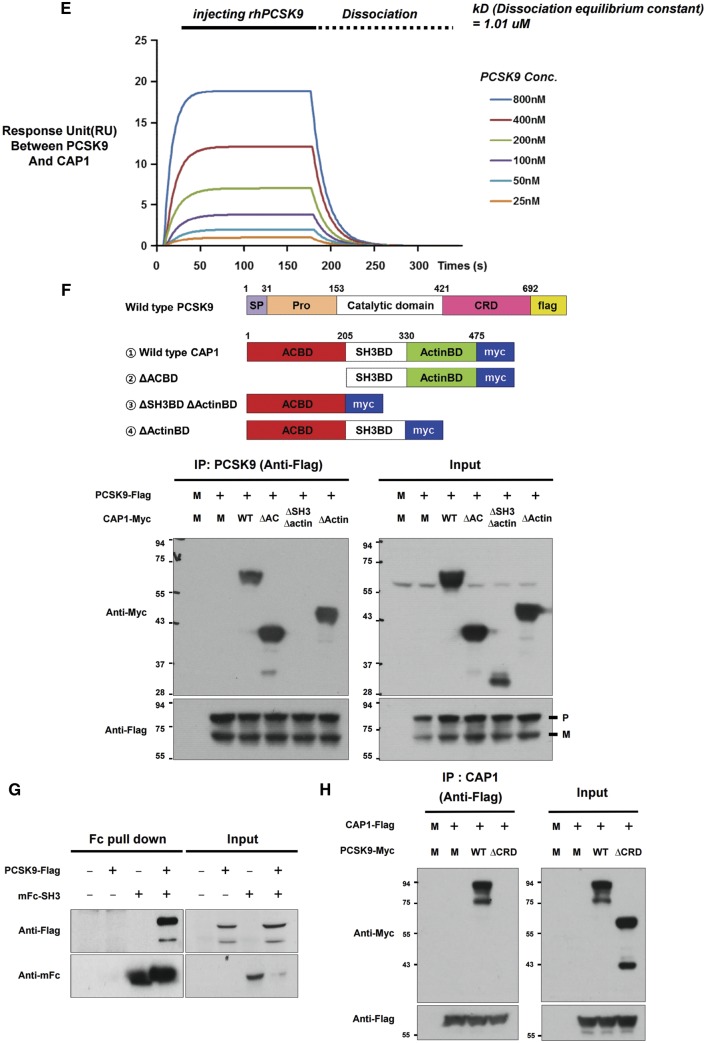
A physical interaction between CAP1 and PCSK9 was shown in mouse liver tissue using immunoprecipitation (Figure 1A). For far-western blot analysis,10 purified mFc-CAP1 or His-PCSK9 was used as prey under the non-reducing condition and probed with His-PCSK9 or mFc-CAP1 as bait, respectively (Figure 1B). To visualize this interaction in living cells, we performed a bimolecular fluorescent complementation assay based on the complementation between two non-fluorescent fragments of fluorescent protein that are brought together by an interaction between proteins fused to each fragment,11 which clearly visualized the direct binding between hCAP1 and hPCSK9 in living cells (Figure 1C). In addition, the immunofluorescent staining of HepG2 cells exogenously treated with recombinant PCSK9 demonstrated that PCSK9 was colocalized with CAP1 and LDLR in the membrane and cytosol (Figure 1D). Finally, a direct binding assay using a surface plasmon resonance-based system yielded an increase of response units between PCSK9 and CAP1 in a dose-dependent manner (Figure 1E).
Figure 1.
CAP1 directly interacts with PCSK9. (A) Immunoprecipitation of endogenous CAP1 or PCSK9 from liver lysates of C57/BL6 wild-type mice. (B) Far-western blot analysis of mFc-hCAP1 and hPCSK9-His. (C) Bimolecular fluorescence complementation assay visualizing the interaction between hCAP1 and hPCSK9 in living cells. Green fluorescence was observed when CAP1 and PCSK9 fused to each fragment (pVC155 and pVN173) and were both expressed (left panel). However, when human CAP1 or PCSK9 was expressed alone, no fluorescence was detected (middle and right panel, respectively). (D) Colocalization of CAP1 and PCSK9 as determined by immunofluorescent staining of HepG2 cells exogenously treated with recombinant hPCSK9. Overlap coefficients: mean ± standard deviation. (E) Direct binding assay between rhPCSK9 and CAP1 using surface plasmon resonance. The equilibrium dissociation constant was 1.01 µM. (F) Coimmunoprecipitation of hPCSK9-Flag and wild-type CAP1 or each CAP1 mutant in HEK 293 cells. Pro-PCSK9 (P) and mature PCSK9 (M) are indicated with bold line. (G) Coimmunoprecipitation of hPCSK9-Flag and the SH3BD of CAP1 in HEK 293 cells showing that the SH3BD is sufficient for binding to CAP1. (H) Coimmunoprecipitation of wild-type hCAP1 with wild-type PCSK9 or the CRD deletion PCSK9 mutant in HEK 293 cells showing that PCSK9 binds CAP1 via the CRD.
Since we previously identified that the Src homology 3 binding domain (SH3BD) of CAP1 is a binding partner of the C-terminal domain of homo-trimer human resistin, which has structural homology with the CRD of PCSK9,8 we tested whether the SH3BD of CAP1 binds the CRD of PCSK9. We carried out an in vitro coimmunoprecipitation assay with wtPCSK9-Flag and the following CAP1 mutants: an adenylyl cyclase-binding domain deletion mutant (▵ACBD mutant), an actin binding deletion mutant (▵ActinBD mutant), and a mutant with a deletion of both the SH3BD and ActinBD (▵SH3BD ▵ActinBD mutant). PCSK9 interacted with wtCAP1, the ▵ACBD mutant, and the ▵ActinBD mutant but not with the ▵SH3BD▵ ActinBD mutant, suggesting that CAP1 binds PCSK9 via the SH3BD (Figure 1F). The SH3BD of CAP1 is sufficient for interacting with PCSK9 (Figure 1G). CAP1 bound wtPCSK9 but not the CRD deletion mutant (Figure 1H). A 3D molecular modelling analysis of the complex using a protein–protein docking simulation and a binding energy score analysis determined the key binding interactions among Asp34B in the SH3BD of CAP and Lys494 in the M1 domain and Arg659 in the M3 domain of PCSK9-CRD (Supplementary material online, Figure S1).
PCSK9 loss-of-function mutants (S668R and G670E in CRD) cannot interact with CAP1
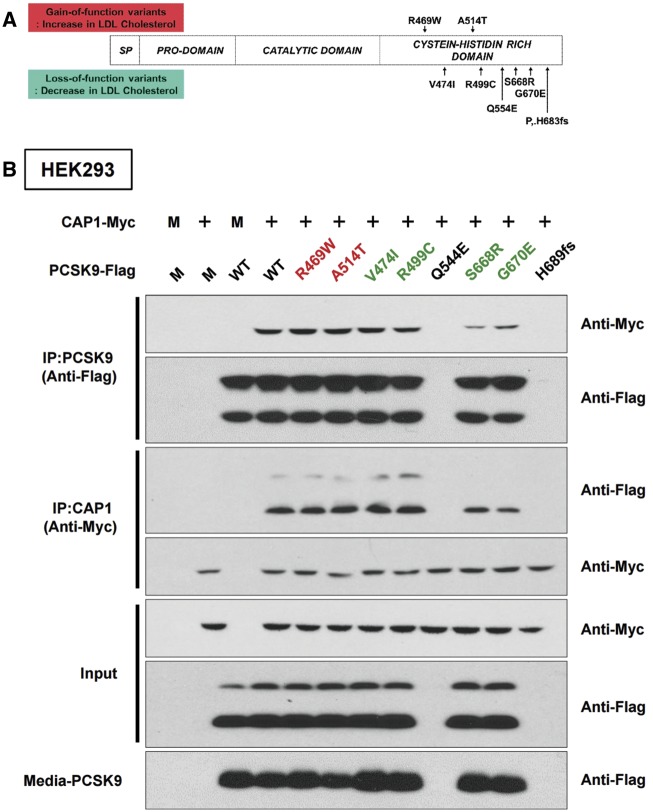
We generated eight PCSK9 point mutants by site-directed mutagenesis of PCSK9-CRD, covering the well-known loss-of-function and gain-of-function variations found in human genetic studies (Figure 2A). Q544E and H683fs mutants of PCSK9 were not expressed in the HepG2 cells, suggesting that these mutants are related to protein expression or stability. S668R and G670E mutants in the CRD of PCSK9 were well expressed, but their binding to CAP1 was severely compromised as assessed by immunoprecipitation (Figure 2B), suggesting that the CRD of PCSK9 is important for binding to CAP1 and for regulating LDLR protein (and, thus, LDL-C levels in humans). To further characterize the direct interaction of PCSK9-CRD mutation with CAP1, we measured the binding affinity of PCSK9 WT, PCSK9 A514T, PCSK9 G670E, and PCSK9 S668RG670E against CAP1 (Supplementary material online, Figure S2, equilibrium dissociation constant, kD: 0.45 µM, 0.11 µM, 0.96 µM, and 2.32 µM, respectively). PCSK9 A514T showed a stronger interaction relative to WT, whereas G670E and G670ES668R mutants showed weak interactions.
Figure 2.
Human PCSK9 loss-of-function mutants have weaker interactions with CAP1 than the wild type. (A) The single-nucleotide variants of PCSK9-CRD reported to be associated with plasma LDL cholesterol levels in humans. (B) The loss-of-function PCSK9 mutants S668R and G679E lose their binding affinity to CAP1. CAP1-Myc and either wild-type PCSK9-Flag or each point mutant of PCSK9-Flag were overexpressed in HEK293 cells. Immunoprecipitation with Flag and immunoblotting with Myc (upper two figures of immunoblot), or immunoprecipitation with Myc and immunoblotting with Flag (immunoblot on the third and fourth lines). The level of PCSK9 in the culture media (the lowest line).
CAP1 induces PCSK9-mediated LDLR degradation and increases the LDL-C level
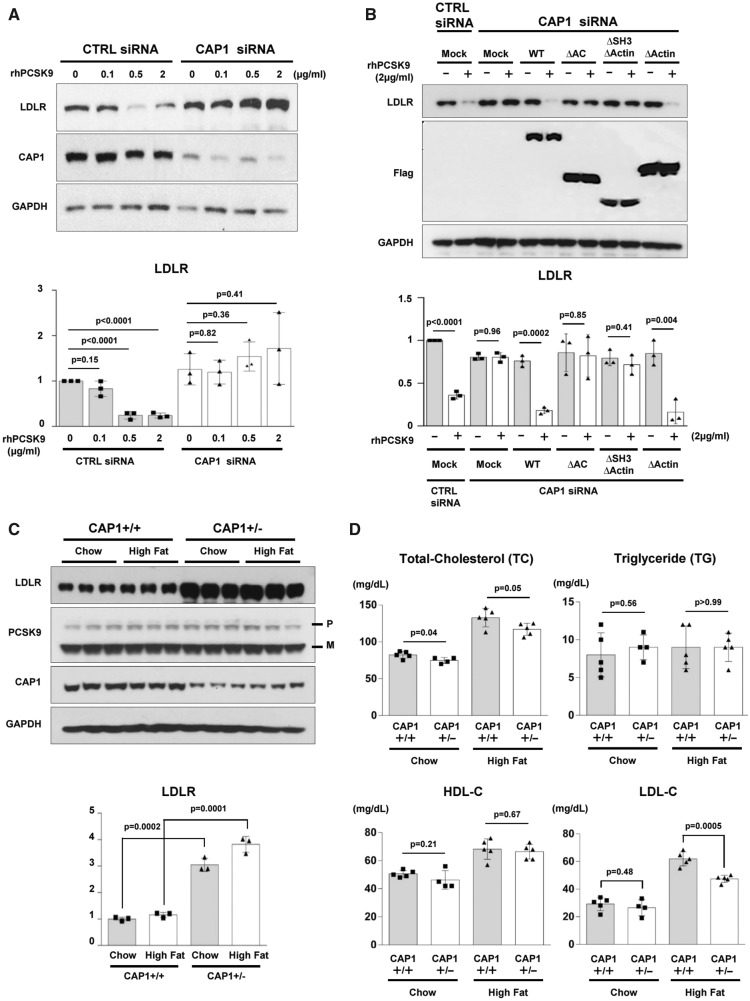
PCSK9 treatment dose-dependently promoted LDLR degradation in HepG2 cells12 (Supplementary material online, Figure S3). Once CAP1 was depleted with siRNA (siCAP1), LDLR degradation by exogenous PCSK9 was significantly attenuated (Figure 3A). In addition, Supplementary material online, Figure S4 demonstrated that LDLR and exogenous His-tagged PCSK9 were less detectable in the cytosol of the CAP1 siRNA-treated cells both 30 and 60 min after His-rhPCSK9 treatment. We rescued the CAP1 deficient cells (siCAP1) with wtCAP1 or each of the CAP1 mutants by overexpression and examined the PCSK9-mediated LDLR degradation. In Figure 3B, only wtCAP1 and the ▵actinBD mutant rescued the attenuated PCSK9-mediated LDLR degradation, which suggests that the SH3BD and ACBD are critical for exogenous PCSK9-mediated LDLR degradation. To examine whether CAP1 regulates PCSK9 interaction with the EGF-A domain of LDLR, we performed immunoprecipitation and proximity ligation assays. The results supported the fact that the interaction between LDLR and PCSK9 is independent of CAP1 (Supplementary material online, Figure S5).
Figure 3.
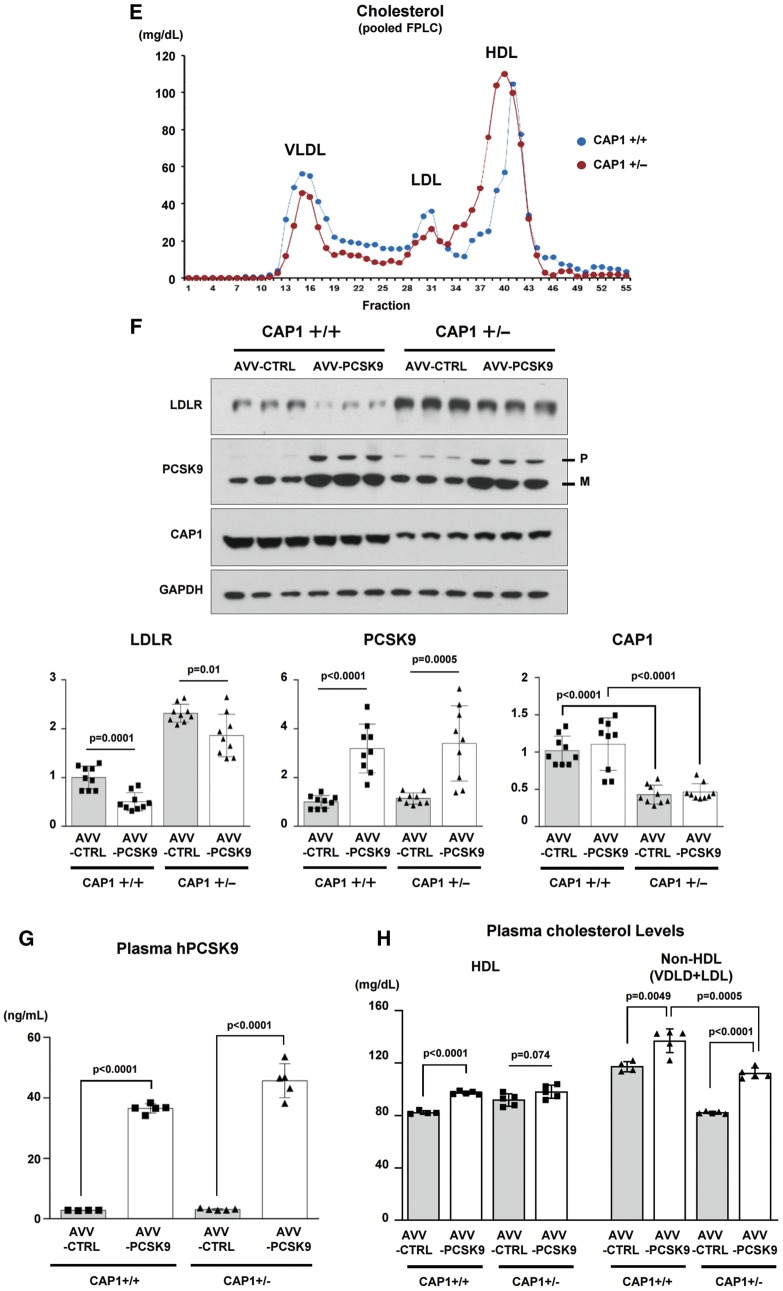
CAP1 is required for PCSK9-induced LDLR degradation. (A) LDLR degradation induced by PCSK9 was attenuated by CAP1 siRNA (upper panel). The representative figure of three independent experiments. Quantification of the expression levels of LDLR (bottom panel). (B) PCSK9-induced LDLR degradation was rescued by wild-type CAP1 and the ▵ACBD mutant (upper panel). Quantification of the expression levels of LDLR (bottom panel). (C) The protein expression levels of LDLR were significantly greater in the CAP1+/− mice than those in the CAP1+/+ mice, regardless of the diet. Pro-PCSK9 (P) and mature PCSK9 (M) are indicated with bold line. Quantification of the expression levels of LDLR/GAPDH in CAP1+/− vs. CAP1+/+ mice (bottom panel). (D) Enzymatic measurement of the plasma cholesterol levels in CAP1+/− and CAP1+/+ mice fed either high fat- or chow diet for 16 weeks (n = 6 per group). (E) Cholesterol levels in FPLC fractions from pooled plasma samples (n = 5 per group) in CAP+/− (red) and CAP1+/+ (blue) mice fed high-fat diet. (F–H) Overexpression of PCSK9 in both CAP1+/− and CAP1+/+ mice using adeno-associated virus. (F) Immunoblot analysis of PCSK9-induced LDLR degradation in CAP1+/− and CAP1+/+ mice liver. Pro-PCSK9 (P) and mature PCSK9 (M) are indicated with bold line. The representative figure of three independent experiments. Quantification of the expression levels of LDLR (bottom left), PCSK9 (bottom middle), and CAP1 (bottom right) (n = 9). (G) Plasma levels of hPCSK9 were measured by ELISA (n = 5 per group). (H) Enzymatic analysis of plasma cholesterol levels in CAP1+/− and CAP1+/+ mice with or without PCSK9 overexpression (n = 5 per group).
We generated a CAP1 knock-out mouse by TALEN-mediated targeting of CAP1 exon 3 that allowed us to determine the role of CAP1 in vivo (Supplementary material online, Figure S6). Heterozygous knock-out mice (CAP1+/− mouse) were used, as the CAP1 homozygous knock-out mice died at embryonic day 16.5 (Supplementary material online, Figure S7). The organs of CAP1+/− mice did not differ from the wild-type organs up to approximately 16 weeks (Supplementary material online, Figure S8), and the mRNA and protein levels of CAP1 were significantly decreased in various organs from the CAP1+/− mouse (Supplementary material online, Figure S9). Thus, we compared the expression levels of LDLR and PCSK9 in the liver of CAP1+/− mice and CAP1+/+ mice with or without a high-fat diet. While there was no significant difference in LDLR mRNA expression via real-time PCR (Supplementary material online, Figure S10), the protein levels of LDLR were significantly higher in the CAP1+/− mice than in the CAP1+/+ mice (Figure 3C). Accordingly, under high-fat diet, the CAP1+/− mice had lower total cholesterol and LDL-C levels than WT mice (Figure 3D). There was no significant difference in the plasma triglyceride (TG) and high-density lipoprotein cholesterol (HDL-C) levels. Then, we fractionated the plasma lipoproteins by fast-performance liquid chromatography, which confirmed the decrease in the LDL-C and VLDL-C levels and the shift of HDL-C towards the large-buoyant form in the CAP1+/− mice compared to the CAP1+/+ mice fed a high-fat diet (Figure 3E). We overexpressed PCSK9 using adeno-associated virus both in CAP1+/− and CAP1+/+ mice and measured the levels of LDLR expression and LDL-C (Figure 3F–H). In CAP1 hetero-knock-out mice, a decrease or degradation of LDLR protein by transduction of PCSK9 was prevented, leading to improved cholesterol profiles compared to wild type animals.
CAP1 guides the PCSK9/LDLR complex to undergo caveolae-dependent endocytosis, leading to degradation of LDLR
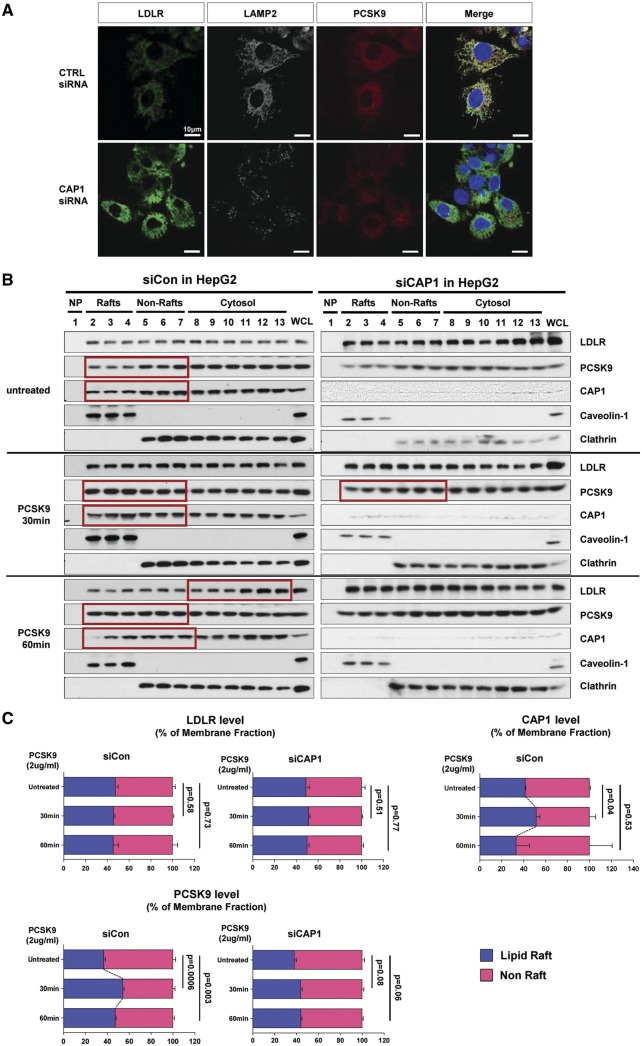
The PCSK9-mediated degradation of LDLR was exclusively blocked by the lysosomal protease inhibitor (E-64d), but not by inhibitors of the proteasome (lactacystin) or autophagy (bafilomycin), suggesting that LDLRs are degraded by the lysosomal pathway as previously reported13,14 (Supplementary material online, Figure S11). This finding was also illustrated in HepG2 cells treated with PCSK9 conjugated with Cy3 dye (PCSK9-Cy3) by tracking LDLRs with PCSK9-Cy3, the endosomal marker EEA1 (early endosome antigen 1), and the lysosomal marker LAMP2 (lysosome-associated membrane protein 2) at various time points (Supplementary material online, Figure S12A). After the PCSK9-Cy3 treatment, EEA1 was colocalized with PCSK9 and LDLR within 30 min (Supplementary material online, Figure S12B and C). Then, LAMP2 appeared, colocalizing with PCSK9 and LDLR within 60 min. It increased until 240 min, at which point PCSK9 and LDLR disappeared (Supplementary material online, Figure S12A, C). This lysosome formation, but not the early endosome formation, was blocked by CAP1 depletion (Figure 4A, Supplementary material online, Figure S13).
Figure 4.
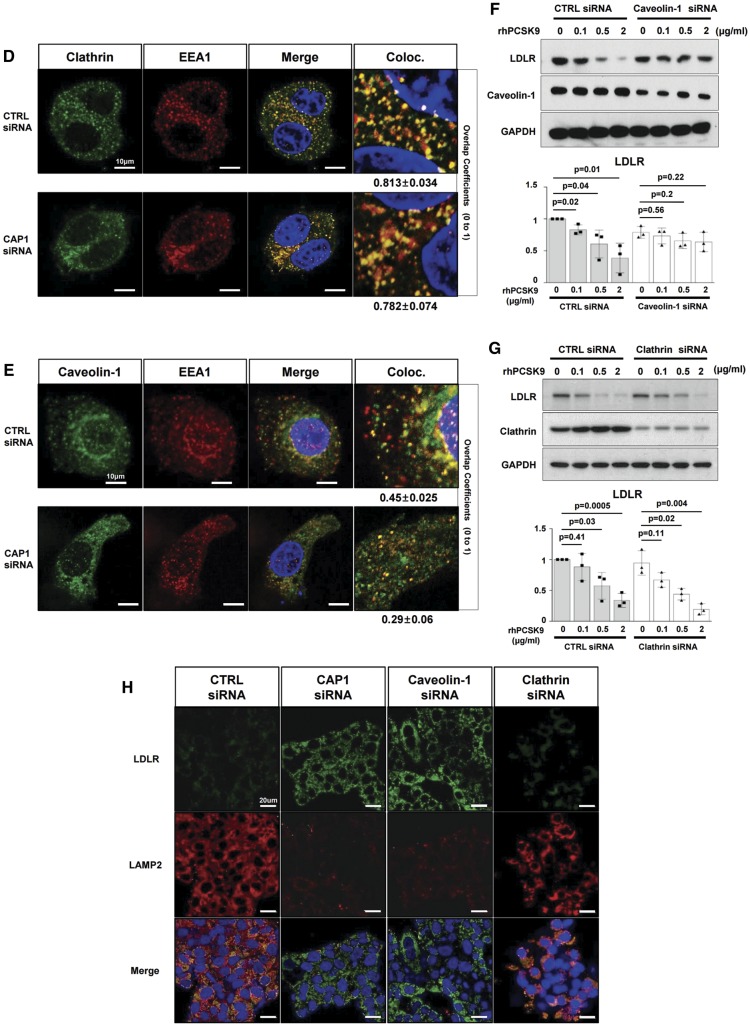
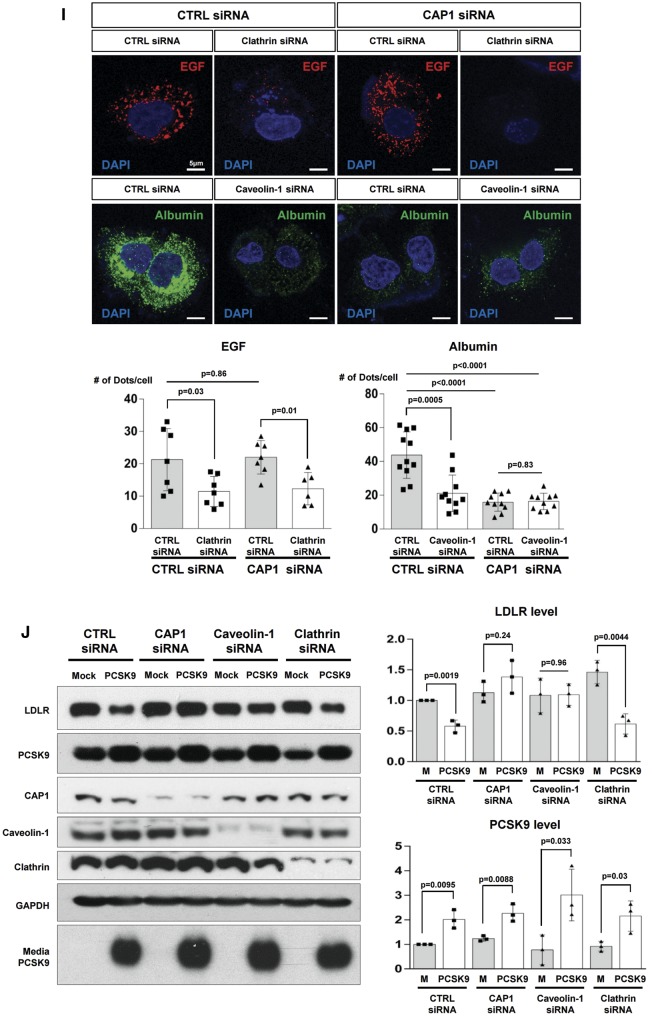
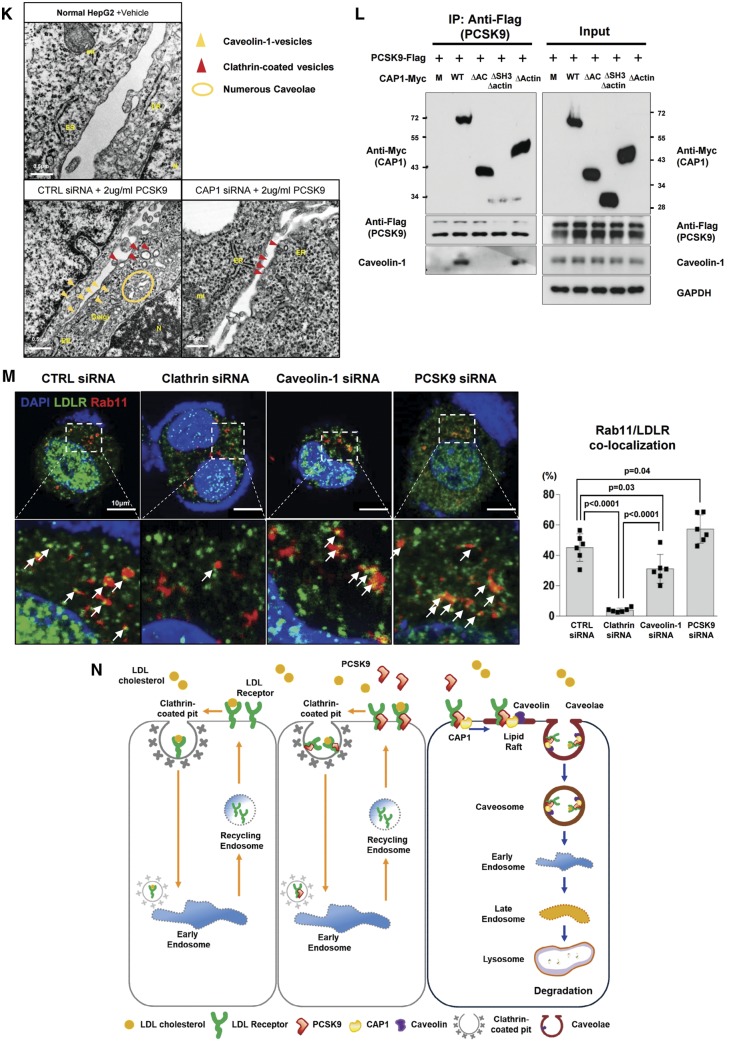
CAP1 mediates the caveolin-dependent endocytosis and lysosomal degradation of LDLR. (A) Immunofluorescent staining of HepG2 cells with LDLR (green), PCSK9 (red), and the lysosomal marker LAMP2 (white) 240 min after treatment with recombinant hPCSK9. (B) Membrane fractionation and immunoblot analysis demonstrating the cellular distribution of LDLR, PCSK9 and CAP1 after PCSK9 treatment in HepG2 cells treated with either a control or CAP1 siRNA. The representative figure of three independent experiments. (C) Quantification of the immunoblot. (D and E) Co-localization of endosomal marker EEA1 with clathrin. . (D) and caveolin-1 (E) in HepG2 cells treated with recombinant rhPCSK9 for 30 min. Overlap coefficients: mean ± standard deviation. (F and G) Immunoblot analysis of LDLR expression demonstrating that PCSK9-induced LDLR degradation was attenuated by caveolin siRNA (F) in HepG2 cells but was not affected by clathrin siRNA (G). Quantification of the expression levels of LDLR (bottom panel, n = 3). (H) Immunofluorescent staining of HepG2 cells with LDLR (green) and the lysosomal marker LAMP2 (red) 240 min after hPCSK9 treatment. HepG2 cells were pre-treated with either the control, CAP1, caveolin1, or clathrin siRNA. (I) Immunofluorescent staining of HepG2 cells with EGF (red) and albumin (green). Bar indicates 10 μm. The representative figure of three independent experiments, and the dots in a cell were counted from ten random fields of each independent experiment (bottom panel). (J) LDLR degradation by the overexpression of PCSK9 was attenuated by CAP1 siRNA and caveolin1 siRNA but not clathrin siRNA. Bar graphs showing the LDLR and PCSK9 level in the lysate. The representative figure of three independent experiments. (K) Transmission electron microscopic images of HepG2 cells without treatment (upper left) or with pretreatment with either a control siRNA (lower left) or CAP1 siRNA (lower right) and exposed to PCSK9 for 15 min. Numerous caveolae (yellow circle), caveolin-vesicles (yellow arrows), and clathrin-coated vesicles (red arrows) are observed in the control siRNA-PCSK9-treated cells, while only clathrin-coated vesicles are observed in the CAP1 siRNA-PCSK9-treated cells. Bar indicates 0.5 μm; ER, endoplasmic reticulum; MT, mitochondrion; N, nucleus. (L) hPCSK9-Flag and either wild-type CAP1-Myc or its mutants were overexpressed in HEK 293 cells. Immunoprecipitation with anti-Flag beads and immunoblotting with Myc or caveolin-1. (M) LDLR localizes to the recycling endosomes in PCSK9-deficient cells but not much in clathrin-deficient cells. Immunofluorescent staining of HepG2 cells ‘at 4 h after LDL-C treatment’ with Rab11 (red), LDLR (green), and DAPI (blue). Colocalization of Rab11 and LDLR was measured and expressed as percent colocalization (bottom graph). (N) Schematic model in which LDLR undergoes caveolae-dependent endocytosis and lysosomal degradation induced by PCSK9. CAP1 mediates this process by interfacing PCSK9 with caveolin through the SH3BD and ACBD.
Take home figure.
CAP1 is a new binding partner of PCSK9 and mediates caveolae-dependent endocytosis and lysosomal degradation of LDLR.
In the raft isolation experiment, endogenous PCSK9 before exogenous application was localized more in non-raft fractions [raft (36.8%)/non-raft (63.2%)]. Within 30 min after application of exogenous PCSK9, it was mainly incorporated in lipid raft fractions [raft (54.3%)/non-raft (45.7%)] (Figure 4B and C), which are closely involved in caveolae formation. LDLR expression in the membrane fraction decreases 60 min after the PCSK9 treatment. However, in the CAP1-deficient cells, there is no significant change in the PCSK9 and LDLR expression patterns, even after PCSK9 treatment (Figure 4B and C). It is reported that early endosome formation by PCSK9 is mediated by the clathrin pathway.14,15 However, PCSK9 treatment increased the numbers of endosomes colocalized with not only clathrin but also caveolin (Supplementary material online, Figure S14). Interestingly, regarding the enhanced endosome formation, either with clathrin or caveolin, after PCSK9 treatment, CAP1 knock-down decreased only the caveolin-endosome but not the clathrin-endosome, suggesting that CAP1 induces caveolin-mediated endocytosis of the PCSK9/LDLR complex but not clathrin-mediated endocytosis (Figure 4D and E).
Therefore, we knocked down caveolin or clathrin to compare caveolin- and clathrin-mediated LDLR endocytosis, respectively, following treatment with PCSK9. In the caveolin-deficient cells, the LDLR was not degraded even with PCSK9 treatment (Figure 4F). In the clathrin-deficient cells, however, the PCSK9 treatment was able to degrade the LDLR in a dose-dependent manner, implying that PCSK9-mediated LDLR degradation is clathrin-independent (Figure 4G). In the absence of caveolin, LAMP2 could not form and LDLR was not degraded by PCSK9, whereas in the absence of clathrin, LAMP2 appeared and LDLR was degraded (Figure 4H). In addition, the knock-down of CAP1 was unable to produce LAMP2, suggesting that CAP1 is closely involved in the caveolin-mediated degradation of LDLR by PCSK9 (Figure 4H). Next, we evaluated the effect of CAP1 deficiency on the endocytosis of EGF or albumin. EGF and its receptor complex is mainly internalized by clathrin-dependent endocytosis,16 whereas albumin uptake has been reported to be dependent on caveolae.17 The caveolin-dependent albumin endocytosis was significantly decreased by CAP1 deficiency, whereas the clathrin-dependent EGF receptor endocytosis was not affected (Figure 4I). These observations suggest that CAP1 might be involved not only in the endocytosis of the PCSK9/LDLR complex but also in caveolin-dependent endocytosis in general. LDLR degradation mediated by endogenously overexpressed PCSK9 was similarly attenuated by siRNA against CAP1 or caveolin (Figure 4J). An electron microscopic analysis (Figure 4K) demonstrated that only caveosome formation, but not clathrin, after PCSK9 treatment was significantly attenuated by CAP1 siRNA. The mechanism by which CAP1 guides the PCSK9/LDLR complex to the caveosome is based on the binding of the AC-domain of CAP1 to caveolin-1. In the immunoprecipitation experiment, PCSK9 (with LDLR) can form a complex with wtCAP1 and caveolin-1, whereas it cannot in the presence of mutant CAP1, such as ▵SH3BD▵actinBD or ▵ACBD mutant CAP1 (Figure 4L), suggesting again that the SH3BD of CAP1 is necessary for binding with the CRD of PCSK9. The ACBD of CAP1 is necessary for binding with caveolin. Immunoprecipitation assay with wild-type mouse liver lysate also showed that caveolin binds to LDLR, CAP1, and PCSK9 (Supplementary material online, Figure S15). In addition, LDLR/PCSK9/CAP1 complexes co-localized with caveolin in mouse liver (Supplementary material online, Figure S16). LDLR endocytosis co-stained with early endosomal marker Rab5,18 1 h after LDL-C treatment significantly decreased by both the clathrin siRNA and caveolin siRNA. When we blocked PCSK9 with the siRNA-attenuating LDLR degradation pathway, caveolin-mediated LDLR endocytosis was significantly reduced, whereas there was no significant change in clathrin-mediated endocytosis (Supplementary material online, Figure S17). Furthermore, when we co-stained LDLR with Rab11, which is related to the recycling mechanism,18 4 h after LDL-C treatment, the amount of recycling LDLR was significantly decreased by clathrin siRNA relative to that with caveolin siRNA (Figure 4M).
Discussion
Previous milestone discoveries in human genetics have established a clear link between PCSK9 function and LDL-C concentrations.19 The gain-of-function missense mutations in PCSK9 cause a rare form of autosomal codominant familial hypercholesterolaemia.20 Conversely, loss-of-function mutations in PCSK9 have been associated with hypocholesterolaemia and protection against atherosclerotic cardiovascular diseases.21 Mechanistic analysis revealed that secreted PCSK9 binds to the LDLR at the hepatocyte cell surface and promotes its endocytosis and lysosomal degradation.10 Molecular mapping of the PCSK9-LDLR binding interface led to the development of therapeutic anti-PCSK9 monoclonal antibodies.22 We have further explored in more detail how PCSK9 directed LDLR towards endocytosis and lysosomal degradation after binding to LDLR. In the present study, we have broadened our understanding of the process by identifying CAP1 as a new binding partner for PCSK9. First, we showed that the SH3BD of CAP1 binds directly with the CRD of PCSK9. Two loss-of-function polymorphisms found in human PCSK9 had a defective interaction with CAP1. We could prevent PCSK9-mediated LDLR degradation not only in cultured liver cells with siRNA against CAP1 but also in heterozygous CAP1 knock-out mice, which had lower plasma LDL-C levels. Finally, our mechanistic study elucidated that CAP1 binds to caveolin-1, subsequently leading the PCSK9-LDLR complex to caveolae-dependent endocytosis and lysosomal degradation.
Internalization of LDLR has been postulated as clathrin-mediated endocytosis. The adaptor protein ARH binds to the FXNPXY motif of the LDLR as well as clathrin and AP-2, which forms clathrin coated pits and leads to the intracellular recycling pathway.23 siRNA against clathrin inhibited LDLR degradation by PCSK9 overexpression in HepG2 cells.14 However, the recycling pathway and endogenously overexpressed PCSK9 might not be related to the endocytosis process that physiologically occurs for the degradation of LDLR. PCSK9-mediated degradation of LDLR could occur under hypertonic conditions, which blocks clathrin-coated pit formation.15 We observed that PCSK9-mediated LDLRs endocytosis occurred both via clathrin- and caveolae-dependent pathways. However, in the caveolin-deficient cells, the LDLR was not degraded with the PCSK9 treatment (Figure 4F), while LDLR in the clathrin-deficient cells were degraded in a dose-dependent manner (Figure 4G). The fate of the LDLR/PCSK9 complex differed depending on the endosome pathway: degradation in the caveolin-pathway vs. recycling in the clathrin-mediated pathway. There is a similar example of cell surface receptors that have two different fates, degradation vs. recycling, depending on the binding partners. TGFβRs present in lipid rafts are integrated into ‘caveolin-1 positive vesicles’, preferentially delivered to the SMAD7-SMURF2-ubiquitin-ligase complex, and degraded.24 However, for TGFβ-induced SMAD2 activation, TGFβRs are internalized through ‘clathrin-mediated endocytosis’ and found in Rab11-positive ‘recycling endosomes’, not in p62-positive late endosomes.25
As summarized in Figure 4N, LDLR enters cells once bound to LDL-C via clathrin-dependent endocytosis, and then the acidic pH of endosomes causes the allosteric dissociation of the LDLR and its recycling to the cell surface.26 During the process, adaptor molecules, such as ARH and DAB2, mediate clathrin-dependent endocytosis.27,28 While PCSK9 also promotes clathrin-dependent endocytosis, it does not lead to lysosomal degradation of LDLR (Figure 4G and H). When the complex of LDLR and PCSK9 interacts with CAP1, however, these proteins enter cells by caveolin-dependent endocytosis because CAP1 can bind to caveolin-1 through the ACBD (Figure 4L). Then, caveolin-coated endosomes containing LDLR/PCSK9/CAP1 are escorted to lysosomes for degradation. The identification of CAP1 as the unknown binding partner of PCSK9 during LDLR degradation clearly delineates caveolin-dependent LDLR endocytosis and lysosomal degradation.
Supplementary Material
Acknowledgements
We thank Jay Horton (UT Southwestern) for the Flag-tagged hPCSK9 and AAV-PCSK9 construct, and Kyoung Tai No for allowing to access the computational resources in his group.
Funding
This study was supported by the Korea Health Technology R&D Project ‘Korea Research-Driven Hospital (HI14C1277)’, ‘Strategic Center of Cell & Bio Therapy (HI17C2085)’, and ‘Medi-star (HI12C1691)’ through the Korea Health Industry Development Institute (KHIDI), funded by the Korea Government (MHW). The research was also supported by a National Research Foundation of Korea (NRF) grant funded by the Korea Government (MSIP) (NRF-2017R1D1A1A02017804).
Conflict of interest: none declared.
See page 253 for the editorial comment on this article (doi: 10.1093/eurheartj/ehz771)
References
- 1. Seidah NG, Awan Z, Chretien M, Mbikay M.. PCSK9: a key modulator of cardiovascular health. Circ Res 2014;114:1022–1036. [DOI] [PubMed] [Google Scholar]
- 2. Burke AC, Dron JS, Hegele RA, Huff MW.. PCSK9: regulation and target for drug development for dyslipidemia. Annu Rev Pharmacol Toxicol 2017;57:223–244. [DOI] [PubMed] [Google Scholar]
- 3. Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, Sever PS, Pedersen TR; FOURIER Steering Committee and Investigators. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med 2017;376:1713–1722. [DOI] [PubMed] [Google Scholar]
- 4. Ridker PM, Revkin J, Amarenco P, Brunell R, Curto M, Civeira F, Flather M, Glynn RJ, Gregoire J, Jukema JW, Karpov Y, Kastelein JJP, Koenig W, Lorenzatti A, Manga P, Masiukiewicz U, Miller M, Mosterd A, Murin J, Nicolau JC, Nissen S, Ponikowski P, Santos RD, Schwartz PF, Soran H, White H, Wright RS, Vrablik M, Yunis C, Shear CL, Tardif JC; SPIRE Cardiovascular Outcome Investigators. Cardiovascular efficacy and safety of bococizumab in high-risk patients. N Engl J Med 2017;376:1527–1539. [DOI] [PubMed] [Google Scholar]
- 5. Hampton EN, Knuth MW, Li J, Harris JL, Lesley SA, Spraggon G.. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc Natl Acad Sci USA 2007;104:14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cho Y, Lee SE, Lee HC, Hur J, Lee S, Youn SW, Lee J, Lee HJ, Lee TK, Park J, Hwang SJ, Kwon YW, Cho HJ, Oh BH, Park YB, Kim HS.. Adipokine resistin is a key player to modulate monocytes, endothelial cells, and smooth muscle cells, leading to progression of atherosclerosis in rabbit carotid artery. J Am Coll Cardiol 2011;57:99–109. [DOI] [PubMed] [Google Scholar]
- 7. Field J, Vojtek A, Ballester R, Bolger G, Colicelli J, Ferguson K, Gerst J, Kataoka T, Michaeli T, Powers S, Riggs L, Rodgers L, Wieland I, Wheland B, Wigler M.. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 1990;61:319–327. [DOI] [PubMed] [Google Scholar]
- 8. Lee S, Lee HC, Kwon YW, Lee SE, Cho Y, Kim J, Lee S, Kim JY, Lee J, Yang HM, Mook-Jung I, Nam KY, Chung J, Lazar MA, Kim HS.. Adenylyl cyclase-associated protein 1 is a receptor for human resistin and mediates inflammatory actions of human monocytes. Cell Metab 2014;19:484–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bertling E, Hotulainen P, Mattila PK, Matilainen T, Salminen M, Lappalainen P.. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol Biol Cell 2004;15:2324–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu Y, Li Q, Chen XZ.. Detecting protein-protein interactions by Far western blotting. Nat Protoc 2007;2:3278–3284. [DOI] [PubMed] [Google Scholar]
- 11. Hu CD, Chinenov Y, Kerppola TK.. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 2002;9:789–798. [DOI] [PubMed] [Google Scholar]
- 12. Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD.. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest 2006;116:2995–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, Trillard M, Abifadel M, Tebon A, Attie AD, Rader DJ, Boileau C, Brissette L, Chretien M, Prat A, Seidah NG.. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem 2004;279:48865–48875. [DOI] [PubMed] [Google Scholar]
- 14. Nassoury N, Blasiole DA, Tebon Oler A, Benjannet S, Hamelin J, Poupon V, McPherson PS, Attie AD, Prat A, Seidah NG.. The cellular trafficking of the secretory proprotein convertase PCSK9 and its dependence on the LDLR. Traffic 2007;8:718–732. [DOI] [PubMed] [Google Scholar]
- 15. Holla OL, Cameron J, Berge KE, Ranheim T, Leren TP.. Degradation of the LDL receptors by PCSK9 is not mediated by a secreted protein acted upon by PCSK9 extracellularly. BMC Cell Biol 2007;8:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sigismund S, Argenzio E, Tosoni D, Cavallaro E, Polo S, Di Fiore PP.. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev Cell 2008;15:209–219. [DOI] [PubMed] [Google Scholar]
- 17. Chatterjee M, Ben-Josef E, Robb R, Vedaie M, Seum S, Thirumoorthy K, Palanichamy K, Harbrecht M, Chakravarti A, Williams TM.. Caveolae-mediated endocytosis is critical for albumin cellular uptake and response to albumin-bound chemotherapy. Cancer Res 2017;77:5925–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Trischler M, Stoorvogel W, Ullrich O.. Biochemical analysis of distinct Rab5- and Rab11-positive endosomes along the transferrin pathway. J Cell Sci 1999;112: 4773–4783. [DOI] [PubMed] [Google Scholar]
- 19. Abifadel M, Rabes JP, Devillers M, Munnich A, Erlich D, Junien C, Varret M, Boileau C.. Mutations and polymorphisms in the proprotein convertase subtilisin kexin 9 (PCSK9) gene in cholesterol metabolism and disease. Hum Mutat 2009;30:520–529. [DOI] [PubMed] [Google Scholar]
- 20. Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C.. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 2003;34:154–156. [DOI] [PubMed] [Google Scholar]
- 21. Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH.. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med 2006;354:1264–1272. [DOI] [PubMed] [Google Scholar]
- 22. Chan JC, Piper DE, Cao Q, Liu D, King C, Wang W, Tang J, Liu Q, Higbee J, Xia Z, Di Y, Shetterly S, Arimura Z, Salomonis H, Romanow WG, Thibault ST, Zhang R, Cao P, Yang XP, Yu T, Lu M, Retter MW, Kwon G, Henne K, Pan O, Tsai MM, Fuchslocher B, Yang E, Zhou L, Lee KJ, Daris M, Sheng J, Wang Y, Shen WD, Yeh WC, Emery M, Walker NP, Shan B, Schwarz M, Jackson SM.. A proprotein convertase subtilisin/kexin type 9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc Natl Acad Sci USA 2009;106:9820–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nagai M, Meerloo T, Takeda T, Farquhar MG.. The adaptor protein ARH escorts megalin to and through endosomes. Mol Biol Cell 2003;14:4984–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Roy C, Wrana JL.. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat Rev Mol Cell Biol 2005;6:112–126. [DOI] [PubMed] [Google Scholar]
- 25. Di Guglielmo GM, Le Roy C, Goodfellow AF, Wrana JL.. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat Cell Biol 2003;5:410–421. [DOI] [PubMed] [Google Scholar]
- 26. Jeon H, Blacklow SC.. Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem 2005;74:535–562. [DOI] [PubMed] [Google Scholar]
- 27. Garuti R, Jones C, Li WP, Michaely P, Herz J, Gerard RD, Cohen JC, Hobbs HH.. The modular adaptor protein autosomal recessive hypercholesterolemia (ARH) promotes low density lipoprotein receptor clustering into clathrin-coated pits. J Biol Chem 2005;280:40996–41004. [DOI] [PubMed] [Google Scholar]
- 28. Maurer ME, Cooper JA.. The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. Journal of Cell Science 2006;119:4235–4246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.