Abstract
Background
Intervertebral disc degeneration (IVDD) is a well-known cause of lower back pain, which is induced by multiple factors including increased apoptosis and decreased survival of nucleus pulposus cells. In this study, we evaluate the effect and potential mechanism of miR-660 on the nucleus pulposus cells apoptosis induced by TNF-α.
Methods
First, we collected tissue of nucleus pulposus from IVDD and healthy controls. General characteristic of the IVDD and healthy control was also collected. And, we also collected nucleus pulposus cells that stimulated by TNF-α or control. miRNA microarray was performed to identify the differentially expressed miRNAs. Apoptosis rate and miR-660 relative expression was measured after stimulated with different concentration of TNF-α to identify the optimal concentration of TNF-α. Second, we successfully constructed antigomiR-660 to block the miR-660 expression in nucleus pulposus cells and then stimulated with TNF-α (100 ng/ml, 12 h). The apoptosis rates and relative protein expression were then measured again. The target association between miR-660 and SAA1 was confirmed by dual-luciferase reporter.
Results
There was no significant difference between the age (IVDD: 39 ± 10 years, healthy controls: 36 ± 7 years), BMI and sex between IVDD and healthy controls. Microarray analysis found that miR-660 was significantly up-regulated in IVDD and TNF-α treated groups, which was further identified by PCR. We found that the rate of apoptosis and miR-660 expression increased with TNF-α concentration increased. Finally, TNF-a with 100 ng/ml was used for further experiment. Compared with TNF-α group, TNF-α + antigomiR-660 could significantly down-regulated the apoptosis rate and relative protein (c-Caspase3 and c-Caspase7). Dual-luciferase reporter revealed that miR-660 could directly binding to the SAA1 at 80–87 sites. Compared with TNF-α alone group, TNF-α + antigomiR-660 significantly up-regulated the SAA1 expression (P < 0.05).
Conclusion
These results indicated that knockdown of miR-660 protected the nucleus pulposus from apoptosis that induced TNF-α via up-regulation of SAA1. Further studies should focus on the role of miR-660 in protecting IVDD in vivo.
Keywords: MicroRNA, Apoptosis, Nucleus pulposus cell, Serum amyloid A1
Background
Intervertebral disc degeneration (IVDD) is a well-known cause of lower back pain and has become the most common orthopedic disease today [1, 2]. Nearly 80% of people will suffer from low back pain in their own life span [3]. Drug therapy (Non-steroidal anti-inflammatory drugs or pregabalin), vertebrectomy and decompression can alleviate the low back pain [4, 5]. However, recurrent pain and disc degeneration of adjacent segments were the weakness of these two therapies [6].
A large of studies shown that the degeneration mainly displays as a large number of the nucleus pulposus cell apoptosis and thus nucleus pulposus cells apoptosis plays a crucial role in IVDD [7, 8]. TNF-α is a key proinflammatory cytokine and could induce the apoptosis of nucleus pulposus cells [9]. Thus, TNF-α was commonly administrated as the model of IVDD [10].
MicroRNAs (miRNAs) are small single-stranded non-coding RNAs with apoptosis-promoting or anti-apoptotic roles, consisting of 20–26 nucleotides [11]. MiRNAs inhibited gene expression through binding to the 3′ untranslated regions (3’UTR) of their target mRNAs [12]. There is growing evidence that miRNAs play key roles in IVDD and miR-199a was associated with clinical grades of degeneration [13].
In this study, we performed microRNA microarray analysis for nucleus pulposus cells between the IVDD and healthy controls. We found that miR-660 was significantly up-regulated in IVDD than healthy control. Moreover, we identify the potential mechanism of miR-660 for the apoptosis of nucleus pulposus cells.
Methods
Sample collection
This study was approved by the Ethics Committee of Huai An Hospital of Chinese Medicine. All included patient signed and dated informed consent form. Human nucleus pulposus specimens were collected from patients with lumbar disc herniation (n = 3; male = 1, female = 2, age = 39 ± 10) and lumbar burst fractures as control [n = 3; male = 2, female = 1, age = 36 ± 7]. Human nucleus pulposus specimens were classified as grade II (lumbar burst fractures) and grade IV (IVDD patients) according to MRI [14]. Human lumbar nucleus pulposus samples were collected from patients with spine burst fractures. There was no significant difference between the IVDD and healthy controls in terms of the BMI (24 ± 2 vs 25 ± 2, P = 0.123). Clinical features of study population can be seen in Table 1.
Table 1.
General characteristic of the included patients
| Variable | Normal | IVDD | P value |
|---|---|---|---|
| Age | 36 ± 7 | 39 ± 10 | 0.153 |
| BMI | 24 ± 2 | 25 ± 2 | 0.123 |
| Sex | |||
| Male | 2 | 1 | 1 |
| Female | 1 | 2 |
miRNAs microarray
The isolated nucleus pulposus tissues in IVDD and healthy controls were immerse into RNA preservation solution and then stored at − 80 °C. Total RNA was extracted and RNA quality was determined by 260/280. RNA labelling and hybridization on miRNA microarray chips were performed as previously described [15]. Following background correction and data normalization with the median method, differentially expressed genes (DEGs) between IVDD and healthy controls was performed using the limma package (version 3.5.1). Volcano plot and heatmap of the differentially expressed genes (DEGs) were also obtained.
Cell culture
Human lumbar nucleus pulposus were kept in PBS contained with 1% penicillin. Nucleus pulposus cells were isolated according to previously described method. In brief, after nucleus pulposus were cut into pieces, the pieces were then digested by 0.2% collagenase type II for 60 min. Then, cell suspension was filtered by strainer and collected the cells by centrifuge in a 1000 r and 5 min. Nucleus pulposus cells were cultured into dulbecco’s modified eagle medium (DMEM) supplemented with 10% FBS and 100 U/mL penicillin in a 37 °C, 5% CO2 environment.
Cell transfection
The lentiviral vector encoding antigomiR-660 and the miR-660 vector were purchased and synthesized by Servicebio (Wuhan, Hubei Province, China). Human nucleus pulposus cells were seeded in a 24-well plate at a density of 3.0 × 105 cells/well. Then, cells were randomly divided into following groups: TNF-α group, TNF-α + vector control group and TNF-α + antagomir-660 group. Following 6 h of incubation, the nucleus pulposus were collected and performed following experiments.
Quantitative real-time RT-PCR
Total RNA was extracted by TRIzol Reagent (Invitrogen). Real-time PCR was performed according to previously described. The PrimeScript RT reagent kit (TaKaRa, Japan) was employed for complementary DNA (cDNA) synthesis, according to the manufacturer’s instructions. RT-PCR analysis was performed with the SYBR Premix Ex Taq II kit (TaKaRa) and detection on a Roche LightCycler 480 sequence detection system. U6 and GAPDH served as an internal reference for quantitation of miRNA and mRNA respectively. Primer for PCR can be seen in Table 2.
Table 2.
Primer information of miRNA for quantitative reverse transcription
| Name | Primer name | Primer sequence (5′ to 3′) |
|---|---|---|
| miR-660 | miR-660-F | TACCCATTGCATATCGG |
| miR-660-R | GTGCAGGGTCCGAGGT | |
| s | U6-F | TGTGTCCGTCGTGGATCTGA |
| U6-R | CCTGCTTCACCACCTTCTTGA |
Apoptosis assay: flow cytometry
Nucleus pulposus cells receiving different treatments were collected and washed with PBS for three times. Apoptosis was detected using an Annexin V-FITC/PI Apoptosis Kit (BD Biosciences, Franklin Lakes, NJ, USA). The collected cells were stained with annexin V-FITC and propidium iodide for 15 min according to the manufacturer’s instructions. Apoptosis was detected with a BD FACS flow cytometer (BD Biosciences).
Western blot
Total proteins were extracted from the nucleus pulposus cells using RIPA buffer (Solarbio, Beijing, China). Protein concentration was determined by BCA Protein Assay Kit (Solarbio, Beijing, China). Equal amount of protein was resolved on a 10% SDS-PAGE gel and transferred to polyvinylidene fluoride membranes (Servicebio, Wuhan, China). The membranes were then washed with 5% nonfat milk in Tris-buffered saline plus 0.1% Tween 20. Subsequently, the membrane was incubated at 4 °C overnight with antibodies specific for c-Caspase3, c-Caspase7, MMP3, MMP13, Collagen II, Aggrecan, SSA1 and GAPDH (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). After the membranes were washed three times with TBST, horseradish peroxidase-conjugated secondary antibody was incubated for 1 h at indoor temperature. The immunoreactivities were detected enhanced chemiluminescence kit (Santa Cruz Biotechnology, Dallas, TX, USA).
Luciferase reporter constructs and assays
The 3′-untranslated region (3′-UTR) of SAA1 fragments were inserted into luciferase vector (Promega, WI, USA). NPCs were seeded in 96-well plates at 8 × 103 cells per well, and co-transfected with the vectors, miR-660 and luciferase vector. The luciferase activity was measured using a luminometer (Promega, WI, USA) after 48 h. The cells were incubated in 5% CO2 at 37 °C for 48 h. The dual luciferase reporter assay system (Promega Corporation) was used to measure luciferase activity. The firefly luciferase activity was normalized.
Statistical analyses
All data are expressed as mean ± SD. Statistical analyses were performed using statistical software programs SPSS 17.0 (IBM, NY, USA) and GraphPad Prism 7 (GraphPad, CA, USA). p < 0.05 was considered statistically significant.
Results
MiR-660 was up-regulated in degenerated nucleus pulposus tissues
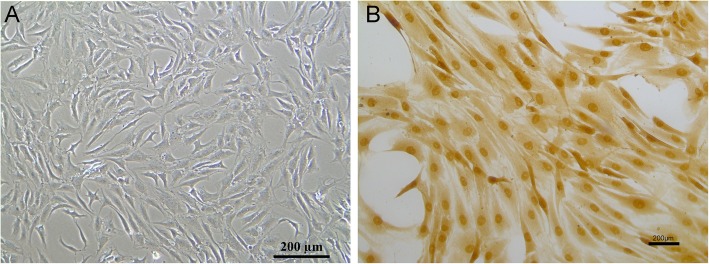
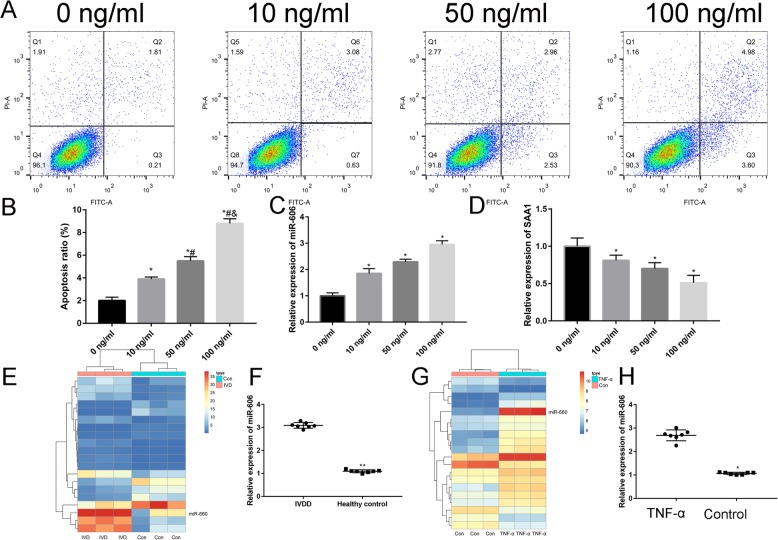
As shown in Fig. 1a, nucleus pulposus cells emerge as long spindle and strong expressed with type II collagen (Fig. 1b). As shown in Fig. 2a and b, TNF-α with 100 ng/ml resulted in the highest apoptosis rate than control group and other concentration of TNF-α. Thus, we administrated TNF-α (100 ng/ml, 12 h) for subsequent experiments. PCR results found that TNF-α increased miR-660 (Fig. 2c) and decreased SAA1 expression (Fig. 2d) in concentration manners.
Fig. 1.
cell morphology of nucleus pulposus cells (a); immunohistochemical of collagen II (b)
Fig. 2.
a Effect of TNF-α on nucleus pulposus cells apoptosis. TNF-α-induced apoptosis in concentration (0 ng/ml, 10 ng/ml, 50 ng/ml, and 100 ng/ml) at 12 h (*p < 0.05); b the percentages of apoptotic cells for all groups; c Relative expression of miR-660 in different concentration of TNF-α groups (0 ng/ml, 10 ng/ml, 50 ng/ml, and 100 ng/ml). d Relative expression of SAA1 in different concentration of TNF-α groups (0 ng/ml, 10 ng/ml, 50 ng/ml, and 100 ng/ml); e heatmap of differentially expressed genes in IVDD and healthy controls; f PCR result to identify the relative expression of miR-660 in IVDD and healthy control; g heatmap of differentially expressed genes in TNF-α and control group; h PCR result to identify the relative expression of miR-660 in TNF-α and control group
Following gene expression data normalization, heatmap of the DEGs can be seen in Fig. 2e, and miR-660 was the most significantly up-regulated miRNA in IVD patients, which verified by PCR (Fig. 2f).
Heatmap of DEGs between TNF-α and control group can be seen in Fig. 2g, and miR-660 was the most significantly up-regulated miRNA in TNF-α treated group, which verified by PCR (Fig. 2g and h).
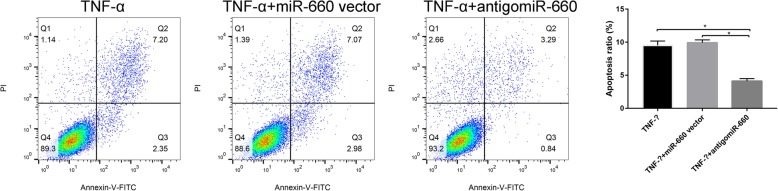
MiR-660 mediates TNF-α induced nucleus pulposus cells apoptosis
The rate of apoptosis was significantly decreased in the antigomiR-660+ TNF-α compared to the TNF-α alone and TNF-α + vector groups (Fig. 3, *P < 0.05). These results indicated that knockdown miR-660 could protect nucleus pulposus cells apoptosis that stimulated by TNF-α.
Fig. 3.
Comparison of the apoptotic cells of the different treated groups (TNF-α vs TNF-α + miR-660 vector vs TNF-α + antigomiR-660, *p < 0.05)
MiR-660 potentially mediates cell apoptosis via targeting SAA1
MiR-600 had potential binding sites in the 3’UTR of SAA1 (Fig. 4a). To identify whether miR-660 can directly target SAA1 3’UTR, luciferase report vectors were generated with wild-type SAA1 3’UTR (pGL3-SAA1-WT) and mutated SAA1 3’UTR (pGL3-SAA1-MUT). The SAA1 luciferase expression vector and activity were measured for describing miR-660 function on luciferase translation. Luciferase activity of SAA1 was inhibited by miR-660 significantly, yet pGL3-SAA1-MUT relieved this effect. Then, we proved that SAA1 is the direct target of miR-660.
Fig. 4.
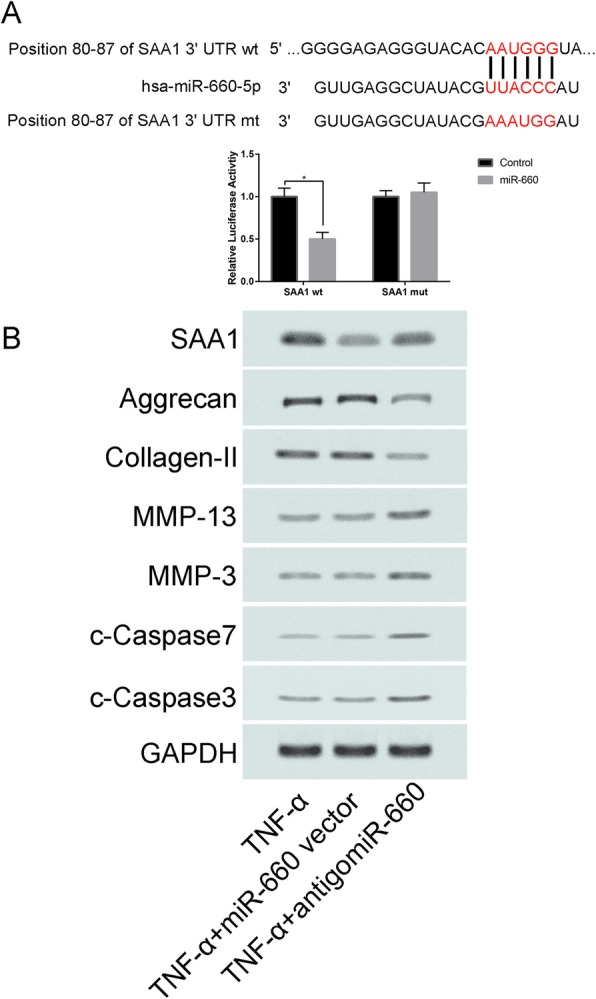
a schematic of SAA1 mRNA showing a potential binding site in the SAA 3′-UTR for miR-660; SAA1-mut indicates the SAA1 with the mutated sites; b Western-blot analyses of different protein expression (c-Caspase3, c-Caspase7, MMP-3, MMP-13, Collagen-II, Aggrecan, SAA1) in TNF-α, TNF-α + miR-660 vector and TNF-α + antigomiR-660 groups
In addition, the expression levels of the apoptotic proteins c-Caspase3 and c-Caspase7 were significantly increased in (Fig. 4b) than TNF-α and TNF-α + miR-660 vector groups. Moreover, the expression of MMP-3, MMP-13, Collagen-II and Aggrecan in TNF-α + antigomiR-660 were significantly increased than TNF-α and TNF-α + miR-660 vector groups (Fig. 4b, P < 0.05).
Discussion
Nucleus pulposus cells apoptosis is the main reason for IVDD [16]. In current study, we administrated with different dose of TNF-α to explore the optimal dose of TNF-α to induce apoptosis of nucleus pulposus cells. Optimal dose of TNF-α for inducing nucleus pulposus cells apoptosis was 100 ng/ml. Wang et al. [17] stimulated nucleus pulposus cells with TNF-α at different concentrations, and they found that 100 ng/ml was the optimal dose to induce apoptosis of nucleus pulposus cells. We then performed miRNAs microarray chip between nucleus pulposus cells in patient with IVDD and healthy controls. We found that miR-660 was significantly up-regulated in the IVDD patients than healthy controls. Furthermore, we identified that the relative expression of miR-660 in TNF-α treated group was higher than control group.
We found that knockdown of miR-660 protects nucleus pulposus cells from TNF-α-induced apoptosis by targeting serum amyloid A1. A major strength of current study was that we firstly identify the role of miR-660 in the process of IVDD.
In the past, some miRNAs, like miR-184 [18] and miR-640 [19], are reported to be involved in regulation of nucleus pulposus cells apoptosis. Dong et al. [19] found that miR-640 aggravates intervertebral disc degeneration via NF-κB and WNT signaling pathway. Cai et al. [20] found that miR-15a could targeting MAP 3 K9 and might be considered as a novel therapeutic target for IVDD treatment.
Firstly, we found that miR-660 was significantly up-regulated in the TNF-α treated group than control group. We used Targetscan and miRDB databases to identify the overlapping target genes of miR-660. According to the cumulative weighted context++ score, we found that SAA1 rank the first, and we further explored the role of SAA1 in the apoptosis of nucleus pulposus cells.
Serum amyloid A1 (SAA1) is a 12.5 kd acute phase protein which could induce inflammation, proliferation and cell death [21]. And previous study has shown that SAA possess an anti-apoptotic role in liver injury and other disease [22]. We found that miR-660 could directly bind to the SAA1 and thus exert an anti-apoptotic role for nucleus pulposus cells.
There are several limitations of our experiment. Firstly, although our results support that knockdown miR-660 could ameliorate nucleus pulposus cells apoptosis by regulating serum amyloid A1, the in vivo effects remain unclear. Secondly, the downstream signaling pathway was not further explored, further investigations are needed to gain deeper understanding of the role of miR-660 in IVDD.
Conclusion
In conclusion, the present study demonstrated that knockdown miR-660 protects nucleus pulposus cells from TNF-α-induced apoptosis by targeting serum amyloid A1. These findings provide a better understanding of the mechanism involved in the pathogenesis of IVDD and miR-660 may be as a potential target for IVDD.
Acknowledgements
Not applicable.
Abbreviations
- 3’UTR
3′ untranslated regions
- DEGs
Differentially expressed genes
- DMEM
Dulbecco’s modified eagle medium
- IVDD
Intervertebral disc degeneration
- miRNAs
MicroRNAs
- SAA1
Serum amyloid A1
Authors’ contributions
HJZ and XHM design the study; SLX and SlQ perform the study; CZL and ZGZ supervise the experiment. All authors prove the final study. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
We declare that the materials described in the manuscript will be freely available to all scientists for non-commercial purposes.
Ethics approval and consent to participate
Ethics approval was approved by Huai An Hospital of Chinese Medicine and all patients were consent to participate into this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu G, Cao P, Chen H, Yuan W, Wang J, Tang X. MiR-27a regulates apoptosis in nucleus pulposus cells by targeting PI3K. PLoS One. 2013;8(9):e75251. doi: 10.1371/journal.pone.0075251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Xunlin, Wu Aimin, Han Chen, Chen Chen, Zhou Tangjun, Zhang Kai, Yang Xiao, Chen Zhiqian, Qin An, Tian Haijun, Zhao Jie. Bone marrow-derived mesenchymal stem cells in three-dimensional co-culture attenuate degeneration of nucleus pulposus cells. Aging. 2019;11(20):9167–9187. doi: 10.18632/aging.102390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi Y, Ma J, Chen Y. PTEN promotes intervertebral disc degeneration by regulating nuclear pulposus cell behaviors. Cell Biol Int. 2019. [epub ahead of print]. [DOI] [PubMed]
- 4.Enke O, New HA, New CH, Mathieson S, AJ ML, Latimer J, Maher CG, Lin CC. Anticonvulsants in the treatment of low back pain and lumbar radicular pain: a systematic review and meta-analysis. CMAJ. 2018;190(26):E786–e793. doi: 10.1503/cmaj.171333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sclafani J, Leong M, Desai MJ, Mehta N, Sayed D, Singh JR. Conventional vs high frequency neuromodulation in the treatment of Low Back Pain following Spine Surgery. PM R : J Injury Function Rehabil. 2019. [epub ahead of print]. [DOI] [PubMed]
- 6.Forozeshfard M, Jahan E, Amirsadat J, Ghorbani R. Incidence and factors contributing to low Back pain in the nonobstetrical patients operated under spinal anesthesia: a prospective 1-year follow-up study. J Perianesth Nurs. 2019. [epub ahead of print]. [DOI] [PubMed]
- 7.Jiao S, Li J, Liu B, Yang M, Xiu J, Qu D. Nucleus pulposus cell apoptosis is attenuated by CDMP-2 through regulating oxidative damage under the hyperosmotic environment. Biosci Rep. 2018;38(5):20181176. [DOI] [PMC free article] [PubMed] [Retracted]
- 8.Li Z, Chen X, Xu D, Li S: Circular RNAs in nucleus pulposus cell function and intervertebral disc degeneration. 2019;52(6):e12704. [DOI] [PMC free article] [PubMed]
- 9.Xie L, Huang W, Fang Z, Ding F, Zou F, Ma X, Tao J, Guo J, Xia X, Wang H, et al. CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis. 2019;10(10):751. doi: 10.1038/s41419-019-1978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Xue-Qiang, Tu Wen-Zhan, Guo Jia-Bao, Song Ge, Zhang Juan, Chen Chang-Cheng, Chen Pei-Jie. A Bioinformatic Analysis of MicroRNAs’ Role in Human Intervertebral Disc Degeneration. Pain Medicine. 2019;20(12):2459–2471. doi: 10.1093/pm/pnz015. [DOI] [PubMed] [Google Scholar]
- 11.Ma QQ, Huang JT, Xiong YG, Yang XY, Han R, Zhu WW. MicroRNA-96 regulates apoptosis by targeting PDCD4 in human Glioma cells. Technol Cancer Res Treat. 2017;16(1):92–98. doi: 10.1177/1533034616629260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Jicheng, Liu Shizhang, Shi Jiyuan, Li Jingyuan, Wang Songbo, Liu Huitong, Zhao Song, Duan Keke, Pan Xuezhen, Yi Zhi. The Role of miRNA in the Diagnosis, Prognosis, and Treatment of Osteosarcoma. Cancer Biotherapy and Radiopharmaceuticals. 2019;34(10):605–613. doi: 10.1089/cbr.2019.2939. [DOI] [PubMed] [Google Scholar]
- 13.Farrokhi MR, Karimi MH, Ghaffarpasand F, Sherafatian M. MicroRNA-199a Upregulation mediates lumbar intervertebral disc degeneration and is associated with clinical grades of degeneration. Turk Neurosurg. 2019. [Epub ahead of print]. [DOI] [PubMed]
- 14.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine. 2001;26(17):1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 15.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101(26):9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qin C, Lv Y, Zhao H, Yang B, Zhang P. MicroRNA-149 suppresses inflammation in nucleus Pulposus cells of intervertebral discs by regulating MyD88. Med Sci Monit. 2019;25:4892–4900. doi: 10.12659/MSM.915858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Li P, Ma X, Tian P, Han C, Zang J, Kong J, Yan H. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-alpha-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. doi: 10.1016/j.biochi.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Wang P, Zhang Z, Wang W, Liu Y, Qi Q. MiR-184 Regulates Proliferation in Nucleus Pulposus Cells by Targeting GAS1. World Neurosurg. 2017;97:710–715.e711. doi: 10.1016/j.wneu.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Dong W, Liu J, Lv Y, Wang F, Liu T, Sun S, Liao B, Shu Z. MiR-640 aggravates intervertebral disc degeneration via NF-kappaB and WNT signalling pathway. Cell Prolif. 2019;52(5):e12664. doi: 10.1111/cpr.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai P, Yang T, Jiang X, Zheng M, Xu G, Xia J. Role of miR-15a in intervertebral disc degeneration through targeting MAP 3K9. Biomed Pharmacother. 2017;87:568–574. doi: 10.1016/j.biopha.2016.12.128. [DOI] [PubMed] [Google Scholar]
- 21.Sun L, Ye RD. Serum amyloid A1: structure, function and gene polymorphism. Gene. 2016;583(1):48–57. doi: 10.1016/j.gene.2016.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siegmund SV, Schlosser M, Schildberg FA, Seki E, De Minicis S, Uchinami H, Kuntzen C, Knolle PA, Strassburg CP, Schwabe RF. Serum amyloid a induces inflammation, proliferation and cell death in activated hepatic stellate cells. PLoS One. 2016;11(3):e0150893. doi: 10.1371/journal.pone.0150893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declare that the materials described in the manuscript will be freely available to all scientists for non-commercial purposes.