Abstract
Background
Growth hormone inducible transmembrane protein (GHITM) is a highly conserved transmembrane protein. This study was conducted to investigate the role of GHITM gene in the apoptosis and growth of the golden apple snail Pomacea canaliculate.
Results
The complete cDNA of this gene was cloned using the rapid amplification of cDNA ends (RACE) method and subjected to bioinformatics analysis. The full-length cDNA was 2242 bp, including an open reading frame of 1021 bp that encoded a protein of 342 amino acid residues. The mRNA expression profiles of GHITM gene in different tissues (liver, kidney, gonad and foot) and different growth phases (6-months old and 2-years old) showed that it was expressed in various tissues and different growth phases. Silencing of the GHITM gene by RNAi (RNA interference) experiments revealed that the GHITM gene possibly plays a role in inhibiting apoptosis through detecting the Caspase (Cysteine-requiring Aspartate Protease)-3 activity. In addition, the aperture width and body whorl length of the snail was significantly affected by RNAi, suggesting that this gene plays a significant role in promoting the growth of the organism.
Conclusions
These results demonstrated that the GHITM gene was involved in apoptosis and growth in golden apple snail.
Keywords: Pomacea canaliculate, GHITM gene, Complete cDNA, RNAi, Cell apoptosis, Growth
Background
Growth hormone inducible transmembrane protein (GHITM) is a highly conserved transmembrane protein that exists widely in plants and animals [1]. This gene plays an important role in the growth and anti-aging mechanisms of the body. Reimers analysis revealed that there was a special structure (UPF005) at the C-terminus that is unique to the BAX inhibitor (BI)-1 superfamily [2]. Therefore, the GHITM gene was classified as a member of the BI-1 superfamily. Other members of this superfamily, responsive to centrifugal force and shear stress gene 1, Golgi anti-apoptotic protein and Lifeguard, all play roles in the growth and development of the body and inhibit apoptosis [3–6]. We speculate that GHITM also has similar functions, but the specific effects need further research and certification.
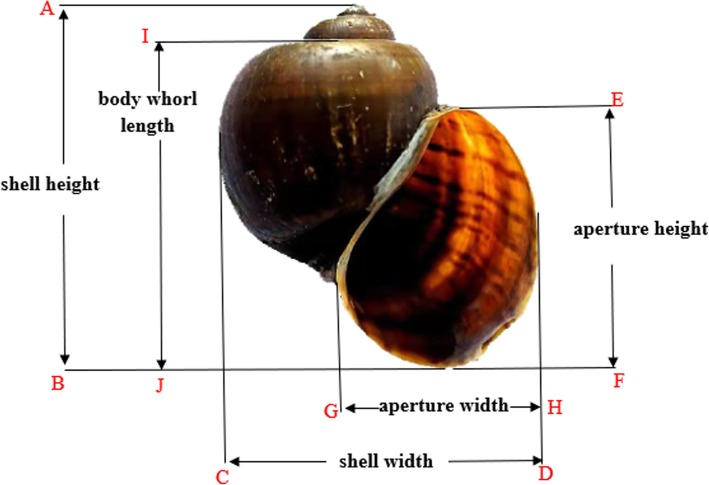
The golden apple snail (its body size traits were shown in Fig. 1.) originated in South America and was introduced into Asia as food in the 1980s [7, 8]. It has been listed as an invasive species owing to its characteristics of feeding on a variety of crops and rapidly reproducing. It also has a low economic value [9, 10] and serious damage to the development of the rice planting industry. In addition to the lack of natural enemies, its strong adaptability to temperature [11–15], food [16, 17] and wet environments [18], as well as its strong ability to grow and reproduce have been keys to its invasiveness. Studies of other species have shown that the GHITM gene functions in regulating the growth of the body and in cell apoptosis. Therefore, improving our knowledge of the GHITM gene of the golden apple snail may elucidate the mechanisms behind the golden apple snail’s strong adaptability and rapid growth and reproduction. In addition, golden apple snails are rich in fat and protein [19]; therefore, studying genes related to their growth may help increase their protein levels, making them feed for some economically important fish [20, 21]. The GHITM gene was first discovered in the brown fat of mice [1], and then was studied in Crassostrea gigas [22], Anthocidaris crassispina [23], Bombyx mori [24], Actiniaria, Pinctada martensii and Apostichopus japonicus [25]. To date, no studies on the GHITM gene in Pomacea canaliculata have been published.
Fig. 1.
The measurement standards of the growth indexes. AB, shell height; CD, shell width;EF, aperture height; GH, aperture width; IJ, body whorl length
Apoptosis is an important mechanism for the preservation of a healthy and balanced immune system in vertebrates [26]. For molluscs, more and more researches showed that apoptosis was related with maintenance of tissue homeostasis, responses to environmental stress, and fights against diseases [27]. Especially, hemocytes can participate in all steps of the immune response pathway as cellular mediators in molluscs [28], and integrins regulate the phagocytic ability of molluscan hemocytes and induce hemocytic apoptosis [29, 30].
In this study, the full-length cDNA sequence of the GHITM gene was cloned successfully, and the structure, properties and subcellular localization of the putative protein were predicted using bioinformatics software. The expression levels of the GHITM gene in different tissues of golden apple snail were assessed by quantitative real-time PCR. After RNAi-mediated knockdown for this gene, the growth and apoptotic indexes of the snail were evaluated and compared. The findings in this study will provide insights into the function of GHITM gene in the golden apple snail.
Results
Characterization of the GHITM cDNA sequence
The full-length cDNA sequence of GHITM included a 5′- UTR (Untranslated Region) of 324 bp, an open reading frame of 1029 bp, encoding a protein of 342 amino acids, and a 3′-UTR of 890 bp. The GenBank accession number for this nucleotide sequence is MN219409. The characterization is shown as (Fig. 2).
Fig. 2.

Nucleotide and deduced amino acid sequences of GHITM cDNA. The start code ATG is in bold, and the stop codon is indicated with an asterisk. In the 3′ UTR, the consensus polyA signal AATTAAA is double underlined. The amino acids of the transmembrane region are underlined
Bioinformatics analysis of GHITM
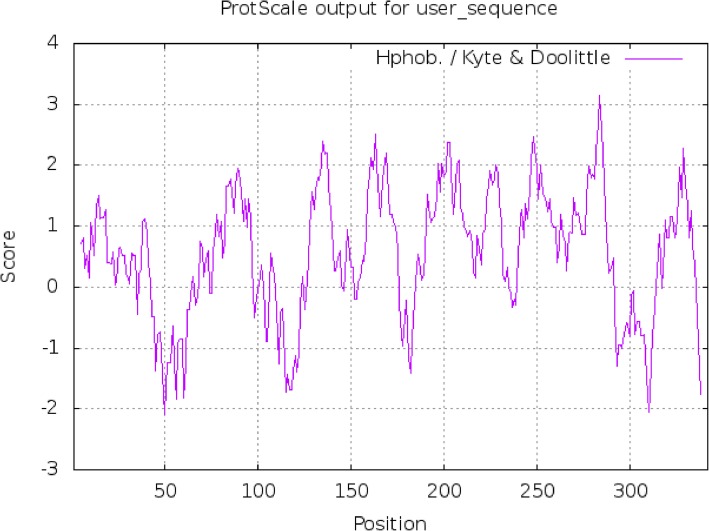
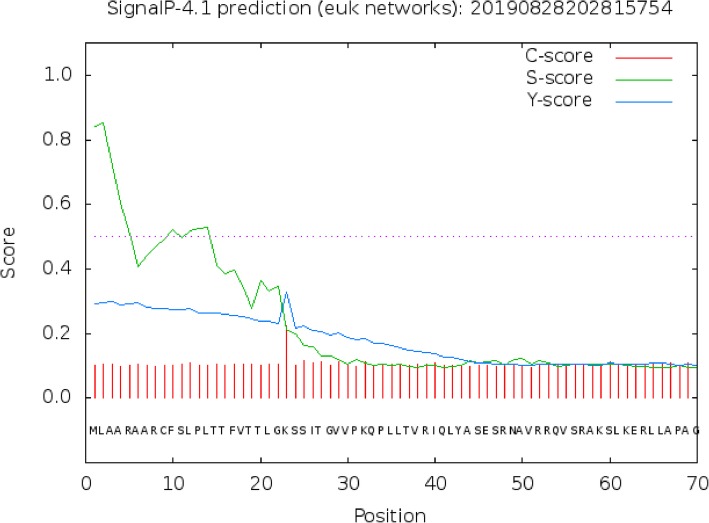
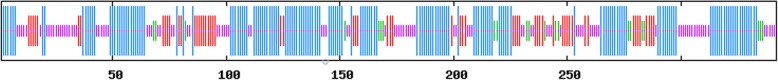
The identified open reading frame of GHITM encoded a putative protein of 342 amino acids with a predicted molecular mass of 36,373.87 Da and theoretical isoelectric point of 9.98. The total number of negatively charged residues (Asp + Glu) was 14, and the total number of positively charged residues (Arg + Lys) was 32. The instability index was calculated to be 44.87, and the adipose index was 109.06. The minimum value of the strongest hydrophilic amino acid was − 2.089, and the maximum value of the strongest hydrophobic amino acid was 3.156. The average hydrophilic coefficient was 0.530, suggesting that this is a hydrophobic protein (Fig. 3). SignalP was used to predict the signal peptide position of the protein, and the average D value was 0.416, which is less than the threshold 0.450. Thus, the existence of signal peptide is not likely (Fig. 4). Using online analysis of SOPMA server, the secondary structure of GHITM protein was predicted to be: 161 alpha helices (Hh), accounting for 47.08%; 59 beta folds (Ee), 17.25%; 21 beta angles (Tt), accounting for 6.14%; There were 101 crimps without regulation (Cc), accounting for 29.53% (Fig. 5).
Fig. 3.
Hydrophilic and hydrophobic profile of the GHITM protein. Notes: The horizontal axis indicates the positions of amino acids in this protein and vertical axis indicates the hydrophobicity
Fig. 4.
Prediction of the golden apple snail GHITM protein’s signal peptide. Note: C-score, raw cleavage site score. S-score, signal peptide score. Y-score, combined cleavage site score.
Fig. 5.
Prediction of the second structure of GHITM protein. Blue, red, green and purple stand for alpha helix, extended strand, beta turn and random coil, respectively
The subcellular localization of the GHITM protein was predicted using the software PSORT II Prediction. In total, 55.6, 33.3 and 11.1% of the GHITM protein was distributed in the endoplasmic reticulum, plasma membrane and vacuole, respectively. Therefore, the GHITM protein appears to be mainly distributed in cytoplasm.
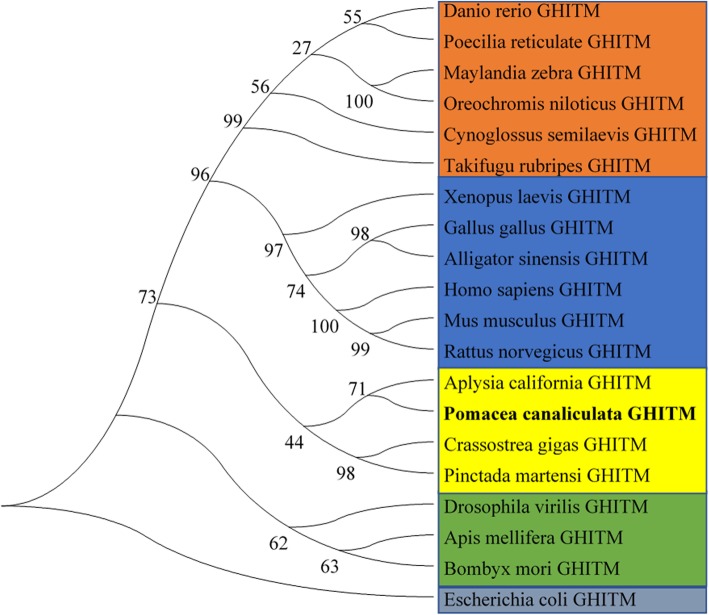
A phylogenetic tree was constructed using the MEGA 7.0 program with the Neighbor-Joining method according to the deduced amino acid sequences of GHITM and other previously reported GHITM proteins. The phylogenetic tree revealed that these GHITMs were divided into three clusters, vertebrates, mollusks, and insects. The GHITM of golden apple snails was grouped into the clade containing GHITMs from molluscs. Moreover, it was most closely related to that of Aplysia california (Fig. 6), and they are both Gastropods.
Fig. 6.
The phylogenetic tree of GHITM amino acid sequences from 20 species
Expression level of GHITM in different tissues and different growth phases of the Golden apple snail
To explore the expression pattern of the GHITM gene in different tissues, qPCR was performed using total RNA from liver, kidney, gonad and foot. GHITM gene is generally expressed in various tissues without significant difference (Fig. 7). In addition, to detect the expression pattern of the GHITM gene during different growth phases, RT-qPCR was performed with total RNA from young (6-months old) and adult (2-years old) golden apple snails. The GHITM transcript levels were not significantly different between 6-month-old and 2-year-old golden apple snails (Fig. 8).
Fig. 7.
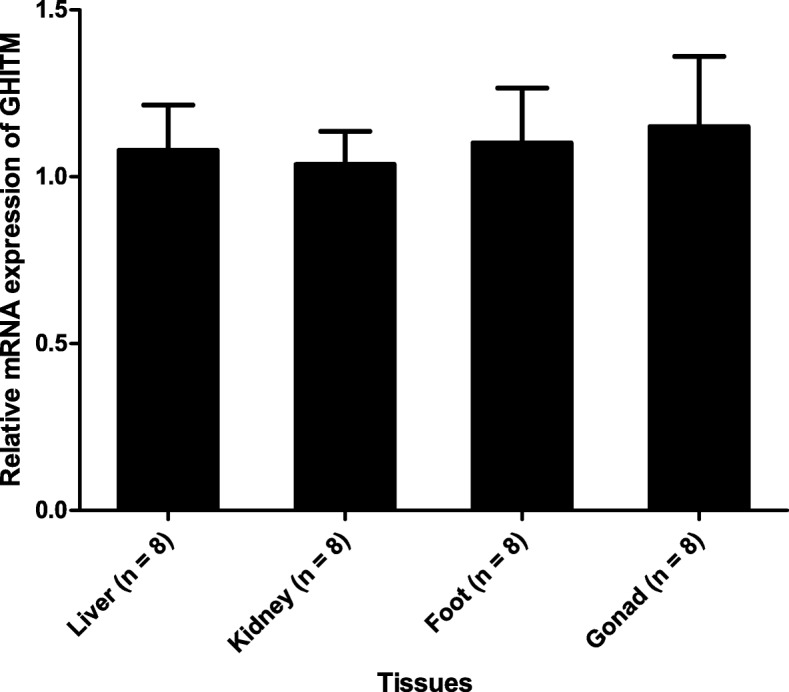
Expression analysis of GHITM in different tissues of the golden apple snail (n = 8). Each bar represents the mean of 8 independent experiments performed in duplicate. The real-time quantitative PCR (qPCR) data were expressed as the mean ± SE
Fig. 8.
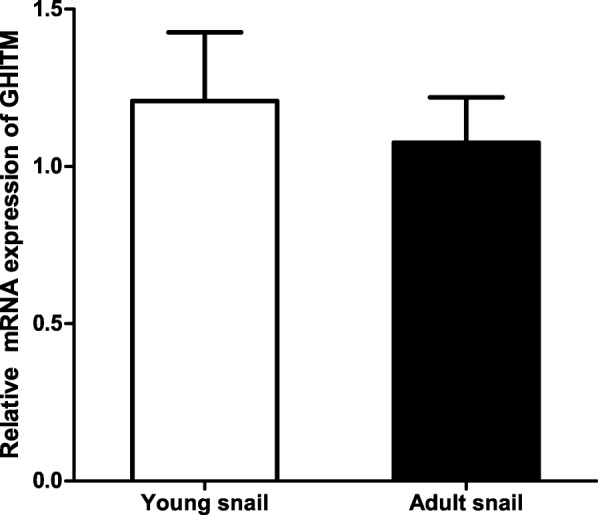
GHITM mRNA expression during different growth phases of the golden apple snail (n = 8). Each bar represents the mean of 8 independent experiments performed in duplicate. The real-time quantitative PCR (qPCR) data were expressed as the mean ± SE
The siRNA-mediated Down-regulation of target genes
To reveal the function of the GHITM gene, RNAi was performed on healthy golden apple snails. Because the foot had the highest GHITM expression level of the tested organs, it was used to determine changes after RNAi. The expression of the GHITM gene was down-regulated by RNAi, with a significantly lower expression level being detected when 10 μg siRNA + 50 μl PBS was used (Fig. 9).
Fig. 9.
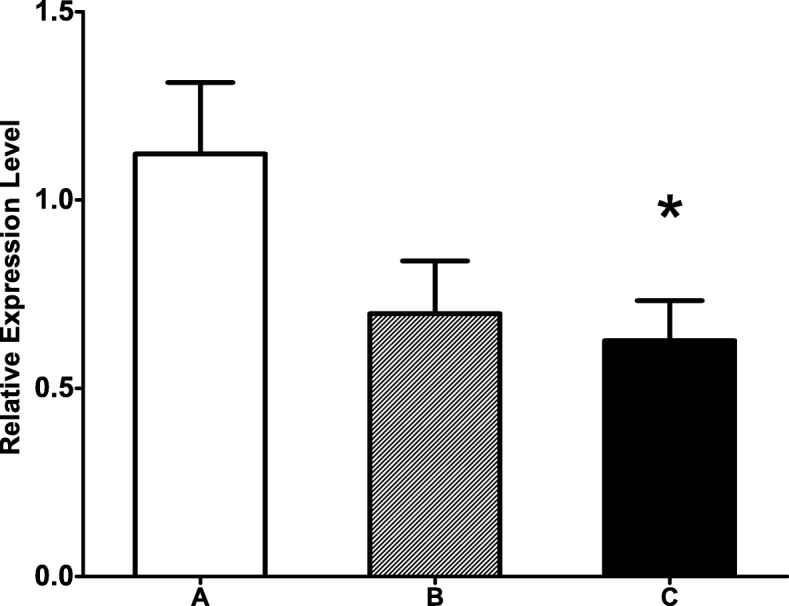
Expression of GHITM gene in foot after RNAi, A was a control group (n = 10), B and C were two experimental groups (n = 8). Each bar represents the mean of 8 independent experiments performed in duplicate. The (*) indicates statistically significant difference (P < 0.05). The real-time quantitative PCR (qPCR) data were expressed as the mean ± SE
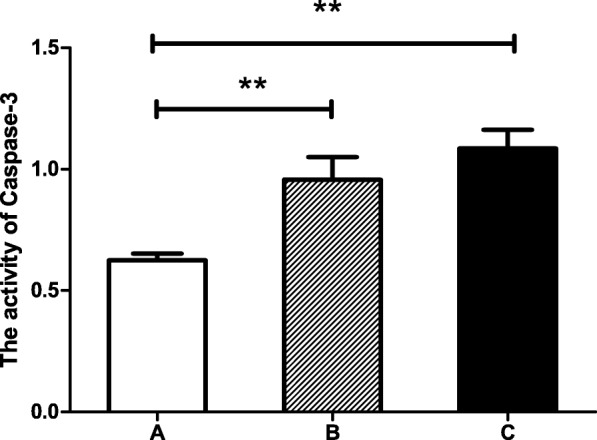
Apoptosis index measurement
The activity levels of caspase-3 in the feet of the three groups were measured after the RNAi experiment. The activity levels of caspase-3 in the two experimental groups were significantly higher than that of the control group (Fig. 10).
Fig. 10.
The activity of Caspase-3 in foot of the 3 groups golden apple snails (n = 8). Each bar represents the mean of 8 independent experiments performed in duplicate. The double-asterisk (**) indicates statistically significant difference (P < 0.01). The data were expressed as the mean ± SE
The effects of RNAi on the growth rate
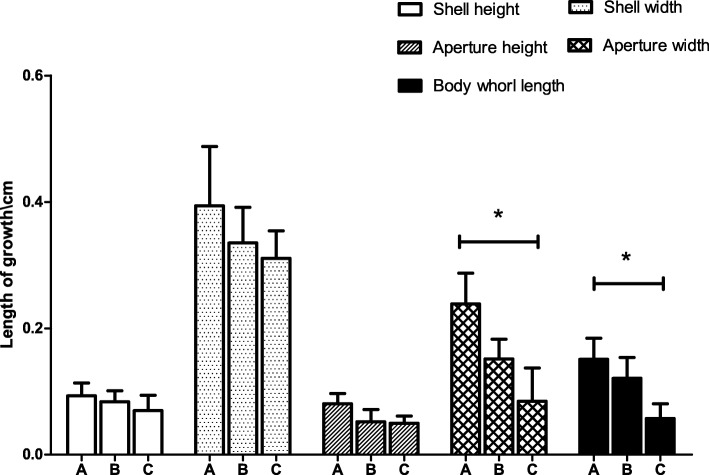
The changes in the growth indexes, including shell height, shell width, aperture height, aperture width and body whorl length, of the control and experimental groups were calculated. The calculation method subtracted the first measurement value from the second measurement value. The change in the growth of the control group was greater than those of the two experimental groups, and the aperture width and body whorl length of the control group were significantly greater than those of the C group (Fig. 11).
Fig. 11.
The change of shell height, shell width, aperture height, aperture width and body whorl length (A is control group, B and C are experimental groups, n = 8. Each bar represents the mean of 8 independent experiments performed in duplicate. The (*) indicates statistically significant difference)
Discussion
Since the first discovery of GHITM in mice in 2001, it has been identified successively in vertebrates and invertebrates, such as human, chicken, zebrafish and oyster [23]. In this study, the GHITM gene was identified for the first time in golden apple snail and this is also the first report of full-length cDNA cloning and sequence analysis of GHITM from the golden apple snail.
Bioinformatics methods, including computer simulations and predictions of gene structure and biological function, have been widely used in pathogen detection, gene information analysis, vaccine development and other fields [31]. In this study, the GHITM gene was cloned and its physical and chemical properties, hydrophilic and hydrophobic properties, signal peptide prediction, subcellular localization prediction and other bioinformatics analyses were also performed to predict the basic structure, properties and distribution of the GHITM gene of the golden apple snail. In addition, a phylogenetic tree was constructed using the GHITM amino acid sequences of 20 species, and it revealed that the GHITM sequence of the golden apple snail was highly homologous with that of A. california, which was consistent with the classification provided by biological evolution. These results provide information for the further study of the gene’s function.
The transcript levels of the GHITM gene in different tissues (liver, kidney, gonad and foot) and different growth phases (6-months old and 2-years old) were assessed using fluorescent qPCR. The results showed that GHITM gene was expressed in different tissues and growth stages, and also widely throughout individuals, which was basically consistent with previous studies on other species [23, 32]. To some extent, it was speculated that the gene may play a role in different tissues and different growth phases of golden apple snail.
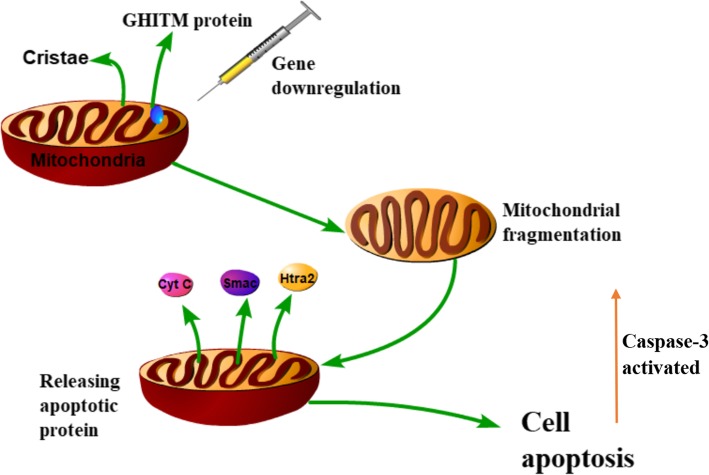
Mitochondria play an important role in the early stage of apoptosis. The release of pro-apoptotic proteins in mitochondria activate downstream caspase proteins and leads to apoptosis [33]. Caspase-3 belongs to the CED (Caenorhabditis elegans death gene)-3 subfamily of the Caspase family [34]. In a normal state, caspase-3 exists in the cytoplasm in the form of zymogen with no activity. However, caspase-3 was activated when apoptosis occurs. We reduced the expression of the GHITM gene in the experimental group by RNAi technology, and then tested the activity of caspase-3 in the control group and the experimental group. The activity level of caspase-3 in the two experimental groups were significantly higher than that in the control group, indicating that apoptosis of the two experimental groups was severe. Therefore, it was speculated that the GHITM gene was involved in inhibiting apoptosis in golden apple snails. GHITM was reported to be a mitochondrial protein that functions to maintain cristae organization [33, 35]. We presumed that down-regulation of the GHITM gene leads to mitochondrial rupture, which leads to the release of apoptotic factors such as cytochrome c, Smac (Second mitochondria-derived activator of caspase), and Htra2 (HtrA serine peptidase 2) [36], and ultimately induces apoptosis and activates Caspse-3 (Fig. 12).
Fig. 12.
Predictive pathway map of apoptosis induced by downregulation of GHITM gene
The effect of the GHITM gene on the growth of the golden apple snail was verified by comparing the changes in several growth indexes between the RNAi experiment groups and the control group after 20 d. Therefore, it was deduced that the growth rates of the golden apple snails in the experimental groups were slow owing to the RNAi-based disrupted expression of the GHITM gene. Thus, the GHITM gene could affect the growth of the golden apple snail. The golden apple snail was introduced into Asia as a human food, but it has become a serious pest of important agricultural crops as well as a threat to aquatic and wetland ecosystems [37]. Currently, the prevention and control of the golden apple snail are concentrated on mechanical methods, biological control and the use of chemical pesticides [38, 39]. The study results provide a theoretical basis and new insights into the prevention and control of this agricultural pest.
As a biological process, apoptosis is carried out during maturation, remodeling, growth, developmental, defense and immunity processes [40, 41]. There were some studies on inhibiting apoptosis and promoting cell growth, for example, transcriptional co-activators with PDZ-binding motif (TAZ) can promote cell growth and inhibits Celastrol-induced cell apoptosis [42], HSP90 (Heat Shock Protein 90) can inhibit apoptosis and promote growth by regulating HIF-1α (Hypoxia Inducible Factor-1α) abundance in hepatocellular carcinoma [43]. In this study, it was verified that the GHITM gene played roles in inhibiting apoptosis and promoting growth in the golden apple snail, which could increase our understanding of apoptosis and growth mechanisms in molluscs.
Conclusions
These results demonstrated that the GHITM gene was involved in apoptosis and growth in golden apple snail, which provides a theoretical basis and new insights into the prevention and control of the growth and reproduction of the golden apple snail as an agricultural pest.
Methods
Animals
Healthy golden apple snails (Adult snail body weight = 25 - 28 g, young snail body weigh = 8 - 10 g) were collected from the Xiamen, Fujian Province, China, and maintained in freshwater in an incubator at 25 ± 2 °C (12-h:12-h light: dark photocycle) before the experiment. The golden apple snails were fed Chinese cabbage daily.
RNA extraction and cDNA synthesis
Total RNA was extracted from the golden apple snails using the TRIzol (TaKaRa Biotechnology, Dalian, China) method. Briefly, samples were dissociated in TRIzol, then extracted using chloroform to eliminate proteins, precipitated with isopropyl alcohol and successively washed with 75% ethanol. RNase inhibitor was added into the extracted RNA to ethanol. RNase inhibitor was added into the extracted RNA to eliminate RNase. The quality, purity and integrity of the RNA were determined using a spectrophotometer (A 260/A 280) and agarose gel electrophoresis. The full-length cDNA was synthesized using SMARTer® RACE 5′/3′ Kit following the instructions in the User Manual (634,858, TaKaRa Biotechnology, Dalian, China). The RNA and cDNA were stored at − 80 °C for further analyses.
GHITM cDNA cloning
A partial sequence (GBZZ01005442.1) was predicted to be the GHITM cDNA of the golden apple snail by a comparison between the transcriptome (GBZZ00000000.1) and GenBank database. Specific primers were designed based on the predicted sequences (Table 1). PCR was conducted at 95 °C for 3 min followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. A final extension step at 72 °C for 5 min was performed, and the PCR products were cloned into the pMD-18 T vector (6011, TaKaRa Biotechnology, Dalian, China) and sequenced. Based on the sequenced cDNA fragment, the 5′- and 3′-ends of the GHITM fragment were cloned using the SMARTer® RACE 5′/3′ Kit (634,858, TaKaRa Biotechnology, Dalian, China).
Table 1.
Primers used in gene cloning and RT-qPCR
| Primer | Sequences (5′-3′) | Purpose |
|---|---|---|
| GHITM-F | ACTTCCTTTTGCTGCTTCC | Conventional PCR |
| GHITM-R | GGAGCACTTGTCTACTATGGGT | |
| 5′- GHITM-R | GATTACGCCAAGCTTCCTGTCTTTGCTCAACGCCCTTAT | RACE |
| 3′- GHITM-F | GATTACGCCAAGCTTCCTGCGCTTAACTTCTTCTGACCAC | |
| Long primer | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | |
| Short primer | CTAATACGACTCACTATAGGGC | |
| qGHITM-F | CGCTGTTTCTCCCTACCAT | qRT-PCR |
| qGHITM-R | AAGCCAGGAATTTCTCATC | |
| q18S RNA-F | AATACATGCAAACCAGCTCC | |
| q18S RNA-R | ATTTTTCGTCACTACCTCCC |
Bioinformatics analysis of GHITM
The nucleic acid sequence was transformed into the amino acid sequence using Editseq of DNAStar (Madison, Wisconsin, USA). The amino acid isoelectric point and molecular mass of the protein were analyzed by ProtParam (http://web.expasy.org/protparam/). A hydrophilic and hydrophobic profile of the GHITM gene was predicted using the software Protscale (https://web.expasy.org/protscale/). The SignalP 4.1 server (http:/www.cbs.dtu.dkP/ services/SignalP-4.1/) was used to predict the signal peptide of GHITM. The subcellular localization and protein function of the GHITM protein were analyzed using PSORT II Prediction (http: r/p sort.hgc. jp/form2.html) and Protfun2.2 (http://www.cbs.dtu.dk/services/prot function/). The secondary structure of GHITM protein was predicted Using online analysis of SOPMA server (https://omictools.com/sopma-tool). A multiple sequence alignment of GHITM with other GHITM protein sequences from mollusk, crustaceans and vertebrates was performed using the ClustalW Multiple Alignment, and a phylogenetic tree was constructed with the Neighbor-Joining method using MEGA 7.0 (Center for Evolutionary Medicine and Informatics, The Biodesign Institute).
GHITM mRNA expression in different tissues and different growth phases
The transcript levels of the GHITM gene in different tissues (liver, kidney, gonad and foot) and various growth phases (6 months and 2 years) were assessed using fluorescent quantitative PCR (qPCR) with the SYBR@ Premix Ex Taq™ (DRR081A, TaKaRa Biotechnology, Dalian, China). A pair of specific primers qGHITM -F/R (Table 1) for qPCR was designed, and the q18S RNA gene was used as an internal control. The qPCR profile was 95 °C for 2 min followed by 39 cycles at 95 °C for 5 s and 60 °C for 30 s. The amplification efficiency (E) of each primer pair was calculated when the curve of the Ct [Log (cDNA dilution)] was generated. The E value of qGHITM-F/q18S RNA-R was 100.6%. The relative transcription levels of the GHITM gene from different samples (n = 8) were analyzed using the 2−ΔΔCt method [44].
The growth index measurement
A total of 30 golden apple snails were randomly divided into 3 groups. A control group (named A) and two experimental groups (named B and C). The growth indexes, including shell height, shell width, aperture height, aperture width and body whorl length, were measured [45].
RNAi experiment
Based on the sequence of GHITM, the target siRNA was designed and synthesized by Sangon Biotech (Shanghai). The RNA interference location for GHITM was labeled as 271TP. The following siRNA sequences were used in this experiment: Sense: GCUCUACGGCCUUGACAUUTT and Antisense: AAUGUCAAGGCCGUAGAGCTT.
The siRNAs of the GHITM gene were dissolved in PBS buffer for foot muscle injection. The experimental groups (B and C) were injected with the following treatments: 5 μg siRNA + 50 μl PBS and 10 μg siRNA + 50 μl PBS, respectively, while the control group (A) was only injected with 50 μl of PBS. Preliminary experiments showed that the effects of a PBS injection were the same as no injection; consequently, the PBS injection group was selected as the control group. The foot muscle injection was conducted once every 4 d, for 20 d.
Verification of RNAi interference effects
After 20 d of siRNA injection, members of the three groups living golden apple snails (n = 8) were sacrificed, and their foot tissues were sampled. Total RNAs were extracted from foot tissues and reverse transcribed into cDNA for real-time qPCR experiments. The mRNA expression levels were assessed using the SYBR@ Premix Ex Taq™ (DRR081A, TaKaRa Biotechnology, Dalian, China) and analyzed using the 2−ΔΔCt method.
Apoptosis index measurement
To verify the possible role of GHITM in apoptosis, the activity levels of caspase-3 in the feet of the three groups of golden apple snails (n = 8) were also measured after the RNAi experiment. After homogenization of the tissue, Ac-DEVD-pNA was added. After the sample turned yellow, the absorbance of A400nm was measured by a microplate reader, and the relative caspase 3 activity was calculated according to the ratio of the absorbance of the apoptosis-induced cells to the absorbance of the blank control cells (Caspase 3 Activity Assay Kit, G015–1-3, Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Further reflects the degree of apoptosis of tissue samples.
The growth index measurement after RNAi experiment
To determine the possible function of GHITM in maintaining normal growth, we measured the growth index and observed the changes between experimental groups and the control group (n = 8) after the RNAi experiment.
Acknowledgements
We would like to thank Dr. Baoyu Huang for the suggestions of animal feeding.
Abbreviations
- BI
BAX inhibitor
- Caspase
Cysteine-requiring Aspartate Protease
- CED
Caenorhabditis elegans death gene
- GHITM
Growth hormone inducible transmembrane protein
- HIF-1α
Hypoxia Inducible Factor-1α
- HSP90
Heat Shock Protein 90
- Htra2
HtrA serine peptidase 2
- qPCR
Real-time quantitative PCR
- RACE
Rapid amplification of cDNA ends
- RNAi
RNA interference
- Smac
Second mitochondria-derived activator of caspase
- TAZ
Transcriptional co-activators with PDZ-binding motif
- UTR
Untranslated Region
Authors’ contributions
WXT conceived and designed the experiments, YWC and ZBL performed the experiments, SHC, ZR and LLL analyzed the data, HC, JQY and WXN contributed materials, and YWC prepared figures and/or tables, WXT, YWC and ZBL wrote the article and WL, ZNN and GW reviewed and edited the manuscript before submission. All authors have read and approved the final manuscript.
Funding
This study was supported by National Key R&D Program of China (No. 2018YFD0901400), the National Natural Science Foundation of China (Nos. 41876193 and 41906088), the Key R & D Program of Shandong Province, China (No. 2018GHY115027), the Special Funds for Taishan Scholars Project of Shandong Province, China (No. tsqn201812094), the Shandong Provincial Natural Science Foundation, China (No. ZR2019MC002), the Fine Agricultural Breeds Project of Shandong Province, China (2019LZGC020), the Modern Agricultural Industry Technology System of Shandong Province, China (SDAIT-14-03), the Plan of Excellent Youth Innovation Team of Colleges and universities in Shandong Province, China (2019KJF004) and the Key R&D Program of Yantai City, China (No. 2017ZH054).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The cDNA sequences generated during the current study are available in the [GenBank] repository, [MN219409].
Ethics approval and consent to participate
Ethical review and approval were not required for the animal study because Pomacea canaliculata is not an endangered or protected species and is not a vertebrate.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenchao Yu and Baolu Zhang contributed equally to this work.
References
- 1.Li Y, Kelder B, Kopchick JJ. Identification, isolation, and cloning of growth hormone (GH)-inducible interscapular brown adipose complementary deoxyribonucleic acid from GH antagonist mice. Endocrinology. 2001;142(7):2937–2945. doi: 10.1210/endo.142.7.8233. [DOI] [PubMed] [Google Scholar]
- 2.Reimers K, Choi CY, Bucan V, Vogt PM. The growth-hormone inducible transmembrane protein (Ghitm) belongs to the Bax inhibitory protein-like family. Int J Biol Sci. 2007;3(7):471–476. doi: 10.7150/ijbs.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Q, Reed JC. Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell. 1998;1(3):337. doi: 10.1016/S1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 4.Holzerlandt R, Orengo C, Kellam P, Albà MM. Identification of new herpesvirus gene homologs in the human genome. Genome Res. 2002;12(11):1739–1748. doi: 10.1101/gr.334302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Somia NV, Schmitt MJ, Vetter DE, Antwerp DV, Heinemann SF, Verma IM. LFG: an anti-apoptotic Gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci U S A. 1999;96(22):12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reimers K, Choi CY, Bucan V, Vogt PM. The Bax Inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med. 2008;8(2):148–156. doi: 10.2174/156652408783769562. [DOI] [PubMed] [Google Scholar]
- 7.Halwart M. The golden apple snail Pomacea canaliculata in Asian rice farming systems: present impact and future threat. Intjpest Manag. 1994;40(2):199–206. [Google Scholar]
- 8.Joshi RC. Problems with the Management of the Golden Apple Snail Pomacea canaliculata: an important exotic Pest of Rice in Asia. Netherlands: Springer; 2007. [Google Scholar]
- 9.Baloch WA, Memon UN, Burdi GH, Soomro AN, Tunio GR, Khatian AA. Invasion of channeled apple snail Pomacea canaliculata, Lamarck (Gastropoda: Ampullariidae) in Haleji Lake, Pakistan. Alzheimers Dementia. 2012;10(4):654. [Google Scholar]
- 10.Brönmark C, Hansson LA. Invading Herbivory: the Golden apple snail alters ecosystem functioning in Asian wetlands. Ecology. 2004;85(6):1575–1580. doi: 10.1890/03-3146. [DOI] [Google Scholar]
- 11.Wada T, Matsukura K. Linkage of cold hardiness with desiccation tolerance in the invasive freshwater apple snail, Pomacea canaliculata (Caenogastropoda: Ampullariidae) J Molluscan Stud. 2011;77(2):149–153. doi: 10.1093/mollus/eyq049. [DOI] [Google Scholar]
- 12.Matsukura K, Tsumuki H, Izumi Y, Wada T. Physiological response to low temperature in the freshwater apple snail, Pomacea canaliculata (Gastropoda: Ampullariidae) J Exp Biol. 2009;212(Pt 16):2558. doi: 10.1242/jeb.031500. [DOI] [PubMed] [Google Scholar]
- 13.Giraudbilloud M, Vega IA, Tosi ME, Abud MA, Calderón ML, Castrovazquez A. Antioxidant and molecular chaperone defences during estivation and arousal in the south American apple snail Pomacea canaliculata. J Exp Biol. 2013;216(4):614–622. doi: 10.1242/jeb.075655. [DOI] [PubMed] [Google Scholar]
- 14.Zheng G, Dong S, Hou Y, Yang K, Yu X. Molecular characteristics of HSC70 gene and its expression in the golden apple snails, Pomacea canaliculata (Mollusca: Gastropoda) Aquaculture. 2012;358-359(1):41–49. doi: 10.1016/j.aquaculture.2012.06.002. [DOI] [Google Scholar]
- 15.Song HM, Mu XD, Gu DE, Luo D, Yang YX, Xu M, Luo JR, Zhang JE, Hu YC. Molecular characteristics of the HSP70 gene and its differential expression in female and male golden apple snails (Pomacea canaliculata) under temperature stimulation. Cell Stress Chaperones. 2014;19(4):579–589. doi: 10.1007/s12192-013-0485-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lach L, Britton DK, Rundell RJ, Cowie RH. Food preference and reproductive plasticity in an invasive freshwater snail. Biol Invasions. 2000;2(4):279–288. doi: 10.1023/A:1011461029986. [DOI] [Google Scholar]
- 17.Tamburi NE, Martín PR. Reaction norms of size and age at maturity of Pomacea canaliculata (Gastropoda: Ampullariidae) under a gradient of food deprivation. J Molluscan Stud. 2009;75(1):19–26. doi: 10.1093/mollus/eyn031. [DOI] [Google Scholar]
- 18.Kruatrachue M, Sumritdee C, Pokethitiyook P, Singhakaew S. Histopathological effects of contaminated sediments on golden apple snail (Pomacea canaliculata, Lamarck 1822) Bull Environ Contamination Toxicology. 2011;86(6):610. doi: 10.1007/s00128-011-0265-4. [DOI] [PubMed] [Google Scholar]
- 19.Subhan A, Yuwanta T, Zuprizal Z, Supadmo S. The use of pomacea canaliculata snails in feed to improve quality of alabio duck (anas plathyrinchos borneo) meat. J Indonesian Tropical Animal Agric. 2015;40(4):238–44.
- 20.Phonekhampheng O, Hung LT, Lindberg JE. Nutritive value of potential feed resources used in Laos for African catfish (Clarias gariepinus) production. Livest Res Rural Dev. 2008;20(12):article #207.
- 21.Phonekhampheng O, Hung LT, Lindberg JE. Ensiling of Golden apple snails (Pomacea canaliculata) and growth performance of African catfish (Clarias gariepinus) fingerlings fed diets with raw and ensiled Golden apple snails as protein source. Livest Res Rural Dev. 2009;21(2):article #18.
- 22.Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490(7418):49–54. doi: 10.1038/nature11413. [DOI] [PubMed] [Google Scholar]
- 23.Hibino T, Loza-Coll M, Messier C, Majeske A, Cohen A, Terwilliger D, Buckley K, Brockton V, Nair S, Berney K. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300(1):349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida T, Nagata S, Kataoka H. Ghitm is an ortholog of the Bombyx mori prothoracic gland-derived receptor ( Pgdr ) that is ubiquitously expressed in mammalian cells and requires an N-terminal signal sequence for expression. Biochem Biophys Res Commun. 2006;341(1):13–18. doi: 10.1016/j.bbrc.2005.12.141. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Wang SN, Shi-Gen YE, Hua LI. Cloning and LPS-induced expression analysis of ghitm gene in sea cucumber Apostichopus japonicus. J Dalian Ocean Univ. 2014;29(6):543–9.
- 26.Osborne BA. Apoptosis and the maintenance of homeostasis in the immune system. 1996. [DOI] [PubMed] [Google Scholar]
- 27.Romero A, Novoa B, Figueras A. The complexity of apoptotic cell death in mollusks: an update. Fish Shellfish Immunol. 2015;46(1):79–87. doi: 10.1016/j.fsi.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 28.Escoubas J-M, Destoumieux-Garzón D, Montagnani C, Gourbal B, Duval D, Green T, Charrière G. Immunity in Molluscs. Encyclopedia Immunobiol. 2016:417–36.
- 29.Sokolova IM. Apoptosis in molluscan immune defense. Invertebr Surviv J. 2009;6(1):49–58. [Google Scholar]
- 30.Terahara K, Takahashi KG. Mechanisms and immunological roles of apoptosis in molluscs. Curr Pharm Des. 2008;14(2):131–137. doi: 10.2174/138161208783378725. [DOI] [PubMed] [Google Scholar]
- 31.Korber B, Labute M, Yusim K. Immunoinformatics comes of age. PLoS Comput Biol. 2006;2(6):e71. doi: 10.1371/journal.pcbi.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317(5834):86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 33.Oka T, Sayano T, Tamai S, Yokota S, Kato H, Fujii G, Mihara K. Identification of a novel protein MICS1 that is involved in maintenance of mitochondrial morphology and apoptotic release of cytochrome c. Mol Biol Cell. 2008;19(6):2597–2608. doi: 10.1091/mbc.e07-12-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Dan L, Zhao W, Zheng X, Jin W, Wang E. A potent Lead induces apoptosis in pancreatic Cancer cells. PLoS One. 2012;7(6):e37841. doi: 10.1371/journal.pone.0037841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lisak DA, Schacht T, Enders V, Habicht J, Kiviluoto S, Schneider J, Henke N, Bultynck G, Methner A. The transmembrane Bax inhibitor motif (TMBIM) containing protein family: tissue expression, intracellular localization and effects on the ER CA 2+ −filling state ☆. Biochim Biophys Acta. 2015;1853(9):2104–2114. doi: 10.1016/j.bbamcr.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Uren RT, Dewson G, Bonzon C, Lithgow T, Newmeyer DD, Kluck RM. Mitochondrial release of pro-apoptotic proteins Electrostatic interactions can hold cytochrome c but not smac/diablo to mitochondrial membranes. J Biol Chem. 2005;280(3):2266. doi: 10.1074/jbc.M411106200. [DOI] [PubMed] [Google Scholar]
- 37.Naylor R. Invasions in agriculture: assessing the cost of the golden apple snail in Asia. Ambio. 1996;25(7):443–448. [Google Scholar]
- 38.Plan MRR, Ivana S, Cagauan AG, Craik DJ. Backbone cyclised peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail) J Agric Food Chem. 2008;56(13):5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- 39.Huang H-C, Liao S-C, Chang F-R, Kuo Y-H, Wu Y-C. Molluscicidal Saponins from Sapindus mukorossi, inhibitory agents of Golden apple snails, Pomacea canaliculata. J Agric Food Chem. 2003;51(17):4916–4919. doi: 10.1021/jf0301910. [DOI] [PubMed] [Google Scholar]
- 40.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinkovics JG. Programmed cell death (apoptosis): its virological and immunological connections (a review) Acta microbiologica Hungarica. 1991;38(3–4):321–334. [PubMed] [Google Scholar]
- 42.Wang S, Ma K, Chen L, Zhu H, Liang S, Liu M, Xu N. TAZ promotes cell growth and inhibits Celastrol-induced cell apoptosis. Biosci Rep. 2016;36(5):e00386–e00386. [DOI] [PMC free article] [PubMed]
- 43.Liu X, Chen S, Tu J, Cai W, Xu Q. HSP90 inhibits apoptosis and promotes growth by regulating HIF-1alpha abundance in hepatocellular carcinoma. Int J Mol Med. 2016;37(3):825–835. doi: 10.3892/ijmm.2016.2482. [DOI] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 45.Harasewych MG. Mathematical modeling of the shells of higher prosobranchs. 1981. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The cDNA sequences generated during the current study are available in the [GenBank] repository, [MN219409].