Abstract
Background
Malaria remains to be one of the deadliest infectious diseases and imposes substantial financial and social costs in the world. Mosquitoes rely on the immune system to control parasite infection. Peptidoglycan recognition proteins (PGRPs), a family of pattern-recognition receptors (PRR), are responsible for initiating and regulating immune signaling pathways. PGRP-LA is involved in the regulation of immune defense against the Plasmodium parasite, however, the underlying mechanism needs to be further elucidated.
Methods
The spatial and temporal expression patterns of pgrp-la in Anopheles stephensi were analyzed by qPCR. The function of PGRP-LA was examined using a dsRNA-based RNA interference strategy. Western blot and periodic acid schiff (PAS) staining were used to assess the structural integrity of peritrophic matrix (PM).
Results
The expression of pgrp-la in An. stephensi was induced in the midgut in response to the rapid proliferating gut microbiota post-blood meal. Knocking down of pgrp-la led to the downregulation of immune effectors that control gut microbiota growth. The decreased expression of these immune genes also facilitated P. berghei infection. However, such dsLA treatment did not influence the structural integrity of PM. When gut microbiota was removed by antibiotic treatment, the regulation of PGRP-LA on immune effectors was abolished and the knock down of pgrp-la failed to increase susceptibility of mosquitoes to parasite infection.
Conclusions
PGRP-LA regulates the immune responses by sensing the dynamics of gut microbiota. A mutual interaction between gut microbiota and PGRP-LA contributes to the immune defense against Plasmodium parasites in An. stephensi.
Keywords: PGRP-LA, Gut microbiota, Peritrophic matrix, Immune effectors, Plasmodium berghei, Anopheles stephensi
Background
Malaria, a mosquito-borne disease (MBD), is caused by parasites of the genus Plasmodium. It remains to be a high concern of the World Health Organization due to the continued emergence and spread of drug-resistant parasites and insecticide-resistant mosquitoes [1]. Malaria prevention and control primarily relies on Anopheles-targeted intervention strategies [2]. Consequently, a further understanding of the interaction between the Anopheles mosquito and Plasmodium is extremely urgent.
The main bottleneck for Plasmodium infection in the mosquito is the traverse of ookinetes across the midgut [3, 4]. During this process, two physical barriers are encountered by Plasmodium. The first barrier, peritrophic matrix (PM), composed of chitin, proteoglycans and proteins, is produced by intestinal epithelial cells in response to blood-feeding [5, 6]. PM poses an indispensable role in the defense of Plasmodium because its maturation time coincides with the ookinete invasion time [7]. When artificially increasing the thickness of PM by feeding mosquitoes with latex particles and animal blood, the number of P. gallinaceum oocysts significantly decreases in Aedes aegypti [8]. Midgut epithelium is the second barrier that inhibits Plasmodium infection [9]. When ookinetes start to traverse the midgut epithelium, epithelial nitration will be activated, promoting thioester-containing protein 1 (TEP1)-mediated lysis of Plasmodium [10, 11]. Once inside the cell cytoplasm, the invaded intestinal epithelial cells tend to undergo apoptosis that extrudes ookinetes from the epithelium [7, 12]. Besides, epithelial cells are also immune competent cells, involved in the production of nitric oxide (NO), antimicrobial peptides (AMPs) and reactive oxygen species (ROS) to limit Plasmodium survival [13, 14].
Mosquito gut microbiota is another important factor that can influence the outcome of Plasmodium infection [15–19]. Oral administration of live or heat-inactivated bacteria isolated from mosquito midgut significantly decreases the infection rate of P. falciparum [20]. Wickerhamomyces anomalus, a yeast involved in symbiotic relationships with Anopheles, is able to directly eliminate Plasmodium through secreting killer toxin [21, 22]. Another stably inherited gut commensal bacteria, Asaia, can be genetically modified and directly inhibits pathogen development by secreting antiplasmodial effectors [23]. In addition, these microbes inhibit parasites indirectly through promoting the formation of PM and stimulating the oxidative defense system and NF-κB dependent immune responses [24–27].
Mosquito immune response plays a pivotal role in protecting the host against pathogen infection. Peptidoglycan recognition proteins (PGRPs) are a family of important pattern-recognition receptors (PRR) that initiate immune signaling pathways [28, 29]. In Anopheles gambiae, PGRP-LC is the receptor of NF-κB transcription factor REL2-mediated signaling pathway. It plays an important role in maintaining the homeostasis of gut microbiota [27]. PGRP-LD protects the mosquito from Plasmodium infection through regulating gut microbiota-mediated PM formation in Anopheles stephensi [30]. PGRP-LB serves as a negative regulator of immune pathways in Aedes and Anopheles mosquitoes [31, 32]. PGRP-LA also participates in antiparasitic immune defenses, but the underlining mechanism needs to be further elucidated [31].
In this study, we demonstrate that the expression of pgrp-la is induced in the midgut in response to a blood meal. Such induction is due to the rapid proliferation of gut microbiota post-feeding. Once gut microbiota is removed by antibiotic treatment, PGRP-LA fails to initiate the synthesis of downstream immune effectors. Knocking down of pgrp-la in antibiotic-treated mosquitoes has no influence on the outcome of infection with P. berghei. These data elucidate that the anti-Plasmodium effect of PGRP-LA depends on the homeostasis of gut microbiota.
Methods
Mosquito rearing and antibiotic treatment
The An. stephensi mosquito (the Hor strain) was reared in the insectary at a temperature of 28 °C, relative humidity of 80% and a 12:12 h light/dark photocycle. Adults were fed on 10% sucrose solution and mouse blood. For antibiotic treatment experiment, newly emerged adult mosquitoes were orally administrated with 10% sucrose solution containing 10 U/ml penicillin, 10 μg/ml streptomycin and 15 μg/ml gentamycin daily for 3 days [20]. Then the antibiotic-treated mosquitoes and untreated mosquitoes were collected and surface sterilized. The homogenates were plated onto LB-agar to test the efficacy of antibiotic treatment.
Plasmodium berghei infection
Six to eight-week-old BALB/c mice were injected intraperitoneally (ip) with 106 infected RBCs with GFP-tagged P. berghei (ANKA). To evaluate parasitemia, tail smears were taken and stained with Giemsa (Baso Diagnostics Inc, Zhuhai, China), the number of parasites per 3000 RBCs were counted [33]. When the parasitemia reached 4–6%, the infected mice were used to feed mosquitoes that had been starved overnight; the mosquitoes were then maintained at 21 °C. Unengorged mosquitoes were removed 24 h post-blood meal. Midguts were dissected and infection intensity (oocyst number) were determined microscopically at day 8 post-infection.
Knockdown by RNA interference
PCR amplification from 606 to 1082 bp of pgrp-la (ASTE016413) was performed using T7-tagged primers. The plasmid eGFP (BD Biosciences, San Jose, USA) was used as a template for control dsRNA amplification. The double-stranded RNA (dsRNA) was synthesized using the MEGAscript RNA kit (Ambio, Invitrogen, Shanghai, China). Two to three-day-old mosquitoes were injected with 69 nl dsRNA (4 μg/μl) intra-thoracically using a nanoject II microinjector (Drummond, Philadelphia, USA). Mosquitoes treated with an equal volume of dsGFP and distilled water were used as controls. For the antibiotic treatment experiment, dsRNAs were injected three days after antibiotic treatment. The midguts of dsRNA-treated mosquitoes were dissected two days post-treatment and knock down efficacy was examined by qPCR as previously described [30]. For the P. berghei infection experiment, the mosquitoes were fed on an infectious blood meal four days after dsRNAs injection.
Total RNA extraction and reverse transcription quantitative PCR (RT-qPCR)
To evaluate transcription of related genes, total RNA was extracted from whole mosquitoes or midguts using TRIzol reagent (Sigma-Aldrich, Shanghai, China) according to the manufacturerʼs protocol. The cDNA was prepared from total RNA using the 5× All-in-One MasterMix (with AccuRT Genomic DNA Removal Kit; ABM, Shanghai, China). Quantitative PCR was performed using SYBR Green qPCR Master Mix (Bimake, Shanghai, China) according to a previously described protocol [30]. Ribosomal gene s7 was used as the internal reference gene.
Gut microbiota measurement
For CFU measurement, midguts were dissected and collected in 0.9% NaCl 5 days after dsRNA treatment and homogenized. The homogenates were plated on LB agar plates. The CFUs were counted 2 days after incubation at 28 °C. For total gut microbiota measurement, total DNA of midguts treated with dsRNA were extracted using the Holmes & Bonner [34] method at 2 time points, 5 days post dsRNA injection and 24 h post-infectious blood meal. Bacterial density was quantified by qPCR using universal 16S rRNA primers [35] (Additional file 1: Table S1).
Peritrophic matrix analysis
For measurement of mRNA levels, 3 PM synthetic genes [peritrophin1 (ASTE010406), peritrophin14 (ASTE009456), chitin synthase (ASTE007145)) and 2 chitinases (chitinaseA (ASTE005630) and chitinaseB (ASTE000328)] were quantified by qPCR using specific primers (Additional file 1: Table S1) 24 h, 36 h and 45 h post-infectious blood meal. The significance was determined using the Student’s t-test. For western blot analysis, 10 midguts were dissected 24 h post-infectious blood meal. Proteins were extracted in cracking buffer (8 M urea, 2% SDS, 5% β-mercaptoethanol and 125 mM Tris-HCl). Immunoblotting was carried out using rabbit anti-per1 antibody (1:1000) and mouse anti-β-actin antibody (1:2000) (Abbkine, Beijing, China). To generate Per1 rabbit polyclonal sera, recombinant Per1 (recPer1) was amplified using specific primers (Additional file 1: Table S1) corresponding to 55–462 bp of peritrophin1, and expressed in pET-42a using One step cloning (C112; Vazyme Biotech, Nanjing, China). The recPer1 was purified through Ni-NTA Superflow resin by the AKTA Explorer system (GE Healthcare, Shanghai, China) and was used to generate the antibody commercially (GL Biochem, Shanghai, China). For PM structure analysis, the abdomen of An. stephensi was fixed at 45 h post-infectious blood meal and sectioned at 5 μm using the paraffin sectioning method and stained with Periodic Acid Schiff (PAS) (Sigma-Aldrich) as previously described [36]. The slides were examined under bright field illumination of a Nikon ECLIPSE IVi microscope connected to a Nikon Digital Sight DS-U3 digital camera.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 6 (GraphPad Software, La Jolla, CA). Statistics of gene expression were tested for using the Student’s t-test. The Mann–Whitney test was used to determine the significance of infection rate and microbiota level of dsRNA-treated mosquitoes.
Results
PGRP-LA is a sensor of gut microbiota
We first analyzed spatial expression pattern of pgrp-la in the midgut and the remaining carcass of An. stephensi 24 h prior to (− 24 h) and 24, 48 and 72 h post-infectious blood meal. It was expressed abundantly in the carcass before blood-feeding. However, pgrp-la was upregulated significantly in the midgut within 48 h after the mosquito took an infectious blood meal (Fig. 1a). To investigate whether the upregulation of pgrp-la in the midgut is due to blood-feeding or the presence of parasite, we compared expression of pgrp-la in the midguts of mosquitoes fed on normal blood and blood containing Plasmodium. A blood meal, no matter whether it contained P. berghei or not, significantly increased the expression of pgrp-la comparing to sugar-fed mosquitoes (Fig. 1b). In addition, the presence of the Plasmodium parasite further induced the expression level of pgrp-la significantly than normal blood did. Because blood-feeding causes an extreme bloom of gut microbiota [9], we next hypothesized that upregulation of pgrp-la in response to a blood meal could be due to the proliferation of gut microbiota. Then we compared pgrp-la expression in the midgut of normal and antibiotic-treated (Abx) mosquitoes. As expected, once the gut microbiota was removed by antibiotic treatment, the expression of pgrp-la in the midgut significantly decreased, compared to control mosquitoes (Fig. 1c). Blood meal alone failed to increase the expression of pgrp-la (Fig. 1c). Taken together, these results indicate that the induction of pgrp-la expression in response to blood-feeding, is a result of the proliferation of gut microbiota in the midgut.
Fig. 1.
Gut microbiota is required for the expression of PGRP-LA. a Relative gene expression of pgrp-la in the midgut and carcass of mosquitoes before (− 24 h IB) and after (+ 24 h IB, + 48 h IB and + 72 h IB) feeding with infectious blood meal. b Relative gene expression of pgrp-la in midgut of sugar-fed (SF), 24 h post normal blood-fed (NB) and infectious blood-fed (IB) mosquitoes. c Relative gene expression of pgrp-la in normal and Abx midguts of sugar-fed (SF), 24 h post normal blood-fed (NB) and infectious blood-fed (IB) mosquitoes. Error bars indicate standard error of the mean (n = 8). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (for details, see Additional file 3: Text S1). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant
PGRP-LA regulates the homeostasis of gut microbiota
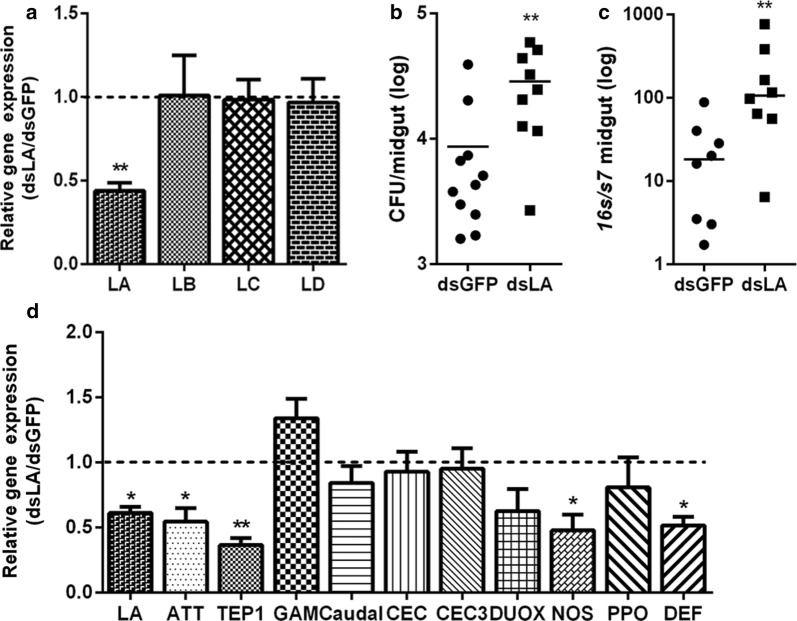
Given that PGRP-LA senses gut microbiota, we next examined whether it could be involved in regulating the homeostasis of gut microbiota. PGRP-LA-specific double-stranded RNA (dsLA) was injected intrathoracically into An. stephensi. In addition to dsGFP controls, we also selected the buffer solution for dsRNA as a blank control to confirm that there was no intrinsic deficit of exogenous RNA (Additional file 2: Figure S1a). The mRNA level of prpg-la reduced by 58% 2 days post-dsLA treatment compared to dsGFP controls. Such treatment did not influence the expression of other long PGRPs (PGRP-LB, PGRP-LC and PGRP-LD) (Fig. 2a). Knock down of PGRP-LA resulted in an around 6-fold increase in bacterial CFU and 16S rRNA gene copy number per midgut, respectively, compared to that in dsGFP controls (Fig. 2b, c). As PGRP-LA is the receptor of the Imd pathway in insects [31, 37, 38], we next analyzed whether the increase of gut microbiota could be due to the downregulation of immune effectors when PGRP-LA was knocked down. A total of 10 immune related genes were analyzed in mosquitoes 4 days post-dsRNA treatment. These genes include 5 antimicrobial peptides (attacin, cecropin, cecropin 3, gambicin and defensin), 1 negative regulator of IMD signaling pathway (caudal) and 4 proteins related to cellular and epithelial immune responses (tep1, ppo, nos and duox). As expected, the expressions of four genes, attacin, tep1, nos and defensin were significantly decreased in dsLA-treated mosquitoes comparing to those in dsGFP (Fig. 2d). Given the bactericidal effect of these antimicrobial peptides, our results indicate that PGRP-LA controls the abundance of gut microbiota through regulating the synthesis of downstream immune effectors.
Fig. 2.
Influence of PGRP-LA on gut microbiota in An. stephensi. a PGRP-LA silencing efficiency and specificity. Relative expression level of pgrp-la in dsLA mosquitoes was normalized to the gene’s expression in dsGFP controls. Error bars indicate standard error of the mean (n = 10). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (t(8) = 3.431, P = 0.0089). b Median culturable gut microbiota in dsRNA-treated mosquitoes. Each dot represents an individual mosquito and horizontal lines represent the medians. Significance was determined by Mann–Whitney test (U = 15.00, P = 0.0074). c Relative expression level of 16S in dsRNA-treated mosquitoes. Each dot represents an individual mosquito and horizontal lines represent the medians. Significance was determined by Mann–Whitney test (U = 7.00, P = 0.0070). d Relative expression levels of immune-related genes in dsRNA-treated, sugar-fed mosquitoes normalized to the gene’s expression in dsGFP controls. Error bars indicate standard error of the mean (n = 8). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (for details, see Additional file 3: Text S1). *P < 0.05, **P < 0.01
PGRP-LA regulates immune defense against P. berghei
Given the role of PGRP-LA in regulating immune activity and its anti-parasitic function in An. coluzzii, we next investigated the influence of PGRP-LA on parasite defense in An. stephensi [31]. Knocking down of pgrp-la (dsLA) resulted in a significant increase in the infection rate of P. berghei from 9 oocysts in dsGFP to 46 oocysts in dsLA mosquitoes (Fig. 3a). Such increasing susceptibility to parasite infection was due to the downregulation of immune genes in the absence of pgrp-la. The four genes, attacin, tep1, nos and defensin were again downregulated in the absence of pgrp-la (Fig. 3b). Similarly, the downregulation of immune effectors in dsLA-treated mosquitoes facilitated the proliferation of gut microbiota as the 16S rRNA gene was significantly higher in dsLA compared to dsGFP mosquitoes (Fig. 3c). To examine the influence of dsRNA treatment on mosquito blood uptake, we compared the weight of dsRNA-treated mosquitoes at 0 h and 24 h post-infectious blood meal, respectively. The weight of dsLA-treated mosquitoes was comparable to that of dsGFP mosquitoes at each time point (Additional file 2: Figure S1b). Taken together, our results suggest that PGRP-LA defends against Plasmodium infection in An. stephensi by positively regulating the immune signaling pathways.
Fig. 3.
PGRP-LA is required for resistance to P. berghei. a Median oocyst number in dsRNA-treated mosquitoes. Each dot represents an individual mosquito and horizontal lines represent the medians. Results from one of three independent experiments are shown. Significance was determined by Mann–Whitney test (U = 132.00, P = 0.0003). b Relative expression levels of immune-related genes in dsRNA-treated mosquitoes 24 h post-infectious blood meal. Error bars indicate standard error of the mean (n = 8). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (for details, see Additional file 3: Text S1). *P < 0.05, **P < 0.01, ***P < 0.001. c Relative expression level of 16S in dsRNA-treated mosquitoes 24 h post-infectious blood meal. Each dot represents an individual mosquito and horizontal lines represent the medians. Significance was determined by Mann–Whitney test (U = 12.00, P = 0.0057)
PGRP-LA is not involved in the regulation of peritrophic matrix synthesis
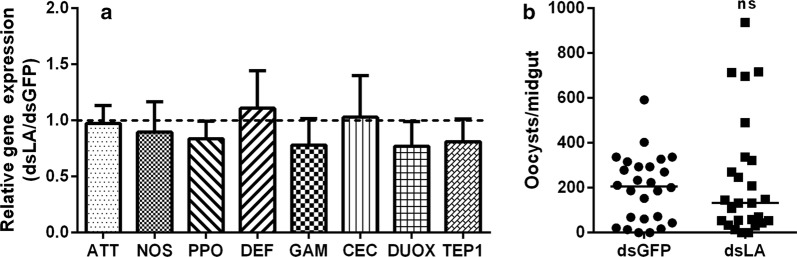
Gut microbiota stimulates the synthesis of peritrophic matrix (PM), thereby inhibiting Plasmodium infection [30, 39, 40]. As we observed an increased level of gut microbiota in dsLA mosquitoes, we next analyzed whether the increased gut microbiota in these mosquitoes could influence the integrity of PM. We first analyzed the expression level of five PM-related genes in dsRNA-treated mosquitoes 24 h, 36 h and 48 h post-infection, respectively. These genes include three PM synthesis genes (peritrophin1, peritrophin14 and chitin synthetase) and two PM digesting chitinases [30]. Except for peritrophin1 (per1) and chitinase A which were differentially regulated 24 h post-infection in dsLA mosquitoes, the expression of most of the genes was comparable to dsGFP mosquitoes (Fig. 4a). Then we examined the protein level of Per1 24 h post-infection by western blot and found silencing pgrp-la did not impact the protein level of Per1 (Fig. 4b). We further analyzed the structural integrity of PM by PAS staining. Again, no significant difference was observed between dsLA and dsGFP mosquitoes. Although knockdown of pgrp-la resulted in the increasing level of gut microbiota, such increase had no influence on PM structure. Neither did PGRP-LA participate in the regulation of PM synthesis directly.
Fig. 4.
The influence of PGRP-LA on PM synthesis. a Relative expression levels of PM-related genes in dsRNA-treated mosquitoes 24 h, 36 h and 48 h post-infectious blood meal. Error bars indicate standard error of the mean (n = 8). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (for details, see Additional file 3: Text S1). *P < 0.05. b Western blot of Per1 in dsGFP-treated and dsLA-treated mosquitoes. Actin was used as a loading control. Results from one of three independent experiments are shown. c PAS staining of PM in dsGFP-treated and dsLA-treated mosquitoes at 100× and 200× magnification. Red arrowheads denote the PM. Results from one of three independent experiments are shown. Scale-bars: c, 100 μm
Gut microbiota is required for PGRP-LA-regulated immune defense against Plasmodium
As the expression level of pgrp-la is correlated with the abundance of gut microbiota, we next investigated whether the regulation of PGRP-LA on immune response could rely on the presence of gut microbiota. The dsRNAs were introduced into An. stephensi of which gut microbiota was removed by antibiotic treatment. Surprisingly, once the gut microbiota was cleared, knock down of pgrp-la had no impact on the expression of the immune effectors, especially attacin, tep1, nos and defensin (Fig. 5a). The infection rate in dsLA mosquitoes was comparable to that in dsGFP mosquitoes (Fig. 5b). Taken together, these results indicate that the presence of gut microbiota is required for PGPR-LA-mediated protection against infection. PGRP-LA and the regulated immune effectors in turn monitor the levels of the gut microbiota.
Fig. 5.
Gut microbiota is necessary for anti-Plasmodium of PGRP-LA. a Fold change of immune-related genes in antibiotic-treated mosquitoes treated with dsRNA 24 h post-infectious blood meal. Error bars indicate standard error of the mean (n = 8). Results from one of three independent experiments are shown. Significance was determined by Student’s t-test (for details, see Additional file 3: Text S1). b Median oocyst number in antibiotics-treated mosquitoes treated with dsRNA. Each dot represents an individual mosquito and horizontal lines represent the medians. Significance was determined by Mann–Whitney test (U = 314.50, P = 0.6729). Abbreviation: ns, not significant
Discussion
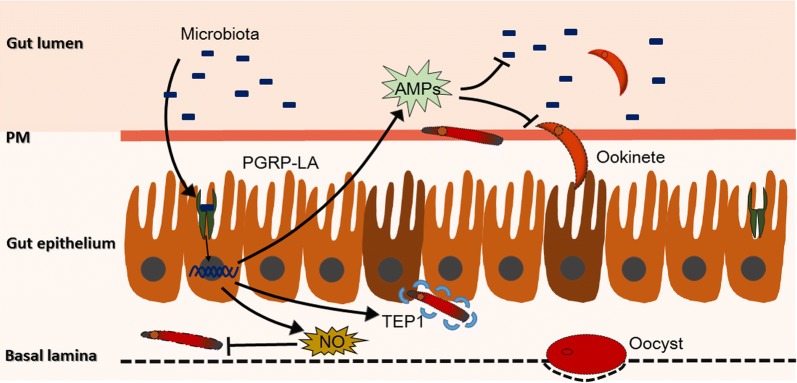
The PGRP family is functionally conserved from insects to mammals, acting as pattern recognition receptors and effectors in innate immunity [29, 41, 42]. In this study, we elucidated that gut microbiota stimulates the expression of pgrp-la in the midgut of An. stephensi, which in turn regulates the microbiota growth and parasite infection through controlling immune activity. Once gut microbiota is removed, such regulation is abolished (Fig. 6).
Fig. 6.
Model of influence of PGRP-LA in An. stephensi. PGRP-LA is enriched in midgut in response to the proliferation of gut microbiota. It protects Anopheles from Plasmodium infection through initiating the synthesis of downstream immune effectors, including AMPs, NOS and TEP1. Such regulation requires the presence of gut microbiota
While PGRP-LA is present in multiple insects, its function in regulating the immune response is not fully understood. In Drosophila, the expression of pgrp-la is enriched in the barrier epithelia, but low in the fat body. It is not required for the production of antimicrobial peptides by the fat body in response to a systemic infection, while it significantly stimulates antimicrobial peptide gene expression in the trachea upon oral bacterial infection [37]. In Tribolium castaneum, PGRP-LA functions as a pivotal sensor of the IMD pathway via sensing both Gram-negative and Gram-positive bacteria. It is mainly responsible for the recognition of Gram-positive bacteria, which is contrary to what has been found in Drosophila and Aedes [32, 37, 43]. Here we show that PGRP-LA is essential in regulating the immune defense against P. berghei. This finding is consistent with the function of PGRP-LA in An. coluzzii [31]. Our study further describes the mutual regulation between PGRP-LA and gut microbiota. The proliferation of gut microbiota after blood-feeding induces the expression of pgrp-la, while PGRP-LA regulates the homeostasis of gut microbiota. In addition, gut microbiota is necessary for PGRP-LA-regulated immune protection against P. berghei.
The interactions between insect immune system and gut microbiota play vital roles in determining infection outcomes. Here we show that the regulation of immune responses by PGRP-LA relies on the presence and abundance of gut microbiota. In agreement with our findings, other PGRP family members are also involved in maintaining the homeostasis of gut microbiota. For example, PGRP-SC2 contributes to the gut immune homeostasis through limiting age-related gene, foxo, that induces commensal dysbiosis in Drosophila [44]. PGRP-LC controls the Imd activity via cooperating with PGRP-LC-interacting inhibitor of Imd signaling (PIMS), which is a negative regulator of Drosophila innate immune signaling. Such interaction helps control the homeostasis of commensal bacteria [45]. PGRP-LE also promotes the growth of the symbiotic bacteria Wolbachia, which in turn induces the expression of PGRP-LE in the carcass of A. aegypti [46]. Additionally, catalytic PGRP-LB acts as a feedback inhibitor of the Imd/Rel innate immune signaling pathway by scavenging peptidoglycan released from bacteria in multiple insects [47–49].
Conclusions
In this study, we show that gut microbiota regulates the expression of pgrp-la, which in turn controls the homeostasis of gut microbiota. Such finely tuned balance is key in regulating the defense against pathogen infection. Our findings will pave the way for further understanding the tripartite interactions between the mosquito, its gut microbiota and the Plasmodium parasite.
Supplementary information
Additional file 1: Table S1. Primers used in this study.
Additional file 2: Figure S1. a PGRP-LA silencing efficiency in control, dsGFP and dsLA mosquitoes. b The weight of dsRNA-treated mosquitoes at 0 h and 24 h post-infectious blood meal.
Additional file 3: Text S1. Significant results from statistical analyses in this study.
Acknowledgements
Not applicable.
Abbreviations
- MBD
mosquito-borne disease
- PM
peritrophic matrix
- TEP1
thioester-containing protein 1
- NO
nitric oxide
- AMPs
antimicrobial peptides
- ROS
reactive oxygen species
- PGRP
peptidoglycan recognition proteins
- PRR
pattern-recognition receptor
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- ip
intraperitoneally
- CFU
colony-forming units
- PAS
periodic acid schiff
- PIMS
PGRP-LC-interacting inhibitor of Imd signaling
Authors’ contributions
LG performed the experiments and drafted the manuscript. LG and XS participated in data analysis. LG and JW revised the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by National Natural Science Foundation of China (31822051) (https://isisn.nsfc.gov.cn/egrantweb/) and National Institutes of Health Grant (R01AI129819) (https://grants.nih.gov/grants).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional files.
Ethics approval and consent to participate
All animals were handled strictly in accordance with the guidelines of the Care and Use of Laboratory Animals of the National Institutes of Health and the Office of Laboratory Animal Welfare, China. The research protocol was approved by the Institutional animal care and use committee, Department of Laboratory Animal Science, Fudan University, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Li Gao, Email: 17210700112@fudan.edu.cn.
Xiumei Song, Email: 17110700026@fudan.edu.cn.
Jingwen Wang, Email: jingwenwang@fudan.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3876-y.
References
- 1.WHO. World malaria report 2018. Geneva: World Health Organization; 2018. https://www.who.int/malaria/publications/world-malaria-report-2018/en/. Accessed 10 May 2019.
- 2.Ramirez AL, van den Hurk AF, Mackay IM, Yang ASP, Hewitson GR, McMahon JL, et al. Malaria surveillance from both ends: concurrent detection of Plasmodium falciparum in saliva and excreta harvested from Anopheles mosquitoes. Parasit Vectors. 2019;12:355. doi: 10.1186/s13071-019-3610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aly AS, Vaughan AM, Kappe SH. Malaria parasite development in the mosquito and infection of the mammalian host. Annu Rev Microbiol. 2009;63:195–221. doi: 10.1146/annurev.micro.091208.073403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlachou D, Schlegelmilch T, Runn E, Mendes A, Kafatos FC. The developmental migration of Plasmodium in mosquitoes. Curr Opin Genet Dev. 2006;16:384–391. doi: 10.1016/j.gde.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Baia-da-Silva DC, Alvarez LCS, Lizcano OV, Costa FTM, Lopes SCP, Orfano AS, et al. The role of the peritrophic matrix and red blood cell concentration in Plasmodium vivax infection of Anopheles aquasalis. Parasit Vectors. 2018;11:148. doi: 10.1186/s13071-018-2752-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegedus D, Erlandson M, Gillott C, Toprak U. New insights into peritrophic matrix synthesis, architecture, and function. Annu Rev Entomol. 2009;54:285–302. doi: 10.1146/annurev.ento.54.110807.090559. [DOI] [PubMed] [Google Scholar]
- 7.Han YS, Thompson J, Kafatos FC, Barillas-Mury C. Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. EMBO J. 2000;19:6030–6040. doi: 10.1093/emboj/19.22.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Billingsley PF, Rudin W. The role of the mosquito peritrophic membrane in bloodmeal digestion and infectivity of Plasmodium species. J Parasitol. 1992;78:430–440. doi: 10.2307/3283640. [DOI] [PubMed] [Google Scholar]
- 9.Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G. Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol. 2010;34:387–395. doi: 10.1016/j.dci.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira Gde A, Lieberman J, Barillas-Mury C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science. 2012;335:856–859. doi: 10.1126/science.1209678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith RC, Vega-Rodriguez J, Jacobs-Lorena M. The Plasmodium bottleneck: malaria parasite losses in the mosquito vector. Mem Inst Oswaldo Cruz. 2014;109:644–661. doi: 10.1590/0074-0276130597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakani P, Suman S, Gupta L, Kumar S. Ambivalent outcomes of cell apoptosis: a barrier or blessing in malaria progression. Front Microbiol. 2016;7:302. doi: 10.3389/fmicb.2016.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marois E. The multifaceted mosquito anti-Plasmodium response. Curr Opin Microbiol. 2011;14:429–435. doi: 10.1016/j.mib.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 14.Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, Zamora RE, et al. The STAT pathway mediates late-phase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe. 2009;5:498–507. doi: 10.1016/j.chom.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirimotich CM, Ramirez JL, Dimopoulos G. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 2011;10:307–310. doi: 10.1016/j.chom.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinparast Djadid N, Jazayeri H, Raz A, Favia G, Ricci I, Zakeri S. Identification of the midgut microbiota of An. stephensi and An. maculipennis for their application as a paratransgenic tool against malaria. PLoS ONE. 2011;6:e28484. doi: 10.1371/journal.pone.0028484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boissiere A, Tchioffo MT, Bachar D, Abate L, Marie A, Nsango SE, et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012;8:e1002742. doi: 10.1371/journal.ppat.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendrin M, Rodgers FH, Yerbanga RS, Ouedraogo JB, Basanez MG, Cohuet A, et al. Antibiotics in ingested human blood affect the mosquito microbiota and capacity to transmit malaria. Nat Commun. 2015;6:5921. doi: 10.1038/ncomms6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahia AC, Dong Y, Blumberg BJ, Mlambo G, Tripathi A, BenMarzouk-Hidalgo OJ, et al. Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol. 2014;16:2980–2994. doi: 10.1111/1462-2920.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Y. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cappelli A, Valzano M, Cecarini V, Bozic J, Rossi P, Mensah P, et al. Killer yeasts exert anti-plasmodial activities against the malaria parasite Plasmodium berghei in the vector mosquito Anopheles stephensi and in mice. Parasit Vectors. 2019;12:329. doi: 10.1186/s13071-019-3587-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker GM. Pichia anomala: cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek. 2011;99:25–34. doi: 10.1007/s10482-010-9491-8. [DOI] [PubMed] [Google Scholar]
- 23.Bongio NJ, Lampe DJ. Inhibition of Plasmodium berghei development in mosquitoes by effector proteins secreted from Asaia sp. bacteria using a novel native secretion signal. PLoS ONE. 2015;10:e0143541. doi: 10.1371/journal.pone.0143541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennison NJ, Saraiva RG, Cirimotich CM, Mlambo G, Mongodin EF, Dimopoulos G. Functional genomic analyses of Enterobacter, Anopheles and Plasmodium reciprocal interactions that impact vector competence. Malar J. 2016;15:425. doi: 10.1186/s12936-016-1468-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, Mulenga M, et al. Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–858. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez JL, Short SM, Bahia AC, Saraiva RG, Dong Y, Kang S, et al. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 2014;10:e1004398. doi: 10.1371/journal.ppat.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song X, Wang M, Dong L, Zhu H, Wang J. PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis. PLoS Pathog. 2018;14:e1006899. doi: 10.1371/journal.ppat.1006899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendrin M, Turlure F, Rodgers FH, Cohuet A, Morlais I, Christophides GK. The peptidoglycan recognition proteins PGRPLA and PGRPLB regulate Anopheles immunity to bacteria and affect infection by Plasmodium. J Innate Immun. 2017;9:333–342. doi: 10.1159/000452797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Beerntsen BT. Functional implications of the peptidoglycan recognition proteins in the immunity of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2015;24:293–310. doi: 10.1111/imb.12159. [DOI] [PubMed] [Google Scholar]
- 33.Shimada M, Hirose Y, Shimizu K, Yamamoto DS, Hayakawa EH, Matsuoka H. Upper gastrointestinal pathophysiology due to mouse malaria Plasmodium berghei ANKA infection. Trop Med Health. 2019;47:18. doi: 10.1186/s41182-019-0146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes DS, Bonner J. Preparation, molecular weight, base composition, and secondary structure of giant nuclear ribonucleic acid. Biochemistry. 1973;12:2330–2338. doi: 10.1021/bi00736a023. [DOI] [PubMed] [Google Scholar]
- 35.Narasimhan S, Rajeevan N, Liu L, Zhao YO, Heisig J, Pan J, et al. Gut microbiota of the tick vector Ixodes scapularis modulate colonization of the Lyme disease spirochete. Cell Host Microbe. 2014;15:58–71. doi: 10.1016/j.chom.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S. Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol. 2008;54:1236–1242. doi: 10.1016/j.jinsphys.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gendrin M, Zaidman-Remy A, Broderick NA, Paredes J, Poidevin M, Roussel A, et al. Functional analysis of PGRP-LA in Drosophila immunity. PLoS ONE. 2013;8:e69742. doi: 10.1371/journal.pone.0069742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc Natl Acad Sci USA. 2000;97:13772–13777. doi: 10.1073/pnas.97.25.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodgers FH, Gendrin M, Wyer CAS, Christophides GK. Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 2017;13:e1006391. doi: 10.1371/journal.ppat.1006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romoli O, Gendrin M. The tripartite interactions between the mosquito, its microbiota and Plasmodium. Parasit Vectors. 2018;11:200. doi: 10.1186/s13071-018-2784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steiner H. Peptidoglycan recognition proteins: on and off switches for innate immunity. Immunol Rev. 2004;198:83–96. doi: 10.1111/j.0105-2896.2004.0120.x. [DOI] [PubMed] [Google Scholar]
- 42.Boneca IG. Mammalian PGRPs in the spotlight. Cell Host Microbe. 2009;5:109–111. doi: 10.1016/j.chom.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Koyama H, Kato D, Minakuchi C, Tanaka T, Yokoi K, Miura K. Peptidoglycan recognition protein genes and their roles in the innate immune pathways of the red flour beetle, Tribolium castaneum. J Invertebr Pathol. 2015;132:86–100. doi: 10.1016/j.jip.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Pan X, Pike A, Joshi D, Bian G, McFadden MJ, Lu P, et al. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 2018;12:277–288. doi: 10.1038/ismej.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106:12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charroux B, Capo F, Kurz CL, Peslier S, Chaduli D, Viallat-Lieutaud A, et al. Cytosolic and secreted peptidoglycan-degrading enzymes in Drosophila respectively control local and systemic immune responses to microbiota. Cell Host Microbe. 2018;23:215–228. doi: 10.1016/j.chom.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–473. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Primers used in this study.
Additional file 2: Figure S1. a PGRP-LA silencing efficiency in control, dsGFP and dsLA mosquitoes. b The weight of dsRNA-treated mosquitoes at 0 h and 24 h post-infectious blood meal.
Additional file 3: Text S1. Significant results from statistical analyses in this study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.