Abstract
Aims/hypothesis
The conserved hypoxia inducible factor 1 α (HIF1α) injury-response pro-survival pathway has recently been implicated in early beta cell dysfunction but slow beta cell loss in type 2 diabetes. We hypothesised that the unexplained prolonged prediabetes phase in type 1 diabetes may also be, in part, due to activation of the HIF1α signalling pathway.
Methods
RNA sequencing (RNA-Seq) data from human islets with type 1 diabetes or after cytokine exposure in vitro was evaluated for activation of HIF1α targets. This was corroborated by immunostaining human pancreases from individuals with type 1 diabetes for 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), the key effector of HIF1α-mediated metabolic remodelling, and by western blotting of islets and INS-1 832/13 cells exposed to cytokines implicated in type 1 diabetes.
Results
HIF1α signalling is activated (p=4.5×10−9) in islets from individuals with type 1 diabetes, and in human islets exposed in vitro to cytokines implicated in type 1 diabetes (p=1.1×10−14). Expression of PFKFB3 is increased fivefold (p<0.01) in beta cells in type 1 diabetes and in human and rat islets exposed to cytokines that induced increased lactate production. HIF1α attenuates cytokine-induced cell death in beta cells.
Conclusions/interpretation
The conserved pro-survival HIF1α-mediated injury-response signalling is activated in beta cells in type 1 diabetes and likely contributes to the relatively slow rate of beta cell loss at the expense of early defective glucose-induced insulin secretion.
Keywords: Cytokines, HIF1α, PFKFB3, Type 1 diabetes
Introduction
Type 1 diabetes is classified as an autoimmune disease based on the associated risk with other endocrine autoimmune diseases, such as Addison’s disease and Hashimoto’s thyroiditis, genetic linkage to HLA subtypes and the presence of autoantibodies with diabetes-predictive value [1, 2]. Prospective studies of individuals at risk of developing type 1 diabetes have established the time course between the initiation of autoimmunity, as defined by detectable autoantibodies, and diabetes onset [3]. This time course is often surprisingly slow, taking as long as 10 years. Studies of human pancreases from adults with recent-onset diabetes find a low or undetectable frequency of beta cell apoptosis and up to 30% of beta cells are still present [4]. Taken together, these data imply that, despite the presence of autoimmunity, beta cell loss in adult-onset type 1 diabetes is remarkably slow and that decreased beta cell responsiveness to hyperglycaemia precedes beta cell loss.
We recently established that the slow time course of beta cell loss in type 2 diabetes, along with a predominant defect in glucose-stimulated insulin secretion, is at least partially attributable to activation of a highly conserved stress-induced pro-survival hypoxia inducible factor 1 α (HIF1α)/ 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) signalling pathway [5]. Deletion of Hif1a increases vulnerability of beta cells to viral infections and toxins and increases the incidence of autoimmune diabetes in the NOD mouse model [6]. A genetic variant of HIF1α that increases activation of HIF1α in response to stress is less common in type 1 and type 2 diabetes than in non-diabetic conditions, consistent with a protective role of HIF1α against beta cell stress [7, 8].
In the present studies we sought to address the questions: (1) is the HIF1α/PFKFB3 signalling pathway activated in beta cells in humans with type 1 diabetes; and, if so, (2) does activation of this signalling pathway protect beta cells against cytokine-induced cell death?
Methods
National Institutes of Health guidelines for rigour in research
In keeping with the National Institutes of Health (NIH) guidelines for reporting of experimental conditions (www.nih.gov/about/reporting-preclinical-research.htm), we report that no formal randomisation was used in studies of human pancreases or RNA sequencing (RNA-Seq) data. RNA-Seq data was used as available in publication and was assigned to groups by authors of those publications. Human pancreases from the JDRF Network for Pancreatic Organ Donors with Diabetes (nPOD) programme were selected based on sample availability from brain-dead organ donors and were assigned to non-diabetic or type 1 diabetic groups based on clinical data available to the nPOD programme. Isolated islets (rat and human) were allocated randomly to treatments, as were INS-1 832/13 cells. Investigators were blinded to pancreas samples during image analysis but were not blinded to RNA-Seq data or during analysis of isolated islets or INS-1 832/13 cells.
Evaluation of RNA sequence transcriptomes of human islets
For the analysis of HIF1α target genes, we examined the genes listed in ESM Table 1 [9]. Due to limited sample sizes in the three datasets used (see below), and the limited coverage in some of the single-cell datasets, we did not evaluate the probabilities of differential gene expression at the single gene level. Instead we calculated the fold change for each gene as the mean expression of the experimental group (type 1 diabetes samples or cytokine-treated samples) divided by the mean expression of the control samples. For each of the three datasets, we then evaluated whether the log-transformed fold changes of the 76 HIF1α targets differed significantly from the fold changes of all other genes in the genome.
The Mastracci et al dataset consists of bulk RNA-Seq data obtained from pancreatic islets isolated from three non-diabetic donors and two donors with type 1 diabetes [10]. We obtained the reads per kb of transcript per million mapped reads (RPKM) values from the Gene Expression Omnibus (GEO) accession no. GSE102371. In our study, we included all genes with detected expression in at least one type 1 diabetes sample and one control sample (15,036 genes). We calculated the fold change for each gene as the mean RPKM in type 1 diabetes samples divided by the mean RPKM in three control samples; this number is >1.0 when the expression is higher in type 1 diabetes, and <1.0 when expression is higher in control samples. Of the 76 known HIF1α targets, 69 were measured in this dataset. We then evaluated whether the fold changes of these 69 genes were different from the fold changes of all other genes. Specifically, we performed a Wilcoxon rank-sum test on the fold change values of the 69 HIF1α targets, compared with the fold change values for all 15,036 genes in the dataset (median values: 1.2 and 0.77, respectively; p = 4.5×10−9).
The dataset from Eizirik et al includes bulk RNA-Seq data from pancreatic islets from individuals without diabetes, which were exposed to cytokines and compared with control samples [11]. We downloaded the RPKM values for the five samples treated with cytokines and five control samples from GEO accession no. GSE35296. We included all genes with expression in at least one treated and one untreated sample (27,131 genes). We then calculated the fold change for each gene as the mean RPKM in the treated samples vs the five control samples. Finally, we compared the fold changes of the 76 confirmed HIF1α targets present in the dataset to the fold changes of all other genes. The comparison was performed using a Wilcoxon rank-sum test (median values: 1.55 and 0.94, respectively; p = 1.10×10−14).
The Wang et al dataset includes single-cell RNA-Seq data from 17 beta cells from non-diabetic individuals and six beta cells from type 1 diabetic individuals [12]. We obtained the counts per million (CpM) data from GEO accession no. GSE83139, and included all genes with detected expression in at least one beta cell from participants with type 1 diabetes and three beta cells from non-diabetic participants (8322 genes). We then calculated the fold change for each gene as the mean CpM measured in the six cells from individuals with type 1 diabetes divided by the mean CpM in the 17 cells from non-diabetic donors. We compared the fold changes of the 31 confirmed HIF1α targets present in the dataset to the fold changes of all 8322 genes (median values: 1.43 and 1.64, respectively; p = 0.81, Wilcoxon rank-sum).
Human pancreas
Pancreases were procured from brain-dead organ donors by the nPOD, administered by the University of Florida (Gainesville, FL. USA). All procedures were in accordance with federal guidelines for organ donation and the University of Florida Institutional Review Board. Five pancreases from individuals with type 1 diabetes (6050, 6070, 6051, 6069, 6175) and five from non-diabetic control donors (6011, 6030, 6015, 6134, 6104), comparable by age (38±12 vs 37±4 years), sex and BMI (23±2 vs 25±2 kg/m2), were evaluated (Table 1).
Table 1.
Characteristics of nPOD donors, the sample from whom were used for immunofluorescence staining
| Type 1 diabetes | Non-diabetic | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| nPOD sample no. | Age (years) | Sex | BMI (kg/m2) | Diabetes duration (years) | nPOD sample no. | Age (years) | Sex | BMI (kg/m2) | |
| 6050 | 82 | M | NA | 50 | 6011 | 46 | F | 26.3 | |
| 6070 | 23 | F | 21.6 | 7 | 6030 | 30 | M | 27.1 | |
| 6051 | 20 | M | 21.5 | 13 | 6015 | 39 | F | 32.2 | |
| 6069 | 23 | M | 28.8 | 7 | 6134 | 27 | M | 20.1 | |
| 6175 | 42 | M | 19.8 | 12 | 6104 | 41 | M | 20.5 | |
| Mean | 38.0 | – | 22.9 | 17.8 | Mean | 36.6 | – | 25.2 | |
| SEM | 11.7 | – | 2.0 | 8.1 | SEM | 3.6 | – | 2.3 | |
F, female; M, male; NA, not available
Immunohistochemistry and morphometric analysis
Paraffin sections of pancreas (4 μm) were exposed to toluene for 10 min and then to 100% (vol./vol.) ethanol for another 10 min, 95% (vol./vol.) ethanol for 5 min, 70% (vol./vol.) ethanol for 5 min and, finally, water. Sections were transferred in heat-induced antigen-retrieval solution (citrate buffer at pH 6.0), using a microwave, and then cooled to room temperature for 1 h. They were then soaked in soaking buffer (Tris-buffered saline [TBS], 0.4% [vol./vol.] Triton X-100) for 30 min on ice, and washed with TBS. After blocking (TBS, 3% [wt/vol.] BSA, 0.2% [vol./vol.] Triton X-100) for 1 h, the slides were incubated with primary antibodies diluted in antibody buffer (TBS, 3% [wt/vol.] BSA, 0.2% [vol./vol.] Tween 20) overnight at 4°C. After washing in TBS–Tween 20 (TBST), slides were incubated with secondary antibodies diluted in antibody buffer for 1 h at room temperature. Slides were then mounted using Vectashield with DAPI (catalogue no. H-1200; RRID: AB_2336790; Vector Laboratories, Burlingame, CA, USA).
The presence of PFKFB3 in the islets was evaluated in pancreatic sections immunostained for PFKFB3, insulin and glucagon. Images of 20–59 islets per sample were taken using a Leica DM6000 fluorescent microscope (Wetzlar, Germany) with a ×20 objective, equipped with OpenLab 5.5 software (Improvision, Coventry, UK). The frequency of nuclear PFKFB3 staining was expressed as a percentage of beta cells expressing PFKFB3 only in the nuclei. Nuclei were considered positive for PFKFB3 staining only if at least two-thirds of their area was occupied by multiple bright puncta. Image analysis was performed blindly by two independent investigators (HN and LP).
To visualise mitochondria, tissue sections were stained with Tom 20 antibody and images were taken under a ×63 objective with the AxioImager.M2a fluorescence microscope equipped with optical sectioning ApoTome.2 and ZEN2 (blue edition) software (ApoTome.2 and ZEN2: Carl Zeiss Microscopy, Thornowood, NY, USA). The mitochondrial area was quantified using the Image-Pro Premier 9.1 software (Rockville, MD, USA) and expressed as the Tom 20+ area inside the insulin+ area per islet divided by the number of beta cells within this area. To corroborate the findings using anti-Tom 20 immunostaining, we also immunostained sections with insulin and anti-glucose-regulated protein 75 (GRP75).
Rat islets
Animal studies were performed in compliance with the guidelines of the UCLA Office of Animal Research Oversight. Wild-type Sprague Dawley (Crl:CD(SD); RS:0000064; Charles Rivers Laboratory, Wilmington, MA, USA) male rats, between 4 and 9 months of age, were housed as described [13]. For rat islet isolation, after an overnight fast, animals were euthanised using isoflurane. The bile duct was cannulated, and a Hanks’ balanced salt solution (HBSS) (Invitrogen, Carlsbad, CA, USA) containing 0.23 mg/ml Liberase (catalogue no. 05401020001; Roche, Basel, Switzerland) and 0.1 mg/ml DNase (Roche; 10104159001) was injected into the pancreatic duct. The pancreas was then removed and transferred into a glass vial containing icecold Liberase solution, digested for 20 min at 37°C, and dispersed by shaking for 30 s. Islets were manually picked and cultured in RPMI 1640 medium (11 mmol/l glucose) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin and 10% (vol./vol.) fetal calf serum (FCS; Invitrogen) at 37°C, in a humidified 5% CO2 atmosphere.
Human islets
Human pancreatic islets were from the Islet Cell Resource Consortium and Prodo Laboratories. The donors, aged 32–67 years, were non-diabetic. Islets were cultured in RPMI 1640 medium (5.5 mmol/l glucose) containing 100 IU/ml penicillin, 100 μg/ml streptomycin and 10% (vol./vol.) fetal bovine serum (Invitrogen). For details of human islet preparations, see human islet checklist in the ESM.
INS-1 823/13 cells
The rat insulinoma cell line INS-1 832/13 was kindly provided by C. Newgard (Duke University, Durham, NC, UK). INS-1 832/13 cells were cultured in RPMI 1640 medium (11 mmol/l glucose) supplemented with 10 mmol/l HEPES, 1 mmol/l sodium pyruvate, 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen), 10% (vol./vol.) FBS (Gemini Bio-Products, West Sacramento, CA, USA), and 50 μmol/l β-mercaptoethanol (Sigma, St Louis, MO, USA), at 37°C, in a humidified 5% CO2 atmosphere. The cell line was free of mycoplasma.
Cytokine treatment
After isolation, rat islets were cultured for 24 h and then treated with a cytokine mix (25 U/ml IL-1β, 500 U/ml TNF-α, 500 U/ml IFN-γ) or vehicle for 24 h. Human islets were exposed to cytokines (50 U/ml IL-1β, 1000 U/ml TNF-α, 1000 U/ml IFN-γ) or vehicle for 48 h. Of note these are lower concentrations of cytokines than concentrations used by Eizirik and colleagues [11] since we required a low level of toxicity to then establish if there was an increase in cell death with inhibition of the putative protection by HIF1α signalling.
Small interfering RNA for PFKFB3 and inhibition of HIF1α
Pfkfb3 small interfering RNAs (siRNAs; L-095107-02-0005) were purchased from Dharmacon, Lafayette, CO, USA. INS-1 832/13 cells were plated at 0.7×106 cells per well in a 6-well plate. Twenty-four hours later, cells were transfected with siRNAs at 75 nmol/l using Lipofectamine RNAiMAX (Invitrogen) and cultured for 48 h before collection. The cytokine mix was added to cells for the last 24 h of culture.
For inhibition of HIF1α we used HIF1α translation and transactivation inhibitor KC7F2 (S7946; Selleckchem, Houston, TX, USA), which was screened for efficacy and toxicity (see ESM Fig. 1). We used concentrations that inhibited HIF1α without toxicity.
Lactate measurements
Medium from cultured islets was sampled at 22 and 24 h following cytokine treatment for rat islets, and 46 and 48 h for human islets. Lactate concentrations were measured using an enzymatic assay (735-10; Trinity Biotech, Wicklow, Ireland).
Western blotting and immunoprecipitation
To prepare whole cell extracts, cells or islets were incubated on ice in RIPA lysis buffer supplemented with protease inhibitors (Sigma P2714), sonicated, and spun at 13,000 g at 4°C for 3 min. Protein concentration was determined using the DC Protein Assay Kit (Bio-Rad, Irvine, CA, USA). Proteins (15–20 μg/lane) were separated by SDS-PAGE (4–12%) and then transferred onto polyvinylidene fluoride membranes by semi-dry electroblotting (Bio-Rad). After blocking with 10% (wt/vol.) milk for 1 h, membranes were probed overnight at 4°C with primary antibodies, then treated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature (Invitrogen). Proteins were visualised using electrochemiluminescence (ECL) reagents from Bio-Rad and expression levels were quantified using LabWorks (version 4.5.00.0) software (UVP, Upland, CA, USA).
To confirm specificity of the three PFKFB3 bands observed by western blot, we performed inversed immunoprecipitations using Protein-A magnetic beads and anti-PFKFB3, anti-phospho-PFKFB3, anti-acetyl lysine and anti-phospho serine antibodies (Invitrogen). PFKFB3-related western blot bands were shown to be specific since their density was reduced after silencing of PFKFB3 and corresponded to partly phosphorylated and acetylated PFKFB3 (ESM Fig. 2).
Antibodies
For detection of PFKFB3, we used rabbit anti-PFKFB3 antibody (Abcam, Cambridge, MA, USA; catalogue no. ab181861; RRID: not available) at 1:400 for immunofluorescence, 1:1000 for western blotting and 1 μg for immunoprecipitation. We have confirmed the specificity of this antibody previously [5]. Other antibodies used in the study were: rabbit anti-HIF1α (Abcam; catalogue no. ab2185; RRID: AB_302883; 1:1000 for western blotting); rabbit anti-lactate dehydrogenase (LDHA; Cell Signaling Technology, Danvers, MA, USA; catalogue no. 2012; RRID: AB_2137173; 1:1000 for western blotting); rabbit anti-hexokinase (HK)1 (Cell Signaling Technology; catalogue no. 2024; RRID: AB_2116996; 1:1000 for western blotting); rabbit anti-cleaved caspase 3 (Cell Signaling Technology; catalogue no. 9664; RRID: AB_2070042; 1:1000 for western blotting); rabbit anti-PFKFB3 (phosphoS461) (Abcam; catalogue no. ab202291; RRID not available; 1:500 for western blotting, 1 μg for immunoprecipitation); rabbit anti-acetyl lysine (Abcam; catalogue no. ab80178; RRID: AB_1640674; 1:500 for western blotting); rabbit anti-GRP75 (Abcam; catalogue no. ab53098; RRID: AB_880311; 1:300 for immunofluorescence); rabbit anti-phosphoserine (Abcam; catalogue no. ab9332; RRID: AB_307184; 1:500 for western blotting); guinea pig anti-insulin (Dako, Glostrup, Denmark; catalogue no. A0564; RRID: AB_10013624; 1:100 for immunofluorescence); mouse anti-glucagon (Sigma-Aldrich, St Louis, MO, USA); Clone K79bB10; catalogue no. G2654; RRID: AB_259852; 1:1000 for immunofluorescence); rat anti-C-peptide (Developmental Studies Hybridoma Bank, Iowa, IA, USA; catalogue no. GN-ID4; RRID: AB_2255626; 1:900 for immunofluorescence); rabbit anti-Tom20 (Santa Cruz Biotechnology, Dallas, TX, USA; catalogue no. sc-11415; RRID: AB_2207533; 1:400 for immunofluorescence); and rabbit anti-GAPDH (Cell Signaling Technology; catalogue no. 2118; RRID: AB_561053; 1:1000 for western blotting). Secondary antibodies for immunofluorescence staining were F(ab′)2 conjugates with Cy3, FITC or Alexa647 and were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA) and used at a dilution of 1:100–1:200.
Chemicals
The chemicals used in this study included: rat IL-1β (IL024; Millipore, Burlington, MA, USA); human IL-1β (I9401, Sigma); rat TNF-α (T5944, Sigma); human TNF-α (T0157, Sigma); rat IFN-γ (I3275, Sigma); human IFN-γ (I17001, Sigma); and the HIF-1α inhibitor KC7F2 (S7946, Selleckchem).
Statistical analysis
Results are expressed as the means ± SEM. Statistical analysis was performed with GraphPad Prism V software (La Jolla, CA, USA), and tests used include the two-tailed t test, Wilcoxon rank-sum test, ANOVA and regression analysis, as stated in the figure legends. Findings were considered significant when p<0.05.
Results
Upregulation of the HIF1α signalling pathway in type 1 diabetes
We examined three RNA-Seq datasets generated from human islets to establish whether the HIF1α signalling pathway is activated in beta cells in type 1 diabetes [10–12]. We expanded the list of 59 HIF1α target genes defined by Ortiz-Barahona et al [9] to include 17 additional genes regulated by HIF1α [14–19] (ESM Table 1).
By performing novel analysis on previously published datasets, we found that the expression of the 76 HIF1α target genes was increased in islets isolated from individuals with type 1 diabetes compared with control islets (median fold change, 1.22). By contrast, non-HIF1α target genes were downregulated in islets from those with type 1 diabetes (median fold change, 0.77; p<10−8 vs HIF1α target genes). To investigate whether cytokines implicated in type 1 diabetes mediate metabolic remodelling of beta cells in a HIF1α-dependent fashion, we analysed expression of HIF1α target genes relative to the rest of the genome using RNA-Seq data from human islets from individuals without diabetes that were treated with cytokines. HIF1α targets were overexpressed in islets treated with cytokines compared with untreated islets (median fold changes 1.6 and 0.94, for HIF1α target genes and non-HIF1α target genes respectively; p<10−14).
Islets from type 1 diabetes donors are relatively depleted of beta cells. To corroborate the findings in isolated beta cells, we investigated the available single-cell RNA-Seq data [12]. HIF1α-activated modulators of metabolism were expressed more highly in beta cells in type 1 diabetes relative to non-diabetic control samples; for example, PFKFB3 increased 37.6-fold, LDHA 22.8-fold and PDK1 14.4-fold (Fig. 1, Table 2). However, statistical power for this single-cell RNA-Seq dataset is limited by the low number of cells (n=6 for type 1 diabetes, n=17 for non-diabetic) and resolution issues.
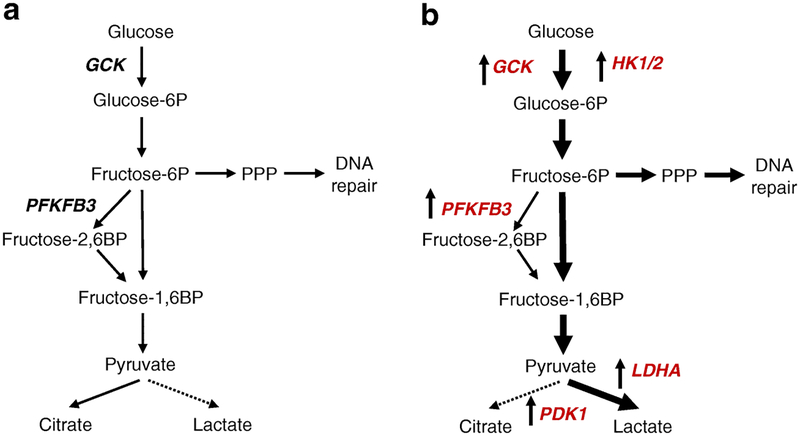
Fig. 1.
The scheme of the functional patterns within the glycolytic pathway in beta cells in health and type 1 diabetes. (a) In health (without diabetes), glucokinase (GCK) limits and regulates glucose flux through glycolysis due to a Km of ~10 mmol/l. (b) Available RNA-Seq data from humans with type 1 diabetes indicates that, apart from GCK, HK1/2, with a Km of ~0.1 mmol/l, are upregulated, facilitating flux through glycolysis. Moreover, the rate-limiting subsequent step in glycolysis (conversion of fructose-6-phosphate to fructose-1,6-bisphosphate) is enhanced in type 1 diabetes by upregulation of PFKFB3, increasing pyruvate production. In type 1 diabetes, the extra pyruvate generated is directed to lactate rather than to the TCA cycle by upregulation of pyruvate dehydrogenase kinase 1 (PDK1) and lactate dehydrogenase (LDHA). In health (a), pyruvate is metabolised to citrate (solid line) since pyruvate conversion to lactate is a disallowed pathway in adult beta cells (dotted line). In contrast, in type 1 diabetes (b), pyruvate is converted to lactate (solid line) rather than to citrate (dotted line). BP, bisphosphate; P, phosphate; PPP, pentose phosphate pathway.
Table 2.
Expression pattern of key aerobic glycolytic genes in three RNA-Seq datasets
| Dataset | Eizirik et al [11] | Mastracci et al [10] | Wang et al [12] |
|---|---|---|---|
| GCK | 1.8 | 0.01 | 10.6 |
| HK1 | 1.9 | 1.5 | NA |
| HK2 | 6.3 | 4.0 | NA |
| PFKFB3 | 3.6 | 2.8 | 37.6 |
| PDK1 | 1.7 | 1.5 | 14.4 |
| LDHA | 4.2 | 1.8 | 22.8 |
Data are presented as fold increase in type 1 diabetes samples vs non-diabetic control samples
NA, not available
Taken together these data suggest that HIF1α-mediated stress signalling is activated in beta cells exposed to cytokines implicated in type 1 diabetes, mirroring the stress response induced by protein misfolding in type 2 diabetes [5]. Given that the most prominent downstream target of HIF1α remodelled metabolism is PFKFB3, we next sought to establish if the PFKFB3 protein levels are increased in beta cells in type 1 diabetes.
PFKFB3 expression in type 1 diabetes
We evaluated pancreas sections from five brain-dead organ donors with type 1 diabetes that have detectable beta cells compared with five non-diabetic control donors. As previously reported, nuclear PFKFB3 immunoreactivity was detectable in approximately 10% of beta cells in non-diabetic individuals [5] (Fig. 2 and Fig. 3a). Of interest, the proportion of beta cells, but not alpha cells, that express PFKFB3 increased with age in non-diabetic individuals (Fig. 3c,d), consistent with accumulating oxidative stress in long-lived cells with a high burden of protein synthesis. In contrast to beta cells, there was minimal PFKFB3 immunoreactivity in the exocrine pancreas at any age (data not shown).
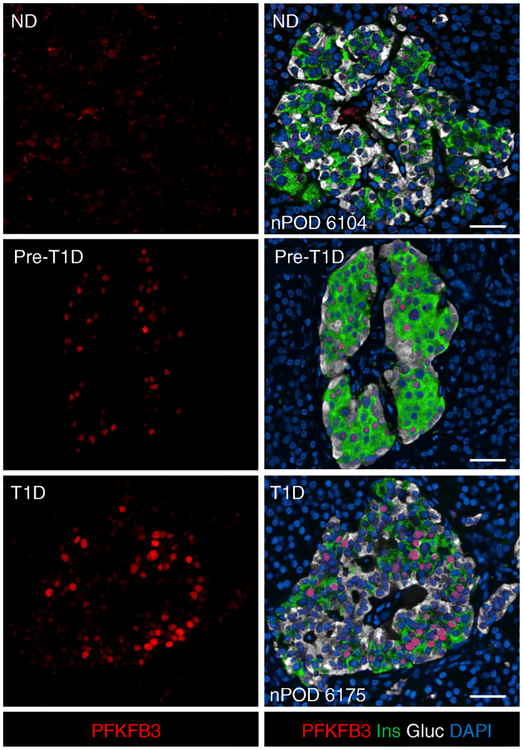
Fig. 2.
Representative immunofluorescent photomicrographs of islets from individuals without diabetes, pre-type 1 diabetes and type 1 diabetes, stained for PFKFB3 (red), insulin (green), glucagon (white) and DAPI (blue). PFKFB3 is weakly present in a minority of beta and alpha cells in islets from individuals without diabetes but is more frequent and intense in beta cells in islets from those with pre-type 1 diabetes and type 1 diabetes. As previously reported, PFKFB3 immunoreactivity was most prominent in the nucleus. Scale bars, 50 μm. Gluc, glucagon; Ins, insulin; ND, non-diabetic; T1D, type 1 diabetic
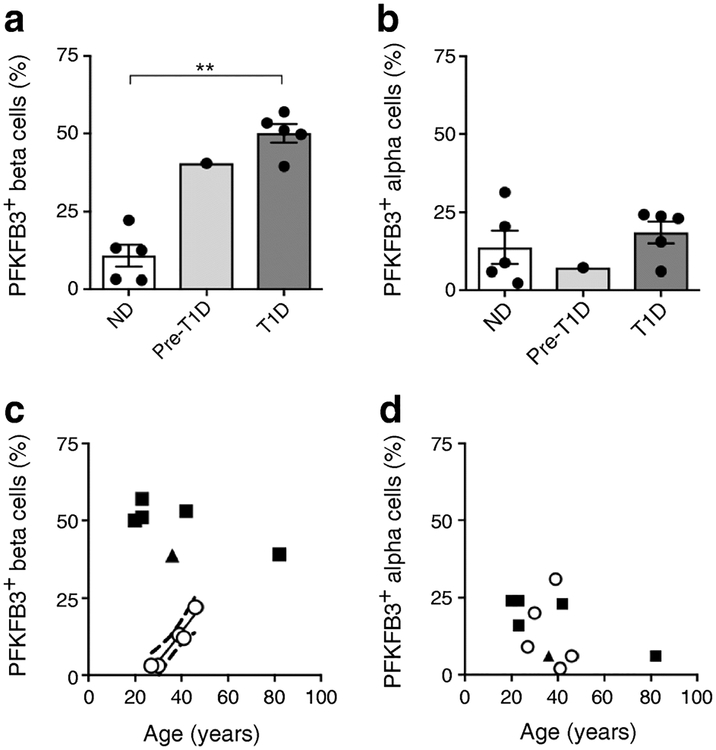
Fig. 3.
(a, b) Quantification of PFKFB3 immunoreactivity in (a) beta cells and (b) alpha cells in non-diabetic (n=5), pre-type 1 diabetes (n=1) and type 1 diabetes (n=5) samples. Data are mean ± SEM. **p<0.01, two-tailed unpaired Student’s t test. (c, d) Relationship between the percentage of (c) beta cells and (d) alpha cells positive for PFKFB3 and age for non-diabetic (white circles), pre-type 1 diabetes (triangles) and type 1 diabetes (squares) samples. The proportion of PFKFB3-positive beta cells increased with age in individuals without diabetes (r=0.96, p<0.01) but was not related to age in type 1 diabetes. The proportion of alpha cells positive for PFKFB3 was not related to age in individuals without diabetes (p=0.73) nor in those with type 1 diabetes (p=0.08), analysed by Pearson’s correlation analysis. ND, non-diabetic; T1D, type 1 diabetic
The proportion of beta cells with immune-detectable PFKFB3 in type 1 diabetes was increased compared with non-diabetic control samples (50.1±3.6% vs 10.8±2.9%; p<0.01; Fig. 3a). In contrast to non-diabetic individuals, in type 1 diabetes the proportion of beta cells that express PFKFB3 was increased irrespective of age. Moreover, the immunoreactivity to PFKFB3 in beta cells in type 1 diabetes was notably more intense than in non-diabetic individuals (ESM Fig. 3). The proportion of alpha cells with immunoreactivity to PFKFB3 was comparable in type 1 diabetes and non-diabetic control samples (18.5±3.5% vs 13.8±5.4%; p=0.5; Fig. 3b) and unrelated to age in either type 1 diabetes or controls (Fig. 3d).
Having established that the protein level of PFKFB3 is increased in beta cells in type 1 diabetes, we next sought to establish if this was also true in the prediabetes phase of type 1 diabetes.
PFKFB3 in pre-type 1 diabetes
We examined pancreases obtained surgically from a 36-year-old donor with pre-type 1 diabetes [20]. Consistent with the finding in type 1 diabetes, there was a 3.7-fold increase in beta cells with nuclear immunoreactivity to PFKFB3 in the individual with pre-type 1 diabetes compared with control samples (Fig. 2 and Fig. 3a). Notably this individual, while prediabetic by fasting glucose values (6.1 mmol/l) and HbA1c (0.06 mmol/mol [5.8%]), had glucose intolerance, as previously reported in the prediabetes phase of type 1 diabetes [21].
Having established, from RNA-Seq analysis, that HIF1α signalling is activated in islets in type 1 diabetes, and that the key HIF1α target PFKFB3 is upregulated in beta cells in type 1 diabetes and pre-type 1 diabetes, we next sought to establish if the cytokines implicated in autoimmunity activate the HIF1α/PFKFB3 pathway and, if so, if this is protective against cytokine-induced beta cell death.
HIF1α signalling is activated by cytokines in human and rat islets
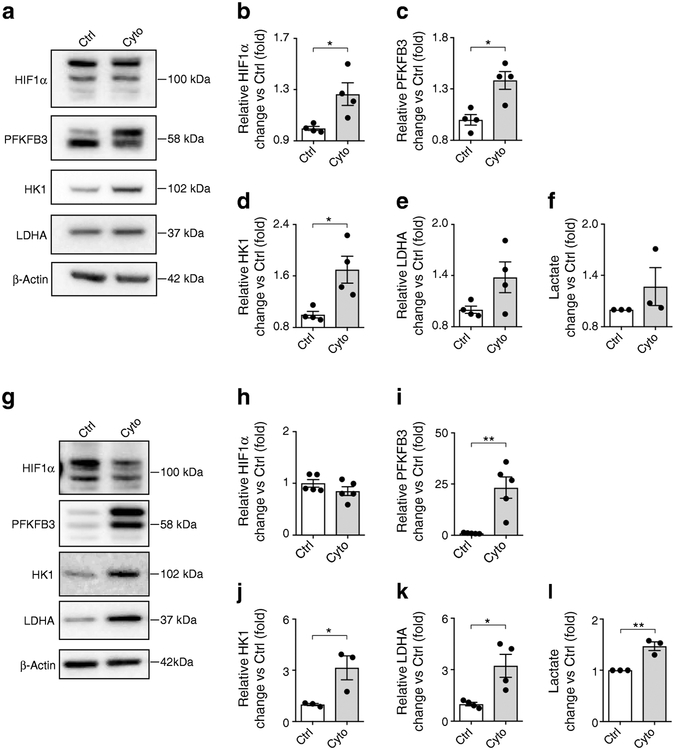
We evaluated human and rat islets for the expression of PFKFB3 and key metabolic targets of HIF1α after cytokine treatment (Fig. 4). Consistent with the RNA-Seq data (Table 2) [11], protein levels of both PFKFB3 and HK1 increased with cytokine treatment of human islets (Fig. 4a,c,d). HIF1α remodels metabolism, diverting the products of glycolysis from oxidative phosphorylation to lactate production, a pathway disallowed in functional beta cells. To establish if cytokine treatment remodels metabolism accordingly, we evaluated lactate production and expression of LDHA in isolated rat and human islets after exposure to cytokines. As expected, there was minimal LDHA expression in unstressed islets, but cytokines increased LDHA expression in rat islets (p<0.05; Fig. 4g,k). Moreover, cytokine treatment also increased lactate production in human (1.27-fold; p=0.30; Fig. 4f) and rat (1.47-fold; p<0.01; Fig. 4l) islets vs untreated controls.
Fig. 4.
(a) Representative western blots of human islets exposed to cytokines vs control. (b–e) Quantification of western blots for (b) HIF1α, (c) PFKFB3, (d) HK1 and (e) LDHA vs control. (f) Lactate production by human islets after cytokine or diluent (control) treatment. (g) Representative western blots of rat islet lysates after cytokine or control treatment. (h–k) Quantification of western blots for (h) HIF1α, (i) PFKFB3, (j) HK1 and (k) LDHA vs control. (l) Lactate production by rat islets after cytokine or diluent (control) treatment. Data are mean ± SEM; n=3–5 experiments; each experiment was performed using islets from an independent islet isolation (single sample per condition). *p<0.05, **p<0.01, two-tailed unpaired Student’s t test. Ctrl, control; Cyto, cytokines
Induction of pro-survival metabolism of aerobic glycolysis from oxidative phosphorylation is associated with remodelling of the mitochondrial network to a perinuclear fragmented form [22]. Therefore, we next investigated the mitochondrial network in beta cells in pancreases from individuals with type 1 diabetes vs control donors.
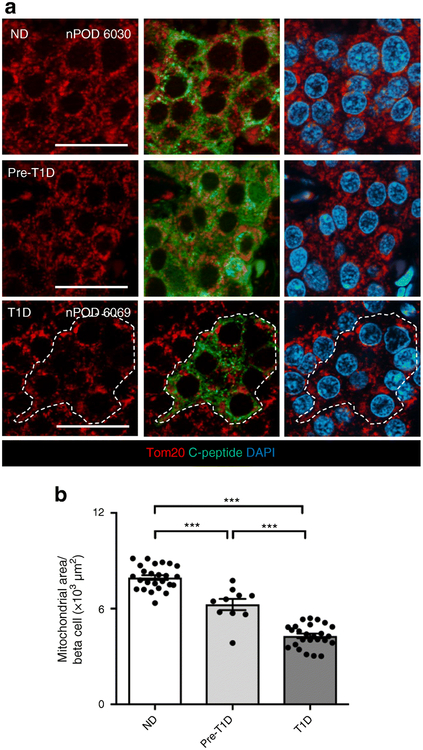
Mitochondrial density is decreased and mitochondrial network fragmented in type 1 diabetes
The mitochondrial network was identified within beta cells by immunostaining for Tom20, as previously described [23]. In non-diabetic control samples, the mitochondrial network extended throughout beta cells, as reported [5]. In contrast, the network was less abundant (p<0.001) and more confined to the perinuclear region in beta cells in type 1 diabetes and pre-type 1 diabetes (Fig. 5). These findings were corroborated by immunostaining of mitochondria with anti-GRP75 antibody [24–27] (ESM Fig. 4). In conclusion, the mitochondrial network adopts a pro-survival format in surviving beta cells in type 1 diabetes, consistent with pro-survival remodelled metabolism induced by HIF1α.
Fig. 5.
(a) Images of pancreatic islets from non-diabetic control, pre-type 1 diabetic and type 1 diabetic donors, stained for Tom20 (to visualise mitochondria; red), C-peptide (green) and DAPI (nuclei; blue). Tom20 immunostaining revealed perinuclear fragmented mitochondrial networks with reduced density in beta cells from a pre-type 1 diabetic individual; this appearance was even more pronounced in type 1 diabetes. Scale bars, 20 μm. (b) Quantification of mitochondrial area in beta cells, as assessed with Image-Pro analysis of pancreatic islets. Data are mean ± SEM. Non-diabetic, n=25; pre-type 1 diabetic, n=10; type 1 diabetic, n=25. ***p<0.001, one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. ND, non-diabetic; T1D, type 1 diabetic
HIF1α signalling is protective against cytokine-induced beta cell death
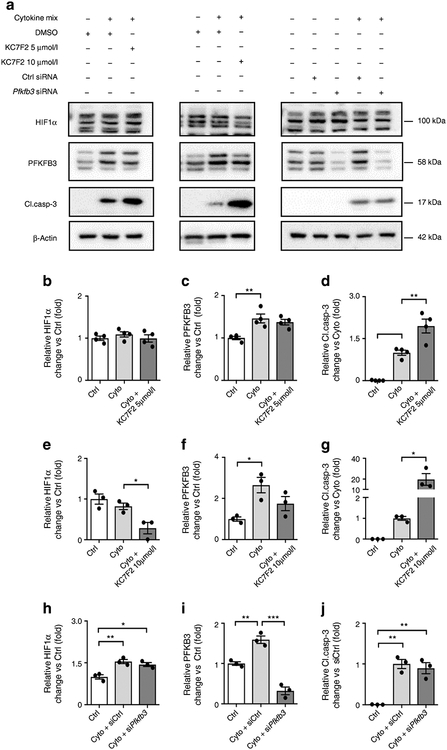
To establish if HIF1α and/or its metabolic effector PFKFB3 are protective against cytokine-induced beta cell death, we manipulated the levels of HIF1α or PFKFB3 in INS-1 832/13 cells in the presence and absence of cytokines.
Inhibition of HIF1α signalling attenuated the upregulation of PFKFB3 by cytokines, as expected (Fig. 6). Inhibition of HIF1α increased cytokine-induced cell death, quantified by activation of caspase-3 (Fig. 6a,d,g). To establish if PFKFB3 inhibition alone is sufficient to impact on cytokine-mediated beta cell death, we exposed INS-1 832/13 to cytokines in the presence and absence of siRNA against PFKFB3. While the Pfkfb3 siRNA successfully suppressed cytokine-induced PFKFB3, this had no impact on cytokine-mediated beta cell death. (Fig. 6a,i,j).
Fig. 6.
(a) Representative western blot analysis of whole cell extracts from INS-1 832/13 cells treated with either HIF1α inhibitor (KC7F2, 5 μmol/l and 10 μmol/l) or Pfkfb3 siRNA, and respective controls, in presence or absence of cytokines. (b–j) Quantification of western blots for (b, e, h) HIF1α, (c, f, i) PFKFB3 and (d, g, j) cleaved caspase-3 in the presence of 5 μmol/l KC7F2, 10 μmol/l KC7F2 or Pfkfb3 siRNA vs control. Data are mean ± SEM; n=3–4 independent experiments-. *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. Cleaved caspase-3, Cl.casp-3; Ctrl, control; Cyto, cytokines
We conclude, under the conditions of cytokines applied to INS-1 832/13 cells in vitro, the numerous targets of HIF1α signalling acting in concert are sufficient to suppress cell death, but PFKFB3 alone is insufficient to impact on cytokine-mediated cell death.
Discussion
We established that the HIF1α pro-survival signalling pathway is activated in beta cells in type 1 diabetes. This finding provides a potential mechanism for the slow rate of beta cell loss and early defects in glucose-mediated insulin secretion in evolving type 1 diabetes.
Analysis of available RNA-Seq data [10–12] revealed increased HIF1α signalling in islets and isolated beta cells from individuals with type 1 diabetes, and in human islets exposed to cytokines implicated in type 1 diabetes. A limitation of this study, and other studies using the RNA-Seq approach in human islets, is the small sample sizes available for evaluation, as well as reliance on RNA expression that is not always reflected by protein levels. Because of this, we corroborated the RNA-Seq finding of increased HIF1α signalling by demonstrating increased protein expression of PFKFB3 in islets exposed to cytokines, which was attenuated by inhibition of HIF1α. We also showed increased protein expression of PFKFB3 in beta cells from individuals with type 1 diabetes. HIF1 is composed of alpha and beta subunits. The HIF1α subunit is degraded following post-translational modification by prolyl-hydroxylases that facilitate ubiquitination and proteasomal degradation [28]. As prolyl-hydroxylase requires oxygen as a co-substrate, the alpha subunit is stabilised under conditions of hypoxia, and the HIF1α complex is mobilised to the nucleus where it activates cell-survival signals [29]. Alternatively, hydroxylation of the alpha subunit of HIF1α is inhibited by increased succinate/fumarate levels that occur when flux through the tricarboxylic acid (TCA) cycle slows, referred to as pseudohypoxia [30]. Mitochondrial dysfunction is an early manifestation of autoimmunity directed at beta cells and, therefore, the most likely inducer of activation of HIF1α signalling in type 1 diabetes [31]. The decreased density and perinuclear distribution of mitochondria noted in beta cells in type 1 diabetes in the current study resembles that observed as a defensive adaptation to stress accomplished by metabolic remodelling to high glycolysis and decreased oxidative phosphorylation [32–34].
The pattern of HIF1α-targeted metabolic remodelling genes in beta cells in type 1 diabetes is consistent with the switch from oxidative phosphorylation to aerobic glycolysis. The genes that encode enzymes that increase flux through glycolysis were increased, including HK, which lowers the Km barrier to initiate glucose metabolism, and PFKFB3, which increases flux through the subsequent rate-limiting step of glycolysis (fructose-6 phosphate to fructose 1,6 biphosphate; Fig. 1). Further, the increased expression of pyruvate dehydrogenase kinase 1 (PDK1) implies that the pyruvate delivered by glycolysis is directed to lactate generation via the LDHA enzyme, rather than to the TCA cycle. The benefit of this reordered metabolism is that it is pro-survival, increasing the resilience of beta cells to cytokine damage. Suppressed mitochondrial respiration implies ATP is generated independently from oxidative phosphorylation, while increased flux through the pentose phosphate pathway generates NADH that buffers against the pro-oxidative actions of cytokines and enhances DNA repair. The expense of the reordered metabolism is that it renders beta cells relatively non-responsive to glucose, consistent with the as-yet-unexplained progressive decline in glucose-mediated insulin secretion and glucose intolerance that precedes diabetes onset in type 1 diabetes [35]. The overlap of transcriptomes of human islets exposed to cytokines and islets isolated from humans with recent-onset type 1 diabetes supports a primary role of cytokines in mediating beta cell dysfunction and loss in type 1 diabetes [10–12].
A recent genome-wide association study of adult-onset type 1 diabetes reported linkage to one of the major targets of HIF1α signalling, PFKFB3 [36]. As yet, it is unknown if this linkage is to variants of PFKFB3 that have increased or decreased activity. Variants of PFKFB3 with enhanced activity might be linked to adult-onset type 1 diabetes by slowing the rate of beta cell loss so that the individuals present later in life, as adults. Alternatively, variants in PFKFB3 that decrease activity might alter the balance of beta cell survival vs loss in favour of developing type 1 diabetes. While PFKFB3 alone was insufficient to suppress cytokine-mediated islet cell death following treatment of INS-1 823/13 cells with cytokines, it is unknown if this is also true of the much less potent conditions present in beta cells in vivo in type 1 diabetes.
We noted that a small subset of beta cells express PFKFB3 in non-diabetic individuals and that their proportion increases with age. From cross-sectional studies, it is not possible to establish if this is a subset of cells that have stable expression of PFKFB3 or if these long-lived endocrine cells transiently express PFKFB3 when undergoing cellular repair. If the latter is the case, then it might be hypothesised that, under conditions of increased beta cell stress (as in type 1 diabetes), beta cells are recruited into the repair pool more rapidly than in health, and that the cells in this pool are relatively protected and so accumulate as a proportion of all beta cells. In support of a DNA repair role for HIF1α/PFKFB3 signalling, remodelled metabolism includes increased flux through the pentose phosphate pathway that is required for DNA repair. Beta cell mass remains remarkably preserved in aged non-diabetic individuals, possibly, in part, as a consequence of the capacity for protection and DNA repair by metabolic remodelling induced by HIF1α/PFKFB3 signalling [37]. In contrast, the exocrine pancreas, which has minimal PFKFB3 expression, undergoes marked atrophy and cell loss with advancing age [38].
From a therapeutic viewpoint, inhibition of the activated HIF1α pathway is not logical since, while it may transiently enhance glucose-mediated insulin secretion, it is likely to do so at the expense of cell survival. The benefit of cytokine-induced HIF1α signalling is that it slows beta cell loss and, therefore, if an effective immune-modulatory therapy is introduced, beta cell function may return to normal.
In summary, cytokines implicated in autoimmunity activate the HIF1α signalling in beta cells resulting in remodelling metabolism towards increased aerobic glycolysis and suppressed cytokine-induced cell death. Activation of this signalling pathway is already present in the period before diabetes onset. Cytokine-induced activation of the HIF1α pathway, therefore, is likely to contribute to the slow rate of beta cell loss but early defective glucose-induced insulin secretion in type 1 diabetes.
Supplementary Material
Research in context.
What is already known about this subject?
Progression from the onset of autoimmunity to the loss of sufficient beta cells to induce diabetes onset is slow in adults with recent-onset type 1 diabetes
The slow progression of beta cell loss in type 2 diabetes is, at least in part, attributable to activation of the conserved hypoxia inducible factor 1 α (HIF1α)/6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3) injury-response signalling programme
Deletion of HIF1α in beta cells increases vulnerability to viral infection and toxins
What is the key question?
Is the HIF1α/PFKFB3 injury-response programme activated in beta cells in type 1 diabetes?
What are the new findings?
The HIF1α signalling pathway is activated in beta cells early in the development of type 1 diabetes
HIF1α delays cytokine-induced beta cell death independently of metabolic remodelling by the HIF1α target PFKFB3
How might this impact on clinical practice in the foreseeable future?
Activation of the HIF1α signalling pathway in beta cells exposed to autoimmunity is likely to preserve beta cell mass during the pre-type 1 diabetes phase that offers the opportunity to allow therapeutic interventions to suppress autoimmunity. Inadvertent suppression of the pathway would likely accelerate diabetes onset
Acknowledgments
This research was performed with the support of the Network for Pancreatic Organ Donors with Diabetes (nPOD), a collaborative type 1 diabetes research project sponsored by the JDRF. Organ Procurement Organizations (OPO) partnering with nPOD to provide research resources are listed at www.jdrfnpod.org/our-partners.php. HN was supported by MSD Life Science Foundation, Public Interest Incorporated Foundation. LP was supported by Department of Endocrinology, Shengjing Hospital of China Medical University, Shenyang, China.
Funding These studies were supported by grant funding from the NIH DK059579 and the Larry L. Hillblom Foundation Network Grant 2014-D-001-NET to PCB; and Larry L. Hillblom Foundation Start-up Grant 2017-D-002-SUP to ST
Abbreviations
- CpM
Counts per million
- GEO
Gene Expression Omnibus
- GRP75
Glucose-regulated protein 75
- HIF1α
Hypoxia inducible factor 1 α
- HK
Hexokinase
- LDHA
Lactate dehydrogenase
- nPOD
JDRF Network for Pancreatic Organ Donors with Diabetes
- PFKFB3
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- RNA-Seq
RNA sequencing
- RPKM
Reads per kb of transcript per million mapped reads
- siRNA
Small interfering RNA
- TBS
Tris-buffered saline
- TCA
Tricarboxylic acid
Footnotes
Data availability All the data presented in this manuscript will be made available upon request.
Duality of interest GC is currently an employee of Regeneron Pharmaceuticals. All other authors declare no duality of interest associated with this manuscript.
References
- [1].Eisenbarth GS (1986) Type I diabetes mellitus. A chronic autoimmune disease. N Engl J Med 314: 1360–1368 [DOI] [PubMed] [Google Scholar]
- [2].Atkinson MA, Eisenbarth GS, Michels AW (2014) Type 1 diabetes. Lancet 383: 69–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Evans-Molina C, Sims EK, DiMeglio LA, et al. (2018) β Cell dysfunction exists more than 5 years before type 1 diabetes diagnosis. JCI insight 3(15):e120877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Butler AE, Galasso R, Meier JJ, Basu R, Rizza RA, Butler PC (2007) Modestly increased beta cell apoptosis but no increased beta cell replication in recent-onset type 1 diabetic patients who died of diabetic ketoacidosis. Diabetologia 50: 2323–2331 [DOI] [PubMed] [Google Scholar]
- [5].Montemurro C, Nomoto H, Pei L, et al. (2019) IAPP toxicity activates HIF1alpha/PFKFB3 signaling delaying beta-cell loss at the expense of beta-cell function. Nature communications 10: 2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lalwani A, Warren J, Liuwantara D, et al. (2019) β Cell hypoxia-inducible factor-1α is required for the prevention of type 1 diabetes. Cell reports 27: 2370–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nagy G, Kovacs-Nagy R, Kereszturi E, et al. (2009) Association of hypoxia inducible factor-1 alpha gene polymorphism with both type 1 and type 2 diabetes in a Caucasian (Hungarian) sample. BMC medical genetics 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yamada N, Horikawa Y, Oda N, et al. (2005) Genetic variation in the hypoxia-inducible factor-1α gene is associated with type 2 diabetes in Japanese. J Clin Endocrinol Metab 90: 5841–5847 [DOI] [PubMed] [Google Scholar]
- [9].Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L (2010) Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res 38: 2332–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mastracci TL, Turatsinze JV, Book BK, et al. (2018) Distinct gene expression pathways in islets from individuals with short- and long-duration type 1 diabetes. Diabetes Obes Metab 20(8):1859–1867 [DOI] [PubMed] [Google Scholar]
- [11].Eizirik DL, Sammeth M, Bouckenooghe T, et al. (2012) The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet 8: e1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang YJ, Schug J, Won KJ, et al. (2016) Single-cell transcriptomics of the human endocrine pancreas. Diabetes 65: 3028–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Butler AE, Jang J, Gurlo T, Carty MD, Soeller WC, Butler PC (2004) Diabetes due to a progressive defect in beta-cell mass in rats transgenic for human islet amyloid polypeptide (HIP rat): a new model for type 2 diabetes. Diabetes 53: 1509–1516 [DOI] [PubMed] [Google Scholar]
- [14].Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ (2009) An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic acids research 37: 4587–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC (2003) Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol Cell Biol 23: 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim JW, Tchernyshyov I, Semenza GL, Dang CV (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- [17].Liu W, Shen SM, Zhao XY, Chen GQ (2012) Targeted genes and interacting proteins of hypoxia inducible factor-1. International journal of biochemistry and molecular biology 3: 165–178 [PMC free article] [PubMed] [Google Scholar]
- [18].Semenza GL (2013) HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 123: 3664–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guimaraes-Camboa N, Stowe J, Aneas I, et al. (2015) HIF1α represses cell stress pathways to allow proliferation of hypoxic fetal cardiomyocytes. Dev Cell 33: 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nomoto H, Gurlo T, Rosenberger M, Girgis MD, Dry S, Butler PC Low grade islet but marked exocrine pancreas inflammation in an adult with autoimmune pre-diabetes. Case Reports in Endocrinology (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ismail HM, Xu P, Libman IM, et al. (2018) The shape of the glucose concentration curve during an oral glucose tolerance test predicts risk for type 1 diabetes. Diabetologia 61: 84–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R (2004) Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 16: 59–68 [DOI] [PubMed] [Google Scholar]
- [23].Neuspiel M, Schauss AC, Braschi E, et al. (2008) Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol 18: 102–108 [DOI] [PubMed] [Google Scholar]
- [24].Trigo D, Goncalves MB, Corcoran JPT (2019) The regulation of mitochondrial dynamics in neurite outgrowth by retinoic acid receptor beta signaling. FASEB journal 33: 7225–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Agnew T, Goldsworthy M, Aguilar C, et al. (2018) A Wars2 mutant mouse model displays OXPHOS deficiencies and activation of tissue-specific stress response pathways. Cell reports 25: 3315–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Erster S, Mihara M, Kim RH, Petrenko O, Moll UM (2004) In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Molecular and cellular biology 24: 6728–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mazkereth N, Rocca F, Schubert JR, et al. (2016) Complement triggers relocation of mortalin/GRP75 from mitochondria to the plasma membrane. Immunobiology 221: 1395–1406 [DOI] [PubMed] [Google Scholar]
- [28].Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92: 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chilov D, Camenisch G, Kvietikova I, Ziegler U, Gassmann M, Wenger RH (1999) Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1α. J Cell Sci 112: 1203–1212 [DOI] [PubMed] [Google Scholar]
- [30].Selak MA, Armour SM, MacKenzie ED, et al. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer cell 7: 77–85 [DOI] [PubMed] [Google Scholar]
- [31].Chen J, Stimpson SE, Fernandez-Bueno GA, Mathews CE (2018) Mitochondrial reactive oxygen species and type 1 diabetes. Antioxidants & redox signaling 29: 1361–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buck MD, O’Sullivan D, Klein Geltink RI, et al. (2016) Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166: 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hall AR, Burke N, Dongworth RK, et al. (2016) Hearts deficient in both Mfn1 and Mfn2 are protected against acute myocardial infarction. Cell Death Dis 7: e2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Coronado M, Fajardo G, Nguyen K, et al. (2018) Physiological mitochondrial fragmentation is a normal cardiac adaptation to increased energy demand. Circ Res 122: 282–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sosenko JM, Skyler JS, Herold KC, Palmer JP, Type 1 Diabetes TrialNet, Diabetes Prevention Trial-Type 1 Study Group (2012) The metabolic progression to type 1 diabetes as indicated by serial oral glucose tolerance testing in the Diabetes Prevention Trial-type 1. Diabetes 61: 1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cousminer DL, Ahlqvist E, Mishra R, et al. (2018) First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. Diabetes Care 41: 2396–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Saisho Y, Butler AE, Manesso E, Elashoff D, Rizza RA, Butler PC (2013) β-Cell mass and turnover in humans: effects of obesity and aging. Diabetes Care 36: 111–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Saisho Y, Butler AE, Meier JJ, et al. (2007) Pancreas volumes in humans from birth to age one hundred taking into account sex, obesity, and presence of type-2 diabetes. Clin Anat 20: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.