Abstract
The tumor suppressor Folliculin (FLCN) enables nutrient-dependent activation of the mechanistic target of rapamycin complex 1 (mTORC1) protein kinase via its guanosine triphosphatase (GTPase) Activating Protein (GAP) activity toward the GTPase RagC. Concomitant with mTORC1 inactivation by starvation, FLCN relocalizes from the cytosol to lysosomes. To determine the lysosomal function of FLCN, we reconstituted the lysosomal FLCN complex (LFC) containing FLCN, its partner FLCN-interacting protein 2 (FNIP2), the RagAGDP:RagCGTP GTPases as they exist in the starved state with their lysosomal anchor Ragulator complex, and determined its cryo-EM structure to 3.6Å. The RagC-GAP activity of FLCN was inhibited within LFC, due to displacement of a catalytically required Arginine in FLCN from the RagC nucleotide. Disassembly of the LFC and release of the RagC-GAP activity of FLCN enabled mTORC1-dependent regulation of the master regulator of lysosomal biogenesis, transcription factor E3, implicating the LFC as a checkpoint in mTORC1 signaling.
One Sentence Summary:
The inactive Rag GTPase dimer is sandwiched between Ragulator and FLCN:FNIP in the cryo-EM structure of a lysosomal complex that controls Rag GTPase activation and modulates mTORC1 signaling output.
The mechanistic Target of Rapamycin Complex 1 (mTORC1) kinase is at the center of a complex biochemical network that relays information about the availability of nutrients, growth factors, oxygen, and energy to downstream signaling programs. When cells are adequately nourished, these signaling programs drive growth and metabolism, and inhibit catabolic pathways such as autophagy (1, 2). Nutrients including amino acids, glucose, and cholesterol activate mTORC1 by triggering its translocation from the cytosol to the lysosomal membrane (3–6). This process is regulated by the heterodimeric Rag guanosine triphosphatases (GTPases)(3, 4, 7), which localize to the lysosomal membrane via binding to the pentameric Ragulator complex (4, 8, 9). Rag heterodimers consist of two functionally equivalent pairs, RagA or RagB in complex with RagC or RagD. Nutrients trigger the transition from the ‘inactive’ combination of guanosine diphosphate (GDP)-bound Rag A and guanosine triphosphate (GTP)-bound Rag C (RagAGDP:RagCGTP) to the ‘active’ RagAGTP:RagCGDP state. The ‘active’ Rag heterodimer binds directly to mTORC1 and recruits it to the lysosome, enabling its subsequent activation (3, 4, 7, 10, 11).
Two GTPase-activating protein (GAP) complexes mediate, in part, the conversion between ‘active’ and ‘inactive’ Rag GTPase states. When amino acid concentrations are low, GATOR1 promotes GTP hydrolysis by RagA or B in response to a well-characterized upstream nutrient-sensing pathway (12–14). Conversely, nucleotide hydrolysis on RagC or D is stimulated by Folliculin (FLCN), the tumor suppressor responsible for Birt-Hogg-Dubé syndrome (BHD) (15), in complex with FLCN-interacting protein 1 or 2 (FNIP1 or 2) (16–18). Consistent with its role in Rag GTPase activation, acute depletion of FLCN in cell culture impairs activation of mTORC1 signaling by amino acids (18, 19). Yet in cellular and mouse models of BHD, the effects of FLCN loss on mTORC1 signaling range from neutral to activating, leading to a controversial and context-dependent interpretation of FLCN function (16, 20, 21). A further conundrum concerns the subcellular localization of FLCN:FNIP. FLCN has been reported to localize to the lysosomal membrane in low nutrient conditions, and to relocalize to the cytosol as nutrient levels increase (18, 19, 22). How these changes in FLCN localization relate to regulation of its GAP activity is not understood. Finally, structural details of FLCN interactions with its partner FNIP, how FLCN:FNIP physically interacts with Rag GTPases, and the key residues for the GAP activity of FLCN:FNIP are unknown.
Assembly of a stable lysosomal FLCN complex lacking GAP activity
Consistent with previous reports (18, 19, 22), FLCN clustered on LAMP2-positive lysosomes in amino acid-depleted conditions and became dispersed in the cytosol upon refeeding (Fig. S1, A and B). FLCN relocalization was controlled by the nucleotide state of the Rag GTPases: expressing Rag mutants locked in the ‘inactive’ nucleotide binding state caused FLCN to be constitutively bound to lysosomes irrespective of amino acid conditions; conversely, the ‘active’ Rag GTPase mutants caused FLCN to become cytosolic under all conditions (Fig. S1, A and B).
To determine the function of lysosomal FLCN in starvation, we monitored the FLCN:FNIP2 GAP reaction in in vitro RagC GTPase assays. To selectively generate Rag heterodimers containing one diphosphate and one triphosphate nucleotide, we used the mutant RagCD181N, which specifically binds xanthine nucleotides (8, 18) (henceforward referred to simply as RagC). RagAGTP:RagCXTP was incubated with a catalytic amount of the appropriate GAP complex (FLCN:FNIP2 or GATOR1) to generate GTP:XDP or GDP:XTP bound heterodimers that were confirmed by high pressure liquid chromatography (HPLC) (Fig. 1A). An intrinsic tryptophan fluorescence GTPase assay (23) was used to monitor in real time the FLCN:FNIP2-catalyzed RagC-GTPase activity (Fig. S2, A and B). FLCN:FNIP2 was added to Rag heterodimers containing a GDP or GTP-bound RagA, with or without the addition of stoichiometric amounts of Ragulator. We observed similar FLCN:FNIP2 GAP activity for all tested substrate permutations, except that the RagAGDP:RagCXTP-Ragulator combination greatly reduced the GAP activity of FLCN:FNIP2 (Fig. 1B). An HPLC-based GAP assay confirmed reduction of FLCN:FNIP2 GAP activity in the presence of GDP-bound RagA and Ragulator (Fig. 1C).
Fig. 1. Absence of FLCN GAP activity in the Lysosomal Folliculin Complex (LFC).
(A) Nucleotide elution profiles from High Performance Liquid Chromatography (HPLC) analysis of Rag nucleotide binding states. Rag heterodimers were loaded with GTP and XTP to generate RagAGTP:RagCXTP, boiled to release bound nucleotides, and the HPLC elution profile is shown (grey). RagAGTP:RagCXTP were incubated with FLCN:FNIP2 (orange) or Gator1 (cyan) then analyzed by HPLC. A nucleotide standard consisting of 50 μM GDP, GTP, XDP, and XTP is shown (black). (B) FLCN:FNIP2 RagC-GAP activity visualized by tryptophan fluorescence signal decay. Rags bound to the specified combinations of nucleotides and Ragulator (black: RagAGDP:RagCXTP-Ragulator, grey: RagAGDP:RagCXTP, cyan: RagAGTP:RagCXTP, blue: RagAGTP:RagCXTP-Ragulator) were incubated with FLCN:FNIP2. Plotted are mean +/− SEM. N=3. (C) HPLC-based GTPase assay comparing FLCN:FNIP2 RagC-GAP activity on the indicated Rag GTPase or Rag GTPase-Ragulator substrates. “No GAP” control was performed on a RagAGTP:RagCXTP substrate. Plotted are mean +/− SD. N=3. “U.” indicates that remaining XTP signal was undetectable by HPLC. (D) SEC profile of RagAGDP:RagCXTPγS-Ragulator (cyan), FLCN:FNIP2 (orange), or the full LFC containing RagAGDP:RagCXTPγS-Ragulator-FLCN:FNIP2 (green). (E) Coomassie stain of eluted SEC fraction containing assembled LFC.
Rag heterodimers containing GDP-bound RagA form a tight complex with Ragulator at the lysosomal membrane (24). To determine whether RagAGDP:RagCXTP and Ragulator form a larger complex that includes FLCN:FNIP2, we combined all three subcomplexes (Fig. S3, A to C) and subjected the mixture to size exclusion chromatography (SEC). We observed an early eluting peak when all components were combined, and confirmed that FLCN:FNIP2, RagA:RagC, and Ragulator co-eluted in this fraction (Fig. 1, D and E). Thus, FLCN:FNIP2 forms a stable complex with RagAGDP:RagCGTP-Ragulator, explaining the localization of FLCN on the lysosomal membrane and suggesting that the FLCN GAP activity is inhibited during starvation. Because this complex corresponds to all the properties expected of the lysosomal form of FLCN, we designate it the “Lysosomal FLCN Complex” (LFC).
Structure and nucleotide state dependence of the LFC
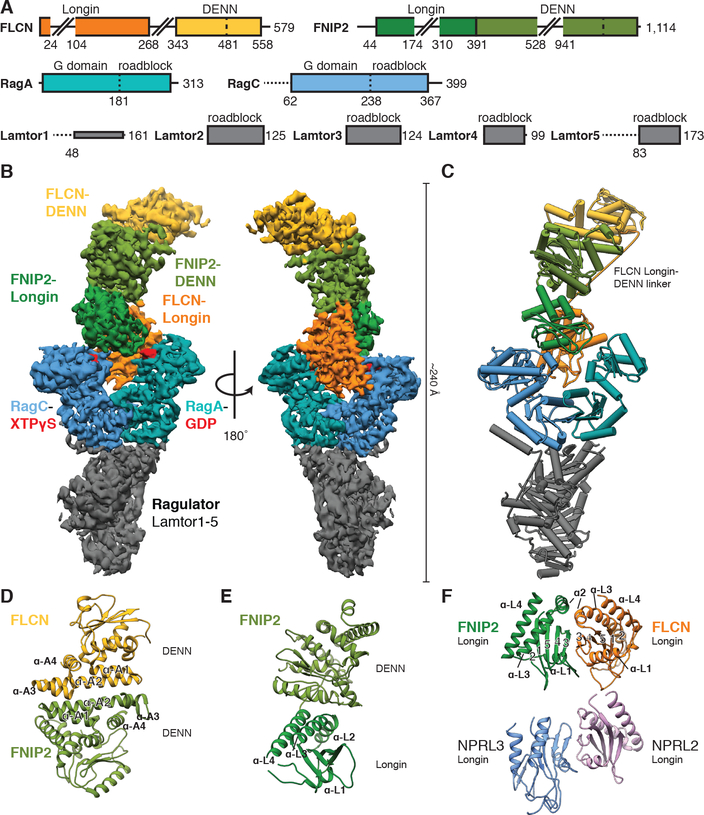
We determined the structure of the LFC at a resolution of 3.6 Å by cryo-EM (Fig. 2, A to C, Figs. S4 to S5 and Table S1). The complex adopts an elongated shape of 240 Å in the longest dimension. The reconstructed cryo-EM density was of sufficient quality to build an atomic coordinate model (Fig. 2C, Figs. S6 and S7 and Table S2) comprising about 60 % of the mass of the complex. The remainder of the molecular mass consists of flexible loops (Fig. 2A) with no corresponding cryo-EM density visible.
Fig. 2. Cryo-EM structure of the LFC.
(A) Domain organization of the LFC proteins. Horizontal dashed lines indicate disordered regions. Extended loops in FLCN and FNIP2 are indicated by skewed lines. (B) Cryo-EM density map of the LFC with model-based amplitude scaling employed (LocScale map, used for all LFC density depictions, contour 0.15). (C) Refined and assembled coordinate model of the LFC represented as pipes (α-helices) and planks (β-sheets). Color scheme as in (A). (D to F) Ribbon representation of FLCN:FNIP2 domain architecture. DENN-DENN interaction (D), FNIP2 DENN module arrangement (E) and longin-longin domain (F, top). The NPRL2:NPRL3 longin-longin domain from GATOR1 is displayed for comparison (F, bottom; pdb 6CES).
FLCN and FNIP1/2 are both DENN module proteins composed of an N-terminal longin and a C-terminal DENN (differentially expressed in normal and neoplastic cells) domain (16–18, 25). The known crystal structures (26, 27) of the Ragulator sub-complex and the FLCN DENN domain are positioned at opposite tips of the complex. The Rag GTPases were built on the basis of crystal and cryo-EM structures (14, 28, 29), and their location was consistent with the crystallographically resolved interaction of their C-terminal roadblock domains with Ragulator (29). The remaining structural elements from FLCN and FNIP2 were built on the basis of newly generated homology models (Supplementary Text, Fig. 2, D to F and Figs. S8 to S10).
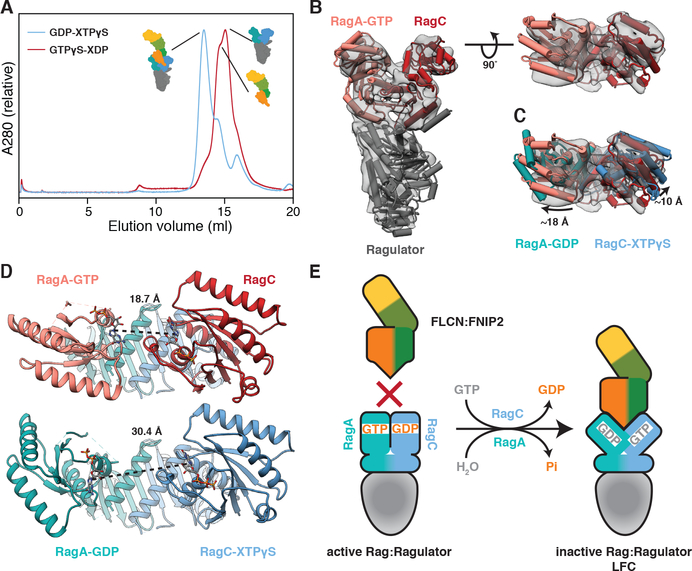
The two longin domains of FLCN:FNIP2 form a heterodimer similar to the NPRL2:NPRL3 longin domains of GATOR1 (Fig. 2F and Fig. S8C) (14). This longin-longin heterodimer is the only FLCN:FNIP2 element to directly interact with the Rag GTPases via their G domains. To understand how the Rag nucleotide state controls LFC formation, we tested whether FLCN:FNIP2 could form a stable complex with Ragulator and Rag GTPases in their active nucleotide binding state (RagAGTP:RagCXDP). Consistent with the cytoplasmic localization of FLCN under nutrient-rich conditions (18, 19, 22), FLCN:FNIP2 did not form a stable complex with active Rag-Ragulator as judged by SEC (Fig. 3A). To gain structural insight into how the Rag GTPase nucleotide states control LFC assembly, we determined an ~8–9 Å cryo-EM structure of the 150-kD Ragulator-RagAGTP:RagCXDP complex (Fig. 3B and Table S1). We did not detect any differences in the RagA and RagC G domain orientations between our active Rag-Ragulator structure and the previously reported GATOR1-RagA:RagC structure in a pseudo-active binding state (RagAGTP:RagCempty) (14) (Fig. 3B). Alignment of RagA:RagC in the active with their counterparts in the inactive nucleotide state within the LFC showed that both G domains reorient relative to the roadblock domains (Figs. 3C, S11A–B and Movie S1). Similar reorientation has been observed for the RagA and RagC orthologs Gtr1 and Gtr2 from S. cerevisiae (28, 30). As a consequence of this reorientation, the cleft between the RagA and RagC G domains is nearly twice as wide in the inactive (RagAGDP:RagCGTP) state as it is in the active (RagAGTP:RagCGDP) state (Fig. 3D). Without this cleft opening, the FLCN:FNIP2 longin heterodimer would be sterically unable to bind RagA:RagC as observed in the LFC (Fig. S11C). This explains structurally why the LFC does not form when nutrient concentrations are high and the Rag GTPases are in the active nucleotide binding state (Fig. 3E).
Fig. 3. A nucleotide-driven conformational change of the Rag GTPases controls LFC assembly.
(A) SEC profile of FLCN:FNIP2 mixed with Rag-Ragulator in the inactive (blue) and active (red) nucleotide binding state. (B) Side (left) and top (right) view of the cryo-EM structure of Ragulator-RagAGTP:RagCXDP (grey, transparent, contour 0.32). Rigid-body fitted atomic models for Ragulator (dark grey, pdb 6B9X) and RagAGTP:RagCempty (light red, red, pdb 6CES) are represented as pipes (α-helices) and planks (β-sheets). (C) Top view of the active Rag-Ragulator structure as in (B) with the atomic model of RagAGDP:RagCXTPγS from the LFC overlaid (cyan, blue). Model alignment was based on the constant roadblock domains of RagA:RagC. Extend of the G domain rotations are indicated. (D) Nucleotide-nucleotide distance in active (top) and inactive (bottom) RagA:RagC. As RagC in the active RagA:RagC structure is nucleotide-free, the respective nucleotide-nucleotide distance is based on a hypothetical model build by aligning the RagA and RagC G domains of the inactive LFC structure to the G domains of the pseudo-active RagAGTP:RagCempty model. (E) Structure-based model for the nucleotide dependent formation of the LFC.
Regulation of Rag nucleotide state by the LFC
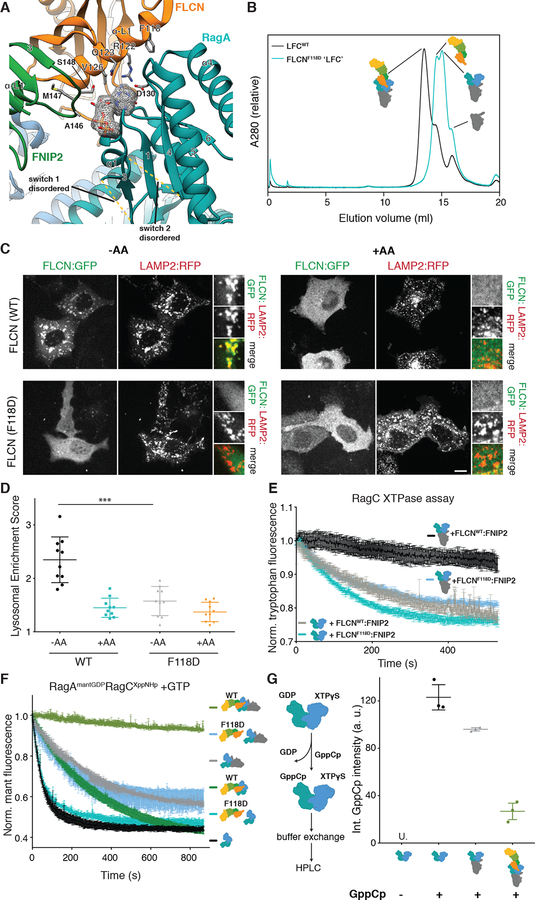
Investigation of the interfaces between FLCN:FNIP2 and Rag G domains revealed that FLCN:FNIP2 interacts more extensively with RagA than with RagC. This is exemplified by its direct interaction with the clearly visible GDP and XTPγS nucleotides, respectively (150 vs. 20 Å2 buried surface area) (Figs. 4A, 5A, S7C–D and Fig. S12). A FLCN:FNIP2 complex containing a FLCNF118D mutation located at the FLCN-RagA interface (Fig. 4A, Fig. S3D and Fig. S13A), did not assemble into a stable LFC as judged by SEC (Fig. 4B). Consistent with a disrupted LFC, GFP-FLCNF118D failed to localize to the lysosomal membrane upon amino acid-starvation, establishing the LFC as the structural basis for FLCN localization under low nutrient conditions (Fig. 4, C and D). Also consistent with its inability to form the LFC, FLCNF118D:FNIP2 had uninhibited GAP activity even in the presence of Ragulator, as judged by both tryptophan fluorescence- and HPLC-based GAP assays (Fig. 4E and Fig. S13B).
Fig. 4. The LFC stabilizes GDP-bound RagA and is necessary for lysosomal FLCN localization during amino acid starvation.
(A) Nucleotide binding pocket and interaction with FLCN:FNIP2 (orange, green) of the RagA G domain (cyan) in the LFC. GDP is highlighted by the according cryo-EM density and stick representation. FLCN:FNIP2 residues in the interface are labeled and side chains are shown as sticks. Disordered switch 1 and switch 2 of RagA absent from the coordinate model are indicated by dashed lines. (B) SEC elution profile of Ragulator-RagAGDP:RagCXppNHp-FLCN:FNIP2 containing FLCNWT (black) or FLCNF118D (cyan). Peak shift indicates the FLCNF118D-containing complex failed to assemble into a stable LFC. (C) Fluorescence images of HEK293A cells expressing the indicated FLCN:GFP construct along with HA:FNIP2 and the lysosomal marker Lamp2:RFP. Cells were starved for one hour for amino acids (-AA) or starved for 50 min then restimulated with amino acids for 10 min (+AA). Scale bar 10 μm. (D) Quantitation of FLCN Lysosomal Enrichment Score for immunofluorescence images in (C) (mean +/− SD, N=10 cells for all conditions). (E) Tryptophan fluorescence XTPase assay with FLCNF118D:FNIP2. RagAGDP:RagCXTP was incubated with FLCNWT:FNIP2 (black, grey) or FLCNF118D:FNIP2 (blue, cyan) in the presence (black, blue) or absence (grey, cyan) of Ragulator as indicated. Plotted are mean +/− SEM. N=3 (F) Mant fluorescence nucleotide exchange assay: intrinsic mant fluorescence intensity decreases by ~50% when released from the GTPase. Mant RagA nucleotide exchange in response to addition of unlabeled GTP visualized by mant fluorescence for RagAmantGDP:RagCXppNHp alone (black), in complex with Ragulator (grey), in complex with FLCNWT:FNIP2 (dark green), Ragulator and FLCNWT:FNIP2 (light green), FLCNF118D:FNIP2 (cyan), or Ragulator and FLCN118D:FNIP2 (blue). Plotted are mean +/− SEM. N=3. (G) HPLC-based nucleotide exchange assay for RagA. GDP replacement by GppCp (non-hydrolyzable GTP analog) for RagAGDP:RagCXTPγS alone (black), in complex with Ragulator (grey), or in the full LFC (light green) is monitored by the GppCp peak in the HPLC elution profile. “U.” indicates that GppCp signal was undetectable. Plotted are individual data points and mean +/− SD. N=3.
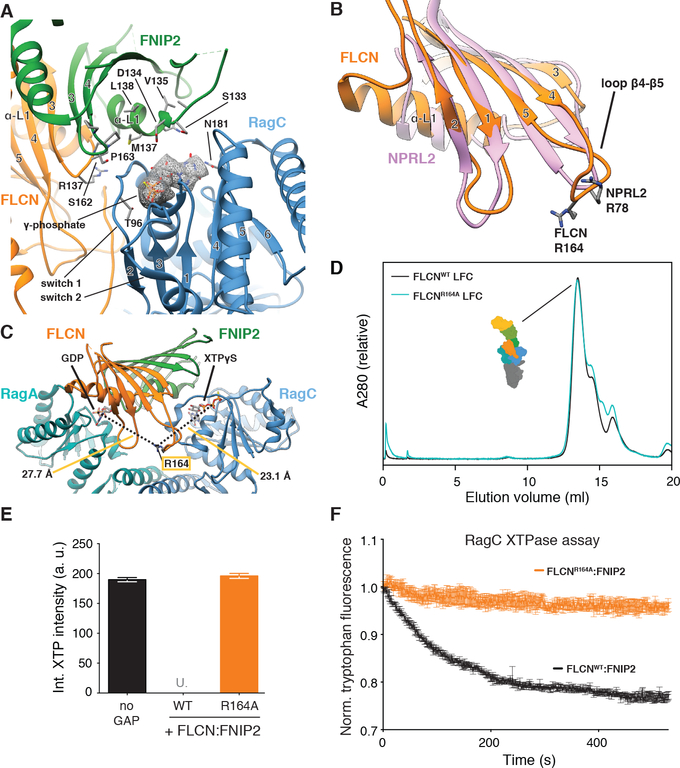
Fig. 5. FLCN Arg164 mediates GAP activity and is in a GAP-incompetent position in the LFC.
(A) Nucleotide-binding pockets and interaction with FLCN:FNIP2 (orange, green) of the RagC G domain (blue) in the LFC. XTPγS is highlighted by the according cryo-EM density and stick representation. FLCN:FNIP2 residues in the interface are labeled and side chains are shown as sticks. The γ-phosphate of XTPγS locking the switch 1 and switch 2 region of RagC is indicated. (B) Overlay of the FLCN (orange) and NPRL2 (rose) longin domains. α-helices α3 and α4 are omitted for clarity. NPRL2 arginine finger (R78) and FLCN R164 in loop β4-β5 are represented as sticks. (C) Position of FLCN R164 in the LFC. Distances to the β-phosphate of GDP (RagA) and XTPγS (RagC) are indicated. (D) SEC elution profile of the LFC (Ragulator-RagAGDP:RagCXTPγS-FLCN:FNIP2) containing FLCNWT (black) or mutant FLCNR164A (cyan). (E) HPLC-based GTPase assay assessing GAP activity of FLCNWT:FNIP2 or FLCNR164A:FNIP2 (orange) on a RagAGTP:RagCXTP substrate by integrating the remaining XTP nucleotide signal. Plotted are mean +/− SD. N=3. “U.” indicates that remaining XTP signal was undetectable by HPLC. (F) Tryptophan fluorescence XTPase assay with FLCN:FNIP2 mutants. RagAGTP:RagCXTP is incubated with FLCNWT:FNIP2 (black) or FLCNR164:FNIP2 (orange). Plotted are mean +/− SEM. N=3.
Compared to other GTPases bound to GDP, e.g. the ER-associated small GTPase Sar1 (31), the RagAGDP switch regions are largely disordered in the LFC and do not contribute stabilizing interactions to the bound GDP (Fig. 4A and Fig. S6C). This prompted us to investigate whether FLCN:FNIP2 binding to the RagA G domain influenced its nucleotide state within the LFC. We monitored RagA nucleotide exchange using both HPLC and a fluorescence-based assay in which the RagA:RagC dimer was first bound to (2’/3’)-O-(N-Methylanthraniloyl) (mant)GTP and non-hydrolyzable XppNHp, then incubated with GATOR1 to generate RagAmantGDP:RagCXppNHp substrate. Nucleotide exchange was monitored as mantGDP fluorescence decrease upon release from RagA in the presence of excess unlabeled GTP. Rapid and complete replacement of mantGDP with GTP occurred in the absence of other proteins and was confirmed by HPLC (Fig. 4, F and G). In contrast, Sar1 showed negligible mantGDP release under identical conditions (Fig. S13C) (32). The addition of either Ragulator or FLCN:FNIP2 slowed the kinetics of mantGDP release (Fig 4F). Strikingly, when the entire LFC was assembled, RagA nucleotide exchange was barely detectable (Fig. 4F and G). Consistent with its inability to form the LFC, mutant FLCNF118D:FNIP2 did not inhibit mantGDP exchange on RagA either by itself or in the presence of Ragulator (Fig. 4F). These data show that RagA in the context of the isolated Rag dimer, unlike most other small GTPases, is competent to spontaneously exchange GDP for GTP, and that the exchange process is blocked in the LFC.
The switch 1 and switch 2 regions of RagC are well defined (Fig. 5A). They adopt a conformation analogous to GTP-bound RagA, Gtr1 and other small GTPases (14, 28, 30, 33). In this conformation, RagC Thr96 and Gly119 stabilize the γ-phosphate of the bound XTPγS nucleotide (Fig. 5A). In line with the finding that FLCN:FNIP2 does not exert GAP activity in the LFC (Fig. 1B), the interface between FLCN:FNIP2 and RagCXTPγS lacks any Arg residue poised to function as the catalytic ‘arginine finger’ characteristic for most GAPs (34) (Fig. 5A). The catalytic arginine of GATOR1 is Arg78 located in loop β4-β5 of the NPRL2 longin domain (35). The longin domains of FLCN and FNIP2 each contain an arginine residue in loop β4-β5 as well, but only Arg164 of FLCN is highly conserved from yeast to human (Fig. 5B and Figs. S9 to S10). In the LFC, FLCN Arg164 is located more than 20 Å away from either nucleotide between the two G domains, with no intersubunit contacts (Fig. 5C). We found that mutant FLCNR164A assembles into an LFC that migrates normally on SEC (Fig. 5D and Fig. S3E), yet GAP activity is abolished (Fig. 5, E and F). Moreover, the RagC-GAP-incompetent FLCNR164A:FNIP2 complex is capable of blocking RagA nucleotide exchange (Fig. S13D), and localizes to the lysosomal membrane upon amino acid-starvation (Fig. S13E). These data establish FLCNR164 as a catalytically required arginine for canonical FLCN RagC-GAP activity and explain structurally why FLCN does not function as an active GAP in the LFC.
Regulation of MiT/TFE factors by FLCN and LFC
FLCN has emerged as a central controller of the master regulators of lysosomal biogenesis and autophagy, the MiT/TFE family transcription factors TFE3 and TFEB (19, 21, 36–39). TFE3 and TFEB are phosphorylated by mTORC1 to prevent their nuclear translocation and to promote retention in the cytoplasm in nutrient replete conditions. Upon nutrient starvation and mTORC1 inactivation, the MiT/TFE factors relocalize to the nucleus and activate their gene expression programs (40–43). In agreement with a key role for FLCN in MiT/TFE regulation, its knockdown using both shRNA and dCas9-mediated silencing led to nuclear localization of TFE3 under full nutrients (Fig. S14, A–D). In contrast, FLCN depletion was largely dispensable for phosphorylation of canonical mTORC1 substrates, S6K1 and 4E-BP1 (Fig. S14, E and F) implicating the LFC as specific mechanisms for mTORC1-dependent control of the MiT/TFE factors.
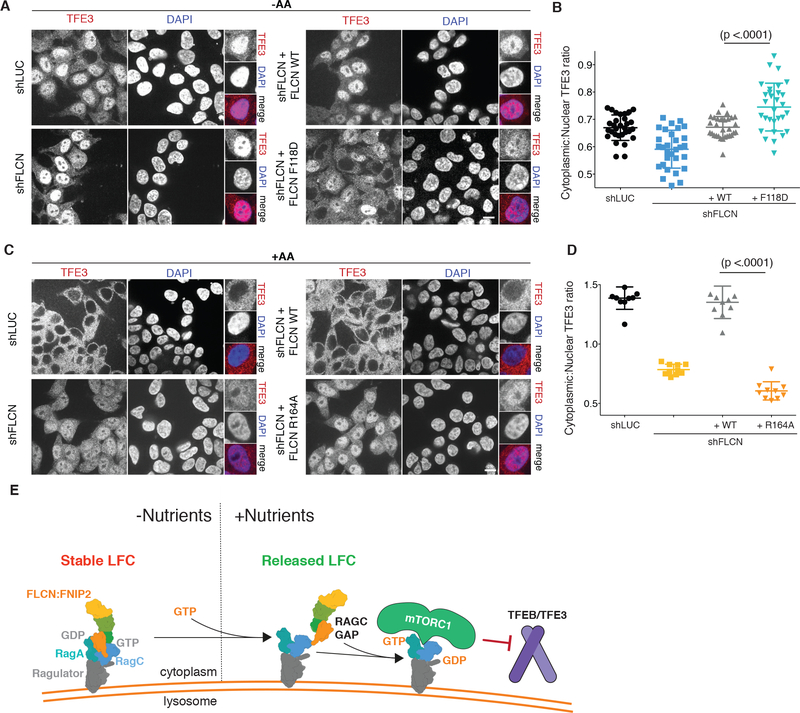
In amino acid-deprived cells in which FLCN was depleted, or depleted and reconstituted with wild-type FLCN, endogenous TFE3 was largely nuclear-localized. Relative to wild-type, reconstituting FLCN-depleted cells with LFC-defective FLCNF118D, which has uninhibited RagC-GAP activity, caused TFE3 to become less nuclear (Fig. 6, A and B). In FLCN-depleted cells that were restimulated with amino acids, TFE3 failed to relocalize from the nucleus to the cytoplasm (Fig. 6, C and D). Cytoplasmic distribution of TFE3 was fully rescued by re-expressing wild-type FLCN. In contrast, expressing GAP-defective FLCNR164A failed to restore cytoplasmic translocation of TFE3, which therefore remained nuclear despite high nutrient concentrations (Fig. 6, C and D).
Fig 6. FLCN GAP activity is necessary for nutrient-induced cytoplasmic TFE3 localization.
(A) Immunofluorescence images of HEK293T cells expressing the indicated shRNA and FLCN rescue constructs. Cells were starved for one hour for amino acids (-AA). Scale bar 10 μm. (B) Quantitation of Cytoplasmic:Nuclear TFE3 Ratio for immunofluorescence images in (A) (mean +/− SD, N=30 cells for all). (C) Immunofluorescence images of HEK293T cells expressing the indicated shRNA and FLCN rescue constructs. Cells were starved for one hour and restimulated with amino acids for two hours (+AA). Scale bar 10 μm. (D) Quantitation of Cytoplasmic:Nuclear TFE3 Ratio for immunofluorescence images in (C) (mean +/− SD, N=10 cells for all). (E) Model for regulation of Rag GTPases activation by the LFC. In low nutrient conditions, binding of FLCN:FNIP2 to a stable RagAGDPR:RagCGTP-Ragulator platform leads to LFC formation. Upon nutrient restimulation, RagA nucleotide exchange and LFC disassembly liberate the RagC-GAP activity of FLCN:FNIP2. The resulting active Rag GTPase complex is competent to activate mTORC1 and promote TFEB/TFE3 cytoplasmic retention.
Together, these data strongly suggest that the RagC-GAP activity of FLCN enables proper regulation of MiT/TFE nuclear translocation by nutrient inputs (Fig. 6E). Under low nutrients, the LFC suppresses the RagC-GAP activity of FLCN, maximizing nuclear translocation of the MiT/TFE factors. Conversely, in high nutrients, disassembly of the LFC liberates the RagC-GAP function of FLCN, which is key for mTORC1-dependent phosphorylation and cytoplasmic retention of the MiT/TFE factors.
Discussion
Our structure of the LFC reveals for the first time how FLCN and FNIP2 assemble into one complex via an intricate domain arrangement, and how they interact with the G domains of the Rag GTPases in their ‘inactive’ state. It serves as the structural basis for FLCN:FNIP2 localization at the lysosomal membrane under low amino acid concentrations, and explains our findings from biochemical reconstitution and in vitro GAP and nucleotide exchange assays. First, we identified Arg164 of FLCN as a catalytically required arginine residue for GAP activity. Second, we find that Arg164 is located far from the RagC nucleotide pocket within the LFC, and thus the LFC is incompatible with FLCN GAP activity. Third, LFC formation strongly depends on RagA in its GDP-bound conformation, explaining the requirement of GATOR1 for lysosomal FLCN localization (22). Fourth, FLCN:FNIP2 interacts with RagAGDP and, together with Ragulator, blocks exchange of GDP with GTP in RagA, possibly stabilizing the LFC. Fifth, by controlling the RagC-GAP activity of FLCN, the LFC regulates a subset of mTORC1 substrates, the MiT/TFE transcription factors.
Mutations in the FLCN tumor suppressor gene are responsible for BHD, which predisposes patients to kidney tumors (44). However, several missense mutations map to the surface of the DENN domain and, based on our structure seem unlikely to disrupt Rag GTPase interaction or LFC stability (Fig. S15). Interestingly, the MiT/TFE factors, TFEB and TFE3, are independently translocated and upregulated in renal cell carcinoma (44–46), providing a plausible mechanism for carcinogenesis downstream of FLCN loss.
The ability of FLCN:FNIP2 to contact its client RagC in two states, the inactive, stable LFC and an active, GAP-compatible but transient interaction, is rare among known GAPs. The other notable exception is GATOR1. The cryo-EM structure of GATOR1 in complex with a Rag dimer revealed a stable, GAP-inhibited state in which the non-catalytic subunit of GATOR1 makes direct contact with the G domain of RagA (14). FLCN:FNIP2 is the mirror image of the GATOR1 complex in the sense that both contain a dual longin domain-based GAP and both complexes have positive and negative regulatory functions. The multiple configurations of this complex with respect to the Rag GTPases are substantially more elaborate than the single reaction carried out by conventional GAP complexes, and suggest that the ability of cells to respond to changing nutrient concentrations may be so important that uniquely stringent mechanisms are called for.
Supplementary Material
Acknowledgments:
We thank H.R. Shin and M.-Y. Su for comments on the manuscript. We thank D. Toso for microscope operational support. Access to the FEI Titan Krios was provided through the BACEM UCB facility. We thank the J. Thorner lab for generous use of their spectrofluorimeter. We thank L. Yuan and D. Melville from the R. Schekman lab for assistance with tryptophan fluorescence experiments and the gift of purified Sar1 GTPase. We thank M. Kampmann for the gift of the 239T CRISPRi cells.
Funding: This work was supported by NIH 1DP2CA195761–01 (R.Z.), GM111730 (J.H.H.), CA223029 (L.N.Y.); the Pew-Stewart Scholarship for Cancer Research and Damon Runyon-Rachleff Innovation Award (R.Z.); a University of California Cancer Research Coordinating Committee Predoctoral Fellowship (R.E.L.), a EMBO Long-Term Fellowship (S.A.F), and a Jane Coffin Childs fellowship (A.L.Y.).
Footnotes
Competing interests: R.Z. is co-founder and stockholder in Frontier Medicines Corp. J.H.H. is a scientific founder of Casma Therapeutics.
Data and materials availability:
EM density maps have been deposited in the EMDB with accession numbers EMD-0554 (LFC) and EMD-0556 (active Rag-Ragulator). Atomic coordinates for the LFC have been deposited in the PDB with accession number 6NZD.
Supplementary Materials:
Materials and Methods
Figures S1-S15
Tables S1-S2
Movie S1
References and Notes:
- 1.Saxton RA, Sabatini DM, mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence RE, Zoncu R, The lysosome as a cellular centre for signalling, metabolism and quality control. Nature cell biology 21, 133–142 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM, The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM, Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM, Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, Zoncu R, Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL, Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology 10, 935–945 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM, Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Teis D, Wunderlich W, Huber LA, Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Developmental cell 3, 803–814 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Rogala KB, Gu X, Kedir JF, Abu-Remaileh M, Bianchi LF, Bottino AMS, Dueholm R, Niehaus A, Overwijn D, Fils AP, Zhou SX, Leary D, Laqtom NN, Brignole EJ, Sabatini DM, Structural basis for the docking of mTORC1 on the lysosomal surface. Science, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anandapadamanaban M, Masson GR, Perisic O, Berndt A, Kaufman J, Johnson CM, Santhanam B, Rogala KB, Sabatini DM, Williams RL, Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 366, 203–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM, A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panchaud N, Peli-Gulli MP, De Virgilio C, Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science signaling 6, ra42 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Shen K, Huang RK, Brignole EJ, Condon KJ, Valenstein ML, Chantranupong L, Bomaliyamu A, Choe A, Hong C, Yu Z, Sabatini DM, Architecture of the human GATOR1 and GATOR1-Rag GTPases complexes. Nature 556, 64–69 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, Sharma N, Walther M, Munroe D, Hill R, Maher E, Greenberg C, Lerman MI, Linehan WM, Zbar B, Schmidt LS, Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer cell 2, 157–164 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF 3rd, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR Jr., Linehan WM, Schmidt LS, Zbar B, Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A 103, 15552–15557 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasumi H, Baba M, Hong SB, Hasumi Y, Huang Y, Yao M, Valera VA, Linehan WM, Schmidt LS, Identification and characterization of a novel folliculin-interacting protein FNIP2. Gene 415, 60–67 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM, The Folliculin Tumor Suppressor Is a GAP for the RagC/D GTPases That Signal Amino Acid Levels to mTORC1. Molecular cell, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petit CS, Roczniak-Ferguson A, Ferguson SM, Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol 202, 1107–1122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasumi Y, Baba M, Ajima R, Hasumi H, Valera VA, Klein ME, Haines DC, Merino MJ, Hong SB, Yamaguchi TP, Schmidt LS, Linehan WM, Homozygous loss of BHD causes early embryonic lethality and kidney tumor development with activation of mTORC1 and mTORC2. Proc Natl Acad Sci U S A 106, 18722–18727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada S, Neinast M, Jang C, Ibrahim YH, Lee G, Babu A, Li J, Hoshino A, Rowe GC, Rhee J, Martina JA, Puertollano R, Blenis J, Morley M, Baur JA, Seale P, Arany Z, The tumor suppressor FLCN mediates an alternate mTOR pathway to regulate browning of adipose tissue. Genes Dev 30, 2551–2564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng J, Ferguson SM, GATOR1-dependent recruitment of FLCN-FNIP to lysosomes coordinates Rag GTPase heterodimer nucleotide status in response to amino acids. J Cell Biol, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmadian MR, Zor T, Vogt D, Kabsch W, Selinger Z, Wittinghofer A, Scheffzek K, Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc Natl Acad Sci U S A 96, 7065–7070 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence RE, Cho KF, Rappold R, Thrun A, Tofaute M, Kim DJ, Moldavski O, Hurley JH, Zoncu R, A nutrient-induced affinity switch controls mTORC1 activation by its Rag GTPase-Ragulator lysosomal scaffold. Nature cell biology 20, 1052–1063 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D, Iyer LM, He F, Aravind L, Discovery of Novel DENN Proteins: Implications for the Evolution of Eukaryotic Intracellular Membrane Structures and Human Disease. Frontiers in genetics 3, 283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su MY, Morris KL, Kim DJ, Fu Y, Lawrence R, Stjepanovic G, Zoncu R, Hurley JH, Hybrid Structure of the RagA/C-Ragulator mTORC1 Activation Complex. Molecular cell 68, 835–846 e833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montano B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL, Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol 2, 120071 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong R, Li L, Liu Y, Wang P, Yang H, Wang L, Cheng J, Guan KL, Xu Y, Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev 25, 1668–1673 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Araujo MEG, Naschberger A, Furnrohr BG, Stasyk T, Dunzendorfer-Matt T, Lechner S, Welti S, Kremser L, Shivalingaiah G, Offterdinger M, Lindner HH, Huber LA, Scheffzek K, Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science 358, 377–381 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Jeong JH, Lee KH, Kim YM, Kim DH, Oh BH, Kim YG, Crystal Structure of the Gtr1p(GTP)-Gtr2p(GDP) Protein Complex Reveals Large Structural Rearrangements Triggered by GTP-to-GDP Conversion. Journal of Biological Chemistry 287, 29648–29653 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE, Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export. J Cell Biol 155, 937–948 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barlowe C, Schekman R, SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349 (1993). [DOI] [PubMed] [Google Scholar]
- 33.Vetter IR, Wittinghofer A, The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A, Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nature structural biology 4, 686–689 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Shen K, Valenstein ML, Gu X, Sabatini DM, Arg78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J Biol Chem, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villegas F, Lehalle D, Mayer D, Rittirsch M, Stadler MB, Zinner M, Olivieri D, Vabres P, Duplomb-Jego L, De Bont E, Duffourd Y, Duijkers F, Avila M, Genevieve D, Houcinat N, Jouan T, Kuentz P, Lichtenbelt KD, Thauvin-Robinet C, St-Onge J, Thevenon J, van Gassen KLI, van Haelst M, van Koningsbruggen S, Hess D, Smallwood SA, Riviere JB, Faivre L, Betschinger J, Lysosomal Signaling Licenses Embryonic Stem Cell Differentiation via Inactivation of Tfe3. Cell stem cell 24, 257–270 e258 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Kennedy JC, Khabibullin D, Hougard T, Nijmeh J, Shi W, Henske EP, Loss of FLCN inhibits canonical WNT signaling via TFE3. Human molecular genetics, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong SB, Oh H, Valera VA, Baba M, Schmidt LS, Linehan WM, Inactivation of the FLCN tumor suppressor gene induces TFE3 transcriptional activity by increasing its nuclear localization. PLoS One 5, e15793 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A, Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153, 335–347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A, A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. The EMBO journal 31, 1095–1108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martina JA, Chen Y, Gucek M, Puertollano R, MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM, The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Science signaling 5, ra42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napolitano G, Esposito A, Choi H, Matarese M, Benedetti V, Di Malta C, Monfregola J, Medina DL, Lippincott-Schwartz J, Ballabio A, mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun 9, 3312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt LS, Linehan WM, FLCN: The causative gene for Birt-Hogg-Dube syndrome. Gene 640, 28–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuiper RP, Schepens M, Thijssen J, van Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF, van Kessel AG, Upregulation of the transcription factor TFEB in t(6;11)(p21;q13)-positive renal cell carcinomas due to promoter substitution. Human molecular genetics 12, 1661–1669 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Davis IJ, Hsi BL, Arroyo JD, Vargas SO, Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi M, Fletcher JA, Fisher DE, Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A 100, 6051–6056 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH, New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Zhang K, Gctf: Real-time CTF determination and correction. Journal of structural biology 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD, Real-space refinement in PHENIX for cryo-EM and crystallography. Acta crystallographica. Section D, Structural biology 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jakobi AJ, Wilmanns M, Sachse C, Model-based local density sharpening of cryo-EM maps. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burnley T, Palmer CM, Winn M, Recent developments in the CCP-EM software suite. Acta crystallographica. Section D, Structural biology 73, 469–477 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallographica Section D-Biological Crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS, Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Notredame C, Higgins DG, Heringa J, T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Webb B, Sali A, Protein structure modeling with MODELLER. Protein Structure Prediction, 1–15 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V, A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. Journal of molecular biology 430, 2237–2243 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Wang Z, Schroder GF, Real-space refinement with DireX: from global fitting to side-chain improvements. Biopolymers 97, 687–697 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS, CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS, Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL, Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol 5, 150174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu X, Bradley MJ, Cai Y, Kummel D, De La Cruz EM, Barr FA, Reinisch KM, Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci U S A 108, 18672–18677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen K, Sabatini DM, Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc Natl Acad Sci U S A 115, 9545–9550 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen K, Choe A, Sabatini DM, Intersubunit Crosstalk in the Rag GTPase Heterodimer Enables mTORC1 to Respond Rapidly to Amino Acid Availability. Molecular cell 68, 552–565 e558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.