Light-activated phytochrome influences alternative splicing by promoting the association of a heterogeneous nuclear ribonucleoprotein with a regulatory cis element on pre-mRNA.
Abstract
Plants perceive environmental light conditions and optimize their growth and development accordingly by regulating gene activity at multiple levels. Photoreceptors are important for light sensing and downstream gene regulation. Phytochromes, red/far-red light receptors, are believed to regulate light-responsive alternative splicing, but little is known about the underlying mechanism. Alternative splicing is primarily regulated by transacting factors, such as splicing regulators, and by cis-acting elements in precursor mRNA. In the moss Physcomitrella patens, we show that phytochrome 4 (PpPHY4) directly interacts with a splicing regulator, heterogeneous nuclear ribonucleoprotein F1 (PphnRNP-F1), in the nucleus to regulate light-responsive alternative splicing. RNA sequencing analysis revealed that PpPHY4 and PphnRNP-F1 coregulate 70% of intron retention (IR) events in response to red light. A repetitive GAA motif was identified to be an exonic splicing silencer that controls red light-responsive IR. Biochemical studies indicated that PphnRNP-F1 is recruited by the GAA motif to form RNA-protein complexes. Finally, red light elevates PphnRNP-F1 protein levels via PpPHY4, increasing levels of IR. We propose that PpPHY4 and PphnRNP-F1 regulate alternative splicing through an exonic splicing silencer to control splicing machinery activity in response to light.
Light is the major source of energy for plant growth and influences developmental processes throughout the life cycle of plants. Light-regulated plant development, or photomorphogenesis, is modulated by sophisticated photoreceptor systems. The photosensory protein phytochrome perceives red (RL) and far-red (FR) light and triggers photomorphogenic responses by regulating multiple steps of gene expression including chromatin modification, transcription, translation, and posttranslational modification (Tu and Lagarias, 2005; Guo et al., 2008; Huq and Quail, 2010; Li et al., 2012; Liu et al., 2012; Wu, 2014). Although phytochrome-mediated regulation is found in almost every stage of gene expression, information about whether phytochrome regulates pre-mRNA splicing is still limited.
Alternative splicing (AS) is a widespread mechanism in eukaryotes in which multiple mRNAs are generated from the same gene by the use of variable splice sites during pre-mRNA splicing. AS largely increases complexity to the transcriptome and therefore plays a key regulatory role during development and in response to environmental changes. In terms of light responses, various studies have suggested that AS potentially modulates photomorphogenesis in plants (Zhou et al., 1998; Mano et al., 1999, 2000; Penfield et al., 2010; Yoshimura et al., 2011; Shikata et al., 2012). For example, overexpressing the alternatively spliced isoforms of light signaling factors CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and PHYTOCHROME INTERACTING FACTOR6 (PIF6) in Arabidopsis (Arabidopsis thaliana) have dominant-negative effects in photomorphogenesis (Zhou et al., 1998; Penfield et al., 2010). Light also promotes AS of SPA1-RELATED3 (SPA3) transcripts to produce truncated proteins that have a dominant-negative effect to the formation of endogenous COP1-SPA3 complex in ubiquitin-dependent protein degradation (Shikata et al., 2014).
During pre-mRNA splicing, spliceosome assembly is guided by intron-defining splicing signals including the 5′ splice site (5′ SS), 3′ SS, polypyrimidine tract, and branching point sequence, which are recognized by spliceosomal components. In addition to core spliceosomal proteins, splicing regulators such as serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) play important roles in SS selection and are usually targeted to the splicing regulatory cis elements (SREs) in pre-mRNA sequences (Wang and Burge, 2008). Numerous SREs have been defined as exonic/intronic splicing enhancers or exonic splicing silencers (ESSs)/intronic splicing silencers based on their location and function in pre-mRNA splicing (Wang et al., 2004). In general, enhancers and silencers recruit SR proteins and hnRNPs, respectively, to promote or suppress the assembly of the spliceosome (Wang et al., 2004). Changes of binding affinities to regulatory cis elements, differential expression, and posttranslational modification of splicing regulators together diversify AS patterns and tremendously increase transcriptome complexity and proteome diversity.
In comparison to trans-acting factors, the function of regulatory cis elements is less addressed in plants. We previously identified an exonic GAA repetitive sequence potentially involved in light-mediated splicing regulation in Physcomitrella patens based on the analysis of RL-responsive intron retention (IR) events (Wu et al., 2014). In human and other vertebrate species, the GAA motif was first identified as an exonic splicing enhancer that recruits AS FACTOR (ASF) and SPLICINGFACTOR2 (SF2; Yeakley et al., 1993; Tanaka et al., 1994; Staffa and Cochrane, 1995; Tacke and Manley, 1995). In plants, a GAA-repetitive motif auto-regulates the intron splicing of SC35-LIKE SPLICING FACTOR33 (SCL33) by recruiting the SR protein SCL33 (Thomas et al., 2012). High-throughput sequencing of RNA immunoprecipitation also revealed the GAA motif as a potential cis element bound by Arabidopsis SER/ARG-RICH45 (SR45; Xing et al., 2015). However, the functional roles of the GAA motif in regulating RL-responsive AS are still unclear.
Large-scale studies have revealed that AS is responsive to light (Shikata et al., 2014; Wu et al., 2014; Hartmann et al., 2016; Mancini et al., 2016). Light-responsive AS has been shown to be mediated by phytochromes in parallel with transcription in Arabidopsis (Shikata et al., 2014). In the bryophyte P. patens, mRNA sequencing (RNA-seq) studies have also revealed a global change in AS in response to RL and a role for phytochromes in regulating this process (Wu et al., 2014). In Arabidopsis, phyB signal transduction leading to AS in response to RL involves the SR-like protein REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND1 (RRC1; Shikata et al., 2012). An Arabidopsis homolog of human Splicing factor 45 protein named SPLICING FACTOR FOR PHYTOCHROME SIGNALING (SFPS) was recently found to be a direct interacting partner of phyB that modulates the pre-mRNA splicing of light signaling genes (Xin et al., 2017). Overall, there is increasing evidence for phytochrome-mediated AS (Cheng and Tu, 2018), but the detailed mechanism is not well understood.
In this study, we showed the Physcomitrella splicing regulator PphnRNP-F1 interacts with phytochrome4 (PpPHY4) in the nucleus under RL. Genome-wide analysis indicated that both PpPHY4 and PphnRNP-F1 are required for the modulation of RL-responsive IR events. The GAA motif was enriched among these PpPHY4- and PphnRNP-F1-mediated RL-responsive IR events. We experimentally validated that this GAA motif functions as an ESS for the RL-responsive IR pattern and recruits protein complexes containing PphnRNP-F1. Together, our results suggest that many IR events that occur in response to RL in P. patens are mediated through an interaction between PpPHY4 and PphnRNP-F1 and that this protein binds to the GAA motif on the pre-mRNA of RL-regulated genes.
RESULTS
PpPHY4 Interacts with PphnRNP-F1 In Vitro and In Vivo
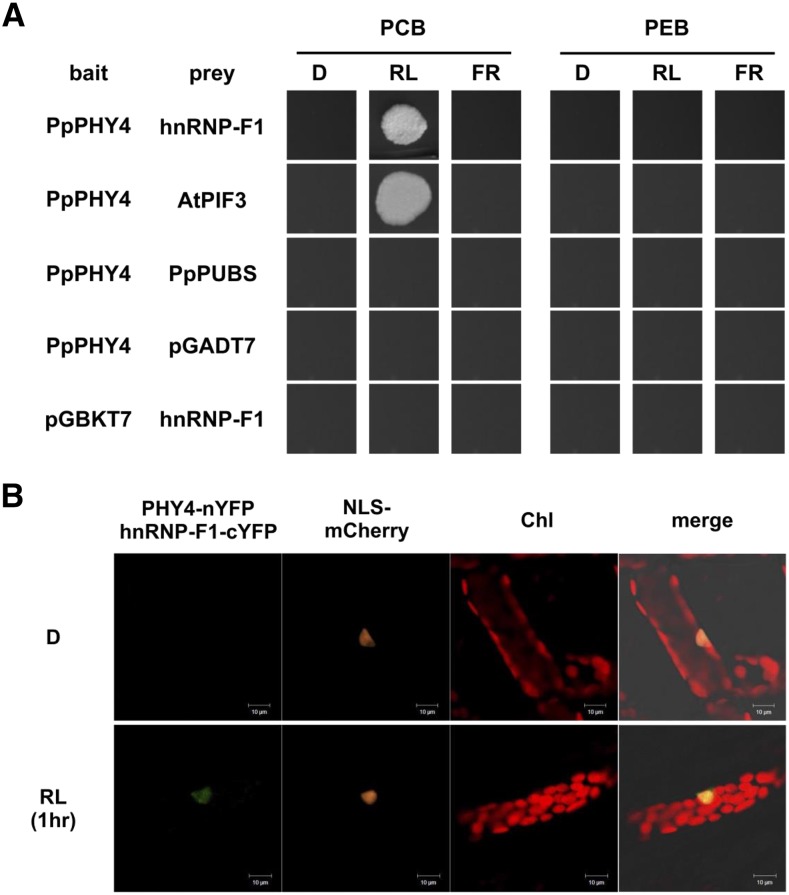
We previously showed that AS is modulated upon RL irradiation in P. patens, which depends on the presence of the RL photoreceptors, phytochromes (Wu et al., 2014). Phytochromes transduce light signals via a direct or indirect physical interaction with downstream regulatory factors. Since auxiliary factors such as SR proteins and hnRNPs function in spliceosome assembly, we tried to identify splicing regulators that interact with phytochromes. By performing yeast two-hybrid (Y2H) targeted screening, we identified a hnRNP-F-type protein named PphnRNP-F1 that interacts with PpPHY4 (Supplemental Fig. S1). In the Y2H assay, yeast cells expressing both PpPHY4 and PphnRNP-F1 were fed with phycocyanobilin (PCB) to generate photo-convertible phytochromes and were incubated under different light conditions. PpPHY4, the ortholog of Arabidopsis phyB, interacted with PphnRNP-F1 only in RL and not in FR or darkness (Fig. 1). This finding points to the importance of RL-activated PpPHY4 for this interaction. To further confirm the RL dependency of the interaction, we performed the same Y2H experiment but used phycoerythrobilin (PEB) instead of PCB as the chromophore to lock phytochrome in the Pr form (Li and Lagarias, 1992). Due to their assembly with PEB, the RL-dependent interaction between PpPHY4 and PphnRNP-F1 no longer occurred (Fig. 1A). These results indicate that only photoactivated PpPHY4 interacts with PphnRNP-F1, with AtPIF3 (Ni et al., 1998) and the chloroplast protein PpPUBS (Chen et al., 2012) serving as the positive and negative control, respectively.
Figure 1.
PpPHY4 interacts with PphnRNP-F1 in vitro and in vivo. A, Yeast cells expressing PpPHY4-pGBKT7 and PphnRNP-F1-pGADT7 were incubated in the dark (D), red light (RL), or far-red light (FR) on dropout medium containing phycocyanobilin (PCB) or phycoerythrobilin (PEB) at 30°C for 3 d. The interaction between PpPHY4 and the Arabidopsis protein AtPIF3 was used as a positive control. The plastid-localized protein PpPUBS was used as a negative control. B, In a BiFC assay, PpPHY4 and PphnRNP-F1 were fused with the N- and C-terminal half of YFP, respectively. Gametophore cells were cobombarded with PpPHY4-nYFP, PphnRNP-F1-cYFP, and NLS-mCherry plasmids and incubated in the dark or RL for 1 h. NLS-mCherry, nucleus marker. The result is the representative one of three biological repeats.
In P. patens, PphnRNP-F1 localized to both the cytoplasm and nucleus regardless of light treatment (Supplemental Fig. S2). PpPHY4 localizes to the cytosol in the dark and translocates to the nucleus once it receives RL, like phytochromes in other plant systems (Sakamoto and Nagatani, 1996; Possart and Hiltbrunner, 2013). Since PphnRNP-F1 and PpPHY4 both localize to the nucleus under RL irradiation, we investigated the interaction between PpPHY4 and PphnRNP-F1 in vivo by performing a bimolecular fluorescence complementation (BiFC) assay in moss cells. After coexpressing PpPHY4 tagged with the N-terminal portion of yellow fluorescent protein (nYFP) and PphnRNP-F1 tagged with the C-terminal portion of YFP (cYFP), we incubated the cells in the dark or RL for 1 h and observed the fluorescence from YFP. Consistent with the results of the Y2H assay, PpPHY4 and PphnRNP-F1 interacted only in the presence of RL in moss nuclei (Fig. 1B). We performed the same BiFC assay using orchid petal cells to monitor RL-dependent interactions within a single cell (Lee et al., 2012). YFP fluorescence was not detected in dark-treated cells, but after RL treatment, their interaction was observed in the nucleus (Supplemental Fig. S3). Taken together, these results suggest that RL is necessary for promoting the interaction between PpPHY4 and PphnRNP-F1.
PpPHY4 and PphnRNP-F1 Control RL-Responsive IR Cooperatively
Increasing evidence indicates that phytochromes are responsible for RL-dependent AS in plants (Shikata et al., 2014; Wu et al., 2014; Xin et al., 2017). Our finding of the RL-dependent interaction of PpPHY4 and PphnRNP-F1 further strengthens the hypothesis that phytochromes and splicing regulators cooperate in the regulation of RL-responsive AS. To dissect the roles of PpPHY4 and PphnRNP-F1 in splicing regulation, we subjected the PpPHY4-deficient mutant phy4 (Wu et al., 2014) and the PphnRNP-F1-deficient mutant hnrnp-f1 (Supplemental Fig. S4) to RNA-seq analysis using dark-adapted protonemal cells as a control (D) and tested the responsiveness of AS to 1 h of RL exposure (R1).
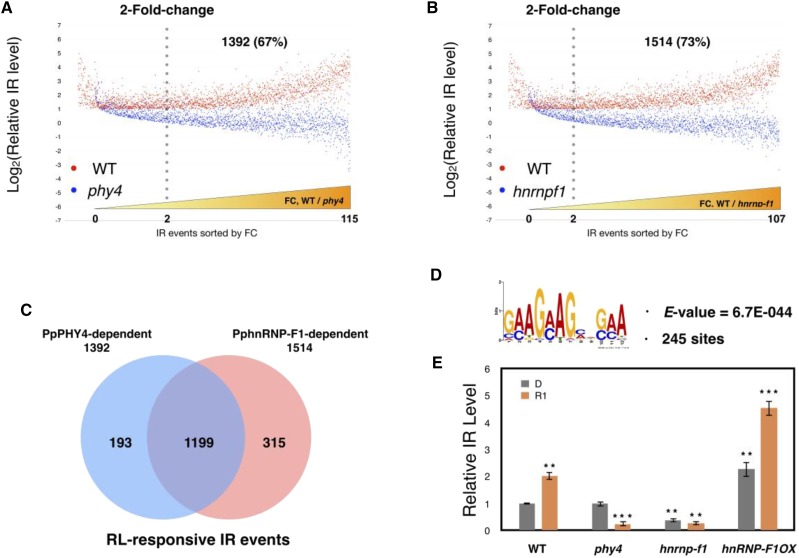
We analyzed the RL-responsive IR events by monitoring relative IR levels as described in our previous study (Wu et al., 2014). First, we measured the levels of the retained intron and total transcripts of the corresponding gene by calculating intron reads per kilobase of retained intron per million mapped reads (IPKM) and reads per kilobase of exon model per million mapped reads (RPKM), respectively. To quantify the level of IR, we normalized the IPKM value with the RPKM value of the corresponding gene. We then calculated the relative IR level by comparing the normalized IPKM values in samples D and R1. In wild-type samples, 2056 RL-induced IR events (relative IR level ≥ 2) were identified as RL-responsive IR events. We compared the relative IR levels of these RL-induced IR events in wild type with those in phy4 or hnrnp-f1, finding that 1392 (67%) and 1514 (73%) events showed less RL responsiveness (fold change, wild type/mutant ≥ 2) in phy4 (Fig. 2A) and hnrnp-f1 (Fig. 2B), respectively, than in wild type (Supplemental Dataset S1). These results indicate that a large proportion of RL- responsive IR events are regulated by PpPHY4 or PphnRNP-F1, suggesting that both PpPHY4 and PphnRNP-F1 are required for the regulation of RL-mediated AS. A comparison of the RL-responsive IR events mediated by PpPHY4 or PphnRNP-F1 revealed 1199 RL-induced IR events (70%) in common (Fig. 2C), indicating that PpPHY4 and PphnRNP-F1 cooperatively regulate RL-responsive IR. For those RL-responsive IR events, we further performed functional enrichment analysis to identify potential signaling processes downstream of RL response mediated by PpPHY4 and PphnRNP-F1. As shown in Supplemental Table S1, genes encoding translation-related protein were enriched. It is worthwhile to further investigate the effect of AS for these gene transcripts to figure out their functions in PpPHY4-mediated RL signaling pathway. We also performed reverse transcription quantitative PCR (RT-qPCR) analysis of several IR events selected from among the 1199 RL-induced IR events found to be cooperatively regulated by PpPHY4 and PphnRNP-F1 in the RNA-seq data and confirmed that these genes were insensitive to RL treatment in phy4 and hnrnp-f1 plants (Supplemental Fig. S5). We further performed analyses for identification of light-responsive exon skipping and alternative donor/acceptor events in wild type and checked splicing pattern of these events in phy4 and hnrnp-f1. As shown in Supplemental Fig. S6, the majority of these events are misregulated in the mutants, further supporting that both PHY4 and hnRNP-F1 are involved in splicing regulation (Supplemental Dataset S2).
Figure 2.
PpPHY4 and PphnRNP-F1 cooperatively control RL-responsive IR. A and B, Analysis of 2056 RL-induced IR events. Profiles of the relative levels of RL-responsive IR events in wild type (WT) and PpPHY4-knockout mutant (phy4; A) or PphnRNP-F1-knockout mutant (hnrnp-f1) plants (B) Log2-transformed relative IR levels of the corresponding events in wild type and phy4 or hnrnp-f1 are shown in the order of fold-change (FC; WT/phy4). C, Venn diagram of PpPHY4-dependent and PphnRNP-F1-dependent RL-induced IR events. D, The repetitive GAA motif is enriched in 1199 RL-responsive IR events cooperatively regulated by PpPHY4 and PphnRNP-F1. The E-value (indicating the statistical significance of the motif) is shown under the motif, and the total number of sites is indicated. E, Relative IR levels of PpRPS8 (Pp3c13 20020) intron 4 in wild type, phy4, hnrnp-f1, and PphnRNP-F1 overexpression lines (PphnRNP-F1-OX) detected by RT-qPCR. Samples were collected from dark-grown protonemata (D) and protonemata treated with RL for 1 h (R1). Relative IR level was calculated according to a previous study and tested in three independent biological replicates with primer sets designed for IR splice variant and total transcripts. Error bars show the SEM (n = 3 biological replicates; *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test).
The GAA Motif Is Enriched in RL-Induced IR Events Cooperatively Regulated by PpPHY4 and PphnRNP-F1
Cis-regulatory elements play important roles in spliceosome assembly by recruiting RNA-binding proteins such as SR proteins and hnRNPs to pre-mRNA (Meyer et al., 2015). We previously identified a purine-rich GAA motif in the adjacent exonic region of the retained intron from RL-responsive IR events (Wu et al., 2014), suggesting a role for the GAA motif in connecting RL signaling and pre-mRNA splicing. In this study, we showed that PpPHY4 controls RL-responsive IR of a number of genes cooperatively with PphnRNP-F1. To identify possible cis elements enriched in RL-induced IR regions, we performed motif searches within the 200-nucleotide sequences of the 5′ and 3′ flanking regions of both the donor and acceptor sites of the 1199 retained introns coregulated by PpPHY4 and PphnRNP-F1 (Chang et al., 2014). A control data set was generated from 1200 randomly selected IR events that were not RL responsive. We compared the occurrence of the motif in the sample and control data sets based on enrichment level with a P value of 0.001 by Fishers exact test. Interestingly, the GAA- repetitive motif was again overrepresented, with an E value of 6.7E-044 (Fig. 2D; Supplemental Dataset S3). These results, together with our previous identification of the enriched GAA motif (Wu et al., 2014), suggest that this motif is a potential cis element for RL-responsive IR events controlled by PpPHY4 and PphnRNP-F1. By contrast, the GAA motif was not enriched among IR events suppressed by RL and coregulated by PpPHY4 and PphnRNP-F1, further confirming the unique role of the GAA motif in silencing splicing.
To experimentally validate the function of the GAA motif, we chose intron 4 of the Physcomitrella Ribosomal Protein S8 (PpRPS8) gene, which undergoes significant RL-responsive IR and contains a GAA motif in the adjacent exon, as a model to study the function of the GAA motif (Wu et al., 2014). We measured the IR level of PpRPS8 intron 4 in wild type, phy4, hnrnp-f1, and a PphnRNP-F1 overexpression line (PphnRNP-F1-OX) by RT-qPCR and normalized with that of wild type in the dark. After 1 h of RL treatment, the relative IR level of PpRPS8 intron 4 increased in wild type (Fig. 2E) but significantly decreased in phy4 and hnrnp-f1 (Fig. 2E), suggesting that both PpPHY4 and PphnRNP-F1 are required for the regulation of RL-responsive IR of PpRPS8 intron 4. Moreover, while the absence of PphnRNP-F1 reduced the RL response, overexpression of PphnRNP-F1 enhanced RL-mediated IR of PpRPS8 intron 4 (Fig. 2E), indicating that PphnRNP-F1 functions as a splicing silencer of PpRPS8.
The GAA Motif Suppresses pre-mRNA Splicing and Recruits RNA-Protein Complexes
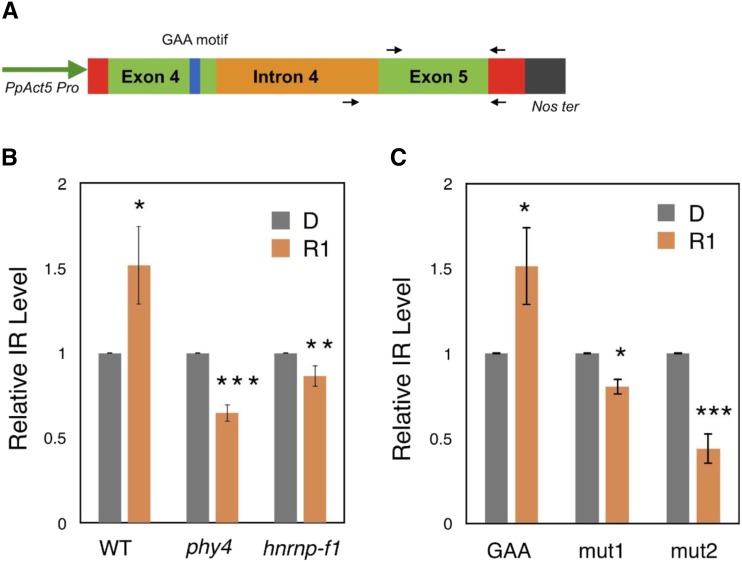
The GAA motif was first identified as a cis element that recruits ASF/SF2 to promote pre-mRNA splicing in humans (Yeakley et al., 1993; Tanaka et al., 1994; Staffa and Cochrane, 1995; Tacke and Manley, 1995). The GAA motif was also predicted to be an exonic cis element in Arabidopsis (Pertea et al., 2007) and was experimentally proven to recruit SCL33, SR45, and SC35-LIKE SPLICING FACTOR30 (SCL30; Thomas et al., 2012; Xing et al., 2015; Yan et al., 2017). To further investigate whether the GAA motif functions as a cis-regulatory element and affect splicing efficiency, we investigated the role of the GAA motif in regulating RL-responsive pre-mRNA splicing. We designed a mini-gene construct composed of exon 4, intron 4, and exon 5 (E4-I4-E5) of PpRPS8 (Fig. 3A) and transiently expressed this mini-gene in wild type, phy4, and hnrnp-f1 protonemata, followed by incubation in the dark or RL. We quantified mini-gene transcripts by RT-qPCR using primer sets designed to detect the splicing efficiency of intron 4. As shown in Figure 3B, wild-type plants expressing the mini-gene construct with an intact GAA motif showed RL-responsive intron 4 retention at a level similar to that of the endogenous gene (Fig. 2E), validating the use of the mini-gene transcript as a reporter for IR level. In the absence of PpPHY4 and PphnRNP-F1, intron 4 retention in the mini-gene transcripts was insensitive to RL (Fig. 3B), confirming the role of PpPHY4 and PphnRNP-F1 in the RL-mediated regulation of splicing.
Figure 3.
The GAA motif in exon 4 of PpRPS8 functions in RL-responsive IR in a mini-gene assay. A, Schematic representation of the PpAct5 promoter-PpRPS8 mini-gene constructs. Nos ter, Nos terminator. The mini-gene construct is shown with E4-I4-E5 of PpRPS8; the blue box shows the location of the GAA motif, and the red box represents a partial sequence (99 bp) from firefly (Photinus pyralis) and Renilla (Renilla reniformis) luciferases used to distinguish endogenous transcripts from transcripts of the mini-gene reporter. Arrows indicate the primer sets used for RT-qPCR. B, RT-qPCR of the PpRPS8 mini-gene in wild-type, phy4, and hnrnp-f1 protonemata. Relative IR levels were calculated according to a previous study. The mini-gene construct was transiently expressed in 7-d-old light-grown protonemata separately using particle bombardment. Samples were collected before RL treatment (dark-grown, D) and at 1 h of RL exposure (R1). C, RT-qPCR of the PpRPS8 mini-gene in wild-type protonemata. Each mini-gene construct was transiently expressed in 7-d-old light-grown protonemata separately using particle bombardment. Samples were collected before RL treatment (dark-grown, D) and at 1 h of RL exposure (R1). Error bars show the SEM (n = 3 biological replicates; *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test).
To explore the role of the GAA motif in splicing regulation, we introduced mini-gene constructs with mutations in the GAA motif into wild-type plants. Mini-gene transcripts with GAA motif mutations showed reduced IR after RL irradiation, especially for the mut2 construct (Fig. 3C). These results confirm in planta that the GAA motif is essential for IR in response to RL, further strengthening the notion that the GAA motif functions as an ESS in Physcomitrella.
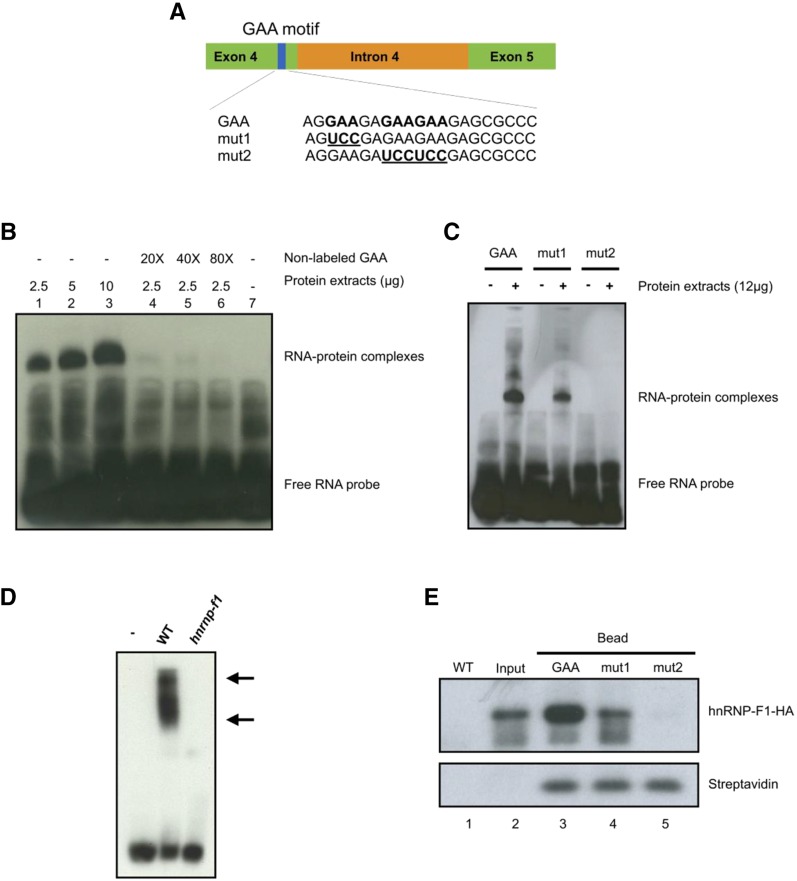
To further investigate whether the GAA motif functions as a cis-regulatory element by recruiting RNA-protein complexes, we performed an RNA-electrophoretic mobility shift assay (EMSA) using plant extracts from light-grown protonemata combined with biotin-labeled RNA oligonucleotides containing the GAA motif. The RNA oligonucleotide was designed based on the flanking sequence of the GAA motif in the adjacent exonic region of PpRPS8 intron 4 (Fig. 4A). As shown in Figure 4B, the plant extracts retarded the mobility of GAA RNA oligonucleotides, indicating that RNA-protein complexes had formed. Moreover, increasing the protein abundance enhanced complex formation, whereas the use of nonlabeled RNA as a competitor significantly decreased this process, pointing to the specificity of the association between GAA-containing RNA oligonucleotides and proteins. To further investigate the importance of the GAA motif in recruiting proteins, we generated two RNA oligonucleotides with GAA mutations (mut1 and mut2), where the GAAs were changed to UCC (Fig. 4A). Notably, mut2 RNA oligonucleotides totally lost the ability to form complexes, whereas mut1 retained partial complex-forming activity (Fig. 4C), indicating that the GAA motif is essential for RNA/protein association. Results from RNA-EMSA competition assay with nonlabeled mut1 and mut2 RNA oligonucleotides further confirmed the specificity of the protein binding activity of GAA motif (Supplemental Fig. S7).
Figure 4.
The GAA motif recruits RNA-protein complexes including PphnRNP-F1. A, Schematic diagram showing E4-I4-E5 of PpRPS8. Blue box illustrates the GAA motif found in the adjacent exon 4. A 21-nucleotide biotin-labeled RNA oligonucleotide used in RNA-EMSA was designed based on the GAA motif of exon 4 of PpRPS8. The GAA motif is shown in bold; GAA-to-UCC mutations are underlined and referred to as mut1 and mut2. B, Increasing amounts of plant extract (lanes 1–3, 2.5, 5, and 10 μg, respectively) were incubated with biotin-labeled GAA RNA oligonucleotides (25 nM) in the RNA-EMSA. For the competition assay (lanes 4–6), a 20-, 40-, and 80-fold excess of nonlabeled GAA RNA oligonucleotides was incubated with 2.5 μg of plant extract and 25 nm biotin-labeled GAA RNA oligonucleotides. Lane 7 shows the shift of free biotin-labeled GAA RNA as a control. C, 25 nm biotin-labeled GAA, mut1, or mut2 RNA oligonucleotides was incubated with or without 12 μg of plant extracts from 7-d-old light-grown protonemata in the RNA-EMSA. The shifted RNA-protein complexes and the free RNA probes were visualized with streptavidin-horseradish peroxidase, followed by chemiluminescent detection. D, RNA-EMSA, 25 nm biotin-labeled GAA RNA oligonucleotides was incubated with or without 12 μg of plant extracts from 7-d-old light-grown wild-type or hnrnp-f1 protonemata. Arrows indicate the RNA-protein complexes. E, RNA pull-down assay using biotin-labeled RNA oligonucleotides (GAA, mut1, and mut2) and 5 mg of plant extracts from 7-d-old light-grown PphnRNP-F1-OX plants. The RNA-bound PphnRNP-F1-OX was precipitated with streptavidin-conjugated magnetic beads and visualized by immunoblotting using anti-HA antibody and horseradish peroxidase-conjugated secondary antibody. Lane 1, wild type (WT); 4 μg of plant extracts from 7-d-old light-grown wild-type plants as a negative control. Lane 2, input, 0.1% of total plant extract was used for the RNA pull-down assay. Lane 35, RNA pull-down assay using biotin-labeled GAA, mut1, and mut2 RNA oligonucleotide-conjugated magnetic beads. Streptavidin is shown as a loading control.
The GAA Motif Is Required for the Recruitment of PphnRNP-F1
Our results suggest that the GAA motif is important for the role of the splicing regulator PphnRNP-F1 in modulating RL-responsive IR in Physcomitrella (Fig. 2, D and E). We therefore investigated the association between this motif and PphnRNP-F1. To determine whether PphnRNP-F1 is involved in the formation of the GAA-mediated RNA-protein complex, we performed an RNA-EMSA in the presence or absence of PphnRNP-F1 using plant extracts from wild type and hnrnp-f1. As shown in Fig. 4D, the formation of RNA-protein complexes was significantly diminished when using PphnRNP-F1-depleted plant extracts, suggesting that PphnRNP-F1 is essential for complex formation and/or stability and that it may interact directly with the GAA motif. In an RNA pull-down assay using plant extracts from the PphnRNP-F1-OX line constitutively expressing an human influenza hemagglutinin (HA)-tagged version of PphnRNP-F1 and biotin-labeled RNA oligonucleotides (GAA, mut1, and mut2), PphnRNP-F1 was pulled down by the GAA motif (Fig. 4E). In the presence of biotin-labeled RNA oligonucleotides corresponding to the mutated versions of the GAA motif (mut1 and mut2), the pull-down of PphnRNP-F1 failed to occur or was strongly reduced (Fig. 4E). These results support the hypothesis that the GAA motif is specifically involved in the recruitment of PphnRNP-F1 for the formation of a RNA-protein complex. We proposed that PphnRNP-F1 is recruited by the GAA motif directly to suppress pre-mRNA splicing.
PpPHY4 Promotes RL-Mediated Accumulation of PphnRNP-F1
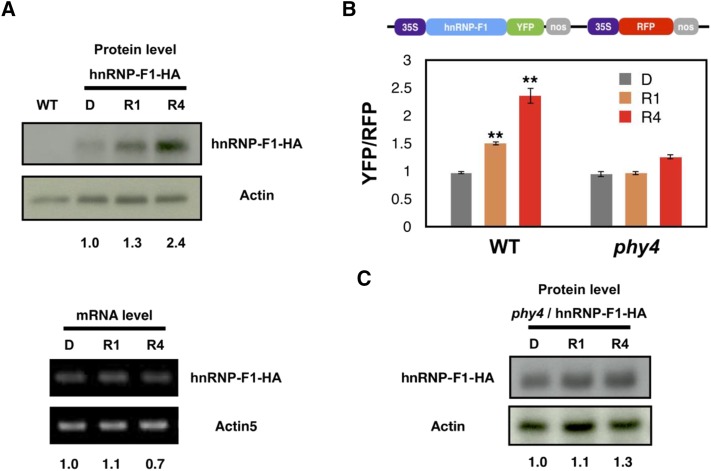
Our data suggest that PphnRNP-F1 interacts with PpPHY4 in an RL-dependent manner associated with the GAA motif and suppresses pre-mRNA splicing. We therefore explored how light-activated phytochrome modulates PphnRNP-F1 activity to control splicing efficiency. Phytochromes generally control the abundance of their interacting protein partners to regulate downstream gene expression (Monte et al., 2004), and the protein level of a tagged version of PphnRNP-F1 increased along with RL exposure (Fig. 5A), although the mRNA level remained constant (Fig. 5A). To examine whether PpPHY4 regulates the PphnRNP-F1 accumulation, we transiently expressed YFP-tagged PphnRNP-F1 and a red fluorescent protein (RFP) reporter gene in wild type and phy4 protonemal cells. We estimated the PphnRNP-F1 protein levels based on YFP fluorescence normalized to the level of RFP fluorescence in each cell. The PphnRNP-F1-YFP protein levels increased after RL irradiation in wild-type but not phy4 cells (Fig. 5B; Supplemental Fig. S8). To confirm PphnRNP-F1 accumulation is PHY4 dependent, we knocked out PpPHY4 in PphnRNP-F1-OX line by using the clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) method (Supplemental Fig. S9). Indeed, we found the protein level of PphnRNP-F1 is not accumulated after RL exposure in the absence of PpPHY4 (Fig. 5C). These findings support the notion that PpPHY4 is involved in regulating PphnRNP-F1 abundance upon RL irradiation. Thus, we propose that RL, through the action of PpPHY4, either stabilizes PphnRNP-F1 protein or promotes its translation through an unknown mechanism, which in turn controls IR in targeted transcripts.
Figure 5.
PpPHY4 is required for PphnRNP-F1 accumulation after RL irradiation. A, Top indicates the immunoblot of plant extracts from wild-type (WT) and PphnRNP-F1- OX plants. Protonemata were grown for 7 d in the light and 3 d in the dark and transferred to RL for 0, 1, and 4 h (D, R1, and R4). Equal amounts of plant extracts were subjected to SDS-PAGE. The numbers below indicate the quantitative results of PphnRNP-F1 protein measurements normalized to actin levels and calculated using ImageJ software. Anti-HA antibody and anti-actin antibody were used to detect the expression of PphnRNP-F1 and actin, respectively. The gel was visualized using HRP-conjugated secondary antibody via the chemiluminescence method. Bottom, indicates the result of the RT-PCR of PphnRNP-F1-OX and PpActin5 mRNA levels. RNA samples were collected from the same materials (D, R1, and R4) used to produce the plant extracts. An equal amount of RNA was subjected to cDNA synthesis. The numbers below indicate the quantitative results of PphnRNP-F1 mRNA level normalized with PpActin5. B, Top indicates the construct of the plasmid used to quantify PphnRNP-F1 accumulation, coexpressing PphnRNP-F1-YFP and RFP. Bottom shows the quantification of PphnRNP-F1 protein level in vivo in response to RL. The plasmid was introduced into wild-type and PpPHY4-knockout mutant (phy4) gametophores by particle bombardment before dark recovery and RL treatment. Regenerated cells (dark grown, D) were irradiated with RL for 1 (R1) and 4 (R4) h prior to confocal microscopy. n ≥ 20. The YFP signal was normalized to the RFP signal using ImageJ software. Error bars show the SEM (*P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test). C, The immunoblot of plant extracts from phy4/PphnRNP-F1-OX plants. Protonemata were grown for 7 d in the light and 3 d in the dark and transferred to RL for 0, 1, and 4 h (D, R1, and R4). Equal amounts of plant extracts were subjected to SDS-PAGE. The numbers below indicate the quantitative results of PphnRNP-F1 protein measurements normalized to actin levels and calculated using ImageJ software. Anti-HA antibody and anti-actin antibody were used to detect the expression of PphnRNP-F1 and actin, respectively. The gel was visualized using horseradish peroxidase-conjugated secondary antibody via the chemiluminescence method. The result is the representative one of three biological repeats.
DISCUSSION
The importance of light in regulating AS has been confirmed in both vascular and nonvascular plants (Petrillo et al., 2014; Shikata et al., 2014; Wu et al., 2014). However, little is known about the molecular mechanism underlying light-regulated AS. Two previous studies showed that splicing factors participate in light-mediated AS (Shikata et al., 2012; Xin et al., 2017). The genetic identification of light signaling components revealed that RRC1, an SR-like splicing regulator, functions as a downstream partner of phyB to control AS and photomorphogenesis (Shikata et al., 2012). More recently, SFPS, an Arabidopsis homolog of human splicing factor45 involved in phytochrome signaling, was shown to directly interact with phyB and regulate photomorphogenesis through pre-mRNA splicing (Xin et al., 2017). A plastid signal induced by light is also thought to modulate AS (Petrillo et al., 2014). Here, we showed that PpPHY4, a phyB-type phytochrome, physically interacts with the splicing regulator PphnRNP-F1 in an RL-dependent manner. Although plant hnRNPs, including the polypyrimidine tract binding proteins Arabidopsis GLY-RICH RNA-BINDING PROTEIN7 (AtGRP7) and AtGRP8, are known to regulate AS, mainly through inhibiting splicing (Staiger et al., 2003; Schöning et al., 2008; Stauffer et al., 2010), they have not been shown to play a role in phytochrome signaling for AS regulation. Here, we demonstrated that most RL-induced IR events are repressed in the absence of PphnRNP-F1, pointing to its silencing role in regulating pre-mRNA splicing, as well as the involvement of this process in RL signaling. Furthermore, more than half of the RL-responsive IR events regulated by PpPHY4 and PphnRNP-F1 overlapped. These results indicate that the two proteins modulate AS cooperatively upon RL irradiation. Our findings thus help to uncover a detailed mechanism underlying how phytochromes regulate AS in conjunction with a potential downstream regulator.
SRE is important for the accuracy and efficiency of intron splicing. The deletion of SRE decreases splicing efficiency (Yeakley et al., 1996). Here, we showed that a repetitive GAA motif was enriched among RL-induced IR events. Interestingly, no GAA repetitive motif was enriched among RL-reduced IR events, suggesting that the GAA motif plays a negative role in RL-responsive AS. In humans, this motif also recruits hnRNP A1 to modulate the splicing of HIV-1 transcripts (Marchand et al., 2002). In plants, only SR proteins such as SCL33, SR45, SCL30, and SER/ARG-RICH SPLICING FACTOR35 (SC35) were previously shown to target the GAA motif (Yoshimura et al., 2011; Thomas et al., 2012; Xing et al., 2015). SR30 is an ASF/SF2-like SR protein that associates with GAA repetitive RNA sequences (Tacke and Manley, 1995). SCL33 also associates with the GAA motif in SCL33 transcripts for autoregulation (Thomas et al., 2012). In this study, we found that the GAA motif functions as a regulatory cis element in RL-induced AS cooperatively regulated by PpPHY4 and PphnRNP-F1. Furthermore, an RNA pull-down assay showed that the GAA motif recruits PphnRNP-F1, suggesting that the GAA-repetitive sequence and PphnRNP-F1 interact. Given that GAA-mediated protein complexes were observed in protein lysates only in the presence of PphnRNP-F1, it is likely that PphnRNP-F1 is required for the formation of GAA-protein complexes. Whether PphnRNP-F1 directly interacts with the GAA motif in pre-mRNA requires further investigation.
We found that the abundance of PphnRNP-F1 protein increased along with RL irradiation, while the mRNA level remained unchanged, perhaps due to the enhancement of protein synthesis or the repression of protein degradation. Light exposure increases the protein level of the bZIP transcription factor ELONGATED HYPOCOTYL5 (HY5) and promotes photomorphogenesis in Arabidopsis (Hardtke et al., 2000). Light-activated phyB also causes SUPPRESSOR OF PHYA-1051 (SPA1) to accumulate in the nucleus via a physical interaction (Saijo et al., 2008; Zheng et al., 2013; Ranjan et al., 2014; Sheerin et al., 2015). Our work shows that the presence of PpPHY4 is required for the accumulation of PphnRNP-F1. It will be important to investigate how the RL-induced PpPHY4-PphnRNP-F1 interaction promotes the accumulation of PphnRNP-F1.
CONCLUSION
AS increases transcriptome complexity as well as proteome diversity, thereby providing alternative ways for plants to rapidly respond to the changing environment. Several studies have demonstrated the importance of AS in photomorphogenesis. For example, AS inhibits the function of COP1, and an AS variant of PIF6 promotes seed germination (Zhou et al., 1998). Moreover, splicing factors such as SFPS and RRC1 regulate AS and photomorphogenesis (Shikata et al., 2012; Xin et al., 2017). However, the exact role and regulatory mechanism of AS in plant light responses requires further investigation. Our understanding of the functional roles of cis-regulatory elements in the regulation of AS is also limited. In this study, we provided evidence that a transacting factor directly participates in light-mediated splicing regulation and identified a cis element in pre-mRNA that functions in this process. By further deciphering the relationship between the splicing regulator and cis-regulatory element and their roles in splicing regulation, we hope to shed light on the detailed mechanism underlying pre-mRNA splicing in plants.
MATERIALS AND METHODS
Plant Growth Conditions and Light Treatment
Physcomitrella patens protonemata were cultured on solid BCDAT medium overlaid with cellophane at 25°C under continuous white light (80–100 μmol m−2 s−1). For propagation, 7-d-old protonemata were collected, blended in sterile water with an SH-48 tissue grinder (Kurabo) at 12,000 rpm for 5 min, and spread onto solid BCDAT medium overlaid with cellophane. Before light treatment, 7-d-old protonemata of the corresponding plants were grown in the dark for 3 d, followed by exposure to RL (660 nm LED, 5 μmol m−2 s−1) at 25°C for 1 and 4 h, as described previously (Chen et al., 2012).
Plasmid Construction
The plasmids used for Y2H and moss transformation were constructed using an In-Fusion HD Cloning kit (Clontech Laboratories). In brief, the complementary DNA (cDNA) sequences of the desired genes were amplified using primers containing 15 nucleotides identical to the gene regions and 15 nucleotide extensions homologous to the vector ends according to the manufacturer’s instructions. PCR-generated sequences were assembled with linearized vectors using In-Fusion enzyme and transformed into Escherichia coli for plasmid amplification. For the mini-gene constructs, fragments of interest were cloned into the pGEM-T-Easy vector (Promega) with partial sequences (99 bp) located both upstream and downstream of the mini-gene from two luciferase genes from firefly (Photinus pyralis) and Renilla (Renilla reniformis) to distinguish endogenous from transiently expressed transcripts while performing the mini-gene reporter assay. To quantify PphnRNP-F1 (Pp3c12_3660) protein level, Cauliflower mosaic virus 35S promoter-driven PphnRNP-F1 was tagged with YFP at its C terminus. The RFP gene driven by another Cauliflower mosaic virus 35S promoter was inserted into the same construct following PphnRNP-F1-YFP.
Transformation of P. patens and Generation of Knockout Mutants
The upstream and downstream regions of the target coding sequences were amplified by PCR and cloned into the pTN80 vector containing an nptII cassette (gift from Mitsuyasu Hasebe, National Institute for Basic Biology, Okazaki, Japan; Supplemental Fig. S4). P. patens was transformed as described, with minor modifications (Chen et al., 2012). Seven-day-old protonemata were harvested for protoplast isolation and polyethylene glycol-mediated transformation. The regenerated protoplasts were selected on BCDAT medium supplemented with the appropriate antibiotic (20 mg/L G418). Resistant colonies were transferred to nonselective BCDAT medium for 1 week, followed by transfer to selection medium. The correct gene-specific insertion in stable transformants was checked by PCR using specific primers and further confirmed by Southern blot analysis (Supplemental Dataset S4).
Y2H Analysis
The coding regions of PphnRNP-F1 (Pp3c12_3660) and PpPUBS (Pp3c6_8700; Lee et al., 2012) from Physcomitrella (Chen et al., 2012) and PIF3 from Arabidopsis (Arabidopsis thaliana; Ni et al., 1998) were cloned into the pGEM-T vector and subcloned into the pGADT7 vector. The coding regions of Physcomitrella phytochromes, including PpPHY1 (Pp3c25_2610), PpPHY2 (Pp3c16_20280), PpPHY3 (Pp3c16_18760), and PpPHY4 (Pp3c27_7830), were cloned into the pGBKT7 vector. Yeast strain Y187 (Clontech) and Y2HGold (Clontech) cells were cultured overnight in liquid yeast extract peptone dextrose adenine medium and transformed with the plasmids according to the manufacturer’s instructions. Two single colonies grown on SD-Leu or SD-Trp medium were mated on solid yeast extract peptone dextrose adenine plates for 24 h, followed by SD/Leu/Trp medium for 3 d. Single transformants were cultured for 3 d on solid SD/Ade/His/Leu/Trp medium supplemented with 5 μm PCB or PEB in the dark, RL (5 μmol m−2 s−1), or FR (1 μmol m−2 s−1).
BiFC
Particle bombardment was performed using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) to transiently coexpress the target proteins fused with N- or C-split YFP in moss gametophores and orchid petal according to the manufacturer’s protocol. Plasmids containing NLS-mCherry were also included as a nuclear localization marker along with the other plasmids. After bombardment, the moss cells and the orchid petal were recovered overnight in the dark and irradiated with RL (5 μmol m−2 s−1) for 1 h. Fluorescence was visualized by confocal microscopy. The plant cells were observed under a Zeiss LSM510 microscope (Zeiss) with the following conditions: YFP and RFP were excited by the 514-nm line of an argon laser and the 543-nm line of a HeNe laser, respectively. YFP fluorescence was detected using a 535- to 560-nm band-pass filter, RFP fluorescence was detected using a 565- to 615-nm band-pass filter, and chloroplast autofluorescence was detected at 650 to 710 nm.
RNA Isolation and RT-qPCR
Plant tissues collected before and after light treatment were frozen in liquid nitrogen for total RNA extraction with a Plant Total RNA Miniprep Purification kit (GeneMark) following the manufacturer’s instructions and quantified using the NanoDrop system (Thermo Fisher Scientific). On-column DNase digestion was performed to remove any genomic DNA contamination prior to cDNA synthesis. First-strand cDNA was synthesized using 1.5 μg of RNA with a SuperScript III RT kit (Invitrogen) according to the standard protocol. Diluted cDNA was subjected to RT-qPCR on a QuantStudio 12K Flex Real-Time PCR System (Thermo Fisher Scientific) using Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). Primers for RT-qPCR analysis were designed based on the sequence of the corresponding region and are listed in Supplemental Dataset S4. The RT-qPCR was performed with three biological replicates. Unpaired Student’s t test was applied for the statistical analysis.
RNA-Seq and Data Analysis
RNA-seq was performed by Yourgene Bioscience on the HiSeq 2000 platform. On average, a total of 50 million 100-nucleotide paired-end reads were obtained for each library. Sequence reads were mapped to the P. patens genome v3.3 (large-scale annotation data in Joint Genome Institute (http://www.phytozome.net/physcomitrella.php) using the BLAT program (Kent, 2002). RPKM values were calculated using the in-house package RACKJ (http://rackj.sourceforge.net/) with a similar algorithm to that described previously (Mortazavi et al., 2008). Calculation of intron reads per kilobase of retained IPKM values and other data-processing steps were performed using Excel (Microsoft). RACKJ was also used to compute light-regulated IR events, as described previously, with minor modifications (Wu et al., 2014). After reads aligned to introns for each sample were counted, events with at least five supported reads were retained. To increase the confidence of calling an IR event, an extra filtering step was included. The read coverage of each retained intron was calculated. Only introns with 100% read coverage in any of the samples were defined as an IR event. χ2 values for goodness-of-fit were computed by comparing the read counts supporting an IR event (intronic read counts) with the read counts of the corresponding gene exons between samples. The corresponding P values were calculated based on χ2 distribution using Microsoft Excel. IR events with P < 0.001 were retained. All RL-responsive IR data were list in Supplemental Dataset S1.
Protein Extraction
Seven-day-old light-grown protonemata were collected and immediately frozen in liquid nitrogen for protein extraction. Frozen samples were ground into a fine powder in liquid nitrogen using a mortar and pestle, resuspended in protein extraction buffer (40 mm Tris-HCl, pH 7.5, 75 mm NaCl, 5 mm EDTA, and 1% [v/v] Triton X-100) containing 1× Proteinase inhibitor (Sigma-Aldrich), and incubated for 20 min on ice. The resuspended mixtures were centrifuged at 16,400g for 1 h at 4°C. The supernatant was collected and used to quantify protein concentrations with Pierce 660 nm Protein Assay Reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
RNA-EMSA
The following single-stranded RNA oligonucleotides were used (mutations are underlined): GAA, 5′-AGGAAGAGAAGAAGAGCGCCC-3′; mut1, 5′-AGUCCGAGAAGAAGAGCGCCC-3′; and mut2, 5′-AGGAAGAUCCUCCGAGCGCCC-3′. The RNA oligonucleotides were synthesized and biotin-labeled at their 3′ ends (PURIGO Biotechnology). The experiment was carried out with a LightShift Chemiluminescent RNA EMSA kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Protein extracts were incubated with biotin-labeled RNA oligonucleotides (25 nm) in a binding reaction (20 μL) containing 10× binding buffer, 5% (v/v) glycerol, and 2 μg tRNA for 20 min at room temperature. For the competition assays, the binding reaction was mixed with a 40-fold excess (1 μm) of unlabeled GAA RNA oligonucleotides. The samples were mixed with loading buffer (30% [v/v] glycerol, 0.5% [w/v] bromophenol blue, and 0.5% [w/v] xylene cyanol) before being separated by electrophoresis in a 6% (v/v) DNA retardation gel (Thermo Fisher Scientific) in 0.5× TBE (Tris-borate-EDTA) buffer (100 V, 1.5 h). The gels were transferred to positively charged nylon membranes (PerkinElmer), UV-cross-linked, and visualized by chemiluminescent detection following the manufacturer’s protocol. RNA-EMSA experiments were repeated at least three times.
RNA Pull-Down Assays and Immunoblotting
Biotin-labeled RNA oligonucleotides (500 pmol) were conjugated with 100 μL of precleaned, streptavidin-coated magnetic beads (Thermo Fisher Scientific) following the manufacturer’s instructions. After washing off the unbound RNA oligonucleotides, 5 mg of protein extracted from light-grown protonemata overexpressing HA-tagged PphnRNP-F1 was incubated with 1 mL RNA-conjugated magnetic beads in an RNA-protein binding reaction containing 10× RNA-protein binding buffer (1 m Tris-HCl, pH 7.5, 4 m NaCl, 1 m MgCl2, and 25% Tween 20) and proteinase inhibitor for 2 h at 4°C. The unbound proteins were washed off by rinsing three times with precooled RNA washing buffer (20 mm Tris-HCl, pH 7.5, 10 mm NaCl, and 0.1% Tween 20) on ice. Protein-bound magnetic beads were resuspended in 100 μL RNA washing buffer for further analysis. One-third of the protein-bound magnetic beads from each biotin-labeled RNA oligonucleotide (GAA, mut1, and mut2) was subjected to immunoblot analysis. HA-tagged PphnRNP-F1 proteins were detected first using mouse anti-HA antibody (BioLegend) and visualized by chemiluminescent detection using horseradish peroxidase-conjugated donkey anti-mouse IgG secondary antibody (Jackson ImmunoResearch). RNA pull-down assays were performed for more than three times.
Transient Expression of the Mini-gene Splicing Reporter by Particle Bombardment
Light-grown 7-d-old protonemata were transiently transformed via particle bombardment with the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) following the manufacturer’s instructions. After mixing 25 μL of gold particles (1 μm in diameter) with 25 μL of plasmid (1 μg), the sample was vortexed with 50 μL of 2.5 m CaCl2 and 20 μL of 0.1 m spermidine for 3 min. The DNA-coated gold particles were pelleted by centrifugation (10,000g) for 10 s and washed twice with 99% (v/v) ethanol. Pelleted gold particles were resuspended in 30 μL of 99% (v/v) ethanol and distributed onto a macro-carrier for particle bombardment. The distance from the rupture disc to the macro-carrier was 1.7 cm, the distance from the macro-carrier to the samples was 6 cm, and the pressure was 1100 psi. After bombardment, the samples were recovered in the dark for 3 d at 25°C and exposed to RL (660 nm LED, 5 μmol m−2 s−1) for 1h.
Quantification of YFP-Fused PphnRNP-F1 Protein
Particle bombardment of moss gametophores was performed using the Biolistic PDS-1000/He Particle Delivery System to transiently express a construct encoding the PphnRNP-F1 protein fused with YFP and RFP driven by two 35S promoters, respectively. RFP protein was used to normalize the transformation efficiency of individual cells. After bombardment, the cells were recovered for 72 h in the dark and irradiated with RL (5 μmol m−2 s−1) for 1 and 4 h. Fluorescence was visualized by confocal microscopy with a LSM510 microscope (Zeiss) under the following conditions: the 514-nm line of an argon laser and the 543-nm line of a HeNe laser were used to excite YFP and RFP, respectively. YFP fluorescence was detected using a 535- to 560-nm band-pass filter, RFP fluorescence was detected using a 565- to 615-nm band-pass filter, and chloroplast autofluorescence was detected at 650 to 710 nm. Twenty to twenty-five cells were observed with both fluorescent signals imaged for further quantification. ImageJ was used to quantify YFP and RFP fluorescence intensity. Relative fluorescence intensity was calculated as YFP fluorescence intensity divided by that of RFP.
Generation of PpPHY4 Mutation in PphnRNP-F1-OX by CRISPR-Cas9
Generation of PpPHY4 mutation in the PphnRNP-F1-OX line was done by using the CRISPR-Cas9 system as previously described (Collonnier et al., 2017). The guide RNA sequence was designed using the CRISPOR program (Haeussler and Concordet, 2016). A fragment containing the 20-nucleotide guide RNA sequence driven by the P. patens U6 promoter was cloned into the pDONR207 vector by Gateway cloning to obtain the Pp-SpgRNA-U6plasmid. A mixture of 7 µg of Pp-SpgRNA-U6 plasmid, 7 µg of pAct-CAS9 (encoding Cas9 nuclease), and 7 µg of pBNRF (provides resistance to G418) was transformed into P. patens protoplasts by polyethylene glycol-mediated transformation (Nishiyama et al., 2000). Fragments containing the mutation site were amplified from genomic DNA obtained from five transformants showing G418 resistance and sequenced to verify the mutation. A mutant line with a 17-bp deletion at the guide RNA target site of the PpPHY4 locus, which encoded a truncated protein with a premature termination codon at the 86th amino acid was selected for further study (Supplemental Fig. S9).
Accession Numbers
RNA-seq data from this publication have been submitted to the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) and assigned the identifier SRP115845. Gene information described in this article can be found in the Phytozome JGI (http://www.phytozome.net/physcomitrella.php) under the following gene locus numbers: PpPHY1 (Pp3c25_2610), PpPHY2 (Pp3c16_20280), PpPHY3 (Pp3c16_18760), PpPHY4 (Pp3c27_7830), PphnRNP-F1 (Pp3c12_3660), PpRPS8 (Pp3c13_20020), PpPUBS (Pp3c6_8700), PpU170k (Pp3c4_6130), PpU2AF-65 (Pp3c17_14650), PpLSM1 (Pp3c12_17600), PpLSM10 (Pp3c11_10290), PphnRNPA1 (Pp3c4_6610), PpRPL13A (Pp3c11_17280), PpRPS24 (Pp3c13_22220), and PpRPL27 (Pp3c8_24410).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Yeast two-hybrid (Y2H) screening for interactions between phytochromes and splicing regulators in Physcomitrella.
Supplemental Figure S2. Subcellular localization of PphnRNP-F1.
Supplemental Figure S3. The RL-dependent interaction between PpPHY4 and PphnRNP-F1 in the same orchid petal cell.
Supplemental Figure S4. Construction of the PphnRNP-F1 KO and OX lines.
Supplemental Figure S5. Validation of the RNA-seq data for RL-induced IR events regulated by PpPHY4 and PphnRNP-F1.
Supplemental Figure S6. PpPHY4 and PphnRNP-F1 are involved in RL-responsive AS.
Supplemental Figure S7. RNA-EMSA competition assay.
Supplemental Figure S8. PpPHY4 is required for the RL-induced accumulation of PphnRNP-F1 protein.
Supplemental Figure S9. CRISPR/Cas9 strategy for generating PpPHY4 mutation in the hnRNP-F1-OX line.
Supplemental Table S1. Functional enrichment of RL-induced IR event coregulated by PpPHY4 and PphnRNP-F1.
Supplemental Dataset S1. RL-responsive IR events regulated by PpPHY4 and PphnRNP-F1.
Supplemental Dataset S2. RL-responsive exon skipping and alternative donor/acceptor site events regulated by PpPHY4 and PphnRNP-F1.
Supplemental Dataset S3. GAA motif enriched from 1199 RL-induced IR events regulated by PpPHY4 and PphnRNP-F1.
Supplemental Dataset S4. Primers used in this study.
ACKNOWLEDGMENTS
We thank Fabien Nogué and Nancy Hofmann for critically reading the manuscript and Shu-Hsing Wu for valuable discussion. Mei-Jane Fang in the Live Cell Imaging Core Laboratory, Wen-Dar Lin in the Bioinformatics Core Laboratory and Shu-Jen Chou in the Genomic Technology Core Laboratory of the Institute of Plant and Microbial Biology, and Choun-Sea Lin at the Agricultural Biotechnology Research Center and Lin-yun Kuang in the Transgenic Plant Core Laboratory of Academia Sinica, Taiwan, for technical assistance.
Footnotes
This work was supported by the Ministry of Science and Technology (grant no. MOST 106-2311-B-001 -033 -MY3 to S.-L.T.) and by the Academia Sinica, Taiwan (grant no. AS-105-TP-B03 to S.-L.T.).
Articles can be viewed without a subscription.
References
- Chang C-Y, Lin W-D, Tu S-L (2014) Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol 165: 826–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-R, Su YS, Tu S-L (2012) Distinct phytochrome actions in nonvascular plants revealed by targeted inactivation of phytobilin biosynthesis. Proc Natl Acad Sci USA 109: 8310–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y-L, Tu S-L (2018) Alternative splicing and cross-talk with light signaling. Plant Cell Physiol 59: 1104–1110 [DOI] [PubMed] [Google Scholar]
- Collonnier C, Epert A, Mara K, Maclot F, Guyon-Debast A, Charlot F, White C, Schaefer DG, Nogué F (2017) CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol J 15: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Zhou J, Elling AA, Charron J-BF, Deng XW (2008) Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Concordet J-P (2016) Genome editing with CRISPR-Cas9: Can it get any better? J Genet Genomics 43: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann L, Drewe-Boß P, Wießner T, Wagner G, Geue S, Lee H-C, Obermüller DM, Kahles A, Behr J, Sinz FH, et al. (2016) Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 28: 2715–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH (2010) Phytochrome signaling In Briggs WR, and Spudich J, eds, Handbook of Photosensory Receptors, Vol Vol. 20 Wiley, Weinheim, Germany, pp 151–170 [Google Scholar]
- Kent WJ. (2002) BLAT--the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L-Y, Wu F-H, Hsu C-T, Shen S-C, Yeh H-Y, Liao D-C, Fang M-J, Liu N-T, Yen Y-C, Dokládal L, et al. (2012) Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24: 1746–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Terzaghi W, Deng XW (2012) Genomic basis for light control of plant development. Protein Cell 3: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lagarias JC (1992) Phytochrome assembly. Defining chromophore structural requirements for covalent attachment and photoreversibility. J Biol Chem 267: 19204–19210 [PubMed] [Google Scholar]
- Liu M-J, Wu S-H, Chen H-M, Wu S-H (2012) Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol 8: 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini E, Sanchez SE, Romanowski A, Schlaen RG, Sanchez-Lamas M, Cerdán PD, Yanovsky MJ (2016) Acute effects of light on alternative splicing in light-grown plants. Photochem Photobiol 92: 126–133 [DOI] [PubMed] [Google Scholar]
- Mano S, Hayashi M, Nishimura M (1999) Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J 17: 309–320 [DOI] [PubMed] [Google Scholar]
- Mano S, Hayashi M, Nishimura M (2000) A leaf-peroxisomal protein, hydroxypyruvate reductase, is produced by light-regulated alternative splicing. Cell Biochem Biophys 32: 147–154 [DOI] [PubMed] [Google Scholar]
- Marchand V, Méreau A, Jacquenet S, Thomas D, Mougin A, Gattoni R, Stévenin J, Branlant C (2002) A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J Mol Biol 323: 629–652 [DOI] [PubMed] [Google Scholar]
- Meyer K, Koester T, Staiger D (2015) Pre-mRNA splicing in plants: In vivo functions of RNA-binding proteins implicated in the splicing process. Biomolecules 5: 1717–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH (2004) The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA 101: 16091–16098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Nishiyama M, Hong K, Mikoshiba K, Poo MM, Kato K (2000) Calcium stores regulate the polarity and input specificity of synaptic modification. Nature 408: 584–588 [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse E-M, Halliday KJ (2010) A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol 73: 89–95 [DOI] [PubMed] [Google Scholar]
- Pertea M, Mount SM, Salzberg SL (2007) A computational survey of candidate exonic splicing enhancer motifs in the model plant Arabidopsis thaliana. BMC Bioinformatics 8: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrillo E, Godoy Herz MA, Fuchs A, Reifer D, Fuller J, Yanovsky MJ, Simpson C, Brown JWS, Barta A, Kalyna M, et al. (2014) A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344: 427–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possart A, Hiltbrunner A (2013) An evolutionarily conserved signaling mechanism mediates far-red light responses in land plants. Plant Cell 25: 102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Dickopf S, Ullrich KK, Rensing SA, Hoecker U (2014) Functional analysis of COP1 and SPA orthologs from Physcomitrella and rice during photomorphogenesis of transgenic Arabidopsis reveals distinct evolutionary conservation. BMC Plant Biol 14: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Zhu D, Li J, Rubio V, Zhou Z, Shen Y, Hoecker U, Wang H, Deng XW (2008) Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell 31: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A (1996) Nuclear localization activity of phytochrome B. Plant J 10: 859–868 [DOI] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D (2008) Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res 36: 6977–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T (2014) Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci USA 111: 18781–18786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, Matsuoka K, Lin C, Matsushita T (2012) The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J 70: 727–738 [DOI] [PubMed] [Google Scholar]
- Staffa A, Cochrane A (1995) Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol 15: 4597–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33: 361–371 [DOI] [PubMed] [Google Scholar]
- Stauffer E, Westermann A, Wagner G, Wachter A (2010) Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J 64: 243–255 [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL (1995) The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J 14: 3540–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Watakabe A, Shimura Y (1994) Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol 14: 1347–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Palusa SG, Prasad KVSK, Ali GS, Surabhi G-K, Ben-Hur A, Abdel-Ghany SE, Reddy ASN (2012) Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J 72: 935–946 [DOI] [PubMed] [Google Scholar]
- Tu S-L, Lagarias C (2005) The Phytochromes In Briggs WR, and Spudich J, eds, Handbook of Photosensory Receptors. Wiley, Weinheim, Germany, pp 121–149 [Google Scholar]
- Wang Z, Burge CB (2008) Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB (2004) Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845 [DOI] [PubMed] [Google Scholar]
- Wu H-P, Su Y-S, Chen H-C, Chen Y-R, Wu C-C, Lin W-D, Tu S-L (2014) Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol 15: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-H. (2014) Gene expression regulation in photomorphogenesis from the perspective of the central dogma. Annu Rev Plant Biol 65: 311–333 [DOI] [PubMed] [Google Scholar]
- Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E (2017) SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc Natl Acad Sci USA 114: E7018–E7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Wang Y, Hamilton M, Ben-Hur A, Reddy ASN (2015) Transcriptome-wide identification of RNA targets of Arabidopsis SERINE/ARGININE-RICH45 uncovers the unexpected roles of this RNA binding protein in RNA processing. Plant Cell 27: 3294–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Xia X, Sun Z, Fang Y (2017) Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet 13: e1006663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Hedjran F, Morfin JP, Merillat N, Rosenfeld MG, Emeson RB (1993) Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol 13: 5999–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley JM, Morfin JP, Rosenfeld MG, Fu XD (1996) A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci USA 93: 7582–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Mori T, Yokoyama K, Koike Y, Tanabe N, Sato N, Takahashi H, Maruta T, Shigeoka S (2011) Identification of alternative splicing events regulated by an Arabidopsis serine/arginine-like protein, atSR45a, in response to high-light stress using a tiling array. Plant Cell Physiol 52: 1786–1805 [DOI] [PubMed] [Google Scholar]
- Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F, et al. (2013) Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell 25: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DX, Kim YJ, Li YF, Carol P, Mache R (1998) COP1b, an isoform of COP1 generated by alternative splicing, has a negative effect on COP1 function in regulating light-dependent seedling development in Arabidopsis. Mol Gen Genet 257: 387–391 [DOI] [PubMed] [Google Scholar]