The main function of the E2FB transcription factor is to restrict cell proliferation and establish quiescence during Arabidopsis leaf development; it acts in complex with RETINOBLASTOMA-RELATED.
Abstract
Cell cycle entry and quiescence are regulated by the E2F transcription factors in association with RETINOBLASTOMA-RELATED (RBR). E2FB is considered to be a transcriptional activator of cell cycle genes, but its function during development remains poorly understood. Here, by studying E2FB-RBR interaction, E2F target gene expression, and epidermal cell number and shape in e2fb mutant and overexpression lines during leaf development in Arabidopsis (Arabidopsis thaliana), we show that E2FB in association with RBR plays a role in the inhibition of cell proliferation to establish quiescence. In young leaves, both RBR and E2FB are abundant and form a repressor complex that is reinforced by an autoregulatory loop. Increased E2FB levels, either by expression driven by its own promoter or ectopically together with DIMERIZATION PARTNER A, further elevate the amount of this repressor complex, leading to reduced leaf cell number. Cell overproliferation in e2fb mutants and in plants overexpressing a truncated form of E2FB lacking the RBR binding domain strongly suggested that RBR repression specifically acts through E2FB. The increased number of small cells below the guard cells and of fully developed stomata indicated that meristemoids preferentially hyperproliferate. As leaf development progresses and cells differentiate, the amount of RBR and E2FB gradually declined. At this stage, elevation of E2FB level can overcome RBR repression, leading to reactivation of cell division in pavement cells. In summary, E2FB in association with RBR is central to regulating cell proliferation during organ development to determine final leaf cell number.
The time window for cell proliferation is the most fundamental determinant for meristem size and has the largest impact on final organ size (Gázquez and Beemster, 2017). This is set by the coordination of cell cycle and exit to differentiation governed through complex regulatory mechanisms culminating in the evolutionarily conserved Retinoblastoma (Rb) repressor protein and E2F transcription factor targets (van den Heuvel and Dyson, 2008). According to the textbook model established in animal systems, cell cycle entry is guarded by cyclin-dependent kinases (CDKs), which, upon activation by mitogenic signals, phosphorylate and thereby inactivate Rb and other related pocket proteins. When released from Rb repression, the so-called activator E2Fs drive the cell cycle by activating the expression of cell cycle genes required for the G1-to-S phase transition. By contrast, the repressor-type E2Fs function together with Rb to instigate quiescence and to allow differentiation (Morgan, 2007).
In Arabidopsis (Arabidopsis thaliana), a single gene codes for the RETINOBLATOMA RELATED (RBR), and this protein acts through three E2F transcription factors, known as E2FA, E2FB, and E2FC. These three E2Fs can only bind to DNA in complex with the DIMERIZATION PARTNER A (DPA) or DPB (De Veylder et al., 2007). Modeling Arabidopsis E2Fs on the animal scenario places E2FA and E2FB as activators and E2FC as a repressor, but similar to animal cells, this subdivision is largely supported by overexpression studies (Magyar et al., 2016). Ectopic co-overexpression of E2FB with DPA allows the continued proliferation of cultured tobacco (Nicotiana tabacum) cells in the absence of the plant growth hormone auxin (Magyar et al., 2005). This is reminiscent of the effect of human E2F1 overexpression, which triggers S-phase entry in growth factor-deprived cultured cells (Johnson et al., 1993). Overexpression of E2FB without the DP also leads to the upregulation of cell cycle genes and surprisingly a much reduced root growth both in Arabidopsis (Sozzani et al., 2006) and in tomato (Solanum lycopersicum; Abraham and del Pozo, 2012), with fruit size increased in the latter. E2FB is expressed throughout the cell cycle phases (Magyar et al., 2000, 2005; Mariconti et al., 2002) and has the ability to drive both the G1-to-S and G2-to-M transitions, leading to shortened cell doubling time and reduced cell sizes (Magyar et al., 2005). The accelerated entry into mitosis was correlated with the induced expression of the G2-M-specific CDKB1;1, following E2FB overexpression (Magyar et al., 2005; Henriques et al., 2013). The activity of E2FB is tightly controlled by RBR phosphorylation in response to Suc availability, overexpression of CYCLIN D3;1 (CYCD3;1), or the counteracting CDK inhibitor KIP-RELATED PROTEIN 2 (KRP2; Magyar et al., 2012).
E2FA differs from E2FB in many respects: (1) E2FA is most abundant in S-phase cells; (2) when overexpressed, it can promote cell proliferation in meristematic cells, whereas in cells that have lost cell division competence, E2FA overexpression supports a modified cell cycle with repeated S-phases, called endoreduplication; and (3) the association of E2FA with RBR is not disrupted, but rather enhanced, when cell proliferation is induced by excess Suc or overexpression of CYCD3;1 (De Veylder et al., 2002; Magyar et al., 2012). Furthermore, E2FA function in endoreduplication does not rely on promoting the transcription of S-phase genes through the transactivation domain, but rather on the ability of E2FA to associate with RBR and to repress genes regulating the entry into endoreduplication and cell differentiation (Magyar et al., 2012). Therefore, it was suggested that E2FA in association with RBR plays a role in maintaining cell proliferation competence in meristems. In addition, E2FA was shown to play roles in maintaining genome integrity and viability in meristematic cells (Horvath et al., 2017).
E2FA and E2FB appear to be redundantly required for cell proliferation, because no viable plants can be generated when predictably null mutants are combined (Li et al., 2017). However, a viable double e2fab mutant plant was generated by combining different loss-of-function mutant alleles for E2FA (e2fa-2) and E2FB (e2fb-1; Heyman et al., 2011), suggesting that at least the C-terminal transactivation function of these E2Fs are dispensable for plant growth and development.
That E2FC is a repressor function is supported by its overexpression suppressing meristematic cell divisions and expression of mitotic CYCB1;1 and its silencing leading to the upregulation of both S-phase-associated HISTONE 4 (H4) and CELL DIVISION CYCLE 6 (CDC6) and the mitotic CYCB1;1 genes (del Pozo et al., 2006). In mammalian cells, the DP, RB-like, E2F4, and Multivulval class B (MuvB) multiprotein complex, known as DREAM, acts as a repressor on cell cycle genes to impose quiescence (Sadasivam and DeCaprio, 2013). In Arabidopsis, E2FC, RBR, and MYB3R3 (a repressor-type MYB3R or Rep-MYB3R) are part of the DREAM complex with a repressive function that establishes quiescence (Kobayashi et al., 2015b). However, unique to plants is that the activator-type E2FB partners the mitosis-specific activator MYB3R4 (an Act-MYB3R) in another DREAM complex (Kobayashi et al., 2015a, 2015b; Harashima and Sugimoto, 2016). This provides additional support for the mitotic role of E2FB.
The leaf is an excellent model to study how the coordinated action between cell proliferation and differentiation is regulated (Andriankaja et al., 2012; Kalve et al., 2014). The leaf has a determinate size, and its growth is the result of two partially overlapping processes: the initial cell proliferation followed by cell expansion, which occurs as cells permanently exit the cell cycle. Cell division is differently regulated in distinct cell populations within the leaf epidermis. The meristematic protodermal cells go through formative cell divisions with a cell proliferation front progressively restricted to the base of the leaf during development. When epidermal leaf cells exit mitosis, they become lobed and enlarged in size, which is coupled with an increase in ploidy through a switch from the mitotic cell cycle to the endoreduplication program (De Veylder et al., 2011). A substantial bulk of pavement cells originate from stomata meristemoids interspersed along the leaf surface, forming a stem cell population that goes through several rounds of asymmetric divisions to produce cells that differentiate into either pavement cells or stomata (Andriankaja et al., 2012). The identity of these meristemoid cells is determined by a set of key regulators, such as SCPEECHLESS, but can also be visually recognized by their characteristic round or square shape and a small size of cells below the stomata guard cells, specifically <100 μm2 (Dong et al., 2009). The temporal and spatial regulation of the cell cycle arrest front in the cell populations originating from protodermal cells or meristemoids are different, but the underlying molecular mechanisms are hitherto unknown (White, 2006).
We investigated how E2FB, which is considered to be an activator of cell proliferation, is regulated by RBR interaction to underpin cell proliferation, the exit to differentiation, and establishment of quiescence during leaf development. In combination, our biochemical and genetic analyses suggest that E2FB regulates organ development as a corepressor complex with RBR.
RESULTS
Elevated E2FB Level Inhibits Cell Proliferation in Association with RBR at Early Stages of Leaf Development, Whereas It Perturbs the Establishment of Quiescence at Later Leaf Developmental Stages When RBR Levels Decline
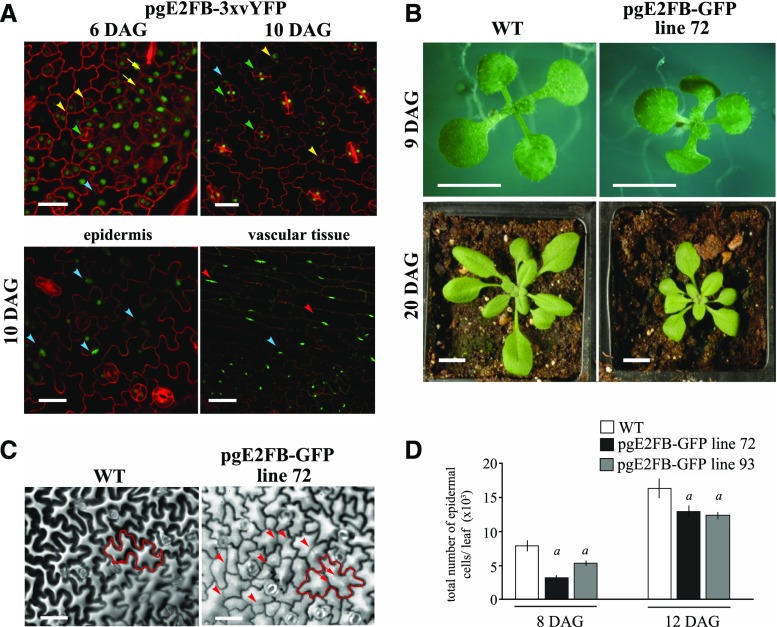
To follow E2FB protein level in its native context during leaf development, we generated Arabidopsis plants carrying the genomic region of E2FB under the control of its own promoter and tagged its C terminus with 3× Venus yellow fluorescent protein (YFP), a modified YFP (pgE2FB-3×vYFP). In young leaves at 6 d after germination (6 DAG), the E2FB-3×vYFP signal was detected in the nuclei both in the proliferating protodermal and meristemoid cells (Fig. 1A, 6 DAG). Interestingly, at a later stage of leaf development, the E2FB-3×vYFP remained present in fully developed stomata as well as in lobed differentiated pavement cells and vascular cells with characteristic elongated nuclei close to the cell wall (Fig. 1A, 10 DAG). By comparing the E2FB-3×vYFP distribution with the localization of E2FA-3×vYFP and RBR-GFP (Magyar et al., 2012), we found that the E2FA-3×vYFP was largely restricted to proliferating epidermal cells and was not detectable in fully differentiated stomata (Supplemental Fig. S1, A and B). The RBR-GFP signal was present in the meristemoids and in the proliferating cells, and also in the differentiated pavement cells (Supplemental Fig. S1C). RBR was also detectable in fully differentiated stomata, although at a lower level (Matos et al., 2014).
Figure 1.
Elevated E2FB level in its own expression domain inhibits cell proliferation in young leaves and disturbs quiescence in older leaves. A, Representative confocal laser scanning microscopy images of the abaxial leaf surface from the first leaf pair of the transgenic line pgE2FB-3×vYFP at 6 and 10 DAG (top), and localization in the epidermis and vascular tissues of the same transgenic line at 10 DAG (bottom). The YFP signal (green) is counterstained for cell membrane with PI (red). Yellow arrows point toward dividing protodermal cells, yellow arrowheads indicate stomatal meristemoids, green arrowheads label fully developed stomata guard cells, blue arrowheads mark elongated pavement cells, and red arrowheads show elongated vascular cells with GFP signal in their nucleus. Scale bars = 20 μm (top) and 25 μm (bottom). B, Images of the wild type (WT) and the transgenic line with high E2FB expression (pgE2FB-GFP line 72) grown for 9 DAG in vitro and for 20 DAG on soil. Scale bars = 0.5 cm. C, Representative images of the abaxial epidermal cell layer of the first leaf pair from wild-type and pgE2FB-GFP line 72 seedlings (12 DAG) taken by differential interference contrast microscopy, for which the imprints were made by the gel casting method. An example of an elongated puzzle-formed pavement cell is outlined in red (left). Arrows indicate straight cell walls inside the cell, whereas arrowheads mark newly formed cell walls inside the elongated pavement cells. Scale bars = 20 μm. D, Quantification of the total number of epidermal cells from the first leaf pair of the wild type and two pgE2FB-GFP transgenic lines (lines 72 and 93). Values represent means and error bars indicate the sd. Significance was determined by Student’s t test; a, P < 0.05. n = 3 and n > 600. The quantifications of cellular parameters are summarized in Supplemental Tables S1 and S2 from 8 and 12 DAG leaves, respectively. Data information, n = biological repeat, n = samples per biological repeat, here and in following figure legends.
To reduce a possible effect of 3×vYFP on the protein function, we also generated transgenic Arabidopsis lines with a single GFP tag (pgE2FB-GFP), and showed that the localizations of both E2FA-GFP and E2FB-GFP were comparable to that observed for E2FA-3×vYFP and E2FB-3×vYFP in the different epidermal cell types (Supplemental Fig. S1, A–F). Although E2FB-GFP expression was driven by the E2FB regulatory region, different expression levels of E2FB-GFP were identified among the 36 independent transformants (low, medium, and high for pgE2FB-GFP lines 61, 93, and 72, respectively; Supplemental Fig. S2A). Despite the difference in the levels, temporal E2FB-GFP expression followed the same declining pattern with leaf development as endogenous E2FB in the wild-type control (Supplemental Fig. S2B). The GFP-tagged E2FB was functional in respect to its ability to interact with RBR as well as to dimerize with and stabilize DPA and DPB proteins (Supplemental Fig. S2, C–E). Its interaction with RBR protein was also regulated as expected; it did not associate with the phosphorylated RBR form (Supplemental Fig. S2C).
Plants of pgE2FB-GFP line 72, with high E2FB-GFP expression driven by the E2FB promoter, showed reduced growth habit compared to the wild type both at the seedling stage and as a full-grown plant. As illustrated in Figure 1B, the leaf area in pgE2FB-GFP line 72 was smaller than that in the wild type. To investigate the cellular basis underlying the growth retardation, we imaged the epidermal layer of the first leaf pair and quantified the leaf area, total cell number, stomata number, cell size, and cell shape at three equal sections of the base, middle, and tip (Supplemental Tables S1 and S2). We took samples from pgE2FB-GFP lines 72 and 93 at two developmental time points representing the young leaf with abundant cell proliferation (8 DAG) and the older leaf when the majority of cells undergo expansion growth (12 DAG; Fig. 1D; Supplemental Fig. S3). Surprisingly, this analysis revealed significantly fewer cells in pgE2FB-GFP lines 72 and 93 compared to the wild type at 8 DAG, whereas this difference became lower at 12 DAG (Fig. 1D; Supplemental Tables S1 and S2). In parallel, flow cytometry analysis of DNA content showed an accumulation of 2C nuclei in pgE2FB-GFP lines 72 and 93, representing the G1 phase at an early developmental stage (8 DAG) of the first leaf pair, in comparison to the wild type, which also indicates a block in cell proliferation (Supplemental Fig. S3C). We also observed a shift toward larger cell size in pgE2FB-GFP line 72 compared to the wild type at 8 DAG (Supplemental Fig. S3B; Supplemental Table S1). However, in spite of the enlarged cell size, the entry into endoreduplication was delayed in both pgE2FB-GFP lines 72 and 93 compared to the wild type, as shown by the reduced 8C nuclei in the first leaf pair at 12 and 15 DAG (Supplemental Fig. S3C). Ploidy level of the cotyledons was also behind that of the wild type in pgE2FB-GFP line 72, as indicated by the reduced 16C nuclei and the complete lack of 32C nuclear DNA content (Supplemental Fig. S3D). In agreement with this, the circularity index of epidermal cells was higher in pgE2FB-GFP lines than in the corresponding wild type, suggesting that cells with elevated E2FB level are more round and thus have delayed cell shape differentiation (Supplemental Tables S1 and S2).
At 12 DAG, the majority of wild-type epidermal cells exited the cell cycle, as indicated by their elongated and lobed outline. In pgE2FB-GFP line 72, we observed numerous straight and less pronounced cell walls in these puzzle-shaped pavement cells, especially in cells located further toward the leaf-tip area (Fig. 1C; Supplemental Fig. S3A; Supplemental Tables S1 and S2). The formation of a new division plane across the differentiated pavement cells was even more frequent and pronounced on the cotyledon surface of pgE2FB-GFP line 72 (Supplemental Fig. S3E). Some of these elongated pavement cells contained more than a single straight cell wall. Similar divisions of enlarged pavement cells have been previously reported in wild-type Arabidopsis leaves (Asl et al., 2011), but the frequency of these divisions was dramatically increased in pgE2FB-GFP line 72 (Supplemental Tables S1 and S2). In agreement, the proportion of middle-sized cells (≤300–1,000 μm2) was elevated at 12 DAG at the expense of the number of larger cells (3,000–6,000 μm2) in pgE2FB-GFP line 72 as compared to the wild type (Supplemental Fig. S3B).
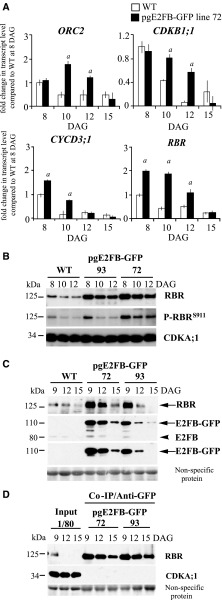
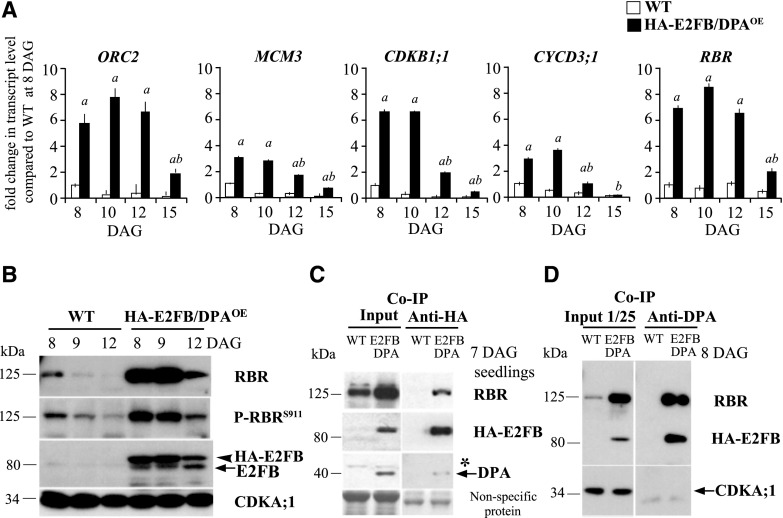
To gain insights into the molecular mechanism leading to the altered cell proliferation when E2FB level is elevated during leaf development, we first determined the expression levels of the S-phase-related ORIGIN RECOGNITION COMPLEX 2 (ORC2) and the mitotic CYCLIN-DEPENDENT KINASE B1;1 (CDKB1;1). In pgE2FB-GFP line 72, the expression levels of ORC2 and CDKB1;1 were comparable to that in the wild type in young leaves (Fig. 2A, 8 DAG). At 10 and 12 DAG, the expression of ORC2 and CDKB1;1 declined in the wild type, whereas expression of these genes in pgE2FB-GFP line 72 remained elevated (Fig. 2A). The transcription of CYCD3;1 and RBR also increased in pgE2FB-GFP line 72, most strikingly at the time point of 10 DAG, when expression of these genes in the wild type was significantly reduced (Fig. 2A). The sustained expression of these cell cycle genes correlated well with the division of enlarged pavement cells.
Figure 2.
RBR efficiently counteracts excess E2FB accumulation in proliferating, but not in differentiating, first leaf pairs. A, Relative expression levels of ORC2, CDKB1;1, CYCD3;1, and RBR in the wild type (WT) and pgE2FB-GFP line 72 from the developing first leaf pair of seedlings at 8, 10, 12, and 15 DAG. Values represent the mean of fold change normalized to the value of the relevant transcript of the wild type at 8 DAG, which was set arbitrarily at 1. Error bars indicate the sd. a, P < 0.05; statistical significance determined using Student’s t test between the wild type and the transgenic line at a given time point (n = 3, n > 50). Abbreviations of genes and primer sequences are listed in Supplemental Table S3. B, The phosphorylation level of RBR on the conserved Ser site at position 911 (P-RBRS911) was followed in the developing first leaf pairs of two independent pgE2FB-GFP-expressing lines (lines 93 and 72) with different E2FB protein levels and compared to the wild type at the indicated time points (DAG) using anti-RBR and P-RBRS911-specific antibody (anti-P-Rb807/811) in immunoblot analysis. C, To follow RBR accumulation in conjunction with E2FB level, anti-RBR, anti-E2FB, and anti-GFP antibodies were used in immunoblot analysis of proteins in the developing first leaf pairs in the same transgenic lines as in B. In the top set of blots, the antibody labels RBR (arrow); in the second set, the anti-E2FB antibody labels both the E2FB-GFP (arrow) and the endogenous E2FB (arrowhead); and in the third set, the anti-GFP antibody marks the accumulation of the E2FB-GFP fusion protein (arrow). D, Co-IP of RBR in the E2FB-GFP pull-down was labeled on the immunoblot with anti-RBR. On the same gel, 1/80 of the IP from the extract of the pgE2FB-GFP 72 line was loaded as input. For comparison, 1/20 of IP was loaded for all genotypes in C. Nonspecific membrane-bound proteins stained by Coomassie-blue were used as loading controls (C and D). Note: The relative intensities of the protein bands in B and C are quantified in Supplemental Figure S4, A and B (B) and C and D (C), and the measurements related to proteins in C and D are quantified in Supplemental Figure S4E.
To understand how E2FB activity is regulated during leaf development, we studied both RBR and its phosphorylation level and the interaction between E2FB and RBR. For this, we utilized the human phosphospecific RbS807/811 antibody that has been shown to recognize the conserved phosphorylation site of RBR proteins in multiple plant species, specifically at the 911 Ser position in Arabidopsis (P-RBRS911; Abraham et al., 2011; Magyar et al., 2012; Wang et al., 2014). In the wild type, both RBR and E2FB protein levels, as well as RBR phosphorylation, were highest at the early stage of leaf development (8 DAG) and displayed a gradual decline afterward when cells exited proliferation (10–12 DAG; Fig. 2, B and C). By comparing RBR protein and phosphorylation levels in pgE2FB-GFP lines 93 and 72 to that in the wild type, we observed clear differences in their kinetics (Fig. 2B). The endogenous RBR level was highly elevated throughout the studied developmental stages in both pgE2FB-GFP transgenic lines, indicating a regulatory loop to counteract the excess E2FB level (Fig. 2, B and C). However, whereas RBR phosphorylation remained high at all studied time points in pgE2FB-GFP line 72, it declined in pgE2FB-GFP line 93 to a level similar to the wild-type level, indicating that RBR is more active as a repressor in pgE2FB-GFP line 93 than in pgE2FB-GFP line 72 (Fig. 2B; quantification in Supplemental Fig. S4, A and B). In agreement, in differentiated epidermal cells at 12 DAG, a considerably greater number of divisions were observed in pgE2FB-GFP line 72 than in pgE2FB-GFP line 93 (Supplemental Table S2).
Next, we compared complex formation between E2FB-GFP and RBR proteins in pgE2FB-GFP lines 93 and 72 (for inputs and co-IP, see Fig. 2, C and D, respectively). Immunoprecipitation (IP) of E2FB-GFP showed that the majority of RBR protein was in complex with E2FB-GFP fusion protein throughout leaf development and that the E2FB-RBR complex was the most abundant in young leaves of both pgE2FB-GFP lines, providing an explanation as to why cell number was decreased in the leaves of these lines (Fig. 2D). The level of E2FB and RBR proteins decreased as leaf development progressed, much more in pgE2FB-GFP line 93 than in line 72 (Fig. 2C; quantification in Supplemental Fig. S4, C and D), whereas the level of E2FB-associated RBR was comparable between the pgE2FB-GFP lines (Fig. 2D; for quantification, see Supplemental Fig. S4E). Based on these data, we concluded that more RBR-bound E2FB-GFP is present in pgE2FB-GFP line 93 than in line 72, whereas RBR-free E2FB might be more prevalent in pgE2FB-GFP line 72 and consequently could promote cell proliferation in lobed differentiated leaf pavement cells.
In summary, in young leaves, the elevated E2FB level together with the abundant presence of RBR represses rather than activates cell proliferation. The cellular and molecular data indicate that excess E2FB can only be liberated from RBR repression at later developmental stages when their levels decline, which leads to extra cell divisions in lobed pavement cells.
The e2fb Mutant Has Increased Number of Cells in Developing Leaves
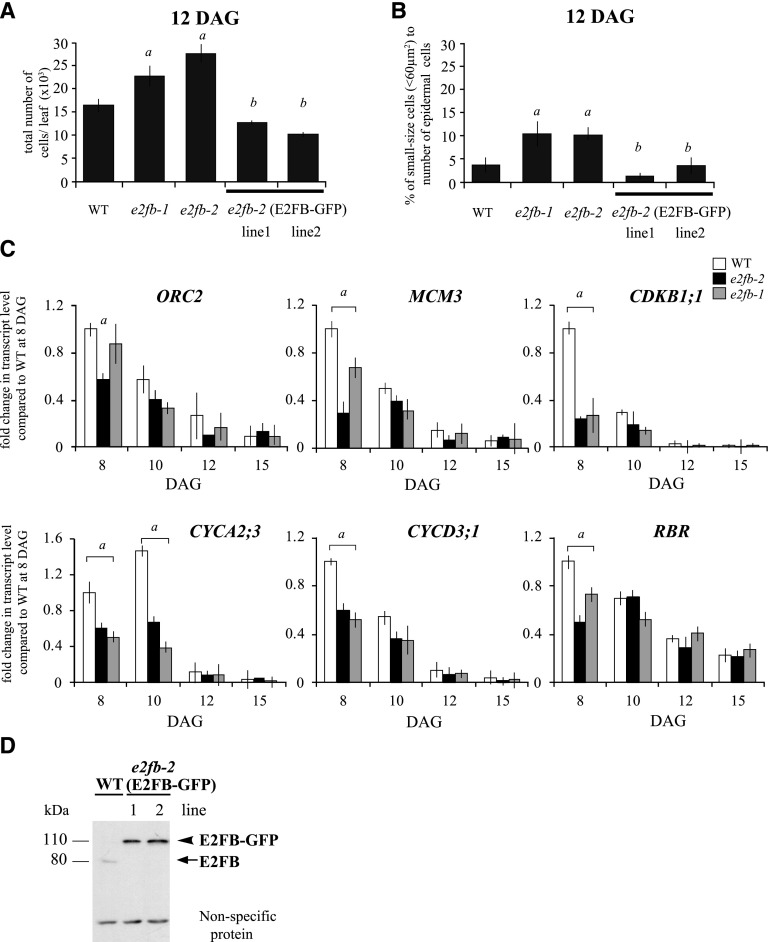
To investigate the effect of E2FB loss of function during leaf development, we analyzed two e2fb T-DNA insertion mutant alleles, e2fb-1 (SALK_103138) and e2fb-2 (SALK_120959; Berckmans et al., 2011a; Kobayashi et al., 2015a, 2015b). The T-DNA insertions in these mutants are located just behind and within the E2FB dimerization domain, respectively (Supplemental Fig. S5A). Based on the position of the T-DNA insertion, it is likely that e2fb-2 is a null mutant, as it lacks the dimerization domain required to form a complex with DP proteins, which is a prerequisite for E2Fs to bind to target promoters. Although no full-length E2FB protein could be detected in either of these mutants (Supplemental Fig. S5B; for e2fb-2, see Berckmans et al., 2011a), the size and morphology of both e2fb-1 and e2fb-2 seedlings were largely comparable to those of wild-type seedlings; however, the area of the first leaf pair was moderately, but significantly, larger than that in the wild type at 8 and 12 DAG (Supplemental Fig. S5C; Supplemental Tables S1 and S2). In young leaves (8 DAG), the cell number in e2fb mutants was comparable to that in the wild type, but cells were found to be enlarged in size (Supplemental Table S1). Flow cytometry analysis revealed that some e2fb mutant leaf cells entered prematurely into the endoreduplication cycle (Supplemental Fig. S5D), thus suggesting that certain cells exit cell proliferation earlier. By contrast, at the later developmental stage of 12 DAG, the number of leaf epidermal cells in both e2fb mutants was significantly increased in comparison to the wild type (Fig. 3A; Supplemental Tables S1 and S2). By introducing pgE2FB-GFP into the e2fb-2 mutant background, we could restore the e2fb leaf epidermal cell number to close to that of the wild type, providing evidence of functional complementation (Fig. 3A; Supplemental Tables S1 and S2).
Figure 3.
E2FB restricts cell proliferation in developing first leaf pairs. A and B, Total cell number (A) and ratio of small-sized cells (<60 μm2; B) in the epidermis of the first leaf pairs from the wild type (WT), the e2fb-1 and e2fb-2 mutants, and the e2fb-2 mutant expressing E2FB-GFP under its own promoter (e2fb-2 E2FB-GFP lines 1 and 2) at 12 DAG (n = 3, n > 600). Error bars indicate the sd. a, P < 0.05, statistical significance determined using Student’s t test between the wild type and the two e2fb mutants; b, P < 0.05, statistical significance between the complemented lines and e2fb mutants. C, Comparison of the ORC2, MCM3, CDKB1;1, CYCA2;3, CYCD3;1, and RBR transcript levels in the first leaf pairs of seedlings of the e2fb-2 and e2fb-1 mutants and the wild type at 8, 10, 12, and 15 DAG. Values represent the mean of fold change normalized to the value of the relevant transcript of the wild type at 8 DAG, which was arbitrarily set at 1 (n = 3, n > 50). a, P < 0.05, statistical significance determined using Student’s t test between the wild type and the mutant lines. Error bars indicate the sd. Abbreviations of genes and primer sequences are listed in Supplemental Table S3. D, Endogenous E2FB and transgenic E2FB-GFP proteins were detected in 1-week-old seedlings from the wild type and the two complemented lines [e2fb-2 (E2FB-GFP) lines 1 and 2]. The arrow indicates the position of E2FB, and the arrowhead indicates E2FB-GFP. Nonspecific, cross-reacting proteins are used as loading control.
It is known that cells with meristemoid identity have a characteristic round or square shape and a small cell size (<100 μm2) below the stomata guard cells (Dong et al., 2009). We measured these cell types on the leaf epidermis at 12 DAG and found them to be distributed below 60 μm2. To reveal whether the increased cell number may result from overproliferation of meristemoids, we counted cells smaller than 60 μm2. We indeed found a much larger increase in both e2fb mutants within this cell population (Fig. 3B). In agreement, the total number of fully developed stomata also increased in the e2fb mutant lines (Supplemental Tables S1 and S2). These phenotypes were also complemented by expressing E2FB-GFP in the e2fb-2 mutant (Fig. 3B), indicating that E2FB represses the proliferation of leaf meristemoid cells. The E2FB-GFP protein accumulated to a much higher level in the pgE2FB-GFP-complemented e2fb-2 lines than did endogenous E2FB protein in the wild type, which explains why there was overcompensation (Fig. 3D).
To study the impact of e2fb mutation on the expression of E2F target genes, we selected the S-phase-specific genes ORC2 and MINICHROMOSOME MAINTENANCE COMPLEX COMPONENT 3 (MCM3), the mitotic CDKB1;1 and CYCLIN A2;3 (CYCA2;3), and the two mitosis upstream regulators CYCD3;1 and RBR. The expression levels of all these genes were reduced in the e2fb mutants, especially in young leaves (8 DAG). The reduction was stronger in the null-mutant e2fb-2 than in e2fb-1 (Fig. 3C). We also investigated how the expression levels of the other two E2F genes were affected in the e2fb mutants. The expression of E2FA did not change, whereas the E2FC transcript level showed a slight elevation from 10 DAG onward (Supplemental Fig. S5E).
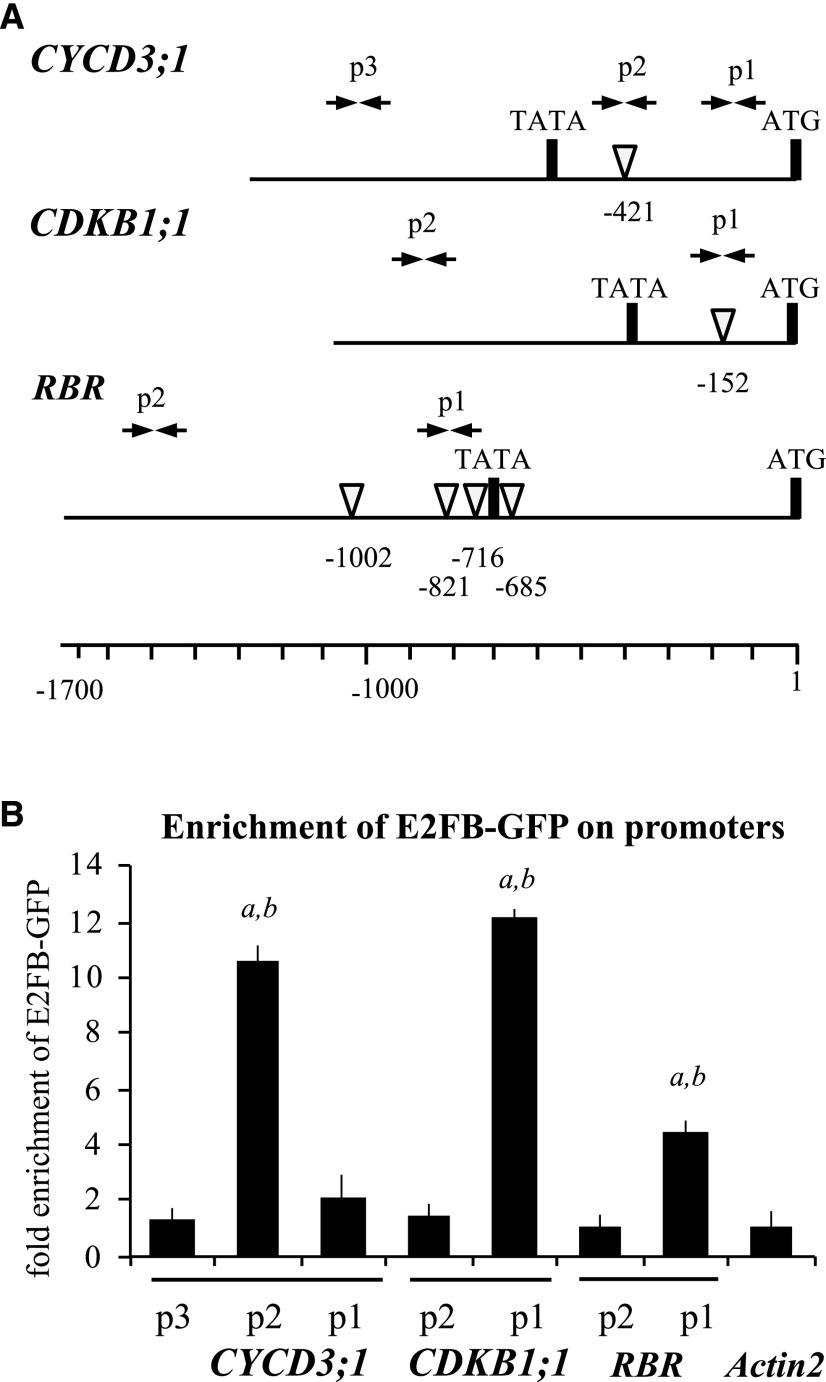
To gather further evidence that the mitotic CDKB1;1, CYCD3;1, and RBR genes are directly regulated through the binding of E2FB to their promoters, we performed chromatin IP (ChIP) experiments using the e2fb-2 mutant complemented with the pgE2FB-GFP construct. There was a significant enrichment of E2FB-GFP protein at the promoter of these genes, specifically in the regions where consensus E2F binding elements were predicted (Fig. 4).
Figure 4.
E2FB directly binds to CYCD3;1, CDKB1;1, and RBR promoters. A, Schematic representation of the CYCD3;1, CDKB1;1, and RBR promoters; arrow pairs labeled p1, p2, and p3 indicate the positions of the primer pairs used for qPCR analysis. The position of the canonical E2F elements (white arrowheads) and their distance from the start codon (ATG) are depicted. Primer sequences are listed in Supplemental Table S3. B, ChIP followed by qPCR was carried out on chromatin isolated from complemented e2fb-2 E2FB-GFP seedlings (7 DAG) using polyclonal antirabbit GFP antibody; the graph shows fold enrichment calculated as the ratio of chromatin bound to the numbered section of the CYCD3;1, CDKB1;1, and RBR promoters with or without antibody. Shown is a representative experiment with three biological replicates. a and b, P < 0.01, statistically significant enrichment between the relevant fragment and the neighboring fragments (a) and between the relevant regulatory region and the negative control (Actin2; b) determined by Student’s t test. The values represent the means of three technical replicates. Error bars indicate the sd. The enrichment on the Actin2 promoter was arbitrarily set to 1. The labels p1, p2, and p3 on the x axis refer to the regions indicated in A.
These results show that whereas E2FB is required for the full activation of cell cycle target genes at early stages of leaf development, its absence does not result in compromised cell proliferation. On the contrary, E2FB has a prevalent importance in inhibiting cell proliferation, though at a later leaf developmental stage. This effect is most pronounced in cells with a small size that likely belong to the stomata meristemoid lineage.
Co-Overexpression of E2FB with DPA Does Not Lead to Hyperproliferation in Developing Leaves
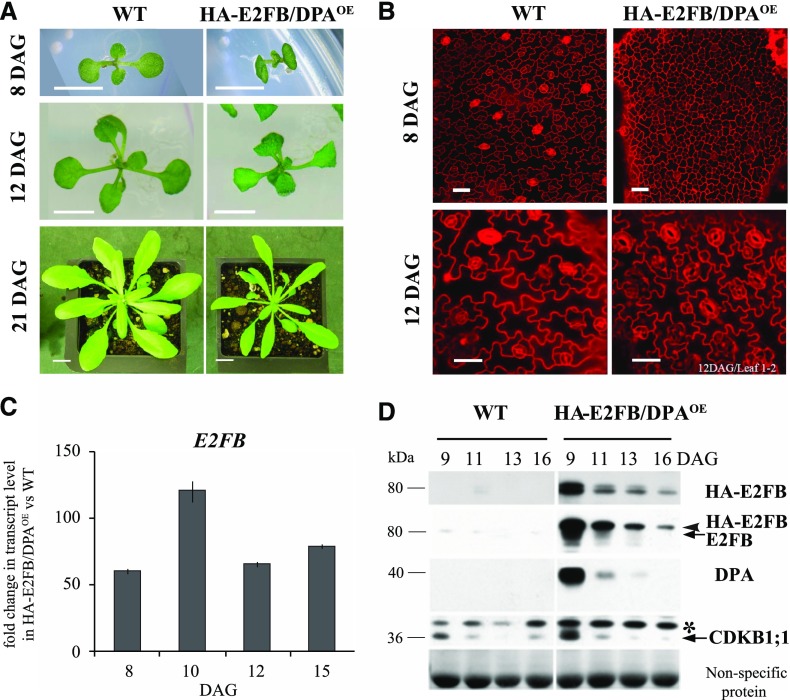
Co-overexpression of E2FB, but not E2FA, with DPA was shown to overcome the requirement of the phytohormone auxin to promote cell proliferation in cultured BY2 tobacco cells (Magyar et al., 2005). In animals, the expression of activator E2Fs is increased in most cancer types and thought to be responsible for uncontrolled cancerous cell proliferation (Chen et al., 2009). To determine whether such overexpression causes cell overproliferation in plants, we studied the Arabidopsis line p35S::HA-E2FB/DPA (E2FB/DPAOE), which overexpresses both E2FB and DPA (De Veylder et al., 2002; Magyar et al., 2012; Horvath et al., 2017). In contrast to the expected deregulation of cell proliferation and disruption of plant development, we did not observe tumorous growth. Leaf initiation proceeded normally; however, E2FB/DPAOE seedlings were smaller, and the total leaf area was reduced to half that of the wild type (Fig. 5A).
Figure 5.
Co-overexpression of E2FB and DPA results in reduced leaf and cell size. A, Representative images of wild-type (WT) and p35S::HA-E2FB/DPAOE (HA-E2FB/DPAOE) seedlings grown in vitro (8 and 12 DAG) and on soil (21 DAG). Scale bars = 0.5 cm at 8 and 12 DAG and 1 cm at 21 DAG. B, Representative confocal microscopy images of PI-stained abaxial leaf surfaces taken from the tip to the base of the first leaf pairs from wild-type and HA-E2FB/DPAOE seedlings (8 and 12 DAG). Scale bars = 20 μm. C, Comparison of E2FB expression levels in the developing first leaf pairs of HA-E2FB/DPAOE and wild-type seedlings at 8, 10, 12, and 15 DAG, where the expression of E2FB was set arbitrarily at 1 at each time point. Values represent fold change. Error bars indicate the sd, referring to technical repeats. The data are from one biological replicate (n < 50), and the transcript level correlates well with the HA-E2FB protein accumulation illustrated in D. D, Detection of protein levels of epitope-tagged (HA-E2FB) and endogenous E2FB, DPA, and CDKB1;1 in the first leaf pairs of wild-type and HA-E2FB/DPAOE seedlings at the indicated time points (DAG) using anti-HA, anti-E2FB, anti-DPA, and anti-CDKB1;1 antibodies. The arrowhead indicates the position of HA-tagged E2FB, whereas arrows indicate endogenous E2FB and CDKB1;1 proteins. The asterisk indicates a nonspecific protein cross reaction with the anti-CDKB1;1 antibody. Nonspecific membrane-bound proteins stained by Coomassie-blue were used as loading control.
To study the cellular basis behind the retarded leaf growth, we imaged the epidermal cell layer of the E2FB/DPAOE line at 8 and 12 DAG (Fig. 5B) and measured cell parameters (Supplemental Tables S1 and S2). At 8 DAG, we observed predominantly small-sized and polygonal shaped cells across the entire leaf surface, but the total calculated cell number was less than in the wild type (Fig. 5B; Supplemental Fig. S6, A and D), indicating that both cell proliferation and cell enlargement are inhibited at early stages of leaf development by the overexpression of E2FB together with DPA. By contrast, at 12 DAG, the calculated leaf epidermal cell number of E2FB/DPAOE was comparable to that of the wild type, whereas cell size remained smaller (Fig. 5B, 12 DAG; Supplemental Fig. S6D; Supplemental Table S2), suggesting that the transition from proliferation to cell elongation is delayed. The reduced stomatal index and the less complex shape of pavement cells (circularity index) at both time points also indicated an inhibition of stomata as well as pavement cell differentiation (Supplemental Tables S1 and S2). E2FB/DPAOE seedlings also displayed down-curling cotyledons (Fig. 5A). In wild-type cotyledons at 6 DAG, cell proliferation ceases and all pavement and stomata cells appear differentiated. By contrast, there were a large number of small cells in the cotyledons of E2FB/DPAOE seedlings (Supplemental Fig. S6B).
In E2FB/DPAOE seedlings, the level of E2FB expression increased from 50- to 100-fold that of the wild-type level throughout leaf development (Fig. 5C). By contrast, the accumulation of E2FB protein did not match the constitutive overexpression of the E2FB transcript; its level was highly elevated at the earliest time point only (9 DAG) and showed diminished accumulation, reaching levels comparable to that of the endogenous E2FB protein at later time points (Fig. 5D). The DPA protein level showed the same kinetics as E2FB (Fig. 5D), suggesting their developmental coregulation at the protein level. The level of the mitotic CDKB1;1 protein was also high in young leaves but diminished toward the 16 DAG time point (Fig. 5D). Coregulation of E2FB and DPA protein with the same kinetics was also observed in cotyledons (Supplemental Fig. S6C).
Surprisingly, there was no excess cell proliferation in the E2FB/DPAOE line, so we looked to see whether there was any deregulation of E2F target genes in this line. We analyzed the expression of two S-phase-specific genes, ORC2 and MCM3, and the mitotic CDKB1;1 (Fig. 6A). These E2F target genes were greatly upregulated throughout leaf development in the E2FB/DPAOE line, although they declined in parallel with the diminishing E2FB and DPA protein levels as leaf development progressed (Figs. 5 D and 6, A and B). Two other cell cycle genes were tested, namely the CDK inhibitor KIP-RELATED PROTEIN 4 (KRP4) and CYCLINA3;1 (CYCA3;1). These genes were also upregulated, but not to the same extent, and their upregulated expression was not observed at every time point (Supplemental Fig. S6E). Expression of the upstream positive and negative regulators of E2FB, CYCD3;1, and RBR was also upregulated in the E2FB/DPAOE line (Fig. 6A), indicating the presence of a regulatory feedback loop. In accordance, we also found an elevated RBR protein level and RBR phosphorylation (P-RBRS911) in E2FB/DPAOE leaves compared to the wild type (Fig. 6B; for quantification, see Supplemental Fig. S6, F and G). RBR was also strongly upregulated in E2FB/DPAOE cotyledons (Supplemental Fig. S6C).
Figure 6.
Ectopic E2FB/DPA functions as transcriptional activator on cell cycle genes. A, The expression levels of ORC2, MCM3, CDKB1;1, CYCD3;1, and RBR were determined in wild-type (WT) and HA-E2FB/DPAOE seedlings by RT-qPCR. Developing first leaf pairs were analyzed at each time point, as indicated. Values represent the mean of fold change normalized to values of the relevant transcript from the wild type at 8 DAG, which was set arbitrarily at 1. Error bars indicate the sd; a, P < 0.05, statistical significance between the wild type and the transgenic line at a given time point; b, P < 0.05, significance between two consecutive time points determined using Student’s t test (n = 3, n > 100). Abbreviations of genes and the list of primers used in this study are listed in Supplemental Table S3. B, Protein level of RBR, P-RBRS911, HA-E2FB, and endogenous E2FB in the developing first leaf pairs of wild-type and HA-E2FB/DPAOE seedlings at 8, 9, and 12 DAG detected using anti-RBR, anti-P-RBRS911 (anti-P-Rb807/811), anti-E2FB, and anti-CDKA;1 antibodies in immunoblot assays. Note, the relative intensities of the RBR and P-RBRS911 protein bands are quantified in Supplemental Figure S6, F and G. C and D, Co-IP of HA-E2FB with RBR and DPA proteins in wild-type and HA-E2FB/DPAOE seedlings at 7 DAG (C) and in first leaf pairs at 8 DAG (D). Co-IP of RBR or HA-E2FB proteins with DPA was determined through immunoblot analysis with anti-RBR or anti-E2FB antibodies. One twenty-fifth of the IP from the extract was loaded as input. The asterisk indicates a nonspecific protein cross-reaction with the anti-DPA antibody in the input. In B and D, anti-CDKA;1 antibody was used as control. In C, nonspecific membrane-bound proteins stained by Coomassie-blue were used as loading control. The arrowhead in B indicates HA-E2FB and arrows mark the positions of endogenous E2FB, DPA, and CDKA;1 in B–D, respectively.
To explore how the overexpression of E2FB/DPA and the consequent change in RBR level and its phosphorylation affected the amount of RBR-associated E2FB, we performed co-IP experiments (Fig. 6, C and D). Utilizing the hemagglutinin (HA) tag on E2FB in the E2FB/DPAOE line, we immunoprecipitated HA-E2FB from seedlings (7 DAG). As Figure 6C shows, only a relatively small amount of DPA was associated with HA-E2FB, and RBR was also not enriched in the complex. However, using the DPA antibody in young leaves (8 DAG), we detected a higher level of immunoprecipitated E2FB, as well as RBR, compared to levels observed in seedlings (Fig. 6, C and D). This shows that RBR effectively binds to the overexpressed E2FB-DPA heterodimer in young leaves, which explains the repression of cell proliferation. However, in some cells, or at some cell cycle stages, active RBR-free E2FB-DPA heterodimer must also be present to cause the high upregulation of E2F target genes.
RBR Recruitment through E2FB Is Important to Halt Cell Proliferation in Developing Leaves
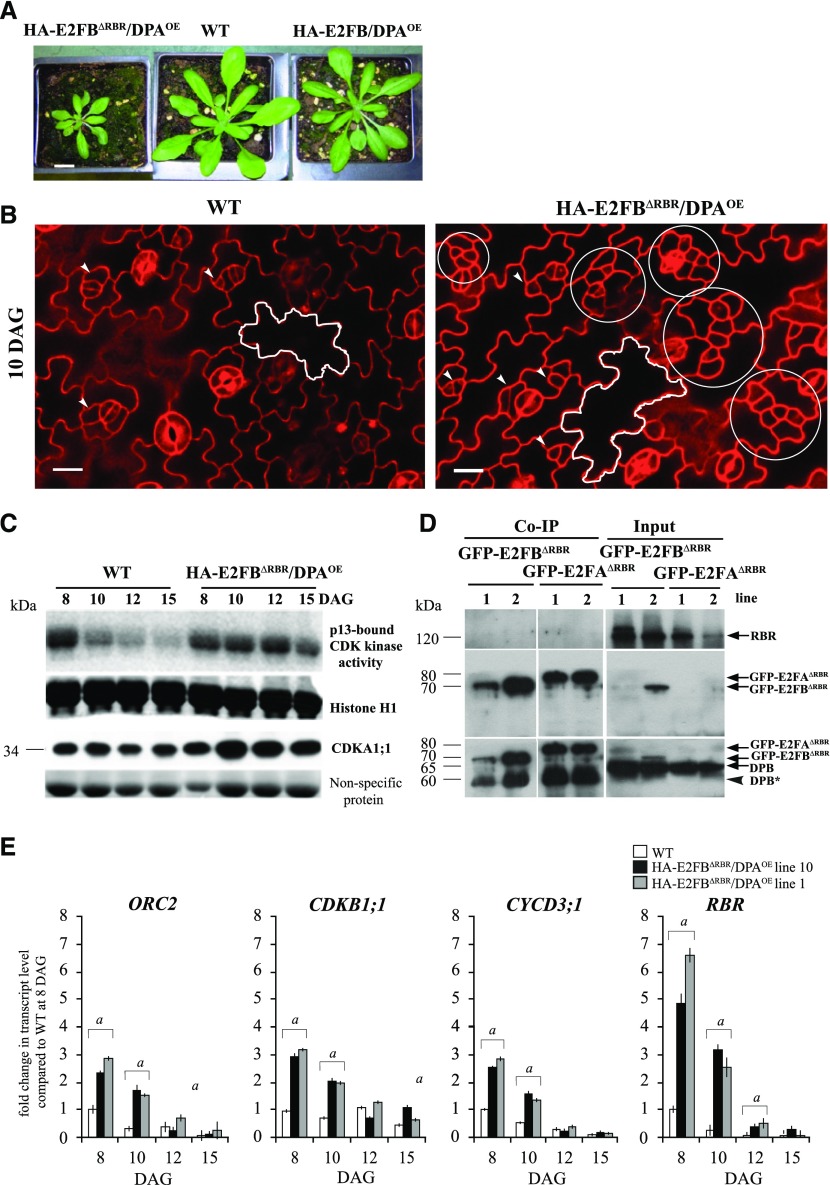
To address how the function of E2FB is dependent on its ability to bind RBR, we constructed a truncated E2FB where we deleted the C-terminal 84-amino acid region containing the conserved RBR-binding and overlapping transactivation domains, as we previously did for E2FA (Magyar et al., 2012), and we co-overexpressed this HA-tagged E2FBΔRBR with DPA (Supplemental Fig. S7A), as we did for the full-length E2FA earlier. Two independent HA-E2FBΔRBR/DPA lines (1 and 10) showed identical developmental abnormalities; their growth was arrested both in vitro and on soil (Fig. 7A; Supplemental Fig. S7, B and C). With high frequency (10% to 15%), we observed abnormally developing seedlings that had three cotyledons and missing or fused organs, indicating abnormal embryo development (Supplemental Fig. S7B). In the HA-E2FBΔRBR/DPA line, we observed clusters of small cells on the leaf epidermis interspersed among large lobed pavement cells (Fig. 7B; Supplemental Fig. S8, A and F). Quantifying epidermal cell sizes over a developmental time series (8, 10, and 12 DAG; Supplemental Fig. S8B; Supplemental Tables S1 and S2) showed that the ratio of small-sized cells (≤300 μm2) diminished gradually in the wild type, but remained high in both independent HA-E2FBΔRBR/DPA lines. On the other hand, large cells (1,000–3,000 μm2) formed earlier in the HA-E2FBΔRBR/DPA lines than in the wild type, and at 8 DAG, the large cells were also more prominent in the middle and the tip region of the leaf (Supplemental Fig. S8C). In agreement, the total cell number in the leaf was also higher in the E2FBΔRBR/DPA lines compared to the wild type at the later developmental stage of 12 DAG (Supplemental Tables S1 and S2). To reveal the proportion of possible stomata meristemoids among the small cells that are prominent at the late leaf developmental stage of 12 DAG, we quantified the number of cells with size smaller than 60 μm2. This cell population showed an even larger increase, specifically >4-fold, in the HA-E2FBΔRBR/DPA lines compared to the wild type (Supplemental Fig. S8D).
Figure 7.
Coexpression of the mutant HA-E2FBΔRBR with DPA, which is unable to transactivate and bind to RBR, hyperactivates meristematic cell divisions in leaf epidermis. A, Representative images of p35S::HA-E2FBΔRBR/DPA (HA-E2FBΔRBR/DPA), wild-type (WT), and p35S::HA-E2FB/DPA (HA-E2FB/DPAOE) plants grown for 20 d on soil. Scale bar = 1 cm. B, Confocal laser scanning microscopy images of PI-stained abaxial leaf surfaces from the first leaf pairs of wild-type and HA-E2FBΔRBR/DPA seedlings at 10 DAG. The white outline shows a typical puzzle-shaped pavement cell. Arrowheads in both images indicate normally dividing meristemoid cells, whereas white circles illustrate clusters of overproliferated meristemoid cells. Scale bars = 20 μm. C, Total CDK histone H1 kinase activity purified by p13suc1-Sepharose beads is shown and compared to Histone H1 from the first leaf pairs at four different developmental time points (8, 10, 12, and 15 DAG). For comparison, the CDKA;1 protein level is also shown in the same leaf samples. Coomassie-stained nonspecific membrane-bound proteins in the range 50–60 kD were used as loading controls. D, Co-IP of RBR and DPB proteins in the GFP-E2FB∆RBR and GFP-E2FA∆RBR pull-down was labeled with anti-RBR and anti-DPB antibodies. On the same gel, 1/12 of the IP from the extracts of the GFP-E2FB∆RBR and GFP-E2FA∆RBR lines was loaded as input. Arrows point toward the specific proteins. The arrowhead indicates a faster-migrating DPB protein. Molecular weight markers are indicated on the left. E, Expression levels of ORC2, CDKB1;1, CYCD3;1, and RBR were followed in two independent HA-E2FBΔRBR/DPA lines (lines 10 and 1) using RT-qPCR. The developing first leaf pairs were analyzed at each time point, as indicated. Values represent the fold change normalized to values of the relevant transcript from the wild type at 8 DAG, which was set arbitrarily at 1. As the two independent lines show the same tendencies, here, n = 2, n > 50. a, P < 0.05, statistical significance between the wild type and the transgenic line at a given time point determined using Student’s t test.
To reveal whether cell size relates to ploidy changes, we measured the DNA content in the first leaf pairs of HA-E2FBΔRBR/DPA, but found no difference compared to the wild type (Supplemental Fig. S8E). Thus, the observed phenotypes of HA-E2FBΔRBR/DPA lines were markedly different from what was observed previously for the HA-E2FAΔRBR/DPA line, which showed a dramatically elevated extent of endoreduplication (Magyar et al., 2012).
To gather molecular evidence behind the sustained proliferation in the cell clusters observed in the HA-E2FBΔRBR/DPA line, we determined CDK activity using p13Suc1 affinity chromatography that pulls down both A- and B-type CDKs (Magyar et al., 2005). As expected, CDK activity declined in the wild type, whereas it remained high throughout leaf development in the HA-E2FBΔRBR/DPA line (Fig. 7C), further supporting the persistence of cell proliferation in this line. To demonstrate that the C-terminally truncated E2FB cannot bind RBR, we utilized transgenic lines where we tagged the N-termini of both E2FA and E2FB deletion constructs with GFP for efficient pull down (Fig. 7D; see details in “Materials and Methods”). By using these transgenic lines in co-IP experiments, we confirmed that neither E2FA nor E2FB could pull RBR down in the absence of the C-terminal RBR-binding domain, but both associated with the DPB protein (Fig. 7D).
We also determined the expression of cell cycle E2F target genes (ORC2, CDKB1;1, CYCD3;1, and RBR) in both HA-E2FBΔRBR/DPA lines during leaf development (Fig. 7E). The transcript levels of all examined genes were upregulated at 8 and 10 DAG compared to the wild type (Fig. 7E). Since HA-E2FBΔRBR lacks the transactivation domain, this upregulation is likely due to the lack of RBR repression on these genes.
In summary, whereas the deletion of the RBR-binding domain in the HA-E2FAΔRBR/DPA lines leads to dramatic over-endoreduplication (Magyar et al., 2012), the same manipulation made to E2FB in HA-E2FBΔRBR/DPA lines results in overproliferation of cell clusters during leaf development.
DISCUSSION
Plant growth is centered on meristem activity, yet surprisingly little is known about how cell proliferation is regulated at the molecular level in a developmental context. E2F transcription factors are the prime candidates for regulating meristematic function in close association with RBR. Previously, we showed that E2FA in complex with RBR is involved in meristem maintenance (Magyar et al., 2012). E2FB was considered as a canonical transcriptional activator, and indeed we found that its overexpression can activate the expression of cell cycle genes, whereas e2fb mutations compromise expression of these same genes. However, the cell proliferation outcome does not follow these molecular changes in the developing leaves. On one hand, elevated or ectopic overexpression of E2FB (pgE2FB-GFP or p35S:HA-E2FB/DPA) causes a decrease in total cell number rather than an increase. On the other hand, the e2fb mutant lines produce more cells during leaf development in comparison to the wild-type control. Furthermore, we demonstrated both biochemically and genetically that the E2FB function of repressing cell proliferation relies on the RBR association, which is reinforced by autoregulatory loops.
In animal cells, Rb level and activity increase as cells exit proliferation and enter differentiation (Zacksenhaus et al., 1996). By contrast, RBR in plants is most abundant in meristematic cells, and its level diminishes as development proceeds (Borghi et al., 2010; Magyar et al., 2012). Thus, RBR coexpresses with E2FA and E2FB in proliferating plant cells and forms repressor complexes. Moreover, we found that elevated and ectopic overexpression of E2FB leads to increased RBR level. This autoregulatory loop enforces the repression, which ensures that cell proliferation is kept under control and thus that increased E2FB level does not lead to tumorous growth. RBR repression of cell proliferation through inhibiting E2FB is suppressed by RBR phosphorylation, and E2FB positively regulates the regulatory cyclin subunit (CYCD3;1) of the RBR-kinase (CDKA;1) as well. It is known that Rb phosphorylation and thus repressor activity is cell cycle regulated; dephosphorylated Rb is active in G1 phase and as cells pass through the G1/S control point the hyperphosphorylated Rb becomes inactive, leading to the expression of cell cycle genes (Morgan, 2007). It is feasible that in plants the elevated E2FB and consequent RBR levels in G1 lead to overabundance of E2FB-RBR repressor complex and thereby inhibition of cell proliferation, whereas after cells pass through the control point, when RBR becomes hyperphosphorylated, the overexpressed and now free E2FB hyperactivates cell cycle target genes. A block in cell proliferation is consistent with increased 2C DNA content when E2FB is elevated.
The protein levels of both E2FB and RBR decline as leaf development proceeds. During this transition phase from cell proliferation to differentiation, the E2FB-RBR complex is important for exiting cell proliferation and to establish quiescence. When E2FB escapes from RBR repression after the transition phase, differentiated cells reenter cell division, which is the case when E2FB level is elevated with expression driven by its own promoter. When E2FB is ectopically overexpressed together with DPA, these extra cell divisions of differentiated pavement cells were not present. Instead, cells are arrested in an undifferentiated state, as indicated by their small size without lobed shape and decreased number of stomata. This suggests that overexpression of E2FB together with DPA prevents the transition from proliferation to differentiation. Thus, the ectopic co-overexpression of E2FB with DPA or elevation of E2FB with expression driven by its own promoter have very different consequences. In the first case, a large amount of E2FB-DPA heterodimer is present that is still kept under the control of RBR to inhibit both cell proliferation and differentiation, leading to growth arrest. The destabilization of E2FB and DPA during leaf development may allow an escape mechanism from this block. By contrast, elevated E2FB with expression driven by its own promoter can form heterodimers with either the endogenously available DPA or DPB. It was suggested that the interaction of DPA with activator E2Fs stimulates nuclear translocation and mediates a higher level of transactivation than interaction with DPB (Kosugi and Ohashi, 2002). This might explain why there is less pronounced growth arrest and cells can exit proliferation when the E2FB level is elevated on its own.
By demonstrating that leaves produce fewer cells when E2FB is overexpressed and more cells when it is mutated, we show that E2FB is required and sufficient to restrain cell proliferation in developing leaves. We also show biochemically that E2FB has strong affinity to associate with RBR in young leaves enriched with proliferating cells. To provide further evidence that RBR acts through E2FB to inhibit cell proliferation, we deleted the C-terminal RBR binding domain of E2FB and overexpressed this mutant form with DPA. Indeed, we observed overproliferation of cells in developing leaves that strongly suggests that the formation of RBR-E2FB repressor complex is important for controlling cell proliferation during leaf development. Based on their small size and shape, proliferation in clusters, and the increased number of fully developed stomata at a later stage, the cell overproliferation is likely within the stomata meristemoid lineage, but this has to be confirmed by cell type-specific markers, such as the expression of SPEECHLESS. Because the C-terminal deletion on E2FB also removed the transactivation domain, the overproliferation of meristemoids must be a consequence of derepression from RBR control. The presence of meristemoid overproliferation in two independent e2fb mutants strongly suggests that this phenotype is E2FB specific.
RBR silencing was shown to upregulate the expression of TOO MANY MOUTH (TMM), the key regulator of stomata meristemoid divisions, leading to their overproliferation (Borghi et al., 2010). At later developmental stages in the stomata lineage, RBR silencing can also interfere with the division arrest of the fully developed guard cells (Borghi et al., 2010; Yang et al., 2014). We did not observe such phenotypes when the truncated E2FB was overexpressed, suggesting that RBR regulates these later steps in stomata differentiation not through E2FB association, but likely through binding and repression of other transcription factors, as shown in the case of FAMA (Xie et al., 2010). Interestingly, SOL1 and SOL2, two Arabidopsis homologs of LIN54, a component with DNA binding activity within the mammalian DREAM complex, were shown to regulate cell fate and division in the stomatal lineage (Simmons et al., 2019). Both SOL1 and SOL2 were found to be upregulated in the E2FA/DPA overexpression line, but only SOL2 was hyperactivated in RBR-silenced RBR-RNAi plants and has the consensus E2F-binding element in its promoter region (Borghi et al., 2010). Accordingly, the E2F-RBR pathway could regulate these transcription factors, but whether these DREAM-related components function in complex with E2Fs and RBR to control cell proliferation in the stomatal lineage is not yet known.
Using GFP-tagged constructs, we found important differences in the expression pattern of these two E2Fs; E2FA is largely restricted to proliferating cells, whereas E2FB and RBR are also present in differentiated pavement and fully developed stomata guard cells. The co-occurrence of E2FB, but not E2FA, with RBR in these differentiated cell types is consistent with the idea that E2FB with RBR is required to repress cell proliferation and impose quiescence to allow differentiation, whereas E2FA acts with RBR to maintain proliferation competence (Magyar et al., 2012). E2FA and E2FB are also distinctly regulated by RBR; excess Suc or overexpression of CYCD3;1 promotes E2FA-RBR interaction, whereas these factors disrupt E2FB-RBR interaction (Magyar et al., 2012). The distinct cellular phenotypes upon the overexpression of C-terminally truncated dominant-negative forms of E2FA or E2FB further underlines the difference in the mode of their action in relation to RBR-repression and transactivation of target genes. The overexpression of E2FAΔRBR resulted in over-endoreduplication due to the inability to repress the expression of endoreduplication genes (Magyar et al., 2012), whereas E2FBΔRBR overexpression had no effect on endoreduplication, but led to the early formation of large pavement cells and clusters of small cells. The fact that overexpression of both the full-length and truncated forms of E2FA and E2FB yields specific phenotypic outcomes suggest that they might have distinct sets of target genes. In agreement, overexpression of E2FA and E2FC also caused very different genes to be deregulated (de Jager et al., 2009).
The functional difference between E2FA and E2FB may rely on their interaction with distinct sets of proteins. As we previously showed, E2FB and E2FC can associate with proteins that are known to be conserved components of the so-called DREAM complex (Kobayashi et al., 2015b). By contrast, though E2FA can interact with RBR and DPs, none of the DREAM components were found in complex with E2FA (Horvath et al., 2017). Both E2FB and E2FC function as part of the DREAM complex to repress cell proliferation. However, our results suggest that E2FB acts at an earlier stage during the transition from proliferation to differentiation, as well as in the immediate establishment of quiescence, possibly as part of the activator MYB3R1-MYB3R4 complex (Kobayashi et al., 2015a, 2015b), whereas E2FC might be required at a later stage to permanently maintain cell cycle repression (del Pozo et al., 2006) as part of the repressor MYB3R1-MYB3R3-MYB3R5 complex (Kobayashi et al., 2015b).
Plants are remarkably resistant to cancerous transformation, but this ability is poorly understood (Doonan and Hunt, 1996). In animals, the activator E2Fs are found to be increased in most cancer types, and they contribute to uncontrolled proliferation (Chen et al., 2009). Here, we show that E2FB, the canonical activator E2F in Arabidopsis, could not drive cancerous divisions even when its level was elevated 50-fold. A potential reason why the large amount of E2FB does not activate tumorous growth is the direct activation of RBR by E2FB and the accumulation of RBR/E2FB repressor complex in proliferating cells. However, CYCD3;1 is also a direct target of E2FB, leading to increased RBR phosphorylation and inactivation of RBR repression. It is likely that the simultaneous activation of positive and negative upstream regulators to E2FB is important to keep cell proliferation under tight control in plant cells.
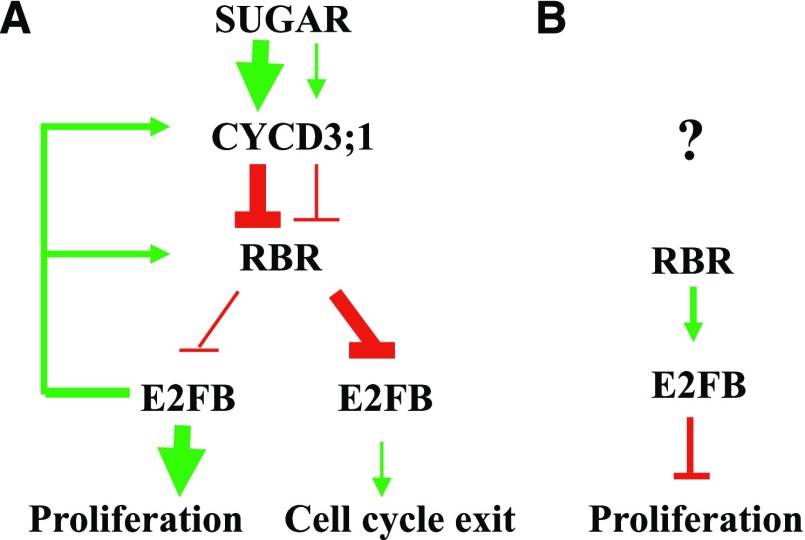
In summary, E2FB-RBR relays meristematic activities to differentiation through the regulation of (1) cell cycle transitions by transcriptional activation of cell cycle genes, (2) cell cycle exit and establishment of quiescence through the repression of cell cycle genes when associated with RBR, and (3) stem cell amplifying divisions through an active repression mechanism together with RBR (Fig. 8). Plant growth is fundamentally determined by the number of cells kept in proliferation in the meristem (Bögre et al., 2008). Meristem size is sensitively responsive to environmental conditions, and we suggest that the interconnected action of the three E2Fs plays a central role in meristem activities, thus providing an entry point to understand and manipulate the growth potential of plants and crops.
Figure 8.
Model explaining the functions of E2FB during leaf development. E2FB has three different activities, and each is dominant at different leaf developmental stages (A) or in different cell types (B). A, Activator E2FB is in its RBR-free form, characteristic of young leaves consisting of mostly proliferating cells. The young meristematic leaf is a nutrient-rich sink tissue, where E2FB is released from the repression of RBR by the CYCD3;1-regulated RBR kinase in a Suc-dependent manner. E2FB controls the activity of RBR by using CYCD3;1 activity to regulate RBR transcriptional and protein level, as well as phosphorylation status. In leaf cells where the growth-promoting signal is weakened, the protein levels of both E2FB and RBR decrease and RBR becomes more active (less phosphorylated) to bind and inhibit E2FB. This repression is important to establish quiescence in leaf cells committed to differentiation. B, In developing leaves, E2FB also forms a repressor complex with RBR in meristemoid leaf cells to corepress their divisions. How this repression is regulated by upstream signal(s) is hitherto unknown.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia wild-type and transgenic seeds were sterilized in commercial bleach, resuspended in sterile water, and cold treated at 4°C in darkness for 2 d (Clough and Bent, 1998). Unless otherwise stated, plants were grown under a 16-h light/8-h dark photoperiod at 22°C in vitro on one-half strength germination medium with 100 μEm−2 s−1 light intensity or on soil mixture of decomposed raised bog peat (Plantobalt; Plantaflor Humus Verkaufs-GmbH) under long-day conditions (16-h light/8-h dark) with 100 μEm−2 s−1 light intensity. The cotyledons and the first leaf pairs of the wild-type or transgenic Arabidopsis lines (p35S:HA-E2FB/DPA, pgE2FB-GFP, and p35S:HA-E2FB∆RBR/DPA) grown in vitro were harvested 8–15 DAG, flash frozen, and stored at −80°C. The T-DNA insertion mutants of E2FB were previously reported (e2fb-1 [SALK_103138] and e2fb-2 [SALK_120959]; Berckmans et al., 2011a; Heyman et al., 2011; Horvath et al., 2017).
Plasmid Construction and Generation of Transgenic Arabidopsis Plants
The construct of the pE2FB:gE2FB-GFP (pgE2FB-GFP) and the pE2FA:gE2FA-GFP (pgE2FA-GFP) translational fusion has been described before (Berckmans et al., 2011a, 2011b; Magyar et al., 2012). Using the pgE2FB-GFP construct, transgenic Arabidopsis lines were generated by Agrobacterium-mediated transformation in the wild-type (ecotype Columbia-0) background, and 36 independent T1 Arabidopsis lines were identified on selection medium containing norflurazon. The pgE2FB-GFP construct was also introduced into the e2fb-2 mutant by Agrobacterium-mediated transformation, and homozygous T2 lines were generated afterward. The genomic sequence of either E2FB or E2FA was also fused in frame with 3×vYFP in a pGreenII-based pGII0125 destination vector (Galinha et al., 2007) by using the Invitrogen 3way Gateway System (Invitrogen). The previously described HA epitope-tagged full-length E2FB and its C-terminal deletion mutant form (HA-E2FB∆RBR) missing the 84-amino acid-long region containing the conserved RBR-binding motif (Magyar et al., 2000) were placed under the control of the constitutive cauliflower mosaic virus 35S promoter in the Gateway vector pK7WG2 (Karimi et al., 2002). These constructs were introduced into the previously established p35S:DPA transgenic Arabidopsis line (De Veylder et al., 2002) using the floral-dip method for Agrobacterium-mediated transformation as described (Zhang et al., 2006). Thirteen p35S:HA-E2FB/DPA co-overexpression transgenic T1 lines were selected based on the presence of the appropriate antibiotic resistance (kanamycin). A strong HA-E2FB expressing single copy T-DNA insertion line was identified and homozygous T2 segregation was selected on kanamycin-containing medium. Twelve p35S:HA-E2FB∆RBR/DPA primary transgenic lines were identified and two homozygous T2 segregations (named 1/10 and 10/X) were selected on medium containing kanamycin for further studies. We generated the GFP-tagged version of E2FA∆RBR and E2FB∆RBR where we cloned the C-terminal deleted version (missing the entire transactivation domains until the Marked box region; deletion of 135- and 160-amino acid-long regions from the C termini of E2FA and E2FB, respectively) into the pK7WGF2 gateway vector adding the GFP tag to the N-terminal position. In each case, 15 independent single-copy T-DNA insertion lines were identified on kanamycin-containing medium.
RT-qPCR
RNA was extracted from leaf samples using the RNeasy Plant Mini Kit (Qiagen). cDNA was synthesized using 1 μg of RNA using the QuantiTect Reverse Transcription Kit (Qiagen). Reverse transcription quantitative PCR (RT-qPCR) in the presence of SYBR Green was carried out using a BioScript PCR kit (Bioline) according to the manufacturer’s instructions in a Rotor-Gene 6000 apparatus (Corbert Life Science). All the data were normalized to housekeeping genes (ACTIN and/or UBIQUITIN) and the calculated efficiency was added to the analysis. Primer sequences are summarized in Supplemental Table S3. All reactions were carried out in triplicate.
Image and Flow Cytometry Analysis, Determining Cellular Parameters of Leaf Samples
To visualize the leaf or cotyledon epidermis, a gel cast was made of the leaf surface, specifically the adaxial side of the first leaf pair, which was then observed under a differential interference contrast light microscope (Optiphot 2, Nikon), as described (Horiguchi et al., 2006).
The first true leaf pairs of wild-type and various transgenic lines were dissected from seedlings at 8 or 12 DAG. Leaves were stained with propidium iodide (PI; 20 mg/mL) and images on the abaxial side of three different zones (the basal, middle, and tip parts) of the leaf were taken and analyzed by confocal laser microscopy (SP5, Leica). Across the three zones, ∼600 cells were counted and measured per leaf sample (n ≥ 3 studied for each transgenic line and the control) using Image J software. Average cell size was calculated and the total cell number was extrapolated to the whole leaf according to previously described methods (Asl et al., 2011). The stomata number and stomatal index were calculated in a similar way. For determining the circularity of epidermal cells using Image J software, guard cells were extracted (Andriankaja et al., 2012). To visualize the distributions of the cell area, only nonguard epidermal cells from the three zones were pooled together and used for calculation at a given time point, unless described otherwise (Asl et al., 2011). The number of elongated pavement cells with newly formed cell wall (described as extra cell division) was counted in all three zones and extrapolated to the whole leaf.
For flow cytometry measurements, the first leaf pairs were collected and chopped with razor blades in nuclei extraction buffer and stained with 4′,6-diamino-phenylindole, as described before (Magyar et al., 2005). Flow cytometry data were obtained using a Partec PAS2 Particle Analyzing system (Partec).
IP, Immunoblotting, and Kinase Assays
IP and immunoblotting assays were carried out as described (Henriques et al., 2010). Briefly, total proteins were extracted from dissected leaves or seedlings in extraction buffer (25 mm Tris-HCl, pH 7.5, 75 mm NaCl, 15 mm MgCl2, 15 mm EGTA, 15 mm p-nitrophenylphosphate, 60 mm β-glycerophosphate, 1 mm dithiothreitol, 0.1% (v/v) IGEPAL CA-630, 0.5 mm NaF, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor cocktail [P9599, Sigma]). Equal amounts of proteins were loaded to SDS-PAGE gel (10% or 12%), and proteins were transferred onto polyvinylidene difluoride (Millipore) membranes. The membranes were blocked in 5% (w/v) milk powder with 0.05% (v/v) Tween 20 in Tris-buffered saline (TBS; 25 mM Tris-Cl, pH 8.0, and 150 mm NaCl; TBS plus Tween 20 [TBST]) buffer for 1 h at room temperature. The membrane was incubated with 5% (w/v) milk-powder TBST containing the primary antibodies and agitated overnight at 4°C. Primary antibodies used in immunoblotting experiments were chicken anti-RBR antibody (1:2,000 dilution; Agrisera), mouse monoclonal anti-PSTAIRE (1:40,000 dilution, CDKA;1 specific; Sigma), rabbit polyclonal antibody anti-CDKB1;1 (1:2,000 dilution; Magyar et al., 2005), antiphospho-specific Rb (Ser-807/811) rabbit polyclonal antibody (1:500 dilution; Cell Signaling Tech), and anti-E2FB polyclonal rabbit antibody (1:400 dilution, Magyar et al., 2005). After the primary antibody reaction, the membrane was washed three times with TBST and incubated with the appropriate secondary antibody conjugated with horseradish peroxidase for another hour at room temperature, followed by three washing steps (TBST). Afterward, chemiluminescence substrate was applied according to the manufacturer description (SuperSignal West Pico Plus [Thermo Fisher Scientific] or Immobilon western horseradish peroxidase [Millipore]). For IP, equal amounts of protein samples (between 500–800 µg) in extraction buffer (see above) were incubated with antibodies or GFP-trap magnetic agarose beads (8–10 μL; ChromoTek) for 40 min to 1 h at 4°C. The following antibodies were used in co-IP experiments: anti-DPA (Magyar et al., 2005) and anti-DPB (Umbrasaite et al., 2010), and anti-GFP monoclonal mouse antibody (Roche) or GFP-Trap coupled to magnetic agarose beads (ChromoTek). Protein A and protein G-Sepharose were used to pull down polyclonal and monoclonal antibodies, respectively, and then the beads were washed three times with extraction buffer and proteins were eluted by adding SDS ample buffer followed by 5 min boiling. Eluted proteins were loaded on SDS-PAGE gels (10% or 12%) and after protein gel electrophoresis they were immunoblotted as described above.
The kinase assay was carried out as described earlier (Magyar et al., 1997). Briefly, total proteins were extracted from frozen leaf samples harvested at 8–15 DAG and equal protein amounts were incubated with p13Suc1-Sepharose beads for 1 h at 4°C on a rotary shaker. Kinase reaction was initiated by the addition of 1 mg/mL histone H1 substrate and 2.5 μCi of γ-32P-ATP.
ChIP
ChIP assay was carried out as described previously (Saleh et al., 2008). Four grams of E2FB-GFP-, E2FA-GFP-, and GFP-expressing seedlings, the latter from a 35S:GFP line, were crosslinked with 1% (w/v) formaldehyde solution at 6 DAG. Chromatin was precipitated using anti-GFP polyclonal rabbit antibody (Invitrogen) and was collected with salmon sperm DNA/protein A-agarose (Sigma). The purified DNA was used in RT-qPCR reactions to amplify promoter regions with specific primers (Supplemental Table S3). Fold DNA enrichment was calculated by dividing the antibody IP signals by the no-antibody signals.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: AT5G22220 (ATE2FB); AT3G36010 (ATE2FA); AT1G47870 (ATE2FC); AT5G02470 (ATDPA); AT5G03415 (ATDPB); AT3G12280 (ATRBR); AT3G48750 (ATCDKA;1); AT5G54180 (ATCDKB1;1); AT4G34160 (ATCYCD3;1); AT1G15570 (ATCYCA2;3); AT5G34080 (ATCYCA3;1); AT2G32710 (KRP4); AT5G46280 (MCM3); and AT2G37560 (ORC2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. E2FB and RBR, but not E2FA, are present in differentiated pavement and fully developed stomata guard cells.
Supplemental Figure S2. The E2FB-GFP protein could make complex with DPs, and the nonphosphorylated form of RBR, with these well-known, major interactors of E2FB.
Supplemental Figure S3. Elevated expression of E2FB with expression driven by its own promoter inhibits cell proliferation in young leaves and disturbs quiescence in older leaves.
Supplemental Figure S4. E2FB-GFP binds less RBR in older leaves of pgE2FB-GFP line 72 than in those of line 93.
Supplemental Figure S5. Lack of E2FB function prematurely switches mitosis to endocycle.
Supplemental Figure S6. Elevated HA-E2FB/DPA heterodimer stimulates the accumulation of RBR and its phosphorylated form, RBRS911.
Supplemental Figure S7. Mutant E2FB protein (HA-E2FB∆RBR) in conjunction with DPA causes drastic phenotypic changes during development.
Supplemental Figure S8. Expression of HA-E2FBΔRBR/DPA hyper-activates cell proliferation of meristemoid cells.
Supplemental Table S1. Cellular parameters quantified from the first leaf pair of wild-type and E2FB-related transgenic lines of leaf development at 8 DAG.
Supplemental Table S2. Cellular parameters of leaf development at 12 DAG quantified from the first leaf pair of wild-type and E2FB-related transgenic lines.
Supplemental Table S3. List of primers and their sequences used for RT-qPCR analysis and in ChIP assays.
Acknowledgments
We thank Ferhan Ayaydin (Biological Research Centre) for helping in microscopy, and Anita Kovács (Biological Research Centre) for her assistance in plant work.
Footnotes
This work was supported by the Hungarian Scientific Research Fund (OTKA K-105816 to E.Ö., A.P.-S., T.L., E.M., and Z.M.), the Ministry for National Economy of Hungary (GINOP-2.3.2-15-2016-00001 to A.P.-S.), the Hungarian Academy of Sciences (Young Scientist Fellowship to T.L.), the Marie Curie IEF (fellowships FP7-PEOPLE-2012-IEF.330713 and FP7-PEOPLE-2012-IEF.330789 to C.P. and B.H., respectively), and the Biotechnology and Biological Sciences Research Council–National Science Foundation (BB/M025047/1 to C.P. and L.B.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Abraham E, Miskolczi P, Ayaydin F, Yu P, Kotogany E, Bako L, Ötvös K, et al. (2011) Immunodetection of retinoblastoma-related protein and its phosphorylated form in interphase and mitotic alfalfa cells. J Exp Bot 62: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham Z, del Pozo JC (2012) Ectopic expression of E2FB, a cell cycle transcription factor, accelerates flowering and increases fruit yield in tomato. J Plant Growth Regul 31: 11–24 [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Asl LK, Dhondt S, Boudolf V, Beemster GT, Beeckman T, Inzé D, Govaerts W, De Veylder L (2011) Model-based analysis of Arabidopsis leaf epidermal cells reveals distinct division and expansion patterns for pavement and guard cells. Plant Physiol 156: 2172–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Lammens T, Van Den Daele H, Magyar Z, Bögre L, De Veylder L (2011a) Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc. Plant Physiol 157: 1440–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, Vassileva V, Schmid SP, Maes S, Parizot B, Naramoto S, Magyar Z, Alvim Kamei CL, Koncz C, Bögre L, et al. (2011b) Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23: 3671–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bögre L, Magyar Z, López-Juez E (2008) New clues to organ size control in plants. Genome Biol 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L, Gutzat R, Fütterer J, Laizet Y, Hennig L, Gruissem W (2010) Arabidopsis RETINOBLASTOMA-RELATED is required for stem cell maintenance, cell differentiation, and lateral organ production. Plant Cell 22: 1792–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HZ, Tsai SY, Leone G (2009) Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer 9: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Scofield S, Huntley RP, Robinson AS, den Boer BG, Murray JA (2009) Dissecting regulatory pathways of G1/S control in Arabidopsis: Common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol Biol 71: 345–365 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, et al. (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C (2006) The balance between cell division and endoreplication depends on E2FC-DPB, transcription factors regulated by the ubiquitin-SCFSKP2A pathway in Arabidopsis. Plant Cell 18: 2224–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, MacAlister CA, Bergmann DC (2009) BASL controls asymmetric cell division in Arabidopsis. Cell 137: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J, Hunt T (1996) Cell cycle. Why don’t plants get cancer? Nature 380: 481–482 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B (2007) PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gázquez A, Beemster GTS (2017) What determines organ size differences between species? A meta-analysis of the cellular basis. New Phytol 215: 299–308 [DOI] [PubMed] [Google Scholar]
- Harashima H, Sugimoto K (2016) Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA-RELATED 1. Curr Opin Plant Biol 29: 95–103 [DOI] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Bögre L (2013) S6K1 and E2FB are in mutually antagonistic regulatory links controlling cell growth and proliferation in Arabidopsis. Plant Signal Behav 8: e24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Alvim Kamei CL, De Jaeger G, Koncz C, De Veylder L (2011) Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J 48: 638–644 [DOI] [PubMed] [Google Scholar]
- Horvath BM, Kourova H, Nagy S, Nemeth E, Magyar Z, Papdi C, Ahmad Z, Sanchez-Perez GF, Perilli S, Blilou I, et al. (2017) Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J 36: 1261–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Schwarz JK, Cress WD, Nevins JR (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349–352 [DOI] [PubMed] [Google Scholar]
- Kalve S, De Vos D, Beemster GT (2014) Leaf development: A cellular perspective. Front Plant Sci 5: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki T, Iwata E, Magyar Z, Bögre L, Ito M (2015a) MYB3Rs, plant homologs of Myb oncoproteins, control cell cycle-regulated transcription and form DREAM-like complexes. Transcription 6: 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Suzuki T, Iwata E, Nakamichi N, Suzuki T, Chen P, Ohtani M, Ishida T, Hosoya H, Müller S, et al. (2015b) Transcriptional repression by MYB3R proteins regulates plant organ growth. EMBO J 34: 1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol 128: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Cai W, Liu Y, Li H, Fu L, Liu Z, Xu L, Liu H, Xu T, Xiong Y (2017) Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc Natl Acad Sci USA 114: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Atanassova A, De Veylder L, Rombauts S, Inzé D (2000) Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett 486: 79–87 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Bögre L, Ito M (2016) DREAMs make plant cells to cycle or to become quiescent. Curr Opin Plant Biol 34: 100–106 [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 31: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Mészáros T, Miskolczi P, Deák M, Fehér A, Brown S, Kondorosi E, Athanasiadis A, Pongor S, Bilgin M, et al. (1997) Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9: 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Matos JL, Lau OS, Hachez C, Cruz-Ramírez A, Scheres B, Bergmann DC (2014) Irreversible fate commitment in the Arabidopsis stomatal lineage requires a FAMA and RETINOBLASTOMA-RELATED module. eLife 3: e02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. (2007) The Cell Cycle: Principles of Control. Oxford University Press, Oxford [Google Scholar]
- Sadasivam S, DeCaprio JA (2013) The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Simmons AR, Davies KA, Wang W, Liu Z, Bergmann DC (2019) SOL1 and SOL2 regulate fate transition and cell divisions in the Arabidopsis stomatal lineage. Development 146: dev171066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Maggio C, Varotto S, Canova S, Bergounioux C, Albani D, Cella R (2006) Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol 140: 1355–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrasaite J, Schweighofer A, Kazanaviciute V, Magyar Z, Ayatollahi Z, Unterwurzacher V, Choopayak C, Boniecka J, Murray JA, Bogre L, et al. (2010) MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS One 5: e15357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ (2008) Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014) A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DW. (2006) PEAPOD regulates lamina size and curvature in Arabidopsis. Proc Natl Acad Sci USA 103: 13238–13243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E (2010) Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang H, Xue S, Qu X, Zou J, Le J (2014) Requirement for A-type cyclin-dependent kinase and cyclins for the terminal division in the stomatal lineage of Arabidopsis. J Exp Bot 65: 2449–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL (1996) pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev 10: 3051–3064 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1: 641–646 [DOI] [PubMed] [Google Scholar]