Short-, medium-, and long-term acclimation of tobacco seedlings to increased light intensities involves only minor adjustments of the chloroplast transcriptome and protein translation.
Abstract
Acclimation to changing light intensities poses major challenges to plant metabolism and has been shown to involve regulatory adjustments in chloroplast gene expression. However, this regulation has not been examined at a plastid genome-wide level and for many genes, it is unknown whether their expression responds to altered light intensities. Here, we applied comparative ribosome profiling and transcriptomic experiments to analyze changes in chloroplast transcript accumulation and translation in leaves of tobacco (Nicotiana tabacum) seedlings after transfer from moderate light to physiological high light. Our time-course data revealed almost unaltered chloroplast transcript levels and only mild changes in ribosome occupancy during 2 d of high light exposure. Ribosome occupancy on the psbA mRNA (encoding the D1 reaction center protein of PSII) increased and that on the petG transcript decreased slightly after high light treatment. Transfer from moderate light to high light did not induce substantial alterations in ribosome pausing. Transfer experiments from low light to high light conditions resulted in strong PSII photoinhibition and revealed the distinct light-induced activation of psbA translation, which was further confirmed by reciprocal shift experiments. In low-light-to-high-light shift experiments, as well as reciprocal treatments, the expression of all other chloroplast genes remained virtually unaltered. Altogether, our data suggest that low light-acclimated plants upregulate the translation of a single chloroplast gene, psbA, during acclimation to high light. Our results indicate that psbA translation activation occurs already at moderate light intensities. Possible reasons for the otherwise mild effects of light intensity changes on gene expression in differentiated chloroplasts are discussed.
Plants are sessile organisms that must cope in situ with ever-changing environmental conditions. Due to the phototrophic lifestyle of plants, light dramatically influences plant metabolism. Light serves as both an energy source and a signal that controls diverse cellular processes. Plants perceive light through different photoreceptors that regulate nuclear gene expression (e.g. Gyula et al., 2003; Kami et al., 2010; Jenkins; 2014). In addition, the photosynthetic reactions within chloroplasts are directly affected by alterations in light conditions and serve as a “metabolic light sensor”. Changes in light quantity and quality cause imbalances in the activities of PSI and PSII, and between the light reactions of photosynthesis and the carbon fixation reactions of the Calvin-Benson-Bassham cycle (CBC). These imbalances can disturb the photosynthetic reduction/oxidation (redox, NADPH/NADP+) balance and/or the energy charge (ATP/ADP), thus reducing the efficiency of photosynthesis or, if extreme, damaging the photosynthetic machinery (e.g. Allen and Pfannschmidt, 2000; Cruz et al., 2005), for example, by producing reactive oxygen species (ROS; Krieger-Liszkay et al., 2008).
Several molecular mechanisms acting at different time scales adjust photosynthetic reactions to the available light and the metabolic requirements (e.g. Foyer et al., 2012; Rochaix, 2014; Tikkanen and Aro, 2014; Dietz, 2015). An immediate consequence of increasing light irradiation is the saturation of the CBC as the major sink for NADPH and ATP. Consequently, key components of linear photosynthetic electron transport become (over-) reduced (leading to the accumulation of plastoquinol and reduced thioredoxin/ferredoxin) and the thylakoid lumen acidifies. To prevent photooxidative damage, excess light energy is dissipated as heat by nonphotochemical quenching (NPQ) and electron flow through the cytochrome b6f complex (Cyt b6f) becomes regulated. Electrons can also cycle around PSI, thus uncoupling ATP synthesis from NADPH production (e.g. Joliot and Johnson, 2011; Yamori and Shikanai, 2016). Furthermore, several enzymes of the CBC are activated by reduced thioredoxin/ferredoxin and/or stromal ATP levels (e.g. Kaiser et al., 2015). Moreover, the thylakoid kinase STN7 (Bellafiore et al., 2005) is inactivated, blocking light harvesting complex II (LHCII) phosphorylation and state transitions (Rintamäki et al., 2000; Ancín et al., 2019). In this way, excitation energy is preferentially dissipated through NPQ.
In the longer term, plants adjust their photosynthetic complex stoichiometry to increased light intensity. Tobacco (Nicotiana tabacum), for instance, substantially increases its PSII reaction center content (at the expense of LHCII) as well as Cyt b6f and chloroplast ATP synthase contents in response to high light (Petersen et al., 2011; Schöttler and Tóth, 2014; Schöttler et al., 2017). These adjustments, together with changes in key enzymes of the CBC ultimately reset photosynthetic capacity (Schöttler et al., 2015).
The mechanisms underlying both short-term responses and long-term acclimation of the photosynthetic apparatus to high light are still not fully understood. Light-dependent chloroplast-derived (redox) signals control the expression of numerous nuclear genes encoding photosynthetic proteins (e.g. Fey et al., 2005; Pogson et al., 2008; Berry et al., 2013). The core subunits of photosynthetic complexes in the thylakoid membrane are encoded in the plastid genome and synthesized by chloroplast 70S-like ribosomes (Bock, 2007). The expression of these core components is redox-regulated and contributes to light acclimation. For example, differential transcription of plastid-encoded PSII (psbA) and PSI (psaA/psaB) core subunits is controlled by the redox state of the plastoquinone pool in response to changes in light quality (Pfannschmidt et al., 1999) and likely contributes to a long-term acclimation of PSII-PSI stoichiometry (Chow et al., 1990; Allen and Pfannschmidt, 2000).
Chloroplast translation has also been shown to be light regulated (Zoschke and Bock, 2018). In de-etiolating barley (Hordeum vulgare) seedlings, light activates translation of numerous plastid transcripts coding for photosynthetic subunits (e.g. Klein and Mullet, 1986, 1987; Klein et al., 1988; Kim and Mullet, 2003). However, energy limitation in etioplasts may contribute to this phenomenon. Moreover, de-etiolation involves etioplast-to-chloroplast differentiation that causes dramatic changes in nuclear and plastid gene expression. In de-etiolating tobacco leaves, only minor changes in chloroplast transcript abundance occur (Armarego-Marriott et al., 2019). Thus, de-etiolation may be very different from light acclimation of differentiated chloroplasts. Also, due to the different patterns of leaf development, data obtained in monocots such as barley may not be transferable to dicots such as tobacco or Arabidopsis (Arabidopsis thaliana).
The PsbA (D1) protein is particularly prone to photodamage. This necessitates the PSII repair cycle, which depends on PSII core protein phosphorylation, dissociation of the PSII core from its LHCII, and diffusion of the core from grana stacks into unstacked thylakoids. Here, the damaged PsbA is replaced by a newly synthesized copy (Herbstová et al., 2012; Järvi et al., 2015). Thus, PsbA synthesis is gradually activated by increasing light (e.g. Ohad et al., 1984, Puthiyaveetil et al., 2014; Chotewutmontri and Barkan, 2018). Although this translational activation is often believed to occur under high light, there is evidence that psbA translation is already substantially (and gradually) induced over a range of rather low light intensities (Sundby et al., 1993). Light-controlled psbA and rbcL translation was suggested to be determined by redox/thiol signals, the ATP status, and/or the transthylakoidal proton gradient (e.g. Blair and Ellis, 1973; Eaglesham and Ellis, 1974; Taniguchi et al., 1993; Mühlbauer and Eichacker, 1998; Zhang et al., 1999, 2000), and occurs, in part, at the elongation level (e.g. Berry et al., 1988; Edhofer et al., 1998; Mühlbauer and Eichacker, 1998). A recent comprehensive study of green maize (Zea mays) and Arabidopsis seedlings suggests that translation elongation of all chloroplast genes is reduced after transferring plants from light to dark and increased after transfer back to the light (Chotewutmontri and Barkan, 2018). Light-induced ribosome recruitment to psbA is superimposed on this global response (Chotewutmontri and Barkan, 2018). However, little is known about the genome-wide regulation of chloroplast translation in response to changes in light intensity that are more typical of physiological conditions. Hence, our knowledge about the magnitude of psbA translational activity at different light intensities is very limited. In addition, for many chloroplast genes, it is unknown whether they are translationally regulated at increased light intensities. Consequently, the relative contributions of translational and physiological adjustments during acclimation to high light have remained elusive.
Here, we used ribosome profiling to comprehensively examine the responses of chloroplast transcript accumulation and translation in tobacco seedlings following a transfer from moderate to high light intensity, a change in the environment that plants frequently experience in nature. Our results revealed only small contributions of translational regulation during short-, medium-, and long-term high light acclimation. Furthermore, although psbA translation is believed to be induced by high light, our data indicate that most of its light-dependent activation occurs at moderate light intensities. Together, our data strongly indicate that other regulatory mechanisms predominate during light acclimation in the analyzed time frame and conditions.
RESULTS
A Shift from Moderate Light to High Light Causes Only Mild Changes in Parameters of Photosynthetic Light Reactions in Tobacco Seedlings
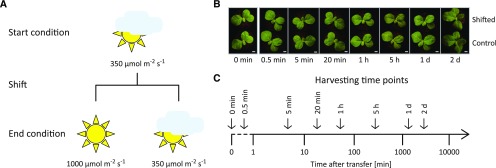
To study acclimation to high light in the model dicotyledonous C3 plant tobacco, plants were grown at 350 µmol m−2 s−1 for 11 d before they were shifted to 1,000 µmol m−2 s−1 (Fig. 1). Seedlings were chosen for this analysis because, in contrast to fully developed plants, photosynthetic complex biogenesis is ongoing in seedlings and thereby flexible in response to environmental stimuli (Cyt b6f and PSI complex biogenesis, for example, are repressed in mature tobacco leaves; Krech et al., 2012; Hojka et al., 2014). Exposure to high light had virtually no effect on leaf temperature and caused only a marginal increase in soil surface temperature (Supplemental Fig. S1). Within the analyzed time frame, the ∼3-fold change in light intensity had almost no effect on the visual appearance of the plants. A slightly paler leaf color (barely visible in Fig. 1B) and slightly thicker leaves were noticed after 2 d in high light in comparison to seedlings kept at control conditions. To assess the physiology of the photosynthetic high light responses, the chlorophyll a/b ratio, chlorophyll content, and chlorophyll-a fluorescence-derived parameters were determined for specific time points after the shift (Supplemental Fig. S2). Throughout the time course, only minor differences in chlorophyll content were observed between control plants and high light-shifted plants. After 2 d, the chlorophyll a/b ratio was increased in high light-shifted plants, indicating a lower content in LHCII antenna proteins (binding both chlorophyll a and b) relative to PSI and PSII reaction centers (which only bind chlorophyll a); a typical initial step of high-light acclimation (Schöttler and Tóth, 2014). Throughout the time course, plants exhibited mild PSII photoinhibition, as indicated by the slightly reduced maximum quantum efficiency of PSII in the dark-adapted state (FV/FM). This suggests an ongoing high light acclimation, which was further substantiated by the unaltered photosynthetic electron transport capacity (suggesting that the typical upregulation of Cyt b6f and ATP synthase content had not yet started; Schöttler and Tóth, 2014; Schöttler et al., 2017). Additionally, chlorophyll a fluorescence emission spectra at 77 K did not reveal substantial alterations (Supplemental Fig. S3). These physiological measurements were complemented by immunoblot analyses that revealed virtually unaltered contents of PSI, PSII, Cyt b6f, chloroplast ATP synthase, and the NADH dehydrogenase-like complex during high light acclimation (Supplemental Fig. S4). Taken together, the applied changes in light intensity were considered to induce a physiological high light acclimation.
Figure 1.
Experimental design for the analysis of high light-induced acclimation processes in tobacco chloroplast gene expression. A, Tobacco plants were grown under moderate light conditions (350 µmol m−2 s−1) for 11 d before they were shifted to high light (1,000 µmol m−2 s−1, in a similar growth chamber with identical growth parameters apart from the light intensity). A control set of plants was kept at 350 µmol m−2 s−1. The comparison of shifted and control plants for each time point intends to exclude circadian and developmental effects from the analyses. For details, see “Materials and Methods”. B, Phenotypes of shifted and control plants at the harvesting time points indicated at the bottom. Scale bars = 1 cm. Note the slightly paler leaf color after 2 d in high light. C, The aerial parts (except the cotyledons) of shifted and control plants were harvested at the indicated time points to identify short-, medium-, and long-term acclimation responses in chloroplast translation. Please note the logarithmic scale of the time axis starting at 1 min. Plants harvested immediately before the shift served as technical controls (0 min). A dashed line connects the time of the shift event with the logarithmic time axis.
Tobacco Chloroplast Translation Is Altered Only Slightly Following a Shift from Moderate Light to High Light
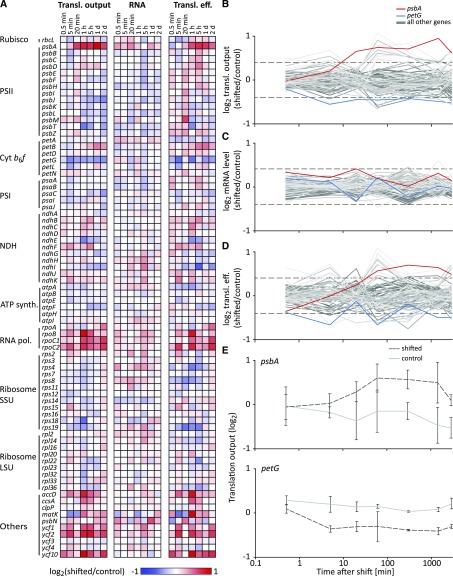
To conduct a transcriptome-wide analysis of chloroplast translational regulation during high light acclimation, true leaves were harvested from treated and control plants at seven time points after the light shift (including the three time points used for photosynthetic measurements; Fig. 1C) and used for analyses of chloroplast translational output and transcript accumulation by microarray-based ribosome and transcript profiling. Results for translational output (represented by ribosome footprint levels), RNA accumulation (represented by transcript levels), and translation efficiencies (the ratio of ribosome footprint abundance to RNA abundance) are reported for all chloroplast genes (Zoschke et al., 2013; Trösch et al., 2018). Relative changes in expression levels (translational output, RNA accumulation, and translation efficiency) between the shifted and control plants harvested at the same time were determined by normalizing total ribosome footprint or RNA signals between samples and calculating signal ratios for each reading frame (RF; see “Materials and Methods”; Supplemental Fig, S5; Supplemental Dataset S1), which were plotted as heat maps, line plots (Fig. 2, A–D), and scatter plots (Supplemental Fig. S6).
Figure 2.
Dynamics in chloroplast translation during acclimation to high light. A, Heat maps representing the average log2-transformed relative translational (Transl.) output and transcript level (RNA) of high light-shifted tobacco plants compared to control tobacco plants (n = 3), and the resulting relative translation efficiency (Transl. eff.; relative ribosome footprint levels normalized to relative transcript abundances) for each plastid gene (labeled on the left) for the analyzed time points after the light shift (indicated on top of the heat maps). Increased and decreased relative expression levels of shifted plants compared to control plants are represented by red and blue colors, respectively (as indicated in the legend below the heat maps; note the limited scale used to visualize the maximum ∼2-fold changes). Genes are grouped in the functional categories PSII, Cyt b6f, PSI, NDH (NADH dehydrogenase-like complex), ATP synth. (ATP synthase), RNA pol. (RNA polymerase), Ribosome SSU (ribosome small subunit), Ribosome LSU (ribosome large subunit), and Others. B to D, Line plots of the log2-transformed relative translational output (B), transcript level (C), and translation efficiency (D) for all chloroplast genes during high light acclimation. psbA and petG, whose translational output is mildly but consistently (i.e. reproducible >1.3-fold change for two or more consecutive time points) up- and downregulated are plotted in red and blue, respectively, to mark the most consistent changes in translational output (for details, see “Results”). All other genes are plotted in different shades of gray. Note the logarithmic scale of the x axis. Individual scatter plot diagrams for each single time point after high light shift are shown in Supplemental Figure S6. E, Representative line plots of log2-transformed relative abundances of ribosome footprint levels of psbA and petG in shifted and control (dark gray dotted and light gray solid lines, respectively) tobacco plants for the analyzed time points, represented as average values for three biological replicates (vertical error bars indicate standard deviations). Note the logarithmic scale of the x axis.
Some mild changes in translational output upon high light exposure were observed, but none of the changes was significant in a Student’s t test (all values adjusted, P > 0.1; Supplemental Dataset S1). Still, the trends for several genes appear consistent (i.e. a reproducible >1.3-fold change for two or more consecutive time points) and may be physiologically meaningful. For example, the data suggested a small increase in ribosome occupancy on psbA mRNA as early as 20 min after high light exposure, and this occupancy remained elevated until the latest analyzed time point (Fig. 2, B and E). Conversely, ribosomes on the petG RF appeared to be reduced slightly already after 5 min of high light treatment, and ribosome occupancy remained below the control level for the remainder of the time course (Fig. 2, B and E). The relative fold change in translational output of some other genes, namely accD, matK, psbJ, psbT, rpoB,rpoC1, rpoC2, ycf1, ycf 2, and ycf 10, showed alterations at a similar magnitude for several consecutive time points (Fig. 2A; Supplemental Dataset S1). However, these very mild effects showed a slightly higher variability between biological replicates (Supplemental Dataset S1) due to the low ribosome footprint coverage in these RFs, and with the exception of psbA, none of the 78 tobacco chloroplast RFs showed a substantial difference in translational output at subsequent time points after high light exposure. This becomes obvious when translational outputs in moderate and high light are plotted against each other (Supplemental Fig. S6). The changes at the RNA level upon high light treatment were even milder than those observed for ribosome footprints (Fig. 2, A–C; Supplemental Fig. S6; Supplemental Dataset S1), together suggesting stable transcript levels and very mild translational regulation, if any (Fig. 2, A–C).
Ribosomes Are Recruited Specifically to the psbA RNA Following a Shift from Low Light to High Light
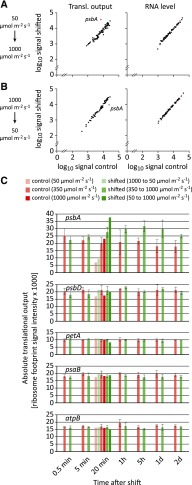
To test whether more drastic changes from low light to high light cause different responses in chloroplast gene expression, we grew seedlings at a light intensity of 50 µmol m−2 s−1 and shifted them to 1,000 µmol m−2 s−1. Again, photosynthetic parameters were determined (Supplemental Fig. S2). The 20-fold increase in light intensity resulted in strong responses of several photosynthetic parameters. The maximum quantum efficiency of PSII decreased drastically within 1 h of high light exposure and did not recover during the 2 d of high light treatment. The underlying substantial photodamage of PSII was also evident in 77 K chlorophyll a fluorescence emission spectra, which after 2 d of high light exposure revealed a shift of the PSII-LHCII emission signal from 687 to 685 nm wavelength and the occurrence of an additional shoulder at 680 nm wavelength (Supplemental Fig. S3). These changes indicate the presence of free, uncoupled LHCII in the thylakoid membrane (Krause and Weis, 1991), likely a consequence of massive photodamage to PSII and its dissociation from LHCII during the repair cycle.
Tissues from shifted seedlings and seedlings maintained at 50 µmol m−2 s−1 were harvested 20 min after the shift and used for microarray-based ribosome profiling. This experiment revealed a >5-fold increase of psbA translational output, whereas all other RFs showed minor changes and transcript levels of all chloroplast genes were virtually unaltered (Fig. 3, A and C; Supplemental Dataset S1). Similarly, we found a specific reduction of psbA translation (>2-fold) when plants were grown at 1,000 µmol m−2 s−1 and shifted to 50 µmol m−2 s−1 (Fig. 3, B and C).
Figure 3.
Specific dynamics in psbA translation in response to extreme changes in light intensity. A, Average ribosome footprint abundances (Transl. output, left plot) and transcript accumulation (RNA level, right plot) for each plastid RF from control samples grown at 50 µmol m−2 s−1, and samples shifted from low light (50 µmol m−2 s−1) for 20 min to high light (1,000 µmol m−2 s−1) are plotted against each other in log10 scale for one biological replicate. psbA, whose translational output substantially changes in response to light intensity, is highlighted in red. B, Same analysis as in A for plants shifted from high light (1,000 µmol m−2 s−1) to low light (50 µmol m−2 s−1). C, From top to bottom, diagrams show average absolute translational output, which was measured by ribosome footprint signal intensities, in the psbA, psbD, petA, psaB, and atpB RFs for control plants grown in constant low, moderate, or high light conditions of 50, 350, or 1,000 µmol m−2 s−1 (light to dark red bars) or plants that were shifted from high light to low light, from moderate light to high light, and from low light to high light (light to dark green bars) as indicated in the legend above the plots. Data were obtained for different harvesting time points, which are specified below the plots. Vertical lines depict standard deviations calculated from three biological replicates. Note that only psbA shows a substantial light intensity-dependent change in the average ribosome footprint signal intensity both in constant light conditions and after alterations in light intensity. psbD, petA, psaB, and atpB are shown as additional examples of genes encoding core subunits of the major photosynthetic complexes.
Interestingly, the difference in the absolute psbA translational output under steady light conditions (control plants) was higher between 50 and 350 µmol m−2 s−1 (∼2.8-fold) than between 350 and 1,000 µmol m−2 s−1 (<1.2-fold; Fig. 3C), indicating a substantial activation of psbA translation at rather low light intensities. No other chloroplast gene exhibited such a substantial light intensity-dependent change in translational output (for selected examples, see Fig. 3C).
Altogether, these results indicate that there is little change in tobacco chloroplast gene expression following a shift from moderate to high light, and that there is a specific promotion of psbA translation upon a shift from low light to high light.
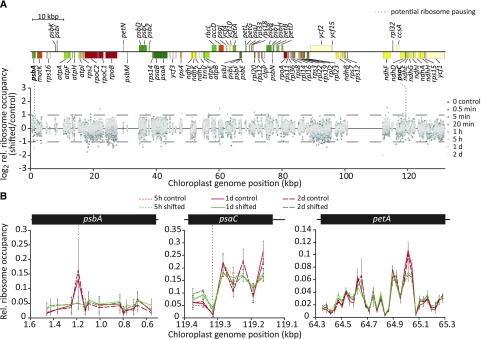
Increased Light Intensity Does Not Induce Substantial Alterations in Chloroplast Ribosome Pausing
In most cases, translation is regulated at the level of initiation. This leads to an increase or decrease in the abundance of ribosomes per RF, a change that can be readily detected by standard ribosome profiling methods (e.g. Ingolia et al., 2019). However, the elongation behavior within specific RFs could be changed during high light acclimation in a way that does not influence the number of ribosomes per RF, for example, through altered ribosome pausing. If this were the case, a change in the distribution of ribosome footprint abundance along an RF should become visible in our ribosome profiles. To test this possibility, we normalized ribosome footprint signal intensities of individual array probes in protein-coding regions to the sum of the footprint signal observed for the respective RF to examine potentially altered ribosome distribution independent of changes in translational output of the whole RF, as previously described (Chotewutmontri and Barkan, 2018). Next, we calculated the ratios of these relative ribosome occupancies between treated (1,000 µmol m−2 s−1) and control (350 µmol m−2 s−1) plants for each time point after the high light shift and plotted these values against the probe position in the tobacco chloroplast genome (Fig. 4A; Supplemental Fig. S7; Supplemental Dataset S1). This analysis showed in almost all cases very similar ribosome distributions between high light-exposed and control plants. However, two probes, located in the psbA and psaC RFs, displayed >2-fold decreased or increased relative ribosome occupancies, respectively, for two or more consecutive time points after the high light shift (Fig. 4A, vertical dashed lines), although in the case of psaC, the overall ribosome occupancy level was relatively low. This may indicate a slightly altered ribosome elongation behavior causing a specific reduction and increase of relative ribosome occupancies in these regions of the psbA and psaC RFs, respectively (Fig. 4B). However, it should be noted that the microarray-based ribosome profiling method has a maximal resolution of ∼30 nucleotides that is sometimes decreased by the exclusion of individual probes with saturated or below-background signal intensities (see “Materials and Methods” for details). This allows the detection of pronounced ribosome pausing events, but it may limit inferences that can be made about weak pausing sites.
Figure 4.
Chloroplast translation elongation is not substantially affected by increased light intensity. A, Quantitative plastome-wide comparison of relative ribosome occupancies within RFs between shifted (1,000 µmol m−2 s−1) and control (350 µmol m−2 s−1) conditions in tobacco. For each probe in protein-coding regions, ribosome footprint signal intensity was normalized to the sum of the signal intensities of all probes in the respective RF for high light-shifted and control samples. Ratios of these relative ribosome occupancy values were calculated for three biological replicates, and their averages were plotted for the indicated time points (different shades of gray as indicated in the legend) against the position in the tobacco chloroplast genome to reveal altered chloroplast ribosome pausing after high light exposure. Only one of the large inverted repeats is shown. Probes with very low signal intensities (<100) were not considered in this analysis. Dashed horizontal lines and bold gene names indicate two probes located in the psbA and psaC RFs with >2-fold changes in relative ribosome occupancy in high light for two or more subsequent time points (detailed magnifications are shown in B). The genome map was drawn with OGDRAW (Lohse et al., 2013). kbp, Kilobase pairs. Individual diagrams for each time point after the high light shift are shown in Supplemental Figure S7. B, Relative ribosome occupancy and the two potential light-dependent ribosome pausing sites in the psbA and psaC RFs. Relative ribosome occupancy within the psbA and psaC RFs is plotted as the fraction of probe signal to the summed RF signal for each of the indicated conditions and time points (see legend; control = 350 µmol m−2 s−1, shifted = 1,000 µmol m−2 s−1) that showed a maximum >2-fold change. A similar representative plot for petA is shown to illustrate unchanged ribosome occupancy in all other chloroplast RFs. Error bars depict standard deviations calculated from three independent biological replicates. Note that for better visual comparison between the shown subsequent time points of the time series, the only probes plotted are those for which data for all three relevant time points were determined in three biological replicates. Due to signal saturation for specific probes in particular replicates, certain data points in the 5′ region and around the genomic position of ∼900 bp within the psbA RF were excluded from this plot (see Supplemental Dataset S1 for data of individual replicates).
Taken together, our data provide evidence against the existence of substantial alterations in ribosome distribution within specific chloroplast RFs in response to high light.
DISCUSSION
In this work, we systematically analyzed changes in chloroplast transcript accumulation and translation in tobacco seedlings following transfer from moderate light to sudden high light. Our results demonstrate that transcript accumulation, ribosome occupancy, and ribosome distribution are virtually unaltered for the vast majority of chloroplast genes. Recently, ribosome profiling analyses in maize and Arabidopsis investigated chloroplast translation in response to sudden darkness and subsequent reillumination during the photoperiod, and came to similar conclusions (Chotewutmontri and Barkan, 2018). The light regimes examined by Chotewutmontri and Barkan (2018) did not include high light (as included in this study), a condition plants regularly experience in natural environments. Furthermore, our ribosome profiling analyses were complemented by examinations of photosynthetic parameters and the accumulation of photosynthetic complexes during high light acclimation, and thereby document physiological effects and changes in photosynthesis during high light acclimation. Overall, our data suggest only relatively minor adjustments in chloroplast translation that occur, however, in a gene-specific manner (Fig. 2). Most strikingly, psbA translation was rapidly upregulated following the shift from moderate light to high light. When tobacco plants were shifted from low light to high light, this effect was much more pronounced (∼5.5-fold change; Fig. 3) and PSII photodamage was massive, as indicated by dramatically decreased FV/FM values and the appearance of emission signals of free, uncoupled LHCII in 77K chlorophyll a fluorescence emission spectra (Supplemental Figs. S2 and S3). This is likely because tobacco has an exceptionally high capacity for long-term light acclimation (Schöttler and Tóth, 2014). Hence, photosynthesis in low light-acclimated plants is easily light-saturated due to low content of Cyt b6f, ATP synthase, and CBC enzymes. Therefore, after the shift from 50 to 1,000 µmol m−2 s−1, additional electrons generated in the light reactions of photosynthesis cannot be efficiently used in the CBC, and both the plastoquinol pool and the PSII acceptor side, and likely also the stromal electron acceptor systems, become rapidly reduced. Consequently, this leads to increased photodamage of PSII, requiring the stimulation of PsbA synthesis (Järvi et al., 2015; Theis and Schroda, 2016). This response has been previously observed in other organisms (e.g. maize) shifted from dark to light (Chotewutmontri and Barkan, 2018). Notably, this response is reversible in that it is reverted by transfer from high light to low light (Fig. 3) or from light to dark (Chotewutmontri and Barkan, 2018). It was shown in cyanobacteria that the extent of photodamage is proportional to light intensity, whereas PsbA repair synthesis peaks, independent of prior growth irradiance, at much lower light intensities in both plants and cyanobacteria (e.g. Sundby et al., 1993; Park et al., 1996; Murata and Nishiyama, 2018). These findings suggest a model in which (1) light intensity-triggered translational activation of psbA occurs only in a limited window of rather low light intensities in a linear manner, and (2) psbA translation reaches a maximum at light intensities below that at which photoinhibition peaks. Indeed, we observed much greater psbA translation in plants shifted from low light to high light than in those shifted from moderate light to high light (Fig. 3C). In addition, control plants that were grown under constant light conditions exhibited a psbA translational output at 350 µmol m−2 s−1 that is close to that at 1,000 µmol m−2 s−1, whereas psbA translation was much less active at 50 µmol m−2 s−1 (with only about one-third of the translational output at 350 or 1,000 µmol m−2 s−1; Fig. 3C). Together, this supports the model that psbA translation is already activated under rather low light intensities and reaches a maximum under light intensities below that which cause substantial photoinhibition (Sundby et al., 1993; Park et al., 1996; Murata and Nishiyama, 2018). Remarkably, this implies that psbA translation should not be considered to be specifically induced by high light.
Apart from psbA, our data suggest that in the time frame analyzed (up to 2 d), adjustments in relative rates of translation among chloroplast genes play only a minor role in plant acclimation to physiological high light. Nevertheless, our results do not exclude the possibility that there is a global change in chloroplast translation following the light shifts we examined. A global increase or decrease in chloroplast translation elongation was recently observed in maize plants shifted from dark to light or vice versa (Chotewutmontri and Barkan, 2018). However, such a global regulation cannot adjust the translational output of specific RFs to changing turnover rates of the respective proteins in high light, as is known for psbA/PsbA, and our data did not reveal any additional gene-specific regulation of translation initiation or elongation. This suggests that among plastid-encoded proteins, the light-induced turnover of PsbA is unique, which is a problem that has not been addressed previously for many small subunits of photosynthetic complexes (due to the difficulties in detecting them in pulse labeling or mass spectrometry experiments). Altogether, this finding may not be surprising when taking into consideration a recent study that demonstrated that dark-to-light shifts in maize and Arabidopsis induce substantial alterations in translation initiation rates only for psbA, presumably to comply with PSII repair demands (Chotewutmontri and Barkan, 2018). However, the herein applied changes in light intensity certainly require acclimation mechanisms to maintain energy and redox homeostasis. Hence, other mechanisms have to be considered to predominate at least in short- and medium-term responses to high light. This raises the question of why the regulation of translation initiation for specific chloroplast transcripts contributes so little to light acclimation in the tested conditions. Due to the kinetics in the gene expression cascade, translational regulation acts potentially much faster than transcriptional regulation, especially in the chloroplast, where transcript half-lives are exceptionally long (Zoschke and Bock, 2018). However, changes in protein synthesis rates that lead to substantial alterations in the abundant photosynthetic complexes that are often comprised of proteins with rather low turnover (e.g. Hojka et al., 2014; Li et al., 2017) are still much slower than many posttranslational or physiological regulation mechanisms, such as the onset of NPQ, cyclic electron flow around PSI, dissipation of excess reducing energy through plastid terminal oxidase and mitochondrial alternative oxidase, or chloroplast movement (to avoid high light; Dietz, 2015; Wada and Kong, 2018). These processes, as well as many redox-dependent posttranslational mechanisms, are triggered within seconds to minutes after high light exposure, thus providing a quasi-immediate mechanism to maintain photosynthetic homeostasis (Dietz, 2015; Wada and Kong, 2018). In line with that argumentation, only very long-term acclimation to different light intensities caused major increases in photosynthetic complexes, especially PSII, the Cyt b6f, and the ATP synthase, whereas PSI levels were largely unaltered (e.g. Schöttler et al., 2017; Albanese et al., 2018). However, these stoichiometric adjustments had not really started after 2 d in high light, as indicated by largely unaltered photosynthetic electron transport capacity and further substantiated by our immunoblot analyses for core subunits of these complexes (Supplemental Fig. S4). Finally, sudden but constant high light exposure may represent a rather minor challenge to plant metabolism, considering that it has evolved to maintain homeostasis even under conditions where light intensities fluctuate by orders of magnitude within seconds (Armbruster et al., 2017).
In high light conditions, the abundance and/or activity of CBC enzymes increase (e.g. Kaiser et al., 2015; Miller et al., 2017). However, the only plastid-encoded component of this pathway, the large subunit of Rubisco (RbcL), did not show translational upregulation (Figs. 2 and 3; Supplemental Fig. S6). rbcL is among the most efficiently expressed chloroplast genes, with exceptionally high transcript levels that are efficiently translated already under moderate light intensities (Chotewutmontri and Barkan, 2016; Trösch et al., 2018). Hence, it can be assumed that the translational output of rbcL is already maximized and cannot be substantially increased further (Salesse-Smith et al., 2018, and references therein).
We cannot completely rule out the possibility that the chosen harvesting intervals skipped specific time points with stronger, but temporarily restricted, responses of chloroplast gene expression to high light (e.g. directly after onset of light). It also should be noted that our analysis averages the response in four leaves from rather young seedlings, which are, however, dominated by source tissue with fully differentiated chloroplasts (as evidenced by different photosynthetic parameters such as maximum quantum efficiency of PSII, linear electron transport capacity, and chlorophyll content; Supplemental Fig. S2). Hence, it is possible that potential stronger responses (that, for example, could occur in chloroplasts of the underrepresented immature sink tissues) were overlooked. However, for several reasons, this appears to be unlikely. Except for the repair of photodamaged PSII, photosynthetic complex biogenesis may not rapidly respond to changes in light intensity, because in natural environments, light intensity (and quality) is fluctuating at timescales of seconds to hours and the photosynthetic metabolism is able to rapidly adjust to these sudden changes in light intensity (Armbruster et al., 2017). To avoid futile responses to these permanently occurring fluctuations at the level of complex biogenesis, plants need to integrate changes in light intensity over a longer time interval of several hours to a few days until acclimation responses are initiated. Full acclimation of the photosynthetic apparatus to a major change in the light environment can take >1 week to be completed (Chow and Anderson, 1987; Anderson et al., 1988).
Taken together, our data strongly indicate that in differentiated chloroplasts, rapid posttranslational and physiological regulatory processes dominate in light acclimation, whereas the regulation of gene expression plays only a minor role. Previous studies have analyzed the effects of light (1) in de-etiolation experiments, (2) in response to extreme high light (i.e. stressful) conditions, or (3) in the single-celled alga Chlamydomonas reinhardtii or in cyanobacteria (reviewed in Dietz, 2015; Sun and Zerges, 2015; Zoschke and Bock, 2018). All these experimental systems are valuable but may not be well suited to examine the adjustments in gene expression of differentiated chloroplasts in vascular plants during acclimation to physiological high light for several reasons. First, during de-etiolation, light-dependent programs and developmental (plastid differentiation) programs operate at the same time, and their distinct effects cannot be clearly disentangled (e.g. Sun and Zerges, 2015; Zoschke and Bock, 2018). Second, extreme high light stress induces massive damage to chloroplasts that can cause degradation of whole organelles and even cell death, and it activates systemic “rescue programs” that may be partially overlapping but are clearly not identical to acclimation responses (e.g. Dietz, 2015; Nakamura and Izumi, 2018). Only a few studies used rather mild, nonstress conditions to study light acclimation, but due to previous technical limitations, they either focused on synthesis of PsbA and very few additional photosynthetic core subunits (Mattoo et al., 1984; Fromm et al., 1985) or were performed in Chlamydomonas (e.g. Mettler et al., 2014). However, third, the control of chloroplast gene expression in Chlamydomonas and vascular plants relies on different mechanisms and factors (e.g. Nickelsen et al., 2014; Trösch et al., 2018; Zoschke and Bock, 2018), and many metabolic, morphological, and physiological features distinguish sessile multicellular plant species from motile unicellular algae. Hence, our study fills a gap and provides comprehensive information on the responses of gene expression in differentiated chloroplasts of seed plants during high light acclimation.
Together with a recent study (Chotewutmontri and Barkan, 2018), our work reported here provides a baseline for the response of gene expression in differentiated chloroplasts to continuous changes in light intensity. It will be interesting to investigate other variations in light quantity and/or quality such as fluctuating light or changes in the spectral quality (that predominantly excite PSI or PSII) and determine their impact on plastid RNA metabolism and translation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Wild-type tobacco (Nicotiana tabacum ‘Petit Havana’) was germinated and grown on soil under fully controlled conditions (16 h light/8 h dark, 100 µmol m−2 s−1, 22°C, 75% humidity) for 8 d, after which seedlings were transplanted. Starting at 10 d after sowing, the seedlings were grown under control conditions (16 h light/8 h dark; 350, 50, or 1,000 µmol m−2 s−1; 24°C/20°C; 60%/55% humidity). The light-shift experiment was performed in duplicate growth chambers with, apart from light, the same growth parameters 21 d after sowing when the seedlings were in a four-true-leaves stage, 5 h after the start of the light period (when photosynthesis runs in steady-state mode and the effects of sudden high light on chloroplast gene expression can be measured with minimal influence from other perturbations or variable parameters such as circadian or diurnal regulation). First, aerial parts of some plants (excluding cotyledons) were harvested directly before the light shift (zero control) and snap frozen in liquid nitrogen. Then, the remaining plants were removed from control conditions and within 1 min, half of the plants were transferred to high light or low light conditions (16 h light/8 h dark, 1,000 or 50 µmol m−2 s−1, 24°C/20°C, 60%/55% humidity), whereas the other half were transferred back to control conditions (see above). After transfer, aerial parts of the plants (excluding cotyledons) were harvested from shifted and control plants in parallel at defined time points (0.5 min, 5 min, 20 min, 1 h, 5 h, 1 d, and 2 d after the shift; for the low light to high light shifts, plants were only harvested 20 min after the shift), snap frozen in liquid nitrogen, and stored together with the zero control tissue at −80°C until use.
Measurements of Photosynthetic Parameters
Chlorophyll was extracted from leaf discs of defined surface area with 80% (v/v) acetone and measured according to Porra et al. (1989). Chlorophyll a fluorescence emission spectra at 77 K were measured using an F-6500 fluorometer (Jasco Inc.). The sample was excited at 430 nm wavelength with a bandwidth of 10 nm, and the emission spectrum was recorded between 655 and 800 nm wavelength in 0.5-nm intervals with a bandwidth of 1 nm. Leaf discs were immediately frozen in liquid nitrogen, homogenized in a mortar, rapidly diluted in 10 mL of ice-cold buffer (5 mm MgCl2, 40 mm KCl, 330 mm sorbitol, and 50 mm HEPES, pH 7.8) to a chlorophyll concentration of <10 µg/mL to avoid reabsorbance of PSII fluorescence emission by PSI-LHCI, and then immediately transferred into the 77 K fluorometer.
In vivo measurements of chlorophyll a fluorescence parameters of intact plants at 24°C were performed using the MAXI version of the Imaging-PAM M-series (Heinz Walz GmbH). Plants were dark adapted for 20 min prior to determination of FV/FM. Then, the actinic light intensity was stepwise increased to 1,200 µmol m−2 s−1, with measuring intervals at each light intensity between 150 s (for light intensities up to the growth light intensity, to allow for the slow activation of the CBC) and 30 s (for saturating light intensities). Linear electron transport rate was calculated from the PSII yield, and corrected for leaf absorptance, which was calculated from leaf transmittance and reflectance spectra as 100% minus transmittance (percent) minus reflectance (percent). Spectra were measured between 400 and 700 nm wavelength using an integrating sphere attached to a photometer (V650, Jasco Inc.) with a spectral bandwidth of 1 nm and a scanning speed of 200 nm min−1. Per plant, photosynthetic parameters of two subsequent leaf generations were analyzed, and no significant leaf-age-related differences were obtained, so that data were averaged.
Protein Methods
Proteins were extracted from 400 mg leaf tissue as described by Cahoon et al. (1992). Of the extracted total protein, 4 µg was separated by SDS-PAGE on Mini-Protean TGX gels (Bio-Rad), blotted onto nitrocellulose membranes, and hybridized with antibodies obtained from Agrisera (except for the actin antibody, which was obtained from Sigma-Aldrich). Luminescence signals were detected using the ECL Prime western blotting detection reagents (GE Healthcare) and the G:Box Chemi XT4 (Syngene). Signal intensity was quantified using Image Lab (version 6.0; Bio-Rad). As loading control, the membranes were stained with Ponceau S prior to hybridization and subject to final hybridization with an actin antibody.
Ribosome Profiling
Microarray-based ribosome profiling of tobacco seedlings was performed as described before (Trösch et al., 2018). Ribosome footprints, as well as total RNA, from shifted and control plants were differentially labeled with Cy3 and Cy5, respectively, and competitively hybridized onto custom microarrays as described (Trösch et al., 2018).
Data processing and analysis were carried out as previously described (Trösch et al., 2018), with the following minor modifications. To reduce background noise to a minimum, local background-subtracted single-channel probe signals (F635-B635 and F532-B532, respectively; Supplemental Dataset S1) <100 were considered as background and were set to zero. Probes that displayed signal saturation were excluded from further analysis. The signals of each dataset were normalized to the average signal of all datasets from the moderate light-to-high light shift experiments. RF signals were calculated as the average of normalized signals from probes whose middle position was located within the respective RF and tested for reproducibility between biological replicates (Supplemental Fig. S5). To minimize potential systematic deviations, e.g. by the different labeling dyes, these values from each RF from zero control samples (collected before shift) were logarithmized and subtracted from the similar value for every time point after the shift (except for extreme shifts). The average values of biological replicates (number indicated in figure legends) of the shifted plants were subsequently normalized to the values of the respective controls to enable the comparative visualization of relative changes over time for genes with very different expression levels. The averages and standard deviations of ribosome footprints and total RNA of each RF were calculated from three biological replicates (except for that of the low light-to-high light shift that was derived from one biological replicate), each including two to four technical replicates (replicate probe spots). Relative ribosome footprint and transcript accumulation were calculated for each RF by subtracting the average logarithmized signal of the control sample from the average logarithmized signal from the shifted sample. Translation efficiencies were calculated for each RF by subtracting the summarized logarithmized signals of the total mRNA from the summarized logarithmized signals of the ribosome footprints. Significance of changes in gene expression was assessed by Student’s t test. Resulting P-values were adjusted for multiple testing according to Hochberg and Benjamini (1990). Differential pausing of elongating ribosomes was evaluated by normalizing ribosome footprint signal intensities of probes located in protein-coding regions to the sum of the signal intensities of all probes in the respective RF and subsequent calculation of ratios of these relative ribosome occupancies between shifted and control plants for each time point after high light shift, as previously described (Chotewutmontri and Barkan, 2018).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NC_001879.2 (tobacco chloroplast genome).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf and soil surface temperatures after high light shift of tobacco plants.
Supplemental Figure S2. Chlorophyll a/b ratio, chlorophyll content, maximum quantum efficiency of PSII in the dark-adapted state, and linear electron transport capacity in high light-shifted and control plants.
Supplemental Figure S3. 77K chlorophyll-a fluorescence emission spectra of high light-shifted and control plants.
Supplemental Figure S4. Immunoblot analysis of core photosynthetic proteins in high light-shifted and control plants.
Supplemental Figure S5. Reproducibility of footprint abundance and transcript accumulation data between representative biological replicates.
Supplemental Figure S6. Dynamics in chloroplast transcript accumulation and translation during acclimation to high light for individual time points after high light shift.
Supplemental Figure S7. Quantitative plastome-wide comparison of relative ribosome occupancies within reading frames between shifted and control conditions for individual time points after high light shift.
Supplemental Dataset S1. Plastid tiling microarray design and ribosome profiling and transcriptome data.
Acknowledgments
We are grateful to Darya Malko, Anita Henze, Md Ahsan Habib Maruf, and Ines Gerlach (all from Max Planck Institute of Molecular Plant Physiology) for excellent technical assistance. We thank Alice Barkan and Prakitchai Chotewutmontri (University of Oregon) for critical discussions on the manuscript.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grants ZO 302/4-1 to R.Z. and SFB-TRR 175 A04 to R.B. and R.Z.) and the Max Planck Society (R.B., M.A.S., and R.Z.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Albanese P, Manfredi M, Re A, Marengo E, Saracco G, Pagliano C (2018) Thylakoid proteome modulation in pea plants grown at different irradiances: Quantitative proteomic profiling in a non-model organism aided by transcriptomic data integration. Plant J 96: 786–800 [DOI] [PubMed] [Google Scholar]
- Allen JF, Pfannschmidt T (2000) Balancing the two photosystems: Photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond B Biol Sci 355: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancín M, Fernández-San Millán A, Larraya L, Morales F, Veramendi J, Aranjuelo I, Farran I (2019) Overexpression of thioredoxin m in tobacco chloroplasts inhibits the protein kinase STN7 and alters photosynthetic performance. J Exp Bot 70: 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Chow W, Goodchild D (1988) Thylakoid membrane organisation in sun/shade acclimation. Funct Plant Biol 15: 11–26 [Google Scholar]
- Armarego-Marriott T, Kowalewska Ł, Burgos A, Fischer A, Thiele W, Erban A, Strand D, Kahlau S, Hertle A, Kopka J, et al. (2019) Highly resolved systems biology to dissect the etioplast-to-chloroplast transition in tobacco leaves. Plant Physiol 180: 654–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Correa Galvis V, Kunz HH, Strand DD (2017) The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr Opin Plant Biol 37: 56–62 [DOI] [PubMed] [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix JD (2005) State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Berry JO, Carr JP, Klessig DF (1988) mRNAs encoding ribulose-1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc Natl Acad Sci USA 85: 4190–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JO, Yerramsetty P, Zielinski AM, Mure CM (2013) Photosynthetic gene expression in higher plants. Photosynth Res 117: 91–120 [DOI] [PubMed] [Google Scholar]
- Blair GE, Ellis RJ (1973) Protein synthesis in chloroplasts. I. Light-driven synthesis of the large subunit of fraction I protein by isolated pea chloroplasts. Biochim Biophys Acta 319: 223–234 [DOI] [PubMed] [Google Scholar]
- Bock R. (2007) Cell and Molecular Biology of Plastids, Vol 19 Springer Verlag, Berlin [Google Scholar]
- Cahoon EB, Shanklin J, Ohlrogge JB (1992) Expression of a coriander desaturase results in petroselinic acid production in transgenic tobacco. Proc Natl Acad Sci USA 89: 11184–11188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Barkan A (2016) Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet 12: e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Barkan A (2018) Multilevel effects of light on ribosome dynamics in chloroplasts program genome-wide and psbA-specific changes in translation. PLoS Genet 14: e1007555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WS, Anderson JM (1987) Photosynthetic responses of Pisum sativum to an increase in irradiance during growth. II. Thylakoid membrane components. Funct Plant Biol 14: 9–19 [Google Scholar]
- Chow WS, Melis A, Anderson JM (1990) Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA 87: 7502–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JA, Avenson TJ, Kanazawa A, Takizawa K, Edwards GE, Kramer DM (2005) Plasticity in light reactions of photosynthesis for energy production and photoprotection. J Exp Bot 56: 395–406 [DOI] [PubMed] [Google Scholar]
- Dietz KJ. (2015) Efficient high light acclimation involves rapid processes at multiple mechanistic levels. J Exp Bot 66: 2401–2414 [DOI] [PubMed] [Google Scholar]
- Eaglesham ARJ, Ellis RJ (1974) Protein synthesis in chloroplasts: II. Light-driven synthesis of membrane proteins by isolated pea chloroplasts. Biochimica et Biophysica Acta 335: 396–407 [DOI] [PubMed] [Google Scholar]
- Edhofer I, Mühlbauer SK, Eichacker LA (1998) Light regulates the rate of translation elongation of chloroplast reaction center protein D1. Eur J Biochem 257: 78–84 [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Bräutigam K, Pfannschmidt T (2005) Photosynthetic redox control of nuclear gene expression. J Exp Bot 56: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Neukermans J, Queval G, Noctor G, Harbinson J (2012) Photosynthetic control of electron transport and the regulation of gene expression. J Exp Bot 63: 1637–1661 [DOI] [PubMed] [Google Scholar]
- Fromm H, Devic M, Fluhr R, Edelman M (1985) Control of psbA gene expression: In mature Spirodela chloroplasts light regulation of 32-kd protein synthesis is independent of transcript level. EMBO J 4: 291–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyula P, Schäfer E, Nagy F (2003) Light perception and signalling in higher plants. Curr Opin Plant Biol 6: 446–452 [DOI] [PubMed] [Google Scholar]
- Herbstová M, Tietz S, Kinzel C, Turkina MV, Kirchhoff H (2012) Architectural switch in plant photosynthetic membranes induced by light stress. Proc Natl Acad Sci USA 109: 20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9: 811–818 [DOI] [PubMed] [Google Scholar]
- Hojka M, Thiele W, Tóth SZ, Lein W, Bock R, Schöttler MA (2014) Inducible repression of nuclear-encoded subunits of the cytochrome b6f complex in tobacco reveals an extraordinarily long lifetime of the complex. Plant Physiol 165: 1632–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Hussmann JA, Weissman JS (2019) Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 11: a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Aro EM (2015) Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim Biophys Acta 1847: 900–909 [DOI] [PubMed] [Google Scholar]
- Jenkins GI. (2014) The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Johnson GN (2011) Regulation of cyclic and linear electron flow in higher plants. Proc Natl Acad Sci USA 108: 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser E, Morales A, Harbinson J, Kromdijk J, Heuvelink E, Marcelis LF (2015) Dynamic photosynthesis in different environmental conditions. J Exp Bot 66: 2415–2426 [DOI] [PubMed] [Google Scholar]
- Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development In Timmermans MCP, ed, Current Topics in Developmental Biology, Vol Vol 91 Academic Press, Cambridge, MA, pp 29–66 [DOI] [PubMed] [Google Scholar]
- Kim J, Mullet JE (2003) A mechanism for light-induced translation of the rbcL mRNA encoding the large subunit of ribulose-1,5-bisphosphate carboxylase in barley chloroplasts. Plant Cell Physiol 44: 491–499 [DOI] [PubMed] [Google Scholar]
- Klein RR, Mason HS, Mullet JE (1988) Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol 106: 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Mullet JE (1986) Regulation of chloroplast-encoded chlorophyll-binding protein translation during higher plant chloroplast biogenesis. J Biol Chem 261: 11138–11145 [PubMed] [Google Scholar]
- Klein RR, Mullet JE (1987) Control of gene expression during higher plant chloroplast biogenesis. Protein synthesis and transcript levels of psbA, psaA-psaB, and rbcL in dark-grown and illuminated barley seedlings. J Biol Chem 262: 4341–4348 [PubMed] [Google Scholar]
- Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: The basics. Annu Rev Plant Physiol Plant Mol Biol 42: 313–349 [Google Scholar]
- Krech K, Ruf S, Masduki FF, Thiele W, Bednarczyk D, Albus CA, Tiller N, Hasse C, Schöttler MA, Bock R (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol 159: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A (2008) Singlet oxygen production in photosystem II and related protection mechanism. Photosynth Res 98: 551–564 [DOI] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell 29: 207–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41: W575–W581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M (1984) Regulation of protein metabolism: Coupling of photosynthetic electron transport to in vivo degradation of the rapidly metabolized 32-kilodalton protein of the chloroplast membranes. Proc Natl Acad Sci USA 81: 1380–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler T, Mühlhaus T, Hemme D, Schöttler MA, Rupprecht J, Idoine A, Veyel D, Pal SK, Yaneva-Roder L, Winck FV, et al. (2014) Systems analysis of the response of photosynthesis, metabolism, and growth to an increase in irradiance in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 2310–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MAE, O’Cualain R, Selley J, Knight D, Karim MF, Hubbard SJ, Johnson GN (2017) Dynamic acclimation to high light in Arabidopsis thaliana involves widespread reengineering of the leaf proteome. Front Plant Sci 8: 1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer SK, Eichacker LA (1998) Light-dependent formation of the photosynthetic proton gradient regulates translation elongation in chloroplasts. J Biol Chem 273: 20935–20940 [DOI] [PubMed] [Google Scholar]
- Murata N, Nishiyama Y (2018) ATP is a driving force in the repair of photosystem II during photoinhibition. Plant Cell Environ 41: 285–299 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Izumi M (2018) Regulation of chlorophagy during photoinhibition and senescence: Lessons from mitophagy. Plant Cell Physiol 59: 1135–1143 [DOI] [PubMed] [Google Scholar]
- Nickelsen J, Bohne A-V, Westhoff P (2014) Chloroplast gene expression—Translation In Theg SM, and Wollman F-A, eds, Plastid Biology, Vol Vol 5 Springer, New York, pp 49–78 [Google Scholar]
- Ohad I, Kyle DJ, Arntzen CJ (1984) Membrane protein damage and repair: Removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J Cell Biol 99: 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-I, Anderson JM, Chow WS (1996) Photoinactivation of functional photosystem II and D1-protein synthesis in vivo are independent of the modulation of the photosynthetic apparatus by growth irradiance. Planta 198: 300–309 [Google Scholar]
- Petersen K, Schöttler MA, Karcher D, Thiele W, Bock R (2011) Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucleic Acids Res 39: 5181–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Allen JF (1999) Photosynthetic control of chloroplast gene expression. Nature 397: 625–628 [Google Scholar]
- Pogson BJ, Woo NS, Förster B, Small ID (2008) Plastid signalling to the nucleus and beyond. Trends Plant Sci 13: 602–609 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Puthiyaveetil S, Tsabari O, Lowry T, Lenhert S, Lewis RR, Reich Z, Kirchhoff H (2014) Compartmentalization of the protein repair machinery in photosynthetic membranes. Proc Natl Acad Sci USA 111: 15839–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamäki E, Martinsuo P, Pursiheimo S, Aro EM (2000) Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc Natl Acad Sci USA 97: 11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2014) Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol 65: 287–309 [DOI] [PubMed] [Google Scholar]
- Salesse-Smith CE, Sharwood RE, Busch FA, Kromdijk J, Bardal V, Stern DB (2018) Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat Plants 4: 802–810 [DOI] [PubMed] [Google Scholar]
- Schöttler MA, Thiele W, Belkius K, Bergner SV, Flügel C, Wittenberg G, Agrawal S, Stegemann S, Ruf S, Bock R (2017) The plastid-encoded PsaI subunit stabilizes photosystem I during leaf senescence in tobacco. J Exp Bot 68: 1137–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler MA, Tóth SZ (2014) Photosynthetic complex stoichiometry dynamics in higher plants: Environmental acclimation and photosynthetic flux control. Front Plant Sci 5: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler MA, Tóth SZ, Boulouis A, Kahlau S (2015) Photosynthetic complex stoichiometry dynamics in higher plants: Biogenesis, function, and turnover of ATP synthase and the cytochrome b6f complex. J Exp Bot 66: 2373–2400 [DOI] [PubMed] [Google Scholar]
- Sun Y, Zerges W (2015) Translational regulation in chloroplasts for development and homeostasis. Biochim Biophys Acta 1847: 809–820 [DOI] [PubMed] [Google Scholar]
- Sundby C, McCaffery S, Anderson JM (1993) Turnover of the photosystem II D1 protein in higher plants under photoinhibitory and nonphotoinhibitory irradiance. J Biol Chem 268: 25476–25482 [PubMed] [Google Scholar]
- Taniguchi M, Kuroda H, Satoh K (1993) ATP-dependent protein synthesis in isolated pea chloroplasts. Evidence for accumulation of a translation intermediate of the D1 protein. FEBS Lett 317: 57–61 [DOI] [PubMed] [Google Scholar]
- Theis J, Schroda M (2016) Revisiting the photosystem II repair cycle. Plant Signal Behav 11: e1218587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM (2014) Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci 19: 10–17 [DOI] [PubMed] [Google Scholar]
- Trösch R, Barahimipour R, Gao Y, Badillo-Corona JA, Gotsmann VL, Zimmer D, Mühlhaus T, Zoschke R, Willmund F (2018) Commonalities and differences of chloroplast translation in a green alga and land plants. Nat Plants 4: 564–575 [DOI] [PubMed] [Google Scholar]
- Wada M, Kong SG (2018) Actin-mediated movement of chloroplasts. J Cell Sci 131: jcs210310. [DOI] [PubMed] [Google Scholar]
- Yamori W, Shikanai T (2016) Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol 67: 81–106 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–16067 [DOI] [PubMed] [Google Scholar]
- Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (2000) Biogenesis of the chloroplast-encoded D1 protein: Regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12: 1769–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Bock R (2018) Chloroplast translation: Structural and functional organization, operational control, and regulation. Plant Cell 30: 745–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Watkins KP, Barkan A (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25: 2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]