Characterizing different small RNA populations reveals the roles of development and genotype in expression variation in maize inbreds and their hybrids.
Abstract
Small RNAs (sRNAs) regulate gene expression, play important roles in epigenetic pathways, and are hypothesized to contribute to hybrid vigor in plants. Prior investigations have provided valuable insights into associations between sRNAs and heterosis, often using a single hybrid genotype or tissue, but our understanding of the role of sRNAs and their potential value to plant breeding are limited by an incomplete picture of sRNA variation between diverse genotypes and development stages. Here, we provide a deep exploration of sRNA variation and inheritance among a panel of 108 maize (Zea mays) samples spanning five tissues from eight inbred parents and 12 hybrid genotypes, covering a spectrum of heterotic groups, genetic variation, and levels of heterosis for various traits. We document substantial developmental and genotypic influences on sRNA expression, with varying patterns for 21-nucleotide (nt), 22-nt, and 24-nt sRNAs. We provide a detailed view of the distribution of sRNAs in the maize genome, revealing a complex makeup that also shows developmental plasticity, particularly for 22-nt sRNAs. sRNAs exhibited substantially more variation between inbreds as compared with observed variation for gene expression. In hybrids, we identify locus-specific examples of nonadditive inheritance, mostly characterized as partial or complete dominance, but rarely outside the parental range. However, the global abundance of 21-nt, 22-nt, and 24-nt sRNAs varies very little between inbreds and hybrids, suggesting that hybridization affects sRNA expression principally at specific loci rather than on a global scale. This study provides a valuable resource for understanding the potential role of sRNAs in hybrid vigor.
Molecular variation is widely studied to understand the basis of plant traits. This can include variation in DNA sequence, DNA methylation, or chromatin modifications, changes in abundance of mRNA or small RNAs (sRNAs), as well as changes in the proteome or metabolome. These types of variation differ substantially in their heritability across generations and stability in different cells or tissues of the same organism. Changes in DNA sequence are highly stable among the cells of an organism, while the abundance of mRNAs or metabolites can vary during development or in response to environmental conditions. This stability is a key factor when considering experimental designs for detecting molecular variation and for attempting to link molecular variation to plant traits. In this study, we monitored variation in sRNA abundance in different tissues and genotypes of maize (Zea mays) to understand the patterns of sRNA variation and inheritance.
Heterosis, or hybrid vigor, occurs when an F1 hybrid outperforms either parent. The contribution of various potential molecular mechanisms to heterosis remains one of the most intriguing and powerful enigmas in plant biology (Birchler et al., 2003; Hochholdinger and Baldauf, 2018). While the molecular basis of heterosis remains uncertain, it is clear that variation between members of the same species is a requirement (Schnable and Springer, 2013). This variation can occur at the genomic, transcriptomic, and epigenetic level (Chen, 2013). Unfortunately, we still lack a coherent molecular mechanism to explain the sources of nonadditive inheritance patterns of gene expression in hybrids (Birchler et al., 2010). However, a variety of studies have found evidence that epigenetic factors, including sRNAs and DNA methylation, play a role in heterosis (reviewed in Groszmann et al., 2013; Ryder et al., 2014; Greaves et al., 2015). sRNAs are promising candidates for modulating the epigenome and gene expression in hybrids. sRNAs can regulate gene expression via posttranscriptional gene silencing and through transcriptional silencing by directing changes in DNA methylation (Borges and Martienssen, 2015). The canonical functional size classes of endogenous sRNAs in plants include 21-nucleotide (nt), 22-nt, and 24-nt sRNAs (Axtell, 2013). The 21-nt sRNAs are mainly composed of microRNAs (miRNAs) that regulate the expression of mRNAs, while the 24-nt size class is predominantly produced by the RNA-directed DNA methylation pathway to control transposable elements (TEs; Borges and Martienssen, 2015). In maize, RNA-directed DNA methylation activity is localized to gene flanks (Gent et al., 2013; Li et al., 2015a) where 24-nt sRNAs are abundant (Wang et al., 2009; Xin et al., 2014) and positively correlate with expression levels of the flanking gene (Gent et al., 2013; Lunardon et al., 2016). In fact, the bulk of the heterochromatic maize genome may be incompatible with sRNA production (Gent et al., 2014). Among the species that have been profiled, the 22-nt size class stands out in maize because of its abundance relative to the other classes of sRNAs. A detailed analysis of a 22-Mb contiguous region of the maize genome found that 22-nt sRNAs are enriched in the highly repetitive regions (Nobuta et al., 2008; Wei et al., 2009; Regulski et al., 2013). Despite our knowledge of sRNA in many species, including maize, there are open questions about the sources of sRNA variation among maize lines, the inheritance pattern of this variation, and how it might contribute to heterosis.
Prior studies have compared sRNA profiles between hybrids and their parents across various species, including in Arabidopsis (Arabidopsis thaliana; Groszmann et al., 2011; Li et al., 2012, 2015b; Shen et al., 2012), rice (Oryza sativa; Chen et al., 2010; He et al., 2010; Chodavarapu et al., 2012), tomato (Solanum lycopersicum and Solanum pennellii; Shivaprasad et al., 2012), wheat (Triticum aestivum; Kenan-Eichler et al., 2011), and maize (Barber et al., 2012; He et al., 2013; Regulski et al., 2013; Xin et al., 2014; Seifert et al., 2018a, 2018b). Collectively, these investigations indicate that, like mRNA expression, sRNA expression can be inherited nonadditively. sRNAs tend to have reduced expression in hybrids relative to parents, particularly 24-nt sRNAs (Groszmann et al., 2011), and this trend is most evident where sRNAs are differentially expressed between parents (Groszmann et al., 2011; Barber et al., 2012; Shen et al., 2012). sRNAs are also associated with changes in DNA methylation and changes in gene expression in hybrids (Chodavarapu et al., 2012; Shen et al., 2012; Greaves et al., 2014; Li et al., 2015b). In maize, associations between sRNA expression and heterotic traits have also been detailed (Seifert et al., 2018a), leading to the identification of hybrid performance-associated sRNAs correlated with hybrid performance for grain yield (Seifert et al., 2018b). Thus, there is potential to incorporate sRNA expression into hybrid performance predictive models and achieve improved accuracy.
Despite these prior investigations, we have an incomplete picture of the genomic nature of sRNA variation. The relative variation among different genotypes or tissues for 21-nt, 22-nt, or 24-nt sRNA abundance has not been investigated. In addition, we have limited understanding of the genomic features that contribute to consistent or variable sRNA abundance. While previous studies have examined sRNA profiles in maize and other plant species, they have often been limited to a single hybrid genotype or a single tissue. In this study, we combine sRNA profiles of five tissues from a diverse set of maize inbreds and hybrids to investigate genotypic and developmental variation in sRNA patterns, the levels and sources of sRNA variation, and the connection between sRNAs and heterosis.
RESULTS
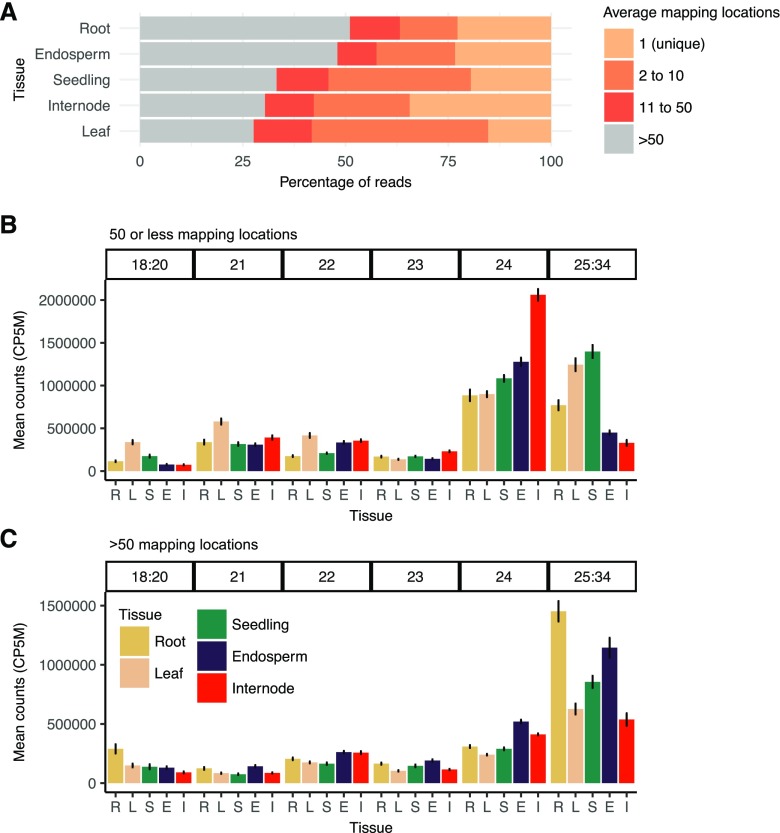
sRNAs were sequenced for five tissues (15-d after pollination endosperm, V7/8 leaf, V1 seedling root, V1 seedling shoot, and V7/V8 internode) of a panel of maize inbreds and hybrids (Supplemental Table S1). After filtering several samples with low sequencing depth, we obtained sRNA profiles for eight inbred and 13 hybrid genotypes for a total of 108 sRNA samples. An average of 5.2 million reads were generated per sample, with a range of 1.9 to 17 million reads (Supplemental Table S2). These sRNA sequences were mapped to the B73 RefGen_v4 genome (Jiao et al., 2017) and then split by size class (bioinformatic approach detailed by Mathioni et al. [2016]). Depending on the tissue, 15% to 34% of the sRNAs could be mapped uniquely to a single best location in the genome (Fig. 1A). There were significant differences in the multimapping rates between tissues; for instance, nearly 50% of sRNAs mapped to highly repetitive sequences in endosperm and root compared with only around 30% in other tissues (Fig. 1A; Supplemental Fig. S1A; ANOVA, Tukey’s honestly significant difference P < 0.001). When sRNAs mapped to multiple genomic locations, the count attributed to each location was scaled proportional to the number of locations to which it mapped.
Figure 1.
Multimapping frequency and length distribution of sRNA reads. A, Average read multimapping frequency of sRNAs. Mapping frequency is shown for genome mapped reads (excluding structural RNAs) of length 18-nt to 34-nt that perfectly aligned with no mismatches to the B73 reference genome for all samples in this study, including inbreds and hybrids. For each sample, mapping rates were categorized into four mapping frequency bins and expressed as a proportion of all mapped reads, and then the distributions were averaged for each tissue. Reads with mapping frequency 1 have a single (unique) high-confidence mapping location. Reads were then categorized into bins with two to 10, 11 to 50, or greater than 50 mapping locations. B, Comparison of the relative abundance of different size classes (bar along top) of sRNAs between different tissues for sequences with fewer than 50 mapping locations. C, Relative abundance of sRNAs mapping to highly repetitive regions (more than 50 mapping locations). CP5M, Clusters with at least 10 counts per five million. Error bars denote se among genotypes. R = seedling root (n = 20), L = leaf (n = 18), S = seedling shoot (n = 21), E = endosperm (n = 28), and I = internode (n = 21).
Substantial Developmental Variation in sRNA Profiles
The size distribution for sRNAs with 50 or fewer mapping locations was assessed for each tissue in the full set of inbred and hybrid genotypes (Fig. 1B). Most work on sRNAs has focused on 21-nt, 22-nt, or 24-nt size classes, but there are also many sRNA reads that are 18-20-nt or 25-34-nt (Supplemental Fig. S2A). The abundance of individual size classes in the range 18-20-nt and 25-34-nt was low compared with 21-nt, 22-nt, or 24-nt sRNAs (Supplemental Fig. S2A); however, collectively, there was a substantial number of RNAs from these size classes when they were summed together (Fig. 1B). There was significant variability in the total level of sRNAs between 25-nt and 34-nt observed in the five tissues. For some tissues (leaf and seedling shoot), these were the most abundant size classes of sRNAs. If we normalized the sRNA expression levels based on the abundance of 25-34-nt sRNAs, it was clear that 21-nt, 22-nt, and 24-nt sRNAs were higher in internodes than in other tissues (Supplemental Fig. S2B). Endosperm also tended to have somewhat elevated levels of 24-nt and 22-nt sRNAs compared with other tissues when normalized in this manner (Supplemental Fig. S2B). The analysis of sRNAs from highly repetitive regions (more than 50 mapped locations) revealed high contributions of 25-34-nt sRNAs and slightly elevated levels of 21-nt, 22-nt, and 24-nt sRNAs in endosperm relative to other tissues (Fig. 1C). We note that the choice about whether to include sRNAs from highly repetitive regions or sRNAs from the 25-34-nt size range in normalization can have significant impacts upon the perceived abundance of 21-nt, 22-nt, and 24-nt sRNAs, especially in comparisons among tissues. For this study, we elected to focus most analyses on sRNAs that have fewer than 50 mapped locations in B73 and to normalize using the total counts for the 18-34-nt set of sRNA size classes.
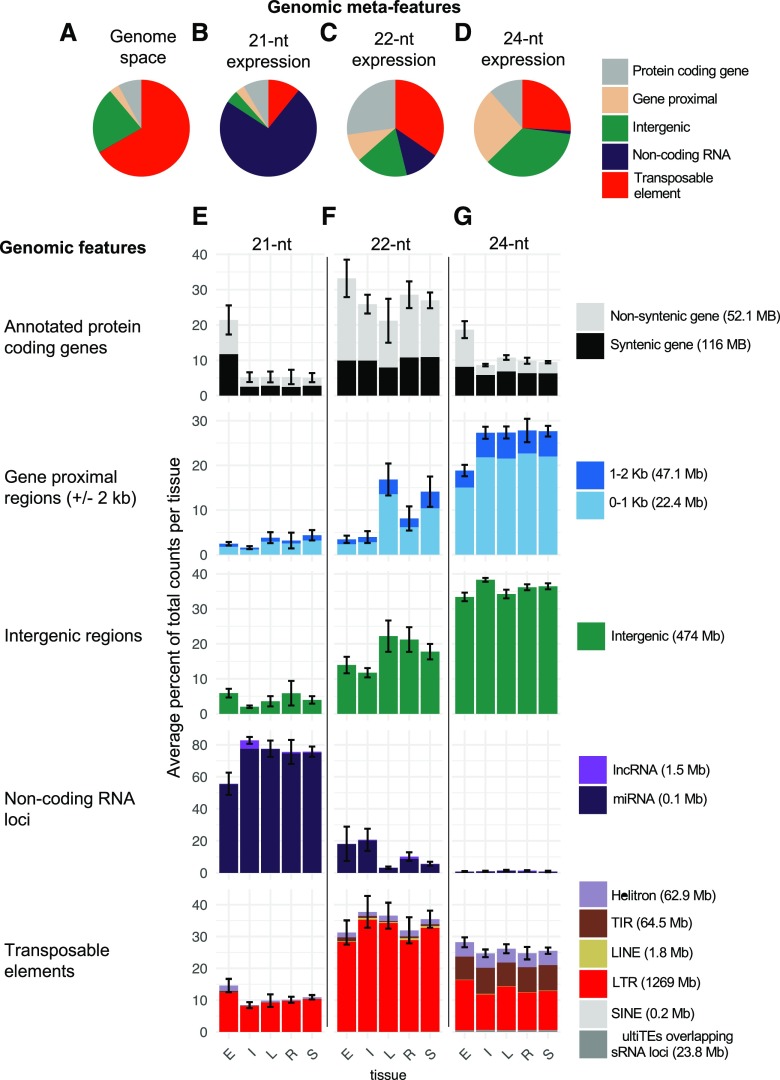
To evaluate variation in locus-specific abundances of sRNAs, we counted the 21-nt, 22-nt, and 24-nt sRNA reads in 100-bp windows of the maize genome that we termed sRNA loci. We examined in detail the genomic locations of expressed sRNA loci, which were defined as CP5M in at least one sample. sRNA loci can occur in both genic and nongenic regions of the genome. We divided the genome into five meta-feature categories by hierarchically classifying the genomic location of sRNA loci (see “Materials and Methods”) as noncoding RNA (e.g. miRNA and long noncoding RNA), genic, gene-proximal (within 2 kb of a gene), TE, or intergenic (greater than 2 kb from a gene). First, we determined the background distribution of each of the genomic features in the genome (Fig. 2A). As expected, TEs accounted for the majority of the maize genome space (∼66.7%) based on the latest TE annotation of B73 (Anderson et al., 2019), followed by intergenic regions (22.1%), genes and gene-proximal regions (11.1%), and a very small fraction was annotated as noncoding RNA loci (0.1%). We then determined the distribution of genomic features represented by each size class of sRNAs (Fig. 2, B–D). For instance, for 21-nt sRNAs, relatively few noncoding RNA loci in the genome accounted for the majority of all 21-nt counts (Fig. 2B). These analyses revealed substantial differences in the distribution of genomic features that contribute to the pool of 21-nt, 22-nt, and 24-nt sRNAs that fit with expectations based on prior research on plant sRNAs. The 21-nt sRNAs were mostly generated from noncoding RNAs (the vast majority from miRNA), 22-nt sRNAs predominantly from genes and TEs, while the largest fraction of 24-nt sRNAs came from intergenic and gene-proximal regions, with significant contributions from TEs as well.
Figure 2.
Distribution of sRNA expression among the genomic features of the genome. A, Background genomic distribution of genomic features determined by annotating each 100-bp tile of the genome as noncoding RNA (e.g. miRNA and long noncoding RNA [lncRNA]), genic, gene-proximal (within 2 kb of a gene), TE, or intergenic using the genome reference B73 RefGen_v4 and annotation based on Gramene version 36 and miRbase release 22. B to D, Distribution of sRNA expression (CP5M) across meta-features for each sample averaged to determine the meta-feature distribution for each size class. E to G, Each meta-feature was then divided into constituent features, and the average distribution of sRNA expression was determined per tissue. In parentheses, the total Mb pairs of each genomic feature category in the genome are listed. The bars in the same vertical column add to 100%. Error bars denote sd among samples for the meta-feature category and represent the variation across the panel of inbred and hybrid genotypes. E = endosperm (n = 28), I = internode (n = 21), L = leaf (n = 18), R = seedling root (n = 20), and S = seedling shoot (n = 21). LINE, Long interspersed nuclear element; LTR, long terminal repeat; SINE, short interspersed nuclear element; TIR, terminal inverted repeat.
It is unclear whether development leads to changes in the distribution of sRNA in the genome in maize. To explore the developmental variation in sRNA profiles, we assessed the relative contributions of distinct types of genomic regions to the sRNA expression profiles in each tissue (Fig. 2, E–G). For each of the 21 genotypes with data across the five tissues, we assessed the independence between tissue and the genomic distribution of sRNA within each size class using Pearson’s χ2 test. For all genotypes, the relationship was highly significant (P < 0.001), indicating significant variation between tissues. To identify the source of the variation, we extracted the residuals to compare the contribution of different genomic elements with the variation observed (Supplemental Fig. S3A). For 21-nt sRNAs, vegetative tissues had a highly consistent profile, mostly composed of miRNAs (Fig. 2E; Supplemental Fig. S3A). However, endosperm had a distinct increase in genic 21-nt abundance from both syntenic and nonsyntenic genes and an increase in long terminal repeat (LTR) retrotransposons. This was the major source of variation between the tissues for 21-nt sRNA (Supplemental Fig. S3A) and was consistently observed for the different genotypes assessed in this study (Supplemental Fig. S4). For 22-nt expression, there was substantial variation between the tissues, particularly for gene-proximal and intergenic regions (Fig. 2F), which contributed the highest proportion to the χ2 statistic (Supplemental Fig. S3A). Both endosperm and internode had substantially fewer 22-nt sRNAs in gene flanks and in intergenic regions but an increase in 22-nt miRNAs. In contrast, leaf and seedling shoot had the highest level of gene-proximal 22-nt sRNAs. This analysis also revealed that for the abundant class of genic 22-nt sRNAs, the majority were in nonsyntenic genes (Fig. 2F), which is in contrast to both 21-nt and 24-nt sRNAs (Fig. 2, E and G), and the ratio of syntenic versus nonsyntenic sRNAs was significantly associated with size class for all genotypes and tissues examined (χ2, P < 0.001). Overall, there was also a significant difference (χ2, P < 0.001) in the genomic distribution of sRNA between 21-nt, 22-nt, and 24-nt sRNAs (Supplemental Fig. S3B). We note that for the purpose of these analyses, we did not differentiate between intron-, exon-, or untranslated region-derived sRNAs, and a portion of genic sRNAs likely represent mRNA decay products. For 24-nt sRNAs, again the vegetative tissues were relatively consistent and endosperm had a distinct increase in genic 24-nt counts. This analysis also highlighted that while 22-nt and 24-nt sRNA were both abundant at TEs, 22-nt were more than 90% LTRs while 24-nt were only around 50% LTRs, with substantial numbers of reads mapping to both Helitron and TIR DNA transposons. Given that the amount of genome space occupied by LTR elements (59.5%) was substantially larger than that of Helitrons (2.9%) or TIR elements (3%), there was a notable enrichment for Helitrons and TIR elements for 24-nt sRNAs.
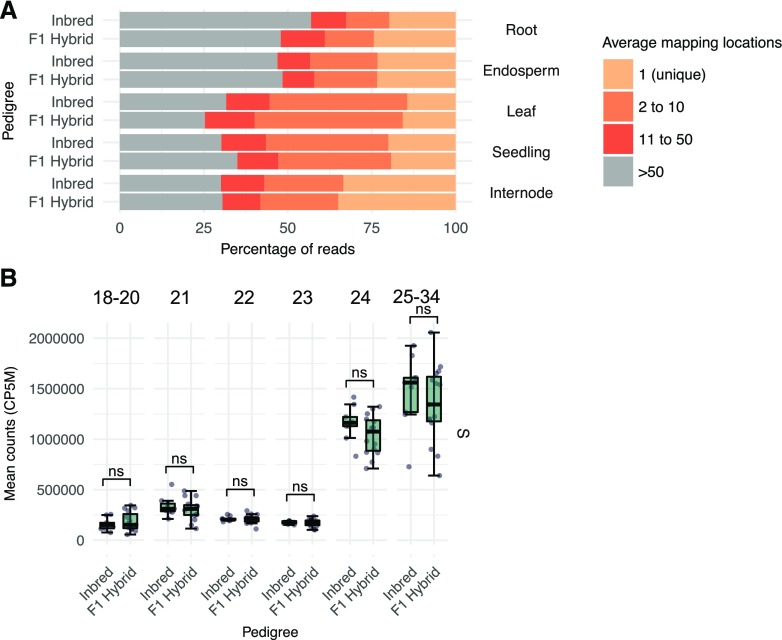
Globally, sRNA Profiles Are Similar between Inbreds and Hybrids
The analyses above included both inbred and hybrid genotypes. Prior work in Arabidopsis (Groszmann et al., 2011) and maize (Barber et al., 2012) have found altered sRNA profiles in heterotic hybrids relative to their inbred parents, suggesting that this may be a general feature of heterosis (Greaves et al., 2015), although prior studies largely focused on a single hybrid genotype relative to the parents. We compared the profiles of the sRNAs in each of the tissues from the full set of inbred and hybrid genotypes used in this study (Fig. 3A). Using a two-way ANOVA interaction model for unbalanced design (type III sum of squares), we found that pedigree (inbred/hybrid) was not a significant factor in sRNA counts for any mapping rate category (ANOVA, pedigree P > 0.05). Likewise, there was limited evidence for any significant global difference in the abundance of the different sRNA size classes, specifically between inbreds and hybrids (false discovery rate-adjusted P < 0.05, Student’s t test), for example in seedling shoot tissue (Fig. 3B) or other tissues (Supplemental Fig. S5). Several inbred/hybrid comparisons were marginally statistically different, including 24-nt abundance in roots, for which 11 of the 13 hybrids had substantially higher 24-nt abundance compared with all inbreds (P = 0.029; Supplemental Fig. S5). In addition, the distribution of the types of genomic loci contributing to sRNA expression was nearly identical between inbreds and hybrids for all size classes (Supplemental Fig. S6). These analyses suggest that maize hybrids exhibiting substantial heterosis do not have globally unique sRNA compositions relative to the inbred parents.
Figure 3.
Comparisons of sRNA profiles in inbreds relative to hybrids. A, Comparisons of multimapping rates in inbreds relative to hybrids. The average read multimapping frequency for all genome-matched sRNAs was summarized into four categories and then divided into averages for inbreds and F1 hybrids. Bars represent the average of all genotypes for each sample. B, Comparison of sRNA abundance between inbreds and hybrids for seedling shoot tissue. Each dot represents a genotype, and whiskers correspond to the first and third quartiles (the 25th and 75th percentiles). S = seedling shoot (n = 21). ns, Nonsignificant false discovery rate-adjusted P < 0.05, Student’s t test.
Size Class-Specific Developmental and Genetic Variation in sRNA Expression
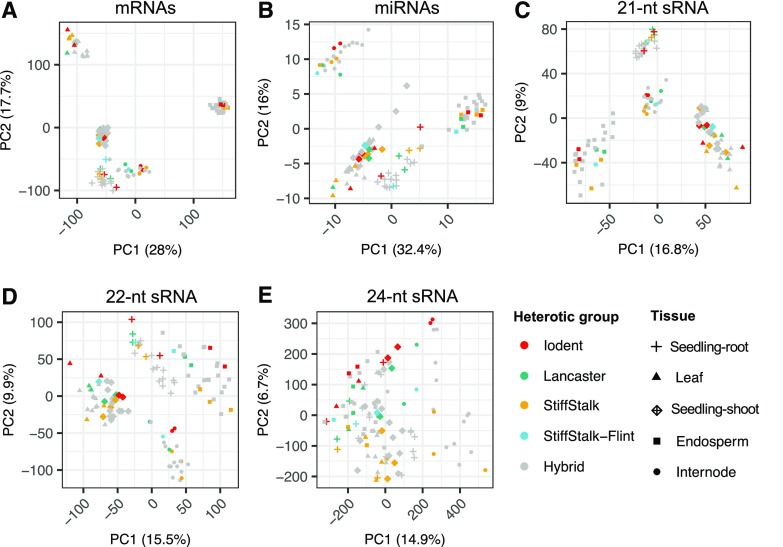
Prior work has suggested that sRNA profiles may capture unique information missed by single-nucleotide polymorphisms or gene (mRNA) expression data (Seifert et al., 2018a, 2018b). For each hybrid and inbred sample, gene expression levels were profiled by RNA sequencing (RNA-seq) using the same RNA sample used for sRNA analysis (Li et al., 2019). To compare and contrast sRNA profiles with gene expression, we summarized RNA-seq reads to counts per million (CPM) per B73v4 gene locus (APGv4 release 36). For all subsequent analyses, we then compared gene expression with sRNA expression using the sRNA data summarized to 100-bp windows. The mRNA and sRNA levels per locus were used to perform principal component analysis (PCA) to assess the relationships of profiles for different tissues and genotypes (Fig. 4). For the mRNA data, variation in the expression of genes was heavily driven by tissue type (Fig. 4A), as has been observed previously (Zhou et al., 2019). Samples clustered by tissue type into discrete groups, with principal component 1 (PC1) explaining 28% of the variation and PC2 another 17%. For miRNA, 21-nt, and 22-nt sRNA loci, samples also clustered by tissue (Fig. 4, B–D). However, seedling shoot and leaf were not separated by the first two principal components, which also accounted for relatively less of the variation compared with mRNA data, capturing ∼16% and 9%, respectively. PC3 separated seedling shoot and leaf for miRNA loci, although not for 21-nt and 22-nt sRNAs (Supplemental Fig. S7). In contrast to mRNAs, miRNAs, 21-nt, and 22-nt sRNAs, 24-nt sRNA loci were not strongly clustered by tissue type; instead, there was evidence for separation by genotype, particularly in PC2. Separate PCAs were performed for each type of molecule (mRNA, 21-nt, 22-nt, or 24-nt) within each tissue (see representative examples in Supplemental Fig. S8, A–D). One hypothesis is that heterosis could lead to a consistent change in sRNA profiles in hybrids relative to their parents. However, there was no evidence for clustering of sRNA profiles of hybrids relative to inbreds, but in most cases the hybrids were intermediate relative to the two parents in both PC1 and PC2; an example is highlighted using dashed lines in Supplemental Figure S8B for the PH207 hybrids. Very similar patterns are seen for the other size classes and tissues (representative examples are provided in Supplemental Fig. S8). This behavior of the hybrids (intermediate to the parents) was less clear for gene expression (Fig. 4E).
Figure 4.
Clustering analysis of sRNAs, miRNAs, and mRNAs. PCA of all tissues on expressed loci for mRNA, miRNA, and 21-nt to 24-nt sRNAs are summarized to 100-bp windows. For each sRNA size class, PCA was performed using expressed loci with greater than 10 CP5M in at least one sample, while mRNA loci with greater than 1 CPM was used as input. Counts were log2 transformed, scaled by unit variance, and clustered using singular value decomposition. Colors represent genotype (heterotic group or hybrid). Symbols represent tissue type. Percentages in parentheses refer to the percentage of variance explained by each principal component.
Inbred-Specific sRNA Expression Drives sRNA Variability among Genotypes
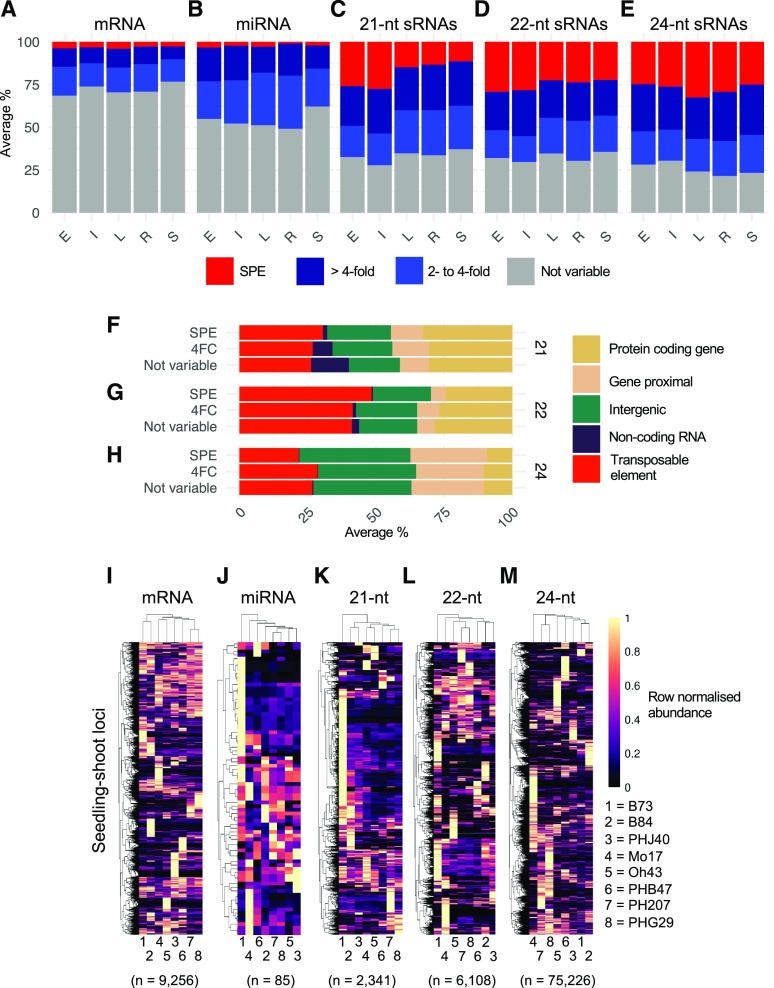
Next, we examined sRNA loci for variable expression between inbred parents. The experimental design did not include biological replicates, limiting our ability to robustly assess differential expression. However, we compared trends in terms of the proportion of sRNA loci with greater than 2-fold and greater than 4-fold variation or single parent expression (SPE) among inbreds to determine whether certain size classes or tissues might have more variation (Fig. 5, A–E). To stringently assess variable expression, only loci with greater than 10 CP5M (or the equivalent 2 CPM for mRNA) in at least one genotype were considered, and SPE was strictly defined as greater than 10 CP5M in one parent and 0 in the other. The proportion of sRNA loci with variation for sRNA expression (Fig. 5, B–E) was much greater than the proportion of genes that exhibit variation (Fig. 5A), although miRNAs were less variable compared with other sRNAs. Across all tissues and sRNA size classes (excluding miRNAs), ∼50% of regions with sRNA expression had at least 4-fold variation, and approximately half of these were SPE in at least one pair of inbreds. In contrast, miRNAs were rarely SPE. In total, more than 60% of all sRNA loci had greater than 2-fold variation; by contrast, many fewer genes exhibited variation (Fig. 5A). The level of variation was slightly higher for internode than for other tissues for 21-nt and 22-nt sRNAs but was slightly lower in this tissue for 24-nt sRNAs. For leaf, root, and seedling shoot tissue, the 24-nt sRNAs seem to have more variable loci than for 21-nt or 22-nt sRNAs, while the relative levels were similar for the size classes for internode and endosperm tissue.
Figure 5.
Frequency and source of expression variation between inbreds. A to E, For each hybrid, the variability in expression between the parents was assessed. Only loci expressed in at least one of the parents were considered (10 CP5M or greater in one parent) by calculating the log2 ratio of high parent (HP) to low parent (LP). The proportions of loci in each variable expression category (not variable, twofold to fourfold, greater than fourfold, and SPE) were collated and then averaged per tissue and size class combination. SPE was strictly defined as greater than 10 CP5M in one parent and 0 in the other parent; not variable loci were defined as less than twofold variable between inbreds. E = endosperm (n = 20 contrasts), I = internode (n = 13 contrasts), L = leaf (n = 11 contrasts), R = seedling root (n = 13 contrasts), and S = seedling shoot (n = 13 contrasts). F to H, Genomic loci contributing variable expression of sRNAs. The distribution of sRNA expression (CP5M) across meta-features for each sample was averaged per tissue and per variable expression category to determine the meta-feature distribution for each size class. n = 70 contrasts averaged per size class. 4FC, Fourfold change. I to M, Expression patterns of loci with variable expression in seedling shoot. Per size class, each locus with variable expression (greater than fourfold or SPE) in at least one contrast was profiled for each hybrid. Expression was normalized to the maximum expression (0–1 scale) for that locus and hierarchically clustered. The number of variable loci per size class is shown in parentheses. For visualization, a maximum of 40,000 loci were included per heat map.
Considering the loci with variable sRNA expression, we hypothesized that certain categories would be more likely to vary between inbreds; for instance, sRNAs in genic regions might increase or decrease in concert with changes in gene expression, while TE-associated sRNAs might be highly consistent between genotypes, given that TEs generally remained repressed in the inbreds profiled. For these analyses, we focused on loci with high variation, more than 4 fold change or SPE, and low variation, less than 2 fold change, and omitted the 2- to 4-fold change category. However, an analysis of the genomic loci contributing variable sRNAs (Fig. 5, F–H) revealed that TEs, gene loci, and intergenic regions were, broadly speaking, equally as likely to be variably expressed (not variable loci, defined as less than 2-fold variable between inbreds). For 21-nt loci, the relatively few noncoding RNA loci, mostly miRNAs, were rarely SPE and relatively less likely to be more than 4-fold variable, which led to increases in the proportion of the other genomic feature categories (Fig. 5F; Supplemental Fig. S6A). The 22-nt loci showed slightly more variation between variable and not variable loci, particularly in genes and TEs (Fig. 5G); this was most evident when comparing tissues (Supplemental Fig. S9B). For instance, in endosperm and internode, variable (greater than 4-fold and SPE) expression patterns were relatively enriched in TEs and depleted in genes (Supplemental Fig. S9B). For 24-nt sRNAs, there was very little difference between the proportion of genomic features of variable (greater than 4-fold and SPE) compared with not variable loci consistently across tissues and the individual inbred comparisons (Fig. 5H; Supplemental Fig. S6).
For each size class and tissue, we identified a set of nonredundant loci exhibiting at least fourfold variation, including SPE, in at least one pairwise comparison. For any given locus, we assessed variation across multiple inbred contrasts both within and between heterotic groups (Fig. 5, I–M; Supplemental Fig. S7). Variable expression could reflect consistent differences between heterotic groups, rare gains, or rare losses of expression. A heat map of the normalized expression levels for seedling shoot loci revealed that many sRNA and mRNA loci have expression in only one of the inbreds (Fig. 5, I–M), such that expression variation was driven by rare gains of expression. While rare gains in expression were the most common type of variation, there were differences in the number of loci, or genes, that exhibited expression in the majority of inbreds. For mRNA, miRNA, and 22-nt sRNAs, there was a set of genes or loci that exhibited expression in the majority of genotypes, but this pattern was quite rare in 21-nt and 24-nt sRNAs. There are a large number of miRNA and 21-nt sRNAs that are expressed only, or primarily, in B73, which might be evidence of mapping bias toward the B73 reference genome. In contrast, there was very little enrichment for B73-specific expression for genes or 24-nt sRNAs. These overall patterns of variation among genotypes were quite similar in other tissues for both mRNA and 21-24-nt sRNAs (Supplemental Fig. S10). Thus, we found that the majority of sRNA loci showed variation in expression, and this was predominantly due to high levels of expression in a single inbred.
There was also a high level of expression variation for sRNAs among genotypes within a single tissue. We were interested in assessing how the variation observed in a single tissue related to expression levels and patterns in other tissues (Supplemental Fig. S11). For each of the heat maps of variation among inbreds (shown in Fig. 5, I–M), we maintained the same order of genes and genotypes and assessed the expression of the same loci in the other tissues (Supplemental Fig. S11). The analysis of 21-nt sRNAs with variable expression in seedling shoot tissue revealed that very few of these loci exhibit similar genotype variation in other tissues. Many of these were expressed at substantially lower levels or exhibited fairly consistent expression among genotypes in other tissues (Supplemental Fig. S11A). For instance, the block of B73-specific loci marked as cluster A in Supplemental Fig. S11A was rarely expressed in other inbreds or in B73 in other tissues (with the exception that these loci were lowly expressed in leaf tissue, which was the most similar tissue type to seedling shoot). By contrast, for both 22-nt and 24-nt sRNA loci, the patterns of variability observed in seedling shoot were frequently seen in other tissues, albeit at slightly lower levels of expression (Supplemental Fig. S11, B and C).
sRNA Loci Exhibit Nonadditive Expression across a Panel of Different Hybrids
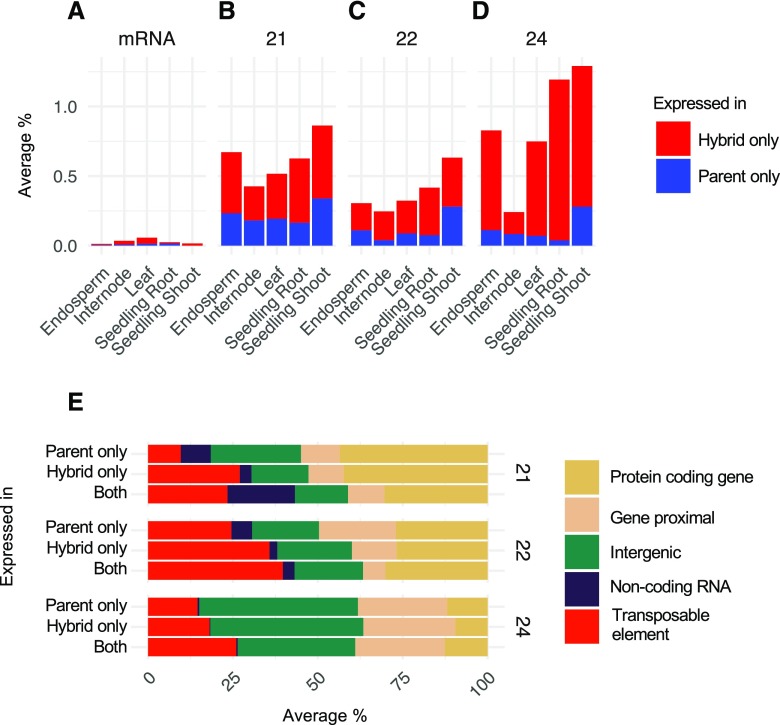
We sought to examine if and how hybrids differ from inbreds for sRNA expression. We focused on two separate types of hybrid-inbred comparisons, with comparisons conducted discretely for each inbred parent triplet in this study and then summarized across triplet comparisons. An initial analysis focused on searching for loci only expressed in the parents or in the hybrid. The majority of loci expressed in parents were also expressed in hybrids, but there was a small subset of less than 1% of loci only expressed in the inbred parents or only in the F1 hybrid (Fig. 6, A–D). Hybrid-specific expression was more common than inbred-specific expression for both genes and sRNA loci; however, hybrid-specific expression was 1 order of magnitude more rare for gene expression compared with sRNA expression (Fig. 6A). sRNA loci expressed only in the hybrid or the parents were located in a variety of genomic features, including TEs, genes, and intergenic regions (Fig. 6E). There were slightly higher levels of inbred-specific or hybrid-specific expression for 24-nt sRNAs compared with other size classes in all tissues except internode (Fig. 6, B–D).
Figure 6.
Loci uniquely expressed in hybrids or inbreds. A to D, Proportion of loci uniquely expressed in hybrids. For each inbred hybrid trio, the proportion of loci expressed only in either hybrids or inbred parents was calculated and then proportions were averaged for all sample groups in each tissue. miRNA loci were excluded due to the low number of loci, as only one to three loci were uniquely expressed in hybrids or parents on average. For sRNAs, only loci that expressed 10 or more CP5M in one of each trio were considered: hybrid only was defined as loci that expressed 10 or more CP5M in the hybrid and less than two CP5M in both parents; parent only was defined as loci that expressed 10 or more CP5M in at least one parent and less than two CP5M in the hybrid. E, The distribution of sRNA expression (CP5M) across meta-features for each sample was averaged per tissue and per expression category to determine the meta-feature distribution for each size class. n = 38 contrasts averaged per size class.
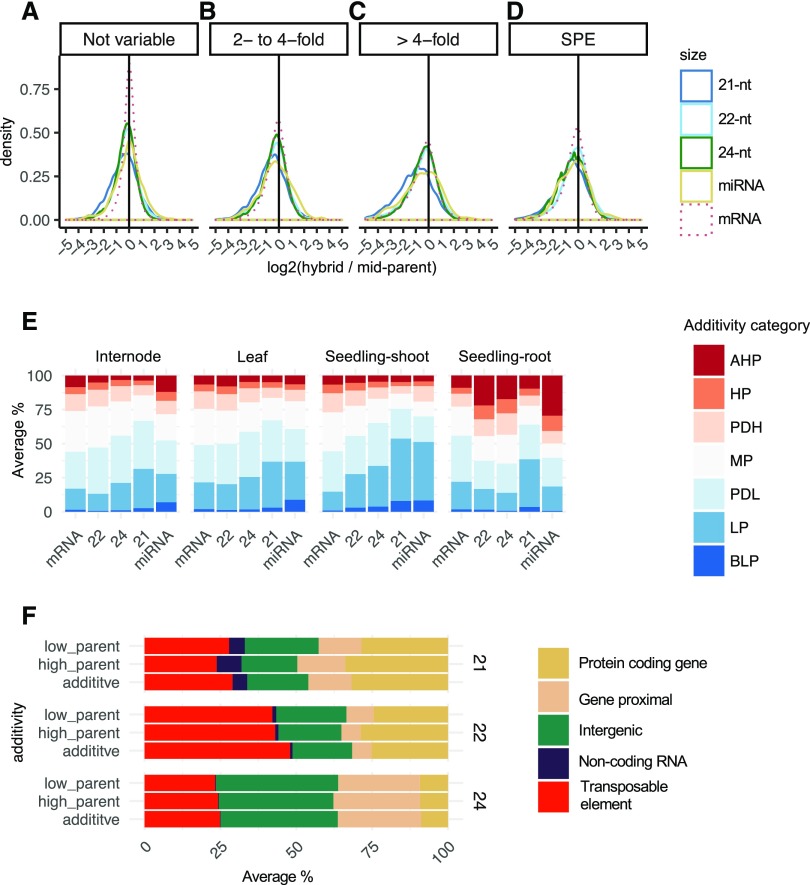
For loci expressed in both inbreds and hybrids, we compared the hybrid expression levels with the average of the two parents (the midparent [MP] level; Fig. 7A). This analysis revealed a normal distribution of hybrid expression values relative to MP levels, with examples of increases or reductions of expression in the hybrids. Overall, for sRNA loci that did not vary between parents, the levels of sRNA abundance exhibited a fairly normal distribution centered at additive levels, similar to the distribution for mRNA, although mRNA values had less variance. This trend was consistent across all tissues and size classes (Supplemental Fig. S12A), although, in root tissues, 21-nt loci shifted toward below parental levels, while 22-nt and 24-nt loci shifted above parental levels. We proceeded to address the additivity of expression levels for sRNA loci with differential abundance in the inbred parents. Prior work on transcript abundance suggests that most genes that are differentially expressed in the two parents exhibit additive, MP patterns in the hybrids, with some examples of expression at the HP or LP levels (Hochholdinger and Hoecker, 2007; Stupar et al., 2007; Zhou et al., 2019), and we found similar results for mRNA data for the parents and hybrids in this study (Fig. 7, B–D, dashed line). Likewise, sRNA loci exhibit a similar frequency of nonadditive expression in all categories: 2- to 4-fold variable, greater than 4-fold, and SPE loci (Fig. 7, B–D, solid lines). In several tissues, the 21-nt sRNAs exhibited some enrichment for expression values lower than the MP values. While the relative frequency of nonadditive expression was similar for gene expression and sRNA expression, given that many more sRNA loci were variably expressed between the parents, a much higher proportion of sRNA loci overall exhibited nonadditive expression.
Figure 7.
Prevalence of nonadditive expression in hybrids. A to D, Overall distribution of additivity in hybrids per sRNA size class compared with mRNA. Additivity was calculated as log2(hybrid/MP) for sRNA and mRNA loci with at least 10 CP5M (two CPM for mRNA) in one parent and divided into categories for the level of expression variation between the inbred parents. For plotting, the tails were concatenated at ±5. E, Distribution of nonadditivity categories per tissue. For sRNA loci that were fourfold or greater variable between inbred parents, the degree of dominance (d/a), calculated as [hybrid – MP/(HP – LP/2)], was determined. d/a values were then divided into either additive MP or six nonadditivity levels: above high parent (AHP) > 1.25; HP 0.75:1.25; partial dominance high parent (PDH) 0.25:0.75; MP −0.25:0.25; partial dominance low parent (PDL) −0.75:0.25; LP −0.75:−1.25; and below low parent (BLP) < −1.25. F, Genomic features of nonadditive loci. For nonadditive HP like (AHP, HP, and PHD) and LP like (PDL, LP, and BLP), the distribution of sRNA expression (CP5M) across meta-features for each sample was averaged per tissue for each size class. n = 30 hybrid parent trio contrasts averaged per size class. Endosperm was excluded owing to the genome imbalance.
Prior analyses have found an enrichment for LP expression for sRNAs (Groszmann et al., 2011; Barber et al., 2012; Shen et al., 2012). In this data set, for both sRNAs and genes, a greater number of loci were below MP levels in the hybrids (Fig. 7E, blue bars). However, for internode, leaf, and seedling shoot tissue, sRNA loci showed a greater level of LP expression compared with mRNA, consistent across 21- to 24-nt size classes. Similar trends were observed in endosperm after taking into consideration the genome imbalance in this tissue (Supplemental Fig. S12B). By contrast, 22-nt and 24-nt loci in root tissue had higher levels of HP expression compared with other tissues and compared with gene expression in root tissue. Next, we examined the genomic locations of nonadditively inherited sRNA loci. This analysis revealed that nonadditive 21-nt, 22-nt, and 24-nt loci occur in all regions of the genome (Fig. 7F), largely reflecting the expression distribution of these sRNAs (Figs. 2 and 5, F–H–H). The patterns were highly consistent between HP-like and LP-like loci. While there was substantial variation between the size classes, overall, there was very little difference in the distribution of genomic features between additive and nonadditive loci (Fig. 7F). Similarly, the different tissues exhibited very consistent genomic feature distributions (Supplemental Fig. S13), with the exception that 22-nt loci in internodes were somewhat enriched for TEs compared with other tissues. Overall, these results suggest that sRNA loci originating from different genomic features have a similar propensity to be nonadditively expressed.
DISCUSSION
sRNAs have been hypothesized to play key roles in plant responses to environmental conditions, transgressive inheritance, and heterosis (Ryder et al., 2014; Greaves et al., 2015; Li et al., 2017). However, deciphering their role in crops such as maize has been limited by our incomplete picture of the sources of sRNA variation. Many studies have focused on a single tissue and/or a limited number of genotypes. We were motivated to undertake a broad survey of sRNA profiles in several distinct tissue types in a panel of diverse inbreds and hybrids in the highly heterotic crop maize. We performed a detailed analysis of sRNA and mRNA expression in a panel of 108 maize samples, including eight inbreds and 14 hybrids across five tissues: endosperm, seedling root, leaf, seedling shoot, and internode. This has provided a more detailed view of the genomic features that contribute sRNAs of different size classes, the developmental variation for sRNA abundance, and the potential connections between sRNAs and heterosis.
Genomic Features Contributing to Consistent or Variable sRNAs
We documented substantial developmental and genotypic influences on sRNA variation; however, we found that the contributions of these factors were different for 21-nt, 22-nt, and 24-nt sRNA expression. This likely reflects established differences in molecular functions and modes of biogenesis of each size class. For instance, 21-nt sRNAs are predominantly miRNAs originating from a relatively small number of noncoding miRNA loci, 24-nt clusters are frequently located near genes, and 22-nt clusters often originate from repetitive sequences (Nobuta et al., 2008; Wei et al., 2009; Barber et al., 2012; Regulski et al., 2013). While these generalizations are supported by our analyses, we also find that each size class of sRNAs has a complex makeup that can vary in different tissue types. For example, while 22-nt sRNAs are enriched for TEs (∼30%–40% of 22-nt sRNA loci), they also are frequently associated with genes, especially nonsyntenic genes (Fig. 2). The 21-nt sRNAs are rarely found associated with genes, except in endosoperm tissue, where ∼20% of 21-nt sRNAs arise from genes. Overall, there was more variability in the features associated with 22-nt sRNAs in different tissues compared with 21-nt or 24-nt sRNAs (Fig. 2, E–G). In particular, we note the reduction in 22-nt sRNAs in gene flanks in endosperm and internode and also the abundant class of genic 22-nt sRNAs mostly occurring in nonsyntenic genes. One possibility is that many of these nonsyntenic genes could be nonfunctional and silenced by the plant, and the enigmatic class of 22-nt small interfering RNAs (siRNAs) in maize might be involved in this silencing mechanism. Previously, Barber et al. (2012) observed differences in sRNA abundance between retrotransposon families. Here, armed with an updated annotation of maize TEs (Anderson et al., 2019; Stitzer et al., 2019), we observed differences in the distribution of 22-nt and 24-nt sRNAs between different TE orders, with 22-nt around more than 90% LTRs. However, 24-nt sRNAs have a high composition from Helitron and TIR DNA transposons far greater than the genome space these TEs occupy: 2.9% and 3%, respectively. This hints at differences in the potential roles of sRNAs in regulating different classes of TEs in maize.
We also performed comparisons of multiple inbreds or of inbreds with hybrids to identify sRNAs that exhibit variable expression between different genotypes or nonadditive expression in hybrids relative to parents. While there were many examples of sRNA loci exhibiting variability in expression levels, we did not find that particular types of sRNAs were more likely to exhibit variation. The distribution of genomic features associated with variable 21-nt, 22-nt, or 24-nt sRNAs was largely similar to the complex distribution observed for sRNA loci without variable expression. This suggests that specific types of sRNAs are not necessarily enriched for variability.
Varying Influence of Tissue and Genotype on Different Size Classes of sRNAs
Prior work on sRNAs and heterosis has often been limited to a single hybrid genotype or a single tissue, for instance in maize (Barber et al., 2012; He et al., 2013; Regulski et al., 2013; Xin et al., 2014) and Arabidopsis (Groszmann et al., 2011; Li et al., 2012, 2015b; Shen et al., 2012). A significant motivation of this study was to investigate whether prior interpretations are representative of other tissues and developmental stages or whether there is variation and uniqueness between tissue types. The analysis of variability for sRNA expression in different tissues and genotypes revealed distinct patterns for different size classes of sRNAs. Gene (mRNA) expression has greater variability among tissues than among genotypes. For example, gene expression profiles for the B73 root were more similar to the root samples of other genotypes than to profiles from B73 leaf; this trend was generally consistent across genotypes and tissues. A very similar pattern was also observed for 21-nt sRNAs and, to a lesser extent, 22-nt sRNAs (Fig. 4). This suggests substantial differences in tissue-specific abundance of 21-nt and 22-nt sRNAs that are largely reproducible in different genotypes. In contrast, 24-nt sRNAs exhibit relatively little clustering by tissue type and instead are clustered by genotype. This indicates a greater role for genetic variation in driving differences in 24-nt sRNA abundance. This is supported by the analysis of the patterns of variable abundances for the expression of sRNA loci in multiple tissues. The 21-nt sRNAs exhibiting variable expression seem to often have tissue-specific expression, while 24-nt sRNA variation is more frequently observed in multiple tissues (Supplemental Fig. S11). Thus, differences in 24-nt sRNA abundance in one tissue are more predictive of variability in other tissues, while observations of 21-nt or 22-nt sRNAs will often be limited to a single tissue type. Thus, developmental variation is a significant factor that should be considered when designing or interpreting analysis of heterosis and sRNA profiles.
Connections between sRNAs and Heterosis
Important early work on the inheritance of sRNA in hybrids in Arabidopsis showed that F1 hybrids have reduced levels of 24-nt siRNAs relative to parents for many loci, with an estimated 25% or greater reduction in production of these siRNAs (Groszmann et al., 2011). Similarly, a study in maize suggested that 24-nt sRNAs often exhibit nonadditive expression in hybrids with expression levels lower than expected (Barber et al., 2012). Here, we find little difference between the global profiles of 21-nt, 22-nt, or 24-nt sRNAs between inbreds and hybrids on a global scale over a diverse panel of heterotic hybrids in maize (Fig. 2B; Supplemental Fig. S5). Likewise, clustering of sRNA profiles did not reveal any underlying characteristic of hybrids that could separate them as a class from inbred parents (Fig. 4). This is consistent with the data underlying other work, noting that there appears to be no general shift in the abundance of the size classes between inbreds and hybrids in maize (Barber et al., 2012) or Arabidopsis (Li et al., 2012). There are many sRNAs with nonadditive expression levels, but the majority of these are expressed at levels expected for partial or complete dominance with rare expression outside of the parental range. Of the size classes, 21-nt sRNA showed the strongest trend toward below MP levels in the hybrids. There was no strong enrichment for sRNA loci with nonadditive expression outside the parental range consistently observed in multiple tissues (Fig. 7E). Collectively, our results and this body of literature suggest that hybridization likely does not substantially alter biogenesis or the accumulation of different sRNA size classes.
In addition to comparing the global profiles of 21-nt, 22-nt, and 24-nt sRNAs in inbreds and hybrids, we also performed targeted searches for sRNAs exhibiting unexpected abundance in hybrids relative to parents. There was an enrichment for sRNAs solely expressed in hybrids or inbreds relative to mRNAs, but these still accounted for only ∼1% of sRNA loci (Fig. 6). In addition, these sRNA loci with unique hybrid expression patterns did not necessarily show strong enrichments for certain genomics features. There was no evidence that genic or TE-derived sRNAs experience unusual accumulation patterns within hybrids relative to parents. Similarly, the analysis of nonadditive expression for sRNAs in hybrids did not find striking enrichments for specific features. These findings suggest that sRNA are unlikely to be the general basis of heterosis. Clearly, quantitative variation in sRNA expression is common, and the nonadditive expression of particular loci could be important for heterotic traits. However, we did not find evidence of whole-scale alterations to sRNA accumulation or hybrid uniqueness suggesting a major upheaval of sRNA regulation and influence in hybrids.
Several prior studies on maize have provided interesting insights into heterosis and sRNA abundance. Barber et al. (2012) reported nonadditive expression, particularly for 24-nt sRNAs from repetitive genome regions, in maize hybrids but also found that mutations that greatly reduced the total abundance of 24-nt sRNAs did not greatly reduce heterosis. Seifert et al. (2018a, 2018b) assessed variation in seedling sRNAs in a panel of 21 inbred lines and found that many sRNAs (often 22-nt or 24-nt sRNAs) were associated with grain-yield heterosis in hybrid lines. We found quite high levels of variation for 22-nt and 24-nt sRNAs among genotypes. Given the large number of sRNA loci and the high proportion of variation, this means that there are many more variable sRNA loci than differentially expressed genes. For a biomarker with high levels of variation, it is not surprising that a subset of variation patterns would match variation for a measured trait. It is unclear whether this association was due to a causal relationship between sRNA abundance and yield heterosis or whether these observations are simply a reflection of the high level of variation.
This study provides a detailed understanding of the genomic regions associated with sRNAs in maize. We confirm substantially different profiles of different size classes of sRNAs in distinct tissues of maize and find differences in the genomic regions that contribute these sRNAs in distinct tissues. Detailed comparisons of inbred and hybrid sRNA profiles help to provide a detailed understanding of the potential roles of sRNAs in heterosis. We fail to find evidence for major shifts in sRNA abundance that would provide clear insights into the core mechanisms of heterosis.
MATERIALS AND METHODS
Plant Material
To represent the maize (Zea mays) heterotic groups, inbred lines were selected from the stiff stalk synthetic group (B73, B84, PHB47, and PHJ40), the nonstiff stalk synthetic group (Mo17 and Oh43), and the iodent group (PH207 and PHG29). This investigation was also part of a larger germplasm sampled for mRNA analysis (Li et al., 2019). Hybrids were generated by crossing each of these selected inbred lines by three male genotypes that included B73 (stiff stalk synthetic), Mo17 (nonstiff stalk synthetic), and PH207 (iodent) in the scheme described in Supplemental Table S1. Five tissues were sampled from the inbred and hybrid genotypes: seedling root at V1 stage (seedling root), seedling shoot at V1 (seedling shoot), the middle of the eighth leaf at V7/8 (leaf), the upper-most elongated internode at V7/V8 (internode), and endosperm at 15 d after pollination (endosperm; Li et al., 2019). Seeds for field-grown plants were planted at the Minnesota Agricultural Experiment Station located in St. Paul on May 16, 2014, with 30-inch row spacing at ∼52,000 plants per hectare, and sampled during the 2014 field season; specific harvest dates for each sample are listed in Supplemental Table S2. For the V1 tissues (root and shoot samples), seeds were planted in Metro-Mix300 (Sun Gro Horticulture) with no additional fertilizer and grown under greenhouse conditions (27°C/24°C day/night and 16 h of light/8 h of dark) at the University of Minnesota Plant Growth Facilities during 2014.
sRNA-Seq Library Construction, Sequencing, and Analysis
Samples were flash frozen in liquid nitrogen, and the sRNA-enriched total RNA fraction was extracted using the miRNAeasy Mini Kit (Qiagen); this preparation was split and was used for both sRNA-seq and RNA-seq. Extracted RNA was DNase treated using the TURBO DNA-free kit (Life Technologies). Sequence libraries were prepared by the Joint Genome Institute following the standard TruSeq Small RNA library preparation protocol (Illumina). Samples were sequenced on an Illumina HiSeq 2500 at the Joint Genome Institute to generate 51-bp single-end reads (Supplemental Table S2).
Preprocessing of sRNA-seq data was performed as previously described (Mathioni et al., 2016). Data were incorporated into an sRNA database and are available for viewing online at https://mpss.danforthcenter.org/dbs/index.php?SITE=maize_sRNA4. Briefly, Trimmomatic version 0.32 was first used to remove the linker adaptor sequences (Bolger et al., 2014). The trimmed reads were then mapped to version 4 of the B73 maize genome (Jiao et al., 2017) using Bowtie (Langmead et al., 2009), allowing zero mismatches and reads of length 18-34-nt retained. Reads mapping to structural RNAs were then removed, and counts were scaled by multimapping rate (e.g. a read mapping to two locations receives a count of 0.5 at each location). Read abundance was then normalized to library size by scaling to CP5M to allow for direct comparison across libraries. For the whole-genome analysis, unmapped reads and reads mapping to greater than 50 locations were excluded from further analysis; for the 20-Mb regional analysis, there was no upper limit on multimapping rate. Counts were then split into the principal sRNA size classes (21-nt, 22-nt, and 24-nt) and counts summarized into 100-bp fixed windows, tiling the genome, based on the position of the 5′ end of the sRNA read. One sample had less than 1 million mapped reads (PH207 × B73 F1 leaf sample) and was omitted from further analysis.
mRNA-Seq Library Construction and Sequencing
RNA-seq samples were as described (Li et al., 2019) and were downloaded from the Sequence Read Archive. The samples analyzed in this study, which were paired with the sRNA samples, are described in Supplemental Table S3. Briefly, as detailed previously (Li et al., 2019), total RNA was extracted using the miRNAeasy Mini Kit (Qiagen). Extracted RNA was DNase treated using the TURBO DNA-free kit (Life Technologies). Sequence libraries were prepared by the Joint Genome Institute following the standard TruSeq Stranded mRNA HT library preparation protocol (Illumina). Samples were sequenced on an Illumina HiSeq 2500 at the Joint Genome Institute to generate 150-bp paired-end reads. For each RNA-seq library, 21 to 52 million reads were sequenced.
Reads were trimmed using Trimmomatic (Bolger et al., 2014) and mapped to the B73v4 genome (Jiao et al., 2017) by alignment software STAR (Dobin et al., 2013). Uniquely mapped reads were assigned to and counted for the 46,117 B73v4 gene models using FeatureCounts (Liao et al., 2014). Raw read counts were then normalized by library size and accounted for the effect of extremely differentially expressed genes using the TMM (trimmed mean of M values) normalization approach to give CPM for each gene model (Robinson and Oshlack, 2010).
Annotation of Genomic Features
The background genomic distribution of genomic features was determined by annotating each 100-bp tile of the genome as noncoding (e.g. miRNA and long noncoding RNA), genic, gene-proximal (within 2 kb of a gene), TE, or intergenic using the genome reference B73 RefGen_v4 and annotation based on Gramene version 36 and miRbase release 22. Synteny classifications (i.e. syntenic and nonsyntenic) and assignment to maize subgenomes were obtained from a previous study (Schnable et al., 2011) based on pairwise whole-genome alignment between maize and sorghum (Sorghum bicolor), downloaded from Figshare (Schnable, 2019). The B73 TE annotation was sourced from Anderson et al. (2019). The genic category includes exons, introns, untranslated regions, and any TEs overlapping a gene locus. Loci annotated as TEs that were within 2 kb of a protein-coding gene were assigned to the TE category rather than the gene-proximal category. The noncoding RNA category excluded the prefiltered structural rRNA, tRNA, small nuclear RNA, and small nucleolar RNA. The intergenic category includes all other regions lacking a specific annotation.
Statistical Analysis
PCA was performed using the R package pcaMethods. Counts were log2 transformed, scaled by unit variance, and clustered using singular value decomposition (flags: scale = uv, center = T, method = svd).
Accession Numbers
sRNA data generated in this study are available in the Sequence Read Archive under accession number SRA793603 or Joint Genome Institute proposal identifier 1810. The specific accession numbers for the samples analyzed are listed in Supplemental Table S2. The sRNA data and a genome browser are also available for viewing online at https://mpss.danforthcenter.org/dbs/index.php?SITE=maize_sRNA4. RNA-seq data used in this study were previously reported (Li et al., 2019), and the SRR numbers for the samples analyzed in this study are listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison of the abundance of highly multimapping sRNAs between tissues.
Supplemental Figure S2. Abundance of sRNA size classes.
Supplemental Figure S3. Relationship between tissue, size, and genomic location of sRNAs.
Supplemental Figure S4. Individual data points for genomic feature proportions.
Supplemental Figure S5. A comparison of sRNA abundance between inbreds and hybrids.
Supplemental Figure S6. Genomic features of sRNA loci in inbreds and hybrids.
Supplemental Figure S7. PCA3 on expressed loci.
Supplemental Figure S8. PCA on expressed loci on individual tissues.
Supplemental Figure S9. Source of expression variation between inbreds per tissue.
Supplemental Figure S10. Expression patterns of loci with variable expression in each tissue.
Supplemental Figure S11. Comparison of variable loci across tissues.
Supplemental Figure S12. Additivity across tissue and size classes.
Supplemental Figure S13. Distribution of genomic features between additive and nonadditive loci across tissues.
Supplemental Table S1. Overview of the crossing scheme for the maize hybrids analyzed in this study.
Supplemental Table S2. Summary of the sRNA-seq samples generated in this study.
Supplemental Table S3. Summary of the RNA-seq samples analyzed in this study.
ACKNOWLEDGMENTS
We thank Katie Heslip (Michigan State University) for assistance with RNA extracts. Computational support was provided by the Minnesota Supercomputing Institute.
Footnotes
This work was funded in part by the U.S. Department of Energy via the Joint Genome Institute and the Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-FC02-07ER64494) and by the National Science Foundation (IOS-1802848 to P.A.C. and N.M.S. and IOS-1754097 to R.H. and B.C.M.).
Articles can be viewed without a subscription.
References
- Anderson SN, Stitzer MC, Brohammer AB, Zhou P, Noshay JM, O’Connor CH, Hirsch CD, Ross-Ibarra J, Hirsch CN, Springer NM (2019) Transposable elements contribute to dynamic genome content in maize. Plant J 100: 1052–1065 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64: 137–159 [DOI] [PubMed] [Google Scholar]
- Barber WT, Zhang W, Win H, Varala KK, Dorweiler JE, Hudson ME, Moose SP (2012) Repeat associated small RNAs vary among parents and following hybridization in maize. Proc Natl Acad Sci USA 109: 10444–10449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Auger DL, Riddle NC (2003) In search of the molecular basis of heterosis. Plant Cell 15: 2236–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA (2010) Heterosis. Plant Cell 22: 2105–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA (2015) The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16: 727–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, He G, He H, Chen W, Zhu X, Liang M, Chen L, Deng XW (2010) Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol 52: 971–980 [DOI] [PubMed] [Google Scholar]
- Chen ZJ. (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14: 471–482 [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Ding B, Simon SA, Lopez D, Jia Y, Wang GL, Meyers BC, Jacobsen SE, Pellegrini M (2012) Transcriptome and methylome interactions in rice hybrids. Proc Natl Acad Sci USA 109: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Ellis NA, Guo L, Harkess AE, Yao Y, Zhang X, Dawe RK (2013) CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res 23: 628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent JI, Madzima TF, Bader R, Kent MR, Zhang X, Stam M, McGinnis KM, Dawe RK (2014) Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26: 4903–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Gonzalez-Bayon R, Wang L, Zhu A, Liu PC, Groszmann M, Peacock WJ, Dennis ES (2015) Epigenetic changes in hybrids. Plant Physiol 168: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Groszmann M, Wang A, Peacock WJ, Dennis ES (2014) Inheritance of trans chromosomal methylation patterns from Arabidopsis F1 hybrids. Proc Natl Acad Sci USA 111: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Albertyn ZI, Scofield GN, Peacock WJ, Dennis ES (2011) Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA 108: 2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M, Greaves IK, Fujimoto R, Peacock WJ, Dennis ES (2013) The role of epigenetics in hybrid vigour. Trends Genet 29: 684–690 [DOI] [PubMed] [Google Scholar]
- He G, Chen B, Wang X, Li X, Li J, He H, Yang M, Lu L, Qi Y, Wang X, et al. (2013) Conservation and divergence of transcriptomic and epigenomic variation in maize hybrids. Genome Biol 14: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, Guo L, Liang M, He H, Zhang H, Chen F, et al. (2010) Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22: 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Baldauf JA (2018) Heterosis in plants. Curr Biol 28: R1089–R1092 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Hoecker N (2007) Towards the molecular basis of heterosis. Trends Plant Sci 12: 427–432 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenan-Eichler M, Leshkowitz D, Tal L, Noor E, Melamed-Bessudo C, Feldman M, Levy AA (2011) Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics 188: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Gent JI, Zynda G, Song J, Makarevitch I, Hirsch CD, Hirsch CN, Dawe RK, Madzima TF, McGinnis KM, et al. (2015a) RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc Natl Acad Sci USA 112: 14728–14733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li Y, Moose SP, Hudson ME (2015b) Transposable elements, mRNA expression level and strand-specificity of small RNAs are associated with non-additive inheritance of gene expression in hybrid plants. BMC Plant Biol 15: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X (2017) The functions of plant small RNAs in development and in stress responses. Plant J 90: 654–670 [DOI] [PubMed] [Google Scholar]
- Li Y, Varala K, Moose SP, Hudson ME (2012) The inheritance pattern of 24 nt siRNA clusters in Arabidopsis hybrids is influenced by proximity to transposable elements. PLoS ONE 7: e47043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhou P, Della Coletta R, Zhang T, Brohammer AB, Vaillancourt B, Lipzen A, Daum C, Barry K, de Leon N, et al. (2019) Highly genotype- and tissue-specific single-parent expression drives dynamic gene expression complementation in maize hybrids. bioRxiv doi:10.1101/668681 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: An efficient general purpose program for assigning sequence reads to/ genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lunardon A, Forestan C, Farinati S, Axtell M, Varotto S (2016) Genome-wide characterization of maize small RNA loci and their regulation in the required to maintain repression6-1 (rmr6-1) mutant and long-term abiotic stresses. Plant Physiol 170: 1535–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathioni SM, Kakrana A, Meyers BC (2016) Characterization of plant small RNAs by next generation sequencing In Stacey G, Birchler J, Ecker J, Martin CR, Stitt M, and Zhou JM, eds, Current Protocols in Plant Biology. John Wiley & Sons, Hoboken, NJ, pp 39–63 [DOI] [PubMed] [Google Scholar]
- Nobuta K, Lu C, Shrivastava R, Pillay M, De Paoli E, Accerbi M, Arteaga-Vazquez M, Sidorenko L, Jeong DH, Yen Y, et al. (2008) Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA 105: 14958–14963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder P, McKeown PC, Fort A, Spillane C (2014) Epigenetics and heterosis in crop plants In Alvarez-Venegas R, de la Peña C, and Casas-Mollano JA, eds, Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications. Springer, Basel, Switzerland, pp 13–31 [Google Scholar]
- Schnable J. (2019) Grass Syntenic Gene List sorghum v3 maize v3/4 with teff and oropetium v2. Figshare. Available at: https://figshare.com/articles/Grass_Syntenic_Gene_List_sorghum_v3_maize_v3_4_with_teff_and_oropetium_v2/7926674/1 [Google Scholar]
- Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Springer NM (2013) Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 64: 71–88 [DOI] [PubMed] [Google Scholar]
- Seifert F, Thiemann A, Grant-Downton R, Edelmann S, Rybka D, Schrag TA, Frisch M, Dickinson HG, Melchinger AE, Scholten S (2018a) Parental expression variation of small RNAs is negatively correlated with grain yield heterosis in a maize breeding population. Front Plant Sci 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert F, Thiemann A, Schrag TA, Rybka D, Melchinger AE, Frisch M, Scholten S (2018b) Small RNA-based prediction of hybrid performance in maize. BMC Genomics 19: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, He H, Li J, Chen W, Wang X, Guo L, Peng Z, He G, Zhong S, Qi Y, et al. (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC (2012) Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J 31: 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J (2019) The genomic ecosystem of transposable elements in maize. bioRxiv doi:10.1101/559922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupar RM, Hermanson PJ, Springer NM (2007) Nonadditive expression and parent-of-origin effects identified by microarray and allele-specific expression profiling of maize endosperm. Plant Physiol 145: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW (2009) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Stein JC, Liang C, Zhang J, Fulton RS, Baucom RS, De Paoli E, Zhou S, Yang L, Han Y, et al. (2009) Detailed analysis of a contiguous 22-Mb region of the maize genome. PLoS Genet 5: e1000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Yang R, Yao Y, Ma C, Peng H, Sun Q, Wang X, Ni Z (2014) Dynamic parent-of-origin effects on small interfering RNA expression in the developing maize endosperm. BMC Plant Biol 14: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Hirsch CN, Briggs SP, Springer NM (2019) Dynamic patterns of gene expression additivity and regulatory variation throughout maize development. Mol Plant 12: 410–425 [DOI] [PubMed] [Google Scholar]