A mutant allows dissection of brassinosteroid signaling-related and noncanonical functions of the receptor-like kinase BRASSINOSTEROID INSENSITIVE 1.
Abstract
Plants depend on various cell surface receptors to integrate extracellular signals with developmental programs. One of the best-studied receptors is BRASSINOSTEROID INSENSITIVE 1 (BRI1) in Arabidopsis (Arabidopsis thaliana). Upon binding of its hormone ligands, BRI1 forms a complex with a shape-complementary coreceptor and initiates a signal transduction cascade, which leads to a variety of responses. At the macroscopic level, brassinosteroid (BR) biosynthetic and receptor mutants have similar growth defects, which initially led to the assumption that the signaling pathways were largely linear. However, recent evidence suggests that BR signaling is interconnected with several other pathways through various mechanisms. We recently described that feedback from the cell wall is integrated at the level of the receptor complex through interaction with RECEPTOR-LIKE PROTEIN 44 (RLP44). Moreover, BRI1 is required for another function of RLP44: the control of procambial cell fate. Here, we report a BRI1 mutant, bri1cnu4, which differentially affects canonical BR signaling and RLP44 function in the vasculature. Although BR signaling is only mildly impaired, bri1cnu4 mutants show ectopic xylem in place of procambium. Mechanistically, this is explained by an increased association between RLP44 and the mutated BRI1 protein, which prevents the former from acting in vascular cell fate maintenance. Consistent with this, the mild BR response phenotype of bri1cnu4 is a recessive trait, whereas the RLP44-mediated xylem phenotype is semidominant. Our results highlight the complexity of plant plasma membrane receptor function and provide a tool to dissect BR signaling-related roles of BRI1 from its noncanonical functions.
Plant cells perceive a multitude of extracellular signals through a battery of plasma membrane-bound receptors that are crucial for the integration of environmental and developmental signals. The response to the growth-regulatory brassinosteroid (BR) phytohormones is mediated by one of the best-characterized plant signaling pathways (Singh and Savaldi-Goldstein, 2015) initiated by a receptor complex containing the Leu-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1; Li and Chory, 1997) and its coreceptors of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) family (Ma et al., 2016; Hohmann et al., 2017). Binding of the brassinosteroid ligand mediates hetero-dimerization of BRI1 and SERK family members such as BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1; Li et al., 2002; Nam and Li, 2002), which in turn triggers extensive auto- and transphosphorylation of the intracellular BAK1 and BRI1 kinase domains (Hohmann et al., 2017). The activated kinases recruit and activate downstream BR signaling components, which eventually leads to vast changes in gene expression mediated by BR signaling-regulated transcription factors such as BRASSINAZOLE-RESISTANT 1 (BZR1; Wang et al., 2002) and BRI1-EMS-SUPPRESSOR 1 (BES1; Yin et al., 2002). Among the transcriptional targets of these transcription factors, cell wall–related genes are strongly overrepresented, consistent with a growth-regulatory function of BR signaling (Sun et al., 2010; Yu et al., 2011; Chaiwanon and Wang, 2015). Recently, we reported that the state of the cell wall is connected to BR signaling through a feedback mechanism mediated by the RECEPTOR-LIKE PROTEIN 44 (RLP44). In plants in which the activity of the important cell wall modifying enzyme PECTIN METHYLESTERASE (PME) is impaired through ectopic expression of a PME inhibitor protein (PMEIox), BR signaling is activated in a compensatory response that includes transcriptional up-regulation of PMEs to prevent cell (wall) rupture (Wolf et al., 2012). RLP44 is sufficient to activate BR signaling, likely by acting as a scaffold to promote association of BRI1 and BAK1 (Wolf et al., 2014), and this interaction is not affected by increasing BR levels. Thus, information from the cell wall is integrated with BR signaling activity at the level of the plasma membrane. Furthermore, RLP44 is under transcriptional control of BRI1 and is able to promote activity of a second LRR-RLK complex, containing the receptor for the phytosulfokine (PSK) peptide, PSK RECEPTOR 1 (PSKR1), through the same scaffolding mechanism as observed for the activation of BR signaling (Holzwart et al., 2018). As a result, both BRI1 and RLP44 are required for full functionality of PSK signaling in the vasculature, demonstrated by the observation that bri1 null mutants, rlp44 mutants, and PSK-related mutants share the same vascular phenotype in the primary Arabidopsis root: ectopic xylem cells in the position of the procambium (Holzwart et al., 2018). Interestingly, hypomorphic mutants of BRI1 with intermediate growth phenotypes and BR biosynthetic mutants with strong growth phenotypes show wild type-like xylem, suggesting that BRI1’s role in BR signaling is independent from its role in procambial maintenance. Here, we further dissect BRI1 function through the characterization of a novel bri1 allele, which is only marginally affected in canonical BR signaling, but strongly affected in RLP44-mediated control of procambial cell fate. These observations demonstrate that the function of BRI1 in BR signaling can be uncoupled from its emerging additional functions.
RESULTS
Two Novel Suppressor Mutants of PMEIox
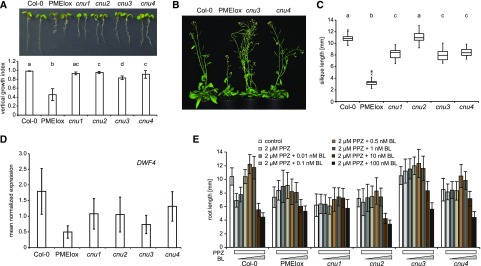
We have previously described that plants overexpressing PECTIN METHYLESTERASE INHIBITOR 5 (PMEIox) show a pleiotropic growth phenotype caused by cell wall feedback signaling. We have used these plants to perform a genetic screen, which identified the comfortably numb1 (cnu1) and cnu2 suppressor mutants affected in the BR receptor BRI1 (Wolf et al., 2012) and RLP44 (Wolf et al., 2014), respectively. Reduced pectin methylesterase activity in PMEIox leads to a compensatory up-regulation of BR signaling, which restores cell wall integrity but causes directional growth phenotypes as a secondary effect (Wolf et al., 2012). RLP44 is required and sufficient for enhancing BR signaling in response to cell wall modification (Wolf et al., 2014), presumably by promoting the interaction between BRI1 and its coreceptor BAK1 (Holzwart et al., 2018). From the cnu suppressor screen we identified two new extragenic suppressor mutants, which we called cnu3 and cnu4 (Fig. 1A). Similar to cnu1 and cnu2, both cnu3 and cnu4 strongly suppressed the macroscopic PMEIox growth phenotype in seedlings, with the exception of a residual root waving phenotype in cnu3, as indicated by measurement of the vertical growth index (vertical distance between hypocotyl junction and root tip divided by root length; Grabov et al., 2005; Fig. 1A). As adult plants, cnu3 and cnu4 appeared similar to wild-type plants, in contrast with their parental line PMEIox (Fig. 1B). Moreover, cnu3 and cnu4 showed suppression of the malformed and short silique phenotype of PMEIox (Fig. 1C). Reverse transcription quantitative PCR (RT-qPCR) analysis revealed that transcript levels of the BR signaling marker gene DWF4 in cnu3 and cnu4 are intermediate between Col-0 and PMEIox, suggesting partial suppression of PMEIox-mediated activation of BR signaling (Fig. 1D). Consistent with this notion, and similar to the cnu1 (mutated in BRI1) and cnu2 (mutated in RLP44) suppressor mutants, cnu3 and cnu4 were more resistant than Col-0 to the depletion of endogenous BR by propiconazole (PPZ; Hartwig et al., 2012), but showed a relatively normal response to exogenous application of epi-brassinolide (BL), in contrast with the largely insensitive cnu1 mutant (Fig. 1E).
Figure 1.
Identification of the PMEIox suppressor mutants cnu3 and cnu4. A, seedling morphology and root vertical growth index (Grabov et al., 2005) of Col-0, PMEIox, the previously published PMEIox suppressor mutants cnu1 (Wolf et al., 2012) and cnu2 (Wolf et al., 2014), and the two novel suppressor mutants, cnu3 and cnu4. Letters indicate statistically significant difference according to one-way ANOVA followed by Tukey’s post hoc test with P < 0.05 (n = 13). B, Adult cnu3 and cnu4 mutants resemble wild-type plants, in contrast with the PMEIox parental line. C, Silique length of Col-0, PMEIox, and the four PMEIox suppressor mutants cnu1 to cnu4. Box plots indicate interquartile range (box), median (bar) and 1.5× interquartile range (whiskers); outliers are indicated with a cross, n = 24–36. Letters indicate statistically significant difference according to one-way ANOVA followed by Tukey’s post hoc test with P < 0.05. D, RT-qPCR analysis of the BR biosynthetic gene DWARF4 (DWF4) in wild type (Col-0), PMEIox, and the cnu1 to cnu4 the suppressor mutants. Bars depict average ± sd; n = 3. E, Root length response of Col-0, PMEIox and the cnu1 to cnu4 suppressor mutants to BR depletion by PPZ and exogenous application of epi-BL. Bars depict average ± sd, n = 19–53.
The cnu3 and cnu4 Suppressor Mutants Carry Two Novel Hypomorphic Alleles of bri1
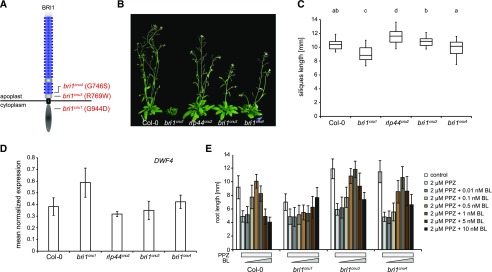
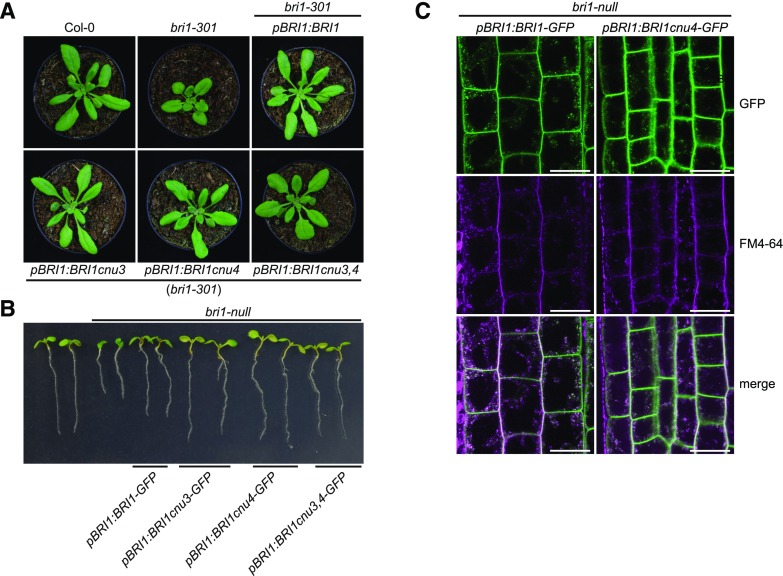
To gain insight into the relationship of cnu3, cnu4, and the previously described rlp44 mutant cnu2, we performed allelism tests by crossing the different suppressor mutants with each other. F1 plants resulting from a cross between cnu3 and cnu4 showed suppression of PMEIox growth defects (Supplemental Fig. S1), whereas F1 plants generated by crossing with cnu2 showed the PMEIox phenotype (Supplemental Fig. S1). This suggests that cnu3 and cnu4 are mutated in the same gene, which is different from RLP44. As we had previously identified a PMEIox suppressor mutation in the BR receptor, we sequenced the BRI1 coding sequence in the novel mutants. We revealed a mutation in cnu3 leading to exchange of Arg 769, located in the extracellular membrane-proximal region, to Trp (R769W). In cnu4, we detected a SNP leading to the exchange of Gly 746, located in the last LRR repeat of the extracellular domain, to Ser (G746S; Fig. 2A). To test whether these variants were causative for the PMEIox suppressor phenotype, we complemented cnu3 and cnu4 by expressing GFP-tagged BRI1 under the control of its native 5′ regulatory sequences. Transgenic BRI1-GFP expression resulted in restoration of the PMEIox phenotype or even a dwarf phenotype (Supplemental Fig. S2), presumably because expression of BRI1 in these hypomorphic mutants in the presence of PMEIox-induced cell wall alterations can lead to excessive BRI1 activity detrimental to growth. Consistent with this assumption, our complementation lines were infertile and reminiscent of plants derived from a cross between PMEIox (Wolf et al., 2012) and BRI1 overexpressing plants (Friedrichsen et al., 2000), which also showed extreme dwarfism and were unable to reproduce (Wolf et al., 2012). To characterize the effect of the mutations in the absence of PMEIox-induced cell wall alterations, we crossed cnu3 and cnu4 to the Col-0 wild-type, and genotyped the F2 population to identify individuals that contained the homozygous bri1 mutations but had lost the PMEIox transgene. We called those mutants derived from cnu3 and cnu4 bri1cnu3 and bri1cnu4, respectively. In sharp contrast with our previously identified PMEIox-suppressing mutant bri1cnu1 (Wolf et al., 2012), both mutants showed relatively normal growth and were not strongly deviating from the wild type with respect to classical BR signaling hallmarks such as fertility, leaf shape, leaf color, silique length, and marker gene expression (Fig. 2, B–D; Supplemental Fig. S3). To assess the capacity of the bri1 mutants to respond to altered BR levels, we grew seedlings on plates under BR-depleting conditions and externally applied varying concentration of BL. Depletion of BRs by PPZ reduced root length of 5-d-old seedlings to ∼5 mm in all genotypes. Cotreatment with 0.5 nm BL completely restored Col-0 root length, whereas 1 nm of BL was required to achieve maximal root length in bri1cnu3 and bri1cnu4 (Fig. 2E). Further increase of BL led to growth depression in wild type and, to slightly lesser degree, in the bri1cnu3 and bri1cnu4 mutants. Thus, in accordance with the subtle growth phenotype, bri1cnu3 and bri1cnu4 were only affected mildly in their response to altered BR levels. In contrast, bri1cnu1 was much less responsive to exogenous BR and did not reach growth depression with the concentrations tested here (up to 10 nm; Fig. 2E), as reported for other bri1 hypomorphic alleles of similar strength (Sun et al., 2017). Consistent with the mild growth phenotype, transformation with constructs encoding the two BRI1 mutant versions alone or the combination of both mutations (BRI1cnu3 and BRI1cnu 4) rescued hypomorphic bri1-301 (Li and Nam, 2002) and the strong, infertile bri1-null (Jaillais et al., 2011) alleles, respectively (Fig. 3, A and B). The subcellular localization of pBRI1-expressed BRI1cnu4-GFP was indistinguishable from pBRI1-expressed BRI1-GFP (Fig. 3C). Taken together, bri1cnu3 and bri1cnu4 are two weak BRI1 mutants with a mild growth phenotype.
Figure 2.
The cnu3 and cnu4 mutants are two novel alleles of BRASSINOSTEROID INSENSITIVE 1 (BRI1). A, Schematic view of BRI1 with indicated position and amino acid substitution of the mutations in bri1cnu1, bri1cnu3 (derived from cnu3, but in the absence of the PMEIox transgene), and bri1cnu4 (derived from cnu4, but in the absence of the PMEIox transgene). B, Comparison of adult plant phenotype of Col-0, bri1cnu1, rlp44cnu2, bri1cnu3, and bri1cnu4. C, Silique length of Col-0, and the mutants derived from the cnu1 to cnu4 suppressor mutants. Box plots indicate interquartile range (box), median (bar) and 1.5× interquartile range (whiskers), n = 25–36. Letters indicate statistically significant difference according to one-way ANOVA followed by Tukey’s post hoc test with P < 0.05. D, RT-qPCR analysis of the BR biosynthetic gene DWF4 in wild type (Col-0), bri1cnu1, rlp44cnu2, bri1cnu3, and bri1cnu4. Bars depict average ± sd; n = 3. E, Root length response of wild type (Col-0), bri1cnu1, bri1cnu3, and bri1cnu4 to BR depletion by PPZ and exogenous application of BL. Bars depict average ± sd; n = 34–70.
Figure 3.
BRI1cnu4 and BRI1cnu3 proteins are functional. A, Mutant BRI1 constructs complement the hypomorphic bri1-301 mutant. B, Constructs encoding mutated BRI1 versions complement the sterile bri1-null mutant. The pBRI1:BRI1cnu3,4-GFP construct combines the mutations of bri1cnu3 and bri1cnu4. C, GFP fluorescence in root meristems of bri1-null mutants complemented with GFP fusion proteins from either the construct pBRI1:BRI1-GFP or pBRI1:BRI1cnu4-GFP shows no apparent difference in subcellular localization. FM4-64 was used as an endocytic membrane tracer dye. Scale bars = 10 µm.
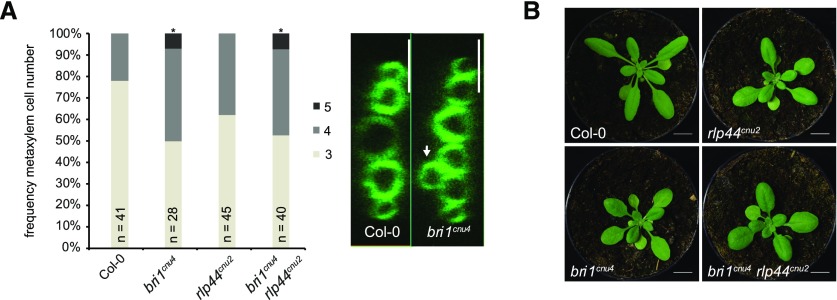
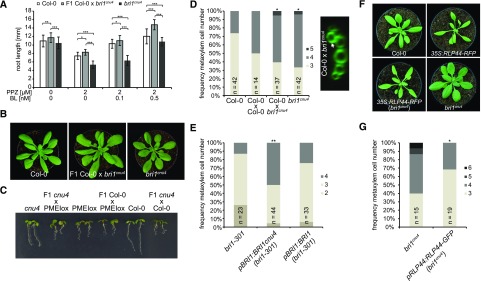
We have previously reported that bri1 null but not bri1 hypomorphic mutants show ectopic xylem cells in place of procambium in the Arabidopsis (Arabidopsis thaliana) primary root. BRI1 controls vascular cell fate through a noncanonical, BR signaling-independent pathway acting through RLP44 and PSK signaling (Holzwart et al., 2018). We therefore tested the xylem phenotype in bri1cnu4, expecting it would behave like other bri1 hypomoprphic mutants such as bri1cnu1, bri1-301, and bri1-5 (Noguchi et al., 1999; Xu et al., 2008; Wolf et al., 2012; Holzwart et al., 2018). In contrast, bri1cnu4 showed a strong increase in xylem cell number, comparable with rlp44 mutants and slightly less pronounced than in bri1-null mutants (Fig. 4A; Holzwart et al., 2018). This clearly distinguishes bri1cnu4 from other BR-related mutants and suggests that the mutation in the BRI1cnu4 protein has a negative effect on RLP44 function, although expression of RLP44 was not decreased in bri1cnu4 (Supplemental Fig. S4, A and B). We reasoned that this could provide valuable insight into the mechanism of xylem cell fate determination by BRI1 and RLP44, concentrating on bri1cnu4 for the remainder of the study. We tested genetic interaction between bri1cnu4 and rlp44cnu2 by generating the double mutant and assessing its xylem phenotype. Simultaneous mutation of rlp44 did not further enhance the bri1cnu4 mutant phenotype, suggesting that bri1cnu4 and rlp44cnu2 are affected in the same pathway with respect to xylem cell fate (Fig. 4A). Likewise, the subtle growth phenotype of rlp44cnu2 and bri1cnu4 was not aggravated in the double mutant (Fig. 4B).
Figure 4.
The mutation in bri1cnu4 negatively affects RECEPTOR-LIKE PROTEIN 44 (RLP44) function. A, Frequency of roots with the indicated number of metaxylem cells in Col-0, rlp44cnu2, bri1cnu4, and the rlp44cnu2 bri1cnu4 double mutant (left) and example of fuchsin-stained xylem cells in Col-0 and bri1cnu4 (right). Asterisks indicate statistically significant difference from Col-0 based on Dunn’s post hoc test with Benjamini-Hochberg correction after Kruskal-Wallis modified U test (*P < 0.05); n = 28–45 as indicated. Scale bars = 10 µm. B, Morphological phenotype of Col-0, rlp44cnu2, bri1cnu4, and the rlp44cnu2 bri1cnu4 double mutant. Scale bar = 1 cm.
The bri1cnu4 Mutant Uncouples BRI1 Roles in BR Signaling and RLP-Mediated Control of Cell Fate
To further test our hypothesis that BRI1cnu4 negatively affects the function of RLP44, we assessed whether the mutation had a dominant effect. We analyzed F1 hybrid seedlings derived from a cross of bri1cnu4 and Col-0 and revealed that the subtle BR insensitivity observed in bri1cnu4 root growth is a recessive trait (Fig. 5A). In line with this, the morphological phenotype of the F1 hybrids appeared closer to the wild type than to that of plants homozygous for the bri1cnu4 mutation (Fig. 5B). In addition, plants heterozygous for the bri1cnu4 mutation were not able to suppress PMEIox-mediated activation of BR signaling (Fig. 5C), indicating that bri1cnu4 rescues PMEIox in the cnu4 mutant through reduced BR signaling strength. Intriguingly, despite the recessive nature of its BR signaling defect, the xylem phenotype of bri1cnu4 was clearly dominant in the F1 seedlings, supporting the idea that the mutation might directly or indirectly impair RLP44 function (Fig. 5D). Consistent with this hypothesis, expression of the BRI1cnu4 transgene in the bri1-301 hypomorphic mutant recapitulated the bri1cnu4 phenotype, whereas expression of wild-type BRI1 did not (Fig. 5E). Interestingly, RLP44-mediated activation of BR signaling was not blocked in bri1cnu4, as the phenotype of plants overexpressing RLP44 in the bri1cnu4 background was intermediate between the overexpressing line and the mutant (Fig. 5F). This is in contrast with what was observed with overexpression of RLP44 in bri1-null (Holzwart et al., 2018) and bri1cnu1, which harbors a mutation in the kinase domain (Wolf et al., 2014). Moreover, increasing the amount of RLP44 through transgenic expression under control of its own promoter rescued the mild BR response phenotype of bri1cnu4 (Supplemental Fig. S4C), and partially rescued xylem cell number (Fig. 5G).
Figure 5.
The bri1cnu4 mutant interferes with RLP44 function. A, Root length of 5-d-old F1 hybrid seedlings of a cross between bri1cnu4 and Col-0 after PPZ treatment and exogenous supply of BL. Bars = mean root length of 5-d-old seedlings ± sd (n = 22–49). Asterisks indicate significance with *P < 0.05, **P < 0.01, and ***P < 0.001 as determined by Tukey’s test after two-way ANOVA. Note that significance is only indicated for comparisons within each treatment. B, Morphological phenotype of Col-0, bri1cnu4, and F1 hybrid plants resulting from crossing the two genotypes. C, Suppression of PMEI5 overexpression phenotype (PMEIox) by the bri1cnu4 allele (cnu4) is a recessive trait, as indicated by the PMEIox-like phenotype of F1 plants from a cross between cnu4 and Col-0. D, Ectopic xylem phenotype in bri1cnu4 and F1 plants from a cross between bri1cnu4 and Col-0. Asterisks indicate statistically significant difference from Col-0 based on Dunn’s post hoc test with Benjamini-Hochberg correction after Kruskal-Wallis modified U test (*P < 0.05); n = 14–42 as indicated. E, Expression of BRI1cnu4, but not of wild-type BRI1 in the bri1-301 mutant results in supernumerary xylem cells. Asterisks depicts significantly altered xylem cell numbers compared with bri1-301 based on Dunn’s post hoc test with Benjamini-Hochberg correction after Kruskal-Wallis modified U test (**P < 0.01); n = 23–44 as indicated. F, RLP44 overexpression can partially rescue the morphological phenotype of bri1cnu4. G, Increased expression of RLP44 can alleviate the bri1cnu4 phenotype. Asterisk indicates statistically significant difference from Col-0 based on two-tailed t test assuming unequal variances (*P < 0.05); n = 15–19 as indicated.
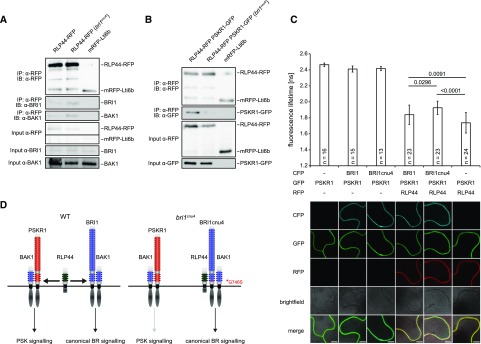
To understand the mechanism by which BRI1cnu4 negatively affects RLP44 function, we analyzed protein-protein interaction. To this end, we compared the association of RLP44 with BRI1 and BRI1cnu4 by immunoprecipitating RLP44 fused to red fluorescent protein (RLP44-RFP) in the Col-0 and bri1cnu4 background, respectively. Interestingly, BRI1cnu4 showed increased abundance in RLP44-containing complexes (Fig. 6A) compared with wild-type BRI1. In addition, BAK1 was slightly more abundant in RLP44-RFP immunoprecipitates in the bri1cnu4 background (Fig. 6A). Increased association of RLP44 with BRI1cnu4 compared with BRI1 was also observed after transient expression in Nicotiana benthamiana (Supplemental Fig. S5A). Furthermore, split-ubiquitin assays in yeast supported a stronger direct interaction between BRI1cnu4 and RLP44 as well as between BRI1cnu4 and BAK1 compared with wild-type BRI1 (Supplemental Fig. S5B). Thus, it is likely that BRI1cnu4 exerts its effect on the maintenance of xylem cell fate by RLP44 sequestration, thereby preventing RLP44 from acting in PSK signaling. To test the hypothesis that BRI1cnu4 negatively affects PSK function in Arabidopsis, we performed coimmunoprecipitation experiments with double transgenic lines expressing RLP44-RFP and PSKR1-GFP under control of the 35S promoter in the Col-0 and bri1cnu4 background, respectively. In the Col-0 background, PSKR1-GFP was readily detected in immunoprecipitates of RLP44-RFP (Fig. 6B), confirming previous results from N. benthamiana (Holzwart et al., 2018). However, the amount of PSKR1-GFP associated with RLP44-RFP was strongly reduced in the bri1cnu4 background (Fig. 6B), supporting the notion that BRI1cnu4 has a negative effect of PSKR1-related functions of RLP44. These results suggest that BRI1 and PSKR1 compete for RLP44. To directly test this hypothesis, we performed FRET-FLIM in N. benthamiana with all three proteins. As previously described (Holzwart et al., 2018), we observed a strong lifetime reduction of PSKR1-GFP when RLP44-RFP was coexpressed (Fig. 6C; Supplemental Fig. S6), reflecting close proximity of the two proteins, likely indicating a direct interaction. Additional expression of BRI1 fused to cyan fluorescent protein (BRI1-CFP) led to an increase in GFP lifetime, suggesting that the presence of BRI1 perturbs the interaction between PSKR1-GFP and RLP44-RFP. Consistent with previous data, this effect was more pronounced when BRI1cnu4-CFP was used instead of BRI1-CFP. Thus, we propose that the PSK and BRI1 receptor complexes are able to compete for RLP44, whereas the BRI1cnu4 mutation sequesters RLP44 in the BR receptor complex, leading to a reduced PSK signaling efficiency (Fig. 6D).
Figure 6.
The BRI1cnu4 protein shows increased interaction with RLP44 and BAK1. A, Coimmunoprecipitation of BRI1-GFP by RLP44-RFP from crude extracts of wild type (Col-0) and bri1cnu4 mutant plants. B, Coimmunoprecipitation of PSKR1-GFP by RLP44-RFP from crude extracts of wild type (Col-0) and bri1cnu4 mutant plants. C, FRET-FLIM analysis of the PSKR1-RLP44 interaction in the absence and presence of BRI1-CFP or BRI1cnu4-CFP. Each bar depicts GFP fluorescence lifetime ± sd. P-values for lifetime comparisons are based on Tukey-Kramer post hoc test following one-way ANOVA; n = 13–24 as indicated. Representative CLSM images demonstrate comparable expression of the proteins across samples. Scale bar = 7 µm. For a box blot representation of the data, see Supplemental Fig. S6. D, model of RLP44 interactions with BRI1 and PSKR1 in the wild type and the bri1cnu4 mutant. The mutation at the base of BRI1´s extracellular domain sequesters RLP44 and prevents it from promoting PSK/PSKR1 signaling.
DISCUSSION
We have previously shown that BRI1 has functions that are independent of classical BR signaling outputs mediated by the canonical BR signaling pathway (Holzwart et al., 2018). Here, we demonstrate that BRI1 mutants, depending on the nature of the allele, differentially affect these functions and can thus serve as a tool to uncouple canonical BR signaling-mediated from noncanonical effects. We isolated a new bri1 allele, bri1cnu4, and compared its impact on classical BR read-outs and the role of BRI1 in the maintenance of procambial cell fate, which depends on RLP44-mediated activation of PSK signaling (Holzwart et al., 2018). These analyses revealed that BR signaling-dependent BRI1 functions are only mildly affected in bri1cnu4, whereas we observed a strong negative effect on RLP44 function in the regulation of vascular cell fate. Interestingly, the same mutation we report here as bri1cnu4, G746A (G2236A on nucleic acid level) has been recently described as bri1-711 in a tilling approach to obtain new bri1 mutants (Sun et al., 2017). Consistent with our results, bri1-711 showed subtle growth defects and mild insensitivity to exogenous application of BL. In addition, the accumulation of nonphosphorylated BES1 as a readout of BR signaling was similar to that of the Col-0 wild-type in response to the hormone (Sun et al., 2017). In contrast with our results obtained with bri1cnu4, other bri1 hypomorphic mutants such as bri1-301, bri1cnu1, and bri1-5 have negligible effects on xylem cell fate in the root, despite their pronounced effect on BR signaling (Holzwart et al., 2018). A possible explanation for the divergent effect of bri1cnu4 is provided by the observation that the BRI1cnu4 protein interacts more strongly with RLP44 than with wild-type BRI1, and that additional RLP44 alleviates the bri1cnu4 xylem phenotype. From these observations we propose that BRI1cnu4 may sequester RLP44, which consequentially has a negative effect on PSK signaling, consistent with reduced association of PSKR1 and RLP44 in the bri1cnu4 background. It has to be noted that BRI1cnu4 also shows increased association with its coreceptor BAK1, corroborating the complexity of receptor associations in the plasma membrane and the challenges associated with deciphering the multilateral interactions observed with many members of the LRR-RLK family (Stegmann et al., 2017; Smakowska-Luzan et al., 2018). Consistent with this, FRET-FLIM experiments demonstrated that the presence of BRI1 or BRI1cnu4 has a negative effect on the interaction between PSKR1 and RLP44. This suggests a scenario in which the PSK and BR receptor complexes compete for RLP44, which might fine-tune the activity of the two pathways. As revealed by the RLP44 interaction pattern, signaling integration and ramification is realized at the level of the receptor complex in the plasma membrane. Additional examples are the interaction of the BRI1-BAK1 complex with G-proteins to mediate sugar-responsive growth (Peng et al., 2018), with the proton pumps of the P-ATPase type to regulate plasma membrane hyperpolarization and wall swelling that precede cell elongation growth (Caesar et al., 2011), and with the BAK1-interacting receptor-like kinase 3 (BIR3) that represses the activity of the complex in the absence of BR (Imkampe et al., 2017; Hohmann et al., 2018; Großeholz et al., 2019). In addition, BRI1 phosphorylates a homolog of the mammalian transforming growth factor-β (TGF-β) receptor interacting protein/eIF3 eukaryotic translation initiation factor subunit TRIP-1 (Ehsan et al., 2005). Although the function of the latter protein is not completely clear at this stage, it seems at least conceivable that it bypasses the canonical BR signaling pathway, even if the morphological defects observed in plants expressing TRIP-1 antisense RNA are reminiscent of BR-deficiency phenotypes (Jiang and Clouse, 2001).
The challenge emerging from the recent discoveries on the example of BRI1 is to understand how distinct responses to extrinsic cues can be generated by the multifaceted network of a receptor in the plasma membrane. Thus, more sophisticated in vivo cell biological approaches in combination with genetic and biochemical tools are required to dissect and understand the function of this important signaling integrator, BRI1.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All mutants and transgenic lines used in this study are in the Col-0 background. The bri1cnu1, rlp44cnu2, bri1-null, and bri1-301 mutants have been described before (Xu et al., 2008; Wolf et al., 2012; Wolf et al., 2014). The 35S:RLP44-RFP (Wolf et al., 2014), pRLP44:RLP44-GFP (Holzwart et al., 2018), and 35S:PSKR1-GFP (Ladwig et al., 2015) lines described previously were used for crossing.
All plants were grown in half-strength Murashige and Skoog (MS) medium supplemented with 1% (w/v) Suc and 0.9% (w/v) plant agar. PPZ (Sigma-Aldrich) and 24-epi-brassinolide (SantaCruz Biotech) were added to the sterilized medium where appropriate.
Plasmid Generation
All constructs beside mbSUS and FRET-FLIM used in this study were generated with GreenGate cloning as previously described (Lampropoulos et al., 2013). For BRI1 (at4g39400) coding sequence GreenGate Cloning, three internal BsaI/Eco31I recognition sites were silently mutagenized via the generation of 4 PCR fragments with the primers BRI1_GGC_1F, BRI1_GGC_1R, BRI1_GGC_2F BRI1_GGC_2R, BRI1_GGC_3F, BRI1_GGC_3R, BRI1_GGC_4F, and BRI1_GGC_4R as previously described (Holzwart et al., 2018). For generating the BRI1cnu4 module the second fragment was amplified with BRI1_GGC_2F BRI1_GGC_2R using gDNA of bri1cnu4 plants as template. For generating the BRI1cnu3 module, the second fragment was amplified with BRI1_GGC_3F BRI1_GGC_3R with gDNA of bri1cnu3 plants. For the combined BRI1-cnu3,4 construct, fragments 1 and 4 from BRI1 wild type were combined with the second fragment of BRI1cnu4 and the third fragment of BRI1cnu3. PCR products of all combinations were gel purified, digested with Eco31I, subsequently ligated, and processed according to the GreenGate protocol (Lampropoulos et al., 2013). For details regarding primers and constructs, please see Supplemental Tables S1 and S2.
Generation of Transgenic Plants
The bri1-301 mutant was transformed with the indicated constructs, and T1 plants were selected on Glufosinate-ammonium containing half-strength MS medium with 0.9% (w/v) plant agar. T1 plants were genotyped for the bri1-301 mutation (Holzwart et al., 2018). Heterozygous bri1-null plants were transformed with the indicated constructs, and T1 plants were selected on sulfadiazine and Glufosinate-ammonium containing half-strength MS medium with 0.9% (w/v) plant agar. T1 plants were genotyped for the bri1-null transfer-DNA insertion (Holzwart et al., 2018).T2 plants were selected to be a single integration line and for survival rate of 75%, and subsequently T3 plants were selected to be homozygote for the respective mutation and transgene.
Genotyping
Genotyping of bri1cnu1, rlp44cnu2, bri1-301, and bri1-null was described previously (Wolf et al., 2012; Wolf et al., 2014; Holzwart et al., 2018). For genotyping of the two new bri1 alleles, we generated CAPS marker using primers bri1cnu3_CAPS_F, bri1cnu3_CAPS_R and restriction enzyme Cfr42I (bri1cnu3) or primers bri1cnu4_CAPS_F, bri1cnu4_CAPS_R with restriction enzyme BseLI (bri1cnu4).
Mating-Based Split Ubiquitin Assays
For mating-based split-ubiquitin assay (mbSUS; Grefen et al., 2009), the coding sequence of RLP44, BAK1, and BRI1 in pDONR207 (Wolf et al., 2014) were cloned into pMetYC-Dest pXNUbA22-Dest, respectively. For generating the BRI1cnu3 Nub construct, primers BRI1_attB1_L + BRI1_attB2_R were used with gDNA of bri1cnu3 plants to create the full-length BRI1cnu3 complementary DNA (cDNA) in pDONR207. For generating the BRI1cnu4 Nub construct, primers BRI1_attB1_L + BRI1_attB2_R were used with gDNA of bri1cnu4 plants to create the full-length BRI1cnu4 cDNA in pDONR207. For generating BRI1cnu3,4 Nub construct, primers BRI1_attB1_L + BRI1_attB2_R were used with the C-Module of BRI1cnu3,4 as a template to create the full-length BRI1cnu3,4 cDNA in pDONR207. The pDONR207 entry modules were recombined with pXNUbA22-Dest. For details regarding primers please see Supplemental Tables S1 and S2. Yeast-based mbSUS assays were performed as described (Grefen et al., 2009; Wolf et al., 2014).
Co-Immunoprecipitation
Material (about 300 mg) from transgenic plants expressing 35S:RLP44-RFP and 35S:RLP44-RFP (bri1cnu4) or 35S:PSKR1-GFP/35S:RLP44-RFP and 35S:PSKR1-GFP/35S:RLP44-RFP (bri1cnu4) or 35S:PSKR1-GFP/35S:RLP44-RFP and 35S:RLP44-RFP at 6 dag were frozen in liquid nitrogen and ground to a fine powder using mortar and pestle. Extraction buffer (100 mm Tris-HCl [pH 8.0], 150 mm NaCl, 10% [v/v] glycerol, 5 mm EDTA [Sigma-Aldrich], 2% [v/v] Igepal CA-630 [Sigma-Aldrich], 5 mm dithiothreitol [Sigma-Aldrich, added immediately before use], 1% [v/v] Protease Inhibitor Cocktail [Bimake, added immediately before use]) was added to the frozen powder (2 mL per 1 g fresh weight), and the homogenate was centrifuged at 12 000g and 4°C after thawing. Input samples were taken from the supernatant, and the remaining supernatant was incubated with 15 µL of RFP-trap or GFP-trap slurry (Chromotek) for 2 h at 4°C on a rotary shaker. The beads were subsequently washed with extraction buffer 4 times and then boiled in 30 µL 2× SDS-PAGE sample buffer at 95°C for 5 min. SDS-PAGE, Western Blotting, and immunological detection of RLP44-RFP, PSKR1-GFP, BRI1, and BAK1 was performed as described (Holzwart et al., 2018).
For coimmunoprecipitation after transient expression in N Nicotiana benthamiana, the coding sequences of BRI1, BRI1cnu4, and RLP44 were expressed with pK7CWG2 (CFP), pK7RWG2 (RFP; Karimi et al., 2002). Constructs were cotransformed in the indicated combinations via Agrobacterium in N. benthamiana leaves. At 2 d after transformation, CFP and RFP fluorescence signal was checked on a Leica SP5 microscope system and subsequently, 500 mg of each combination were harvested and frozen in liquid nitrogen. Coimmunoprecipitation was performed as already described with transgenic plants above.
Confocal Microscopy
GFP, FM4-64, and basic fuchsin fluorescence were analyzed on a Leica SP5 microscope system equipped with a 63× water immersion objective using laser lines of 488 nm (GFP), 514 nm (basic fuchsin), and 543 nm (FM4-64). Fluorescence was recorded between 490 and 525 nm for GFP, between 530 and 600 nm for basic fuchsin, and between 600 nm and 720 nm for FM4-64. Images were analyzed with Fiji.
Xylem Imaging
Basic fuchsin staining of seedling roots was performed as described (Holzwart et al., 2018).
RT-qPCR
Total RNA was extracted from 100 mg of tissue harvested from 5-d-old seedlings using the GeneMATRIX Universal RNA Purification Kit (EURx/Roboklon). AMV Reverse Transcriptase Native according the manufacturer’s protocol (Roboklon E1372) with RiboLock RNase Inhibitor (Thermo Fisher Scientific EO0381) was used for generating cDNA. PCR reactions were performed in a Rotor Gene Q 2plex cycler (Qiagen) using 1:40 diluted cDNA template, JumpStart Taq DNA polymerase (Sigma-Aldrich), and SYBR-GreenI (Sigma-Aldrich). Expression of DWF4, SAUR15, and RLP44 was normalized against at5g46630, respectively (see Supplemental Table S1 for oligonucleotide sequences).
FRET-FLIM
For FRET‐FLIM analysis, the coding sequences of BRI1, BRI1cnu4, PSKR1, and RLP44 were expressed as C‐terminal fluorophore fusions pK7CWG2 (CFP), pH7FWG2 (GFP), pB7RWG2 (RFP; Karimi et al., 2002). These binary vectors and p19 as gene silencing suppressor were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into N. benthamiana leaves. The measurements were performed 2 to 3 d after infiltration using the SP8 laser scanning microscope (Leica Microsystems) with LAS AF and SymPhoTime software as described (Veerabagu et al., 2012). Before performing the FRET‐FLIM measurement, the presence of the fluorophores was detected by using 458, 488, or 561 nm lasers for CFP, GFP, or RFP excitation, respectively.
The fluorescence lifetime τ in nanoseconds of either the donor only expressing cells or the cells expressing the indicated combinations was measured with a pulsed laser as an excitation light source of 470 nm and a repetition rate of 40 MHz (PicoQuant Sepia Multichannel Picosecond Diode Laser, PicoQuant Timeharp 260 TCSPC Module and Picosecond Event Timer). The acquisition was performed until 1,000 photons in the brightest pixel were reached. To obtain the GFP fluorescence lifetime, data processing was performed with SymPhoTime software and bi‐exponential curve fitting, correction for the instrument response function and a fitting range from channel 90 to 1,400. FRET‐FLIM data are derived from three different biological replicates. Statistical analysis was carried out with JMP 14. To test for homogeneity of variance, a Brown-Forsythe test was applied. As the variances are homogenous a one-way ANOVA was permissible followed by a Tukey-Kramer post hoc analysis.
Statistical Analysis
Sample numbers used for quantifications are indicated in the figures and figure legends as (n). Statistical significance of difference between samples was determined by one-way ANOVA combined with Tukey’s post hoc test for root length, root vertical growth index, silique length, and fluorescence lifetime. For xylem cell numbers, statistically significant difference for multiple comparisons was determined by Kruskal-Wallis modified U test followed by Dunn’s post hoc test with Benjamini-Hochberg correction; for pairwise comparison, a two-tailed t test was used.
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database under the following accession numbers: AT4G39400 (BRI1), At3G49750 (RLP44), (AT2G02220 (PSKR1), AT3G50660 (DWF4), AT2G31430 (PMEI5), AT3G05890 (Lti6b), AT4G38850 (SAUR15).
Supplemental Data
The following materials are available.
Supplemental Figure S1. The Arabidopsis (Arabidopsis thaliana) mutants comfortably numb3 (cnu3) and comfortably numb4 (cnu4) are allelic mutants.
Supplemental Figure S2. Mutant BRASSINOSTEROID INSENSITIVE 1 (BRI1) constructs complement the cnu3 and cnu4 mutants.
Supplemental Figure S3. Reverse transcription quantitative PCR (RT-qPCR) analysis of SAUR15 reveals a wild type-like response in bri1cnu3 and bri1cnu4.
Supplemental Figure S4. Expression of RLP44 is not affected in bri1cnu4, whereas enhanced RLP44 expression rescues the bri1cnu4 BR deficiency phenotype.
Supplemental Figure S5. The bri1cnu4 mutation results in increased association with RECEPTOR-LIKE PROTEIN 44 (RLP44).
Supplemental Figure S6. PHYTOSULFOKINE RECEPTOR1 (PSKR1) and BRI1 compete for RLP44 association.
Supplemental Table S1. Oligonucleotides used in this study.
Supplemental Table S2. GreenGate Cloning modules and destination constructs.
Acknowledgments
We thank Michael Hothorn for antiserum against BRI1 and BAK1, as well as Karin Schumacher for antiserum against RFP. We benefited from the Plant Observatory facilities at Institute Jean-Pierre Bourgin, which benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
Footnotes
This work was supported by the German Research Foundation (DFG) (grants WO 1660/6-1 to S.W. and HA2146/22-1 and CRC 1101-D02 to K.H.; the DFG through the Emmy Noether Programme (grant WO 1660/2 to S.W.); and an Agence Nationale de la Recherche grant “Pectosign” (to H.H.).
Articles can be viewed without a subscription.
References
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K (2011) A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. Plant J 66: 528–540 [DOI] [PubMed] [Google Scholar]
- Chaiwanon J, Wang ZY (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehsan H, Ray WK, Phinney B, Wang X, Huber SC, Clouse SD (2005) Interaction of Arabidopsis BRASSINOSTEROID-INSENSITIVE 1 receptor kinase with a homolog of mammalian TGF-beta receptor interacting protein. Plant J 43: 251–261 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J (2000) Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol 123: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Ashley MK, Rigas S, Hatzopoulos P, Dolan L, Vicente-Agullo F (2005) Morphometric analysis of root shape. New Phytol 165: 641–651 [DOI] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K (2009) The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS). Methods Mol Biol 479: 217–233 [DOI] [PubMed] [Google Scholar]
- Großeholz R, Feldman-Salit A, Wanke F, Schulze S, Glöckner N, Kemmerling B, Harter K,Kummer U (2019) Specifying the role of BAK1-interacting receptor-like kinase 3 in brassinosteroid signaling. J Integr Plant Biol doi:10.1111/jipb.12803 [DOI] [PubMed] [Google Scholar]
- Hartwig T, Corvalan C, Best NB, Budka JS, Zhu JY, Choe S, Schulz B (2012) Propiconazole is a specific and accessible brassinosteroid (BR) biosynthesis inhibitor for Arabidopsis and maize. PLoS One 7: e36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M (2017) The structural basis of ligand perception and signal activation by receptor kinases. Annu Rev Plant Biol 68: 109–137 [DOI] [PubMed] [Google Scholar]
- Hohmann U, Nicolet J, Moretti A, Hothorn LA, Hothorn M (2018) The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat Plants 4: 345–351 [DOI] [PubMed] [Google Scholar]
- Holzwart E, Huerta AI, Glöckner N, Garnelo Gómez B, Wanke F, Augustin S, Askani JC, Schürholz AK, Harter K, Wolf S (2018) BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proc Natl Acad Sci USA 115: 11838–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imkampe J, Halter T, Huang S, Schulze S, Mazzotta S, Schmidt N, Manstretta R, Postel S, Wierzba M, Yang Y, et al. (2017) The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 29: 2285–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão-Pires E, Dangl JL, Chory J (2011) Extracellular leucine-rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Clouse SD (2001) Expression of a plant gene with sequence similarity to animal TGF-beta receptor interacting protein is regulated by brassinosteroids and required for normal plant development. Plant J 26: 35–45 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Ladwig F, Dahlke RI, Stührwohldt N, Hartmann J, Harter K, Sauter M (2015) Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 27: 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate---a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8: e83043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Ma X, Xu G, He P, Shan L (2016) SERKing coreceptors for receptors. Trends Plant Sci 21: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Chen L, Li S, Zhang Y, Xu R, Liu Z, Liu W, Kong J, Huang X, Wang Y, et al. (2018) BRI1 and BAK1 interact with G proteins and regulate sugar-responsive growth and development in Arabidopsis. Nat Commun 9: 1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Savaldi-Goldstein S (2015) Growth control: Brassinosteroid activity gets context. J Exp Bot 66: 1123–1132 [DOI] [PubMed] [Google Scholar]
- Smakowska-Luzan E, Mott GA, Parys K, Stegmann M, Howton TC, Layeghifard M, Neuhold J, Lehner A, Kong J, Grünwald K, et al. (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature 553: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Sun C, Yan K, Han JT, Tao L, Lv MH, Shi T, He YX, Wierzba M, Tax FE, Li J (2017) Scanning for new BRI1 mutations via TILLING analysis. Plant Physiol 174: 1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerabagu M, Elgass K, Kirchler T, Huppenberger P, Harter K, Chaban C, Mira-Rodado V (2012) The Arabidopsis B-type response regulator 18 homomerizes and positively regulates cytokinin responses. Plant J 72: 721–731 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Höfte H (2012) Plant cell wall homeostasis is mediated by brassinosteroid feedback signaling. Curr Biol 22: 1732–1737 [DOI] [PubMed] [Google Scholar]
- Wolf S, van der Does D, Ladwig F, Sticht C, Kolbeck A, Schürholz AK, Augustin S, Keinath N, Rausch T, Greiner S, et al. (2014) A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc Natl Acad Sci USA 111: 15261–15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Li B, Li J, Wang Y (2008) Is kinase activity essential for biological functions of BRI1? Cell Res 18: 472–478 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, Rodermel S, Yin Y (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]