Nitrogen nutrient can trigger changes in chromatin modifications, thus affecting RNA processing, metabolism, and growth in plants.
Abstract
Chromatin modification has gained increased attention for its role in the regulation of plant responses to environmental changes, but the specific mechanisms and molecular players remain elusive. Here, we show that the Arabidopsis (Arabidopsis thaliana) histone methyltransferase SET DOMAIN GROUP8 (SDG8) mediates genome-wide changes in H3K36 methylation at specific genomic loci functionally relevant to nitrate treatments. Moreover, we show that the specific H3K36 methyltransferase encoded by SDG8 is required for canonical RNA processing, and that RNA isoform switching is more prominent in the sdg8-5 deletion mutant than in the wild type. To demonstrate that SDG8-mediated regulation of RNA isoform expression is functionally relevant, we examined a putative regulatory gene, CONSTANS, CO-like, and TOC1 101 (CCT101), whose nitrogen-responsive isoform-specific RNA expression is mediated by SDG8. We show by functional expression in shoot cells that the different RNA isoforms of CCT101 encode distinct regulatory proteins with different effects on genome-wide transcription. We conclude that SDG8 is involved in plant responses to environmental nitrogen supply, affecting multiple gene regulatory processes including genome-wide histone modification, transcriptional regulation, and RNA processing, and thereby mediating developmental and metabolic processes related to nitrogen use.
Chromatin modifications have gained increasing attention for their roles in modulating developmental and environmental responses across plant and animal kingdoms (Greer and Shi, 2012; Khan and Zinta, 2016; Sirohi et al., 2016). In plants, environmental signals such as light (Charron et al., 2009), drought (Chinnusamy and Zhu, 2009), and temperature (Lamke et al., 2016) have been shown to affect histone modifications or DNA methylation at functionally relevant gene loci. While it is well known that nitrogen (N)—especially nitrate—acts as a nutrient signal to trigger global reprogramming of gene expression to optimize N uptake and assimilation (Vidal et al., 2015; Varala et al., 2018), little is known about the role of epigenetic regulation in N signaling. One report in Arabidopsis (Arabidopsis thaliana) showed that high levels of N increased the level of H3K27me3 at the Nitrate Transporter2.1 (NRT2.1) gene locus, and this increase was dependent on a Pol II complex protein HIGH NITROGEN INSENSITIVE9 (HNI9; Widiez et al., 2011). However, at the genome scale, our knowledge of the role of chromatin modifications in nitrate or N signaling is limited. Here, we present data that implicates the SET DOMAIN GROUP8 (SDG8) protein as a specific histone methyltransferase involved in mediating genome-wide responses to a changing nitrate environment.
We initially uncovered genes involved in regulation of the N-assimilatory pathway in Arabidopsis using a positive genetic selection for mutants impaired in the regulation of the promoter of Asn synthetase1 (ASN1), a key gene involved in the assimilation of N into Asn (Thum et al., 2008). This genetic selection uncovered a mutant called sdg8-5 (formerly known as cli186 e.g. in Thum et al., 2008), which we later showed was deleted in a gene encoding the histone methyltransferase SDG8 (Li et al., 2015). Despite the existence of 32 SDG proteins encoded in the Arabidopsis genome (Springer et al., 2003), genetic studies show that SDG8 plays a nonredundant role in histone methylation. Specifically, mutant analysis has associated SDG8 with pleiotropic effects on plant growth and development, including early flowering (Soppe et al., 1999; Kim et al., 2005; Zhao et al., 2005; Xu et al., 2008), impaired pigment synthesis (Cazzonelli et al., 2009), enhanced shoot branching (Dong et al., 2008; Cazzonelli et al., 2009), defects in pathogen defense (Berr et al., 2012; Lee et al., 2016), and an impaired touch response (Cazzonelli et al., 2014). Our previous study of the sdg8-5 deletion mutant revealed that SDG8 is also associated with gene responses to carbon and light signals (Li et al., 2015). Our genome-wide transcriptome and epigenome profiling revealed that only a subset of genes lose H3K36me3 modification in the sdg8-5 mutant, and this set of hypomethylated genes is enriched in specific functional categories (photosynthesis, defense, N metabolism, development, etc; Li et al., 2015). Therefore, SDG8 is likely involved in regulating multiple processes with a certain level of target specificity. Mechanistically, we showed that SDG8 is required for depositing the H3K36me3 histone mark on the gene body, which is required to maintain active expression of its target genes (Li et al., 2015).
In addition, several lines of evidence have linked SDG8 with N uptake and assimilation: (1) SDG8 was identified as a regulator of a key N assimilation gene, ASN1, in a mutant screen (i.e. the cli186 mutant; Thum et al., 2008). (2) N metabolism genes are significantly overrepresented among the validated genome-wide targets of SDG8 (Li et al., 2015). (3) SDG8 is required to maintain the high expression level of genes in the N uptake and assimilation pathways including the nitrate transporter (NRT1.5) and Gln synthetase genes (GLN1;1, GLN1;3, and GLN1;4; Li et al., 2015). (4) SDG8 physically binds to the GLN1;1 gene locus (Li et al., 2015). (5) SDG8 is required to maintain wild-type H3K36me3 levels at gene loci encoding nitrate and nitrite reductases (NIA1, NIA2, and NIR1; Li et al., 2015). (6) The mRNA level of SDG8 is induced by nitrate treatment, specifically in the lateral root cap and pericycle in roots (Gifford et al., 2008).
Despite this supportive evidence, it still remains unclear whether SDG8 has a direct role in mediating genome-wide changes in response to fluctuations in environmental N supply. Moreover, how and whether the H3K36me3 marks made by SDG8 in the gene body regulate gene expression remain unclear.
We previously showed that SDG8 has a preference for H3K36me3 in the gene body, suggesting its role in histone methylation that mediates transcriptional elongation or cotranscriptional RNA processing (Li et al., 2015). This is consistent with the role of SETD2, the Arabidopsis SDG8 homolog in mammals, which prevents transcripts from starting in the middle of the gene body (Simon et al., 2014). In Arabidopsis, H3K36me3 and SDG8 were reported to affect RNA splicing of genes during responses to temperature (Pajoro et al., 2017). Alternative splicing of RNAs promotes the diversity of the transcriptome, which was recently shown to be involved in mineral nutrient signaling such as phosphate signaling (Dong et al., 2018). Therefore, these findings beg the questions whether and how SDG8 mediates RNA processing during nitrate signaling.
In this study, we investigated whether the response to nitrate treatment at a genome-wide scale is mediated through chromatin modifications, especially through histone H3K36me3 modification by SDG8, and what impacts this may have on gene expression, especially RNA processing. We hypothesized that SDG8, and the H3K36me3 marks it makes on histones associated with specific N-related genes, plays a role in regulating gene responses to N signals, thus affecting physiological responses to N treatment. To test this hypothesis, we compared the N-induced genome-wide H3K36me3 patterns, transcriptome responses including RNA isoform specific gene expression, and physiological responses, between the sdg8-5 deletion mutant (Thum et al., 2008; Li et al., 2015) and its corresponding wild type, in the presence or absence of a N-treatment. Our results show that the sdg8-5 mutant displays impaired H3K36me3 responses at genomic loci functionally relevant to nitrate uptake and assimilation, as well as reduced metabolic and developmental responses to N treatments. We also show that N-responsive transcriptional reprogramming and RNA isoform switching (i.e. differential production of distinct transcript isoforms under different N conditions) are altered in the sdg8-5 mutant. Finally, we investigated the relevance of SDG8-mediated RNA isoform switching of CCT101, a putative CCT motif regulatory gene, which we functionally validated in shoot cells. We also demonstrated physiological relevance of SDG8 to N responses in planta. Overall, our results support the idea that the histone methyltransferase encoded by SDG8 is required for plants to respond to N signals, affecting genome-wide histone modification, transcriptomic response, and RNA isoform processing, as well as modulating root development, nitrate uptake, and primary metabolism.
RESULTS
SDG8 Mediates Changes in Histone Modifications In Response to N Treatments
SDG8, a histone methyltransferase required for deposition of the histone mark H3K36me3 (Li et al., 2015), was initially uncovered in a screen for mutants impaired in the regulation of ASN1, a key gene involved in N-assimilation (Thum et al., 2008). We thus tested the hypothesis that SDG8 mediates genome-wide changes of H3K36me3 in genes functionally relevant to response to nitrate treatments. To do this, we exposed the sdg8-5 mutant and its wild-type control to nitrate treatments, as described in Wang et al. (2001, 2003). Briefly, seeds were sown on nitrate-free media (0.5 mm of NH4 succinate) for two weeks, followed by N starvation for 24 h, before transferring to either 5 mm of KNO3 or 5 mm of KCl treatments. After 5 h of nitrate versus control treatments, the shoots were harvested for histone chromatin immunoprecipitation sequencing (ChIP-Seq) to profile the global pattern of H3K36me3 (Supplemental Fig. S1). Two independent biological replicates were performed for ChIP-Seq. The raw sequencing reads were first trimmed and mapped to the Arabidopsis genome using the software BowTie (Langmead et al., 2009). The mapped reads were used to call significant H3K36me3 islands and significantly differentially methylated islands using SICER (Zang et al., 2009). To synthesize the output from the two biological replicates, a nitrate-responsive differentially methylated gene had to be identified as significantly different (false discovery rate [FDR] < 0.05) in both replicates, and the mean fold change of enrichment of the two replicates had to be >1.2, a cutoff that was chosen based on our previous study (Li et al., 2015) to reflect the dynamic range and stability of the H3K36me3 mark (see “Materials and Methods” for details).
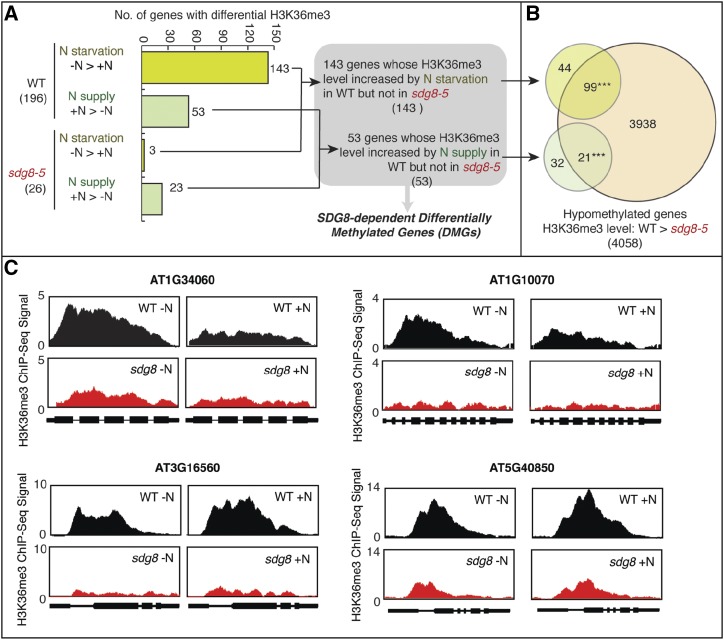
Our results showed that nitrate treatment caused changes in the H3K36me3 levels of 196 genes in wild type, but only 26 genes in the sdg8-5 mutant (Fig. 1). In detail, in wild type, 143 genes show increased H3K36me3 in the N-starvation group compared to the N-supply group (Fig. 1; Supplemental Table S1). These 143 genes are significantly enriched for GO terms “response to nitrogen starvation,” “defense response,” and “response to stress” (FDR < 0.05; Supplemental Table S2). This group of genes includes WRKY transcription factors and a nuclear transcription factor Y family gene, which were previously reported to control N starvation (Krapp et al., 2011) and stress responses (Phukan et al., 2016). In wild type, we also identified 53 genes with increased H3K36me3 in the N-supply group compared to the N-starvation group (Fig. 1; Supplemental Table S1). This group includes genes involved in nitrate reduction (G6PD3 and UPM1), N assimilation (ASN2), regulators of N signaling including HHO2 (Medici et al., 2015) and CIPK3 (Canales et al., 2014), and genes responsive to cytokinin (Arabidopsis Response Regulator7 [ARR7], ARR8, and ARR9)—a downstream signal in the N response (Sakakibara et al., 2006). Significantly enriched GO terms identified among this group of genes include “response to circadian rhythm,” “cytokinin stimulus,” “phosphorelay,” and “sulfate assimilation” (FDR < 0.05; Supplemental Table S3). These GO terms have functional relevance to N responses in plants, as cytokinin (Sakakibara et al., 2006) and phosphorelay signaling (Engelsberger and Schulze, 2012) are known to mediate N signaling, and N was previously reported to regulate circadian rhythm (Gutiérrez et al., 2008) and sulfate assimilation (Wang et al., 2003). Overall, these results support that genes relevant to N signaling, reduction, and assimilation are regulated at the chromatin level through H3K36me3 modification in response to changes of N levels in wild-type plants. In contrast, the N-mediated modification of these N-relevant target genes in wild type are abrogated in the sdg8-5 mutant. Instead, in the sdg8-5 mutant the changes of H3K36me3 occur at a different set of genes in response to N starvation (three genes) or N supply (23 genes), respectively (Fig. 1; Supplemental Table S4), with no significant enrichment of GO terms.
Figure 1.
N-responsive H3K36 methylation is attenuated in the sdg8-5 deletion mutant. A, The comparison of genome-wide H3K36me3 patterns between N-supplied (+N) and N-depleted (−N) wild-type (WT)plants identified 196 genes with differentially methylated H3K36, including 143 genes that have higher H3K36me3 in −N than in +N, and 53 genes that have higher H3K36me3 in +N than in −N. In the sdg8-5 mutant, only three genes were identified to have higher H3K36me3 in −N than in +N, and 23 genes were identified to have higher H3K36me3 in +N than in −N. There is no overlap between the genes identified in the wild type and that identified in the sdg8-5 mutant. Therefore, we identified 143 genes that gain H3K36me3 in N-starvation condition in the wild type but not in the sdg8-5 mutant, and 53 genes that gain H3K36me3 in N-supplied condition in the wild type but not in the sdg8-5 mutant. B, Overlaps of the two groups of differentially methylated genes with the H3K36 hypomethylated genes (i.e. genes with less H3K36me3 in the sdg8-5 mutant compared to wild type) are shown in the Venn diagram. Asterisks indicate a significant overlap: ***P < 0.001 determined using the software Genesect. C, Exemplary genes show H3K36me3 responses to nitrate level change in the wild type but not in the sdg8-5 mutant.
In summary, our comparative analyses identified 143 genes whose H3K36me3 levels are increased by N starvation in wild type, and this response is abrogated in the sdg8-5 mutant (Fig. 1). Similarly, we identified 53 genes whose H3K36me3 levels are increased by N supply in wild type, but this response is abrogated in the sdg8-5 mutant (Fig. 1). We collectively call this group of 196 genes (143 and 53 genes, Fig. 1) SDG8-dependent Differentially Methylated Genes (DMG).
To determine whether these SDG8-dependent DMGs are directly or indirectly regulated by SDG8, we compared the list of 196 SDG8-dependent DMGs with the set of genes shown to be directly bound targets of SDG8, as previously determined under highly comparable experimental conditions (Li et al., 2015). This analysis showed that a modest, but statistically significant portion of the 196 SDG8-dependent DMGs were indeed directly bound by SDG8 (Li et al., 2015; 28/196, 14% overlap with an enrichment P value < 0.001, Supplemental Fig. S2). This significant enrichment provides support that the differential H3K36me3 in response to N is at least partially attributed to a direct role of SDG8. It is likely that some of the “unbound” DMGs may also be direct targets of SDG8, but they could not be identified by the SDG8 binding assay, which requires stable binding. These false negatives may occur, as the ChIP-Seq assay—performed at a given timepoint—identifies only a subset of the authentic targets, because the interaction of a regulatory protein (in this case an HMT enzyme) to its target genes is a dynamic process (Swift and Coruzzi, 2016). Lastly, a portion of the SDG8-dependent DMGs could be regulated by SDG8 indirectly, i.e. through an intermediate partner. Moreover, we also observed a significant overlap between the 196 SDG8-dependent DMGs and a list of 4,058 genes that display significantly less H3K36me3 in the sdg8-5 mutant compared to wild type (overlap P < 0.001; Fig. 1; Supplemental Results; Supplemental Table S5). In summary, our results suggest that SDG8 controls N-responsive changes in H3K36me3 at a set of functionally relevant genomic loci.
SDG8 Affects Transcriptomic Responses to N Treatments
To determine whether and how histone methylation mediated by SDG8 affects the transcription of N-responsive genes, we performed RNA sequencing (RNA-Seq) to compare these responses in the sdg8-5 mutant and wild type exposed to nitrate treatments (Supplemental Fig. S1). Specifically, we identified two types of SDG8-mediated regulation of gene transcription: (1) differentially expressed genes (DEGs) that are regulated by SDG8 in the context of nitrate treatment (i.e. genotype × treatment); and (2) genes whose RNA isoform switching is affected by the deletion of SDG8 in the sdg8-5 mutant.
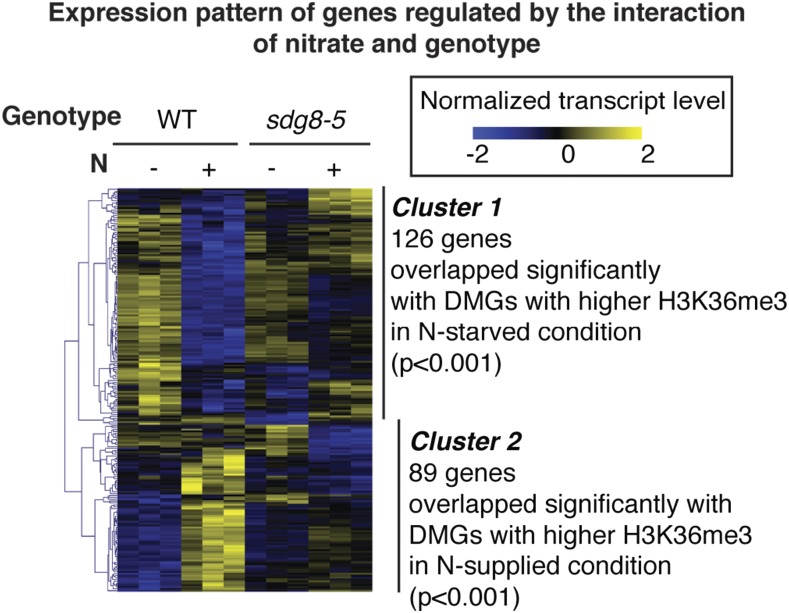
Overall, our RNA-Seq analysis supports the notion that SDG8 mediates N-triggered reprogramming of gene expression. Specifically, the RNA-Seq reads were first trimmed and mapped to the Arabidopsis genome (from The Arabidopsis Information Resource 10 [TAIR10], www.arabidopsis.org) using the TopHat suite (Trapnell et al., 2012). The gene counts were generated by HTSeq (Anders et al., 2015), and then processed by DESeq2 (Love et al., 2014) with the design (∼Genotype+Treatment+Genotype:Treatment) to detect DEGs. Genes that are regulated by the interaction of N and genotype were determined based on P value for Genotype:Treatment interaction, which was adjusted for multitesting error using FDR and controlled at 5%. In total, we identified 215 DEGs regulated by the interaction of SDG8 genotype and N treatment using the two-factor design by DESeq2 (FDR < 0.05; Fig. 2; Supplemental Table S6). This analysis identified genes whose regulation by nitrate is different in wild type compared to the sdg8-5 deletion mutant. Among this group of misregulated genes are ASN1—whose promoter was used to isolate the original sdg8-5 mutant (i.e. cli186; Thum et al., 2008)—and genes encoding NRT2.1 and NRT1.5 and GLN1;4. Next, using hierarchical clustering, we identified two major clusters of the 215 DEGs. Cluster 1 comprised 126 genes that are highly induced by N starvation in wild type, but not in the sdg8-5 mutant (Fig. 2; Supplemental Table S6). The enriched GO terms for this group include “cellular amino acid derivative metabolic process” (FDR < 0.0019), “response to nutrient level” (FDR < 0.039), and “cellular nitrogen compound metabolic process” (FDR < 0.047; Supplemental Table S7). These cluster-1 genes whose N response is misregulated in sdg8-5 share a significant overlap (P < 0.001, six genes) with the genes that display higher H3K36me3 under the N-starved condition than the N-supplied condition (Fig. 2). This finding for the 126 Cluster-1 genes suggests that SDG8 affects the expression of these N-responsive genes through regulating the histone mark H3K36me3, which is known to be associated with actively transcribed genes (Bannister et al., 2005). Cluster 2 contains 89 genes induced by N supply in wild type, which is abolished or attenuated in the sdg8-5 mutant (Fig. 2; Supplemental Table S6). Overrepresented GO terms in cluster 2 include “cellular amine metabolic process” (FDR < 0.0056) and “amino acid transport” (FDR < 0.016; Supplemental Table S8). Similarly, the cluster-2 genes share a significant overlap (P < 0.001, five genes) with genes that exhibit higher H3K36me3 under N-supplied conditions than N-starved conditions (Fig. 2). Overall, the 215 DEGs have a significant overlap with genes directly bound by SDG8 (Li et al., 2015; 30/215, overlap P < 0.024), supporting a direct role of SDG8 in regulating this set of SDG8-dependent DEGs. The 215 DEGs also have a significant overlap with the 4,058 H3K36 hypomethylated genes (74/215, overlap P < 0.001). Together, these results support that histone methylation by SDG8 regulates transcriptional reprogramming of functionally relevant genes during nitrate treatments.
Figure 2.
N-responsive reprogramming of gene expression is altered in the sdg8-5 deletion mutant. Expression levels of 215 genes significantly regulated by the interaction of nitrate and SDG8 genotype are shown as a heatmap. Two major clusters of the 215 DEGs were identified: Cluster 1 comprises 126 genes that are more highly induced by N starvation in wild type (WT), compared to the sdg8-5 mutant; the cluster-1 genes share a significant overlap with the genes that display higher H3K36me3 under the N-starved condition than the N-supplied condition. Cluster 2 contains 89 genes induced by N supply in wild type, which is abolished or attenuated in the sdg8-5 mutant; the cluster-2 genes share a significant overlap with genes that exhibit higher H3K36me3 under the N-supplied condition than the N-starved condition.
SDG8 Affects RNA Isoform Switching in Response to N-Treatments
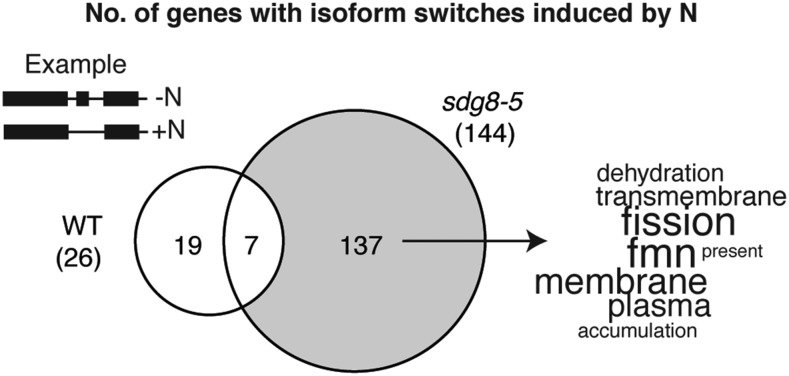
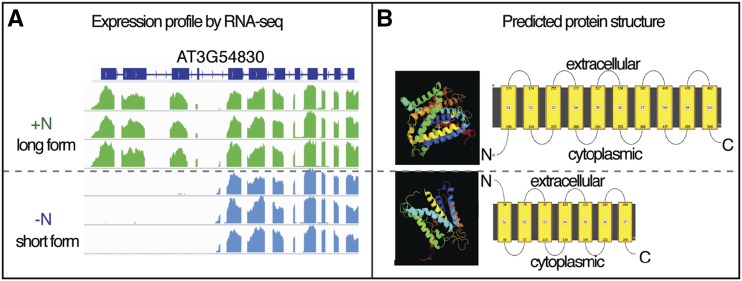
Next, because SDG8 mediates H3K36me3 preferentially in the gene body (Li et al., 2015), we investigated the role that SDG8 might play in regulating the preferential expression of transcript isoforms in responses to nitrate treatments. To do this, we first tested whether nitrate treatments preferentially regulate expression of specific RNA isoforms in wild-type plants. We found 26 genes that switched isoform preference between N-supplied and N-starved samples (Fig. 3; Supplemental Table S9). Notably, under the N-starved condition, the AT3G54830 gene locus is transcribed as a shorter transcript isoform that skips the first four exons, while the longer transcript isoform (with 12 exons) is produced in response to N supply (Fig. 4). This longer isoform encodes a protein that shares high structural similarity with an amino acid/polyamine/organocation transmembrane transporter with 10 transmembrane α-helices (Shaffer et al., 2009; Fig. 4; confidence level 99.9% determined by the software Phyre2; Kelley et al., 2015). Under the N-starved condition, the shorter RNA isoform encodes a truncated putative transmembrane protein that is missing the four N-terminal exons, and the predicted protein structure contains only seven transmembrane α-helices (Fig. 4). Further studies are required to elucidate if there is any functional difference between the putative transmembrane proteins encoded by these two mRNA isoforms of a putative amino acid transporter.
Figure 3.
Deletion of the histone methyltransferase SDG8 increases N-responsive RNA isoform switching. We identified 144 genes that show N-responsive isoform switching in the sdg8-5 mutant, and only 26 genes that show isoform switching in wild type (WT). The comparison between the two gene lists identified 137 genes that are dependent on SDG8 for wild-type–like RNA processing. The functional enrichment within these 137 genes were determined and visualized using the program GeneCloud.
Figure 4.
A putative transporter displays RNA isoform switching in response to N. A, Expression profile of AT3G54830 (encoding a putative transmembrane transporter) shows a distinct preference for RNA isoforms under the N-supplied condition (+N, green, long RNA isoform) compared to the N-starved condition (−N, blue, short RNA isoform). Specifically, a gene-centric view of RNA-Seq sequencing depth is shown with three biological replicates for each condition. B, Predicted structure of proteins encoded by the long or short mRNA isoform suggested that the encoded protein is likely a putative transmembrane transporter, with distinct structures under N-supplied versus N-starved conditions. Structures were predicted using the program Phyre2.
Interestingly, deletion of SDG8 greatly increases the number of genes with N-triggered isoform switches, from 26 in wild type to 144 in the sdg8-5 deletion mutant (Fig. 3; Supplemental Table S9). This suggests that SDG8 normally plays a role in maintaining the stability of RNA isoform expression from specific gene loci when plants experience a change in the N environment. In contrast, in the sdg8-5 mutant, the RNA isoform expression is more likely to alter when plants experience a change in the N environment. Comparison of these two gene lists leads to the identification of 137 genes with RNA isoform switches in the sdg8-5 mutant, but not in wild type in response to N supply (Fig. 3; Supplemental Table S9). Overall, we refer to this group of 137 genes as “sdg8-dependent isoform-switched genes.” As a group, the sdg8-dependent isoform-switched genes are enriched with functional annotations such as “nucleotide binding” (FDR < 0.0011 determined by AgriGO; Tian et al., 2017) and “membrane/transmembrane” (determined by GeneCloud; Krouk et al., 2015; Fig. 3). Transmembrane transport and nucleotide metabolism were also identified as enriched functional groups among alternatively spliced genes involved in maintaining nutrient homeostasis in rice (Dong et al., 2018). In summary, our results indicate that the histone methyltransferase SDG8 affects differential RNA isoform production in response to changes in nutrient levels.
The Histone Methyltransferase SDG8 Affects RNA Isoform Switching of CCT101, Which Influences Its Regulatory Function
We next investigated whether sdg8-dependent RNA isoform switching is related to SDG8-dependent H3K36 methylation. To do this, we compared the gene list of sdg8-dependent isoform-switched genes with H3K36 hypomethylated genes in the sdg8-5 deletion mutant (Supplemental Table S5). We found a significant overlap between these SDG8-dependent genes (overlap P < 0.002, 32/137 genes), which suggests that SDG8-mediated H3K36me3 in the gene body is associated with the RNA isoform switch of its target genes. An example downstream target gene, which encodes a CCT motif protein (CONSTANS, CO-like, and TOC1 motif; AT5G53420; Brambilla and Fornara, 2017), is referred to as “CCT101” hereinafter. To study the functional relevance of RNA isoform switches associated with SDG8 targets, we performed functional studies on the CCT101 RNA isoforms as a case study example.
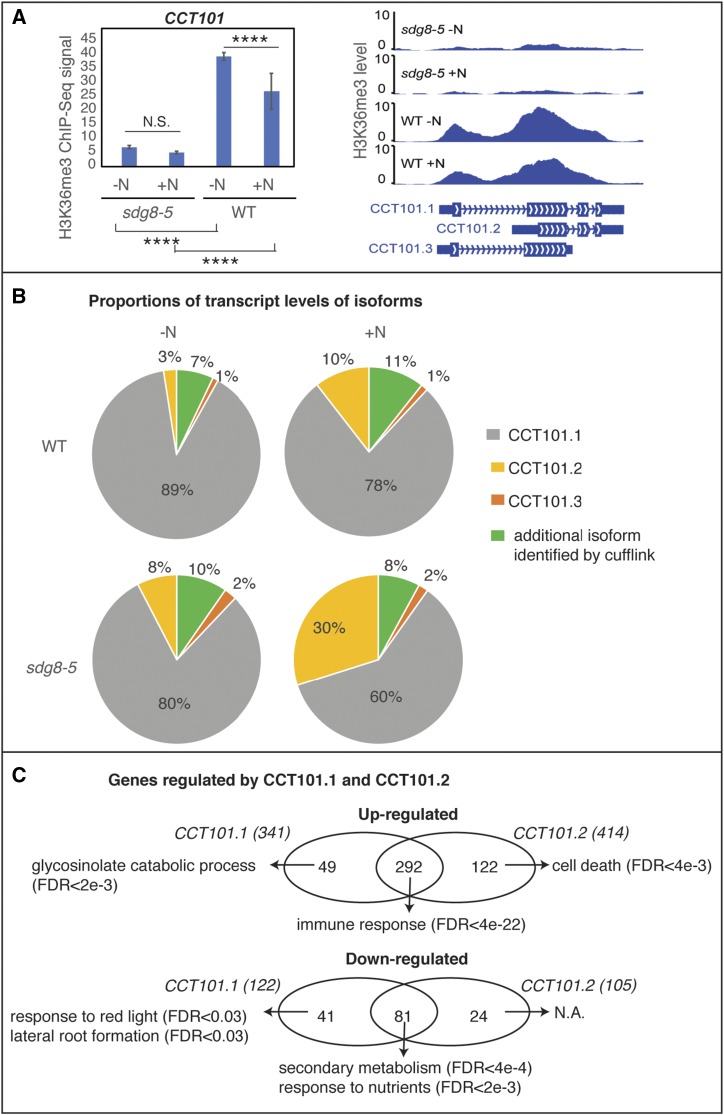
CCT101 belongs to the Activator of Spomin::LUC (ASML2) family of CCT domain proteins (Brambilla and Fornara, 2017). The best-studied member of this family is ASML2, which is a regulatory protein that activates expression of sugar-responsive genes (Masaki et al., 2005). In the sdg8-5 mutant, the H3K36me3 marks associated with the CCT101 locus are significantly lower, compared to wild type (FDR < 0.05; Fig. 5). Moreover, the H3K36me3 level associated with the CCT101 gene locus, which increases in response to N starvation in the wild type, is abrogated in the sdg8-5 mutant (FDR < 0.05; Fig. 5). CCT101 has three isoforms based on the TAIR10 annotation (Fig. 5). In our RNA-Seq analysis, we captured all three CCT101 RNA isoforms and also found support for an additional isoform that is expressed at very low levels (Fig. 5). CCT101.1, the longest transcript isoform, is the dominantly expressed isoform, while the RNA isoform CCT101.2 is truncated at the 5′ end and is expressed at a lower level (Fig. 5). Interestingly, in the sdg8-5 mutant, the proportions of CCT101.1 and CCT101.2 RNA isoforms change significantly in response to nitrate treatment (FDR = 0.02; Fig. 5). In wild type, the change in the proportions of CCT101.1 and CCT101.2 RNA isoform due to N treatment is not statistically significant. These data support the notion that SDG8 affects the preferential accumulation of RNA isoforms CCT101.1 and CCT101.2 in response to N-treatment.
Figure 5.
Histone methylation by SDG8 regulates RNA isoform preference of CCT101 (AT5G53420), which impacts transcriptional regulation. A, H3K36me3 levels at the CCT101 locus are affected by the sdg8-5 deletion and nitrate levels. In wild type (WT), the H3K36me3 mark at the CCT101 locus is induced by N starvation. By contrast, in the sdg8-5 mutant, the H3K36me3 mark at the CCT101 locus is greatly reduced and not responsive to N-level changes. Two biological replicates of histone ChIP-Seq, each sampled as a pool of 200 to 400 seedlings, were used to calculate the H3K36me3 levels. The error bars represent mean ± se. ****P < 0.001. The P values were determined using the program SICER. B, CCT101 has four isoforms identified by the software Cufflinks. Three of them are also annotated by TAIR10 (CCT101.1, CCT101.2, and CCT 101.3, as displayed in A). The proportion of transcripts for each CCT101 mRNA isoform is shown as pie chart for the genotype and condition tested. In the sdg8-5 mutant but not in the wild type, the change of N level induces a significant isoform switch. C, Overlapping yet different genes are regulated by proteins encoded by isoforms CCT101.1 and CCT101.2. Numbers of genes regulated by each isoform are shown within parentheses. Significantly enriched GO terms are displayed for shared targets between the two isoforms, or unique targets of either isoform.
To understand the functional implications of RNA isoform switching of CCT101, we analyzed the regulatory role of the proteins encoded by the two RNA isoforms CCT101.1 and CCT101.2. To do this, we tested their functionality using a plant cell-based transient overexpression system called TARGET (Bargmann et al., 2013). In detail, the CCT protein encoded by each of the RNA isoforms was transiently overexpressed in Arabidopsis shoot protoplasts, and after controlled nuclear import, the resulting effect of the CCT protein on genome-wide expression was assayed by RNA-Seq in three treatment replicates (see “Materials and Methods”). The experiments were performed on the same day using the same batch of protoplasts to ensure that the effect of different CCT isoforms can be directly compared. The overexpression of the protein encoded by the CCT101.1 RNA isoform led to the upregulation of 341 genes and downregulation of 122 genes in shoot protoplasts (FDR < 0.05, fold-change > 2; Fig. 5; Supplemental Table S10). By contrast, the protein encoded by the CCT101.2 RNA isoform upregulates 414 genes and downregulates 105 genes (FDR < 0.05, fold-change > 2; Fig. 5; Supplemental Table S10). Comparisons of these gene lists indicate that the two CCT101 RNA isoforms encode proteins that regulate overlapping, yet distinct gene targets (Fig. 5; Supplemental Table S10). Moreover, genes regulated by one CCT isoform but not by the other isoform have different enriched GO terms compared to the genes regulated by both isoforms (Fig. 5; Supplemental Table S10), suggesting that the proteins encoded by the two CCT isoforms perform related, yet different, functions.
Physiological Responses to Nitrate Are Abrogated in the Histone Methyltransferase Mutant sdg8-5
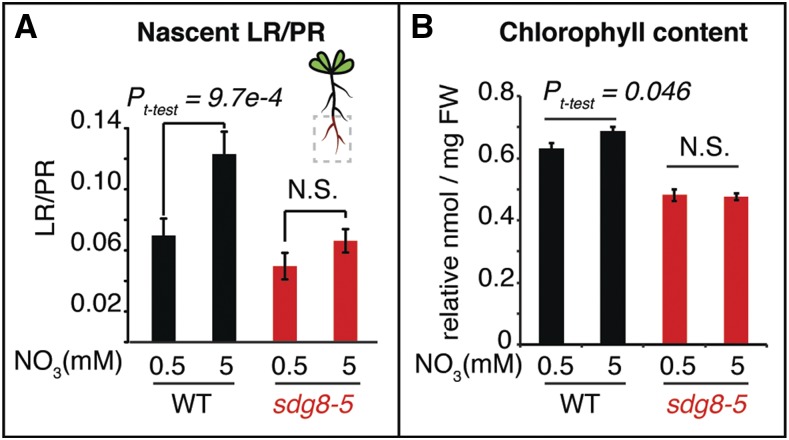
We next set out to test whether the SDG8-mediated changes in histone methylation and RNA isoforms in response to N-treatment have an impact on N-related phenotypic responses in planta. To do this, we first examined whether the sdg8-5 deletion mutant shows altered physiological responses to nitrate treatments. The sdg8-5 mutant and its corresponding wild-type strain (Li et al., 2015) were grown on ammonium succinate for 10 d and then transferred to either low N (0.5 mm of KNO3) or high N (5 mm of KNO3) medium (see “Materials and Methods”). Five to six days after the transfer, we measured chlorophyll content, root architecture, N uptake, and free amino acids. We found that wild-type plants grown under high-N conditions showed significantly enhanced lateral root growth on the nascent primary root (Fig. 6), as well as elevated chlorophyll levels in the shoots (Fig. 6), compared to low-N treatment conditions. By contrast, these responses to high N seen in wild type were abrogated in the sdg8-5 deletion mutant (Fig. 6). The sdg8-5 mutant also showed altered N acquisition and free amino acids compared to wild type (Supplemental Fig. S3). The sdg8-5 mutant had lower nitrate acquisition than wild-type plants under the high-N condition (Supplemental Fig. S3). Although the total amount of free amino acids was comparable between wild type and the sdg8-5 deletion mutant (Supplemental Fig. S3), free Leu was significantly higher in the sdg8-5 mutant compared to wild type under the low-N condition (Supplemental Fig. S3), and gamma-aminobutyric acid (GABA), a nonproteinogenic amino acid, was significantly induced by N supply in the sdg8-5 mutant but not in wild type (Supplemental Fig. S3). In summary, our data support the idea that deletion of the specific histone methyltransferase encoded by SDG8 affects plant physiological responses to nitrate at many different levels.
Figure 6.
Physiological N responses are attenuated in the histone methyltransferase deletion mutant sdg8-5. Root architecture, specifically nascent lateral root growth (A) and chlorophyll content (B), is regulated by N in the wild-type (WT) plants but not in the sdg8-5 mutant. The sdg8-5 mutants and the corresponding wild-type seedlings were first grown on ammonium succinate for 10 d and then transferred to either low N (0.5 mm of KNO3) or high N (5 mm of KNO3) medium. Chlorophyll content (n = 6, individual plants) and root architecture (n = 12–18, individual plants) were measured 5–6 d after the transfer. The P values were calculated by Student’s t test, and the error bars represent mean ± se. FW, fresh weight; LR, lateral roots; PR, primary roots.
DISCUSSION
Chromatin modifications including histone modifications have been reported as an additional regulatory layer of plant responses to environmental fluctuations (Charron et al., 2009; Kim et al., 2010). However, the mechanisms through which a specific histone-modifying protein affects gene transcription and/or RNA processing in response to environmental change is largely unknown. In this study, we addressed the role of epigenetics in the regulation of RNA isoforms in response to the nutrient environment by demonstrating that the sdg8-5 mutant, with a deletion of a specific histone methyltransferase, displays alterations in the genome-wide H3K36me3 landscape, transcriptome, and RNA isoform expression in response to nitrate treatments, as well as impaired physiological and metabolic responses to N-nutrient treatments. Overall, our study links a specific chromatin modification and histone methyltransferase with RNA processing and nitrate responses in plants.
Our previous studies showed that loss of H3K36 methylation in the gene body of SDG8 targets in the sdg8-5 mutant is associated with downregulation of gene expression (Li et al., 2015). Our study has now tested the hypothesis that the histone methylation marks made by SDG8 in the gene body are also able to impact qualitative transcriptomic changes via affecting RNA isoform switches. This hypothesis was inspired by our previous observation that SDG8 is associated with H3K36me3 in the gene body (Li et al., 2015), indicating a potential role for SDG8 in transcriptional elongation or cotranscriptional splicing. This notion was also supported by a recent study showing that H3K36me3 alters alternative RNA splicing of genes in the temperature response (Pajoro et al., 2017), and by findings that the yeast SET2 protein, a homolog of Arabidopsis SDG8, prevents spurious transcripts from starting in the middle of a gene (Carrozza et al., 2005). Moreover, SETD2, the mammalian homolog of Arabidopsis SDG8, is associated with proper RNA processing including the choice of transcription start site and exon-intron splicing (Simon et al., 2014). We thus tested the hypothesis that SDG8 plays a role in RNA processing in nutrient signaling in Arabidopsis. Our results indeed suggest that the Arabidopsis SDG8 protein affects RNA processing in transcriptional responses to N treatments.
How do SDG8 and H3K36me3 affect the production of RNA isoforms? In yeast, the H3K36 methylation deposited in the gene body by SET2 (homolog of SDG8) suppresses histone acetylation in the middle of a gene, thus preventing transcription from alternative start sites (Carrozza et al., 2005). It is possible that Arabidopsis SDG8 functions similarly to its yeast homolog, and this hypothesis could be tested by performing ChIP-Seq for histone acetylation in the sdg8-5 mutant. Alternatively, H3K36me3 might differentially “mark” histones associated with exons and introns, thus guiding spliceosome machinery for proper splicing. In line with this hypothesis, we found that there is a minor, but significant, enrichment of H3K36me3 deposited by SDG8 in the exons compared to the introns (P value < 0.005), based on our epigenome data (Li et al., 2015). In addition, we speculate that the H3K36me3 marks mediated by SDG8 could change the rate of Pol-II mediated transcription, which was shown to affect the fidelity of splicing in yeast (Aslanzadeh et al., 2018).
To further investigate how SDG8-mediated RNA isoform preference can affect protein function, we focused our functional validation on one putative regulatory protein, CCT101. We found that SDG8 affects the level of H3K36me3 associated with the CCT101 locus. Specifically, a reduced H3K36me3 mark at the CCT101 locus favors the transcription of the short RNA isoform (CCT101.2), while high levels of H3K36me3 are associated with the transcription of the long RNA isoform (CCT101.1). The missing N-terminal polypeptide in the short RNA isoform CCT101.2—relative to the longer isoform CCT101.1—is not predicted as a signal peptide by SignalP (Nielsen, 2017). Therefore, the biochemical function of the missing N-terminal polypeptide requires further investigation. To this end, we tested whether the protein encoded by the two CCT101 RNA isoforms have different regulated genome-wide targets using a transient overexpression system in plant cells called TARGET (Bargmann et al., 2013). Our TARGET studies show that both CCT101 isoforms can regulate hundreds of genes. Specifically, the proteins encoded by the CCT101.1 and CCT10 1.2 RNA isoforms regulate overlapping, as well as distinct downstream genes. Therefore, the RNA isoform switch between CCT101.1 and CCT101.2 that is affected by SDG8 results in two distinct functional CCT regulatory proteins that appear to regulate different sets of downstream genes.
How do the genome-wide changes of histone marks that we attribute to SDG8 affect the nitrate response in plants at a physiological level? Previous studies showed that levels of SDG8 mRNA are specifically induced in the root pericycle cells by a supply of nitrate (Gifford et al., 2008). Because lateral root initiation occurs in the pericycle, we tested if SDG8 is required for lateral root growth in response to a nitrate signal. We observed that wild-type plants display an enhanced lateral root growth in the newly grown primary root under high N relative to low N, and this morphological response to N treatments is abolished in the sdg8-5 mutant (Fig. 6). Therefore, SDG8 indeed has a role in mediating lateral root growth in response to N-level changes. However, because the H3K36me3 and transcriptome data in our study were collected from the shoots, an examination of H3K36 status of genes in roots could thus help to elucidate the role of SDG8-mediated histone methylation in sensing and foraging for nitrate.
One of the many possible roles for SDG8 is that it might serve as an integration point across inputs from photosynthesis, carbon metabolism, and N metabolism (Thum et al., 2008; Li et al., 2015). Indeed, among the misregulated N-responsive genes in the sdg8-5 mutant, GO terms related to response to sugar stimulus are significantly enriched (Supplemental Table S7). In agreement with this role in nutrient crosstalk, we observed that the chlorophyll level is increased by N supply in wild type, but not in the sdg8-5 mutant (Fig. 6). Therefore, the sdg8-5 mutant might not be able to provide enough carbon skeletons and reducing power for nitrate assimilation when nitrate level increases. This is supported by the observation that the sdg8-5 mutant has lower nitrate acquisition than wild-type plants under the high nitrate condition (Supplemental Fig. S3). Another piece of evidence comes from the free amino acid measurements comparing the sdg8-5 mutant and wild-type plants. GABA, a nonproteinogenic amino acid, has a central position in the interface between carbon and N metabolism, and is reported to regulate genes involved in N metabolism and carbon metabolism (Barbosa et al., 2010). GABA is synthesized from the N donor Glu and degraded into succinate to enter the citric acid cycle to generate carbon molecules (Michaeli and Fromm, 2015). In our study, GABA is significantly induced by N supply in the sdg8-5 mutant, but not in wild type (Supplemental Fig. S3). Collectively, our results support the notion of altered carbon/N balance in the sdg8-5 mutant deleted in the H3K36 methyltransferase, with impacts on gene expression, RNA processing, and physiological responses to N treatment.
CONCLUSION
Our study shows that nitrate treatments induce genome-wide changes in the histone mark H3K36me3 mediated by SDG8, with impacts on gene expression, RNA processing, and physiological responses. This has revealed a previously overlooked layer of chromatin regulation involved in nutrient signaling. To develop a comprehensive understanding of the scope and function of chromatin regulation in nutrient signaling, it is important that future studies expand to include more epigenetic marks, such as other histone modifications (e.g. histone acetylation), chromatin landscape, and DNA methylation. Such knowledge will reveal how chromatin modifiers could be harnessed and engineered to enable a holistic N response in plants to increase nutrient use efficiency.
MATERIALS AND METHODS
Plant Growth for ChIP-Seq and RNA-Seq
Arabidopsis (Arabidopsis thaliana) sdg8-5 mutant (aka cli186) and corresponding wild-type (Li et al., 2015) seeds were surface-sterilized and grown hydroponically in Phytatrays (Sigma-Aldrich). In detail, ∼200 seeds were sown onto a nylon mesh supported by a plastic platform in a sterile Phytatray filled with 200 mL of plant growth media (1× basal MS medium [cat. no. 97-5068EC, GIBCO], 3 mm of Suc, 0.5 mm of NH4 succinate, and 0.5 g/L of MES hydrate at pH 5.8). The Phytatrays were kept under white light (50 μM s−1 m−2) in a long-day cycle (16-h light/8-h dark) at 22°C for 16 d. Next, plants were starved for N for 24 h in media that is identical to the growth media, except that the 0.5 mm of NH4 succinate was removed. Next, half of the plants used were exposed to nitrate-supplied condition (5 mm of KNO3 added to the starvation solution). As controls, the other half of the plants used were exposed to the starvation medium with 5 mm of KCl added. After 5 h, the plants were harvested for ChIP-Seq or RNA-Seq (Supplemental Fig. S1).
ChIP-Seq Experiment
The H3K36me3 ChIP-Seq was performed as described in Li et al. (2015). Two independent biological replicates were collected from shoot tissues. The ChIP-Seq libraries for the first biological replicate were prepared as described in Li et al. (2015) and sequenced on a GAII platform (Illumina) with paired-end 75-bp format at the Cold Spring Harbor Laboratory. The ChIP-Seq libraries for the second replicate were barcoded with NEXTflex DNA Barcodes (Bioo Scientific) and pooled to sequence on a HiSeq platform (Illumina) with paired-end 100-bp format at the Cold Spring Harbor Laboratory.
ChIP-Seq Analysis
For the first replicate of H3K36me3 ChIP-seq, 20–34 million raw reads were generated for each sample. The raw reads were trimmed for adaptors and low quality using in-house Python and Perl scripts, and then mapped to the Arabidopsis genome (TAIR10) using the software BowTie (Langmead et al., 2009). On average, 89% of raw reads were mapped to genome, while 78% raw reads mapped to chromosomes. The chromosome-mapped reads were used to call significant H3K36me3 islands and significantly differentially methylated islands with H3K36me3 with the tool SICER (Zang et al., 2009), as described in Li et al. (2015). For the second replicate, 40–47 million raw reads were generated for each library and 65% were mapped to the genome, while 57% were mapped to chromosomes. The data analysis was performed in the same way as that for replicates. The ChIP-Seq signal of the two replicates share a high similarity (correlation coefficient = 0.9 across all the identified islands). To combine the results from the two replicates, a nitrate-responsive DMG (differentially methylated gene) has to pass the following criteria: identified as significantly different (FDR < 0.05) for both replicates; and the mean fold change (N-supplied versus N-starved) of Enrichment (Enrichment = ChIP/Input) of the two replicates has to be >1.2 for the transient N treatment. We used a relatively low cutoff for fold change that is similar to a previous study (Li et al., 2015) to detect H3K36me3 changes caused by N, because: (1) compared to RNA-Seq analysis that samples multiple copies of a transcript in a cell, histone ChIP-Seq samples only one copy of genomic DNA from a cell, therefore a smaller dynamic range is expected; (2) the nitrate treatment is a transient treatment that lasts for only a few hours; and (3) histone methylation compared to histone acetylation is relatively stable during the time frame of our treatment (in hours; Barth and Imhof, 2010). Nonetheless, changing the fold change cutoff from 1 to 2 revealed the same trend, which is that wild type shows more H3K36me3 changes in response to N treatment compared to the sdg8-5 mutant (Supplemental Fig. S4).
To identify hypomethylated genes, wild type and sdg8-5 mutants were compared for N-supplied and N-starved conditions separately, and then the overlap (∼90% overlap) between two conditions was identified. The analysis regarding biological replicates and statistical cutoff was similar to determining the nitrate-responsive DMGs described earlier in "ChIP-Seq Analysis", with fold change of enrichment greater than two as the cutoff, because the difference between wild type and mutant is a long-term effect compared to the transient N treatment.
Gene Overlap Analysis
The significance level of overlap between two gene lists was calculated by the tool Genesect within the VirtualPlant platform (Katari et al., 2010). Genesect is a nonparametric randomization test to determine whether an overlap between two gene lists is significantly higher or lower than by chance.
RNA-Seq Experiments
Three biological replicates of plant shoot tissues were harvested for each genotype and treatment, while each biological replicate is a pool of ∼200 seedlings. Total RNA samples were extracted using RNeasy Plant Mini kit (Qiagen) following the manufacturer’s instructions. The quality and quantity of RNA were determined using a Bioanalyzer (Agilent Genomics). Nine milligrams total RNA of each sample was used to prepare an RNA-Seq library using a home-made protocol (Wang et al., 2011). The RNA-Seq libraries were barcoded with the NEXTflex DNA Barcodes (Perkin Elmer). Twelve libraries were pooled and sequenced in one lane of the Illumina Hi-Seq platform with the paired-end 100-bp format at the Cold Spring Harbor Laboratory.
RNA-Seq Analysis
Fifteen million to 20 million read pairs were generated for each RNA-Seq library. The reads were trimmed for quality and adaptor using an in-house Python script. The trimmed reads were then mapped to the Arabidopsis TAIR10 genome using the TopHat suite (Trapnell et al., 2012). On average, 81% of the raw read pairs were mapped to the genome concordantly. Next, two analysis pipelines were performed to answer two different questions: (1) to identify DEGs using DESeq2 (Love et al., 2014); and (2) to identify alternative splicing genes using the cuffdiff function in the Cufflinks package (Trapnell et al., 2012). For the first pipeline, the gene counts were generated by HTSeq (Anders et al., 2015). The raw gene counts were then processed by DESeq2 with the design (∼Genotype+Treatment+Genotype:Treatment). Note that DESeq2 deals with normalization and low counts filtering internally (Love et al., 2014). To identify genes that are regulated by the interaction of N and genotype, P value for N×G interaction was adjusted for multitesting with FDR controlled at 5%. For the second pipeline to detect alternative spliced isoforms, the mapped reads were assembled into transcriptome (to detect new splicing events), and gene counts were generated using the program Cufflinks (Trapnell et al., 2012). The cuffdiff function in the Cufflinks package was used to call genes with alternative splicing between N-supplied groups and N-starved groups, in three categories (alternative promoter use, alternative splicing from the same primary transcript, and alternative coding sequence) with significance cutoff FDR < 0.1 (Trapnell et al., 2012). The conclusion remains the same if a different statistical cutoff such as FDR 5% is used. Note that cuffdiff only considers genes with high read counts for the alternative splicing analysis; therefore, the number of genes considered for the alternative splicing analysis is ∼2,000 for alternative promoter use, ∼5,800 for alternative splicing from the same primary transcript, and ∼1,800 for alternative coding-sequence output. The actual number of significant alternative splicing events is likely to be higher. To confirm the alternative splicing analysis by Cufflinks, we also analyzed the same RNA-Seq data with the tool DEXSeq (Anders et al., 2012) for differential exon usage. The conclusion was also supported by DEXSeq analysis.
Plant Growth for N-Response Phenotyping
The sdg8-5 mutant and the corresponding wild type were surface-sterilized and planted on growth medium as follows (unless indicated otherwise): 1× nitrate-free Murashige and Skoog medium (cat. no. MSP10; Caisson Labs), 3 mm of Suc, 0.5 mm of ammonium succinate, 0.5 g/L of sodium MES (sodium 2-[n-Morpholino]ethanesulfonic acid), 100 mg/L of I-inositol, 0.5 mg/L of niacin, 0.5 mg/L of pyridoxine HCl, 0.1 mg/L of thiamine HCl, and 1% (w/v) agar, at pH 5.8. The seeds were stratified at 4°C in dark for 4 d and then moved to a Percival growth chamber at 22°C, with a light cycle of 16-h light/8-h dark with 50 μM of s−1 m−2 light. The plants were grown vertically on plates. After 10 d, the plants were transferred to two groups of vertical plates, with high-N and low-N treatments, separately. The high-N and low-N plates are identical to the growth plates, except for that the 0.5 mm of ammonium succinate was replaced by either 5 mm (for high N) or 0.5 mm of KNO3 (for low N, with an additional 4.5 mm KCl to balance). For the 15N analysis, the KNO3 was spiked with 1% (v/v) K15NO3 to allow analysis of N uptake. The roots were scanned 5 d after the treatment for root architecture analysis, while the chlorophyll measurement (n = 6, individual plants), 15N measurement (n = 6, individual plants), and amino acid profiling (n = 3–5, pooled 150 seedlings) were performed 6 d after the treatment. For the amino acid profiling experiment, the plants were first grown with the following nutrients: 1× nitrate-free Murashige and Skoog medium (cat. no. MSP10; Caisson Labs), 3 mm of Suc, 0.5 mm of ammonium succinate, 0.5 g/L of NaMES, and 1% (w/v) agar at pH 5.8, and then transferred to high-N or low-N plates where 0.5 mm of ammonium succinate was replaced by either 5 mm of KNO3 or 0.5 mm of KNO3 supplemented with 4.5 mm of KCl.
15N Analysis
After 6 d of N treatments (with 5 or 0.5 mm of KNO3 spiked with 1% v/v of K15NO3), plants were analyzed for net 15NO3− uptake. Individual seedlings were first rinsed in 0.1 mm of CaSO4, and then placed in preweighed tin capsules (CE Elantech) and left in a 70°C oven for 48 h. Dry weight was determined and the total N and atom % 15N were analyzed by a Continuous-Flow Isotope Ratio Mass Spectrometer, using a Euro-EA Euro Vector Elemental Analyzer coupled with an IsoPrime Mass Spectrometer (GV Instruments).
Chlorophyll Analysis
Each individual seedling was weighed, flash-frozen in 1.5-mL tubes, and ground to powder using a Tissuelyzer (Qiagen). Next, 300 μL of methanol was added to each sample and tubes were rotated for 10 min at room temperature. The lysate was then centrifuged at 14,000 rpm for 5 min, and 200 μL of supernatant was transferred and measured with an Infinite M200 Plate Reader for absorbance (Life Sciences/Tecan) at 750, 665, and 652 nm, with methanol as the background controls. The chlorophyll content was calculated as described in Porra (2002). For the high-N versus low-N experiment, six individual seedlings were analyzed for each genotype/condition.
Root Architecture Analysis
After transferring the seedlings to high-N and low-N plates, the end of each primary root was marked with a permanent marker on the plate, to allow measurement of newly grown roots. After 5 d in high-N versus low-N condition, the roots were scanned, and the resulting images were analyzed using the program ImageJ (imagej.nih.gov/ij/). The root measurements generated by ImageJ were summarized and analyzed using in-house Perl and R scripts.
Amino Acid Profiling
Approximately 150 seedlings were pooled for each replicate per genotype and per N condition. In total, three to five replicates were analyzed per genotype and per condition. Amino acid profiling was performed as described in McCoy et al. (2018). Briefly, shoots were extracted in 10 mL of methanol. The neutral and basic amino acids were separated from acidic amino acids to measure Glu and Asp separately from Gln and Asn. Free amino acids were then derivatized with heptafluorobutyric acid and analyzed via gas chromatography mass spectrometry. Pools were quantified in reference to external standards using ⍺-amino-butyrate for neutral and basic amino acids and ⍺-amino-adipate for Glu and Asp. Pyro-Glu was identified and added to the Gln abundance. B-cyano-Ala was also identified and added to the Asn abundance. The resulting measurements were normalized to fresh weights. The significance of difference between nitrate treatments and genotypes was determined by Student’s t test as well as two-way ANOVA (genotype × nitrate). Only amino acids that show significant P value for genotype difference in both Student’s t test and ANOVA are reported (P < 0.05).
Determining the Genome-Wide Targets of Isoforms of CCT 101
The cell-based TARGET system for transcription factor perturbation (Bargmann et al., 2013) was used to identify the genome-wide targets of different isoforms of CCT101. Plants were grown on vertical petri plates containing half-strength Murashige and Skoog medium with 1 mm of KNO3 for 10 d before the experiment. The shoot protoplast preparation, transient transformation, and cell-sorting were performed as described in Bargmann et al. (2013). Protoplasts isolated from shoots were treated with 20 mm of KNO3 and 20 mm of NH4NO3 before the nuclear import of transcription factor induced by dexamethasone. Cells overexpressing either CCT101 isoforms or empty vector controls were collected in triplicate in the same batch of experiment and RNA-Seq libraries were prepared from mRNA using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs). All three isoforms are stably expressed at comparable levels based on RNA-Seq readout. The libraries were pooled and sequenced on the NextSeq 500 platform (Illumina) for 75 cycles. The RNA-Seq reads were aligned to the TAIR10 genome assembly using the software suite TopHat and gene expression was estimated using the GenomicFeatures/GenomicAlignments packages (Lawrence et al., 2013). Genes showing differential expression between the samples with an overexpressed isoform and the samples with empty vector were identified using the software package DESeq2 (Love et al., 2014) at a significance level of FDR < 0.05 and fold change > 2.
Accession Numbers
The ChIP-Seq and RNA-Seq data for this study can be accessed at the National Center for Biotechnology Sequence Read Archive (Accession no. SRP149810). SDG8 is encoded by AT1G77300.
Supplemental Data
The following materials are available.
Supplemental Figure S1. Experimental design for the ChIP-Seq and RNA-Seq experiments.
Supplemental Figure S2. Overlaps of the two groups of differentially methylated genes with the SDG8 bound genes.
Supplemental Figure S3. N uptake and assimilation in wild-type and the sdg8-5 mutant.
Supplemental Figure S4. The number of differentially methylated genes between N-supplied and N-starved conditions identified using different fold change cutoffs.
Supplemental Table S1. Genes with increased H3K36me3 level in N-supplied group compared to N-starved group, or vice versa, in wild-type plants
Supplemental Table S2. Significantly enriched GO terms (Biological process) among the 143 genes whose H3K36me3 increases in response to N-starvation in wild-type, determined by the Biomaps tool in the VirtualPlant platform.
Supplemental Table S3. Significantly enriched GO terms (Biological process) among the 53 genes whose H3K36me3 increases in response to N-supply in wild-type, determined by the Biomaps tool in the VirtualPlant platform.
Supplemental Table S4. Genes with increased H3K36me3 level in N-supplied group compared to N-starved group, or vice versa, in the sdg8-5 mutant.
Supplemental Table S5. 4,058 H3K36 hypomethylated genes that show significantly higher H3K36me3 in WT compared to the sdg8-5 mutant in N-supplied and N-starved groups.
Supplemental Table S6. Genes that are regulated by the interaction of genotype and N.
Supplemental Table S7. Significantly enriched GO terms among the cluster-1 genes regulated by the interaction of genotype and N, determined by AgriGO.
Supplemental Table S8. Significantly enriched GO terms of cluster-2 genes that are regulated by the interaction of genotype and N, determined by AgriGO.
Supplemental Table S9. Genes with isoform switch between N and KCl in wild-type and sdg8-5, determined by cuffdiff in Cufflinks.
Supplemental Table S10. The target genes of CCT101 and their functional enrichments.
Supplemental Results. Supplemental results describing how SDG8 dependent H3K36 methylation is detected to determine the hypomethylated genes.
Acknowledgments
We thank Dr. David Rhodes (Purdue University) for insightful discussion about amino acid analysis. We also thank two undergraduates, Ellen Cho and Teresa Qi, at New York University for their help in root analysis, and Pascal Tillard at the French National Institute for Agricultural Research (Montpellier, France) for 15N measurements.
Footnotes
This work was supported by the U.S. Department of Energy (DE-FG02-92ER20071 to G.M.C.), the National Science Foundation (MCB 1412232 to G.M.C.), the U.S. Department of Agriculture’s USDA National Institute of Food and Agriculture (Hatch project numbers 1013620 to Y.L., and 177845 to J.R.W.), Purdue University (startup funds to Y.L. and J.R.W.), and Centre National de la Recherche Scientifique (CNRS LIA-CoopNet; to G.M.C. and S.R.).
Articles can be viewed without a subscription.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W (2012) Detecting differential usage of exons from RNA-seq data. Genome Res 22: 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanzadeh V, Huang Y, Sanguinetti G, Beggs JD (2018) Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res 28: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T (2005) Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 280: 17732–17736 [DOI] [PubMed] [Google Scholar]
- Barbosa JM, Singh NK, Cherry JH, Locy RD (2010) Nitrate uptake and utilization is modulated by exogenous gamma-aminobutyric acid in Arabidopsis thaliana seedlings. Plant Physiol Biochem 48: 443–450 [DOI] [PubMed] [Google Scholar]
- Bargmann BOR, Marshall-Colon A, Efroni I, Ruffel S, Birnbaum KD, Coruzzi GM, Krouk G (2013) TARGET: A transient transformation system for genome-wide transcription factor target discovery. Mol Plant 6: 978–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth TK, Imhof A (2010) Fast signals and slow marks: The dynamics of histone modifications. Trends Biochem Sci 35: 618–626 [DOI] [PubMed] [Google Scholar]
- Berr A, Ménard R, Heitz T, Shen W-H (2012) Chromatin modification and remodelling: A regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell Microbiol 14: 829–839 [DOI] [PubMed] [Google Scholar]
- Brambilla V, Fornara F (2017) Y flowering? Regulation and activity of CONSTANS and CCT-domain proteins in Arabidopsis and crop species. Biochim Biophys Acta Gene Regul Mech 1860: 655–660 [DOI] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA (2014) Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Cuttriss AJ, Cossetto SB, Pye W, Crisp P, Whelan J, Finnegan EJ, Turnbull C, Pogson BJ (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli CI, Nisar N, Roberts AC, Murray KD, Borevitz JO, Pogson BJ (2014) A chromatin modifying enzyme, SDG8, is involved in morphological, gene expression, and epigenetic responses to mechanical stimulation. Front Plant Sci 5: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron J-BF, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J-K (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, He F, Berkowitz O, Liu J, Cao P, Tang M, Shi H, Wang W, Li Q, Shen Z, et al. (2018) Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 30: 2267–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Ma D-P, Li J (2008) The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem Biophys Res Commun 373: 659–664 [DOI] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX (2012) Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. Plant J 69: 978–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y (2012) Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet 13: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, Tanurdzic M, Dean A, Nero DC, McClung CR, et al. (2008) Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA 105: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, et al. (2010) VirtualPlant: A software platform to support systems biology research. Plant Physiol 152: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Zinta G (2016) Drought stress and chromatin: An epigenetic perspective In Hossain MA, Wani SH, Bhattacharjee S, Burritt DJ, and Tran L-SP, eds, Drought Stress Tolerant Plants, Vol 2. Springer International Publishing, Cham, Switzerland, pp 571–586 [Google Scholar]
- Kim J-M, To TK, Nishioka T, Seki M (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Kim SY, He Y, Jacob Y, Noh Y-S, Michaels S, Amasino R (2005) Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17: 3301–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou J-P, Daniel-Vedele F (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Carré C, Fizames C, Gojon A, Ruffel S, Lacombe B (2015) GeneCloud reveals semantic enrichment in lists of gene descriptions. Mol Plant 8: 971–973 [DOI] [PubMed] [Google Scholar]
- Lamke J, Brzezinka K, Altmann S, Baurle I (2016) A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J 35: 162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ (2013) Software for computing and annotating genomic ranges. PLOS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Fu F, Xu S, Lee SY, Yun D-J, Mengiste T (2016) Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28: 1640–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mukherjee I, Thum KE, Tanurdzic M, Katari MS, Obertello M, Edwards MB, McCombie WR, Martienssen RA, Coruzzi GM (2015) The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol 16: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Tsukagoshi H, Mitsui N, Nishii T, Hattori T, Morikami A, Nakamura K (2005) Activation tagging of a gene for a protein with novel class of CCT-domain activates expression of a subset of sugar-inducible genes in Arabidopsis thaliana. Plant J 43: 142–152 [DOI] [PubMed] [Google Scholar]
- McCoy RM, Utturkar SM, Crook JW, Thimmapuram J, Widhalm JR (2018) The origin and biosynthesis of the naphthalenoid moiety of juglone in black walnut. Hortic Res 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G (2015) AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun 6: 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli S, Fromm H (2015) Closing the loop on the GABA shunt in plants: Are GABA metabolism and signaling entwined? Front Plant Sci 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol Biol 1611: 59–73 [DOI] [PubMed] [Google Scholar]
- Pajoro A, Severing E, Angenent GC, Immink RGH (2017) Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol 18: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: Molecular regulation and stress responses in plants. Front Plant Sci 7: 760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ. (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73: 149–156 [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takei K, Hirose N (2006) Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci 11: 440–448 [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E (2009) Structure and mechanism of a Na+-independent amino acid transporter. Science 325: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan P-F, Jonasch E, Furey TS, et al. (2014) Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 24: 241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi G, Pandey BK, Deveshwar P, Giri J (2016) Emerging trends in epigenetic regulation of nutrient deficiency response in plants. Mol Biotechnol 58: 159–171 [DOI] [PubMed] [Google Scholar]
- Soppe WJ, Bentsink L, Koornneef M (1999) The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana. Development 126: 4763–4770 [DOI] [PubMed] [Google Scholar]
- Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM (2003) Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol 132: 907–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Coruzzi GM (2016) A matter of time—how transient transcription factor interactions create dynamic gene regulatory networks. Biochim Biophys Acta Gene Reg Mech 1860: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum KE, Shin MJ, Gutiérrez RA, Mukherjee I, Katari MS, Nero D, Shasha D, Coruzzi GM (2008) An integrated genetic, genomic and systems approach defines gene networks regulated by the interaction of light and carbon signaling pathways in Arabidopsis. BMC Syst Biol 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z (2017) agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res 45(W1): W122–W129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varala K, Marshall-Colón A, Cirrone J, Brooks MD, Pasquino AV, Léran S, Mittal S, Rock TM, Edwards MB, Kim GJ, Ruffel S, et al. (2018) Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc Natl Acad Sci USA 115: 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Álvarez JM, Moyano TC, Gutiérrez RA (2015) Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr Opin Plant Biol 27: 125–132 [DOI] [PubMed] [Google Scholar]
- Wang L, Si Y, Dedow LK, Shao Y, Liu P, Brutnell TP (2011) A low-cost library construction protocol and data analysis pipeline for Illumina-based strand-specific multiplex RNA-seq. PLoS One 6: e26426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-H, Garvin DF, Kochian LV (2001) Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol 127: 345–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widiez T, El Kafafi S, Girin T, Berr A, Ruffel S, Krouk G, Vayssières A, Shen W-H, Coruzzi GM, Gojon A, et al. (2011) High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3− uptake is associated with changes in histone methylation. Proc Natl Acad Sci USA 108: 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou J-P, Steinmetz A, Shen W-H (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28: 1348–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen W-H (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]