HUB-mediated H2Bub1 regulates the NADPH oxidase RbohD-dependent H2O2 signaling pathway, which plays an important role in the defense response to Vd-toxins.
Abstract
Histone H2B monoubiquitination (H2Bub1) plays critical roles in regulating growth and development as well as stress responses in Arabidopsis (Arabidopsis thaliana). In this study, we used wild-type and HUB1 and HUB2 loss-of-function Arabidopsis plants to elucidate the mechanisms involved in the regulation of the plant’s defense responses to Verticillium dahliae toxins (Vd-toxins). We demonstrated that HUB-mediated H2Bub1 regulates the expression of the NADPH oxidase RbohD by enhancing the enrichment of histone H3 trimethylated on Lys-4 in response to Vd-toxins. RbohD-dependent hydrogen peroxide (H2O2) signaling is a critical modulator in the defense response against Vd-toxins. Moreover, H2Bub1 also affects posttranscriptional mitogen-activated protein kinase (or MPK) signaling. H2Bub1 was required for the activation of MPK3 and MPK6. MPK3 and MPK6 are involved in regulating RbohD-mediated H2O2 production. MPK3 and MPK6 are associated with protein tyrosine phosphatases (PTPs), such as Tyr-specific phosphatase1 and mitogen-activated protein kinases phosphatase1, which negatively regulated H2O2 production. In addition, H2Bub1 is involved in regulating the expression of WRKY33. WRKY33 directly binds to RbohD promoter and functions as a transcription factor to regulate the expression of RbohD. Collectively, our results indicate that H2Bub1 regulates the NADPH oxidase RbohD-dependent H2O2 production and that the PTP-MPK3/6-WRKY pathway plays an important role in the regulation of RbohD-dependent H2O2 signaling in defense responses to Vd-toxins in Arabidopsis.
Post-translational modifications of histones play key roles in regulating chromatin dynamics, gene transcription, and DNA repair. Histone H2B monoubiquitination (H2Bub1) is a key modification that has significant effects, mainly associated with transcriptional activation and elongation, on genes (Pavri et al., 2006; Weake and Workman, 2008). H2Bub1 regulates multiple developmental processes in plants, including seed dormancy, leaf and root growth (Fleury et al., 2007; Liu et al., 2007), control of flowering time, plant development (Cao et al., 2008, 2015; Gu et al., 2009; Schmitz et al., 2009; Xu et al., 2009), photomorphogenesis, and circadian rhythms (Bourbousse et al., 2012; Himanen et al., 2012). The roles of H2Bub1in flowering time and circadian rhythm regulation are associated with histone H3 trimethylated on Lys-4 (H3K4me3) and/or with H3K36me3 (Cao et al., 2008; Himanen et al., 2012; Malapeira et al., 2012; Feng and Shen, 2014; Zhao et al., 2019). Moreover, H2Bub1 is involved in the regulation of defense responses and cuticle composition, which is a defensive barrier that protects plants against biotic stresses (Dhawan et al., 2009; Ménard et al., 2014; Zou et al., 2014). However, the mechanisms of H2Bub1 in plant defense responses are not fully understood.
Verticillium dahliae is a soil-borne pathogen that causes Verticillium wilt in a range of important plant species worldwide (Bhat and Subbarao, 1999). Although the physiology of plant defenses against Verticillium infection is well established (Fradin and Thomma, 2006; Shaban et al., 2018), the molecular mechanisms and regulatory pathways involved in the defense response to Verticillium remain largely unknown. Hydrogen peroxide (H2O2) may act as an upstream signal to modulate the expression of defense genes against V. dahliae toxins (Vd-toxins) in Arabidopsis (Arabidopsis thaliana; Yao et al., 2011). H2Bub1 plays an important role in the defense response to Vd-toxins, and HUB1 and HUB2 loss-of-function plants have reduced levels of Vd-toxins-induced H2O2 production (Hu et al., 2014). However, the molecular mechanisms of H2Bub1 in the regulation of the defense response to Vd-toxins in plants remain unknown.
The recognition of pathogens by plants triggers several early defense responses, including the production of reactive oxygen species (ROS) and the activation of mitogen-activated protein kinases (MAPKs; Lamb and Dixon, 1997; Torres et al., 2005; Boller and Felix, 2009). Plant NADPH oxidases, also known as respiratory burst oxidase homologs (RBOHs), are major sources of ROS during plant-pathogen interactions. RbohD and RbohF mediate diverse physiological processes, including responses to pathogens, in Arabidopsis. Both AtRbohD and AtRbohF were initially described as key components of plant defense (Torres et al., 2002; Torres and Dangl, 2005). However, they have contrasting functions in the regulation of cell death in response to pathogen attack (Torres and Dangl, 2005). RbohD is responsible for the generation of ROS in the response of Arabidopsis to all pathogens tested (Miller et al., 2009; Suzuki et al., 2011; Marino et al., 2012), while AtRbohF is a crucial modulator in the regulation of metabolic responses and resistance in Arabidopsis (Chaouch et al., 2012).
MAPK cascades play pivotal roles in defense-related signaling pathways of plants (Pedley and Martin, 2005). In Arabidopsis, three MAPKs (MPK3, MPK4, and MPK6) have been implicated in the defense against pathogens (Asai et al., 2002; Ichimura et al., 2002). MPK3 and MPK6 play positive roles in the activation of defense responses in Arabidopsis (Beckers et al., 2009; Rodriguez et al., 2010). However, the literature on the connection between MAPK activation and ROS production is contradictory. Some reports indicate that the MAPKs function downstream of the ROS burst during the triggering of plant immunity (Apel and Hirt, 2004; Pitzschke and Hirt, 2009). In contrast, other studies suggest that MAPKs act upstream of ROS production in the plant immunity-related signaling pathway (Zhang et al., 2007; Asai et al., 2008). Recently, studies have indicated that MAPK activation and the ROS burst are two independent early-signaling events in plant immunity (Segonzac et al., 2011; Xu et al., 2014).
MAPKs can be inactivated fully by MAPK phosphatases that dephosphorylate both Ser/Thr and Tyr residues (Chen et al., 2001). Tyr phosphatases play important roles in the regulation of MAPK cascades (Ichimura et al., 2002). Tyr phosphorylation plays a key role in regulating defense signaling in plants (Lin et al., 2014; Macho et al., 2014). Furthermore, protein Tyrosine Phosphatase1 (PTP1) and MAPK Phosphatase1 (MKP1), two main Tyr phosphatases, act as repressors of MPK3/MPK6-dependent stress-related signaling (Bartels et al., 2009). MKP1-mediated deactivation of MPK3/MPK6 in the stomatal signaling pathway has been reported, and MKP1 regulates MAPK signaling specificity and cell fate decisions during stomatal development (Tamnanloo et al., 2018). H2Bub1 modulates the expression levels of AtPTP1 and AtMKP1 genes and affects the activation of MPK3 and MPK6 in salt-stress responses (Zhou et al., 2017).
MAPK cascades also perform roles in transcriptional reprogramming through WRKY transcription factors (Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012). Some WRKY transcription factors may be regulated by MAPKs at the transcriptional and posttranscriptional levels in defense-related signaling pathways (Eulgem and Somssich, 2007; Pandey and Somssich, 2009; Ishihama and Yoshioka, 2012; Adachi et al., 2015). WRKY33 plays a vital role in Arabidopsis defenses against necrotrophic fungi (Zheng et al., 2006; Birkenbihl et al., 2012; Liu et al., 2017). WRKY33 may function as a downstream component of the MPK3/MPK6-mediated signaling pathway, and WRKY33 expression is regulated by the MPK3/MPK6 cascade (Mao et al., 2011). However, the upstream events in the MPK3/MPK6 cascade and the roles of the MPK3/MPK6-dependent signaling pathway in the regulation of defense responses to Vd-toxins are currently unknown.
Here, we show that HUB-mediated H2Bub1 plays an important role in regulating the H2O2-signaling pathway in defense responses to Vd-toxins. H2Bub1 regulated the expression of the NADPH oxidase AtRbohD and WRKY33 by enhancing the enrichment of H3K4me3 in response to Vd-toxins. The active transcription of WRKY33 regulates AtRbohD expression. In addition, H2Bub1 also affects posttranscriptional MAPK signaling. H2Bub1 is required for the activation of MPK3 and MPK6. Moreover, MPK3 and MPK6 associate with PTPs, which negatively regulated H2O2 production. The PTP-MPK3/6-WRKY pathway regulates H2O2 signals in the response against Vd-toxins in Arabidopsis.
RESULTS
HUB1 and HUB2 Are Involved in Regulating the Defense Response to Vd-Toxins
To explore the possible functions of H2Bub1 in the defense response to Vd-toxins, wild-type and HUB1 and HUB2 loss-of-function Arabidopsis plants were used in this study. First, we observed root growth after exposure to Vd-toxins in the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants of Arabidopsis. Root growth was suppressed by exposure to Vd-toxins in a concentration-dependent manner. Root lengths of the mutants were significantly shorter than those of the wild type after a 4-d Vd-toxins treatment (Fig. 1, A and B). Thus, the HUB1 and HUB2 loss-of-function Arabidopsis plants were more susceptible to Vd-toxins. We also found that Vd-toxins could induce global changes in H2Bub1 in wild-type plants (Supplemental Fig. S1).
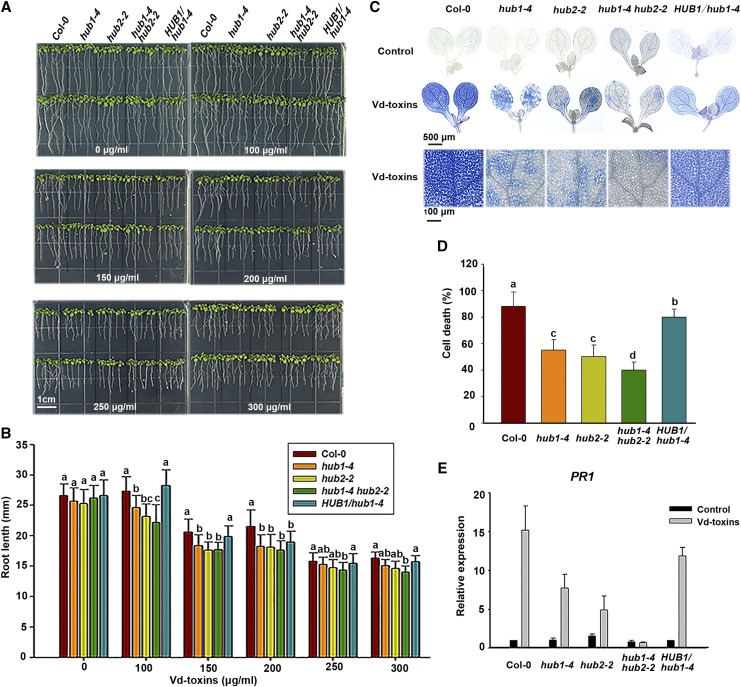
Figure 1.
HUB1 and HUB2 are involved in regulating defense responses to Vd-toxins in Arabidopsis. A, Root growth of wild-type Columbia-0 (Col-0), hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and HUB1/hub1-4 complementation line seedlings treated with Vd-toxins. Seedlings (4 d old) of the wild type and mutants were transferred from one-half-strength Murashige and Skoog (MS) medium to one-half-strength MS medium without or with Vd-toxins (100–300 μg mL−1). Photographs were taken 4 d after transfer. B, Quantification of root lengths in A. Error bars indicate sd; n = 30. C, Vd-toxins-induced cell death in cotyledons of the Arabidopsis wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line. Cotyledons of 7-d-old plants were treated with 150 μg mL−1 Vd-toxins and stained with Trypan Blue. Leaves treated with double distilled water were used as controls. D, Quantification of the cell death rates induced by Vd-toxins in C using ImageJ software. Error bars indicate sd; n = 8. E, Relative expression levels of the defense gene PR1 in the wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line were determined after treatment with 150 μg mL−1 Vd-toxins. Seedlings treated with double distilled water were used as controls. Total RNA was extracted 24 h after the Vd-toxins treatment for RT-qPCR analysis. Error bars indicate sd; n = 15. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. All experiments were repeated at least three times.
The central features of plant defense responses are rapid cell death and the activation of defense-related genes, and this combination is known as the hypersensitive response (HR; Dangl and Jones, 2001; Greenberg and Yao, 2004). Next, we analyzed the rapid cell death and Pathogenesis-Related Protein1 (PR1) gene expression after exposure to Vd-toxins in the wild type and the mutants. We used the Trypan Blue staining method to detect cell death. The extensive cell death was dramatically reduced in the leaves of the hub1-4, hub2-2, and hub1-4 hub2-2 mutants 14 h after the Vd-toxins treatment compared with the wild type (Fig. 1, C and D). Furthermore, the relative expression levels of PR1 were monitored by reverse transcription quantitative PCR (RT-qPCR). The expression of PR1 was strongly increased in the wild type after the Vd-toxins treatment, whereas the expression of PR1 was substantially reduced in the mutants, especially in the hub1-4 hub2-2 mutant (Fig. 1E).
To corroborate that the phenotypes of the hub mutants resulted from the disruption of HUB, the hub1-4-complemented line HUB1/hub1-4 was used to analyze the above phenotypes and defense responses. The phenotype of the HUB1/hub1-4 complementation line was restored to that of the wild type (Fig. 1).
Thus, the mutants showed the suppression of root growth, reduction of Vd-toxins-induced cell death, and expression of defense genes, suggesting that HUB1 and HUB2 play important roles in regulating defense responses to Vd-toxins in Arabidopsis.
HUB1 and HUB2 Loss-of-Function Plants Have Reduced Vd-Toxins-Induced H2O2 Production Levels
Following successful pathogen recognition, ROS are proposed to orchestrate the establishment of plant defenses (Torres et al., 2006). H2O2 accumulation is a marker of the cellular defense response (Sang and Macho, 2017). We next explored whether HUB1 and HUB2 are involved in regulating H2O2 production in defense responses to Vd-toxins. H2O2 in Arabidopsis leaves was labeled using a 2′,7′-dichlorofluorescein diacetate (H2DCF-DA) fluorescent probe. The fluorescence intensity observed in the wild type substantially increased after the Vd-toxins treatment, whereas the fluorescence intensity was only slightly increased in hub1-4, hub2-2, and hub1-4 hub2-2 mutants. The intensity of the HUB1/hub1-4 complementation line was similar to that of the wild type (Fig. 2, A and B). Furthermore, we measured the H2O2 content in leaves using a chromogenic peroxidase-coupled assay, and, similar to the above result, the level of H2O2 accumulation induced by the Vd-toxins was dramatically reduced in the mutant compared with the wild type (Fig. 2C).
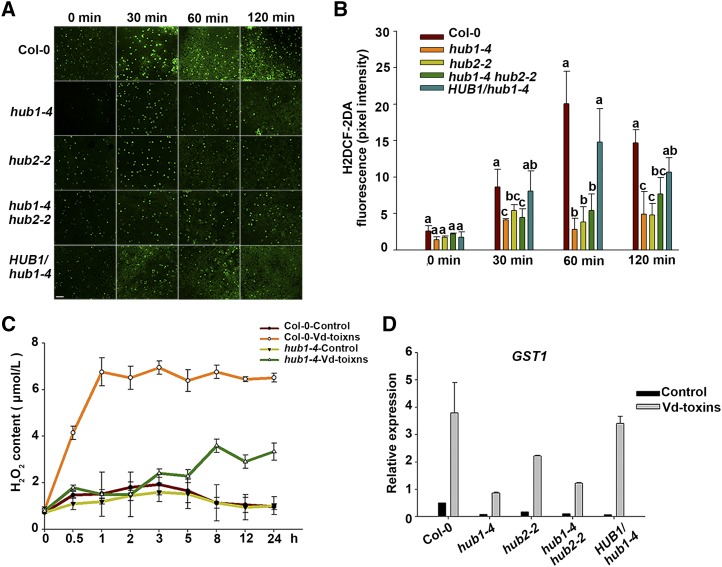
Figure 2.
H2O2 production induced by Vd-toxins in the leaves of wild-type Arabidopsis Columbia-0 (Col-0) and mutants. A, H2O2 was detected by fluorescence resulting from H2DCF-DA, as described in “Materials and Methods.” Bar = 100 μm. B, Quantification of the H2DCF-DA fluorescence intensities in A. Error bars indicate sd; n = 8. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. C, Quantitative measurements of H2O2 concentrations in the leaves using the chromogenic peroxidase-coupled assay. Error bars indicate sd; n = 3. D, Relative expression levels of GST1 after treatment with 150 μg mL−1 Vd-toxins in the wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line. Seedlings treated with double distilled water were used as controls. Total RNA was extracted 24 h after the Vd-toxins treatment for RT-qPCR analysis. Error bars indicate sd; n = 3. All experiments were repeated at least three times.
The Glutathione-S-Transferase1 (GST1) gene can function as a marker gene for ROS (Love et al., 2005). To determine whether HUB1 and HUB2 loss-of-function plants affect H2O2 production at the transcriptional level, we measured the relative expression levels of GST1 using RT-qPCR. The expression of GST1 was strongly increased in the wild type after the Vd-toxins treatment, whereas the expression of GST1 was substantially reduced in the mutants. The expression level in the HUB1/hub1-4 complementation line was partially restored to that of the wild type (Fig. 2D). Thus, HUB1 and HUB2 play functional roles in regulating H2O2 production in Arabidopsis defense responses to Vd-toxins.
HUB1 and HUB2 Regulate NADPH Oxidase AtRbohD Expression
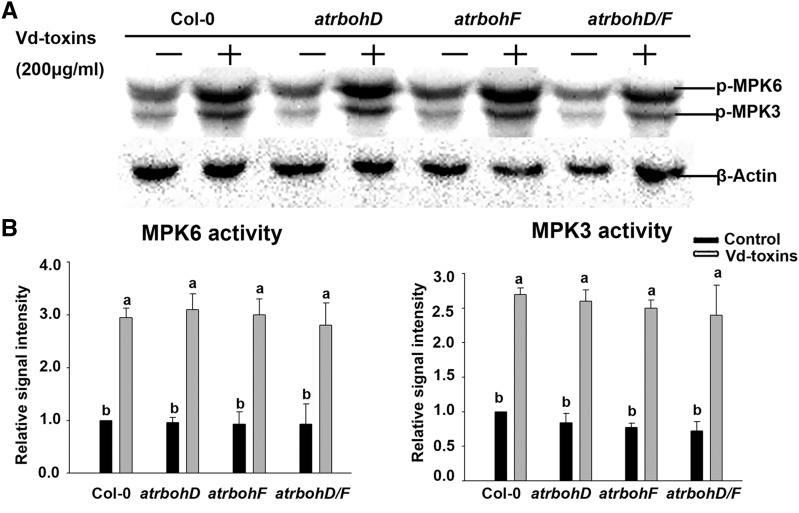
NADPH oxidases are necessary for the H2O2 production induced by Vd-toxins in Arabidopsis (Yao et al., 2011). To further dissect the effect of H2Bub1 on H2O2 production, we first examined H2O2 production induced by Vd-toxins in the wild type, the NADPH oxidase loss-of-function mutants atrbohD and atrbohF, as well as the atrbohD/F double mutant, using the H2DCF-DA staining assay. H2O2 production was induced by Vd-toxins, and the H2O2 level displayed a time-dependent increase in both the wild type and mutants. However, H2O2 production was significantly lower in atrbohD, atrbohF, and atrbohD/F than in the wild type (Fig. 3, A and B). Interestingly, H2O2 production was significantly greater in the atrbohF mutant, especially later in the treatment (25–30 min), than in the atrbohD or atrbohD/F mutant.
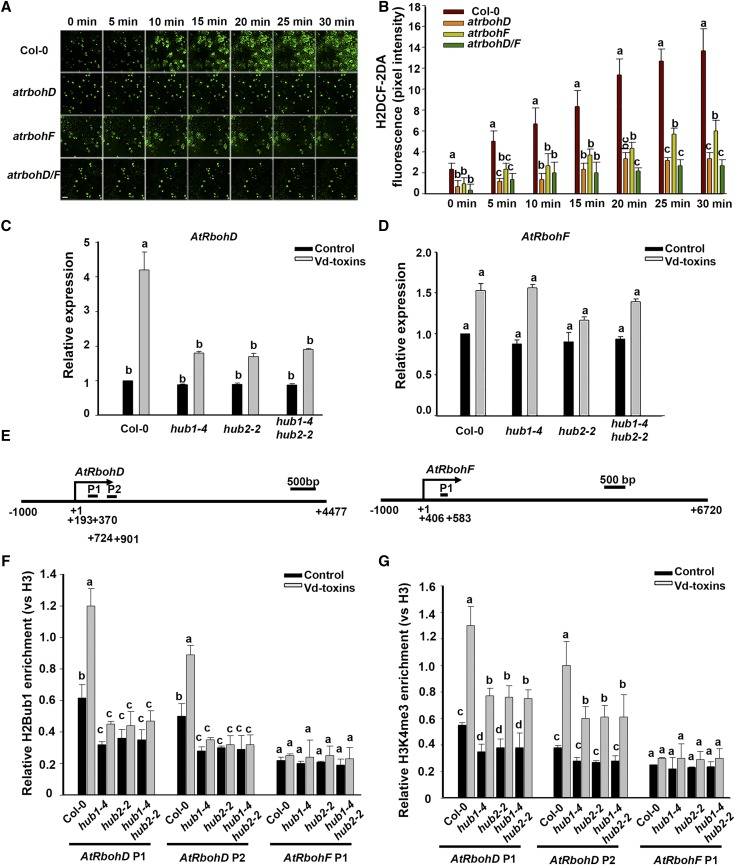
Figure 3.
AtRbohD and AtRbohF expression affect H2O2 production and the HUB1/HUB2-regulated AtRbohD expression level in response to the Vd-toxins treatment in Arabidopsis. A, H2O2 was detected in the leaves of wild-type Columbia-0 (Col-0) and the atrbohD, atrbohF, and atrbohD/F mutants by assessing the fluorescence resulting from H2DCF-DA, as described in “Materials and Methods.” Bar = 20 μm. B, Quantification of the H2DCF-DA fluorescence intensities in A. Error bars indicate sd; n = 8. C and D, Relative expression levels of AtRbohD and AtRbohF after treatment with 150 μg mL−1 Vd-toxins in the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants. Total RNA was extracted 24 h after the Vd-toxins treatment for RT-qPCR analysis. Seedlings treated with double distilled water were used as controls. Error bars indicate sd; n = 3. E, Schematic diagram of the AtRbohD and AtRbohF genes. AtRbohD P1, AtRbohD P2, and AtRbohF P1 are gene body regions; arrows indicate transcription start sites. F and G, Relative enrichments of H2Bub1 and H3K4me3 in the AtRbohD locus after treatment with Vd-toxins in the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants. Chromatin was extracted 12 h after the Vd-toxins treatment, and immunoprecipitated DNA was analyzed using qPCR. Data were determined as percentages of H2Bub1/H3 and H3K4me3/H3 for each individual gene position. Relative enrichments of H2Bub1 and H3K4me3 in the AtRbohF P1 locus were used as negative controls. Error bars indicate sd; n = 3. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. All experiments were repeated three times.
Next, we examined the expression levels of AtRbohD and AtRbohF in the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants after Vd-toxins treatment. The expression of AtRbohD was significantly up-regulated in the wild type, whereas it was slightly up-regulated in hub1-4, hub2-2, and hub1-4 hub2-2 mutants. In contrast, no marked differences in the expression levels of AtRbohF were detected between the hub1-4, hub2-2, and hub1-4 hub2-2 mutants and the wild type (Fig. 3, C and D).
To further investigate how HUB1 and HUB2 influenced the transcription of AtRbohD, we performed a chromatin immunoprecipitation (ChIP) assay using an anti-H2Bub1 antibody against chromatin derived from the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and we designed two fragments based on the chromatin of AtRbohD for ChIP-qPCR, AtRbohD P1 (193–370) and AtRbohD P2 (724–901; Fig. 3E). qPCR was performed to detect the relative H2Bub1 enrichment in the chromatin of AtRbohD, while the relative H2Bub1 enrichment in the chromatin of AtRbohF P1 (406–583) was used as the negative control. H2Bub1 was enriched intensively in the chromatin of the AtRbohD locus of the wild type, whereas it was enriched weakly in the hub mutants. Moreover, H2Bub1 was obviously increased in the chromatin of AtRbohD of the wild type after the Vd-toxins treatment, while it was barely increased in the hub mutants (Fig. 3F).
In general, histone H3K4me3 is necessary for gene activation while H3K27me is associated with transcriptional repression (Li et al., 2007). H2Bub1 is required for H3K4 and H3K79 methylation in yeast (Lee et al., 2007). Therefore, we performed ChIP assays using an anti-H3K4me3 antibody to detect the level of H3K4me3 enrichment in the chromatin of AtRbohD. H3K4me3 was increased in the chromatin of AtRbohD after Vd-toxins treatment in both the wild type and hub mutants; however, it was lower in the hub mutants (Fig. 3G). These results indicated that H2Bub1 regulated the expression of AtRbohD by enhancing the level of H3K4me3 in the AtRbohD locus after the Vd-toxins treatment.
Thus, AtRbohD may play a more important role than AtRbohF in H2O2 production induced by Vd-toxins. Additionally, H2Bub1 regulates the expression of the NADPH oxidase AtRbohD, which is associated with the enhancement of H3K4me3 in the chromatin of AtRbohD.
HUB1 and HUB2 Are Required for the Activation of MPK3 and MPK6 during Defense Responses to Vd-Toxins
MPK3 and MPK6 have emerged as central players in MAPK-mediated stress signaling (Bartels et al., 2009; Pitzschke, 2015), while H2Bub1 is required for the activation of MPK3 and MPK6 in modulating salt-stress tolerance in Arabidopsis (Zhou et al., 2017). Therefore, we examined whether MAPK is involved in the Vd-toxins-induced responses and whether HUB1 and HUB2 are required for the activation of MPK3 and MPK6 during defense responses. To investigate the latter, we confirmed the activation of MPK3 and MPK6 in wild-type and mutant seedlings. Seedlings of the 7-d-old wild type and the hub1-4 mutant were independently treated with 200 μg mL−1 Vd-toxins, and samples were taken at various times. MPK3 and MPK6 were rapidly activated, and activities were significantly elevated in the wild type within 30 min, whereas their activity levels were partially elevated in the hub1-4 mutant compared with the wild type (Fig. 4, A and B; Supplemental Fig. S2). Furthermore, seedlings of hub2-2 and hub1-4 hub2-2 mutants were also independently treated with 200 μg mL−1 Vd-toxins for 30 min, and the results were consistent with those of the hub1-4 mutant (Fig. 4, C and D). Thus, HUB1 and HUB2 appear to be required for the activation of MPK3 and MPK6 during defense responses to Vd-toxins.
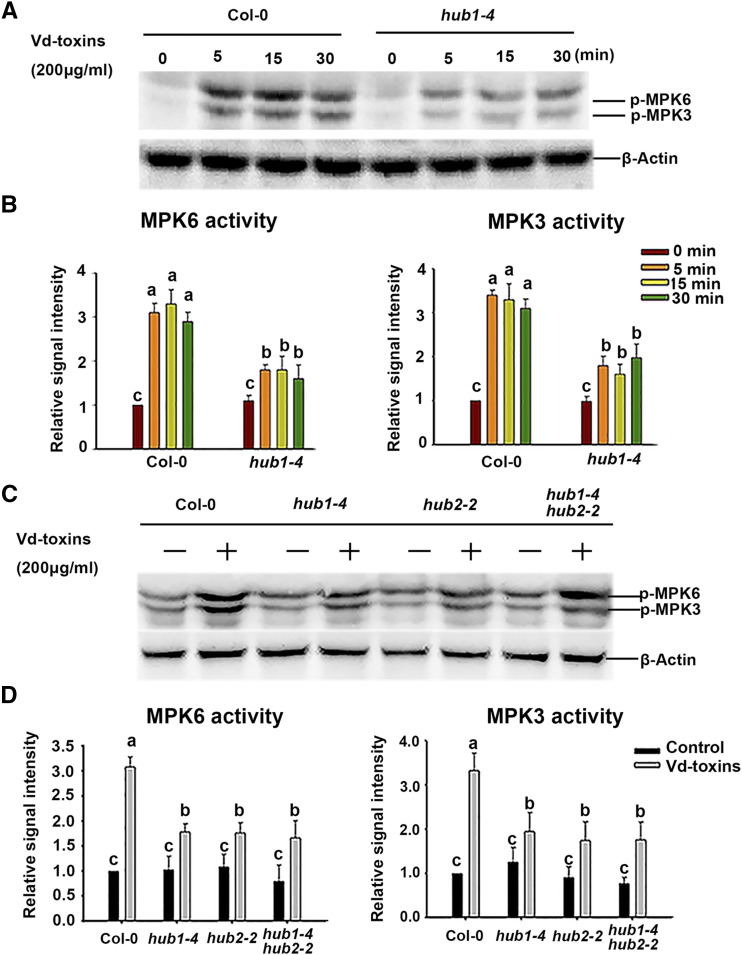
Figure 4.
HUB1 and HUB2 affect the activation of MPK3 and MPK6 in response to the Vd-toxins treatment in Arabidopsis. A, The kinase activities of MPK3 and MPK6 were detected by immunoblotting using anti-phospho-p44/42 MAPK antibodies (p-MPK6 and p-MPK3). Seedlings (7 d old) of wild-type Columbia-0 (Col-0) and the hub1-4 mutant were treated with 200 μg mL−1 Vd-toxins, and then total proteins were extracted after various times for immunoblot analysis. β-Actin was used as the loading control. Results are presented three independent biological replicates. B, Quantification of the kinase activities of MPK3 and MPK6 in A using ImageJ software. Error bars indicate sd; n = 3. C, The kinase activities of MPK3 and MPK6 were measured in the wild type and the hub1-4, hub2-2, and hub1-4 hub2-2 mutants. Seedlings (7 d old) of the wild type and mutants were treated with 200 μg mL−1 Vd-toxins, and then total proteins were extracted after 30 min for immunoblot analysis. Results are presented three independent biological replicates. D, Quantification of the kinase activities of MPK3 and MPK6 in C using ImageJ software. Error bars indicate sd; n = 3. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests.
MPK3 and MPK6 Are Involved in Regulating AtRbohD-Mediated H2O2 Production
To determine whether H2O2 production affected the activation of MPK3 and MPK6 in response to Vd-toxins or whether the activation of MPK3 and MPK6 affected the H2O2 production induced by Vd-toxins, we examined the activation of MPK3/MPK6 and H2O2 production in the wild type, atrbohD, atrbohF, atrbohD/F, mpk3, and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and MKK5DD, a constitutively active MKK5 kinase mutant, which leads to the rapid activation of MPK3 and MPK6 during dexamethasone (DEX) treatment.
We first examined the activation of MPK3 and MPK6 after the Vd-toxins treatment in the wild type and the atrbohD, atrbohF, and atrbohD/F mutants. The activation states of MPK3 and MPK6 were induced by Vd-toxins in the mutants and the wild type; however, there were no significant differences in the activation levels of MPK3 and MPK6 among the tested lines (Fig. 5). H2O2 production was significantly lower in atrbohD, atrbohF, and atrbohD/F mutants compared with the wild type (Fig. 3, A and B). Thus, the activation states of MPK3 and MPK6 are not affected by H2O2 production triggered by Vd-toxins.
Figure 5.
Activation states of MPK3 and MPK6 induced by Vd-toxins in wild-type Arabidopsis Columbia-0 (Col-0) and atrbohD, atrbohF, and atrbohD/F mutants. A, The kinase activities of MPK3 and MPK6 were measured in the wild type and the atrbohD, atrbohF, and atrbohD/F mutants. Seedlings (7 d old) of the wild type and mutants were treated with 200 μg mL−1 Vd-toxins, and total proteins were then extracted after 30 min for immunoblot analysis. β-Actin was used as the loading control. Results are presented three independent biological replicates. B, Quantification of the kinase activities of MPK3 and MPK6 in A using ImageJ software. Error bars indicate sd; n = 3. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests.
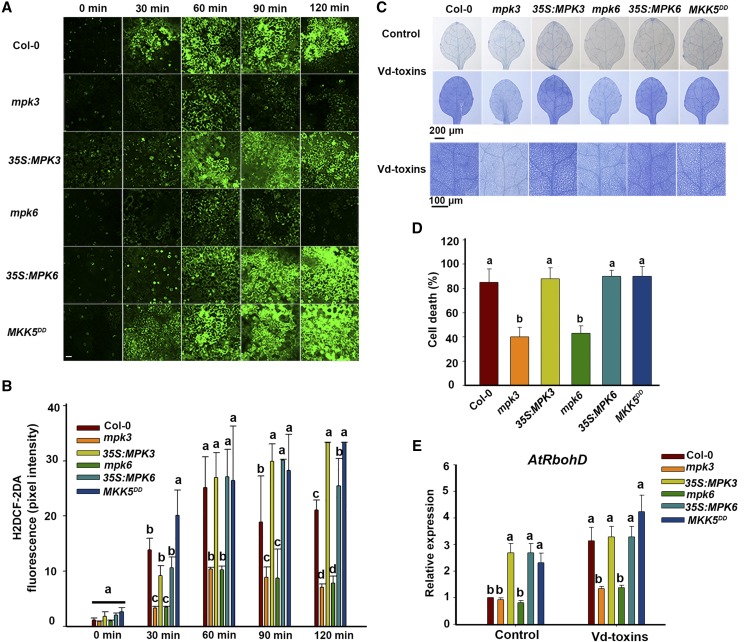
To further determine whether MPK3 and MPK6 affected H2O2 production induced by Vd-toxins, we examined H2O2 production in the wild type, the mpk3 and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and MKK5DD. H2O2 production increased dramatically after the Vd-toxins treatment in the wild type, 35S:MPK3, 35S:MPK6, and MKK5DD, whereas the induced H2O2 production was significantly reduced in the mpk3 and mpk6 mutants compared with the wild type (Fig. 6, A and B). Trypan Blue staining was used to determine whether the mpk3 and mpk6 mutations affected the HR cell death-related phenotypes. As shown in Figure 6, C and D, the mpk3 and mpk6 mutations reduced the extensive cell death, indicating that MPK3 and MPK6 are associated with HR-related cell death in response to Vd-toxins. Furthermore, we examined the expression of AtRbohD in the wild type, the mpk3 and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and MKK5DD after exposure to Vd-toxins. As shown in Figure 6E, the expression levels of AtRbohD in mpk3 and mpk6 mutants were much lower than those in the wild type, 35S:MPK3 and 35S:MPK6 lines, and MKK5DD, indicating that the expression of AtRbohD is positively regulated by MPK3 and MPK6. Thus, MPK3 and MPK6 putatively regulate H2O2 production, and MPK3 and MPK6 are involved in regulating the expression of RbohD during the defense response to Vd-toxins.
Figure 6.
MPK3 and MPK6 regulate H2O2 production and RbohD expression in Arabidopsis. A, H2O2 was detected in the leaves of wild-type Columbia-0 (Col-0), the mpk3 and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and the MKK5DD mutant using fluorescence resulting from H2DCF-DA, as described in “Materials and Methods.” Leaves were treated with Vd-toxins plus 15 μm DEX. Bar = 20 μm. B, Quantification of the H2DCF-DA fluorescence intensities in A. Error bars indicate sd; n = 8. C, Cell death in cotyledons of wild-type, the mpk3 and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and MKK5DD Arabidopsis seedlings. Cotyledons of 14-d-old seedlings were treated 14 h with 150 μg mL−1 Vd-toxins plus 15 μm DEX and stained with Trypan Blue. Seedlings treated with 15 μm DEX were used as controls. D, Quantification of the cell death rates induced by Vd-toxins in C using ImageJ software. Error bars indicate sd; n = 15. E, Relative expression levels of AtRbohD after treatment in wild-type, the mpk3 and mpk6 mutants, the 35S:MPK3 and 35S:MPK6 lines, and MKK5DD seedlings. Total RNA was extracted 12 h after the Vd-toxins treatment for RT-qPCR analysis. Error bars indicate sd; n = 9. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. All experiments were repeated three times.
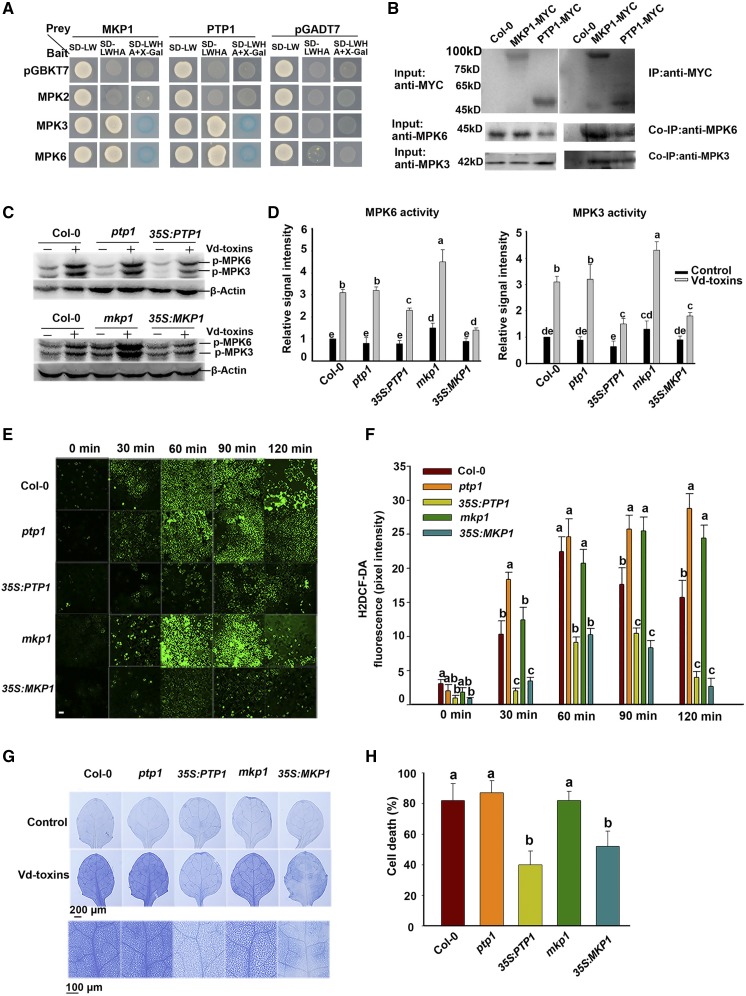
MPK3 and MPK6 Interact with PTP1 and MKP1, and PTP1 and MKP1 Negatively Regulate H2O2 Production
PTP1 and MKP1 are important regulators of MPK3/MPK6-dependent stress signaling (Bartels et al., 2009). To examine whether PTP1 and MKP1 affect MPK3 and MPK6 signaling involved in the defense response to Vd-toxins, we used yeast two-hybrid and coimmunoprecipitation (Co-IP) assays to detect whether PTP1 or MKP1 interacts with MPK3 or MPK6. PTP1 and MKP1 interacted with MPK3 and MPK6 (Fig. 7, A and B). To further determine whether PTP1 and MKP1 affected the activation states of MPK3 and MPK6, kinase activity assays were performed by immunoblotting using an anti-phospho-p44/42 antibody. The wild-type plants were independently transformed with vectors expressing PTP1 and MKP1 cDNA driven by the strong 35S promoter of Cauliflower mosaic virus (35S:PTP1 and 35S:MKP1). The MPK3 and MPK6 activity levels were significantly elevated by exposure to Vd-toxins in the wild type and the ptp1 and mkp1 mutants, whereas the MPK3 and MPK6 activity levels were partially increased in the 35S:PTP1 and 35S:MKP1 lines (Fig. 7, C and D). Thus, PTP1 and MKP1 act as negative regulators of MPK3 and MPK6 activities.
Figure 7.
MPK3 and MPK6 interact with PTP1 and MKP1, which negatively regulate H2O2 production in Arabidopsis. A, Interactions of PTP1 and MKP1 with MPK3 and MPK6 in the yeast two-hybrid system. The experiments were performed three times with similar results. B, Co-IP of MPK3 and MPK6 interactions with PTP1 and MKP1. Total proteins were extracted from 15-d-old wild-type Columbia-0 (Col-0) and transgenic 35S:PTP1 and 35S:MKP1 lines. Input and immunoprecipitated proteins were analyzed by independently immunoblotting with anti-MYC, anti-MPK3, and anti-MPK6 antibodies. C, The kinase activities of MPK3 and MPK6 were detected by immunoblotting using anti-phospho-p44/42 MAPK antibodies (p-MPK6 and p-MPK3). Seedlings (7 d old) of the wild type, the ptp1 and mkp1 mutants, and the 35S:PTP1 and 35S:MKP1 lines were treated with 200 μg mL−1 Vd-toxins, and the total proteins were then extracted at various times for immunoblot analysis. β-Actin was used as the loading control. D, Quantification of the kinase activity levels of MPK3 and MPK6 in C using ImageJ software. Results are presented three independent biological replicates. Error bars indicate sd; n = 3. E, H2O2 was detected in the leaves of the wild type, the ptp1 and mkp1 mutants, and the 35S:PTP1 and 35S:MKP1 lines using a fluorescence assay with H2DCF-DA, as described in “Materials and Methods.” Bar = 20 μm. F, Quantification of the H2DCF-DA fluorescence intensities in E. Error bars indicate sd; n = 8. G, Cell death induced by Vd-toxins in cotyledons of the wild type, the ptp1 and mkp1 mutants, and the 35S:PTP1 and 35S:MKP1 lines. Cotyledons of 14-d-old seedlings were treated 14 h with 150 μg mL−1 Vd-toxins and stained with Trypan Blue. H, Quantification of the cell death rates induced by Vd-toxins in G using ImageJ software. Error bars indicate sd; n = 9. Different letters represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. All experiments were repeated three times.
To further investigate whether PTP1 and MKP1 affect H2O2 production, we examined H2O2 production induced by Vd-toxins in the wild type, the ptp1 and mkp1 mutants, and the 35S:PTP1 and 35S:MKP1 lines. The H2O2 production levels were significantly increased by the Vd-toxins treatment in ptp1 and mkp1 mutants compared with the wild type, whereas H2O2 production levels were markedly decreased in 35S:PTP1 and 35S:MKP1 lines (Fig. 7, E and F). Moreover, we examined whether PTP1 and MKP1 affected the defense response to Vd-toxins. Trypan Blue staining indicated that the 35S:PTP1 and 35S:MKP1 lines reduced the extensive cell death (Fig. 7, G and H). Thus, PTP1 and MKP1 act as negative regulators of defense responses to Vd-toxins. These data suggest that PTP1 and MKP1 negatively regulated H2O2 production and inhibited the activity levels of MPK3 and MPK6 during defense responses to Vd-toxins.
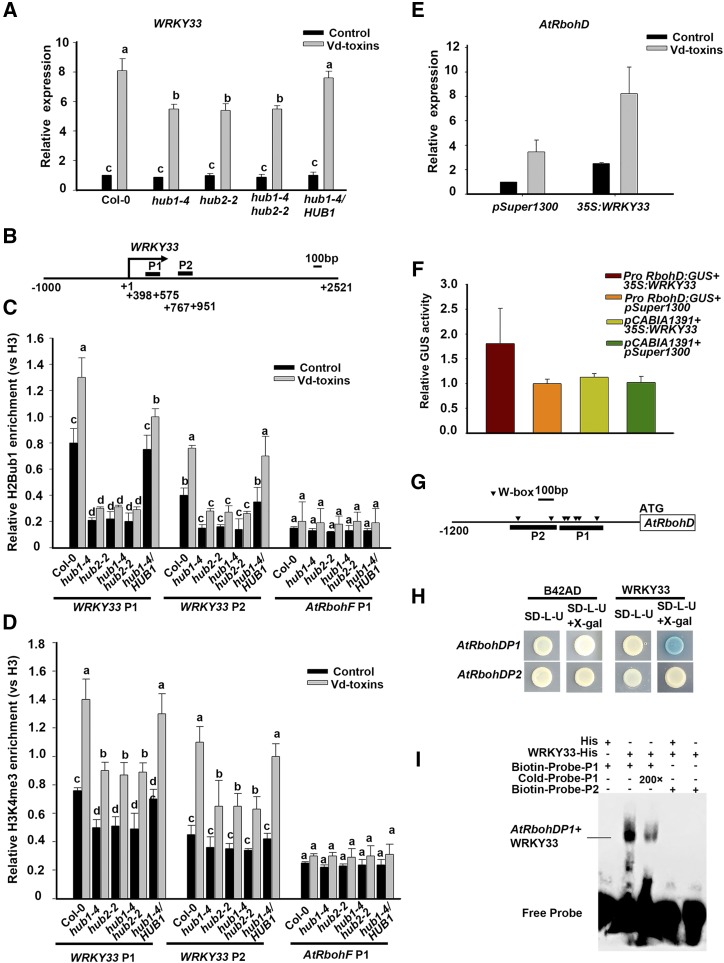
HUB1 and HUB2 Regulate WRKY33 Expression and WRKY33 Regulates AtRbohD Expression
WRKY33 is a pathogen-inducible transcription factor, and its expression is regulated by the MPK3/MPK6 cascade (Mao et al., 2011). Therefore, we hypothesized that HUB1 and HUB2 are involved in regulating the expression of WRKY33 in response to Vd-toxins. To determine whether HUB1 and HUB2 loss-of-function plants affected WRKY33 expression, the relative expression levels of WRKY33 were analyzed after exposure to Vd-toxins in the wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line using RT-qPCR. The expression of WRKY33 was strongly up-regulated in the wild type, while it was reduced in the mutants compared with the wild type. The expression level in the HUB1/hub1-4 complementation line was restored to that of the wild type (Fig. 8A). ChIP assays were performed to examine the levels of H2Bub1and H3K4me3 enrichment in the chromatin of WRKY33 in the wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line. Two fragments on the chromatin of WRKY33 were targeted for ChIP-qPCR (Fig. 8B). H2Bub1 enrichment was strongly detected in the chromatin of WRKY33 in the wild type, but only a weak enrichment was detected in the hub mutants. H2Bub1 was obviously increased in the chromatin of WRKY33 of the wild type after the Vd-toxins treatment, while it was only slightly increased in the hub mutants (Fig. 8C). Moreover, the level of H3K4me3 was greater in the chromatin of WRKY33 after Vd-toxins treatment in the wild type than in the hub mutant (Fig. 8D). These results suggested that HUB1 and HUB2 regulated the expression of WRKY33 by enhancing the enrichment of H3K4me3 after Vd-toxins treatment.
Figure 8.
HUB1 and HUB2 regulated the expression of WRKY33 in response to Vd-toxins, and WRKY33 regulated the expression of AtRbohD in Arabidopsis. A, The relative expression levels of WRKY33 induced by 150 μg mL−1 Vd-toxins in wild-type Columbia-0 (Col-0), the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line. Error bars indicate sd. B, Schematic diagram of the WRKY33 gene. P1 and P2 are gene body regions; the arrow indicates the transcription start site. C and D, Relative enrichments of H2Bub1 and H3K4me3 in the WRKY33 locus after treatment with 150 μg mL−1 Vd-toxins in the wild type, the hub1-4, hub2-2, and hub1-4 hub2-2 mutants, and the HUB1/hub1-4 complementation line. Chromatin was extracted 12 h after the Vd-toxins treatment, and immunoprecipitated DNA was analyzed by qPCR. Data were determined as the percentages of H2Bub1/H3 and H3K4me3/H3 for each individual gene position. Relative enrichments of H2Bub1 and H3K4me3 in the AtRbohF P1 locus were used as negative controls. Error bars indicate sd. Different letters in A, C, and D represent significant differences at P < 0.05 by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. E, WRKY33 regulates the expression of AtRbohD. The protoplasts of Arabidopsis harboring 35S:WRKY33 were used. The expression of AtRbohD was examined after the protoplasts were exposed to Vd-toxins for 4 h. pSuper1300 was used as the negative control. Error bars indicate sd. F, GUS activity measurement in N. benthamiana leaves after the transient expression of ProRbohD:GUS and 35S:WRKY33. pCAMBIA1391 and pSuper1300 were used as negative controls. LUC was used as an internal control. G, Schematic diagram showing the promoter structure of the AtRbohD gene. The upstream regions and part of the coding region are indicated by black wide lines and a white box, respectively. The solid arrowheads indicate the sites containing W-boxes in the AtRbohD promoter. The two fragments (P1 and P2) used for the yeast one-hybrid assay and EMSA are indicated. H, Yeast one-hybrid assay showing that WRKY33 binds to the AtRbohD promoter. I, EMSA showing WRKY33 binding to the AtRbohD promoter. Biotin-labeled fragments of the AtRbohD promoter that contained W-boxes were used as probes. Each experiment was repeated three times with similar results.
Next, we determined whether WRKY33 regulated the expression of the AtRbohD gene. We detected the gene expression of AtRbohD in a WRKY33 overexpression line. The protoplasts of Arabidopsis harboring 35S:WRKY33 were used. AtRbohD was up-regulated in the 35S:WRKY33 line compared with the control, which was protoplasts harboring the empty vector (Fig. 8E).
To further substantiate the role of WRKY33 in the regulation of AtRbohD expression, we performed transient transactivation assays in Nicotiana benthamiana leaves to determine whether WRKY33 regulates the expression of AtRbohD. The leaves were inoculated with Agrobacterium tumefaciens containing ProRbohD:GUS and 35S:WRKY33. pCAMBIA1391 and pSuper1300 were used as negative controls. Luciferase (LUC) was used as the loading control. The expression of AtRbohD was greater in leaves that were cotransformed with 35S:WRKY33 than in the control (Fig. 8F). Next, we performed yeast one-hybrid assays and electrophoresis mobility shift assays (EMSAs) to determine whether WRKY33 bound directly to the promoter of AtRbohD. We analyzed the sequences of AtRbohD promoter regions and found that the P1 and P2 fragments of the AtRbohD promoter contain W-box structures (Fig. 8G). The P1 and P2 fragments were independently inserted into the pLacZi2μ vector, and the coding sequence of WRKY33 was inserted into the pB42AD vector. Only the strain carrying both the AD-WRKY33 and AtRbohD P1:LacZ plasmids turned blue, indicating an interaction (Fig. 8H).
To perform the EMSA, the recombinant WRKY33 fused with a His tag (WRKY33-His) was purified from a prokaryotic system. The P1 and P2 fragments of the AtRbohD promoter were each labeled with biotin. WRKY33-His could bind to the AtRbohD P1, while no binding was observed between the P2 fragment and the WRKY33-His. The negative control and the addition of the competitive probe (Cold-Probe-P1) also resulted in no binding and weak binding, respectively (Fig. 8I).
Thus, HUB1 and HUB2 regulate the expression of WRKY33 by enhancing the enrichment of H3K4me3 in response to Vd-toxins. WRKY33 directly binds to AtRbohD promoter and functions as a transcription factor to regulate the expression of AtRbohD.
DISCUSSION
In this study, we demonstrated that HUB-mediated H2Bub1 plays an important role in regulating the H2O2 signaling pathway in the defense response to Vd-toxins. H2Bub1 regulated the expression of the NADPH oxidase AtRbohD and WRKY33 genes. H2Bub1 also affects posttranscriptional MAPK signaling. H2Bub1 is required for the activation of MPK3 and MPK6. Moreover, MPK3 and MPK6 associate with PTPs to affect H2O2 signal accumulation. The WRKY33 transcription factor is necessary for regulating AtRbohD’s expression. Thus, we have increased our knowledge of the molecular regulatory mechanisms of H2Bub1 that are involved in the defense response against Vd-toxins.
H2O2 signals induce large transcriptional changes and cellular reprogramming to control a large array of biological processes, ranging from the regulation of growth and development to responses to biotic and abiotic stimuli (Gadjev et al., 2006; Mittler et al., 2011). NADPH oxidase has a pivotal role in H2O2 production under biotic and abiotic stress conditions (Suzuki et al., 2011). In Arabidopsis, there are 10 different Rboh genes and their expression is mainly transcriptionally controlled in a tissue-specific manner. The RbohD and RbohF genes belong to the group expressed throughout the whole plant, and RbohD and RbohF are capable of generating H2O2 in response to pathogen attacks (Torres et al., 2002; Torres and Dangl, 2005). However, the expression levels of AtRbohD and AtRbohF are differentially modulated by pathogen-associated molecular patterns, and this could account for their action specificities (Morales et al., 2016). In this study, we found that AtRbohD plays a more important role than AtRbohF in the H2O2 accumulation induced by Vd-toxins. Moreover, HUB1 and HUB2 are involved in regulating the expression of AtRbohD.
H2Bub1 controls flowering time primarily through the transcriptional activation of FLC, and H2Bub1 is required for the enhancement of H3K4me3 in the chromatin of FLC and other FLC clade genes in Arabidopsis (Cao et al., 2008). H2Bub1 also regulates the expression levels of circadian clock genes (e.g. CCA1 and TOC1) by increasing the H3K4me3 deposition in their chromatin (Himanen et al., 2012; Malapeira et al., 2012). However, H2Bub1 and H3K4me3 deposition were only connected at some gene-specific chromatin regions. SET DOMAIN GROUP8 (SDG8; which encodes a histone methyl transferase) and SDG25 affect distinct H3K4 and H3K36 methylations and regulate the expression of plant immunity genes directly through histone Lys methylation or indirectly through H2Bub1 (Lee et al., 2016). In addition, SA2Lb (a euchromatic protein that functionally associates with an AtCOMPASS-like complex) mediates H3K4me3 deposition independent of H2Bub1 (Fiorucci et al., 2019). The interaction between H2Bub1 and H3K4me3 deposition in plant defense responses is poorly understood. Our results showed that HUB1 and HUB2 regulate the expression of AtRbohD by enhancing the level of H3K4me3 in the chromatin of AtRbohD. Therefore, we concluded that AtRbohD is responsible for H2O2 accumulation during responses to Vd-toxins in Arabidopsis and that H2Bub1 regulates the expression of AtRbohD, which is associated with H3K4me3.
MAPK activation may lead to ROS production, and MPK3/MPK6 may regulate the ROS bursts from NADPH oxidases (Zhang et al., 2007; Asai et al., 2008). Conversely, MAPKs may function downstream of ROS bursts when triggering plant immunity (Apel and Hirt, 2004; Pitzschke and Hirt, 2009). In this study, we demonstrated that MPK3 and MPK6 act as positive regulators of defense responses to Vd-toxins. Additionally, MPK3/MPK6 played important roles upstream of H2O2 accumulation, and they regulated the AtRbohD-dependent H2O2 production in the defense response to Vd-toxins. The results indicate that AtRbohD-dependent H2O2 production is required for the activation of MPK3 and MPK6 after exposure to Vd-toxins. Thus, MAPK activation occurs upstream of H2O2 production in this pathway. This is consistent with an earlier study (Zhang et al., 2007). Thus, the mechanisms of MAPK-controlled H2O2 production and H2O2-controlled MAPK activity should be discerned in the near future.
Protein Tyr phosphorylation plays an important role in the regulation of MAPK signaling (Tena et al., 2001; Zhang and Klessig 2001; Ghelis et al., 2008; Nemoto et al., 2015). The two main Tyr phosphatases that dephosphorylate activated MAPKs are MKP1 and PTP1 (Bartels et al., 2009). For example, PTP1 and MKP1 act as repressors of MPK3/MPK6-dependent stress signaling (Bartels et al., 2009), and MKP1 acts as a negative regulator of MPK6-mediated pathogen-associated molecular pattern responses and resistance against bacteria (Anderson et al., 2011). Therefore, it is likely that PTP1 and MKP1 have negative functions in defense signaling pathways. In this study, we demonstrated that PTP1 and MKP1 interact with MPK3 and MPK6 and negatively regulate H2O2 production. A previous study showed that H2Bub1 modulates the expression levels of AtPTP1 and AtMKP1 genes and affects the activation states of MPK3 and MPK6 in salt-stress responses (Zhou et al., 2017).
WRKY transcription factors are transcriptional regulators of signaling pathways involved in biotic and abiotic stress responses, and they act by specifically binding to W-boxes (TTGACC/T) in the promoter regions of target genes (Rushton et al., 1996; Yu et al., 2001; Eulgem and Somssich, 2007; Lippok et al., 2007). The WRKY family transcription factor member WRKY33, a pathogen-inducible transcription factor, is a direct phosphorylation target of MPK3 and MPK6 (Mao et al., 2011). In N. benthamiana, RbohA and RbohB, the putative orthologs of Arabidopsis RbohF and RbohD, are essential for H2O2 production in the response against the oomycete pathogen Phytophthora infestans (Yoshioka et al., 2003). The RbohB promoter contains a functional W-box, and W-boxes are well known cis-regulatory elements recognized by WRKY transcription factors (Eulgem et al., 2000; Adachi et al., 2015). The expression of WRKY33 was rapidly induced by Vd-toxins, and the transcription factor was directly involved in the activation of AtRbohD expression. In addition, it is required for H2Bub1 to regulate AtRbohD-dependent H2O2 signaling in the defense response to Vd-toxins. A positive regulatory loop, consisting of MPK3/6-WRKY33-ALD1-pipecolic acid, is a critical regulatory mechanism of systemic acquired resistance induction in Arabidopsis (Wang et al., 2018).
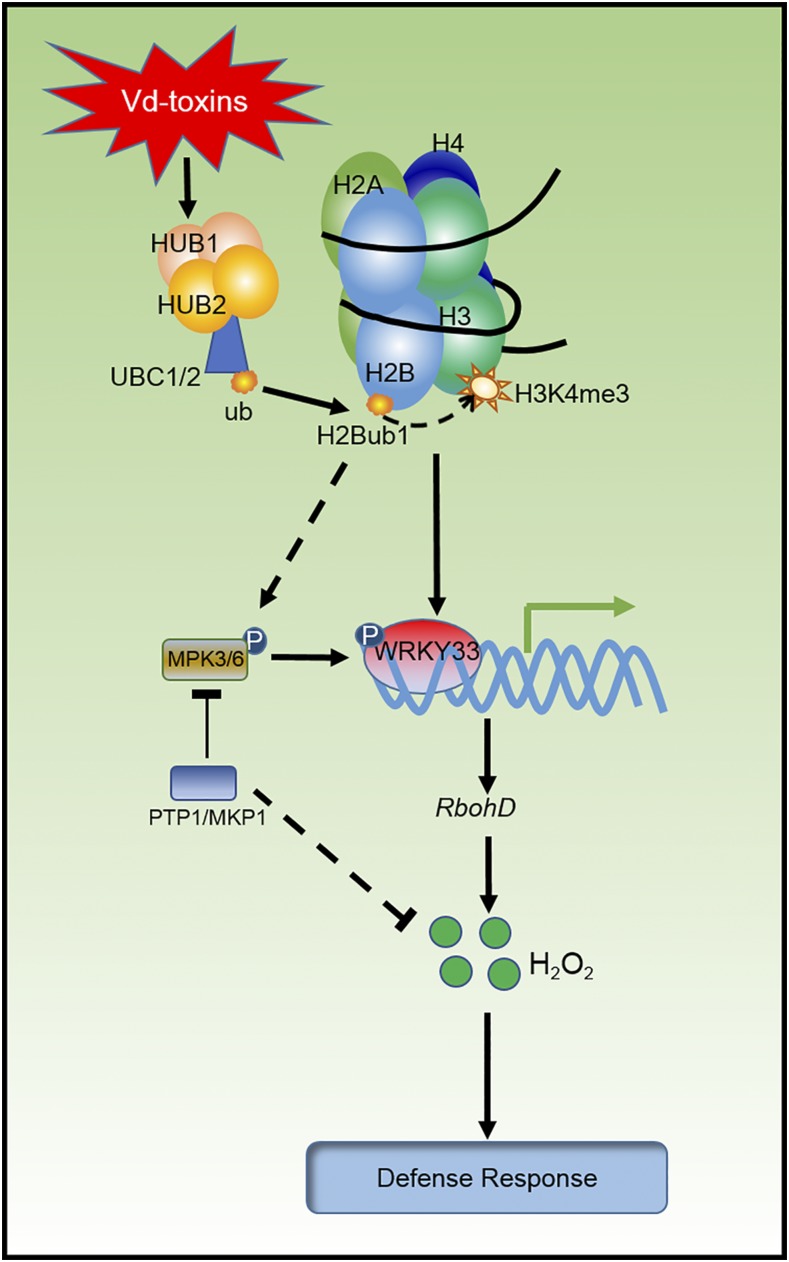
In conclusion, our results demonstrate that HUB-mediated H2Bub1 is involved in the regulation of defense responses against Vd-toxins through the transcriptional activation of the NADPH oxidase AtRbohD and WRKY33. H2Bub1 is associated with H3K4me3 deposition during the transcription induction of AtRbohD and WRKY33. The active transcription of WRKY33 regulates AtRbohD expression, and AtRbohD-dependent H2O2 signaling is a critical modulator in the defense response against Vd-toxins. In addition, H2Bub1 also affects posttranscriptional MAPK signaling. H2Bub1 is required for the activation of MPK3 and MPK6. Moreover, MPK3 and MPK6 associate with PTPs, which negatively regulated H2O2 production. The PTP-MPK3/6-WRKY pathway regulates H2O2 signaling in the response against Vd-toxins in Arabidopsis (Fig. 9).
Figure 9.
Model of the H2Bub1 mechanism used to regulate defense responses to Vd-toxins in Arabidopsis. In this model, HUB-mediated H2Bub1 regulates the expression of the NADPH oxidase RbohD and WRKY33 by enhancing the enrichment of H3K4me3 in response to Vd-toxins. H2Bub1 also affects posttranscriptional MAPK signaling. H2Bub1 is required for the activation of MPK3 and MPK6. Phosphorylation of WRKY33 by MPK3 and MPK6 regulates the expression of RbohD. Moreover, MPK3 and MPK6 associate with PTPs, which negatively regulate H2O2 production. The PTP-MPK3/6-WRKY pathway regulates H2O2 signals in the responses against Vd-toxins in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 was used as the wild-type. Arabidopsis mutants and transgenic lines, hub1-4, hub2-2, hub1-4 hub2-2, HUB1/hub1-4 (Cao et al., 2008), rbohd, rbohf, rbohd/f (Torres et al., 2002), mpk3, mpk6, 35S:MPK3, 35S:MPK6, MKK5DD (Liu et al., 2008), ptp1, mkp1, 35S:PTP1, and 35S:MKP1 (Zhou et al., 2017), were previously described.
The seeds were sterilized with 0.2% (v/v) sodium hypochlorite for 20 min, rinsed with water, and grown on one-half-strength MS medium supplemented with 1% (w/v) Suc and 0.8% (w/v) agar at 16°C or 22°C with a 16-h-light/8-h-dark cycle and 70% relative humidity in a growth cabinet.
Preparation of Crude Vd-Toxins
A highly infectious and defoliating strain of Verticillium dahliae (Vd991) was used to extract Vd-toxins. The Verticillium spp. culture filtrate was purified as described previously (Jia et al., 2007; Shi and Li, 2008). The fungal culture was filtered through filter paper, and the filtrate was centrifuged at 10,000g for 30 min to remove the spores. The supernatant was frozen (−20°C) for 24 h, lyophilized for 36 h, and dissolved in distilled water to make a 0.5 mg mL−1 solution. The solution was dialyzed using 1-kD dialysis membranes (MWCO) in 4°C for 24 h. The concentrated solution was frozen and lyophilized again, and the powder was dissolved in distilled water. The solution was then refiltered through a 0.45-μm Millipore filter. The resulting filtrate was used as crude Vd-toxins extract for further experiments.
Vd-Toxin Treatment Assays
For the phenotypic analysis, seeds of the wild type and the hub1-4, hub2-2, hub1-4 hub2-2, and HUB1/hub1-4 mutants were germinated on one-half-strength MS medium for 4 d and then transplanted onto plates supplemented with Vd-toxins (100–300 μg mL−1) and grown for another 4 d. For root length determinations after exposure to Vd-toxins, the root length of untreated seedlings of each genotype was set at 100%, and the mean values ± sd were shown as percentages of root length of the respective controls. Three independent experiments were performed, and at least 30 seedlings of each genotype were measured in each experiment.
H2O2 Assays
For the H2DCF-DA staining assay, 7-d-old seedlings were incubated in 10 mm MES-KCl buffer (pH 6) supplemented with 200 μg mL−1 Vd-toxins and 20 μm H2DCF-DA for 20 min at room temperature. Then they were washed three times with 10 mm MES-KCl buffer (pH 6) to remove the excess H2DCF-DA. Seedlings incubated in double distilled water were used as controls. Fluorescence was detected with a confocal laser scanning microscope (Zeiss LSM 710) using the following conditions: power 70%, excitation at 488 nm, and emission at 505 to 530 nm. This experiment was independently repeated at least three times.
H2O2 content was measured using the chromogenic peroxidase-coupled assay (Veljovic-Jovanovic et al., 2002). The reaction mixture consisted of 0.1 m phosphate buffer (pH 6.5), 3.3 mm 3-dimethylaminobenzoic acid, 0.07 mm 3-methyl-2-benzothiazolinonehydrazone, and 0.1 unit mL−1 peroxidase (Sigma). The 7-d-old seedlings were treated with 200 μg mL−1 Vd-toxins, then 0.1 g of the leaves was ground to a fine powder in liquid nitrogen, and the powder was extracted in 2 mL of 1 m HClO4. When required, the extractions were performed in the presence of insoluble 5% (w/v) polyvinylpyrrolidone. Homogenates were centrifuged at 12,000g for 10 min, and the supernatants were neutralized with 5 m K2CO3 to pH 5.6 in the presence of 100 μL of 0.3 m phosphate buffer (pH 5.6). The homogenates were then centrifuged at 12,000g for 1 min to remove KClO4. When required, the sample was incubated prior to the assay for 10 min with 1 unit of ascorbate oxidase (Sigma) to oxidize ascorbate. The reaction was initiated by the addition of 200 μL of the sample and 2,700 μL of the reaction mixture. The absorbance change at 590 nm was monitored at 25°C. Three independent experiments were performed, each with three replicates, and similar results were obtained.
Detection of Cell Death in Leaves
Cell death in leaves was determined by staining them with Trypan Blue according to the method of Bowling et al. (1997). The experiments were repeated at least three times.
RT-qPCR
Arabidopsis total RNA was isolated using TRIzol reagent (Invitrogen) and treated with DNaseI according to the manufacturer’s specifications (Invitrogen). For the RT-qPCR analysis, first-strand cDNA was synthesized from 1 μg of total RNA using PrimeScript reverse transcription reagents kit (TaKaRa). RT-qPCR was performed on an ABI 7500 Sequence Detector (Applied Biosystems) using the SYBR Premix Ex Taq Kit (TaKaRa) with gene-specific primers and the internal control (ACTIN2). The gene-specific primer pairs are listed in Supplemental Table S1. cDNA aliquots of 2 μL were amplified in a 25-μL reaction volume containing SYBR Premix Ex Taq, PCR forward primer, PCR reverse primer, and the ROX reference dye in accordance with the manufacturer’s instructions (TaKaRa). PCR was conducted at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s (denaturation) and annealing at 60°C for 34 s. Data analysis, including the determination of the threshold cycle that represents the starting point of the exponential phase of PCR, and graphic presentation were conducted using Sequence Detection Software version 1.07 (Applied Biosystems). The quantification of the transcript levels of the cDNA fragments was normalized to the expression of the ACTIN2 gene in Arabidopsis at several points during the Vd-toxins treatment. Three independent experiments were performed, each with three replicates.
Yeast Two-Hybrid Assays
To create bait and prey plasmids, fragments were amplified by different primers and cloned into pGBKT7 and pGADT7, respectively. All the primer sequences are listed in Supplemental Table S1. MPK2-, MPK3-, and MPK6-coding sequences were independently cloned into the pGBKT7 vector (Clontech) as the bait constructs, while PTP1 and MKP1 were independently cloned into the pGADT7 vector (Clontech) as the prey constructs. Empty pGBKT7 and pGADT7 vectors were used as controls. Bait and prey constructs were cotransformed into Saccharomyces cerevisiae strain AH109. The transformants were tested on synthetic defined (SD) screening medium. Positive clones were identified by the ability to grow on the SD medium minus Leu/Trp/His/adenine and containing 6 mm 3-aminotriazole and 20 μg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galacto-pyranoside.
Yeast One-Hybrid Assays
The yeast one-hybrid assay was performed as described by Miao et al. (2004) and Li et al. (2010). The full-length cDNA of WRKY33 and DNA fragments of the AtRbohD promoter were amplified and independently cloned into pB42AD and pLacZi, respectively. All the primer sequences are listed in Supplemental Table S1. Empty pB42AD and pLacZi vectors were used as controls. The fusion constructs were cotransformed into S. cerevisiae strain EGY48. The transformants were tested on SD medium minus uracil and Leu and containing 20 μg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galacto-pyranoside.
GUS/LUC Assays
The transient GUS/LUC assay was performed as described by Zhao et al. (2016). The pCAMBIA1391 vector containing the Pro-35S:WRKY33 or ProRbohD:GUS construct was transfected into Agrobacterium tumefaciens strain GV3101. All the primer sequences are listed in Supplemental Table S1. Then, disposable sterilized syringes were used to inject these two bacterial solutions into different positions on the same Nicotiana benthamiana leaf. Empty vectors were used as controls. Three days after injection, the treated leaves were collected to measure the GUS and LUC activity levels. Methyl umbelliferyl glucuronide (Sigma) was used in the test of the GUS activity. The binding activity of WRKY33 to the AtRbohD promoter was determined by the GUS-to-LUC ratio.
EMSA
EMSA was performed as described in the protocol of the LightShift EMSA Optimization Control Kit (Thermo). The EMSA protocol of Zhao et al. (2016) was also referenced. All the primer sequences are listed in Supplemental Table S1. The recombinant proteins WRKY33-His and His were expressed and purified from Escherichia coli (BL21). Biotin-labeled or unlabeled primers were used to obtain two fragments (P1 and P2) of the RbohD promoter by PCR amplification. The unlabeled fragments and His protein were used as competitors and negative control, respectively.
ChIP Assays
ChIP was performed as described by Zhao et al. (2019) using 3-week-old seedlings. The anti-H2Bub1, anti-H3K4me3, and anti-H3 antibodies were purchased from Cell Signaling Technology. ChIP-qPCR was performed to analyze the enrichment of H2Bub1 and H3K4me3 in DNA fragments with anti-H2Bub1 anti-H3K4me3 antibodies, and anti-H3 antibodies were used to normalize H2Bub1 and H3K4me3 levels to nucleosome occupancy. As the H2Bub1 and H3K4me3 levels in ACTIN2 chromatin were similar among hub1-4, hub2-2, and the wild type, ACTIN2 was used as an internal control. The primers are provided in Supplemental Table S1.
Immunoblotting and MAPK Analyses
The 7-d-old Arabidopsis seedlings were treated with 200 μg mL−1 Vd-toxins at several time points and carefully placed into centrifuge tubes. Protein extraction buffer (100 mm HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 2 mm DTT, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerolphosphate, 1 mm phenylmethylsulfonyl fluoride, 1 mm proteinase inhibitor cocktail and phosphatase inhibitor cocktail, 10% [w/v] glycerol, and 1% [w/v] polyvinylpolypyrrolidone) was added to resuspend each sample. After centrifugation at 10,000g for 30 min at 4°C, supernatants were transferred into clean tubes. The protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad) with BSA as the standard.
For western-blot analysis, 30 µg of total protein per lane was separated on an 8% SDS-PAGE gel. After electrophoresis, the proteins were transferred onto polyvinylidene difluoride membranes. After blocking for 2 h in Tris-buffered saline plus Tween 20 buffer with 5% (w/v) BSA at room temperature, the membranes were incubated with anti-phospho-p44/42 MAPK (1:1,000; Cell Signaling Technology), anti-AtMPK3 (1:2,000; Sigma), and anti-AtMPK6 (1:2,000; Sigma) as primary antibodies and peroxidase-conjugated goat anti-rabbit IgG (1:2,000; Cell Signaling Technology) as the secondary antibody. Then the membranes were visualized using enhanced chemiluminescence substrate (ThermoFisher).
Co-IP Assays
Co-IP assays were performed as described previously (Ding et al., 2015). Transgenic plants independently harboring 35S:PTP1 and 35S:MKP1 and expressing PTP1-MYC and MKP1-MYC, respectively, were used to detect the interactions of MPK3/6 and PTP1/MKP1. Wild-type plants were used as the negative control. The total proteins extracted from wild-type and stable transgenic plants were independently immunoprecipitated with anti-Myc agarose (Sigma), anti-AtMPK3 (Sigma), and anti-AtMPK6 (Sigma). The immunoprecipitates were separated on a 10% SDS-PAGE gel and detected with corresponding antibodies. Then, the membranes were visualized using an enhanced chemiluminescence substrate (ThermoFisher).
Statistical Analyses
Statistical data were analyzed by SPSS statistics software. Data were analyzed by one-way ANOVA with Tukey’s honestly significant difference posthoc tests. Values of P < 0.05 are noted in the figure legends, and significant differences are indicated by different letters.
Accession Numbers
Sequence data for the genes described in this article can be found in TAIR (https://www.arabidopsis.org/) with the following accession numbers: AT2G44950 for HUB1, AT1G55250 for HUB2, AT5G47910 for AtRbohD, AT1G64060 for AtRbohF, AT2G14610 for PR1, AT2G38470 for WRKY33, AT1G02930 for GST1, AT3G45640 for MPK3, AT2G43790 for MPK6, AT1G71860 for PTP1, and AT3G55270 for MKP1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Global change in H2Bub1 induced by the Vd-toxins treatment in wild-type Arabidopsis.
Supplemental Figure S2. Kinase activity levels of MPK3 and MPK6 detected in wild-type, mpk3, and mpk6 Arabidopsis plants.
Supplemental Table S1. Primers used in this work.
Acknowledgments
We thank Dr. Ligeng Ma (Capital Normal University, Beijing) for the hub1-4, hub2-2, and hub1-4 hub2-2 mutants and the HUB1/hub1-4 complementation line, Dr. Miguel A. Torres (University of North Carolina, Chapel Hill) for the rbohD, rbohF, and rbohD/F mutants, and Dr. Dongtao Ren (China Agricultural University, Beijing) for the mpk3, mpk6, 35S:MPK3, 35S:MPK6, and MKK5DD mutants.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31370292 and 31670252) and the National Transgenic Research Project (grant no. 2015ZX08005-002) to Y.L.
References
- Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H (2015) WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27: 2645–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Bartels S, González Besteiro MA, Shahollari B, Ulm R, Peck SC (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6-mediated PAMP responses and resistance against bacteria. Plant J 67: 258–268 [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, González Besteiro MA, Carreri A, Hirt H, Buchala A, Métraux JP, Peck SC, Ulm R (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21: 2884–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, Conrath U (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RG, Subbarao KV (1999) Host range specificity in Verticillium dahliae. Phytopathology 89: 1218–1225 [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159: 266–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F (2012) Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet 8: e1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Li X, Wang Z, Ding M, Sun Y, Dong F, Chen F, Liu L, Doughty J, Li Y, et al. (2015) Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol 168: 1389–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G (2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Chen P, Hutter D, Yang X, Gorospe M, Davis RJ, Liu Y (2001) Discordance between the binding affinity of mitogen-activated protein kinase subfamily members for MAP kinase phosphatase-2 and their ability to activate the phosphatase catalytically. J Biol Chem 276: 29440–29449 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Mittelsten Scheid O, Mengiste T (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21: 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Feng J, Shen WH (2014) Dynamic regulation and function of histone monoubiquitination in plants. Front Plant Sci 5: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci AS, Bourbousse C, Concia L, Rougée M, Deton-Cabanillas AF, Zabulon G, Layat E, Latrasse D, Kim SK, Chaumont N, et al. (2019) Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination. Genome Biol 20: 100–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Himanen K, Cnops G, Nelissen H, Boccardi TM, Maere S, Beemster GT, Neyt P, Anami S, Robles P, et al. (2007) The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BP (2006) Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol Plant Pathol 7: 71–86 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelis T, Bolbach G, Clodic G, Habricot Y, Miginiac E, Sotta B, Jeannette E (2008) Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol 148: 1668–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Yao N (2004) The role and regulation of programmed cell death in plant-pathogen interactions. Cell Microbiol 6: 201–211 [DOI] [PubMed] [Google Scholar]
- Gu X, Jiang D, Wang Y, Bachmair A, He Y (2009) Repression of the floral transition via histone H2B monoubiquitination. Plant J 57: 522–533 [DOI] [PubMed] [Google Scholar]
- Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, Vuylsteke M, Van Lijsebettens M (2012) Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant J 72: 249–260 [DOI] [PubMed] [Google Scholar]
- Hu M, Pei BL, Zhang LF, Li YZ (2014) Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in Arabidopsis. Plant Physiol 164: 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, et al. (2002) Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H (2012) Post-translational regulation of WRKY transcription factors in plant immunity. Curr Opin Plant Biol 15: 431–437 [DOI] [PubMed] [Google Scholar]
- Jia ZQ, Yuan HY, Li YZ (2007) NO and H2O2 induced by Verticillium dahliae toxins and its influence on expression of GST gene in cotton suspension cells. Chin Sci Bull 52: 1347–1354 [Google Scholar]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A (2007) Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131: 1084–1096 [DOI] [PubMed] [Google Scholar]
- Lee S, Fu F, Xu S, Lee SY, Yun DJ, Mengiste T (2016) Global regulation of plant immunity by histone lysine methyl transferases. Plant Cell 28: 1640–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci USA 111: 3632–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok B, Birkenbihl RP, Rivory G, Brümmer J, Schmelzer E, Logemann E, Somssich IE (2007) Expression of AtWRKY33 encoding a pathogen- or PAMP-responsive WRKY transcription factor is regulated by a composite DNA motif containing W box elements. Mol Plant Microbe Interact 20: 420–429 [DOI] [PubMed] [Google Scholar]
- Liu H, Wang Y, Xu J, Su T, Liu G, Ren D (2008) Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res 18: 422–432 [DOI] [PubMed] [Google Scholar]
- Liu S, Ziegler J, Zeier J, Birkenbihl RP, Somssich IE (2017) Botrytis cinerea B05.10 promotes disease development in Arabidopsis by suppressing WRKY33-mediated host immunity. Plant Cell Environ 40: 2189–2206 [DOI] [PubMed] [Google Scholar]
- Liu Y, Koornneef M, Soppe WJ (2007) The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love AJ, Yun BW, Laval V, Loake GJ, Milner JJ (2005) Cauliflower mosaic virus, a compatible pathogen of Arabidopsis, engages three distinct defense-signaling pathways and activates rapid systemic generation of reactive oxygen species. Plant Physiol 139: 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Schwessinger B, Ntoukakis V, Brutus A, Segonzac C, Roy S, Kadota Y, Oh MH, Sklenar J, Derbyshire P, et al. (2014) A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 343: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Malapeira J, Khaitova LC, Mas P (2012) Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci USA 109: 21540–21545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23: 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Dunand C, Puppo A, Pauly N (2012) A burst of plant NADPH oxidases. Trends Plant Sci 17: 9–15 [DOI] [PubMed] [Google Scholar]
- Ménard R, Verdier G, Ors M, Erhardt M, Beisson F, Shen WH (2014) Histone H2B monoubiquitination is involved in the regulation of cutin and wax composition in Arabidopsis thaliana. Plant Cell Physiol 55: 455–466 [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal 2: ra45. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F (2011) ROS signaling: The new wave? Trends Plant Sci 16: 300–309 [DOI] [PubMed] [Google Scholar]
- Morales J, Kadota Y, Zipfel C, Molina A, Torres MA (2016) The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J Exp Bot 67: 1663–1676 [DOI] [PubMed] [Google Scholar]
- Nemoto K, Takemori N, Seki M, Shinozaki K, Sawasaki T (2015) Members of the plant CRK superfamily are capable of trans- and autophosphorylation of tyrosine residues. J Biol Chem 290: 16665–16677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D (2006) Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 125: 703–717 [DOI] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2005) Role of mitogen-activated protein kinases in plant immunity. Curr Opin Plant Biol 8: 541–547 [DOI] [PubMed] [Google Scholar]
- Pitzschke A. (2015) Modes of MAPK substrate recognition and control. Trends Plant Sci 20: 49–55 [DOI] [PubMed] [Google Scholar]
- Pitzschke A, Hirt H (2009) Disentangling the complexity of mitogen-activated protein kinases and reactive oxygen species signaling. Plant Physiol 149: 606–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Macho AP (2017) Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol Biol 1578: 143–153 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C, Feike D, Gimenez-Ibanez S, Hann DR, Zipfel C, Rathjen JP (2011) Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol 156: 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban M, Miao Y, Ullah A, Khan AQ, Menghwar H, Khan AH, Ahmed MM, Tabassum MA, Zhu L (2018) Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol Biochem 125: 193–204 [DOI] [PubMed] [Google Scholar]
- Shi FM, Li YZ (2008) Verticillium dahliae toxins-induced nitric oxide production in Arabidopsis is major dependent on nitrate reductase. BMB Rep 41: 79–85 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Morales J, Shulaev V, Torres MA, Mittler R (2011) Respiratory burst oxidases: The engines of ROS signaling. Curr Opin Plant Biol 14: 691–699 [DOI] [PubMed] [Google Scholar]
- Tamnanloo F, Damen H, Jangra R, Lee JS (2018) MAP KINASE PHOSPHATASE1 controls cell fate transition during stomatal development. Plant Physiol 178: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena G, Asai T, Chiu WL, Sheen J (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4: 392–400 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2005) Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet 37: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljovic-Jovanovic S, Novtor G, Foyer CH (2002) Are leaf hydrogen peroxide concentrations commonly overestimated? The potential influence of artefactual interference by tissue phenolics and ascorbate. Plant Physiol Biochem 40: 501–507 [Google Scholar]
- Wang Y, Schuck S, Wu J, Yang P, Döring AC, Zeier J, Tsuda K (2018) A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell 30: 2480–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: Triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH (2009) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J 57: 279–288 [DOI] [PubMed] [Google Scholar]
- Yao LL, Zhou Q, Pei BL, Li YZ (2011) Hydrogen peroxide modulates the dynamic microtubule cytoskeleton during the defence responses to Verticillium dahliae toxins in Arabidopsis. Plant Cell Environ 34: 1586–1598 [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Numata N, Nakajima K, Katou S, Kawakita K, Rowland O, Jones JD, Doke N (2003) Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15: 706–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6: 520–527 [DOI] [PubMed] [Google Scholar]
- Zhao S, Zhang ML, Ma TL, Wang Y (2016) Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28: 3005–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Neyt P, Van Lijsebettens M, Shen WH, Berr A (2019) Interactive and noninteractive roles of histone H2B monoubiquitination and H3K36 methylation in the regulation of active gene transcription and control of plant growth and development. New Phytol 221: 1101–1116 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 48: 592–605 [DOI] [PubMed] [Google Scholar]
- Zhou S, Chen Q, Sun Y, Li Y (2017) Histone H2B monoubiquitination regulates salt stress-induced microtubule depolymerization in Arabidopsis. Plant Cell Environ 40: 1512–1530 [DOI] [PubMed] [Google Scholar]
- Zou B, Yang DL, Shi Z, Dong H, Hua J (2014) Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol 165: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]