Disease resistance to the pathogen Pseudomonas syringae in Arabidopsis is enhanced by a lower temperature via up-regulating multiple genes in the salicylic acid pathway that are repressed by ethylene.
Abstract
Temperature has a large impact on plant immune responses. Earlier studies identified intracellular immune receptor nucleotide-binding leucine-rich repeat (NLR) genes and salicylic acid (SA) as targets of high-temperature inhibition of plant immunity. Here, we report that moderately low temperature enhances immunity to the bacterial pathogen Pseudomonas syringae in Arabidopsis (Arabidopsis thaliana). This enhancement is dependent on SA signaling and is accompanied by up-regulation of multiple SA biosynthesis and signaling genes at lower temperature. SA signaling is repressed by jasmonic acid and ethylene at both normal and low temperatures. The inhibition of SA biosynthesis by ethylene, while mainly through ISOCHORISMATE SYNTHASE1/SALICYLIC ACID-INDUCTION DEFICIENT2 (ICS1/SID2) at normal temperature, is through ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5)/SID1, ICS2, and ICS1/SID2 at lower temperature. The repression by ethylene is mediated by a direct regulation of the ethylene response transcription factor ETHYLENE INSENSITIVE3 (EIN3) on multiple SA biosynthesis and signaling genes. Thus, low temperature enhances the SA pathway to promote immunity and at the same time uses ethylene to repress multiple SA regulators to achieve fine-tuned immune responses.
Temperature has a large impact on plant growth, development, and defense responses. Temperature sensitivity was reported for plant disease resistance as early as 1969 (Dropkin, 1969) and has been observed in various plant-pathogen interactions (Wang et al., 2009). Ambient temperature within the normal range of plant growth influences both pathogen virulence and host immune response (Colhoun, 1973; Browder, 1985; Huot et al., 2017).
Plants rely on multilayered and multibranched immune systems to fight off pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). The first branch, named pathogen-triggered immunity (PTI), uses pattern recognition receptors to detect microbe- or pathogen-associated molecular patterns to trigger basal resistance (Boller and Felix, 2009). The second immunity branch, named effector-triggered immunity (ETI), recognizes effectors by RESISTANCE (R) proteins, mostly nod-like receptor, or nucleotide-binding leucine-rich repeat (NLR) proteins and mount a more robust response (Cui et al., 2015).
Plant immune responses are regulated by multiple plant hormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). SA is a major hormone for both ETI and PTI (Vlot et al., 2009). It is synthesized mainly through the isochorismate pathway and partially through the Phe pathway. ISOCHORISMATE SYNTHASE1 (ICS1), also known as SALICYLIC ACID-INDUCTION DEFICIENT2 (SID2), is the key enzyme in the isochorismate pathway and is thought to play a major role for SA production induced by pathogen infection (Wildermuth et al., 2001). ICS2, with the same biochemical function as ICS1, has a minimal role in SA accumulation (Garcion et al., 2008). The Phe pathway of SA biosynthesis is mediated by PHENYLALANINE AMMONIA LYASE (PAL) that catalyzes Phe to trans-cinnamic acid, a precursor of diverse phenolic compounds (Dempsey et al., 2011). In Arabidopsis (Arabidopsis thaliana), pathogen induction of SA biosynthesis occurs predominantly through the isochorismate pathway, which contributes to 90% of the SA content (Rekhter et al., 2019). SA accumulation upon pathogen infection is shown to be dependent on SID1/ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5; Nawrath et al., 2002). SID1/EDS5 encodes a multidrug and toxin extrusion-like transporter and functions in the export of isochorismate, the precursor of SA, from the chloroplast to the cytoplasm (Serrano et al., 2013; Rekhter et al., 2019). The blocking of SA export in the sid1 mutant results in low SA, probably through an inhibition of SA biosynthesis from an overaccumulation of SA in the chloroplast. The SA defense signal is potentiated by positive feedback loops involving signaling components of PTI and ETI. In particular, EDS1 and PHYTOALEXIN DEFICIENT4 (PDA4), as essential components for NLR-mediated ETI, are important for pathogen-induced SA accumulation (Jirage et al., 1999; Feys et al., 2001). Besides SA, plant hormones JA and ET also play critical roles in plant immunity (Shigenaga and Argueso, 2016). These hormones have antagonistic or synergistic interactions among themselves, thus generating complex signaling networks (Tsuda and Katagiri, 2010). This is thought to confer tunable immune regulation to respond to developmental and environmental variations. SA has negative or positive interactions with JA and ET in regulating plant defenses against microbes (Robert-Seilaniantz et al., 2011). JA induces the expression of SID1/EDS5 but inhibits the expression of PAD4, which is likely responsible for the mitigation of SA accumulation in plant immunity by JA (Mine et al., 2017). ET in general acts antagonistically to SA signaling. The ET signaling transcription factors EIN3 and EIN3-LIKE1 (EIL1) bind directly to the promoter of SID2 and repress its expression (Chen et al., 2009). The interaction of plant hormones in plant immunity was extensively investigated using combinations of mutants defective in DELAYED-DEHISCENCE2 (DDE2), EIN2, PAD4, and SID2. PTI induced by a peptide from flagellin (flg22) and ETI induced by the pathogen effector AvrRpt2 were mostly abolished in the dde2 ein2 pad4 sid2 quadruple mutant but were largely intact in plants with mutations in any one of these genes. This suggests that SA, PAD4, JA, and ET signaling sectors together contribute positively to PTI and ETI (Tsuda et al., 2009).
Variations in ambient temperature have a pronounced effect on PTI and ETI in plant immunity. Temperature sensitivity of disease resistance has been found in multiple host-pathogen interactions (Hua, 2014). Resistance of Arabidopsis plants to virulent and avirulent Pseudomonas syringae pv tomato (Pst) strain DC3000 is reduced at a moderately elevated temperature of 28°C, which is accompanied by a reduced expression of the SA signaling molecules PAD4 and EDS1 compared with the normal growth temperature of 22°C (Wang et al., 2009). Temperature modulation of NLR gene activity, exemplified by NLR protein subcellular localization, is one of the key causes for high-temperature inhibition of disease resistance (Zhu et al., 2010). Different variants of the NLR gene Suppressor of npr1, constitutive1 (SNC1) confer immune responses with differential temperature sensitivity, indicating that SNC1 itself is a temperature-sensitive component of plant immune responses (Zhu et al., 2010). In addition, SA biosynthesis is inhibited at 30°C compared with 22°C and application of the SA analog benzothiadiazole potentiates disease resistance to virulent pathogens at 30°C, indicating that inhibition of SA is critical for high-temperature inhibition of basal resistance (Huot et al., 2017). Studies also suggest that high temperature shifts resistance from ETI to PTI, as expression of PTI-induced genes is elevated at moderately high temperatures (Cheng et al., 2013).
Here, we examined the propagation of the bacterial pathogen Pst DC3000 in Arabidopsis at different ambient temperatures: moderately low (16°C; referred to as low), normal (22°C), and moderately high (28°C; referred to as high). We found that low temperature enhances plant immunity and does not compromise the virulence of the pathogen. Furthermore, we utilized mutants deficient in single and multiple signaling sectors of SA, JA, ET, and PAD4 to reveal a potential different genetic requirement for resistance at different temperatures. We found that SA signaling is a major sector mediating the enhancement of resistance at low temperature. In addition, ET and SA have different genetic interactions at low and normal temperatures. Further transcriptome analysis revealed that ET antagonizes the SA sector through the SA biosynthesis gene ICS1/SID2 at normal temperature but through multiple SA biosynthesis regulators, including ICS1/SID2, EDS5/SID1, and ICS2, at low temperature. Therefore, low temperature potentiates a higher number of SA biosynthesis genes for higher disease resistance, which is balanced with direct repression by the EIN3 transcription factor.
RESULTS
Lower Temperature Reduces Pathogen Propagation in Arabidopsis
Prior studies showed that basal resistance to the virulent pathogen Pst DC3000 is inhibited at a moderately high temperature of 28°C compared with the normal growth temperature of 22°C in Arabidopsis. Here, we examined resistance to this bacterial pathogen at a moderately low temperature of 16°C in comparison with 22°C and 28°C in the reference accession Columbia-0 (Col-0) of Arabidopsis.
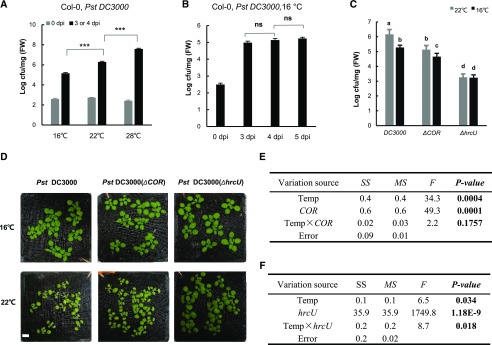
Consistent with previous findings, propagation of Pst DC3000 was higher by more than 10-fold at 28°C compared with 22°C at 3 d post inoculation (dpi; Fig. 1A). In contrast, propagation was lowest at 16°C, with more than 10-fold reduction compared with 22°C at 3 dpi (Fig. 1A). To assess the effect of a lower temperature on bacterial growth in plants, we monitored pathogen propagation at 3, 4, and 5 dpi. No significant difference was found for pathogen growth at 4 dpi compared with 5 or 3 dpi, although there was a slight increase from 3 to 4 dpi at 16°C (Fig. 1B). To minimize the potential effects of lower temperature on pathogen growth and plant growth, we used 4 dpi at 16°C and 3 dpi at 22°C for further analysis.
Figure 1.
Lower temperature reduces pathogen propagation in Arabidopsis. A, Growth of the virulent pathogen Pst DC3000 in wild-type Col-0 plants at 0 and 3 dpi for 22°C and 28°C or at 4 dpi for 16°C. B, Growth of Pst DC3000 in the wild-type Col-0 at 16°C at 3, 4, and 5 dpi. C, Growth of Pst DC3000, Pst DC3000 ΔCOR (coronatine deficient), and Pst DC3000 ΔhrcU (T3SS deficient) in the wild-type Col-0 at 4 dpi at 16°C and 3 dpi at 22°C. For A to C, values represent means ± sd for three biological repeats (n = 3). Asterisks indicate statistically significant differences between samples determined by Student’s t test (***, P < 0.001; ns, not significant). Letters indicate statistically significant differences by ANOVA (P < 0.05). Similar results were obtained from three independent experiments (each with three biological repeats). cfu, colony-forming units; FW, fresh weight. D, disease symptoms after infection with Pst DC3000, Pst DC3000 ΔCOR, or Pst DC3000 ΔhrcU. E and F, MANOVA for growth of Pst DC3000 and Pst DC3000 ΔCOR (E) and Pst DC3000 and Pst DC3000 ΔhrcU (F) at different temperatures. F, F test; MS, mean square; SS, square sum; Temp, temperature.
We next examined the requirement of pathogen virulence factors for the reduced pathogen growth at 16°C compared with 22°C using the Pst DC3000 ΔhrcU strain, which is defective in the type III secretion system (T3SS; Roine et al., 1997) and the Pst DC3000 ΔCOR strain, which is defective in COR production (Ma et al., 1991). Similar to Pst DC3000, Pst DC3000 ΔCOR grew less at 16°C than at 22°C (Fig. 1, C and D). Multivariate ANOVA (MANOVA) analysis revealed no significant influence of COR over the temperature effect on pathogen growth (Fig. 1E). In contrast, the Pst DC3000 ΔhrcU strain had no detectable decrease in growth at 16°C compared with 22°C (Fig. 1C). The interaction between temperature and hrcU on pathogen growth is significant (Fig. 1F). These data indicate that a lower growth of Pst DC3000 in plants at 16°C is dependent on a functional T3SS but not on COR production from the pathogen.
Low Temperature Does Not Reduce Pathogen Virulence
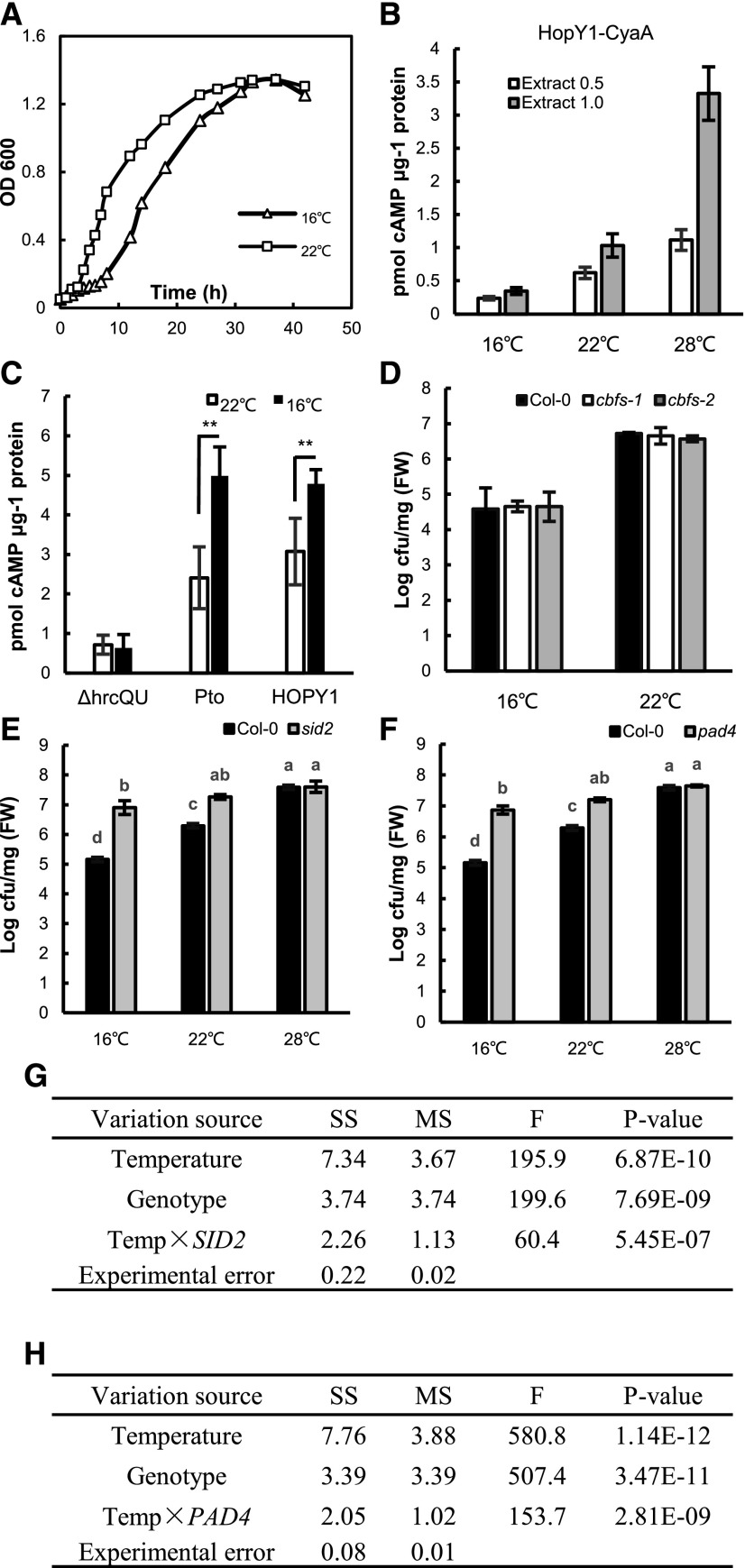
We next examined whether the reduced pathogen propagation in plants at 16°C compared with 22°C is due to an increase of plant immunity or a reduction of pathogen growth or virulence at lower temperature. First, the growth of Pst DC3000 at 16°C and 22°C was analyzed in liquid rich medium where the maximum growth rate could be achieved. From 6 to 30 h post inoculation (hpi), Pst DC3000 became slightly denser at 22°C than at 16°C, but after 30 hpi, they grew to a similar density (Fig. 2A). We expect that the growth rate of the pathogen in plants is unlikely to exceed that in rich medium, and the difference of growth rates at the two temperatures will likely be reduced in plants. Because pathogen growth in liquid medium differed only in the early phase and the maximum difference was 3-fold, the decrease of pathogen propagation in plants by almost 10-fold at 16°C compared with 22°C could not be explained by a lower intrinsic pathogen growth rate at 16°C.
Figure 2.
SA and PAD4 are critical for low-temperature enhancement of plant immunity. A, Growth curve of Pst DC3000 grown in Luria-Bertani liquid medium at 16°C and 22°C. Shown is the result from one experiment, and similar growth curves were observed for each temperature in a second experiment. B, Effect of temperature on the enzymatic activity of HopY1-CyaA. Shown are means ± sd of the amount of cAMP produced in reactions containing the lysate of Pst DC3000 carrying HopY1-CyaA with plant extracts at 16°C, 22°C, and 28°C. The plant extract was provided at a concentration of 0.5 or 1 μg μL−1. Data are means from seven biological repeats, and sd values are indicated by error bars. C, Effector translation at different temperatures. Shown is the amount of cAMP in leaves of temperature-acclimated plants syringe infiltrated with Pst DC3000 with AvrPto-CyaA, Pst DC3000 with HopY1-CyaA, or Pst DC3000 ΔhrcQ-hrcU with AvrPto-CyaA. Values were normalized by the total protein amount and the relative CyaA enzyme activity at the two temperatures. Data are means from seven biological repeats, and sd values are indicated by error bars. Asterisks indicate statistical significance based on Student’s t test (**, P < 0.01) of pairwise comparisons for each individual effector strain at 16°C versus 22°C. D, Growth of Pst DC3000 in wild-type Col-0 and cbf-1 and cbf-2 mutants measured at 4 dpi for 16°C and 3 dpi for 22°C. Values represent means ± sd (n = 3). cfu, Colony-forming units; FW, fresh weight. E and F, Growth of Pst DC3000 in the pad4 (E) and sid2 (F) mutants at 16°C, 22°C, and 28°C. The bacterial growth was measured at 4, 3, and 3 dpi at 16°C, 22°C, and 28°C, respectively. Values represent means ± sd (n = 3). Letters indicate statistically significant differences by ANOVA (P < 0.05). Similar results were obtained in three independent experiments, and shown is the result from one experiment with three biological repeats. G and H, MANOVA for pathogen growth in the mutant plants pad4 (G) and sid2 (H) compared with the wild-type plants at different temperatures. F, F test; MS, mean square; SS, square sum; Temp, temperature.
Second, translocation of two effector proteins from the pathogen was analyzed by cyclic AMP (cAMP) produced by their fusions with the reporter protein adenylate cyclase A (CyaA). CyaA converts ATP into cAMP only when they are present in plant cells but not bacteria where calmodulin is not available for its activity (Schechter et al., 2004). Arabidopsis plants grown at 16°C and 22°C were inoculated by infiltration with Pst DC3000 strains carrying the fusions between the effectors (AvrPto or HopY1) and the reporter CyaA (named AvrPto-CyaA or HopY1-CyaA; as described by Schechter et al. [2004, 2006]), and the amount of cAMP produced by CyaA in leaves at 4 hpi was measured. The cAMP amount had an apparent increase of 1- to 2-fold for both effectors at 22°C compared with that at 16°C (Supplemental Fig. S1). Early biochemical studies indicate that the enzymatic activity of CyaA is positively correlated with temperature, and its activity at 22°C was found to be 1.8-fold and 3-fold that at 16°C in the two studies (Murayama et al., 1994; Raffelberg et al., 2013). To verify this temperature dependence of enzymatic activity from the CyaA reporter protein, we mixed plant extracts with lysate of Pst DC3000 strains carrying HopY1-CyaA or AvrPto-CyaA and incubated the reactions at 16°C, 22°C, and 28°C. With the plant extract provided at the protein concentration of 1 μg μL−1 in the reaction, the CyaA activity was found to increase from 16°C to 22°C by 2.9- to 2.7-fold and from 22°C to 28°C by 3-fold (Fig. 2B; Supplemental Fig. S1B). With the plant extract provided at a lower protein concentration of 0.5 μg μL−1, there was also an increase of cAMP production from 16°C to 22°C and from 22°C to 28°C, but the amount of cAMP was lower, probably due to insufficient calmodulin provided from the plant extract (Fig. 2B; Supplemental Fig. S1B). No cAMP, product of CyaA, could be detected from reactions containing only the plant extracts without the lysate from the bacterial strain carrying AvrPto-CyaA (Supplemental Fig. S1C), indicating that the activity detected came from the bacteria carrying the CyaA reporter protein. We therefore adjusted the measured cAMP amount at 22°C relative to that at 16°C by a factor of 2.4-fold for direct comparison of the amount of the effector-CyaA fusion protein. This factor is the average of those from the two previous biochemical studies and is close to the 2.7- to 3-fold found in this study and therefore is a conservative factor for decreased activity at 16°C. The normalized cAMP concentration was significantly higher in infected plants at 16°C compared with 22°C for Pst DC3000 strains carrying AvrPto-CyaA or HopY1-CyaA (Fig. 2C). As a control, a Pst DC3000 ΔhrcQ-hrcU mutant strain where T3SS is defective (Cunnac et al., 2011) carrying the same AvrPto-CyaA induced little cAMP production in plants after infection, and there was no difference of the cAMP amount between 16°C and 22°C at 4 hpi (Fig. 2C). Therefore, effector secretion from the pathogen is not reduced at 16°C compared with 22°C. This lack of decrease of virulence at low temperature, combined with no drastic reduction of intrinsic pathogen growth, indicates that the lower propagation of the pathogen in plants is mainly due to an enhancement of plant immunity at low temperatures.
Involvement of the SID2 and PAD4 Pathways But Not the C-Repeat-Binding Factor Genes in Low Temperature Enhances Plant Immunity
An early study found that a short-term treatment of 4°C enhances plant immunity (Wu et al., 2019). Therefore, we investigated whether the C-repeat-binding factor (CBF) genes that mediate cold acclimation could be involved in the enhancement of plant immunity by moderately low temperature. The cbf mutants, where all three CBF genes are knocked out (Jia et al., 2016), were analyzed for disease resistance at 16°C and 22°C. Both cbf-1 and cbf-2 mutants supported a similar pathogen growth to the wild type, with a lower pathogen growth at 16°C than 22°C (Fig. 2D). This suggests that the CBF pathway does not play a major role in mediating low-temperature enhancement of disease resistance to Pst DC3000.
Because SA signaling is critical for high temperature-conferred inhibition of disease resistance, we asked if it is critical for the effects of both high and low temperatures on plant immunity. To this end, pathogen growth was analyzed in the SA biosynthesis mutant sid2 and the SA signaling mutant pad4 at 16°C, 22°C, and 28°C. The sid2 and pad4 mutants both exhibited the same susceptibility (slightly more but not significant) at 28°C compared with 22°C, in contrast to that in the wild-type Col-0 (Fig. 2, E and F). The pad4 and sid2 mutants were more susceptible than the wild type at 16°C, indicating an important role of the SA signaling sector in resistance at 16°C. Most strikingly, pathogen growth in the pad4 and sid2 mutants was similar between 16°C and 22°C (Fig. 2, E and F), indicating that the loss of the SA sector abolishes the low-temperature enhancement of disease resistance. Therefore, PAD4 and SID2 play an important role in enhancing plant immunity at lower temperatures. Further MANOVA revealed a significant interaction between temperature and genotypes on disease resistance (Fig. 2, G and H), indicating a strong influence of PAD4 and SID2 over the temperature effect on basal resistance.
ET and JA Repress Resistance through SA and PAD4 at 22°C
Because temperature affects the production and signaling of plant hormones (Scott et al., 2004), we investigated whether varying temperatures could alter the contribution from and interaction among hormones in plant immunity. To do this, we conducted pathogen growth assays at 16°C, 22°C, and 28°C on the well-characterized hormone sector mutant set including the single and combined mutants of sid2, dde2, ein2, and pad4.
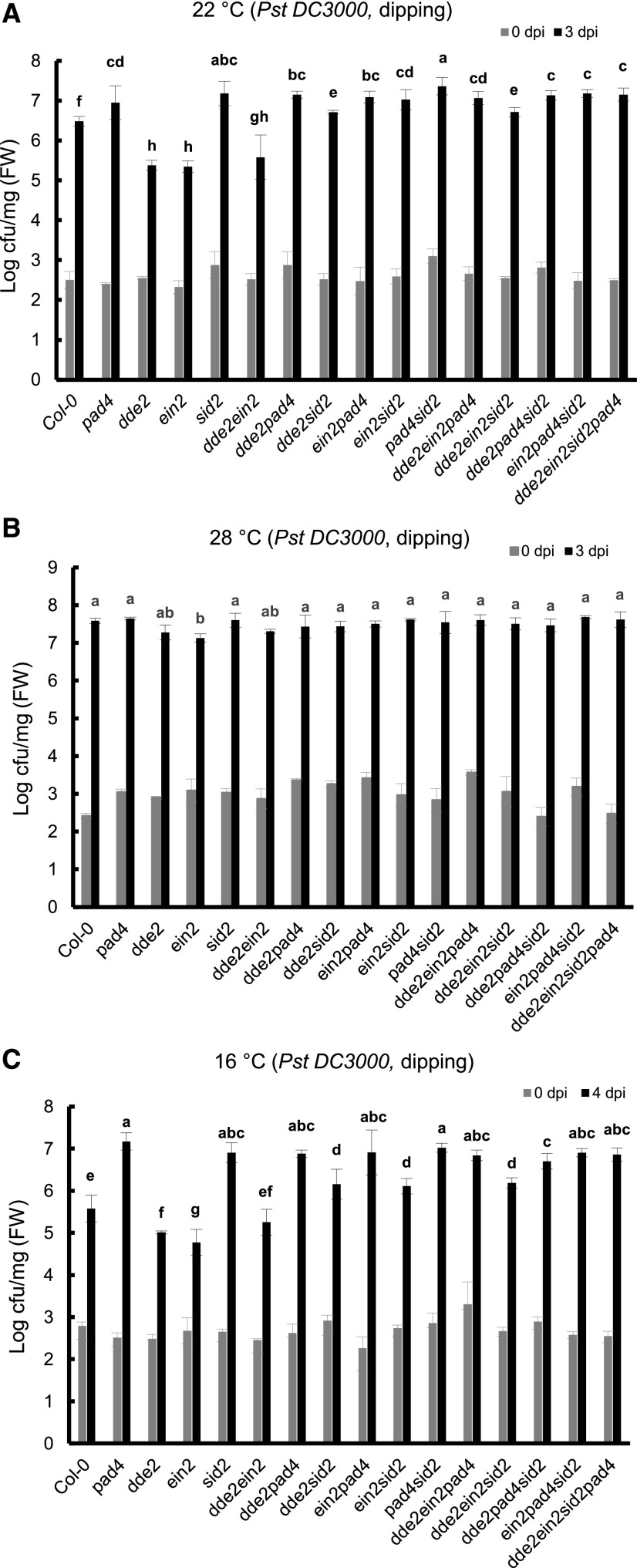
At 22°C, the quadruple mutant dde2 ein2 sid2 pad4 was more susceptible to Pst DC3000 than the wild type assayed by either dipping or infiltration inoculation (Fig. 3A; Supplemental Fig. S2). This indicates that these genes collectively make a positive contribution to plant immunity. Among the single mutants, dde2 and ein2 were more resistant than the wild type while sid2 and pad4 were more susceptible than the wild type, indicating a positive role of SA and a negative role of JA and ET in resistance to Pst DC3000. The inhibition of resistance by JA and ET is mediated by SID2 and PAD4. The double mutants between the more resistant dde2 or ein2 mutants and the more susceptible sid2 or pad4 mutants (dde2 sid2, dde2 pad4, ein2 sid2, and ein2 pad4) were all more susceptible to Pst DC3000 than the wild type. Furthermore, the dde2 pad4 and ein2 pad4 double mutants were as susceptible as the pad4 mutant, while dde2 sid2 was more susceptible than dde2 although more resistant than the sid2 mutant (Fig. 3A; Supplemental Fig. S2). This suggests that the effect of JA and ET is mainly through their inhibition of SID2 and PAD4, although the JA suppression of resistance is not entirely through SID2.
Figure 3.
ET and JA repress disease resistance through SA and PAD4. Growth is shown for Pst DC3000 in the wild-type Col-0 and single and combined mutants of dde2, ein2, sid2, and pad4. The bacterial growth was measured at 3, 3, and 4 dpi for 22°C (A), 28°C (B), and 16°C (C), respectively. Values represent means ± sd (n = 3). Letters indicate statistically significant differences among different genotypes determined by ANOVA (P < 0.05). Similar results were obtained in three independent experiments, and shown is the result from one experiment with three biological repeats. cfu, colony-forming units; FW, fresh weight.
PAD4 appears to be the most important player among the four genes in basal resistance to Pst DC3000 at 22°C. No significant difference in susceptibility was observed between the sid2, pad4, and sid2 pad4 mutants (Fig. 3A; Supplemental Fig. S2). All mutants containing pad4, including dde2 ein2 pad4, had a similar susceptibility to the pad4 single mutant (Fig. 3A), indicating a dependence of resistance inhibition on PAD4 from the combined action of JA and ET. The dde2 ein2 sid2 pad4 mutant, like other combinations containing both sid2 and pad4, was only slightly less susceptible than the sid2 pad4 double mutant (Fig. 3A), and the difference was very small. The dde2 ein2 sid2 mutant, although more susceptible than the dde2 ein2 mutant, was more resistant to Pst DC3000 than the sid2 mutant (Fig. 3A). These data indicate that SID2 has a similar but smaller role compared with PAD4 in resistance to Pst DC3000 and that JA and ET work through PAD4/SID2 in plant immunity at 22°C.
The Contributions from Four Signaling Sectors to Disease Resistance Are Minor at 28°C
The growth of Pst DC3000 was measured in the wild type and the sector mutants at 28°C by dipping inoculation (Fig. 3B) or infiltration inoculation (Supplemental Fig. S3). The pad4 and sid2 single mutants were as susceptible as the wild type while the ein2 mutant exhibited less susceptibility compared with the wild type. A slight decrease of susceptibility was observed in the dde2 mutant but it did not appear to be significant. The double mutants, ein2 pad4, ein2 sid2, dde2 pad4, and dde2 sid2, were as susceptible as the single pad4 and sid2 mutants or the wild type. Therefore, the loss of PAD4 or SID2 function alone does not affect disease susceptibility but their function is revealed in the ein2 and perhaps the dde2 mutant.
PAD4 and SID2 Are Critical for Disease Resistance at 16°C
The growth of Pst DC3000 in the single sector mutants was analyzed at 16°C and the patterns of growth were similar between using dipping and infiltration inoculation methods (Fig. 3C; Supplemental Fig. S4). The dde2 and ein2 mutants were both more resistant than the wild type at 16°C, but the dde2 pad4 and ein2 pad4 double mutants were as susceptible as the pad4 mutant. In addition, the ein2 sid2 pad4 and dde2 sid2 pad4 triple mutants were as susceptible as the sid2 and pad4 single mutants, indicating that the repression of resistance by JA and ET is through PAD4 and SID2. Overall, relative pathogen growth in single and combined mutants at 16°C was also similar to that at 22°C (Fig. 3, A and C), suggesting a similar contribution and interaction from the four signaling sectors at these two temperatures. The only exception is the ein2 sid2 mutant, which exhibited an enhanced resistance compared with the sid2 single mutant at 16°C but had the same resistance as sid2 at 22°C (Fig. 3, A and C; Supplemental Figs. S2 and S4). This suggests that enhanced resistance in the ein2 mutant is fully dependent on SID2 at 22°C but not at 16°C.
Transcriptome Analysis Reveals a Module Associated with Differential Contributions of SID2 to Immunity at 16°C and 22°C
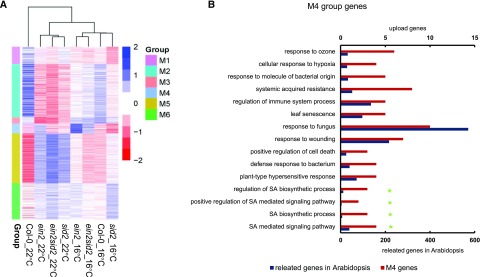
To reveal the underlying mechanism of this temperature dependence of the ein2 and sid2 interaction, we performed transcriptome analysis using 3′ RNA sequencing (RNA-seq; Tandonnet and Torres, 2016) on seedlings of Col-0, ein2, sid2, and ein2 sid2 grown at 16°C and 22°C. A total of 8,400 genes were differentially expressed between the mutants and the wild type or between 16°C and 22°C (Supplemental Fig. S5A). Cluster analysis based on these differentially expressed genes (DEGs) revealed that sid2 at 22°C and ein2 sid2 at 22°C were closely related while ein2 at 16°C and ein2 sid2 at 16°C were closely related (Fig. 4A; Supplemental Fig. S5B). This is in agreement with the pathogen resistance phenotype, where the ein2 sid2 double mutant was similar to sid2 at 22°C but was more similar to ein2 at 16°C (Fig. 3, A and C).
Figure 4.
Transcriptome analysis reveals differential contributions of SID2 to resistance at low and normal temperatures. A, Heat map of gene expression in six modules defined by WGCNA. Blue color indicates relatively high expression, and red color indicates relatively low expression. B, Gene Ontology term analysis of genes in module 4. Shown are the results with Bonferroni-corrected significance (P < 0.05). The green star makers indicate genes involved in SA biosynthesis and signaling.
We carried out Weighted Gene Coexpression Network Analysis (WGCNA; Stuart et al., 2003) on the DEGs to reveal potential gene networks that are differentially associated with ein2 sid2 at 22°C and 16°C. Six modules (M1–M6) were identified for all DEGs (Fig. 4A; Supplemental Table S1; Supplemental Fig. S5B). Genes in the ein2 sid2 mutant had different relative expression levels at 22°C and 16°C in all modules except for M1. Among the modules between M2 and M6, only genes in the M4 module had expression in the ein2 sid2 mutant, resembling ein2 more at 16°C but resembling sid2 at 22°C. In addition, genes in M4 had higher expression in the ein2 mutant but lower expression in the sid2 mutant.
Because the expression pattern of the M4 module is highly correlated with the disease resistance properties of the ein2 sid2 mutant at 16°C and 22°C (Fig. 3, A and C), we examined the genes in this module in detail. Gene Ontology term analysis of the M4 module revealed an enrichment of defense-related genes (Fig. 4B). More specifically, terms related to SA biosynthesis were enriched, including SA biosynthetic processes and signaling pathway (Fig. 4B). This indicates that the SA biosynthesis process might be affected by the ein2 sid2 mutations in a temperature-dependent manner. Consistent with this hypothesis, multiple SA-inducible genes such as PATHOGENESIS RELATED PROTEIN1 (PR1) and PR2 are present in the M4 module. PR2 had a higher expression at 16°C compared with 22°C. PR1 had the same expression at 16°C compared with 22°C (although there was a slight but not significant increase at 16°C), and both expression levels were very low, which is consistent with its induced nature by stress conditions. Their expressions were much higher in the ein2 mutant than in Col-0 at both temperatures and were lower in the sid2 mutant compared with the wild type (Supplemental Fig. S6, A and B). Similar to other genes in the M4 module, both had higher expression in the ein2 sid2 mutant than in the sid2 mutant at 16°C but not at 22°C (Fig. 4A; Supplemental Fig. S5B).
EDS1 and PAD4, regulators of SA signaling and biosynthesis, are also found in the M4 module. EDS1, but not PAD4, had a slightly higher expression at 16°C than at 22°C (Supplemental Fig. S6, C and D). Their expression was not drastically affected by the sid2 mutation but was significantly higher in the ein2 mutant than in the wild type at both 16°C and 22°C (Supplemental Fig. S6, C and D). Similar to other genes in the M4 module, their expression in the ein2 sid2 mutant was significantly higher at 16°C than at 22°C (Supplemental Fig. S6, C and D). In summary, genes in M4 are involved in both SA biosynthesis and signaling, and they likely contribute to the temperature-dependent interaction between ein2 and sid2.
SA Biosynthesis Genes in the ein2 sid2 Mutant Have Higher Expression at 16°C Than at 22°C
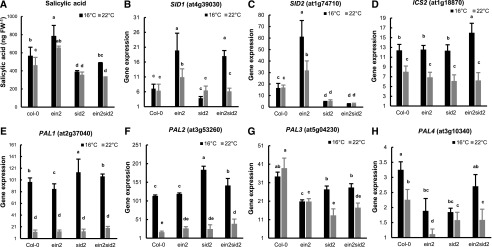
The expression pattern of SA-induced defense genes suggests that SA biosynthesis and/or signaling are affected by the ein2 mutation and that the involvement of SID2 in these SA processes is temperature dependent. To test this hypothesis, we measured SA content in Col-0, ein2, sid2, and ein2 sid2 plants at 22°C and 16°C. The wild-type Col-0 plants accumulated more SA at 16°C than at 22°C (Fig. 5A), which could explain the enhanced disease resistance at 16°C (Fig. 1A). As expected, the sid2 mutant contained less SA than the wild type at both 16°C and 22°C. In contrast, the ein2 mutant accumulated more SA than the wild type at both temperatures, which is consistent with a higher disease resistance in the ein2 mutant than in the wild type (Fig. 3). Interestingly, the ein2 sid2 double mutant had a reduced SA level compared with the ein2 mutant, to a level similar to sid2 at 22°C, but it had a higher SA level compared with the ein2 sid2 mutant, to a level similar to the wild type at 16°C. Therefore, the SA content is closely correlated with disease resistance at the two temperatures for the ein2 sid2 mutant.
Figure 5.
SA biosynthesis genes in the ein2 sid2 mutant have higher expression at 16°C than at 22°C. A, Total SA in 3-week-old plants grown at 22°C or 4-week-old plants grown at 16°C. FW, Fresh weight. B to H, Normalized expression levels as counts per million reads of SID1 (B), SID2 (C), ICS2 (D), PAL1 (E), PAL2 (F), PAL3 (G), and PAL4 (H) analyzed by 3′ RNA-seq. Values are means ± sd from three biological replicates. Letters indicate statistically significant differences among different genotypes determined by ANOVA (P < 0.05).
To better understand the molecular basis for a higher SA content in the ein2 sid2 mutant compared with the sid2 mutant at 16°C but not 22°C, we examined the expression of genes involved in SA biosynthesis. SID2/ICS1 had a higher expression in the ein2 mutant than in the wild type (Fig. 5C), which is associated with a higher SA content in the ein2 mutant than in the wild type. Meanwhile, SID1 was also expressed at a higher level in the ein2 mutant than in the wild type and in the ein2 sid2 mutant than in the sid2 mutant at 16°C (Fig. 5B), indicating an inhibition of SID1 expression by EIN2. At 22°C, SID1 was expressed at a slightly higher level in the ein2 mutant than in the wild type but to a similar level in the ein2 sid2 and sid2 mutants. For the alternative PAL pathway, PAL1 and PAL2, but not PAL3 and PAL4, exhibited a 10-fold increase of expression at 16°C compared with 22°C, and this increase was observed in the wild type as well as in the ein2 and sid2 mutants (Fig. 5, E–H). However, PAL1, PAL2, and PAL3 did not have a higher expression in the ein2 sid2 mutant compared with the sid2 mutant at 16°C (Fig. 5, E–G). PAL4 had an increased expression in the ein2 sid2 mutant compared with the sid2 mutant at 16°C (Fig. 5H), but the contribution of this increase to a higher SA level in the ein2 sid2 mutant might be minimal because its expression is extremely low compared with the other three PAL genes.
Interestingly, ICS2, the SID2/ICS1 homolog, had a higher expression at 16°C compared with 22°C in all genotypes tested. It is worth noting that it is more highly expressed in the ein2 sid2 mutant compared with the sid2 mutant at 16°C (Fig. 5D); the relative expression level of ICS2 in the ein2 sid2 and sid2 mutants was correlated with the SA content at 16°C (Fig. 5A).
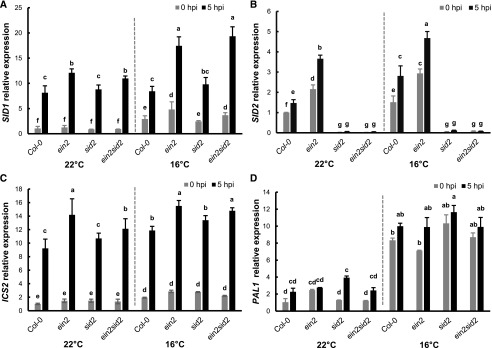
Pst DC3000 Induces SID1 and ICS2 Expression in ein2 sid2 at 16°C
Because SID1 and ICS2 expression under nonpathogenic conditions is correlated with SA accumulation in the ein2 sid2 mutant at 16°C (Fig. 5, A and B), we examined its expression after pathogen infection to determine whether its expression level could contribute to disease resistance. Seedlings of the wild type, ein2, sid2, and ein2 sid2 were dipping inoculated by Pst DC3000, and the expression of SID1 and ICS2 was assayed by reverse transcription quantitative PCR (RT-qPCR; Fig. 6A). In all these wild-type and mutant plants, the SID1 gene was expressed at a higher level by 8- to 12-fold at 5 hpi compared with 0 hpi. In addition, it was expressed at a significantly higher level in the ein2 and ein2 sid2 mutants than in the wild type and the sid2 mutant at 5 hpi, and this difference was more drastic at 16°C than at 22°C. Therefore, SID1 expression is higher in plants containing the ein2 mutation compared with EIN2 wild-type plants, and the higher expression is more pronounced at lower temperature. In contrast to the previous notion, ICS2 expression was induced by the pathogen by 5- to 7-fold in the wild-type and mutant plants at both 16°C and 22°C. In addition, it was expressed at a slightly higher level in the ein2 mutant than in the wild type and in the ein2 sid2 mutant than in the sid2 mutant at 5 hpi (Fig. 6C). These results suggest that ICS2 plays a role in SA induction after pathogen infection.
Figure 6.
Pst DC3000 induces SID1 and ICS2 expression in the ein2 sid2 mutant at low temperature. Relative expression levels of SID1 (A), SID2 (B), ICS2 (C), and PAL1 (D) were analyzed by RT-qPCR in leaves dipping inoculated by Pst DC3000 at 0 and 5 hpi. Expression levels are normalized by gene expression at 22°C in Col-0 at 0 hpi. Values represent means ± sd from two independent experiments. Letters indicate statistically significant differences among different genotypes determined by ANOVA (P < 0.05).
The expression of the other SA biosynthesis genes SID2 and PAL1 was also examined in the wild type and mutants after pathogen infection (Fig. 6, B and D). SID2 was not expressed in the sid2 and ein2 sid2 mutant plants, as expected. It was induced by pathogen infection, although the induction fold was smaller than that of SID1. Unlike SID1, SID2, or ICS2, PAL1 was not significantly induced in expression after pathogen infection except in the sid2 mutant at 22°C (Fig. 6D). However, it had a significantly higher expression at 16°C than at 22°C in all genotypes with or without infection (Figs. 5D and 6D).
We observed an inconsistency in the relative expression levels of SID1 and SID2 at 16°C versus 22°C in RNA-seq data (Fig. 5, B and C) and in the RT-qPCR data without infection (Fig. 6, A and B, 0 hpi). It turns out that the expression of SID1 and SID2 is impacted by the circadian clock, and both had higher expression at 16°C compared with 22°C at the end of the night but similar expression at the two temperatures at the end of the day (Supplemental Fig. S7). This is consistent with earlier findings that the SA pathway is circadian clock regulated (Zheng et al., 2015).
EIN3 Directly Regulates the Expression of SID1 and ICS2
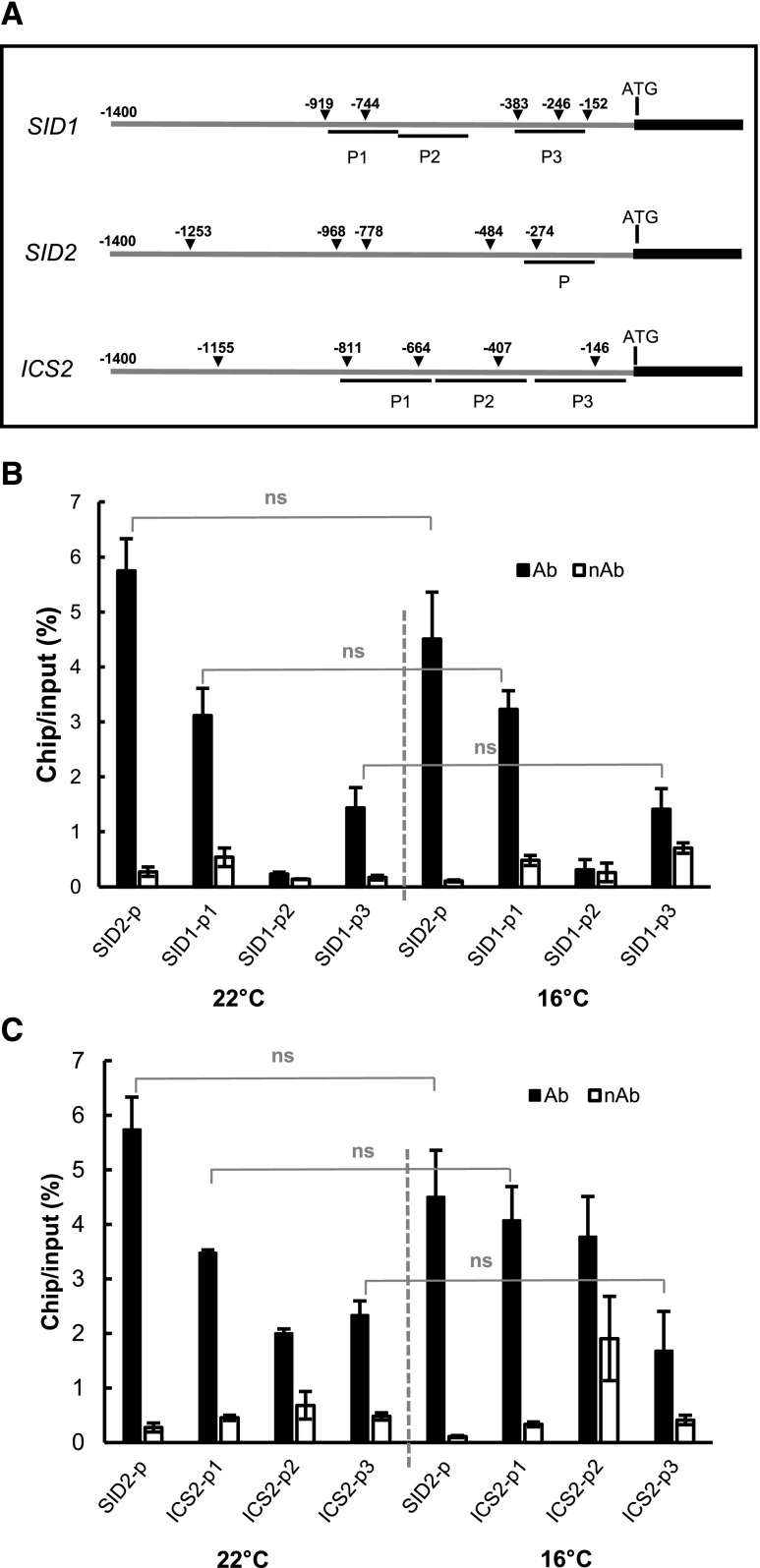
The up-regulation of SA biosynthesis and signaling genes, including SID2, SID1, ICS2, PAD4, and EDS1, likely plays an important role in the enhancement of disease resistance by low temperature. Because early studies revealed a direct transcriptional repression of SID2 by EIN3 (Chen et al., 2009), we explored the possibility that EIN3 acts as a direct transcription factor of these genes. EIN3-binding sites were experimentally identified by DNA affinity purification (DAP)-seq and chromatin immunoprecipitation (ChIP)-seq (Supplemental Fig. S8, A and B; Chang et al., 2013; O’Malley et al., 2016), and a consensus sequence of A(C/T)G(A/T)A(C/T)CT was reported to be the binding motif of the EIN3/EIL family protein (Kosugi and Ohashi, 2000; Chang et al., 2013). We searched in the promoters of these genes for EIN3-binding consensus motifs and EIN3/EIL-binding sites using DAP-seq and ChIP-seq data. EDS1 is a strong candidate for a direct target of EIN3 based on the presence of five perfect binding motifs (Supplemental Fig. S8A), and EIN3 indeed showed binding footprints in its promoter region (Supplemental Fig. S8C; O’Malley et al., 2016). SID2 was demonstrated to be directly bound by EIN3 via a number of in vivo and in vitro methods (Chen et al., 2009), and five EIN3-binding motifs were also found in its promoter (Fig. 7A). However, SID2 was not considered as a strong target gene of EIN3 based on DAP-seq or ChIP-seq (Supplemental Fig. S8C), suggesting a low sensitivity of -seq detection or a low binding activity of EIN3 to the SID2 promoter. For ICS2 and SID1, EIN3-binding motifs were found in their promoter regions (Fig. 7A). EIN3-binding signals were stronger for ICS2 and SID1 than for SID2 in the ChIP-seq experiment (Supplemental Fig. S8C), suggesting that ICS2 and SID1 are direct target genes of EIN3/EILs.
Figure 7.
EIN3 is associated with the promoter regions of SID1, SID2, and ICS2. A, Schematic diagram of DNA fragments used for ChIP experiments and the EIN3-binding motifs found in the promoters of SID1, SID2, EDS1, and ICS2. Triangles indicate the positions of the EIN3-binding motifs relative to the translation start site. B and C, ChIP assays of EIN3 binding to the promoter regions of SID1 (B) and ICS2 (C). Shown are the qPCR results from samples precipitated with antibody (Ab) or without antibody (nAb) of GFP. SID2 was used as a positive control. Significant differences between samples were determined by Student’s t test; ns, no significant difference. Values represent means ± sd from two independent experiments.
We next tested the direct association of EIN3 with the predicted binding sites in the promoters of the SID1 and ICS2 genes using ChIP. A GFP fusion of EIN3 (EIN3-GFP) was expressed under the control of the 35S promoter in Arabidopsis protoplasts. Chromatins associated with EIN3-GFP were immunoprecipitated (IPed) with the anti-GFP antibody, and the precipitated chromatins were analyzed by qPCR for enrichment of the chosen promoter regions. The P region of the SID2 promoter (−115 to −324 bp relative to the transcription start site; Fig. 7A), shown to be bound by EIN3 in an earlier study (Chen et al., 2009), was indeed enriched in the EIN3-GFP ChIPed chromatin compared with the no-antibody control. The P2 region of SID1 (−392 to −618 bp), with no predicted binding motifs of EIN3, did not show any enrichment in the ChIP samples (Fig. 7B). In contrast, predicted EIN3-binding sites, including P1 (−128 to −331 bp) and P3 (−574 to −782 bp) of SID1 as well as P1 (−109 to −358 bp), P2 (−358 to −621 bp), and P3 (−621 to −818 bp) of ICS2, were all found to have higher ChIP output versus the genomic DNA input in GFP-antibody ChIPed chromatins over no-antibody ChIPed controls (Fig. 7, B and C). The association of EIN3 with SID1 and ICS2 was observed at both 22°C and 16°C, with no significant difference in output/input percentage.
DISCUSSION
Temperature is one of the most important climatic factors that impact the interactions between plants and pathogenic bacteria, fungi, and insects. In the model plant-pathogen interaction system, we found a reduced propagation of Pst DC3000 at a moderately low temperature of 16°C compared with the normal growth temperature of 22°C in Arabidopsis. This is due to an increase of basal resistance and NLR-triggered immunity in Arabidopsis at low temperature (Figs. 1 and 3), while the virulence of Pst DC3000 was not reduced at low temperature (Fig. 2, B and C). This enhancement of plant immunity is accompanied by a higher SA content at low temperature even without pathogen infection (Fig. 5A). We further demonstrate that the SA pathway is the major contributor to the enhanced resistance at 16°C. Mutants defective in SA biosynthesis and signaling, sid2 and pad4, not only have higher susceptibility compared with the wild type at 16°C but also exhibit no enhancement of disease resistance at 16°C compared with 22°C (Fig. 2, E and F). Therefore, SA is a key target for temperature regulation of plant immunity, with lower temperature increasing SA signaling and enhancing plant immunity.
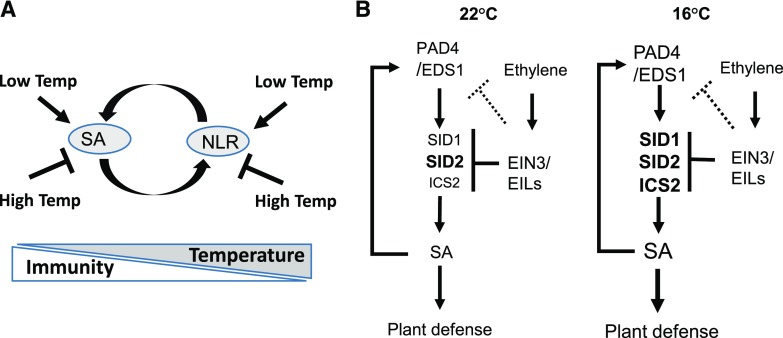
Earlier studies have found that NLR proteins have a higher activity at lower temperatures. Hybrid necrosis mediated by activation of NLR proteins was present at 16°C but was suppressed at 23°C (Bomblies et al., 2007). Chilling-sensitive mutants containing active forms of NLR proteins exhibit low temperature-induced cell death (Wang et al., 2013). Because NLR activation can induce SA biosynthesis and SA can induce the expression of some NLR genes, it is likely that NLR forms an amplification loop with SA to enhance resistance at low temperature (Fig. 8A). Similar feedback regulation likely occurs at moderately high temperatures as well. High temperature inhibits plant immune receptor NLR genes and the plant hormone SA (Huot et al., 2017). The suppression of one or multiple components in the loop could lead to amplification of the effect of high temperature (Fig. 8A).
Figure 8.
Models of low-temperature enhancement of plant immunity. A, Modulation of plant immunity by temperature. SA and NLR form a positive amplification loop. Low temperature promotes the SA pathway and NLR activities, while high temperature inhibits the SA pathway and NLR activities. B, Interaction of ET with the SA pathway at 22°C and 16°C. SA and EDS1/PAD4 form a positive feedback loop, and the up-regulation of biosynthesis genes including SID1, ICS2, and SID2 contributes to the higher SA level at 16°C. SID2 plays a major role in SA biosynthesis at both temperatures. ET represses multiple target genes to influence the SA pathway. SID2 is the ET target that is a major player in SA biosynthesis at 22°C, while SID1 and ICS2 are additional targets that contribute to SA biosynthesis at 16°C. ICS2 has a minor role in SA biosynthesis compared with SID2, but it could replace the role of SID2 at low temperature when its repression by ET is released.
Low temperature induces a higher production of SA mainly due to an up-regulation of SA biosynthesis genes (Fig. 5, A and C). The increase of expression of SID2, the key gene in the isochorismate pathway, is essential for the increased SA content at 16°C because the loss of SID2 function abolishes an increase of SA at 16°C. The production of SA upon pathogen infection at low temperature is not yet determined. A higher production is expected under pathogen infection conditions compared with no infection because the extent of induction of SID2 by infection is much higher at 16°C compared with 22°C (Fig. 6B).
JA and ET repress plant defenses through SA at a broad temperature range. The loss of their biosynthesis or signaling (in dde2 and ein2 mutants) confers higher basal resistance at both 22°C and 16°C and to some extent at 28°C (Fig. 3). This negative regulation is through their inhibition of the SA biosynthesis and signaling pathway because the loss of PAD4 abolishes their effects (Fig. 3). At 28°C, the suppression of the SA pathway by high temperature renders almost all the signaling sector mutants as susceptible as the wild type, removing the differences of basal resistance across the genotypes. Despite the overall enhancement of plant immunity at low temperature, the four signaling sectors have similar relative contributions to plant immunity at low and normal temperatures. However, the interaction between ET and SA is different at 16°C and 22°C. The repression of immunity by ET is totally dependent on SID2 at 22°C but is partially dependent on SID2 at 16°C (Fig. 3, A and B; Supplemental Figs. S2 and S4). Nevertheless, the repression of immunity by ET at 16°C is through the SA pathway, despite a reduced role of SID2 in SA biosynthesis under ET repression. This is supported by the total dependence of PAD4 for ET repression at 16°C (Fig. 3, A and C).
The repression of immunity by ET at low temperature is through multiple SA biosynthesis genes, including SID1 and ICS2 besides SID2. At normal temperature, the SID2 gene is directly regulated by the transcription factor EIN3 (Chen et al., 2009). At low temperature, SID2 is also directly associated with EIN3, and it has a higher expression in the ein2 mutant (Figs. 5C, 6B, and 7). SID1 is another EIN3 target gene, as indicated by its increased expression in the ein2 mutant and an association of the EIN3 protein with its promoter (Figs. 5B, 6A, and 7). Similarly, the SID2 homologous gene ICS2 is also directly repressed by EIN3, as EIN3 is associated with its promoter and represses its expression. In addition, the SA signaling gene EDS1 is another regulatory target of ET (O’Malley et al., 2016), and the up-regulation of EDS1 in the ein2 mutant may amplify SA signaling and thus influence SA biosynthesis at low temperature. Thus, ET represses multiple target genes to influence the SA pathway.
SID2 is the ET target that is a major player in SA biosynthesis at 22°C, while SID1 and ICS2 are additional targets that contribute to SA biosynthesis at 16°C (Fig. 8). ICS2 has a minor role in SA biosynthesis compared with SID2, but it could replace the role of SID2 at low temperature when its repression by ET is released. Expressing EIN3 (EIN3-GFP) under the control of the 35S promoter did not reveal differences of its association with the promoters of SID1 and ICS2 at 16°C and 22°C (Fig. 7, B and C). It has yet to be determined quantitatively the level of endogenous EIN3 protein and its relative binding to target genes at normal and low temperatures. In addition to the potential differential EIN3 activity at the two temperatures, the differential activities of SA biosynthesis proteins at the two temperatures could also contribute to an increased SA level at low temperature.
In sum, ET likely exerts an effective repression on the SA pathway through repressing multiple genes in SA biosynthesis, transport, and signaling. This repression could be more critical for balancing growth and immunity at lower temperature when immunity is enhanced. The temperature-dependent interaction of hormone biosynthesis and signaling might provide versatility to achieve optimal defense responses for a broad range of pathogen interaction and environmental conditions.
MATERIALS AND METHODS
Plants and Growth Conditions
The single and combined Arabidopsis (Arabidopsis thaliana) mutants of dde2, ein2, sid2, and pad4 were as previously described (Tsuda et al., 2009). Primers and restriction enzymes used for mutant analyses are listed in Supplemental Table S2.
The Arabidopsis plants were grown in chambers with a light intensity of 100 μmol m−2 s−1 and relative humidity at 50% to 70%. Plants for the pathogen growth assay were grown under a 12-h-light/12-h-dark photoperiod unless specified.
RNA Expression Analysis
Total RNAs were extracted from soil-grown 14-d-old plants (22°C) and 21-d-old plants (16°C) with TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. For expression analysis of individual genes, cDNAs were synthesized from total RNAs by using the AffinityScript QPCR cDNA Synthesis Kit (Agilent Technologies). qPCR was performed on the Bio-Rad PCR System using iQSYBR GREEN SuperMix (Bio-Rad). Primers used in the qPCR are listed in Supplemental Table S2. ACTIN2 was used as an internal control.
For whole-transcriptome analysis, total RNAs were processed at the Cornell Genomic Facility for 3′ RNA-seq analysis (Tandonnet and Torres, 2016) on Illumina NextSeq500. The read count for each gene was obtained from the mapping results and normalized to counts per million reads. DEGs were defined by false discovery rate ≤ 0.001 and absolute value of log2 ratio ≥ 1.
WGCNA
DEGs were identified by DESeq2 (moderated estimation of fold change) and dispersion for RNA-seq data with DESeq2 based on pairwise comparison between the mutants (ein2, sid2, and ein2 sid2) and the wild type (Col-0), respectively, setting q < 0.05 and log2 fold change > 1. Subsequently, the expression levels of these DEGs were subjected to WGCNA (an R package for weighted correlation network analysis) for gene coexpression analysis. To render the network scale free, a soft thresholding power (softPower = 26) was chosen to transform the Pearson similarity matrix into an adjacency matrix. Modules, in which genes exhibited similar expression patterns, were determined by the dynamic tree-cut method, and modules with highly correlated genes (e.g. Pearson correlation ≥ 0.8) were merged. The heat map of genes assigned to the modules was plot by pheatmap (R package).
ChIP Analysis
ChIP analysis was performed in Arabidopsis protoplasts as previously described (He et al., 2013; Bao et al., 2014). The coding regions of EIN3 were cloned into destination vector pSATN1-GW (Zhang et al., 2017) using LR clonase (Invitrogen). Protoplasts were collected at 16 h after transformation. Immunoprecipitation experiments were carried out with anti-GFP monoclonal antibodies (Life Technologies).
Pathogen Growth Assay
Pathogen growth assays were performed by dipping inoculation (Gou et al., 2015) unless otherwise specified. Seedlings used for infection were grown for 21 d at 16°C, 14 d at 22°C, or 12 d at 28°C under a 12-h-light/12-h-dark photoperiod so that they reached a similar developmental stage. The amounts of bacteria in plants were analyzed at 1 h (0 dpi), 3 d (3 dpi), or 4 d (4 dpi) after dipping. For infiltration infection (Liu et al., 2015), seedlings were grown for 5 weeks at 16°C, 4 weeks at 22°C, or 18 d at 28°C before they were leaf infiltrated with a bacterial solution. The amounts of bacteria in plants were analyzed at 1 h (0 dpi), 2 d (2 dpi), or 3 d (3 dpi) after infiltration. Pathogen growth was determined by log10-transformed bacterial number in infected leaves.
SA Measurement
Total SA contents were assayed as previously described (Lou et al., 2016). Plants were grown for 15 d at 22°C or 21 d at 16°C under a 12-h-light/12-h-dark photoperiod before analysis.
Effector Translocation Assay
To determine the temperature dependence of the CyaA enzymatic activity, Pseudomonas syringae pv tomato DC3000 or the Pst DC3000 ΔhrcQ-hrcU strains carrying the AvrPto-CyaA or HopY1-CyaA effector (as described by Schechter et al. [2004, 2006]) were cultured overnight on a King's B plate. Bacteria were washed from the King's B plate by 10 mm MgCl2, and its concentration was adjusted to an OD600 of 2. The cultures were centrifuged, and the pellets were washed and resuspended in sonication buffer (20 mm Tris-HCl [pH 8] and 10 mm MgCl2). The bacteria were disrupted by sonication with a microtip for 2 min, the cellular debris was pelleted by centrifugation, and the supernatant was collected as cell lysate. Plant extracts were made by grinding the leaf tissues with liquid nitrogen and resuspending the tissue powder in sonication buffer. The reaction was carried out by mixing 10 µL of lysate with plant extracts at 0.5 or 1 µg µL−1 in a 100-µL volume at the designated temperature.
For translocation analysis, plants were infiltrated using a needless syringe with an inoculum (OD = 0.002) of Pst DC3000 or the Pst DC3000ΔhrcQ-hrcU mutant carrying the AvrPto-CyaA or HopY1-CyaA effector. Leaf discs were harvested using a biopsy punch for cAMP quantification.
The CyaA product cAMP was extracted and quantified using the Direct cAMP ELISA kit (ENZO) according to the manufacturer’s instructions, as previously described (Schechter et al., 2004). The measurement was normalized by total plant protein and the relative CyaA enzyme activity at the two temperatures.
Accession Numbers
Accession numbers of the major genes in this research are listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The absolute amount of cAMPs produced by the CyaA reporter protein.
Supplemental Figure S2. Pathogen growth after infiltration inoculation in hormone sector mutants at 22°C.
Supplemental Figure S3. Pathogen growth after infiltration inoculation in hormone sector mutants at 28°C.
Supplemental Figure S4. Pathogen growth after infiltration inoculation in hormone sector mutants at 16°C.
Supplemental Figure S5. DEGs and coexpression modules identified by WGCNA.
Supplemental Figure S6. SA signaling genes have higher expression at lower temperature.
Supplemental Figure S7. Expression of SID1 and SID2 at different times of the day.
Supplemental Figure S8. Binding of EIN3 to the promoters of SA-related genes.
Supplemental Table S1. Expression levels of DEGs used in WGCNA.
Supplemental Table S2. Primers for genotyping, gene expression analysis, and ChIP assays.
Supplemental Table S3. Short description of genes used in this study.
Acknowledgments
We thank the Arabidopsis Resource Center for Arabidopsis mutant seeds. We thank Dr. Allan Collmer for the CyaA reporter strains.
Footnotes
This work was supported by grants from the U.S. National Science Foundation (IOS 1353738) to J. Hua, the National Natural Science Foundation of China (31670269 to J. Hua and 31671774 and 31201279 to J. Hu and Y.G.), the National Key Research and Development Project (2018YFD0100902) to Y.G., the Fundamental Research Funds for the Central Universities (2019QNA6015) to R.P., the Jiangsu Collaborative Innovation Center for Modern Crop Production to J. Hu and J. Hua, and the Dabeinong Funds for Discipline Development and Talent Training in Zhejiang University to J. Hu.
Articles can be viewed without a subscription.
References
- Bao Z, Zhang N, Hua J (2014) Endopolyploidization and flowering time are antagonistically regulated by checkpoint component MAD1 and immunity modulator MOS1. Nat Commun 5: 5628. [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder LE. (1985) Parasite:host:environment specificity in the cereal rusts. Annu Rev Phytopathol 23: 201–222 [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H, Huang SS, Schmitz RJ, Urich MA, Kuo D, et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xue L, Chintamanani S, Germain H, Lin H, Cui H, Cai R, Zuo J, Tang X, Li X, et al. (2009) ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Gao X, Feng B, Sheen J, Shan L, He P (2013) Plant immune response to pathogens differs with changing temperatures. Nat Commun 4: 2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Colhoun J. (1973) Effects of environmental factors on plant disease. Annu Rev Phytopathol 11: 343–364 [Google Scholar]
- Cui H, Tsuda K, Parker JE (2015) Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A (2011) Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA 108: 2975–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9: e0156, doi:10.1199/tab.0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dropkin V. (1969) The necrotic reaction of tomatoes and other hosts resistant to Meloidogyne: Reversal by temperature. Phytopathology 59: 1632–1637 [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C, Lohmann A, Lamodière E, Catinot J, Buchala A, Doermann P, Métraux JP (2008) Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol 147: 1279–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Zhang Z, Zhang N, Huang Q, Monaghan J, Yang H, Shi Z, Zipfel C, Hua J (2015) Opposing effects on two phases of defense responses from concerted actions of HEAT SHOCK COGNATE70 and BONZAI1 in Arabidopsis. Plant Physiol 169: 2304–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Sidhu G, Pawlowski WP (2013) Chromatin immunoprecipitation for studying chromosomal localization of meiotic proteins in maize. Methods Mol Biol 990: 191–201 [DOI] [PubMed] [Google Scholar]
- Hua J. (2014) Temperature and plant immunity In Franklin KA, and Wigge PA, eds, Temperature and Plant Development. John Wiley & Sons, Hoboken, New Jersey, pp 163–180 [Google Scholar]
- Huot B, Castroverde CDM, Velásquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY (2017) Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nat Commun 8: 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212: 345–353 [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28: 960–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Kørner CJ, Du X, Vollmer ME, Pajerowska-Mukhtar KM (2015) Bacterial leaf infiltration assay for fine characterization of plant defense responses using the Arabidopsis thaliana-Pseudomonas syringae pathosystem. J Vis Exp e53364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YR, Bor M, Yan J, Preuss AS, Jander G (2016) Arabidopsis NATA1 acetylates putrescine and decreases defense-related hydrogen peroxide accumulation. Plant Physiol 171: 1443–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Morris V, Cuppels D (1991) Characterization of a DNA region required for production of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 4: 69–74 [Google Scholar]
- Mine A, Nobori T, Salazar-Rondon MC, Winkelmüller TM, Anver S, Becker D, Tsuda K (2017) An incoherent feed-forward loop mediates robustness and tunability in a plant immune network. EMBO Rep 18: 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama T, Hewlett EL, Maloney NJ, Justice JM, Moss J (1994) Effect of temperature and host factors on the activities of pertussis toxin and Bordetella adenylate cyclase. Biochemistry 33: 15293–15297 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffelberg S, Wang L, Gao S, Losi A, Gärtner W, Nagel G (2013) A LOV-domain-mediated blue-light-activated adenylate (adenylyl) cyclase from the cyanobacterium Microcoleus chthonoplastes PCC 7420. Biochem J 455: 359–365 [DOI] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I (2019) Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365: 498–502 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD (2011) Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY (1997) Hrp pilus: An hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 94: 3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J Bacteriol 186: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Vencato M, Jordan KL, Schneider SE, Schneider DJ, Collmer A (2006) Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol Plant Microbe Interact 19: 1180–1192 [DOI] [PubMed] [Google Scholar]
- Scott IM, Clarke SM, Wood JE, Mur LA (2004) Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol 135: 1040–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Wang B, Aryal B, Garcion C, Abou-Mansour E, Heck S, Geisler M, Mauch F, Nawrath C, Métraux JP (2013) Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol 162: 1815–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenaga AM, Argueso CT (2016) No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin Cell Dev Biol 56: 174–189 [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Tandonnet S, Torres TT (2016) Traditional versus 3′ RNA-seq in a non-model species. Genom Data 11: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Katagiri F (2010) Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr Opin Plant Biol 13: 459–465 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bao Z, Zhu Y, Hua J (2009) Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact 22: 498–506 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Wang Z, Zhang X, Yang S (2013) A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J 75: 553–565 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Wu Z, Han S, Zhou H, Tuang ZK, Wang Y, Jin Y, Shi H, Yang W (2019) Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant Cell Environ 42: 2645–2663 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. (2017) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, Dong X (2015) Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci USA 112: 9166–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]