mTERF8 functions as a transcription terminator of chloroplast gene psbJ in Arabidopsis.
Abstract
Members of the mitochondrial transcription terminator factor (mTERF) family, originally identified in vertebrate mitochondria, are involved in the termination of organellular transcription. In plants, mTERF proteins are mainly localized in chloroplasts and mitochondria. In Arabidopsis (Arabidopsis thaliana), mTERF8/pTAC15 was identified in the plastid-encoded RNA polymerase (PEP) complex, the major RNA polymerase of chloroplasts. In this work, we demonstrate that mTERF8 is associated with the PEP complex. An mTERF8 knockout line displayed a wild-type–like phenotype under standard growth conditions, but showed impaired efficiency of photosystem II electron flow. Transcription of most chloroplast genes was not substantially affected in the mterf8 mutant; however, the level of the psbJ transcript from the psbEFLJ polycistron was increased. RNA blot analysis showed that a larger transcript accumulates in mterf8 than in the wild type. Thus, abnormal transcription and/or RNA processing occur for the psbEFLJ polycistron. Circular reverse transcription PCR and sequence analysis showed that the psbJ transcript terminates 95 nucleotides downstream of the translation stop codon in the wild type, whereas its termination is aberrant in mterf8. Both electrophoresis mobility shift assays and chloroplast chromatin immunoprecipitation analysis showed that mTERF8 specifically binds to the 3′ terminal region of psbJ. Transcription analysis using the in vitro T7 RNA polymerase system showed that mTERF8 terminates psbJ transcription. Together, these results suggest that mTERF8 is specifically involved in the transcription termination of the chloroplast gene psbJ.
Plastids in plant cells originated from a single endosymbiotic event, in which the ancestor of the plant lineage engulfed a green photosynthetic cyanobacterium more than one billion years ago. This progenitor retained a small-scale genome during plant evolution (Raven and Allen, 2003; Timmis et al., 2004; Yagi and Shiina, 2012). Although plastid genomes of land plants contain only 120–135 genes (López-Juez, 2007; http://megasun.bch.umontreal.ca/ogmp), their products are essential for chloroplast development and plant survival. Transcription of plastid genomes is mediated by at least two different types of RNA polymerases: a multisubunit Plastid-Encoded RNA Polymerase (PEP) and a single subunit Nucleus-Encoded RNA Polymerase (NEP; Lerbs-Mache, 2011; Börner et al., 2015). PEP is the major RNA polymerase in chloroplasts. It consists of the plastid-encoded core subunits and of numerous nucleus-encoded accessory proteins (Hajdukiewicz et al., 1997; Lopez-Juez and Pyke 2005; Schweer et al., 2010). The core components of the transcriptional machinery share common features with those of prokaryotic organisms (Liere et al., 2011). Numerous nucleus-encoded accessory proteins have been identified from the PEP RNA polymerase complex in plants (Pfannschmidt and Link, 1994; Pfalz et al., 2006; Schweer et al., 2010; Pfalz and Pfannschmidt, 2013; Yu et al., 2014). Thus, the chloroplast transcriptional machinery is a unique hybrid transcription system, which is composed of the remnant prokaryotic subunits as well as of nucleus-encoded eukaryotic components, and is therefore much more complex in plants than in bacteria.
The mitochondrial transcription terminator factor (mTERF) family is characterized by tandem repetition of a 30-amino acid motif called ‘mTERF’. So far, four subfamilies have been identified in vertebrates (Linder et al., 2005). Structure analysis suggested that mTERF proteins have evolved to bind nucleic acids (Byrnes and Garcia-Diaz, 2011). Human (Homo sapiens) mTERF1 was the first identified member of this protein family. It was proposed to promote transcription termination of human mitochondrial genes (Kruse et al., 1989), which gave rise to the mTERF nomenclature. Recent investigations suggest that human mTERF1 is also involved in the termination of antisense transcripts, which would prevent light-strand transcripts from proceeding around the mitochondrial DNA circle and avoid transcriptional interference at the light-strand promoter (Terzioglu et al., 2013). MOC1, an mTERF-like protein in Chlamydomonas reinhardtii, binds specifically to a sequence within the mitochondrial ribosomal RNA (rRNA)-coding module S3 to terminate the antisense transcript as observed for human mTERF1 in vivo (Wobbe and Nixon, 2013). The mTERF family has been characterized in terrestrial plants, but it is absent in fungi and prokaryotes. In Arabidopsis (Arabidopsis thaliana), 35 genes encode mTERF members, 12 of which are localized in chloroplasts (Linder et al., 2005; Babiychuk et al., 2011). Arabidopsis mutants defective in the mTERF proteins SINGLET OXYGEN-LINKED DEATH ACTIVATOR10 (SOLDAT10)/mTERF1 (Meskauskiene et al., 2009) and BELAYA SMERT (BSM)/RUGOSA2 (RUG2)/mTERF4 (Babiychuk et al., 2011; Quesada et al., 2011; Sun et al., 2016) display arrested development of the embryo, and the knockout of mTERF6 (Romani et al., 2015; Zhang et al., 2018b) has serious defects in chloroplast development. mTERFs are also involved in responses to abiotic stress (Meskauskiene et al., 2009; Kim et al., 2012; Quesada, 2016; Xu et al., 2017). However, the molecular functions of mTERFs in plants remain poorly understood. In maize (Zea mays), Zm-mTERF4 is involved in group-II intron splicing, and this function in chloroplast RNA splicing might be conserved in its putative ortholog BSM/RUG2 in Arabidopsis (Hammani and Barkan, 2014). Similar to the role of mTERF4 in splicing in chloroplasts, mTERF15 was shown to be involved in the processing of nad2 pre-RNA in Arabidopsis mitochondria (Hsu et al., 2014). mTERF6 interacts specifically with a sequence within the chloroplast Ile tRNA gene (trnI.2) located in the rRNA operon, and is required for the maturation of this RNA. In vitro, recombinant mTERF6 binds to its plastid DNA target site and can terminate transcription (Romani et al., 2015). mTERF6 was also shown to be involved in the transcription termination of rpoA encoding one core subunit of PEP (Zhang et al., 2018b). mTERF5 was recently characterized as a transcriptional pausing factor to positively regulate transcription of chloroplast psbEFLJ (Ding et al., 2019). The molecular functions of the other mTERFs in plants remain to be elucidated.
Transcription termination includes arrest of RNA biosynthesis, release of the transcript, and dissociation of the RNA polymerase from the DNA template. Proper transcription termination may assist in avoiding interference with transcription of downstream genes and preventing formation of antisense RNAs that can interfere with normal pre-RNA production. Also, it ensures that a pool of RNA polymerases is available for recycling (Greger et al., 2000; Richard and Manley, 2009). Several regulatory mechanisms for transcription termination have been elucidated in bacteria and eukaryotes. In prokaryotes, two different processes exist, namely Rho-independent and Rho-dependent transcription termination. In the former, the termination signal is mainly located on the mRNA that forms a guanine + cytosine-rich stem loop followed by a string of Us, whereas the latter relies on both mRNA elements and transacting factors (Richardson, 2003). In eukaryotes, the mechanisms for terminating transcription are very complex. RNA polymerase II transcription termination is associated with 3′-end processing of the pre-mRNA (Birse et al., 1998; Hirose and Manley 2000; Yonaha and Proudfoot, 2000; Proudfoot, 2004; Buratowski, 2005), and an intact polyadenylation signal is required for transcription termination of protein-coding genes in human and yeast cells (Whitelaw and Proudfoot, 1986; Logan et al., 1987; Connelly and Manley, 1988). By comparison, termination with RNA polymerases III and I is simpler. RNA polymerase III terminates transcription at T-rich sequences located near the mature RNA 3′-end together with a few auxiliary factors (Arimbasseri et al., 2013), whereas RNA polymerase I latter terminates at a major terminator located downstream from the rRNA precursor sequence and requires terminator recognition by specific factors (Kuhn and Grummt, 1989; Lang and Reeder, 1995).
Plastids are specific organelles in plant cells, and the mechanisms of their transcription termination remain obscure. In earlier studies, biochemical assays showed that termination of chloroplast transcription occurs at intrinsic bacterial-like terminators in vitro (Chen and Orozco, 1988). Plastid RNA polymerase is likely to recognize bacterial terminator sequences with specific features under in vivo conditions. Chi et al. (2014) found that a RNA binding protein RHON1 participates in transcriptional termination of rbcL (encoding the large subunit of Rubisco) in Arabidopsis. RHON1 can bind to the mRNA as well as to single-stranded DNA of rbcL, displays RNA-dependent ATPase activity, and terminates transcription of rbcL in vitro (Chi et al., 2014). RHON1 terminates rbcL transcription through an ATP-driven mechanism similar to that of Rho of Escherichia coli.
pTAC15 is one component of the plastid transcriptionally active complex (Pfalz et al., 2006). It was characterized as a member of mTERF family, named mTERF8 (Babiychuk et al., 2011) in Arabidopsis. Our results show that mTERF8 is localized in chloroplast nucleoids, and cofractionates with the core RNA polymerase subunit RpoB. An mterf8 knockout line is affected in plastid gene expression, and displays impaired efficiency of photosystem II electron flow. mTERF8 binds to the DNA of psbJ. The recombinant protein MBP:mTERF8 displays transcription termination activity for psbJ. These data suggest that mTERF8 is involved in transcription termination by specifically binding to the 3′-terminal end of the chloroplast gene psbJ in Arabidopsis.
RESULTS
mTERF8 Is Associated with the PEP Complex in Chloroplasts
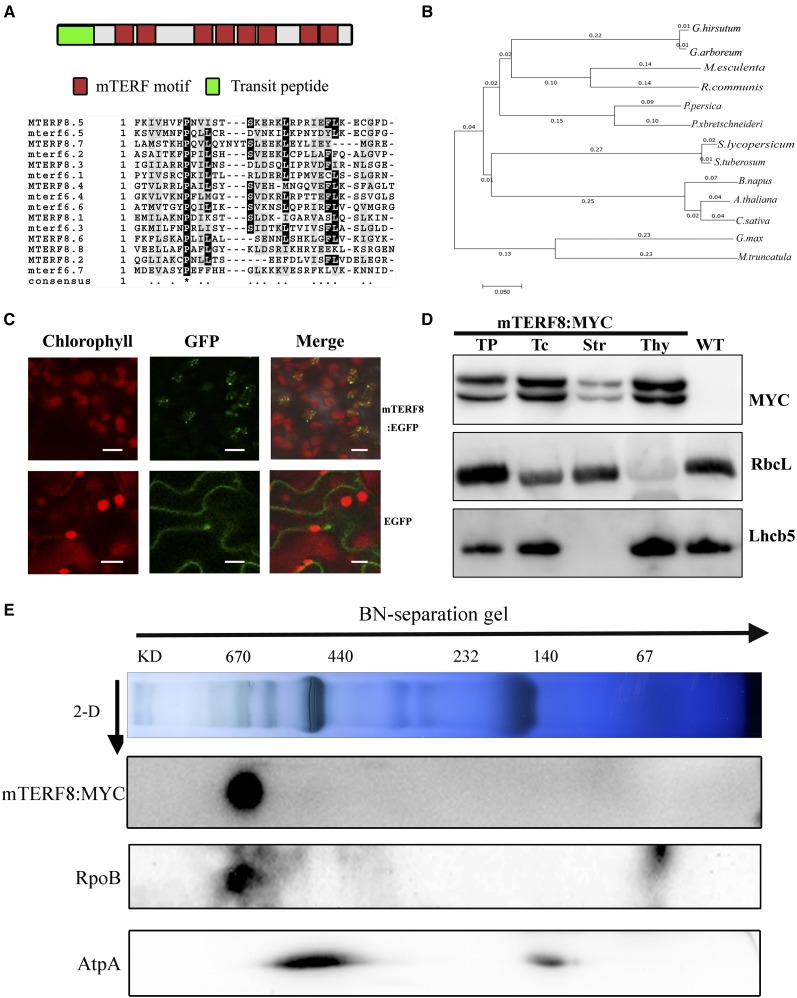
pTAC15 was initially identified in the chloroplast PEP RNA polymerase complex of plants (Pfalz et al., 2006). This protein contains a putative transit peptide of 59 residues at its N terminus and eight mTERF motifs (PPDB; http://ppdb.tc.cornell.edu/). pTAC15 was, thus, also named mTERF8 (Babiychuk et al., 2011). Alignment analysis of the amino acid sequences of the mTERF motifs in mTERF8 with those of other mTERFs revealed the conservation of a Pro residue at the 8th position of the motifs (Fig. 1A). BLAST (BLAST) searching revealed that putative ortholog of mTERF8 exist in angiosperms, gymnosperms, and photosynthetic bacteria (Supplemental Fig. S1). Available putative ortholog of mTERF8 from different species, which represent different plant families, including Brassicaceae (XP_013689181.1), Malvaceae (XP_016726590.1 and XP_017612337.1), Euphorbiaceae (XP_002515871.2 and OAY51230.1), Rosaceae (XP_010482826.1, XP_006347515.1, XP_009361283.1, and XP_020412334.1), Leguminosae (XP_003602380.1 and KRH64303.1), and Solanaceae (XP_004235022.1), were used to construct a phylogenetic tree. This tree revealed that the proteins in the same family form distinct subclades (Fig. 1B), which is consistent with their evolutionary relationship.
Figure 1.
Sequence analysis of mTERF8 and its subcellular localization. A, Scheme of mTERF8 protein, and alignment of eight mTERF motifs. Red boxes represent the mTERF motifs. Green box represents the transit peptide. B, A neighbor-joining phylogenetic analysis of the mTERF8 protein and its putative orthologs in different species, including Camelina sativa (XP_010482826.1), Brassica napus (XP_013689181.1), Gossypium hirsutum (XP_016726590.1), Gossypium arboreum (XP_017612337.1), Manihot esculenta (OAY51230.1), Ricinus communis (XP_002515871.2), Prunus persica (XP_020412334.1), Pyrus xbretschneideri (XP_009361283.1), Glycine max (KRH64303.1), Medicago truncatula (XP_003602380.1), Solanum lycopersicum (XP_004235022.1), and Solanum tuberosum (XP_006347515.1). These species represent different plant families, including Brassicaceae, Malvaceae, Euphorbiaceae, Rosaceae, Leguminosae, and Solanaceae. Scale bar below the tree = branch length. C, Localization of eGFP, and mTERF8:eGFP in tobacco mesophyll cells. Green fluorescence shows GFP fluorescence, red fluorescence indicates chlorophyll autofluorescence, and orange/yellow fluorescence (Merge) shows colocalization. Scale bars = 5 μm. D, mTERF8:MYC is present in both stroma and thylakoids. Proteins of different fractions of chloroplasts from transgenic Arabidopsis seedlings and from wild type were separated by PAGE and immunoblotted with anti-MYC, anti-Lhcb5, and anti-RbcL antibodies. TP, Total protein from the transgenic Arabidopsis seedlings; Tc, Total chloroplast protein; Str, stroma; Thy, thylakoid fraction; WT, total protein from wild type. E, Immunoblot detection of mTERF8, AtpA, and RpoB after two-dimensional gel electrophoresis. AtpA was used as a negative control. Thylakoid membrane proteins from 3-week-old mTERF8:MYC seedlings were fractionated by BN-PAGE in the first dimension and by SDS-PAGE in the second dimension. The approximate molecular masses of the labeled protein complexes are indicated at the top.
Babiychuk et al. (2011) have determined that mTERF8 is targeted to chloroplasts through analysis of GFP fluorescence patterns resulting from mTERF8:GFP expression. In this study, we produced a construct consisting of the full-length mTERF8 coding sequence fused to enhanced GFP (eGFP), and transiently expressed this fusion in tobacco (Nicotiana tabacum) leaf mesophyll cells. Fluorescence microscopy revealed that the fluorescence signal from mTERF8:eGFP colocalized with chlorophyll autofluorescence (Fig. 1C), which confirms its chloroplast localization. GFP fluorescence displayed a punctate pattern in chloroplasts (Fig. 1C). We further produced genomic complementation mTERF8:MYC transgenic plants (see “Materials and Methods”) to investigate mTERF8 subcellular localization (Fig. 1D). Featured proteins of each compartment, such as the light-harvesting complex of PSII (Lhcb5) in the thylakoid and the Rubisco large subunit (RbcL) in the stroma, were examined by immunoblotting. As shown in Figure 1D, as expected, Lhcb5 was mainly present in the thylakoids, whereas RbcL was mainly present in the chloroplast stroma. Immunoblot analysis was performed using an anti-MYC antibody on chloroplast fractions isolated from the transgenic plants. The mTERF8:MYC fusion protein was mainly detected in the thylakoid membrane fraction. mTERF8 was originally identified in the plastid transcription activation complex in Arabidopsis (Pfalz et al., 2006). However, it was not included in the twelve PEP-associated proteins (PAPs) that are tightly associated with the PEP complex (Steiner et al., 2011; Pfalz and Pfannschmidt, 2013). To further examine the association of mTERF8 with the PEP complex, thylakoid membrane proteins from 3-week-old mTERF8:MYC transgenic seedlings were first fractionated by blue native (BN) gel electrophoresis followed by SDS-PAGE in the second dimension. Immunoblotting revealed that mTERF8:MYC comigrates with RpoB, a core component of PEP, in a ∼670 kD complex. To confirm that mTERF8 is specifically associated with the PEP complex, we used ATP synthase as a negative control. Immunoblotting with an antibody against AtpA, a subunit of ATP synthase, showed that this complex migrates at 440 kD where there is no signal corresponding to mTERF8:MYC (Fig. 1E). Taken together these data indicate that mTERF8 is associated with the PEP complex.
Electron Transfer Rate Is Slightly Impaired in the Knockout Mutant of mTERF8
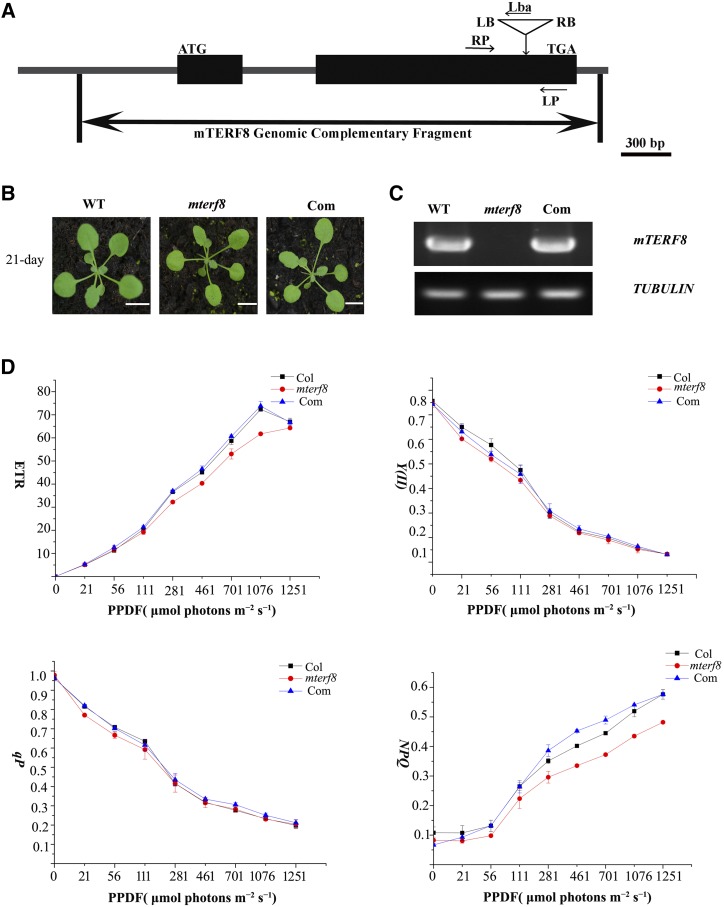
To investigate the function of the mTERF8 gene, we obtained one homozygous transfer DNA (T-DNA) insertion line (SALK_021988) from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus; http://www.arabidopsis.org). The T-DNA is inserted in the first exon (position 17964170 in chromosome 5) in the SALK_021988 line (Fig. 2A). The homozygous plants displayed a wild-type phenotype under standard growth conditions (Fig. 2B). RT-PCR analysis showed that mTERF8 transcripts are not present in the homozygous lines, but are present in the wild type (Fig. 2C). Thus, the mTERF8 gene in the T-DNA homozygous line is not expressed, and this line was named mterf8. To test whether the loss of mTERF8 affects photosynthetic activity, we investigated chlorophyll a fluorescence transients under growth light conditions (90 μmol photons·m−2·s−1). The ratio (Fv/Fm) of variable fluorescence (Fv = Fm − Fo; where Fv is variable fluorescence, Fm is maximum fluorescence, and Fo is minimal fluorescence) to maximum fluorescence in the mterf8 mutant (Fv/Fm = 0.777 ± 0.016) was similar to that in wild type (Fv/Fm = 0.779 ± 0.007). This indicated the presence of an active PSII in the mterf8 mutant. We also measured the light intensity dependence of four chlorophyll fluorescence parameters: light-response curves of nonphotochemical quenching (NPQ), PSII quantum yield, photochemical quenching, and electron transport rate. Nonphotochemical quenching (NPQ), PSII quantum yield, photochemical quenching, and electron transport rate were slightly lower in the mterf8 mutants than in the wild type (Fig. 2D). These data indicate a slightly decreased efficiency of photosynthetic electron flow in the mterf8 mutant. We then investigated whether the absence of mTERF8 affects the accumulation of photosynthetic complexes. Thylakoid membranes were solubilized with 2% n-Dodecyl b-d-maltoside, and membrane protein complexes were separated by BN-PAGE. Our results show that the amount of PSII supercomplexes is slightly decreased in the mterf8 mutant, compared with wild type (Supplemental Fig. S2, A and B). Accumulation of the PSII proteins PsbA and PsbB was slightly reduced (Supplemental Fig. S2C). The other photosynthetic proteins PsaA, PsaD, AtpA, AtpC, CytF, and NdhO accumulated to a similar level as in wild type.
Figure 2.
Characterization of the knockout line of mTERF8. A, Schematic illustrating the genomic structure of mTERF8 and the location of the T-DNA insertion. Black boxes and lines indicate exons and introns, respectively. The T-DNA insertion site is indicated by a triangle. Both left border (LB) and right border (RB) are shown. Lba represents the left border primer of the T-DNA insertion. LP and RP represent the left and right genomic primers, respectively. B, Phenotypes of 21-d-old plants of wild type (WT), mterf8, and the complemented line (Com). Bars = 1 cm. C, RT-PCR analysis of mTERF8 expression in wild type, mterf8, and the complemented line (Com). TUBLIN was used as a control. D, Light-response curves of PSII quantum yield, photochemical quenching (qP), electron transport rate (ETR), and nonphotochemical quenching (NPQ) in the wild type (Col), mterf8 mutant, and the complemented line (Com). Measurements were performed at the following light intensities: 0, 21, 56, 111, 281, 461, 701, 1076, and 1251 μmol photons m−2 s−1. PPDF, photosynthetic photon flux density. Data were presented as mean ± sd. Each data point represents at least 9 independent plants.
To further confirm that the loss of mTERF8 causes the observed mterf8 phenotype, the AT5G54180 genomic sequence, including its 1500-bp upstream and 232-bp downstream regions, was introduced into homozygous mterf8 plants by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). Five out of 41 independent T1 transgenic plants were identified as complemented lines (Fig. 2B). RT-PCR analysis showed that the transcripts of mTERF8 are present in the complemented lines (Fig. 2B). Furthermore, analysis of induction of NPQ and chlorophyll fluorescence kinetics showed that the complemented lines exhibit similar responses as the wild type (Supplemental Fig. S3). Taken together, these results show that the loss of mTERF8 slightly affects the electron transfer rate under normal growth conditions.
Chloroplast Gene Expression Profile in the mterf8 Mutant
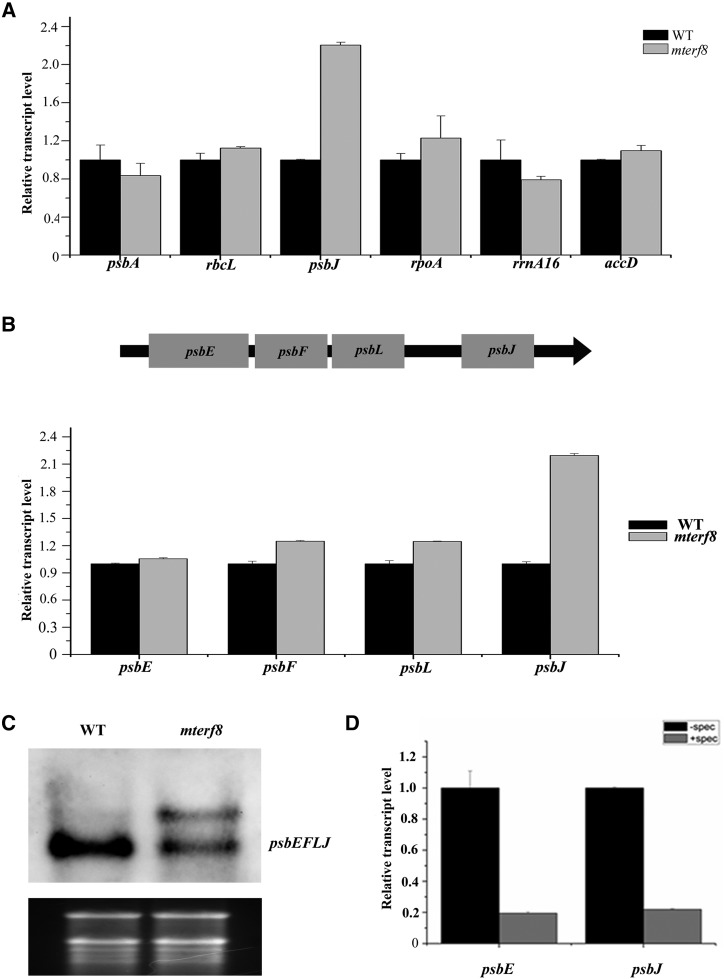
Because mTERF8 is associated with the PEP complex, we analyzed whether the loss of mTERF8 affects the expression profile of plastid genes through reverse transcription quantitative PCR (RT-qPCR) analysis. Our results show that the expression profile of some plastid genes is aberrant in the mterf8 mutant (Fig. 3A; Supplemental Fig. S4). The expression levels of typical PEP-transcribed genes, except rbcL, are reduced. For example, the amounts of the transcripts of two PEP-transcribed genes psbA and psbB were ∼80% of the wild-type level, whereas the transcript abundance of another PEP-transcribed gene rbcL was ∼1.1-fold compared with wild type (Fig. 3A). The abundance of transcripts of NEP-dependent plastid genes, for example, accD and rpoA, was slightly increased (Fig. 3A).The expression of genes transcribed by both NEP and PEP including atpB, atpE, and 16S rRNA were ∼60% to 80% of the wild-type level (Supplemental Fig. S4). Among these plastid transcripts, the most pronounced change was observed for psbJ in the mutant with a 2.4-fold increase compared with wild type (Fig. 3A). In Arabidopsis chloroplasts, the psbE, psbF, psbL, and psbJ genes are part of a large polycistron (Fig. 3B). RT-qPCR analysis showed that the transcripts of psbE, psbF, psbL, and psbJ were increased 1.1-, 1.4-, 1.4-, and 2.4-fold compared with wild type, respectively (Fig. 3B; Supplemental Fig. S4). We also checked the transcript of the psbEFLJ polycistron by RNA blot analysis with a probe spanning the three transcripts. A specific transcript could be observed in the wild type as previously reported (Jin et al., 2018). However, an extra larger transcript was observed in the mterf8 mutant. These data suggest that read-through transcription of the psbJ gene cluster and/or changes in RNA processing occur in the mterf8 mutant (Fig. 3C). It is not clear whether transcription of these genes is performed by PEP polymerase. Spectinomycin is an inhibitor of plastid protein synthesis (Moazed and Noller, 1987). Because the core subunits of PEP are synthesized on plastid ribosomes, growth of Arabidopsis on spectinomycin-containing medium should lead to PEP-deficiency (Zubko and Day, 1998; Swiatecka-Hagenbruch et al., 2007; Chi et al., 2014). Accordingly, we analyzed the transcripts of psbE and psbJ in wild-type plants after spectinomycin treatment and found that their levels were about 20% of those in plants without treatment (Fig. 3D). These results indicate that the psbEFLJ polycistron is transcribed by PEP polymerase. Taken together, our data show that aberrant expression of plastid genes occurs in the mterf8 mutant, especially for the psbJ gene.
Figure 3.
Expression pattern of the chloroplast polycistron psbJ-psbL-psbF-psbE in the knockout line of mTERF8. A, RT-qPCR analysis of chloroplast gene expression in wild type (WT) and the mterf8 mutant. TUBLIN4 was used as an internal control. Relative expression patterns of plastid-encoded genes, including psbA, rbcL, psbJ, rpoA, rrna16, and accD of wild type and mterf8 are shown. B, Schematic illustration of the chloroplast polycistron psbJ-psbL-psbF-psbE in Arabidopsis, and relative expression levels of the chloroplast genes psbJ, psbI, psbF, and psbE in wild type and mterf8. C, Profile of the psbJ-psbL-psbF-psbE polycistron in wild type and mterf8 by RNA blot analysis. The gray values of the transcripts in wild type and mterf8 mutant are 135.8 and 146.5, respectively. The values were measured by Image J software. D, Expression of the chloroplast genes psbJ and psbE in wild type treated with spectinomycin or not. The tublin4 gene was used as control. For the RT-qPCR results, the data represent the mean of triplicate experiments for each gene. Bars represent standard deviations calculated by ABI7300 system SDS software. Two independent biological replicates showed similar results.
psbJ Transcription Termination Is Defective in the mterf8 Mutant
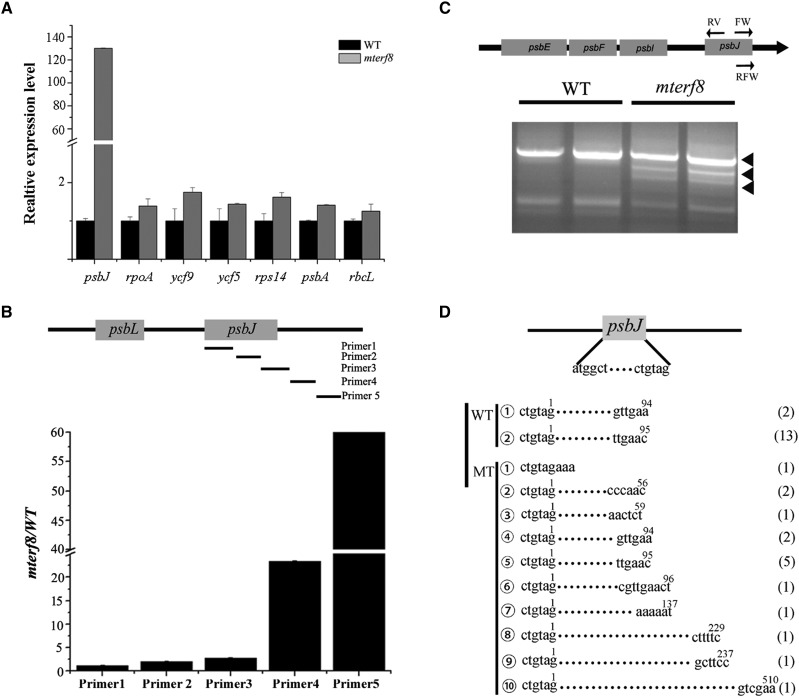
mTERF8 shows structural similarity with human mTERF1 that acts as a transcription termination factor (Supplemental Fig. S5; Zhang, 2008; Byrnes and Garcia-Diaz, 2011). We analyzed the potential transcription read-through in the mutant with specific primers as described (Zhang et al., 2018b). Transcripts from the spacer regions of the ycf9, rbcL, psbA, rps14, ycf5, and psbJ genes were only slightly changed for five of these genes (ycf9, psbA, rpoA, ycf5, and rbcL) in mterf8 relative to the wild type (Fig. 4A). By contrast, the transcripts from the spacer regions of the psbJ gene in the mterf8 mutant were much higher than that in wild type (Fig. 4A). This suggests a potential transcription read-through for the psbJ gene in mterf8. Because the exact transcription termination site of psbJ gene is unknown, we designed five pairs of primers for different regions of psbJ (Fig. 4B) to analyze its transcription termination. Our results showed that the relative levels of the detected transcripts in the mterf8 mutant are close to those of the wild type when three primer sets (primers 1–3) located inside the coding region or within the 3′ termination region were used (Fig. 4C). However, transcript levels of psbJ in the mterf8 mutant were about 30- to 50-fold higher than in wild type when the two primer sets (primers 4 and primers 5) located downstream of the psbJ gene were used (Fig. 4B). These data indicate that transcription of the psbJ polycistron terminates in the region between the primer set 3 and 4, and its transcription termination is defective in the mterf8 mutant.
Figure 4.
Analysis of the defect in psbJ Transcription Termination in the mterf8 mutant. A, Comparison of the expression of genes with the selected primer pairs by RT-qPCR in wild type (WT) and the mterf8 mutant. Error bars indicate standard deviations for triplicate replicates. B, Distribution of the five pairs of primers in the psbJ gene. The names of the five primers are shown on the right. Relative expression level of psbJ transcripts in wild type and mterf8, with the different five sets of primers. C, Schematic illustration of the chloroplast polycistron, and the primers (FW, RV, and RFW) used for cRT-PCR are listed in the Supplemental Table S1. cRT-PCR of chloroplast polycistron psbJ-psbI-psbF-psbE in both wild type and mterf8. The bands of the cRT-PCR products in the mterf8 mutant are indicated by black triangles. D, Schematic illustration of the psbJ genes. Both the 5′-and 3′-end nucleotides of the psbJ transcript are shown. The 3′-end nucleotide sequences (termination sites) of the mature psbJ RNA in both wild type and mterf8 are shown. The numbers in the brackets at the right of the sequences represent the number of sequenced clones that contained each of the sequence variant.
We further identified the termination site of the psbJ gene through a circular RT-PCR (cRT-PCR) analysis. cRT-PCR has been applied to identify the 5′ and 3′ extremities of organelle transcripts (Perrin et al., 2004; Barkan, 2011). Total RNA or polyadenylated RNA are circularized with T4 RNA ligase and reverse transcribed with gene-specific primers. We used cRT-PCR to analyze transcription termination of psbA in wild type and the mterf8 mutant to test the reliability of the method for transcription termination analysis. Transcription termination of psbA was not changed in mterf8 (Supplemental Fig. S6A). Further sequence analysis showed that the psbA transcripts from wild type and the mterf8 mutant terminated at residue 90 or 91 downstream of the psbA stop codon (Supplemental Fig. S6, B and C). This indicated that cRT-PCR can reliably determine transcription termination sites. Subsequently, we analyzed the psbJ termination site in both wild type and the mterf8 mutant through cRT-PCR with the corresponding primers (Fig. 4C). Our results showed the 5′–3′-end ligation products were different between wild-type and mterf8 plants (Fig. 4B). We further determined the termination sites of psbJ through sequence analysis. In the 15 sequenced clones containing the psbJ cRT-PCR product from wild type, the termination site of the psbJ transcript is located at residue 94 or 96 downstream of its stop codon (Fig. 4, C and D). By contrast, among the 16 sequenced clones containing the corresponding product from mterf8, 7 clones contain psbJ transcripts with the same ends as in wild type (Fig. 4, C and D). The remaining clones were variants, with psbJ transcript termination sites at nucleotides 3, 56, 59, 137, 229, 237, or 510 downstream of its stop codon (Fig. 4D). These results indicate that termination of psbJ transcripts is altered in the mterf8 mutant.
mTERF8 Binds to the Chloroplast psbJ Gene
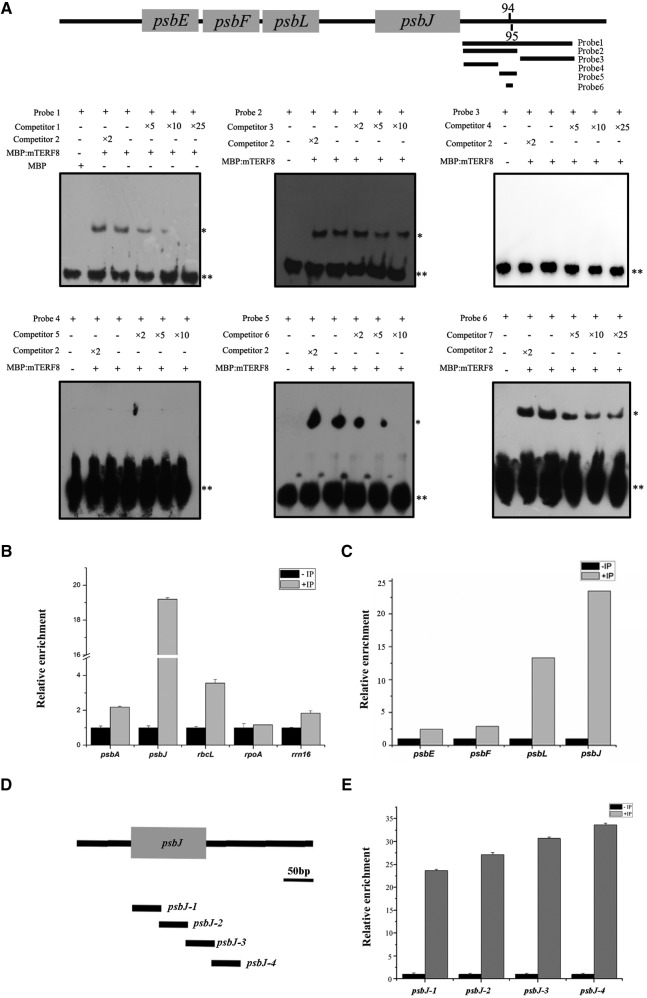
mTERF8 is structurally analogous to human mTERF1 that binds to specific organelle DNA sequences (Terzioglu et al., 2013). Thus, we analyzed whether the mTERF8 protein binds specifically to the psbJ gene. The purified recombinant MBP:mTERF8 protein was incubated with a double stranded DNA (dsDNA) fragment containing the 3′ terminal region of psbJ to perform electrophoretic mobility-shift assays (EMSA). A shifted band that migrated slower than the free DNA probe was detected. The signal of the retarded band became weaker upon addition of increasing amounts of the corresponding unlabeled competitor probe. By contrast, MBP alone did not bind the DNA probe (Fig. 5A, probe 2). To further examine the specificity of binding of mTERF8 to the abovementioned probe, we also test the terminal region of another chloroplast gene, psbA, as a competitor in the EMSA (Fig. 5A). This psbA probe did not compete with the psbJ terminal region, indicating that the recombinant MBP:mTERF8 protein binds specifically to the psbJ probe. Subsequently, we further determined which region of the psbJ 3′-end is bound by the recombinant MBP:mTERF8 protein by testing probes that cover different parts of this region. Our EMSA results show that MBP:mTERF8 binds to the 100-bp fragment downstream of the stop codon of psbJ (Fig. 5A, probe 2), but not to the 100-bp fragment that is further downstream (Fig. 5A, probe 3). In the rice (Oryza sativa) plastid genome, the psbE, psbF, psbL, and psbJ genes are also arranged in a large polycistron, similar to that of Arabidopsis. We checked whether mTERF8 is able to bind to the corresponding region of the rice psbJ terminal region. Our results indicated that this is not the case (Supplemental Fig. S7). Sequence alignments showed that the psbJ terminal region of maize and rice is conserved, but it is different in Arabidopsis (Supplemental Fig. S8). To further determine the binding motif of mTERF8, we split the fragment (Fig. 5A, probe 2) into two fragments, a 55-bp fragment (probe 4) and a 39-bp fragment (probe 5), to perform an EMSA (Fig. 5A). MBP:mTERF8 is able to bind to the 39-bp fragment but not to the 55-bp fragment (Fig. 5A). Based on the above data, we focused on the divergent nucleotides in the 39-bp fragment among Arabidopsis, maize, and rice and prepared the probe 6 with the double tandem nonconserved sequence “TGtcTgattCGagggGG” (lowercase letters represent the nonconserved nucleotides; Supplemental Fig. S8). EMSA shows that MBP:mTERF8 is able to bind to this probe. This data suggest that mTERF8 binds the psbJ 3′ terminal region through this motif. We further tested whether mTERF8 binds to other regions of the polycistron. Three probes corresponding to the coding region of psbJ, psbL, and the intergenic region of psbJ-psbL were designed (Supplemental Fig. S7). EMSA showed that the mTERF8 protein did not bind to any of these probes. This confirms that mTERF8 binds preferentially to the psbJ 3′ terminal region. We further examined whether MBP:mTERF8 binds to the terminal region of another plastid gene, psbA. The psbA probe corresponding to its 3′ terminal region was incubated with the MBP:mTERF8. However, no binding was detected (Supplemental Fig. S7), which is in agreement with the inability of psbA to compete with the binding of MBP-mTERF8 to psbJ (Fig. 5A). Taken together, these data indicate that MBP:mTERF8 binds to the psbJ gene within the 39-bp dsDNA fragment of the psbJ terminal region.
Figure 5.
mTERF8 binds to the chloroplast gene psbJ. A, EMSAs using six DNA probes from psbJ of Arabidopsis. Labeled DNA probes were incubated with recombinant MBP:mTERF8 protein, whereas free biotinylated DNA probes only (Mock) or incubated with empty MBP protein (MBP) served as negative controls. Increasing concentrations of the corresponding competitor probes (competitor 1, 3, 4, 5, 6, and 7) were used, respectively. The terminal region of the psbA gene was used as a competitor probe (competitor 2) in the analysis. The location of the six probes is shown in the schematic of the polycistron. **Bands of free probes; *bands of shifted probes. B, Chloroplast chromatin immunoprecipitation (cpCHIP) analysis of the relative enrichment of the four representative chloroplast genes including psbA, psbJ, rbcL, rpoA, and rrn16. C, cpCHIP analysis of the relative enrichment of the four chloroplast genes psbE, psbF, psbL, and psbJ. D, Distribution of the four different fragments in the psbJ gene. The names of the four fragments (psbJ-1, psbJ-2, psbJ-3, and psbJ-4) are indicated on the right. E, cpCHIP analysis of the relative enrichment of the four different fragments of the psbJ gene. Bars = standard deviations calculated by ABI7300 system SDS software.
To confirm the binding of mTERF8 to the terminal region of psbJ, we further performed a coimmunoprecipitation ChIP analysis with anti-MYC antibody using extracts from the mTERF8:MYC complemented plants. The immunoprecipitated DNA was analyzed by RT-qPCR. Chloroplast genes whose expression is PEP dependent such as psbA, rbcL, and psbJ were enriched in these assays but not rpoA that is not transcribed by PEP (Fig. 5B). The enrichment in these PEP-dependent genes is probably due to the association of mTERF8 with the PEP complex (Fig. 1E). Among these PEP-dependent enriched genes, the highest enrichment was found for the coding region of psbJ (Fig. 5B). We also examined the distribution of mTERF8 along the psbEFLJ polycistron. Here also, the association with mTERF8 with psbJ was the highest (Fig. 5C). Additionally, we used four pairs of primers spanning different regions of psbJ (Fig. 5D). As expected, mTERF8 was enriched with each of the four different segments, and the peak association occurred with fragments of the 3′ terminal part of psbJ (Fig. 5E). Taken together, these data show that mTERF8 binds preferentially near the psbJ termination site.
mTERF8 can Terminate Transcription of the psbJ Gene in vitro
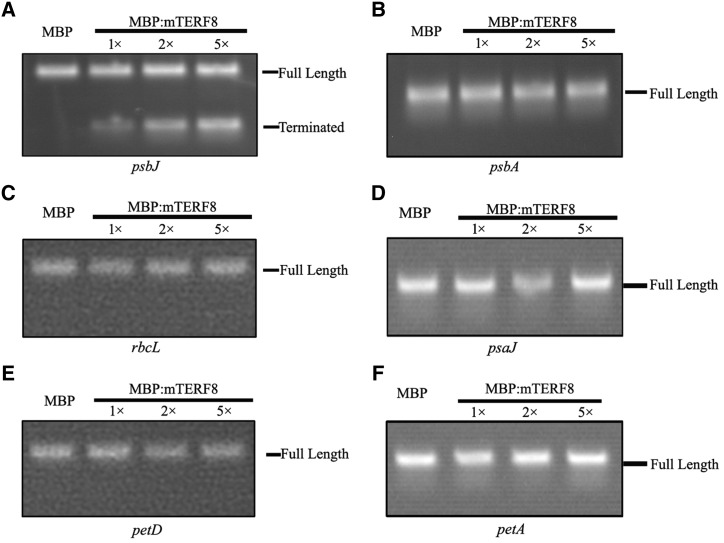
We further investigated whether mTERF8 has transcription termination activity. For this purpose, an assay for transcription termination activity was performed (Supplemental Fig. S9) following previous reports (Prieto-Martín et al., 2004; Romani et al., 2015). We cloned the 100-bp genomic psbJ fragment containing the binding site of mTERF8 into the pGEM vector. After linearization of the plasmid, the construct was transcribed with T7 RNA polymerase in the presence of recombinant MBP:mTERF8. A run-off long transcript (Full-length) was generated with the T7 RNA polymerase. When recombinant MBP:mTERF8 was added to the reaction, an additional shorter transcript corresponding to the psbJ transcript was generated (Fig. 6A). The abundance of this transcript increased gradually with increasing amounts of MBP:mTERF8 in the reaction (Fig. 6A). We also analyzed whether the recombinant MBP:mTERF8 has termination activity for other chloroplast genes. We cloned five chloroplast genes (psbA, rbcL, psaJ, petD, and petA) including their down-stream sequences into the pGEM plasmid. They were linearized as indicated above, and transcribed with T7 RNA polymerase in the presence of MBP:mTERF8 as described above. Only a longer run-off transcript was observed for each gene (Fig. 6, B–F). These data suggest that mTERF8 acts specifically on the 3′ terminal region of psbJ.
Figure 6.
Transcription termination activity of MBP:mTERF8 in vitro. A, Transcription termination activity of MBP:mTERF8 in vitro. MBP:mTERF8 was added to the T7 RNA polymerase transcription system in vitro with the Arabidopsis psbJ gene as a template. B to F, Transcription termination activity of MBP:mTERF8 in vitro. MBP:mTERF8 was added to the T7 RNA polymerase transcription system in vitro with the Arabidopsis psbA, rbcL, psaJ, petD, and petA genes as a template. All the above transcripts were separated by electrophoresis on a 2% agarose gel, then stained with ethidium bromide.
DISCUSSION
mTERF8 Is Associated with the PEP Complex and Is Able to Terminate Transcription
PEP polymerase is a multisubunit bacterial-type enzyme with plastid-encoded core subunits (α2ββ´ß´´) and numerous nucleus-encoded accessory proteins (Schweer et al., 2010). Similar to the case of bacterial RNA polymerase, the nucleus-encoded sigma factors confer promoter specificity on PEP, whereas the PEP core enzyme is responsible for transcription elongation. The accessory proteins include the elongation factor EF-Tu; ribosomal proteins; proteins with DNA/RNA binding domains, such as the PPR, SMR, SAP, OB fold, S1, KOW, NGN, mTERF; or with single-stranded DNA binding motifs (Pfalz et al., 2006; Yu et al., 2014). These proteins are involved in transcription and posttranscriptional processes (Yu et al., 2014). Recent studies on plant mTERF genes have revealed that they are involved in organelle gene expression (Hammani and Barkan, 2014; Hsu et al., 2014; Romani et al., 2015). mTERF8, a member of the mTERF family, was initially named pTAC15 because it was first isolated from the chloroplast PEP RNA polymerase complex in plants (Pfalz et al., 2006). Our results confirm that mTERF8 is localized in chloroplasts (Fig. 1B), consistent with previous proteomics data (Pfalz et al., 2006) and fluorescence microscopy observations (Babiychuk et al., 2011). The fluorescence of the mTERF8:eGFP displayed a punctate pattern (Fig. 1B), as found for PEP-associated proteins such as pTAC3 (Steiner et al., 2011; Yagi et al., 2012), FLN1 (Arsova et al., 2010; Steiner et al., 2011), and FSD3 (Myouga et al., 2008; Steiner et al., 2011). The plastid transcriptional apparatus in chloroplast nucleoids has been proposed to be associated with the thylakoid membranes in mature chloroplasts (Sato, 2001; Jeong et al., 2003; Sato et al., 2003; Karcher et al., 2009; Schweer et al., 2010). Our chloroplast fractionation analysis indicating that most of mTERF8 is associated with thylakoids is in agreement with this proposal (Fig. 2). Our two-dimensional BN-SDS-PAGE electrophoresis and immunoblotting analysis also shows that mTERF8:MYC is present in a supermolecular complex with a molecular mass of 670 kD, which comigrates with RpoB (Zhong et al., 2013), one core subunit of the PEP complex (Fig. 2). Combined, these data indicate that mTERF8/pTAC15 is a component of the PEP complex, which is associated with thylakoids.
Transcription termination is the final step in RNA synthesis, wherein the nascent transcript is released from RNA polymerase. Efficient and timely transcription termination ensures proper gene expression. In human mitochondria, mTERF1, a founder of the mTERF family, is able to bind the organelle genome and is involved in promoting termination of mitochondrial light-strand antisense transcription (Terzioglu et al., 2013). In the unicellular green alga Chlamydomonas reinhardtii, the mTERF protein MOC1 was reported to terminate mitochondrial DNA transcription like human mTERF1. MOC1 binds specifically to an octanucleotide sequence within the mitochondrial rRNA coding module S3, and loss of MOC1 increases read-through transcription at the S3-binding site, thereby causing elevated amounts of antisense RNA in the mutant stm6 (Wobbe and Nixon, 2013). In plants, there are over 30 members of the mTERF family, which are mainly localized in chloroplasts and mitochondria (Babiychuk et al., 2011; Robles et al., 2012; Zhao et al., 2014). The available evidence shows that plant mTERF proteins are involved in posttranscriptional processes, such as organelle RNA splicing (Hammani and Barkan, 2014; Hsu et al., 2014) and chloroplast Ile tRNA maturation (Romani et al., 2015). In vitro assays show that recombinant mTERF6 displays transcription termination activity (Romani et al., 2015). In agreement with this finding, mTERF6 was recently suggested be involved in transcription termination of rpoA (Zhang et al., 2018b). In this study, we report that mTERF8 shares structural features with human mTERF1 (Supplemental Fig. S6). Genetic analysis demonstrates that mTERF8 is involved in transcription termination of psbJ (Figs. 3 and 5). Additionally, recombinant mTERF8 was able to terminate psbJ transcription in vitro (Fig. 6). Taken together, these results indicate that mTERF8, a homolog of human mitochondrial mTERF1, is likely to be involved in transcription termination of psbJ. Although we cannot fully rule out that the loss of mTERF8 promotes initiation of transcription within the terminal region of psbJ, this possibility seems unlikely.
The psbEFLJ operon encodes low-molecular-weight proteins of photosystem II. In tobacco, ΔpsbJ plants grew photoautotrophically, and displayed impaired PSII activity in young leaves. Photosynthetic electron flow was decreased in the absence of PsbJ protein (Regel et al., 2001; Ohad et al., 2004) suggesting a role of PsbJ protein for optimal electron flow. The mterf8 mutant displayed aberrant transcription termination of psbJ (Figs. 3 and 4). Nevertheless, this mutant grows photoautotrophically similar to wild type (Fig. 2). Measurements of several photosynthetic parameters indicate that photosynthetic electron flow is only slightly impaired in mterf8 (Fig. 2). Consistent with these observations, accumulation of the PSII complexes is only weakly reduced in the mutant (Supplemental Fig. S5).
mTERF8 Mediates Preferential Transcription Termination of the Chloroplast Gene psbJ in Arabidopsis
The chloroplast transcription machinery is a unique chimeric system with remnant prokaryotic components and nucleus-encoded eukaryotic components (Yagi et al., 2012). As for the bacterial RNA polymerase, the PEP complex in plants recognizes chloroplast promoters through sigma factors. The association of PEP with sigma factors is enriched at promoter-proximal regions, and declines gradually along the transcribed region, similar to the distribution patterns in bacteria. This suggests that PEP-dependent transcription initiation, elongation, and termination steps are regulated by mechanisms similar to those of bacteria (Yagi et al., 2012). A Rho-like factor, Rhon1, is involved in transcription termination of rbcL in Arabidopsis chloroplasts through an ATP-driven mechanism similar to E. coli Rho factor (Chi et al., 2014). In this work, we found that the eukaryotic-type plastid nucleoid protein mTERF8, which is a component of the PEP complex, can directly bind to the termination region of the psbJ gene as a termination factor (Fig. 5). This region is guanine + cytosine rich and is able to form a stem-loop structure, similar to the terminators of Rho-independent transcription termination in E. coli. Thus, it is likely that mTERF8-mediated transcription termination of psbJ is similar to Rho-independent transcription termination in E. coli.
PEP is the major RNA polymerase responsible for most plastid gene transcription. The psbA gene is transcribed only by the PEP RNA polymerase in Arabidopsis. Knockout of core members of the PEP complex substantially reduced the abundance of psbA transcripts (Pfalz et al., 2006, 2015; Garcia et al., 2008; Myouga et al., 2008; Arsova et al., 2010; Gao et al., 2011; Yagi et al., 2012; Yu et al., 2013, 2018). Although mTERF8 is associated with the PEP complex (Fig. 1) transcription termination of psbA is not affected, although psbA transcript abundance is slightly decreased in mterf8 (Fig. 6; Supplemental Fig. S5). These data suggest that mTERF8 is not responsible for psbA transcription termination. Similarly, MBP:mTERF8 does not terminate transcription of other PEP-dependent genes including rbcL, psaJ, petD, and petA (Fig. 6). Combined, these data suggest that mTERF8 is not a general terminator for PEP-transcribed plastid genes in Arabidopsis. In a similar way, RHON1 only terminates transcription of rbcL but not of other PEP-dependent genes. As in Arabidopsis, the psbE, psbF, psbL, and psbJ genes are also arranged in a large polycistron in the rice plastid genome. However, the sequences of the psbJ terminal regions differ between Arabidopsis and rice (Supplemental Fig. S9). We found that mTERF8 is not able to bind the rice terminal region of the OspsbJ gene (Supplemental Fig. S7), and in vitro analysis showed that the MBP:mTERF8 is not able to terminate OspsbJ transcription. Further analysis revealed that mTERF8 binds to a nonconserved segment of the psbJ terminal sequence region. These data suggest that transcription termination mechanisms, at least in the case of psbJ, have diverged in dicotyledons and monocotyledons. The picture that is emerging from all these studies is that termination of PEP-transcription in chloroplasts is mediated by nucleus-encoded factors that act in a gene-specific manner, at least in the case of psbJ and rbcL. It is noteworthy that several posttranscriptional steps of chloroplast gene expression including RNA turn-over and processing and translation are also modulated by gene-specific factors. Taken together, these results reveal a surprisingly large number of accessory factors involved both in chloroplast transcriptional and posttranscriptional processes.
Molecular Functions of mTERF Proteins in Plants
In mammals, the mTERF family plays important roles in organelle gene expression (Kruse et al., 1989; Park et al., 2007; Cámara et al., 2011; Spåhr et al., 2012). However these mTERF proteins do not actually terminate transcription, as their designation suggests, but appear to function in antisense transcription termination and ribosome biogenesis (Kleine, 2012; Kleine and Leister, 2015). In Chlamydomonas reinhardtii, MOC1, an mTERF-like protein, can bind to the mitochondrial DNA specifically like human mTERF1 in vivo for the regulation of transcription of the mitochondrial genome (Wobbe and Nixon, 2013). The conserved function shared by mTERF genes in Chlamydomonas reinhardtii and humans probably underlies similar mechanisms for mitochondrial transcription regulation (Wobbe and Nixon, 2013). In land plants, the mTERF family has expanded to about 30 members. The additional mTERF members in plants may account for the complicated regulation of the mitochondrial and chloroplast genomes. Plant mTERF proteins share a conserved mTERF motif and a similar three-dimensional structure with human mTERF proteins (Zhao et al., 2014). So far, molecular functions of only a few mTERF proteins in flowering plants have been examined including organellular splicing (Hammani and Barkan, 2014; Hsu et al., 2014), chloroplast Ile tRNA maturation (Romani et al., 2015), and transcriptional pausing (Ding et al., 2019). These mTERF proteins in plants display DNA-binding activity (Romani et al., 2015) and are also able to bind RNA (Hsu et al., 2014). It remains to be seen whether mTERF proteins in plants have similar roles as human mTERFs and MOC1 in Chlamydomonas. The mTERF6 protein shares 25%/46% identity/similarity over a stretch of 194 amino acids with human mTERF1 (Romani et al., 2015), whereas the predicted structure of mTERF8 shares similarity with the human mTERF1 (Supplemental Fig. S6). Both mTERF8 and mTERF6 (Zhang et al., 2018b) are involved in transcription termination of chloroplast genes. Consistent with their functions, recombinant mTERF8 (Fig. 6) and mTERF6 mediate transcription termination in vitro (Romani et al., 2015). It is possible that plant mTERF proteins have an evolutionarily conserved function in transcription termination as in mammals.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Columbia) plants were grown in soil in a greenhouse under a 16-h/8-h light/dark cycle with a light intensity of 120 µmol photons m-2s−1 at 22°C. The mterf8 mutant (SALK_021988) was obtained from the Arabidopsis Biological Resource Center and identified by PCR using genomic DNA and T-DNA specific primers in combination with gene-specific primers (Alonso et al., 2003). The T-DNA insertion site was determined by sequencing of PCR products.
Nucleic Acid Isolation, Complementary DNA Synthesis, RT-qPCR Analysis, and RNA Hybridization
For DNA extraction, leaf tissue was homogenized in lysis buffer containing 200 mm Tris-HCl, pH 7.5; 25 mm NaCl; 25 mm EDTA; and 0.5% (w/v) SDS. After centrifugation, DNA was precipitated from the supernatant with cold isopropyl alcohol. After washing with 70% (v/v) ethanol, the DNA was dissolved in distilled water. For total RNA isolation, frozen tissue was ground in liquid nitrogen; then total RNA isolation was performed with Trizol reagent (Invitrogen, https://www.thermofisher.com/) according to the manufacturer’s instructions. Purification of RNA was performed with QIAGEN RNeasy Mini kit after DNAse I treatment. Complementary DNA (cDNA) synthesis, RT-qPCR analysis, and RNA hybridization were performed as described previously (Yu et al., 2013). All qPCR analysis reactions were performed in triplicate on at least two biological replicates (for oligonucleotide information, see Supplemental Tables S1 and S2).
Genomic Complementation and Subcellular Localization Analysis
For mterf8 mutant complementation, the 3,141-bp genomic sequence without the stop codon was amplified by KOD plus polymerase from the Columbia wild-type genome with specific primers (Supplemental Table S1) and cloned into the modified pCAMBIA1300 vector in frame with four tandem MYC coding sequences. The construct was transformed into Agrobacterium tumefaciens strain GV3101, and introduced into mterf8 mutant plants by the floral-dip method (Clough and Bent, 1998). Transgenic plants were selected for hygromycin resistance and analyzed using PCR analysis. For the mTERF8:eGFP fluorescence analysis, the full-length coding sequence of the mTERF8 gene was amplified by KOD plus polymerase (TOYOBO, http://www.toyobo.co.jp) with gene-specific primers (Supplemental Table S1).The coding sequence of the mTERF8 gene was subcloned into the modified vector pRIN101eGFP (TOYOBO, http://www.toyobo.co.jp) and to produce a fusion encoding mTERF8:eGFP. The construct was transformed into Agrobacterium tumefaciens strain GV3101, and introduced into Nicotiana tabacum by injection. The GFP fluorescence was imaged by laser confocal microscope (Zeiss).
Chlorophyll Fluorescence Analysis
Chlorophyll Fluorescence measurements were performed using an IMAGING-PAM fluorimeter or a MINI-PAM portable chlorophyll fluorimeter (Walz) as described in Duan et al. (2016).
cRT-PCR
For the determination of the psbJ and the psbA transcript ends, circular RT-PCR was performed as described previously (Perrin et al., 2004). Total RNA (5 µg) was circularized using T4 RNA ligase (New England Biolabs). cDNA spanning the junction of the 5′ and 3′ ligated ends was synthesized with the corresponding psbJ and psbA reverse gene primers, respectively (Supplemental Table S1), using SuperScript II RNaseH reverse transcriptase (Invitrogen). The region of circularized psbJ and psbA containing the junction between the original 5′ and 3′ ends was then amplified by RT-PCR using specific primers (Supplemental Table S1), cloned into pMD19-T vector (TAKARA), and sequenced.
MBP:mTERF8 Expression in Escherichia coli and Purification
The sequence encoding the putative mature mTERF8 was amplified from wild-type cDNA with gene-specific primers (Supplemental Table S1), and the PCR product was cloned into the pMALc5X vector (NEB, http://www.neb.com). E. coli BL21-CodonPlus (DE3)-RIPL cells harboring the MBP:mTERF8 construct were induced with 1 mm IPTG, incubated in a shaker overnight at 18°C, and harvested at 7,000 g for 10 min. Purification of MBP:mTERF8 was performed following the manufacturer’s instruction (NEB, http://www.neb.com).
EMSA
The interactions between MBP:mTERF8 and the dsDNA were detected using the LightShift Chemiluminescent EMSA kit (Pierce, http://www.pierce.com). Briefly, the target DNA was amplified with gene-specific primers that were biotin-labeled at the 5′ end (Supplemental Table S1) and then incubated with the purified MBP:mTERF8 in the provided binding reaction system. After incubation for 20 min at 25°C, the samples were resolved on a 6% (w/v) Tris-borate gel in Tris-borate/EDTA buffer and transferred to a nylon membrane. The biotin end-labeled DNA was detected using a streptavidin-horseradish peroxidase conjugate and chemiluminescent substrate. The images were caught by TANON-5500 Chemiluminescent Imaging System (TANON, http://www.bio-tanon.com.cn).
cpChIP
cpChIP assay was performed following the protocol described in Yagi et al. (2012). One gram of 6-d-old transgenic mTERF8:MYC Arabidopsis seedlings was used for cpChIP assays with MYC antibody (Millipore, http://www.merckmillipore.com/).
Transcription Termination Assay in vitro
The sequences of the intact psbJ, rbcL, psaJ, petD, and petA genes were amplified from the wild-type genome (for oligonucleotide information, see Supplemental Table S1) and cloned individually into the pGEM-T easy vector (Promega, http://www.promega.org). In vitro transcription was performed in 20-µL reactions containing 100 ng of each vector; 1 µm mTERF8; ATP, CTP, UTP, and GTP (0.5 mm each); 20 units of T7 RNA polymerase (Fermentas); 4 µL of 5× transcription buffer; and 20 units of RNase Inhibitor (Fermentas). Reactions were incubated for 1 h at 25°C, and the transcripts were subjected to 2% agarose gel electrophoresis.
Chloroplast Subfractionation, Immunoblotting, SDS-PAGE, and BN-PAGE Analyses
Two- to three-week-old seedlings were homogenized in ice-cold buffer I (0.33 m sorbitol; 0.02 m Tricine/KOH, pH 8.4; 5 mm EGTA, pH 8.35; 5 mm EDTA, pH 8.0; 10 mm NaHCO3). The homogenate was successively filtered through a double layer of Miracloth then 100- and 40-µm sieves. The filtered solution was centrifuged in a swing-out rotor at 2000 g for 5 min at 4°C. Intact chloroplasts were further resuspended in buffer II (0.33 m sorbitol; 5 mm MgCl2; 2.5 m EDTA, pH 8.0; 20 mm HEPES/KOH, pH 7.6). Thylakoid membranes and stromal proteins were prepared from isolated intact chloroplasts. The intact chloroplasts were resuspended in buffer III (5 mm MgCl2-6H2O; 25 m EDTA, pH 8.0; 20 mm HEPES/KOH, pH 7.6). After centrifugation at 4°C, the supernatant was used as stroma and the sediment as thylakoid fraction. Total protein preparation and immunoblot analyses were performed as described previously (Yu et al., 2013). BN-PAGE analysis was performed as described previously (Peng et al., 2006; Zhang et al., 2018a). One gram of 2- to 3-week-old seedlings was homogenized in ice-cold HMSN buffer (0.4 m Suc, 10 mm NaCl, 5 mm MgCl2-6H2O, 10 mm HEPES) and then filtered through multilayer microcloth. The isolated thylakoid pellets were suspended in resuspension buffer (25 mm Bis-Tris-HCl, pH 7.0; 1% [w/v] n-Dodecyl b-d-maltoside, and 20% [w/v] glycerol) at 1.0 mg chlorophyll/mL. After incubation at 4°C for 5 min and centrifugation at 12,000 g for 10 min, one-tenth volume of loading buffer (100 mm Bis-Tris-HCl, pH 7.0, 0.5 m 6-amino-n-caproic acid, 5% [w/v] Serva blue G, and 30% [w/v] glycerol) was added to the supernatant and applied to 0.75-mm 4% to 12% acrylamide gradient gels in a Tannon vertical electrophoresis at 4°C. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer for 30 min and layered onto 1-mm 10% SDS polyacrylamide gels containing 6 m urea. After electrophoresis, the proteins were transferred to nitrocellulose membranes, probed with antibodies against MYC (cwbio, http://www.cwbiotech.com/) and RpoB, and visualized by ECL. Polyclonal antibodies against proteins involved in photosynthesis used in this study were obtained from Agrisera (http://www.agrisera.com).
Accession Numbers
The accession numbers of the genes mentioned in this article are given below, and their sequence data can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Arabidopsis thaliana (mTERF8/AT5G54180, NP_200229.1), Camelina sativa (XP_010482826.1), Brassica napus (XP_013689181.1), Gossypium hirsutum (XP_016726590.1), Gossypium arboreum (XP_017612337.1), Manihot esculenta (OAY51230.1), Ricinus communis (XP_002515871.2), Prunus persica (XP_020412334.1), Pyrus xbretschneideri (XP_009361283.1), Glycine max (KRH64303.1), Medicago truncatula (XP_003602380.1), Solanum lycopersicum (XP_004235022.1), Solanum tuberosum (XP_006347515.1), psbA (AtCG00020), rbcL(AtCG00490), psaJ(AtCG00630), petD(AtCG00730), petA(AtCG00540), Oryza sativa (OspsbJ, NC_031333), Zea mays (ZmpsbJ, NP_043038.1).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Sequence alignment of mTERF8 with its putative orthologs.
Supplemental Figure S2. Accumulation of photosynthetic complexes and photosynthetic proteins in wild type and the mterf8 mutant.
Supplemental Figure S3. Photosynthetic parameters of wild type, mterf8, and the complemented line (Com).
Supplemental Figure S4. Plastid gene expression profile of wild type and mterf8.
Supplemental Figure S5. Predicted structure of mTERF8 and protein purification.
Supplemental Figure S6. Circularized RT-PCR of chloroplast psbA transcripts in wild type and mterf8.
Supplemental Figure S7. EMSA with psbA and OspsbJ probes.
Supplemental Figure S8. Sequence alignment of psbJ terminal sequences.
Supplemental Figure S9. Schematic of transcription termination activity analysis in vitro.
Supplemental Table S1. Primers used for mTERF8 functional analysis in this study.
Supplemental Table S2. Primers for RT-qPCR analysis of plastid gene expression used in this study.
Acknowledgments
We thank Arabidopsis Biological Resource Center (The Ohio State University, Columbus), which kindly offered the transgenic Arabidopsis lines (SALK_021988).
Footnotes
This work was supported by the National Natural Scientific Foundation of China (NSFC) (grant nos. 31570232 and 31370271) and the Shanghai Science and Technology Committee (18DZ2260500, 17DZ2252700).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ (2013) Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta 1829: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustün S, Melzer M, Petersen K, Lein W, Börnke F (2010) Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: Evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22: 1498–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011) Studying the structure and processing of chloroplast transcripts. Methods Mol Biol 774: 183–197 [DOI] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ (1998) Coupling termination of transcription to messenger RNA maturation in yeast. Science 280: 298–301 [DOI] [PubMed] [Google Scholar]
- Börner T, Aleynikova AY, Zubo YO, Kusnetsov VV (2015) Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim Biophys Acta 1847: 761–769 [DOI] [PubMed] [Google Scholar]
- Buratowski S. (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Byrnes J, Garcia-Diaz M (2011) Mitochondrial transcription: How does it end? Transcription 2: 32–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara Y, Asin-Cayuela J, Park CB, Metodiev MD, Shi Y, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. (2011) MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab 13: 527–539 [DOI] [PubMed] [Google Scholar]
- Chen LJ, Orozco EM Jr. (1988) Recognition of prokaryotic transcription terminators by spinach chloroplast RNA polymerase. Nucleic Acids Res 16: 8411–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, He B, Manavski N, Mao J, Ji D, Lu C, Rochaix JD, Meurer J, Zhang L (2014) RHON1 mediates a Rho-like activity for transcription termination in plastids of Arabidopsis thaliana. Plant Cell 26: 4918–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connelly S, Manley JL (1988) A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev 2: 440–452 [DOI] [PubMed] [Google Scholar]
- Ding S, Zhang Y, Hu Z, Huang X, Zhang B, Lu Q, Wen X, Wang Y, Lu C (2019) mTERF5 acts as a transcriptional pausing factor to positively regulate transcription of chloroplast psbEFLJ. Mol Plant 12: 1259–1277 [DOI] [PubMed] [Google Scholar]
- Duan Z, Kong F, Zhang L, Li W, Zhang J, Peng L (2016) A bestrophin-like protein modulates the proton motive force across the thylakoid membrane in Arabidopsis. J Integr Plant Biol 58: 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, Yang ZN (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H (2008) An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J 53: 924–934 [DOI] [PubMed] [Google Scholar]
- Greger IH, Aranda A, Proudfoot N (2000) Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 97: 8415–8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Barkan A (2014) An mTERF domain protein functions in group II intron splicing in maize chloroplasts. Nucleic Acids Res 42: 5033–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY (2014) Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS One 9: e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SY, Rose A, Meier I (2003) MFP1 is a thylakoid-associated, nucleoid-binding protein with a coiled-coil structure. Nucleic Acids Res 31: 5175–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Fu M, Duan Z, Duan S, Li M, Dong X, Liu B, Feng D, Wang J, Peng L, Wang HB (2018) LOW PHOTOSYNTHETIC EFFICIENCY 1 is required for light-regulated photosystem II biogenesis in Arabidopsis. Proc Natl Acad Sci USA 115: E6075–E6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D, Köster D, Schadach A, Klevesath A, Bock R (2009) The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant 2: 1223–1232 [DOI] [PubMed] [Google Scholar]
- Kim M, Lee U, Small I, des Francs-Small CC, Vierling E (2012) Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell 24: 3349–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T. (2012) Arabidopsis thaliana mTERF proteins: Evolution and functional classification. Front Plant Sci 3: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D (2015) Emerging functions of mammalian and plant mTERFs. Biochim Biophys Acta 1847: 786–797 [DOI] [PubMed] [Google Scholar]
- Kruse B, Narasimhan N, Attardi G (1989) Termination of transcription in human mitochondria: identification and purification of a DNA binding protein factor that promotes termination. Cell 58: 391–397 [DOI] [PubMed] [Google Scholar]
- Kuhn A, Grummt I (1989) 3′-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev 3: 224–231 [DOI] [PubMed] [Google Scholar]
- Lang WH, Reeder RH (1995) Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc Natl Acad Sci USA 92: 9781–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: Regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Liere K, Weihe A, Börner T (2011) The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J Plant Physiol 168: 1345–1360 [DOI] [PubMed] [Google Scholar]
- Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM (2005) A family of putative transcription termination factors shared amongst metazoans and plants. Curr Genet 48: 265–269 [DOI] [PubMed] [Google Scholar]
- Logan J, Falck-Pedersen E, Darnell JE Jr., Shenk T (1987) A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci USA 84: 8306–8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Pyke KA (2005) Plastids unleashed: Their development and their integration in plant development. Int J Dev Biol 49: 557–577 [DOI] [PubMed] [Google Scholar]
- López-Juez E. (2007) Plastid biogenesis, between light and shadows. J Exp Bot 58: 11–26 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K (2009) A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1987) Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327: 389–394 [DOI] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K (2008) A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20: 3148–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I, Dal Bosco C, Herrmann RG, Meurer J (2004) Photosystem II proteins PsbL and PsbJ regulate electron flow to the plastoquinone pool. Biochemistry 43: 2297–2308 [DOI] [PubMed] [Google Scholar]
- Park CB, Asin-Cayuela J, Cámara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. (2007) MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 130: 273–285 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Lange H, Grienenberger JM, Gagliardi D (2004) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res 32: 5174–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T (2015) ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol 206: 1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T (2013) Essential nucleoid proteins in early chloroplast development. Trends Plant Sci 18: 186–194 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Link G (1994) Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol Biol 25: 69–81 [DOI] [PubMed] [Google Scholar]
- Prieto-Martín A, Montoya J, Martínez-Azorín F (2004) Phosphorylation of rat mitochondrial transcription termination factor (mTERF) is required for transcription termination but not for binding to DNA. Nucleic Acids Res 32: 2059–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Quesada V. (2016) The roles of mitochondrial transcription termination factors (MTERFs) in plants. Physiol Plant 157: 389–399 [DOI] [PubMed] [Google Scholar]
- Quesada V, Sarmiento-Mañús R, González-Bayón R, Hricová A, Pérez-Marcos R, Graciá-Martínez E, Medina-Ruiz L, Leyva-Díaz E, Ponce MR, Micol JL (2011) Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J 68: 738–753 [DOI] [PubMed] [Google Scholar]
- Raven JA, Allen JF (2003) Genomics and chloroplast evolution: What did cyanobacteria do for plants? Genome Biol 4: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regel RE, Ivleva NB, Zer H, Meurer J, Shestakov SV, Herrmann RG, Pakrasi HB, Ohad I (2001) Deregulation of electron flow within photosystem II in the absence of the PsbJ protein. J Biol Chem 276: 41473–41478 [DOI] [PubMed] [Google Scholar]
- Richard P, Manley JL (2009) Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JP. (2003) Loading Rho to terminate transcription. Cell 114: 157–159 [DOI] [PubMed] [Google Scholar]
- Robles P, Micol JL, Quesada V (2012) Unveiling plant mTERF functions. Mol Plant 5: 294–296 [DOI] [PubMed] [Google Scholar]
- Romani I, Manavski N, Morosetti A, Tadini L, Maier S, Kühn K, Ruwe H, Schmitz-Linneweber C, Wanner G, Leister D, Kleine T (2015) A member of the arabidopsis mitochondrial transcription termination factor family is required for maturation of chloroplast transfer RNAIle(GAU). Plant Physiol 169: 627–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. (2001) Was the evolution of plastid genetic machinery discontinuous? Trends Plant Sci 6: 151–155 [DOI] [PubMed] [Google Scholar]
- Sato N, Terasawa K, Miyajima K, Kabeya Y (2003) Organization, developmental dynamics, and evolution of plastid nucleoids. Int Rev Cytol 232: 217–262 [DOI] [PubMed] [Google Scholar]
- Schweer J, Türkeri H, Kolpack A, Link G (2010) Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription - recent lessons from Arabidopsis thaliana. Eur J Cell Biol 89: 940–946 [DOI] [PubMed] [Google Scholar]
- Spåhr H, Habermann B, Gustafsson CM, Larsson NG, Hallberg BM (2012) Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc Natl Acad Sci USA 109: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner S, Schröter Y, Pfalz J, Pfannschmidt T (2011) Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol 157: 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xu D, Liu Z, Kleine T, Leister D (2016) Functional relationship between mTERF4 and GUN1 in retrograde signaling. J Exp Bot 67: 3909–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatecka-Hagenbruch M, Liere K, Börner T (2007) High diversity of plastidial promoters in Arabidopsis thaliana. Mol Genet Genomics 277: 725–734 [DOI] [PubMed] [Google Scholar]
- Terzioglu M, Ruzzenente B, Harmel J, Mourier A, Jemt E, López MD, Kukat C, Stewart JB, Wibom R, Meharg C, et al. (2013) MTERF1 binds mtDNA to prevent transcriptional interference at the light-strand promoter but is dispensable for rRNA gene transcription regulation. Cell Metab 17: 618–626 [DOI] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Whitelaw E, Proudfoot N (1986) α-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human α2 globin gene. EMBO J 5: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe L, Nixon PJ (2013) The mTERF protein MOC1 terminates mitochondrial DNA transcription in the unicellular green alga Chlamydomonas reinhardtii. Nucleic Acids Res 41: 6553–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Leister D, Kleine T (2017) Arabidopsis thaliana mTERF10 and mTERF11, but not mTERF12, are involved in the response to salt stress. Front Plant Sci 8: 1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T (2012) Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc Natl Acad Sci USA 109: 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Shiina T (2012) Evolutionary aspects of plastid proteins involved in transcription: the transcription of a tiny genome is mediated by a complicated machinery. Transcription 3: 290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonaha M, Proudfoot NJ (2000) Transcriptional termination and coupled polyadenylation in vitro. EMBO J 19: 3770–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Huang C, Yang ZN (2014) Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front Plant Sci 5: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN (2013) TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol Plant 148: 408–421 [DOI] [PubMed] [Google Scholar]
- Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Zhao TT, Ye LS, Cheng L, Wu YQ, Huang C, Yang ZN (2018) pTAC10, an S1-domain-containing component of the transcriptionally active chromosome complex, is essential for plastid gene expression in Arabidopsis thaliana and is phosphorylated by chloroplast-targeted casein kinase II. Photosynth Res 137: 69–83 [DOI] [PubMed] [Google Scholar]
- Zhang L, Pu H, Duan Z, Li Y, Liu B, Zhang Q, Li W, Rochaix JD, Liu L, Peng L (2018a) Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor. Plant Cell 30: 1770–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Cui YL, Zhang XL, Yu QB, Wang X, Yuan XB, Qin XM, He XF, Huang C, Yang ZN (2018b) A nuclear-encoded protein, mTERF6, mediates transcription termination of rpoA polycistron for plastid-encoded RNA polymerase-dependent chloroplast gene expression and chloroplast development. Sci Rep 8: 11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Cai M, Zhang X, Li Y, Zhang J, Zhao H, Kong F, Zheng Y, Qiu F (2014) Genome-wide identification, evolution and expression analysis of mTERF gene family in maize. PLoS One 9: e94126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C (2013) Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25: 2925–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Day A (1998) Stable albinism induced without mutagenesis: A model for ribosome-free plastid inheritance. Plant J 15: 265–271 [DOI] [PubMed] [Google Scholar]