GA signaling is required for FR-induced shoot elongation in P. tabuliformis seedlings, and there are different regulatory targets for FR-mediated GA biosynthesis between conifers and angiosperms.
Abstract
Gibberellin (GA) is known to play an important role in low red/far-red (R:FR) light ratio-mediated hypocotyl and petiole elongation in Arabidopsis (Arabidopsis thaliana). However, the regulatory relationship between low R:FR and GAs remains unclear, especially in gymnosperms. To increase our understanding of the molecular basis of low R:FR-mediated shoot elongation in pines and to determine whether there is an association between low R:FR and GAs action, we explored the morphological and transcriptomic changes triggered by low R:FR, GAs, and paclobutrazol (PAC), a GAs biosynthesis inhibitor, in Pinus tabuliformis seedlings. Transcriptome profiles revealed that low R:FR conditions and GAs have a common set of transcriptional targets in P. tabuliformis. We provide evidence that the effect of low R:FR on shoot elongation in P. tabuliformis is at least partially modulated by GAs accumulation, which can be largely attenuated by PAC. GAs are also involved in the cross talk between different phytohormones in the low R:FR response. A GA biosynthesis gene, encoding ent-kaurenoic acid oxidase (KAO), was strongly stimulated by low R:FR without being affected by GAs feedback regulation or the photoperiod. We show that GA signaling is required for low R:FR-induced shoot elongation in P. tabuliformis seedlings, and that there are different regulatory targets for low R:FR-mediated GA biosynthesis between conifers and angiosperms.
Light, an essential energy source and informational signal, is one of the most important environmental factors for plants (Jiao et al., 2007). As sessile organisms, shade-intolerant plants have evolved the ability to sense shading or surrounding light competitors and respond rapidly with shade-avoidance responses (SARs; e.g. elongation of the stem, hypocotyl, or petiole; Jiao et al., 2007). Far-red light (FR) is known to be an important light cue during this process (Ballaré et al., 1990). In the past few decades, significant progress has been made toward understanding the molecular regulatory network of FR-mediated SARs (Carriedo et al., 2016). A small subset of basic helix-loop-helix transcription factors, known as phytochrome-interacting factors (PIFs), act as a signaling hub in this process (Leivar and Quail, 2011).
A variety of plant hormones are known to be involved in the morphological adaptation response to low R:FR (de Wit et al., 2016). Gibberellin (GA) is known to be the dominant hormone in the promotion of stem elongation in plants. Therefore, it not surprising that GA plays an important role in the plant light response (Lau and Deng, 2010). Interestingly, as opposed to auxin, few genes involved in the GA signaling pathway are directly regulated by PIFs (Zhang et al., 2013). A recent study showed that DELLA proteins (the central negative regulators of GA signaling) can directly promote the degradation of PIFs (Li et al., 2016). Therefore, GA plays a role upstream of PIFs and auxin in FR-mediated SAR.
Over the past ten years, a molecular framework of the coordinated regulation of hypocotyl elongation by light and GA has been explored. Red light (R) and FR are both sensed by the phytochrome (phy) light receptor in plants. FR or low R:FR releases PIFs by inactivating phy, eventually leading to growth enhancement. In addition, the activity of PIFs is suppressed by DELLA proteins through a direct protein-protein interaction (de Lucas et al., 2008; Feng et al., 2008), demonstrating that the DELLA proteins block PIF transcriptional regulation activity and also mediate their degradation (Li et al., 2016). Thus, GAs regulate hypocotyl elongation by suppressing the suppressor through the GA-GID1-DELLA module (Harberd et al., 2009).
The current model integrates phy and GA signals with the regulation of hypocotyl elongation through PIFs, which is the central hub of the signaling network. PIFs function as negative regulators of phy-mediated light responses (Shin et al., 2009; Stephenson et al., 2009) and accumulate in the nucleus in darkness to promote hypocotyl elongation (Leivar et al., 2008). Light stimulates DELLA proteins accumulation by reducing GA levels (Achard et al., 2007). DELLA proteins can function as transcriptional repressors by blocking the activity of PIFs, resulting in inhibition of PIF-mediated gene expression and hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008). Under low R:FR conditions, the biosynthesis of GA is stimulated mainly through transcript levels of GA20ox and GA3ox genes in angiosperms (García-Martinez and Gil, 2001; Hisamatsu et al., 2005), targeting the DELLA proteins for degradation via the 26S proteasome pathway (Djakovic-Petrovic et al., 2007; Leone et al., 2014). This process releases PIF3/4 from the negative effect of DELLA proteins in the nucleus, leading to modulation genes involved in cell elongation to promote internode or hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008). These changes in light quality trigger a series of responses known as the shade-avoidance syndrome (SAS; Pierik and de Wit, 2014; Fraser et al., 2016; Ballaré and Pierik, 2017).
Previous studies have shown that a completely functional GA system is required for phyB-mediated stem elongation (Peng and Harberd, 1997) and that active GA level was upregulated by FR-enriched light, which has been observed in many species (García-Martinez and Gil, 2001). However, the underlying molecular mechanism of FR regulation of GA biosynthesis in shoot elongation remains unclear. Two studies have shown that the GA biosynthetic genes GA20ox1 and GA20ox2 in Arabidopsis (Arabidopsis thaliana; Hisamatsu et al., 2005) and GA20ox and GA3ox in Rumex palustris (Pierik et al., 2011) are upregulated by low R:FR. Nevertheless, this effect may be restricted to petioles. When compared with Arabidopsis and model crop species where considerable progress has been made in elucidating the molecular basis of the low R:FR response (García-Martinez and Gil, 2001), details of the molecular mechanism underlying low R:FR-GA interactions in conifers remain poorly understood. Several studies by physiological ecologists have clearly demonstrated a typical SAS induced by supplemental FR in conifers (de la Rosa et al., 1998; Razzak et al., 2017). Recently, transcriptome analysis has shown that the underlying mechanisms of shade avoidance may be diverse in gymnosperms in contrast with that in angiosperms (Ranade et al., 2019).
To understand the effect of low R:FR on the response to GA in conifers, here we investigated the molecular basis of low R:FR-mediated shoot elongation in pines and the role of GAs in low R:FR-stimulated shoot elongation. We explored the morphological and transcriptomic changes triggered by FR, GAs, and paclobutrazol (PAC), a GA biosynthesis inhibitor (Rademacher, 2000), in Pinus tabuliformis seedlings. To account for errors caused by slow growth and the physiological response to light in conifers (Burgin et al., 1999), we analyzed shoots for an extended time (>100 d) rather than using a hypocotyl model system for 1 week (Fernbach and Mohr, 1990; Burgin et al., 1999; Ranade and García-Gil, 2013). We found that low R:FR action on shoot elongation in P. tabuliformis is at least partially modulated by GA accumulation. Furthermore, we show that the GA biosynthesis gene PtKAO2 is regulated by low R:FR in P. tabuliformis without being affected by GA feedback regulation or the photoperiod. These results show that GA signaling is required in conifers for FR-induced shoot elongation, and increases our understanding of the connection between FR and GA in plants.
RESULTS
Both Low R:FR and GA Promote Shoot Elongation in P. tabuliformis Seedlings
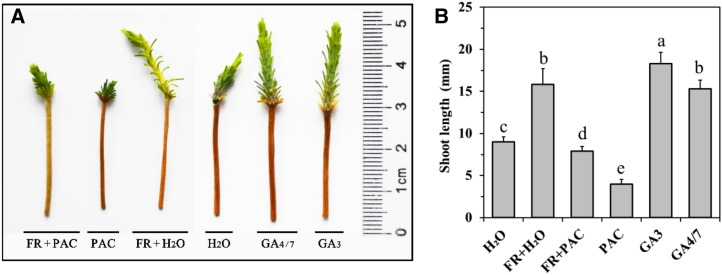
We examined the effects of low R:FR, GA3, GA4/7, and PAC on the regulation of shoot elongation in fully developed P. tabuliformis seedlings. GA3 and GA4/7 weekly irrigation caused a similar growth stimulation effect, suggesting that both are active GAs in pines. Low R:FR significantly promoted shoot growth in P. tabuliformis seedlings under control conditions over 15 weeks (Fig. 1). GA and low R:FR caused shoot elongation through an increase in stem unit length rather than stem unit number (Fig. 1). This result supports the occurrence of FR-mediated SAR in pines (Razzak et al., 2017).
Figure 1.
Effect of low R:FR, GA3, GA4/7, and PAC on shoot elongation in P. tabuliformis seedlings. A, Morphological changes of neoformed shoot elongation treated with low R:FR, GA3, GA4/7, and PAC for 15 weeks. FR, low R:FR light conditions. Needles were cut off before taking pictures; (A) was assembled from two photos which were taken at same time. B, Average shoot length of seedlings with different treatment for 15 weeks. This experiment was repeated at least four times, and data represent the means with SE of more than 16 seedlings. Significant differences between treatments are indicated with different letters.
Low R:FR Mediated Shoot Elongation Associated with GAs Synthesis Pathway
Under normal light conditions, PAC treatment resulted in dwarfing and a dark-green phenotype (Fig. 1), similar to other GA-deficient syndromes in angiosperm plants. Low R:FR rescued growth inhibition by PAC, whereas PAC was seen to partially antagonize low R:FR-induced shoot elongation (Fig. 1). We measured the gibberellin content of the seedlings using a liquid chromatography–mass spectrometry system. The results show that bioactive GA4 and GA1 are the highest abundance GAs in the P. tabuliformis seedlings, and the low R:FR treatment plants have a higher level of both GA4 and GA1 than control seedlings (Table 1). This suggests that the low R:FR and GA response networks are at least partially overlapping and that the GA signaling pathway is required for low R:FR-mediated shoot growth enhancement.
Table 1. Concentrations of Bioactive GAs in P. tabuliformis.
The GA concentration is given in nanograms GAs per gram of fresh needle. The values are the average of two biological replicates, with each sample containing all the needles of five seedlings.
| Treatment | GA Concentration (ng/g) | |
|---|---|---|
| GA1 | GA4 | |
| Control | 0.18 ± 0.02 | 0.30 ± 0.09 |
| Low R:FR | 0.28 ± 0.09 | 0.52 ± 0.02 |
Low R:FR and GAs Have a Common Set of Transcriptional Targets in P. tabuliformis Seedlings
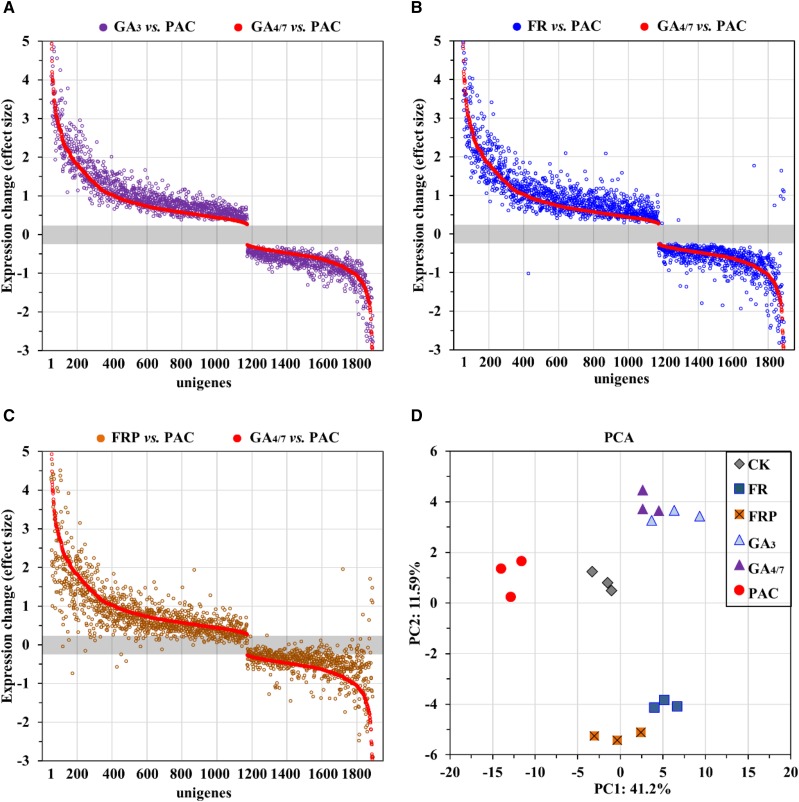
To explore whether there was overlap between the low R:FR and GA response networks, we analyzed global gene expression in P. tabuliformis in response to low R:FR, GAs, and PAC. We found that 26.7% of the differentially expressed genes (P < 0.01) between low R:FR versus Control overlapped with those in GA3 versus Control and GA4 versus Control. However, because most GA-regulated genes have a weak expression response (Nemhauser et al., 2006; Goda et al., 2008; Kakei et al., 2015), we used a high-sensitivity threshold for screening differentially expressed genes as fold change > 1.2 and P < 0.05 compared with the GA-deficient conditions (PAC treatment). We identified 1,842 genes that responded to GAs and were differently expressed between GA3 versus PAC, GA4/7 versus PAC, and low R:FR versus PAC (Supplemental Fig. S1; Supplemental Table S1). We found that 60.9% of these genes were up-regulated by GAs and, at the same concentration, GA3 treatment induced greater gene expression changes than GA4/7 (Fig. 2A). Expression changes of these genes were in the same direction and exhibited a very similar profile in the presence of GA3 or GA4/7 (Fig. 2A), and 99.57% of genes were affected in the same direction under low R:FR conditions (Fig. 2B), supporting that they are GA-regulated genes. In addition, low R:FR conditions combined with PAC resulted in a GA-regulatory like gene expression shift, and the majority of these genes (98.53%) changed in the same direction as that observed under GA regulation (Fig. 2, C and D; Supplemental Fig. S2, heatmap). These results suggest that low R:FR shares a common transcriptional module with GA response networks.
Figure 2.
Transcriptomic changes in response to low R:FR, GAs, and PAC in P. tabuliformis seedlings. A total of 1,842 unigenes were shown, which were differently expressed between GA3 versus PAC, GA4/7 versus PAC, and low R:FR versus PAC with an expression fold change > 1.2, P < 0.05. FR, low R:FR light conditions; FRP, simultaneous low R:FR conditions and PAC irrigation; CK, water control. Unigenes were rearranged in descending order according to the expression fold changes between GA3 and PAC treatment. Data are mean values of three biological replicates. Corresponding expression changes of these unigenes between GA4/7 versus PAC (A), low R:FR versus FRP (B), and FRP versus PAC (C) are shown. D, Principal component analysis based on each of the 1,842 differently expressed genes. Each symbol represents a single sample (n = 3 replicate samples per treatment).
GA-Mediated Plant Hormone Cross Talk in the Low R:FR Response in P. tabuliformis Seedlings
Previous reports have suggested that many plant hormones are involved in the FR response (Kebrom and Mullet, 2016). We analyzed genes involved in hormone responses that are affected by both GA and low R:FR (Fig. 3). Auxin is a key player activated by FR (Procko et al., 2016). Based on our results, auxin-response genes were primarily up-regulated by low R:FR and regulated by GAs, suggesting that GA plays a role upstream of auxin in FR-mediated SAR. Meanwhile, brassinosteroid (BR)- and salicylic acid–response genes were enhanced, whereas ethylene (ET)-response genes were repressed by both GAs and low R:FR. These results suggest that GA plays an important role in plant hormone cross talk in FR-mediated SAR.
Figure 3.
Differential expression of genes involved in the phytohormone response. Among the 1,842 unigenes regulated by low R:FR and GA, 53 genes with functions related to the response of phytohormones were selected and shown. FR, low R:FR light conditions; FRP, simultaneous low R:FR conditions and PAC irrigation; CK, cytokinin; ABA, abscisic acid; JA, jasmonic acid; SA, salicylic acid. The brown histogram (right) indicates genes that are expressed at higher levels under GA or low R:FR conditions and the stellblue histogram (left) indicates genes repressed by GA or low R:FR. The effect size (analogous to fold change) values were obtained from the sleuth software (Pimentel et al., 2017). Values represent the mean of three biological replicates.
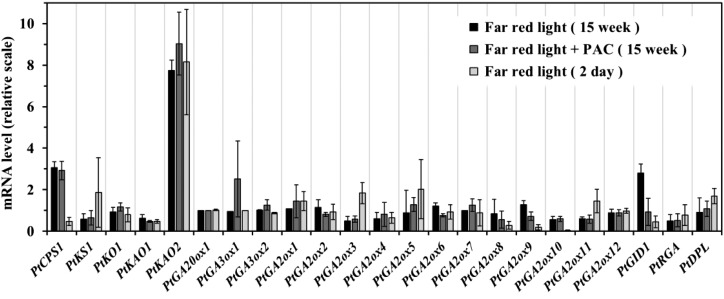
A Putative GA-Biosynthesis Gene was Strongly Stimulated by Low R:FR in P. tabuliformis Seedlings
To explore the role of low R:FR in the GA signaling pathway, we analyzed the expression changes of 20 previously identified putative GA-metabolism genes (Niu et al., 2014), as well as genes encoding the GA receptor PtGID1 and two DELLA proteins (Du et al., 2017). We found that a putative ent-kaurenoic acid oxidase-encoding gene, PtKAO2, was strongly stimulated by low R:FR (Fig. 4). Moreover, the stimulation effect of low R:FR was not inhibited by PAC and showed a stable response over a relatively short period (Fig. 4).
Figure 4.
The expression response of genes involved in GA metabolism and signaling pathways to low R:FR in P. tabuliformis seedlings. The expression levels of each gene are relative to the value of the water control group (set at 1). Seedlings were grown under a 14:10 h photoperiod. FR, low R:FR light conditions. The mean of three biological replicates is plotted with SE.
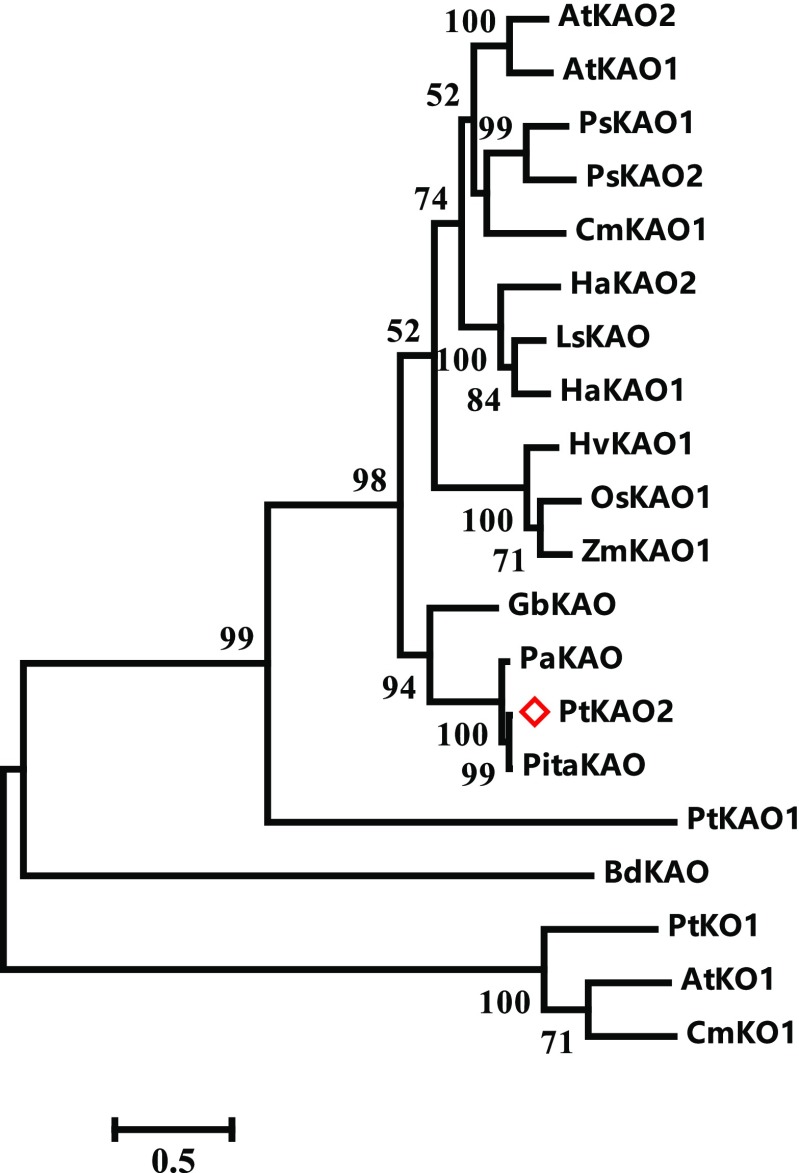
The phylogenetic relationship between putative KAOs in P. tabuliformis and functionally identified KAOs in other plants was analyzed with the recently identified bacterial BdKAO (Nett et al., 2017) as outgroup. We found that KAOs from monocotyledons and dicotyledons were separated into different subclusters, and PtKAO2 is more similar to the angiosperm KAO than PtKAO1 (Fig. 5). The amino acid sequence alignment suggests that KAO was conserved during the evolution of conifers and angiosperms. PtKAO2 shares 81% amino acid identity with at least one of the identified angiosperm KAOs in this study (Supplemental Fig. S3).
Figure 5.
Phylogenetic relationship of PtKAOs in P. tabuliformis and KAOs identified from angiosperms. PtKAO1, -2, and ZmKAO1 (Winkler and Helentjaris, 1995), OsKAO1 (Sakamoto et al., 2004), HvKAO1 (Helliwell et al., 2001), LsKAO (Sawada et al., 2008), HaKAO1 and 2 (Fambrini et al., 2011), PsKAO1 (Davidson et al., 2003), CmKAO1 (Helliwell et al., 2000), AtKAO1 and 2 (Helliwell et al., 2001), and the putative KAOs from other gymnosperms as Ginkgo biloba, Picea abies, and Pinus taeda were used to build the maximum likelihood tree. The bacteria BdKAO (Nett et al., 2017) and the related cytochrome P450 monooxygenase (CYP701A) ent-kaurene oxidases (KOs) were used as outgroup. The red diamond symbol indicates the P. tabuliformis gene that is identified in this study. The horizontal branch lengths are proportional to the estimated number of amino acid substitutions per residue. Bootstrap values were obtained from 1,000 bootstrap replicates.
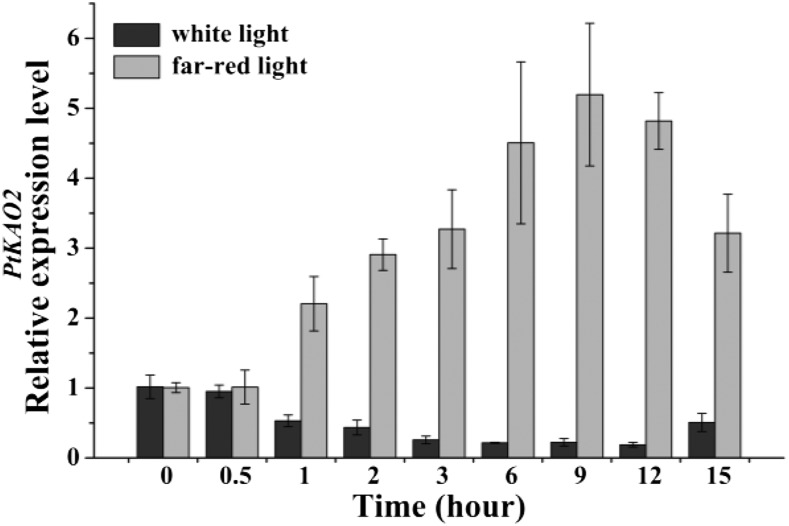
To explore the response pattern of PtKAO2 to low R:FR, the expression of PtKAO2 was detected by reverse transcription quantitative PCR at 0, 0.5, 1, 2, 3, 6, 9, 12, and 15 h (in darkness for 1 h) from the beginning of the light period during a 14:10–h photoperiod (Fig. 6). Expression of PtKAO2 did not show a violent oscillation pattern under regular conditions, and white light repressed this pattern throughout the day. PtKAO2 showed a rapid response to both white light and FR, and the effect was significantly attenuated following 1 h of darkness. However, the stimulation effect of low R:FR was gradually enhanced and peaked within 9 h. This result suggests that 7 h after exposed to low R:FR is an appropriate time for sample collection for RNA-sequencing (RNA-seq) analysis in this study.
Figure 6.
Expression pattern of PtKAO2 under white light and low R:FR in P. tabuliformis. The expression level of each sample was relative to the value of the samples at 0 h, which were collected at the beginning of light treatment (set to 1). The mean of three biological replicates is plotted with SE.
Expression Profile of PtKAO2 in P. tabuliformis
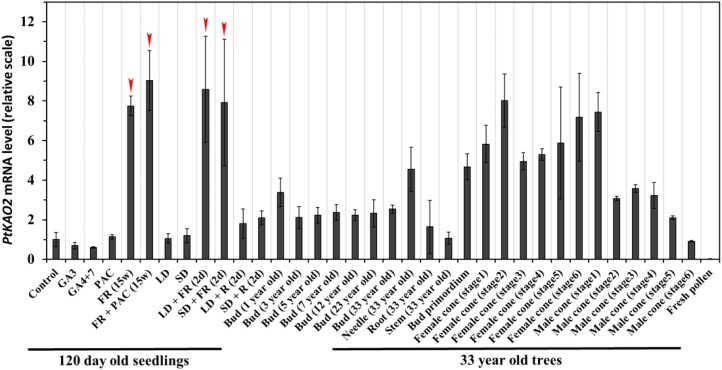
To explore the regulatory and functional roles of PtKAO2 in P. tabuliformis, we explored its expression under different conditions and during development. With the exception of pollen, it was ubiquitously expressed in all tissues (Fig. 7). Similar to AtKAO in Arabidopsis (Regnault et al., 2014), PtKAO2 is more strongly expressed in reproductive organs. Interestingly, low R:FR induced PtKAO2 expression in seedling needles to a high level, similar to that in female cones (Fig. 7). This activation was not affected by short-day or long-day photoperiods. When compared with the effect of low R:FR, GAs, and PAC, R only showed a minor regulatory role on the expression of PtKAO2 (Fig. 7). These data confirm that low R:FR is an important regulator of PtKAO2 expression in P. tabuliformis.
Figure 7.
Expression profile of PtKAO2 in P. tabuliformis under different conditions and during development. The expression level of each gene relative to the water control group (set to 1) is shown. The mean of three biological replicates is plotted with SE. FR,low R:FR light conditions; LD, long-day conditions; SD, short-day conditions; R: high R:FR light conditions. The arrows indicate the specific stimulation of PtKAO2 expression by low R:FR conditions.
Overexpression of PtKAO2 Complements Dwarfing in kao1 kao2 Arabidopsis Plants
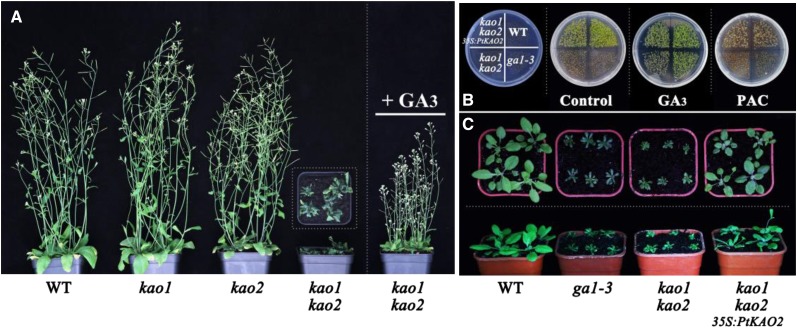
In order to confirm the biochemical function of PtKAO2, we overexpressed this gene in Arabidopsis because comparable genetic tools are still not available in P. tabuliformis. In Arabidopsis, ent-kaurenoic acid oxidase is encoded by two paralogous genes with functional redundancy, designated KAO1 and KAO2 (Regnault et al., 2014). Because kao1 and kao2 single mutants are indistinguishable from wild-type plants (Fig. 8A), we generated the kao1 kao2 double mutant by crossing the corresponding single mutant lines. kao1 kao2 plants exhibited typical nongerminating dwarf phenotypes, which were rescued by exogenous application of GA3 (Fig. 8A). In the kao1 kao2 genetic background, overexpression of PtKAO2 also rescued the growth and germination defects of kao1 kao2 plants and seeds (Fig. 8, B and C). These results suggest that PtKAO2 plays a similar role as KAOs in Arabidopsis in the GA biosynthesis pathway.
Figure 8.
Overexpression of PtKAO2 rescue the growth and germination defects of kao1 kao2 double mutant in Arabidopsis. A, Arabidopsis kao1 kao2 double mutant shows a severe dwarf phenotype that is partially rescued by 5 μm GA3. B, PtKAO2 transgenic lines in a kao1 kao2 double mutant background display a normal germination phenotype similar to that of wild type (WT) under control, 5 μm GA3, and 10 μm PAC treatment. C, Arabidopsis kao1 kao2 double mutant exhibits a dark-green dwarf phenotype typical of ga1-3, which is partially complemented by PtKAO2 overexpression. The parts of the figure were assembled from more than one photo, and the outlines of each original photo are indicate by dotted lines.
DISCUSSION
GAs are pivotal growth-promoting regulators in plants. Therefore, it is not surprising that GAs play an important role in light-regulated growth (Kamiya and García-Martinez, 1999). Indeed, a completely functional GA system is required for phyB-mediated stem elongation (Peng and Harberd, 1997), and the active GA level is up-regulated by low F:FR conditions, which has been observed in many species (García-Martinez and Gil, 2001). However, the underlying molecular mechanism of low F:FR regulation of GA biosynthesis in shoot elongation remains unclear. Two studies have shown that the GA biosynthetic genes GA20ox1 and GA20ox2 in Arabidopsis (Hisamatsu et al., 2005) and GA20ox and GA3ox in Rumex palustris (Pierik et al., 2011) are up-regulated by low R:FR; however, this effect may be restricted to petioles. Genes associated with GA metabolism were analyzed based on transcriptome data in Arabidopsis (Supplemental Table S2), and no consistent differences were observed between the data sets. For example, no GA-biosynthesis genes were differently expressed more than 2-fold after 1, 3, or 24 h of low F:FR treatment (Leivar et al., 2012). By contrast, AtGA20ox1 was significantly down-regulated in seedlings exposed to low F:FR for 4 h in the AtGenExpress data set (http://jsp.weigelworld.org). AtGA20ox1 and AtGA20ox2 showed no differential expression in cotyledons, hypocotyls, or roots under dark conditions (Ma et al., 2005). In another experiment, only AtGA3ox1 showed a 2-fold change after exposure to low F:FR for 1 h, but was subsequently down-regulated after prolonged FR-rich treatment (Sessa et al., 2005). Under the monochromatic FR treatment, some GA2oxs, which contribute to bioactive GA reduction, were significantly up-regulated in both hypocotyls and cotyledons in Arabidopsis (Kirchenbauer et al., 2016; Supplemental Table S2). Recently, one study indicated that inactive GA12 (synthesized by KAO) rather than active GAs is the major mobile GA in vivo (Regnault et al., 2015). These apparently contradictory results could be explained by the regulation of GA20ox and GA3ox genes downstream of GA12 in a highly tissue-specific manner for the local synthesis of active GAs, which are sensitive to the feedback regulation of GAs (Hedden and Thomas, 2012). Moreover, the transcription of GA20ox and GA3ox are under circadian control and are very sensitive to environment cues (Wu et al., 1996; Xu et al., 1997), which results in complex observations. Overall, the association between FR and GA biosynthesis remains poorly understood.
In this study, we found that a putative GA biosynthesis gene, PtKAO2, was strongly stimulated by both short- and long-term low R:FR treatment in the conifer P. tabuliformis, and that this regulation was not affected by feedback regulation of GAs. We confirmed the biological functions of PtKAO2 in Arabidopsis; its ectopic expression complemented the GA-defective phenotype of kao1 kao2 plants, indicating that it encodes a functional ent-kaurenoic acid oxidase that functions in the GA biosynthesis pathway. Interestingly, in wild-type Arabidopsis, neither KAO1 nor KAO2 was consistently regulated by low R:FR in hypocotyls and cotyledons. However, KAO and normal GA synthesis are required for SAS in Arabidopsis, since the low R:FR response was significantly attenuated in kao1 kao2 plant. This suggests that GAs probably play an important role in both gymnosperms and angiosperms, but the regulatory mechanisms may differ.
In Arabidopsis, an annual plant with a very short life cycle, the response to low R:FR is a very rapid process (Tao et al., 2008; Leivar et al., 2012). After the start of low R:FR treatment, DELLA protein degradation and the corresponding petiole elongation were observed within 2 h (Djakovic-Petrovic et al., 2007). The expression changes of PIF3-like 1 (PIL1) in response to low R:FR were detectable within 5 min and increased more than 30-fold within 1 h (Salter et al., 2003). Indeed, the phy response to light is significantly slower in conifers than angiosperms (Burgin et al., 1999). However, although phenotypic changes were barely detectable, the expression of PtKAO2 was fully induced by low R:FR within 2 d in P. tabuliformis (Fig. 3). We further shortened the analysis time after the start of low R:FR treatment. Surprisingly, PtKAO2 was rapidly up-regulated within 1 h and peaked within 9 h (Fig. 6), which is indicative of a highly responsive transcriptional network in conifers.
Many phytohormones play a role in SAR through interactions between different hormone pathways (Carabelli et al., 2007; Kebrom and Mullet, 2016). Under shade conditions, the biosynthesis of auxin, BRs, ET, and GAs are regulated by the interactions of phyB with PIFs in SAR (Ballaré and Pierik, 2017). Auxin is required for shade-induced growth in different plants (Procko et al., 2014; Müller-Moulé et al., 2016), and phyB plays a key role in the regulation of auxin signaling (Morelli and Ruberti, 2000). Previous studies have shown that auxin plays a major role in promoting hypocotyl elongation. In low R:FR shade, auxin is stimulated via the TAA1-dependent pathway (Tao et al., 2008) and the Pr form of phyB-mediated PIFs accumulation and targeted transcription of YUCCA genes that catalyze the rate-limiting step in auxin biosynthesis (Hornitschek et al., 2012; Li et al., 2012; Müller-Moulé et al., 2016). Auxin-responsive defective mutations can suppress the constitutive SAR phenotype of phyB, leading to reduced hypocotyl growth (Tao et al., 2008). In a phyB mutant of sorghum (Sorghum bicolor), the auxin response was enhanced (Kebrom and Mullet, 2016). BR, another growth-promoting hormone, was shown to play a role in SAR in a nonredundant manner with auxin (Keuskamp et al., 2011). Interestingly, we show that the response of both auxin and BR was enhanced by GA in P. tabuliformis, indicating that low R:FR linked the actions of auxin and BR at least partly through the GA pathway. In tobacco (Nicotiana tabacum) and Rumex palustris, ET functions as a positive regulator in SAR, acting upstream of GA (Pierik et al., 2004; Pierik et al., 2011). However, in the sorghum phyB mutant, several ET-biosynthesis genes are down-regulated (Kebrom and Mullet, 2016). GA-induced hypocotyl or stem elongation is associated with regulation of PIFs activity independent of DELLA degradation (Djakovic-Petrovic et al., 2007; Leone et al., 2014). In this study, we identified ET-response genes that are repressed by low R:FR and GA. In contrast with auxin, ET signaling may not be a common factor in SAR across different species. In addition, past studies have shown that the phytohormone jasmonic acid plays a key role in the activation of plant immunity (Ballaré et al., 2012; Cerrudo et al., 2012; Cerrudo et al., 2017; Liu et al., 2019). In this study, several genes involved in cytokinin, abscisic acid, jasmonic acid, and salicylic acid were regulated by both low R:FR and GA (Fig. 3), indicating that GA plays a role in the cross talk between different phytohormones during shade signaling.
SAR is undesirable in agricultural production because carbon resources are diverted to stem elongation rather than agronomically important tissues (Carriedo et al., 2016). However, SAR may be beneficial for tree breeding, specifically for trees with a very slow growth rate. In P. tabuliformis, a conifer tree, the stimulation of PtKAO2 by low R:FR was not affected by photoperiod or GA-feedback regulation. Therefore, low R:FR could easily be applied under any conditions without strict control of illumination, and could be combined with exogenous GA application to promote seedling growth.
MATERIALS AND METHODS
Plant Material and Treatments
The seeds of Pinus tabuliformis were obtained from a primary clonal seed orchard located in Pingquan City, Hebei Province, China (40°99’ N, 118°45’ E, 560 m above sea level). Seeds were sown on sphagnum moss soaked with water and then germinated for 15 d in a growth chamber under conditions of 22°C 14-h/10-h light/dark (controls photosynthetically active radiation [PAR] = 30.438 µmol m−2 s−1, light-emitting diode 50 W). Light quality was measured using an AvaSpec-ULS2048XL-EVO spectrometer (Avantes, http://www.avantes.com), which is able to measure wavelengths 200–1160 nm with a back-thinned CCD detector. The detector was held at the same height as needles to record the quality of light from the light-emitting diode source. Seedlings were then transferred in 8-cm diameter plastic pots with one individual and cultured in a mixture of prefertilized peat moss, perlite, and vermiculite (2:1:1, v/v) at 22°C with 14:10–h (normal), 16:8–h (long-day), or 10:14–h (short-day) photoperiods. The full developmental seedlings (Ranade and García-Gil, 2013) of P. tabuliformis were irrigated weekly with water, 50 μm PAC, 50 μm GA3, and 50 μm GA4+7 as well as low R:FR treatment (FR, peak wavelength at 723.21 nm, PAR = 15.440 µmol m−2 s−1). In our previous experiments, we found that if the monochromatic far-red light treatment area was completely delimited by surrounding it with aluminum foil, the seedlings grew poorly or even died. Therefore, we provided supplementary white light (PAR = 5.707 µmol m−2 s−1) in subsequent far-red light treatments to ensure normal growth of the seedlings. The PAR of FR supplemented with white light was 21.147 µmol m−2 s−1. Therefore, FR treatment refers to low R:FR light conditions in this study for all experiments. Each treatment contained 16 seedlings with similar growth rates. The exact growth chamber and light conditions that were adopted in this study are show in detail in Supplemental Figure S4.
The Arabidopsis kao1 (Stock CS835691) and kao2 (Stock CS827299) mutants were obtained from the The Arabidopsis Information Resource (http://arabidopsis.org). We generated the kao1 kao2 double mutants through crossing of the corresponding single mutant lines. PCR-based screening and sequencing were used for confirmation of double mutant progeny. For proliferation of nongerminating severe dwarf kao1 kao2 double mutants, 5 μm GA3 was applied to promote germination and growth.
Reverse Transcription Quantitative PCR Assays
The seedlings were grown for 3 weeks under normal conditions and were then transferred to low R:FR light conditions. Needles were sampled from the beginning of the light period at regular intervals. Total RNA was extracted using TRIzol reagent (Invitrogen,) for gene expression analysis. Two micrograms of total RNA was used to synthesize single-strand complementary DNA using a M-MLV Reverse Transcriptase kit (Promega). qPCR assays were performed using a QuantiTect SYBR Green PCR Kit (Qiagen). PCR primers are listed in Supplemental Table S3.
RNA-Seq Analysis
Total RNA quantity and purity were assessed using the Nano Photometer spectrophotometer (Implen), and RNA concentration was measured using the Qubit RNA Assay kit in Qubit 2.0 Fluorometer (Life Technologies). RNA integrity was assessed using the RNA Nano 6000 Assay kit of the Bioanalyzer 2100 system (Agilent Technologies). mRNA was fragmented into small pieces using divalent cations under increased temperatures. The cleaved RNA fragments were then reverse-transcribed to create the final cDNA library in accordance with the protocol for the mRNA-Seq sample preparation kit (Illumina). The average insert size for the paired-end libraries was 200–300 bp. The pooled libraries were sequenced on the Illumina Hiseq Xtenplatform (2 × 150 bp) using the paired-end module. The transcript abundances were estimated using kallisto software (Bray et al., 2016), and the differential expression analysis was performed using sleuth software (Pimentel et al., 2017).
Phylogenetic Analysis
The software ProtTest3 (Darriba et al., 2011) was used to determine the best-fit model of evolution. The “LG+G” model was finally selected, and the MAFFT tool (Katoh and Standley, 2013) was used for multiple sequence alignment. The maximum likelihood tree based on the LG+G model was obtained using PhyML 3.0 software (Guindon et al., 2010). Bootstrap values were obtained by 1,000 bootstrap replicates.
Plasmid Construction and Plant Transformation
The complete CDS of PtKAO2 (GenBank: KJ158983) was amplified from the complementary DNA of P. tabuliformis needles by PCR. The products were cloned into pBI121, which contained the nptII gene as a selectable marker, which generated the p35S::PtKAO2 construct. The construct was transformed into Agrobacterium GV3101 strain via the freeze/thaw method. kao1 kao2 double mutant plants were transformed by floral dip. The transgenic lines were screened by kanamycin, and then confirmed by PCR and sequencing using primers that are listed in Supplemental Table S4.
Data Availability
The authors declare that all data supporting the findings of this study are available within the article and Supplementary Material online or are available upon request from the corresponding author.
Accession Numbers
The GenBank accession numbers of proteins mentioned in this paper are AHW42467 (PtKAO2), AHW42466 (PtKAO1), NP_001320198 (AtKAO1), NP_001189657 (AtKAO2), AAO23063 (PsKAO1), AAO23064 (PsKAO2), AAG41777 (CmKAO1), CBV36748 (HaKAO2), BAG71199 (LsKAO), XP_022022633 (HaKAO1), Q9AXH9 (HvKAO1), XP_015643774 (OsKAO1), NP_001105586 (ZmKAO1), AAT28221 (GbKAO), AAC28890 (BdKAO), AHW42468 (PtKO1), NP_197962 (AtKO1), and AAG41776 (CmKO1). The RNA-seq data that support the findings of this study have been deposited in the China National GeneBank Sequence Archive (https://db.cngb.org/cnsa/) of China National GeneBank Database with accession number CNP0000737.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Overlap of genes between GA3 vs. PAC, GA4/7 vs. PAC, and low R:FR vs. PAC.
Supplemental Figure S2. Heatmap of 1,842 GA-response genes under different treatments.
Supplemental Figure S3. Amino acid sequence alignment of PtKAO2 and ent-kaurenoic acid oxidases (KAOs) from angiosperms.
Supplemental Figure S4. The irradiance system and light qualities used in this study.
Supplemental Table S1. Expression level (TPM) and functional description of 1,842 GA-response genes under different treatments.
Supplemental Table S2. Expression data of GA-metabolism genes in response to low far/far-red light in Arabidopsis.
Supplemental Table S3. Primers used in reverse transcription quantitative PCR analysis.
Supplemental Table S4. Primers of PCR-based screening and sequencing for genetic background confirmation.
Footnotes
This work was supported by Central Universities Fundamental Research Funds (2019ZY27) and the National Natural Science Foundation of China (NSFC) (31600535, 31770713, 31870651).
References
- Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, Harberd NP (2007) DELLAs contribute to plant photomorphogenesis. Plant Physiol 143: 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Mazza CA, Austin AT, Pierik R (2012) Canopy light and plant health. Plant Physiol 160: 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R (2017) The shade-avoidance syndrome: Multiple signals and ecological consequences. Plant Cell Environ 40: 2530–2543 [DOI] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Sánchez RA (1990) Far-red radiation reflected from adjacent leaves: An early signal of competition in plant canopies. Science 247: 329–332 [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527 [DOI] [PubMed] [Google Scholar]
- Burgin M, Whitelam G, Sanchez RJ (1999) A light-regulated pool of phytochrome and rudimentary high-irradiance responses under far-red light in Pinus elliottii and Pseudotsuga menziesii. J Exp Bot 50: 831–836 [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriedo LG, Maloof JN, Brady SM (2016) Molecular control of crop shade avoidance. Curr Opin Plant Biol 30: 151–158 [DOI] [PubMed] [Google Scholar]
- Cerrudo I, Caliri-Ortiz ME, Keller MM, Degano ME, Demkura PV, Ballaré CL (2017) Exploring growth-defence trade-offs in Arabidopsis: Phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shade-avoidance responses. Plant Cell Environ 40: 635–644 [DOI] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D (2011) ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 27: 1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Rosa TM, Aphalo PJ, Lehto T (1998) Effects of far-red light on the growth, mycorrhizas and mineral nutrition of Scots pine seedlings. Plant Soil 201: 17–25 [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- de Wit M, Galvão VC, Fankhauser C (2016) Light-mediated hormonal regulation of plant growth and development. Annu Rev Plant Biol 67: 513–537 [DOI] [PubMed] [Google Scholar]
- Davidson SE, Elliott RC, Helliwell CA, Poole AT, Reid JB (2003) The pea gene NA encodes ent-kaurenoic acid oxidase. Plant Physiol 131: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic T, de Wit M, Voesenek LACJ, Pierik R (2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Du R, Niu S, Liu Y, Sun X, Porth I, El-Kassaby YA, Li W (2017) The gibberellin GID1-DELLA signalling module exists in evolutionarily ancient conifers. Sci Rep 7: 16637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrini M, Mariotti L, Parlanti S, Picciarelli P, Salvini M, Ceccarelli N, Pugliesi C (2011) The extreme dwarf phenotype of the GA-sensitive mutant of sunflower, dwarf2, is generated by a deletion in the ent-kaurenoic acid oxidase1 (HaKAO1) gene sequence. Plant Mol Biol 75: 431–450 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernbach E, Mohr H (1990) Coaction of blue/ultraviolet-A light and light absorbed by phytochrome in controlling growth of pine (Pinus sylestris L.) seedlings. Planta 180: 212–216 [DOI] [PubMed] [Google Scholar]
- Fraser DP, Hayes S, Franklin KA (2016) Photoreceptor crosstalk in shade avoidance. Curr Opin Plant Biol 33: 1–7 [DOI] [PubMed] [Google Scholar]
- García-Martinez JL, Gil J (2001) Light regulation of gibberellin biosynthesis and mode of action. J Plant Growth Regul 20: 354–368 [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochem J 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Gebbie L, Peacock WJ, Dennis ` ES, Olive MR (2000) solation of an ent-kaurene oxidase cDNA from Cucurbita maxima. Aust J Plant Physiol 27: 1141–1149 [Google Scholar]
- Hisamatsu T, King RW, Helliwell CA, Koshioka M (2005) The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol 138: 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, Xenarios I, Fankhauser C (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Kakei Y, Mochida K, Sakurai T, Yoshida T, Shinozaki K, Shimada Y (2015) Transcriptome analysis of hormone-induced gene expression in Brachypodium distachyon. Sci Rep 5: 14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, García-Martínez JL (1999) Regulation of gibberellin biosynthesis by light. Curr Opin Plant Biol 2: 398–403 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE (2016) Transcriptome profiling of tiller buds provides new insights into phyB regulation of tillering and indeterminate growth in sorghum. Plant Physiol 170: 2232–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217 [DOI] [PubMed] [Google Scholar]
- Kirchenbauer D, Viczián A, Ádám É, Hegedűs Z, Klose C, Leppert M, Hiltbrunner A, Kircher S, Schäfer E, Nagy F (2016) Characterization of photomorphogenic responses and signaling cascades controlled by phytochrome-A expressed in different tissues. New Phytol 211: 584–598 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2010) Plant hormone signaling lightens up: Integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL (2014) To grow or defend? Low red:far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol 204: 355–367 [DOI] [PubMed] [Google Scholar]
- Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW (2016) DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun 7: 11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wei H, Ma M, Li Q, Kong D, Sun J, Ma X, Wang B, Chen C, Xie Y, Wang H (2019) Arabidopsis FHY3 and FAR1 proteins regulate the balance between growth and defense responses under shade conditions. Plant Cell 31: 2089–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng XW (2005) Organ-specific expression of Arabidopsis genome during development. Plant Physiol 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli G, Ruberti I (2000) Shade avoidance responses. Driving auxin along lateral routes. Plant Physiol 122: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Moulé P, Nozue K, Pytlak ML, Palmer CM, Covington MF, Wallace AD, Harmer SL, Maloof JN (2016) YUCCA auxin biosynthetic genes are required for Arabidopsis shade avoidance. PeerJ 4: e2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nett RS, Montanares M, Marcassa A, Lu X, Nagel R, Charles TC, Hedden P, Rojas MC, Peters RJ (2017) Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat Chem Biol 13: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu S, Yuan L, Zhang Y, Chen X, Li W (2014) Isolation and expression profiles of gibberellin metabolism genes in developing male and female cones of Pinus tabuliformis. Funct Integr Genomics 14: 697–705 [DOI] [PubMed] [Google Scholar]
- Peng J, Harberd NP (1997) Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113: 1051–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Cuppens ML, Voesenek LA, Visser EJ (2004) Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol 136: 2928–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, de Wit M (2014) Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J Exp Bot 65: 2815–2824 [DOI] [PubMed] [Google Scholar]
- Pierik R, De Wit M, Voesenek LA (2011) Growth-mediated stress escape: Convergence of signal transduction pathways activated upon exposure to two different environmental stresses. New Phytol 189: 122–134 [DOI] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Procko C, Burko Y, Jaillais Y, Ljung K, Long JA, Chory J (2016) The epidermis coordinates auxin-induced stem growth in response to shade. Genes Dev 30: 1529–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J (2014) Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol 165: 1285–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W. (2000) GROWTH RETARDANTS: Effects on gibberellin biosynthesis and other metabolic pathways. Annu Rev Plant Physiol Plant Mol Biol 51: 501–531 [DOI] [PubMed] [Google Scholar]
- Ranade SS, Delhomme N, García-Gil MR (2019) Transcriptome analysis of shade avoidance and shade tolerance in conifers. Planta 250: 299–318 [DOI] [PubMed] [Google Scholar]
- Ranade SS, García-Gil MR (2013) Ecotypic variation in response to light spectra in Scots pine (Pinus sylvestris L.). Tree Physiol 33: 195–201 [DOI] [PubMed] [Google Scholar]
- Razzak A, Ranade SS, Strand Å, García-Gil MR (2017) Differential response of Scots pine seedlings to variable intensity and ratio of red and far-red light. Plant Cell Environ 40: 1332–1340 [DOI] [PubMed] [Google Scholar]
- Regnault T, Davière JM, Heintz D, Lange T, Achard P (2014) The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J 80: 462–474 [DOI] [PubMed] [Google Scholar]
- Regnault T, Davière JM, Wild M, Sakvarelidze-Achard L, Heintz D, Carrera Bergua E, Lopez Diaz I, Gong F, Hedden P, Achard P (2015) The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat Plants 1: 15073. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al. (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sawada Y, Katsumata T, Kitamura J, Kawaide H, Nakajima M, Asami T, Nakaminami K, Kurahashi T, Mitsuhashi W, Inoue Y, et al. (2008) Germination of photoblastic lettuce seeds is regulated via the control of endogenous physiologically active gibberellin content, rather than of gibberellin responsiveness. J Exp Bot 59: 3383–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C-H, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci USA 106: 7654–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Helentjaris T (1995) The maize Dwarf3 gene encodes a cytochrome P450-mediated early step in Gibberellin biosynthesis. Plant Cell 7: 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Li L, Gage DA, Zeevaart JA (1996) Molecular cloning and photoperiod-regulated expression of gibberellin 20-oxidase from the long-day plant spinach. Plant Physiol 110: 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YL, Gage DA, Zeevaart JA (1997) Gibberellins and stem growth in Arabidopsis thaliana. Effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiol 114: 1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and Supplementary Material online or are available upon request from the corresponding author.