A mixed-inoculum, “select and resequence” approach efficiently assays for strain-specific effects of host mutations in nodule-specific genes.
Abstract
Genetic studies of legume symbiosis with nitrogen-fixing rhizobial bacteria have traditionally focused on nodule and nitrogen-fixation phenotypes when hosts are inoculated with a single rhizobial strain. These approaches overlook the potential effect of host genes on rhizobial fitness (i.e. how many rhizobia are released from host nodules) and strain-specific effects of host genes (i.e. genome × genome interactions). Using Medicago truncatula mutants in the recently described nodule-specific PLAT domain (NPD) gene family, we show how inoculating plants with a mixed inoculum of 68 rhizobial strains (Ensifer meliloti) via a select-and-resequence approach can be used to efficiently assay host mutants for strain-specific effects of late-acting host genes on interacting bacteria. The deletion of a single NPD gene (npd2) or all five members of the NPD gene family (npd1–5) differentially altered the frequency of rhizobial strains in nodules even though npd2 mutants had no visible nodule morphology or N-fixation phenotype. Also, npd1–5 nodules were less diverse and had larger populations of colony-forming rhizobia despite their smaller size. Lastly, NPD mutations disrupt a positive correlation between strain fitness and wild-type host biomass. These changes indicate that the effects of NPD proteins are strain dependent and that NPD family members are not redundant with regard to their effects on rhizobial strains. Association analyses of the rhizobial strains in the mixed inoculation indicate that rhizobial genes involved in chromosome segregation, cell division, GABA metabolism, efflux systems, and stress tolerance play an important role in the strain-specific effects of NPD genes.
The mutualism between leguminous plants and rhizobial bacteria is both ecologically and economically important. Rhizobial bacteria living inside host-formed nodules convert atmospheric N2 into a plant-accessible form (Herridge et al., 2008). Functional genetic studies of these interactions have traditionally paired single host genotypes with single rhizobial strains and screened for disruptions in binary traits such as nodule formation (Nod+/−) and nitrogen fixation (Fix+/−; e.g. Mitra et al., 2004; Domonkos et al., 2013). Such studies have greatly advanced our understanding of the plant and rhizobial genes underlying this interaction—particularly those that occur early in the establishment of nodules. However, the focus on binary traits, knockout mutations, and the use of single-strain inoculations are likely to miss potentially important quantitative variation in the benefits legumes and rhizobia receive from each other (Burghardt, 2019).
The reliance on a few rhizobial strains in genetic experiments has hindered the identification of genes or alleles with effects that are rhizobial strain specific (Gourion and Alunni, 2018). Such variation may be more common than previously thought (e.g. Burghardt et al., 2017), and the genes responsible are starting to be identified. For instance, two nodule-specific Cys-rich peptides have been identified as regulators of strain specificity after bacteria enter host cells (Tirichine et al., 2000; Wang et al., 2018), and a late-acting nodulin in Lotus japonicus, an aspartic peptidase, is necessary for infection with Mesorhizobium loti strain TONO, but not M. loti strain MAFF303099 (Yamaya-Ito et al., 2018). Recently, the phenotypic effects of NPD1, a member of the nodule-specific polycystin-1, lipoxygenase, alpha toxin (PLAT) domain (NPD) gene family studied here, were shown to be strain specific in Medicago truncatula (Pislariu et al., 2019). Assaying strain-specific phenotypic effects, when surveying many strains, is empirically difficult because pairwise host and strain experiments (i.e. those that rely on single-strain inoculations) quickly become intractably large, and avoiding cross contamination is a challenge. High-throughput screening assays such as the “select and resequence” (S&R) approach that quantify the relative frequencies of strains in nodules avoid these problems (Burghardt et al., 2018). The S&R approach uses pooled sequencing to estimate strain frequency in the initial mixture and in nodules after strains have competed to form nodules and reproduce inside them. These assays have the potential to transform our understanding of host genotype by rhizobial strain interactions (G × G) by measuring how disruption of specific host genes affect dozens of strains simultaneously.
We applied S&R to explore rhizobia (Ensifer meliloti) strain-specific effects of the M. truncatula NPD genes. NPDs comprise a family of five genes that are expressed late in nodule development: expression of NPD genes is not detectable 3 d postinoculation, but by 6 d postinoculation, NPD1, NPD3, and NPD5 peak in expression, and after 10 d, NPD2 and NPD4 peak (Carvalho et al. data hosted on MtGEA v3; Benedito et al., 2008; Supplemental Fig. S1). Characterization of sequential NPD knockouts, conducted with a single rhizobia strain (E. meliloti Sm1021), revealed that NPD disruption can produce quantitative changes in symbiotic phenotypes, including plant size, number of pink nodules, time of nodule senescence, and the area of the fixation zone in nodules (Trujillo et al., 2019). To date, only the NPD1 gene has been functionally characterized in detail (Pislariu et al., 2019). In npd1 hosts, root infection and nodule initiation appear to progress normally, but rhizobia fail to differentiate into nitrogen-fixing bacteroids once inside host cells, leading to early onset of nodule senescence. Pislariu et al. (2019) also found the effects of npd1 to be strain specific. Two of four strains used for phenotyping produced small white ineffective nodules, one strain formed brown nodules hypothesized to result from a buildup of phenolics due to a host immune response, and one formed large, pink nodules that led to bigger plants than the wild type. Strain-specific effects of NPD genes are also supported by Burghardt et al. (2017), who found that host NPD gene expression depends on rhizobial species.
We assayed relative strain frequencies in the wild type and a series of NPD mutant genotypes generated by Trujillo et al. (2019). Here, we focus on the effects of just NPD2 mutation alone (npd2) or disruption of all five NPD genes on strain frequencies, nodule traits, and plant growth. The quintuple NPD mutant npd1 npd2 npd3 npd4 npd5 is abbreviated npd1–5. These two mutant genotypes are representative of the range of effects the mutant series had on rhizobial strain frequency (Supplemental Fig. S2) and also had contrasting effects on nodule phenotypes in previous single-strain experiments (Trujillo et al., 2019). We show that (1) S&R assays can be used to quickly screen for strain-specific effects of late nodulation genes; (2) host NPD genes that affect late-stage nodule development and host biomass have strain-specific effects on rhizobial fitness in competition; and (3) genome-wide association studies (GWAS) can be used to pinpoint candidate genes likely to contribute to among-strain variation in the effects of host NPD genes on rhizobia.
RESULTS
NPD Genes Have Strain-Specific Effects on Plant and Nodule Phenotypes
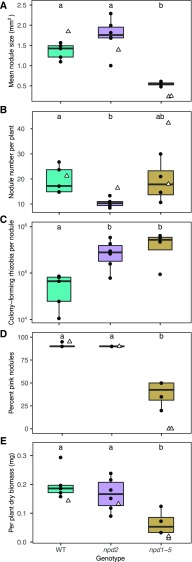
Inoculation of plants with a mixed community of 68 rhizobial strains revealed that knocking out NPD2 (npd2) did not have a strong effect on nodule size (Fig. 1A; P = 0.10), plant biomass (Fig. 1E; P = 0.53), or pink nodules (Fig. 1D; P = 0.97) compared to the wild type, which was consistent with results from single-strain inoculations (Trujillo et al., 2019). However, npd2 hosts produced fewer nodules per plant than wild-type hosts (Fig. 1B; P = 0.04). By contrast, community-inoculated plants with NPD1 to NPD5 knocked out (npd1–5) were smaller than community-inoculated wild-type plants (Fig. 1E; P = 0.006) and produced smaller nodules (Fig. 1A; P = 0.003). The effect of knocking out all five NPD genes is strain specific. When the npd1–5 mutant is inoculated with E. meliloti Sm1021, only white, ineffective nodules are formed. By contrast, in community-inoculated npd1–5 mutants, a subset of nodules are pink (39%; Fig. 1D), and average nodule size is larger than those formed when plants were inoculated with strain Sm1021 only (Fig. 1A; t test P < 0.001). See Supplemental Table S1 for additional information on phenotypes and Supplemental Table S2 for details on statistical tests. The phenotypic differences between the mixed- and single-strain-inoculated plants indicate that the effects of mutating NPD genes are rhizobial strain-specific. Further, there are strains in our mixture that are able to form functional nodules even when the entire nodule-specific NPD family is disrupted. These results demonstrate that multi-strain inoculation experiments can be an efficient way to assess whether mutations in plant genes have strain-specific effects on nodulation.
Figure 1.
Phenotypes of plants with all five NPD genes knocked out (npd1–5) differed from wild-type (WT) M. truncatula plants more than plants with the NPD2 gene knocked out differed from wild type. Measured phenotypes are as follows: mean nodule size (A), nodule number per plant (B) colony-forming rhizobia per nodule (C), percent pink nodules (D), and plant biomass (E). Letters indicate between-group differences in the multi-strain inoculation experiment based on ANOVA (P < 0.05; P adjusted for three contrasts via Tukey method). Solid points indicate average values in multi-strain inoculations for each pot replicate (median of 5.5 plants per pot). Hollow triangles indicate single-strain (Sm1021-only) inoculations.
Quantitative Effects of NPD Genes on Rhizobial Communities
An implicit assumption in many studies of the legume-rhizobia symbiosis is that, relative to white nodules, elongated pink nodules are more beneficial to hosts via increased N fixation and support greater reproduction of the bacterial strains inside those nodules. Dilution plating and rhizobial colony counts revealed that NPD deletions affected the number of reproductive rhizobia in nodules—but not in the direction we anticipated. In the mixed-inoculum experiment, the npd2 and npd1–5 mutants released a greater number of colony-forming rhizobia per nodule (Fig. 1C; P = 0.01, P = 0.006) and per plant (Supplemental Fig. S3) than wild-type plants, despite npd1–5 plants producing smaller (Fig. 1A; P = 0.01) and whiter nodules (Fig. 1D; P < 0.001). A single-strain inoculation of npd1–5 and wild-type hosts with Sm1021 showed that white npd1–5 nodules produced a greater number of reproductively capable rhizobia than pink wild-type nodules (Supplemental Fig. S4A). These results are surprising but show that rhizobial fitness cannot always be inferred from measurement of nodule phenotypes or plant biomass. While directly measuring colony-forming rhizobia released from nodules as we do here can provide novel insights, we caution that it is possible that the rhizobia released from the white nodules have low levels of storage resources that could lead to poor survivorship outside of the plant (Ratcliff et al., 2012; Muller and Denison, 2018).
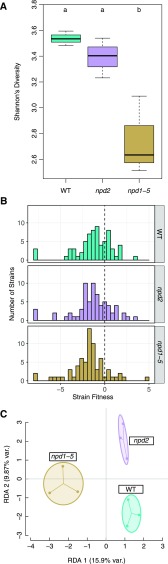
Knocking out NPD2 or NPD1 to NPD5 not only affected the number of reproductively capable rhizobia, but also affected the diversity of strains found in nodules. Strain diversity was slightly lower in npd2 than wild-type nodules and considerably lower in npd1–5 than either wild-type or npd2 nodules (both Pdf = 1 < 0.05; Fig. 2A). This decrease in diversity was not simply due to a reduced number of nodules being formed—npd2 nodule pools had fewer nodules than those from npd1–5 mutants. The same pattern emerges when we examine strain fitness (i.e. the change in the frequency of a strain in pooled nodules relative to the frequency in the initial inoculum). Fewer strains had high fitness with npd1–5 than with either wild-type or npd2 hosts—in other words, the npd1–5 mutant is more selective than the npd2 mutant or wild-type plants (Fig. 2B). The effects of host mutations also are seen in the results of a redundancy analysis (RDA) that revealed that communities in nodules quantitatively and significantly shift when NPD genes are disrupted (Fig. 2C). Moreover, knocking out all five of the NPD genes shifts the community in a different way (RDA1 axis, 15.9% variation) than knocking out NPD2 alone (RDA2 axis, 9.9% variation). Together, these results show that NPD genes can affect the composition of the community of rhizobia that associate with host plants.
Figure 2.
Characterization of shifts in E. meliloti strain communities in nodules of M. truncatula wild-type (WT) plants, plants with NPD2 knocked out (npd2), and plants with all five NPD genes knocked out (npd1–5). A, Nodule strain diversity was greater in wild-type hosts and npd2 mutants than npd1–5 mutants. Higher diversity communities have more strains that are at similar frequencies. Different lowercase letters indicate genotypes that are significantly different from each other (P < 0.05). B, Similarly, the number of strains with fitness greater than zero (i.e. those that increased in frequency in nodules as compared to the initial inoculum) were greater in wild-type hosts and npd2 mutants than npd1–5 mutants. C, Redundancy analysis of relative strain fitness shows distinct effects of npd2 and npd1–5 mutants (host genotype explains 17.2% of strain fitness variation [adj. r2], more than expected by chance [P = 0.005]). ANOVA test results from A, wild type versus npd1–5 (P = 0.011), npd2 versus npd1–5 (P = 0.03), and wild type versus npd2 (P = 0.20).
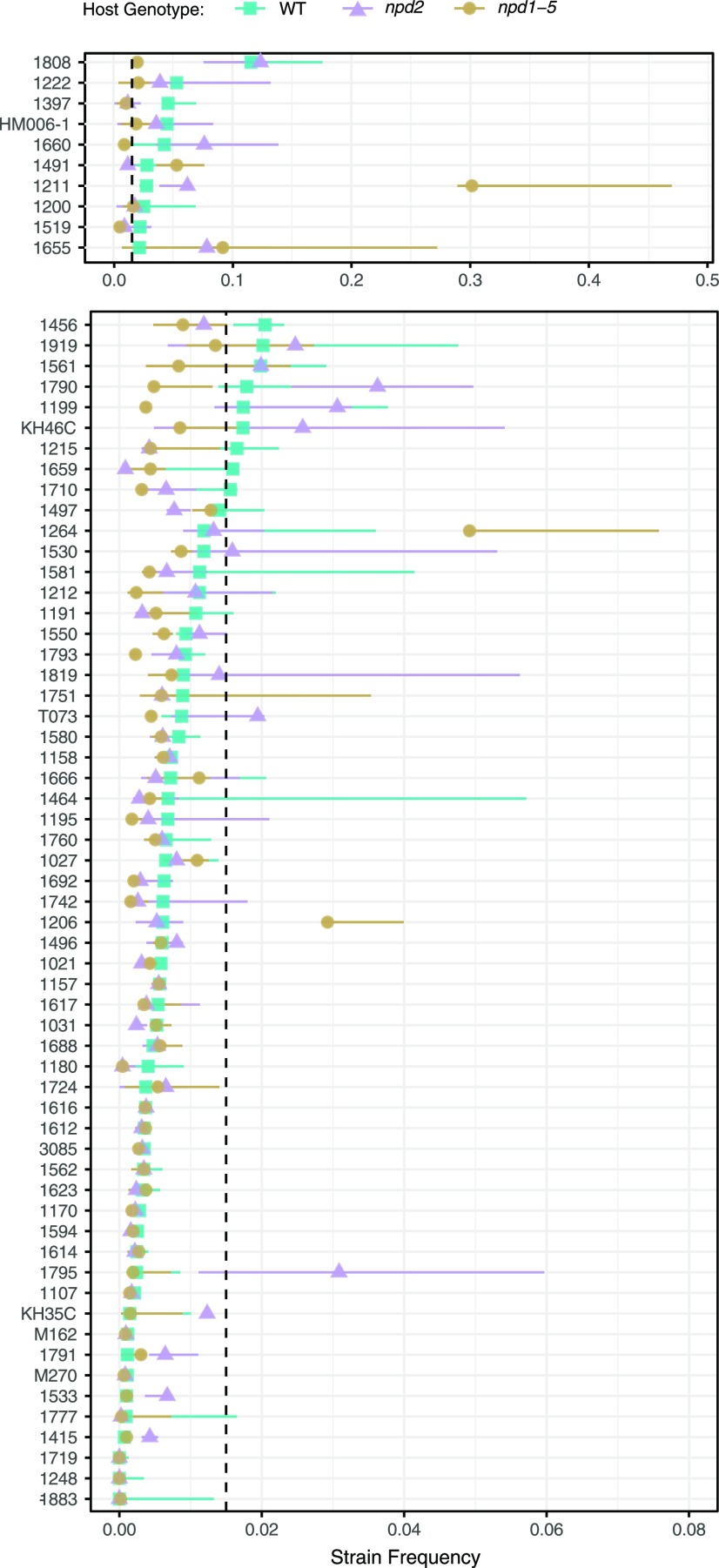
The differential effects of npd2 and npd1–5 on strain composition in nodules can be seen not only by examining community metrics (i.e. Fig. 2), but also by examining the frequency of the individual strains (Fig. 3). Disruption of NPD2 results in increases in the frequency of some strains that were common in wild-type communities (e.g. strain USDA 1660) and others that were extremely rare in wild type (USDA 1795). In npd1–5 mutants, disruption of the other four NPD genes in addition to NPD2 led to even larger shifts in strain frequencies. Similar examination of the entire mutant series suggests that to observe the majority of this shift in strain frequency requires disruption of NPD1, NPD4, and NPD5—but not NPD3. Strain frequencies in npd1/2/4/5 nodules were most similar to npd1–5, while npd2/4, npd1/2/4, and npd2/4/5 nodules were more similar to npd2 and wild type (Supplemental Fig. S2). The biggest drivers of the community shifts between wild type and npd1–5 mutants were large increases in the frequency of five strains (e.g. USDA 1211, USDA 1206, USDA 1264, USDA 1491, USDA 1655) and decreases in the five strains that had been most common in wild-type nodules (including the most frequent strain, USDA 1808). Changes in strain frequencies could be caused either by shifts in nodule initiation or bacterial replication in nodules. However, the observation that NPDs are not expressed until 6 d postinfection suggests the latter (Supplemental Fig. S1).
Figure 3.
Strain frequency estimates for 68 E. meliloti strains in nodules of wild-type M. truncatula (WT) plants, plants with the NPD2 gene knocked out (npd2), and plants with all five NPD genes knocked out (npd1–5) . Points indicate median frequency across replicates, and lines span from minimum to maximum values. Scales have been split for readability, and strains are ordered based on frequency in wild-type nodules. Dashed line indicates median overall strain frequency in the initial inoculum.
Strains with the most dramatic changes in frequency can be used for follow-up experiments to identify the specific mechanisms responsible for changes in strain fitness. For instance, single inoculation of three strains on npd1–5 hosts revealed the low-fitness strain USDA 1808 forms white nodules that released the fewest undifferentiated bacteria (among tested strains), while the high-fitness strain USDA 1206 forms large pink nodules that released 100 times more bacteria (Supplemental Fig. S4B). USDA 1264 formed large exclusively brown nodules (possibly filled with phenolics, see Pislariu et al. [2019]) that released an intermediate number of colony-forming bacteria.
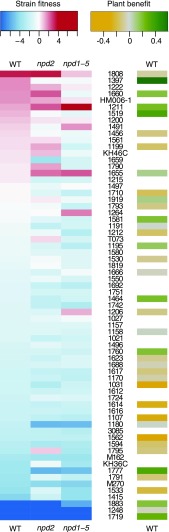
Shifts in strain frequencies could feed back to alter plant benefits. We previously reported (Burghardt et al., 2018) that with plant host R108 (wild type), strain-relative fitness and plant benefit are only weakly correlated. The same is true in this experiment (Fig. 4); rhizobial strains that have high fitness in wild-type nodules tend to provide more host benefit, as measured by vegetative biomass, but this relationship is weak (r2 = 0.12; P = 0.02). The changes in strain nodule communities in the NPD knockouts disturb this already weak correlation between plant weight and strain fitness (P = 0.63 and 0.32 in npd2 and npd1–5, respectively). NPD knockouts can both undermine and strengthen associations with beneficial strains (Fig. 4). For instance, strain USDA 1397 is the most beneficial strain in single-strain inoculation and is the third most common strain in wild-type nodules (5%) but has dramatically reduced representation in both npd2 and npd1–5 communities. By contrast, USDA 1211, a strain that also provides large growth benefits to wild type, is at an intermediate frequency (2.5%) in wild type and becomes more common in both npd2 (6%) and npd1–5 (30%). Disrupting NPD genes also can reduce associations with unhelpful strains; USDA 1808, which provides only a mild benefit to wild type, is the most common strain in both wild-type plant (11%) and npd2 mutant (12%) nodules but is far less common in nodules where all five NPD genes are knocked out (2%).
Figure 4.
Heatmaps (left) show that NPD mutations in M. truncatula have differential effects on relative fitness of E. meliloti strains in nodules. Strain fitness in wild-type M. truncatula (WT) plants are compared to plants with the NPD2 gene knocked out (npd2) and plants with all five NPD genes knocked out (npd1–5). The effects of NPD disruption on strain fitness are not clearly related to the effects of strains on plant biomass (right). Strains are ordered from high relative fitness in wild type (WT; red) to low (blue), and values are the median of three replicates). Right shows plant benefit in wild-type R108 background in terms of plant dry weight (green is high, gray is median, yellow is low, and white is N/A). Plant benefit estimates are from single-strain inoculation experiments (Burghardt et al., 2018).
Rhizobial Genes Associated with Shifts in Bacterial Fitness
GWAS is commonly used to find associations between allelic variants and phenotypic variation. Recently, researchers have begun to extend GWAS to bacteria, but GWAS in bacteria can be complicated by strong linkage disequilibrium (LD) between genomic regions that are noncontiguous and distantly located in the genome. Because of this long-distance LD, before conducting the association analyses, we identified sets of highly correlated single-nucleotide polymorphisms (SNPs), which we refer to as linkage groups, and then used these linkage groups rather than individual SNPs in the analyses (as in Epstein et al., 2018). The 26,332 linkage groups (minor allele frequency > 10%) segregating among the 68 strains were unequally distributed across the Ensifer genome. The ∼3 Mb main chromosome harbored 1969 groups, 60% of which contain SNPs in only a single gene. The ∼1.4 Mb symplasmid pSymA harbored 9880 groups, 86% of which contain SNPs in only a single gene. The ∼1.7 Mb symplasmid pSymB harbored 14,483 groups, 89% of which contain SNPs in only a single gene.
To identify rhizobial alleles responsible for host genotype × strain interactions, we tested for associations on the between-host genotype difference in strain fitness: wild type versus npd2, wild type versus npd1–5, and npd2 versus npd1–5. By performing the association on fitness differences, we focus on rhizobial variants associated with altered strain fitness in nodules of different genotypes rather than nodule fitness that is independent of host identity (variants associated with high nodule fitness in wild type [R108] were reported in Burghardt et al. [2018]). The genomic locations of variants identified by GWAS differed among the genotypic contrasts (Supplemental Fig. S5). Almost all (90%) of the top 50 linkage groups associated with fitness shifts in the wild type versus npd2 contrast were on the main chromosome. In the wild type versus npd1–5 and npd2 versus npd1–5 contrasts, most candidates were on pSymA and pSymB, where many of the traditional symbiosis genes are located.
The GWAS identified bacterial linkage groups that contain genes that are potentially responsible for strain-specific responses to the disruption of host NPD genes. We focus on the 10 most strongly associated linkage groups (Supplemental Tables S3–S5). The largest of these groups was among the largest linkage groups in our sample and contained 425 genes. While the size of this group makes it difficult to identify specific causative genes, it does contain genes known be important for strain fitness in nodules (e.g. intracellular poly-3-hydroxybutyrate depolymerase). Most of the candidate groups, however, contain far fewer genes; 19 of the top 30 groups contain only one or two genes, several of which are promising candidates for future functional validation, including several involved in chromosome segregation, cell division, gamma-aminobutyrate (GABA) metabolism, efflux systems, intercellular signaling, and stress responses (see Supplemental Appendix S1 for discussion of candidates).
DISCUSSION
Reliance on single-strain experiments for functional analyses of genes that mediate interactions between hosts and symbiotic microbes can miss important aspects of gene function (Gourion and Alunni, 2018; Burghardt, 2019). For instance, Yamaya-Ito et al. (2018) discovered the role of the ASPARTIC PEPTIDASE NODULE‐INDUCED1 (APN1) gene on nodulation only by testing multiple strains—many strains did not exhibit a strong phenotype. We have shown how mixed-strain inoculations together with S&R analyses can be used to efficiently screen plant mutants for bacteria strain-specific effects (i.e. genome × genome interactions). In single-strain inoculations with E. meliloti Sm1021, deletion of the whole NPD gene family (npd1 npd2 npd3 npd4 npd5 [npd1–5]) resulted in plants that were small and produced only white nodules (Trujillo et al., 2019; Supplemental Table S1). When inoculated with a community of 68 rhizobia strains, npd1–5 plants were nearly four times larger, and more than a third of the nodules they produced were pink. Thus, even without sequencing, the community inoculations reveal evidence for plant gene × strain interactions and that the NPD family is essential for effective nodule formation with some strains, but not others. By contrast, phenotypes of wild-type and npd2 plants were similar regardless of whether those plants were inoculated with a single strain or a 68-strain community.
Although evidence of strain-specific effects on plant phenotype can be revealed by comparing single-strain and community-inoculated plants, far more information is revealed by sequencing the community of bacteria found inside nodules. In our experiment, sequencing the npd2 nodules revealed clear changes in the relative frequency of strains despite the lack of an observable nodulation or plant phenotype. Further, abolishing all NPD genes leads to a pronounced reduction of strain diversity and has an even greater effect on strain frequencies than disrupting only NPD2. This suggests that NPD genes function in nonredundant ways and broaden the number of strains that have high fitness inside host nodules.
Our finding of strain-specific effects of NPD2 is consistent with the observation of strain-specific effects of NPD1 detected based on nodule phenotypes in four single-strain inoculations (Pislariu et al., 2019). The npd1 mutant forms small white nodules with Sm1021 but pink nodules with Rm41. Nonetheless, incompatibility with Sm1021 appears to be reversible if other NPD genes are knocked out, as npd1/2/4 and npd1/2/4/5 mutants are able to form small pink nodules with this strain (Trujillo et al., 2019). While we did not measure strain specificity in the npd1 single-gene knockout used by Pislariu et al. (2019), examination of strain frequencies in other multigene knockouts do not support the idea that NPD1 alone is responsible for the large shifts in strain frequency between npd2 and npd1–5 nodules. It will be interesting to extend experiments to single-gene knockouts of the other NPD family members, or even to genes encoding nodule-specific Cys-rich (NCR) peptides (e.g. Kim et al., 2015) and other late-acting host genes (e.g. DNF2; Bourcy et al., 2013).
Sequencing the community of rhizobia found inside plant nodules also enables a GWAS to identify bacterial genes responsible for the strain-specific effects of NPD genes. One of the key events during bacteroid differentiation in Medicago nodules involves endoreduplication of bacterial chromosomes, where the chromosome duplicates repeatedly but the cell does not divide (Mergaert et al., 2006; Haag et al., 2013). Interestingly, two of the top candidate groups from the npd2 contrasts contain genes known to be involved in cellular division in multiple prokaryotes. The chromosome partition protein Smc—along with segregation and condensation proteins ScpA and ScpB—forms a structural maintenance of chromosome (SMC) protein complex. This condensin complex is vital for chromosomal compaction, partitioning, and segregation during cell division (Wang et al., 2013). The second candidate, the actin-like FtsA (filamenting temperature-sensitive A) protein, is essential for the localization and completion of cell wall biosynthesis in many bacteria (e.g. Escherichia coli and Streptococcus pneumonia; Mura et al., 2017). Chromosome replication and cell division genes, including FtsA and Smc, are upregulated by E. meliloti in nodule zones undergoing host infection and bacteroid differentiation as compared to the nitrogen fixation zone (Roux et al., 2014). Cell division genes are also upregulated in bacteria growing in host mutant nodules (e.g. dnf1 and dnf5) that arrest during the symbiosome formation stage (Lang and Long, 2015). Thus, it is tempting to speculate that NPD2 either directly or indirectly alters the environment in symbiosomes during bacteroid differentiation and that natural genetic variants in these bacterial genes make strains more or less susceptible to those changes.
Conclusion
The S&R approach provides a high-throughput screen for strain-specific effects of genes that mediate interactions between eukaryotic hosts and bacterial communities. Our application of S&R to legume NPD genes, a small gene family that affects the legume-rhizobia symbiosis after nodules have formed, revealed how sequencing of the host-associated community can identify strain-specific effects that are not readily apparent based on phenotype or single-strain data alone. S&R also provides the resources needed to conduct a bacterial GWAS, thereby identifying candidate genes showing a statistical signal of being responsible for the strain-specific responses to host gene mutations. The utility of S&R for characterizing gene function is not limited to the legume-rhizobia system but should be broadly applicable to other host-microbe systems. This includes host-pathogen systems (e.g. Psuedomonas; Baltrus et al., 2017), in which identification of gene function has mostly relied on single-strain inoculations.
MATERIALS AND METHODS
We grew 68 Ensifer meliloti strains primarily from the USDA core collection (listed in Fig. 3). The 68 strains were chosen from 160 previously sequenced strains (Epstein et al., 2012, 2018; Nelson et al., 2018) to maximize our ability to accurately estimate strain frequency and minimize population structure, a complicating factor in GWAS. In brief, we used dnadiff from MUMmer3 (v3.23; Kurtz et al., 2004) to estimate the pairwise genome-wide average nucleotide identity among the 160 strains, then eliminated one strain from each pair with > 99.6% pairwise identity. Removing these strains results in a set of 68 strains with a mean pairwise nucleotide identity of 99%.
Selected strains were grown in 3 mL of Tryptone Yeast medium (6 g tryptone, 3 g yeast extract, 0.38 g CaCl2 per L) for 3 d at 28°C on a rotary shaker. An equal volume of each strain was then combined to form a community for inoculation and diluted to ∼106 rhizobia per mL with 0.85% (w/v) NaCl. We did not adjust the volume of each strain based on OD600. Preliminary experiments showed that, while OD provided a good estimate of bacterial cell density within a strain over time, OD provided little predictive power when comparing between strains at a single time point (correlation of OD and colony-forming units < 0.05). By sequencing the initial community, we directly estimate the variance in initial strain frequency (median = 0.014; range, 0.009–0.02) and can control for those differences in our estimate of fitness. The initial strain frequency estimate might include some dead cells. However, even if this is the case, the differences we observe among hosts are robust, since all hosts were inoculated with the same initial mixture. Moreover, correlations between initial and final frequencies are weak and do not differ from chance expectations (wild type, Pdf = 66 = 0.73; npd2, Pdf = 66 = 0.81; npd1–5, Pdf = 66 = 0.84). E. meliloti Sm1021 was grown as described above but was not included in the rhizobial community.
We grew plants from Medicago truncatula wild-type (R108) and mutant seeds (Trujillo et al., 2019) that were scarified with a razor blade, sterilized for 3 min with a 33% commercial bleach solution with 0.2% (v/v) Tween 20, well rinsed with sterile deionized water, and incubated at 4°C for 3 d. Seeds were then germinated at room temperature overnight, and 7 to 10 seeds were planted in 6-inch square pots containing a sterile 1:1 mix of turface and vermiculite. Three days after planting, we inoculated five to six pots of each genotype with the rhizobia community and one to two pots of each genotype with E. meliloti Sm1021. Pots were randomized and placed in a single growth chamber set at 22°C to 24°C with 75% humidity on a 16 h:8 h, light (200–350 μmol m−2 s−1):dark photoperiod. Plants were fertilized with N-free Fahraeus solution (Barker et al., 2006) once per week and watered with sterile deionized water twice per week. Seven weeks postinoculation, plant shoots and roots were collected, dry biomass was measured, nodules were counted, and pooled nodules from each pot were photographed for nodule area assessment as described in Trujillo et al. (2019).
As described in Burghardt et al. (2018), nodules from all plants in each pot were pooled. We sequenced nodule pools from the first three replicate blocks. These pools contained a mean of 137 nodules for wild-type plants, 64 nodules for npd2, and 85 nodules for npd1–5. Nodules were surface-sterilized with 10% (v/v) bleach for 15 s, rinsed with sterile deionized water, and crushed in a 1.5 mL microcentrifuge tube with 1 mL 0.85% (w/v) NaCl solution. The suspension was centrifuged at 400g for 10 min to form a pellet enriched for heavier, endo-reduplicated bacteroids and cellular debris, as compared to the supernatant, which is enriched for undifferentiated rhizobia. Note, this does not mean that the bacteroid pellet is devoid of undifferentiated rhizobia. The supernatant was separated and the pellet was resuspended, crushed, and centrifuged again at 400g. The supernatant fractions from the two rounds of spinning were combined, and an aliquot was taken for dilution plating to count the number of colony-forming rhizobia released from the nodules. The supernatant fraction was spun at 10,000g for 2 min to pellet undifferentiated bacteria. Both bacteroid and undifferentiated rhizobia-enriched pellets were frozen prior to extracting DNA using a DNeasy Plant Mini Kit (Qiagen).
DNA from each of the nodule samples and from the initial community was used to generate NexteraXT libraries and sequenced on two runs of an Illumina HiSeq 2500 (125-bp paired-end reads; 3.7–7.1 million read pairs library−1). Reads were trimmed with TrimGalore! (v0.4.1) using default settings, except the minimum read length was set to 100 bp, quality threshold was set to 30, and minimum adapter match length was set to 3. The trimmed reads were aligned to the E. meliloti USDA1106 genome (Nelson et al., 2018) using bwa mem (v0.7.17; Li and Durbin, 2009) with default settings. Mean genome coverage per sample was 116× (range 68×–161×; see Dryad repository for additional statistics). The frequency of each strain was estimated using HARP (haplotype analysis of reads in pools; Kessner et al., 2013) as described in Burghardt et al. (2018). In brief, HARP calculates for each read the probability that it came from each strain and then determines the combination of strain frequencies that maximize the likelihood across reads. We called SNPs for the 68 individual strains in the community using FreeBayes (v1.2.0-2-g29c4002; Garrison and Marth, 2012) with a minimum read-mapping quality of 30. For estimating strain frequency, we used only SNPs for which every strain had an unambiguous base call.
We sequenced both bacteroid- and undifferentiated rhizobia-enriched pellets for three wild-type and npd1–5 replicates. Estimates of the strain representation from these two fractions from the same replicate were nearly identical (mean Rn = 68 = 0.98, for six replicate pairs). Thus, we sequenced only the bacteroid-enriched fraction for all remaining replicates and genotypes and only report strain frequencies inferred from the bacteroid fraction.
Quantification of Changes in Strain Communities
We used two related measures of strain success: raw strain frequency estimated from HARP and relativized strain fitness calculated as log2(freqnodule/freqinitial). The relativized strain fitness controls for slight differences in strain frequency in the initial strain community and normalizes the fitness distribution for downstream analysis. We performed a series of analyses to determine how strain communities in nodules differed among the three focal host genotypes (wild-type, npd2, npd1–5). First, we determined whether there were changes in the diversity of communities by calculating a Shannon diversity index on strain frequencies from nodules in each replicate pot (more diverse communities have a higher index). To determine if host genotype significantly affected Shannon diversity, we ran linear models (“lm” in base R) on each pairwise host contrast. Second, we performed a RDA on relative strain fitness (“rda” function in the vegan R package [Oskanen et al., 2017]) to collapse the dimensionality of the relative fitness data and visualize changes in strain fitness between host genotypes. RDA fits a multivariate linear regression to centered and scaled data and uses principal component analysis to decompose the fitted parameters into the major axes of variation. The adjusted R2 of each RDA model provides an estimate of the proportion of variance in strain relative fitness explained by the predictor(s) (host genotype) in the model. To determine the probability that fitness differences occurred by chance, we reran the analyses on permuted data (ANOVA function set to 999 permutations).
Identification of Bacterial Genes Associated with Host-Genotype-Specific Colonization
We conducted GWAS using 26,332 linkage groups that contained 85,017 SNPs with a minor allele frequency >10%. The linkage groups were formed by grouping SNPs with pairwise LD measured as R2 > 0.95 (Epstein et al., 2018). Initially, we performed a GWAS to find bacterial genetic variants associated with strain fitness in wild-type, npd2, and npd1–5 nodules. The majority of the top 10 variants identified in any one of these analyses were also associated with rhizobia fitness in the other two host genotypes (27 of 30 are in the top 2.5% of P values). To find variants associated with changes in strain fitness between wild-type and NPD mutants, we conducted three additional analyses on fitness differences between each of the three pairwise host combinations (fitness in host 1 − fitness in host 2). We used the linear mixed-model option (−lmm 4) in GEMMA (v0.96; (Zhou and Stephens, 2012) with a standardized K-matrix (−gk 2) to perform the association mapping.
Accession Numbers
The M. truncatula NPD genes (Mt4.0 v2) are as follows: Medtr2g103303 (NPD1); Medtr2g103307 (NPD2); Medtr2g103313 (NPD3); Medtr2g103330 (NPD4); Medtr2g103360 (NPD5). The 68 Ensifer strains used were primarily obtained from the ARS culture collection (https://nrrl.ncaur.usda.gov/). Raw sequencing data has been deposited on NCBI under BioProject number PRJNA401437 (SRX4557823–SRX4557965). All estimated strain frequencies, strain details, phenotypic datasets, and R code are available in a Dryad repository (https://doi.org/10.5061/dryad.pnvx0k6hb).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NPD expression time course from Carvalho et al. data hosted on MtGEAv3.
Supplemental Figure S2. RDA of strain fitness in all six sequential NPD knockouts in Trujillo et al. (2019).
Supplemental Figure S3. Colony-forming rhizobia released per plant for M. truncatula wild type plants, plants with NPD2 knocked out (npd2), and plants with all five NPD genes knocked out (npd1–5).
Supplemental Figure S4. The number of undifferentiated rhizobia within nodules is host, nodule morphology, and strain dependent.
Supplemental Figure S5. The proportion of the top 50 linkage groups from each GWAS contrast found on each of the rhizobial replicons (chromosome, PsymA, and PsymB).
Supplemental Table S1. Summary of M. truncatula wild-type, npd2, and npd1–5 plant phenotypes when grown with a mixture of 68 strains or with Sm1021 alone.
Supplemental Table S2. Statistical test results for pairwise contrasts of wild-type, npd2, and npd1–5 nodules and plant phenotypes when grown with a mixture of 68 strains.
Supplemental Table S3. Summary of GWAS analysis of strain fitness difference between wild-type and npd2 nodules with corresponding ranks of these LD groups in wild-type nodules and other genotype contrasts.
Supplemental Table S4. Summary of GWAS analysis of strain fitness difference between wild-type and npd1–5 nodules with corresponding ranks of these LD groups in wild-type nodules and other genotype contrasts.
Supplemental Table S5. Summary of GWAS analysis of strain fitness difference between npd2 and npd1–5 nodules with corresponding ranks of these LD groups in wild-type nodules and other genotype contrasts.
Supplemental Table S6. Number of genes tagged in each linkage group for all 26,000 LD groups and divided by Ensifer replicon.
Supplemental Appendix S1. Further discussion of bacterial gene candidates.
Acknowledgments
We thank Roxanne Denny and Michelle Hoge for assistance with experimental setup and harvesting. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author and do not necessarily reflect the views of the National Science Foundation. Computing resources were provided by the Minnesota Supercomputing Institute (MSI) at the University of Minnesota.
Footnotes
This work was supported by the National Science Foundation under grant numbers IOS-1724993 and IOS-1856744.
Articles can be viewed without a subscription.
References
- Baltrus DA, McCann HC, Guttman DS (2017) Evolution, genomics and epidemiology of Pseudomonas syringae: Challenges in bacterial molecular plant pathology. Mol Plant Pathol 18: 152–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DG, Pfaff T, Moreau D, Groves E, Ruffel S, Lepetit M, Whitehand S, Maillet F, Nair RM, Journet E-P (2006) Growing M. truncatula: Choice of substrates and growth conditions In Mathesius U, Journet EP, Sumner LW, eds, Medicago truncatula Handbook. Noble Research Institute, Ardmore, OK, https://www.noble.org/medicago-handbook/ [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bourcy M, Brocard L, Pislariu CI, Cosson V, Mergaert P, Tadege M, Mysore KS, Udvardi MK, Gourion B, Ratet P (2013) Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol 197: 1250–1261 [DOI] [PubMed] [Google Scholar]
- Burghardt LT. (2019) Evolving together, evolving apart: Measuring the fitness of rhizobial bacteria in and out of symbiosis with leguminous plants. New Phytol doi:10.1111/nph.16045 [DOI] [PubMed] [Google Scholar]
- Burghardt LT, Epstein B, Guhlin J, Nelson MS, Taylor MR, Young ND, Sadowsky MJ, Tiffin P (2018) Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc Natl Acad Sci USA 115: 2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghardt LT, Guhlin J, Chun CL, Liu J, Sadowsky MJ, Stupar RM, Young ND, Tiffin P (2017) Transcriptomic basis of genome by genome variation in a legume-rhizobia mutualism. Mol Ecol 26: 6122–6135 [DOI] [PubMed] [Google Scholar]
- Domonkos A, Horvath B, Marsh JF, Halasz G, Ayaydin F, Oldroyd GE, Kalo P (2013) The identification of novel loci required for appropriate nodule development in Medicago truncatula. BMC Plant Biol 13: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein B, Abou-Shanab RAI, Shamseldin A, Taylor MR, Guhlin J, Burghardt LT, Nelson M, Sadowsky MJ, Tiffin P (2018) Genome wide association analyses in the model rhizobium Ensifer meliloti. MSphere 3: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein B, Branca A, Mudge J, Bharti AK, Briskine R, Farmer AD, Sugawara M, Young ND, Sadowsky MJ, Tiffin P (2012) Population genomics of the facultatively mutualistic bacteria Sinorhizobium meliloti and S. medicae. PLoS Genet 8: e1002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G (2012) Haplotype-Based Variant Detection From Short-Read Sequencing. https://arxiv.org/abs/1207.3907
- Gourion B, Alunni B (2018) Strain-specific symbiotic genes: A new level of control in the intracellular accommodation of rhizobia within Legume nodule cells. Mol Plant Microbe Interact 31: 287–288 [DOI] [PubMed] [Google Scholar]
- Haag AF, Arnold MFF, Myka KK, Kerscher B, Dall’Angelo S, Zanda M, Mergaert P, Ferguson GP (2013) Molecular insights into bacteroid development during Rhizobium-legume symbiosis. FEMS Microbiol Rev 37: 364–383 [DOI] [PubMed] [Google Scholar]
- Herridge DF, Peoples MB, Boddey RM (2008) Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 311: 1–18 [Google Scholar]
- Kessner D, Turner TL, Novembre J (2013) Maximum likelihood estimation of frequencies of known haplotypes from pooled sequence data. Mol Biol Evol 30: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Chen Y, Xi J, Waters C, Chen R, Wang D (2015) An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci USA 112: 15238–15243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL (2004) Versatile and open software for comparing large genomes. Genome Biol 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C, Long SR (2015) Transcriptomic analysis of Sinorhizobium meliloti and Medicago truncatula symbiosis using nitrogen fixation-deficient nodules. Mol Plant Microbe Interact 28: 856–868 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert P, Uchiumi T, Alunni B, Evanno G, Cheron A, Catrice O, Mausset A-E, Barloy-Hubler F, Galibert F, Kondorosi A, et al. (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA 103: 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra RM, Shaw SL, Long SR (2004) Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA 101: 10217–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller KE, Denison RF (2018) Resource acquisition and allocation traits in symbiotic rhizobia with implications for life-history outside of legume hosts. R Soc Open Sci 5: 181124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura A, Fadda D, Perez AJ, Danforth ML, Musu D, Rico AI, Krupka M, Denapaite D, Tsui HT, Winkler ME, et al. (2017) Roles of the essential protein FtsA in cell growth and division in Streptococcus pneumoniae. J Bacteriol 199: e00608–e00616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M, Guhlin J, Epstein B, Tiffin P, Sadowsky MJ (2018) The complete replicons of 16 Ensifer meliloti strains offer insights into intra- and inter-replicon gene transfer, transposon-associated loci, and repeat elements. Microb Genom 4: e000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskanen J, Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O’Hara R, Simpson G, Solymos P, et al. (2017) vegan: Community ecology package. Version 2.4.2 https://cran.r-project.org/web/packages/vegan/index.html
- Pislariu CI, Sinharoy S, Torres-Jerez I, Nakashima J, Blancaflor EB, Udvardi MK (2019) The nodule-specific PLAT domain protein NPD1 is required for nitrogen-fixing symbiosis. Plant Physiol 180: 1480–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Underbakke K, Denison RF (2012) Measuring the fitness of symbiotic rhizobia. Symbiosis 55: 85–90 [Google Scholar]
- Roux B, Rodde N, Jardinaud MF, Timmers T, Sauviac L, Cottret L, Carrère S, Sallet E, Courcelle E, Moreau S, et al. (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J 77: 817–837 [DOI] [PubMed] [Google Scholar]
- Tirichine L, de Billy F, Huguet T (2000) Mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol 123: 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo DI, Silverstein KAT, Young ND (2019) Nodule-specific PLAT domain proteins are expanded in the Medicago lineage and required for nodulation. New Phytol 222: 1538–1550 [DOI] [PubMed] [Google Scholar]
- Wang Q, Liu J, Li H, Yang S, Körmöczi P, Kereszt A, Zhu H (2018) Nodule-specific cysteine-rich peptides negatively regulate nitrogen-fixing symbiosis in a strain-specific manner in Medicago truncatula. Mol Plant Microbe Interact 31: 240–248 [DOI] [PubMed] [Google Scholar]
- Wang X, Montero Llopis P, Rudner DZ (2013) Organization and segregation of bacterial chromosomes. Nat Rev Genet 14: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya-Ito H, Shimoda Y, Hakoyama T, Sato S, Kaneko T, Hossain MS, Shibata S, Kawaguchi M, Hayashi M, Kouchi H, et al. (2018) Loss-of-function of ASPARTIC PEPTIDASE NODULE-INDUCED 1 (APN1) in Lotus japonicus restricts efficient nitrogen-fixing symbiosis with specific Mesorhizobium loti strains. Plant J 93: 5–16 [DOI] [PubMed] [Google Scholar]
- Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44: 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]