The Evening Complex interacts with the complex responsible for the deposition of the histone variant H2A.Z, creating repressive chromatin domains to repress a cohort of target genes in Arabidopsis.
Abstract
The Evening Complex (EC) is a core component of the Arabidopsis (Arabidopsis thaliana) circadian clock, which represses target gene expression at the end of the day and integrates temperature information to coordinate environmental and endogenous signals. Here we show that the EC induces repressive chromatin structure to regulate the evening transcriptome. The EC component ELF3 directly interacts with a protein from the SWI2/SNF2-RELATED (SWR1) complex to control deposition of H2A.Z-nucleosomes at the EC target genes. SWR1 components display circadian oscillation in gene expression with a peak at dusk. In turn, SWR1 is required for the circadian clockwork, as defects in SWR1 activity alter morning-expressed genes. The EC-SWR1 complex binds to the loci of the core clock genes PSEUDO-RESPONSE REGULATOR7 (PRR7) and PRR9 and catalyzes deposition of nucleosomes containing the histone variant H2A.Z coincident with the repression of these genes at dusk. This provides a mechanism by which the circadian clock temporally establishes repressive chromatin domains to shape oscillatory gene expression around dusk.
The circadian clock generates biological rhythms with a period of ∼24 h to coordinate plant growth and development with environmental cycles (Greenham and McClung, 2015). A large fraction of the Arabidopsis (Arabidopsis thaliana) transcriptome is circadian-regulated (Staiger and Green, 2011). Circadian transcription allows the molecular anticipation of the environmental cycles, which improves plant fitness and adaptation (Yerushalmi et al., 2011). Consistent with its adaptive function, the circadian clock is subject to multiple layers of regulation, which contribute toward accurate oscillations (Seo and Mas, 2014).
Transcriptional regulation of circadian genes is a basic framework of clock architecture (Carré and Kim, 2002; Salomé and McClung, 2004). The Arabidopsis central oscillator consists of a series of sequential regulatory loops composed of genes expressed at different times during the diurnal cycle. Morning-expressed genes such as CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) repress the expression of evening-expressed genes like TIMING OF CAB EXPRESSION1/PSEUDO-RESPONSE REGULATOR1 (Alabadí et al., 2001), while in turn the TOC1 protein represses CCA1 and LHY during the night (Alabadí et al., 2001; Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2013). Repression of CCA1 and LHY throughout the day also occurs by the sequential action of additional members of the PRR family, including PRR9, PRR7, and PRR5 (Nakamichi et al., 2010; Salomé et al., 2010). Additional repressors of morning gene expression include the components of the Evening Complex (EC), EARLY FLOWERING3 (ELF3), ELF4, and LUX ARRYTHMO/PHYTOCLOCK1 (Nusinow et al., 2011; Chow et al., 2012; Herrero et al., 2012).
The EC is a core component of the circadian clock, and elf3 mutants are arrhythmic under continuous light (Hicks et al., 1996; Lu et al., 2012). By binding to the promoters of hundreds of key regulators of circadian clock, photosynthesis, and phytohormone signaling, the EC is able to repress their expression (Ezer et al., 2017). Because the activity of the EC is reduced at warmer temperatures, and environmental sensing phytochromes cobind target promoters, the EC is able to integrate environmental information into endogenous developmental programs (Ezer et al., 2017). However, the molecular mechanisms by which the EC represses gene expression are not known.
Histone variants influence chromatin structure and therefore transcription. H2A.Z is the most well-conserved histone variant, enriched near transcription start sites (TSSs; Raisner et al., 2005; Raisner and Madhani, 2006) and influencing transcriptional activities of associated genes (Raisner et al., 2005). Effects of H2A.Z deposition are likely variable depending on chromatin context: H2A.Z deposition at promoters prevents the spread of heterochromatin in yeast and is associated with transcriptional inducibility (Guillemette et al., 2005), whereas in metazoans, H2A.Z has been shown to play a role in heterochromatin formation and maintenance (Rangasamy et al., 2003; Swaminathan et al., 2005). In plants, H2A.Z-nucleosomes confer transcriptional competence (Deal et al., 2007; To and Kim, 2014), and also appear to wrap DNA more tightly, facilitating inducible gene expression (Kumar and Wigge, 2010; Coleman-Derr and Zilberman, 2012).
The Arabidopsis genome encodes putative homologs of catalytic subunits of the Swi2/Snf2-Related (SWR1)/Swi2/Snf2-Related CBP Activator Protein complex responsible for H2A.Z deposition, including PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), ACTIN-RELATED PROTEIN6 (ARP6), and SERRATED LEAVES AND EARLY FLOWERING (SEF; March-Díaz and Reyes, 2009). The SWR1 complex associates extensively with chromatin and catalyzes H2A.Z exchange at genomic levels. Consistently, SWR1-mediated chromatin remodeling is involved in diverse aspects of plant physiology and development, such as the floral transition, immune responses, and temperature sensing (Noh and Amasino, 2003; Deal et al., 2005; Kumar and Wigge, 2010). Here, we report that the EC associates with the SWR1 complex to repress the evening transcriptome. As a part of its biological impact, this complex shapes circadian oscillations by targeting clock genes such as PRR7 and PRR9 for H2A.Z deposition and gene repression. These results indicate that diurnal H2A.Z deposition provides a mechanism contributing to circadian gene expression in Arabidopsis.
RESULTS
ELF3 Stabilizes Nucleosome Architecture at EC Target Genes
To understand how the EC functions, we created a stringent list of direct EC targets (Supplemental Table S1), defined as genes whose promoters are bound by at least two EC proteins and which are misexpressed at the end of the day in elf3-1 (Ezer et al., 2017). Previously, we have seen that EC targets show the same pattern of misexpression in both elf3-1 and lux-4, when compared to wild type over a 24-h time course (Ezer et al., 2017). In both cases, there is minimal deviation from wild-type gene expression during the day, but maximal deviation during the evening and night-time, coinciding with the activity of the EC (Huang and Nusinow, 2016). Furthermore, the fold-increase in expression in elf3-1 and lux-4 is also conserved between target genes (Ezer et al., 2017). This indicates that the EC components control target gene expression possibly altogether. Thus, we next asked how the EC globally controls endogenous gene expression programs.
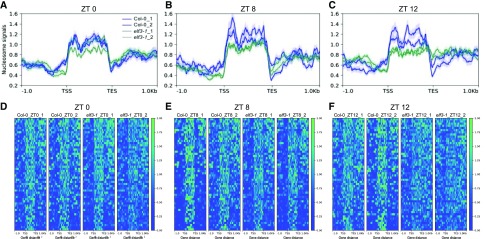
Because chromatin accessibility, which is related to gene responsiveness, is usually coordinated with transcriptional regulation, we investigated if EC targets have a distinctive nucleosome structure, and if this is perturbed in elf3-1. Micrococcal nuclease (MNase) produces double-stranded cuts between nucleosomes, thus providing a simple method for obtaining information on the locations and arrangements of nucleosomes (Shu et al., 2013). We investigated the genome accessibility of the EC target loci at a range of time points using MNase digestion coupled with sequencing (MNase-seq). We observed high nucleosome occupancy for EC target genes (Fig. 1), and this increased at Zeitgeber Time (ZT)8 and ZT12 (Fig. 1, B, C, E, and F), coinciding with maximal EC activity. Notably, a marked increase in chromatin accessibility was observed in the elf3-1 mutant (Fig. 1, B, C, E, and F), consistent with the EC repression of gene expression. The nucleosome occupancy in elf3-1 was most similar to wild type at ZT0 (Fig. 1, A and D), but decreased as the day progresses, showing the greatest loss at ZT12, by which time the gene body nucleosome occupancy was barely detectable (Fig. 1, C and F). These results suggest that EC-dependent gene repression is induced in part by temporal stabilization of repressive chromatin structures.
Figure 1.
Nucleosome occupancy in EC target genes in Col-0 and elf3-1 at ZT0, 8, and 12. Average nucleosome signals (normalized MNase-seq reads) on 52 EC target genes (see also Supplemental Table S1) at ZT0, ZT8, and ZT12 (A–C) in Col-0 and elf3-1 grown under long day conditions. Heat map visualization of nucleosome signals over 52 EC target genes on Col-0 and elf3-1 at ZT0, ZT8, and ZT12 (d–F). Data are plotted from 1 kb upstream of TSSs to 1 kb downstream of transcription end site (TES) of EC target genes. Graphs show the results of two replicates.
Components of the SWR1 Complex Are Necessary for EC Function
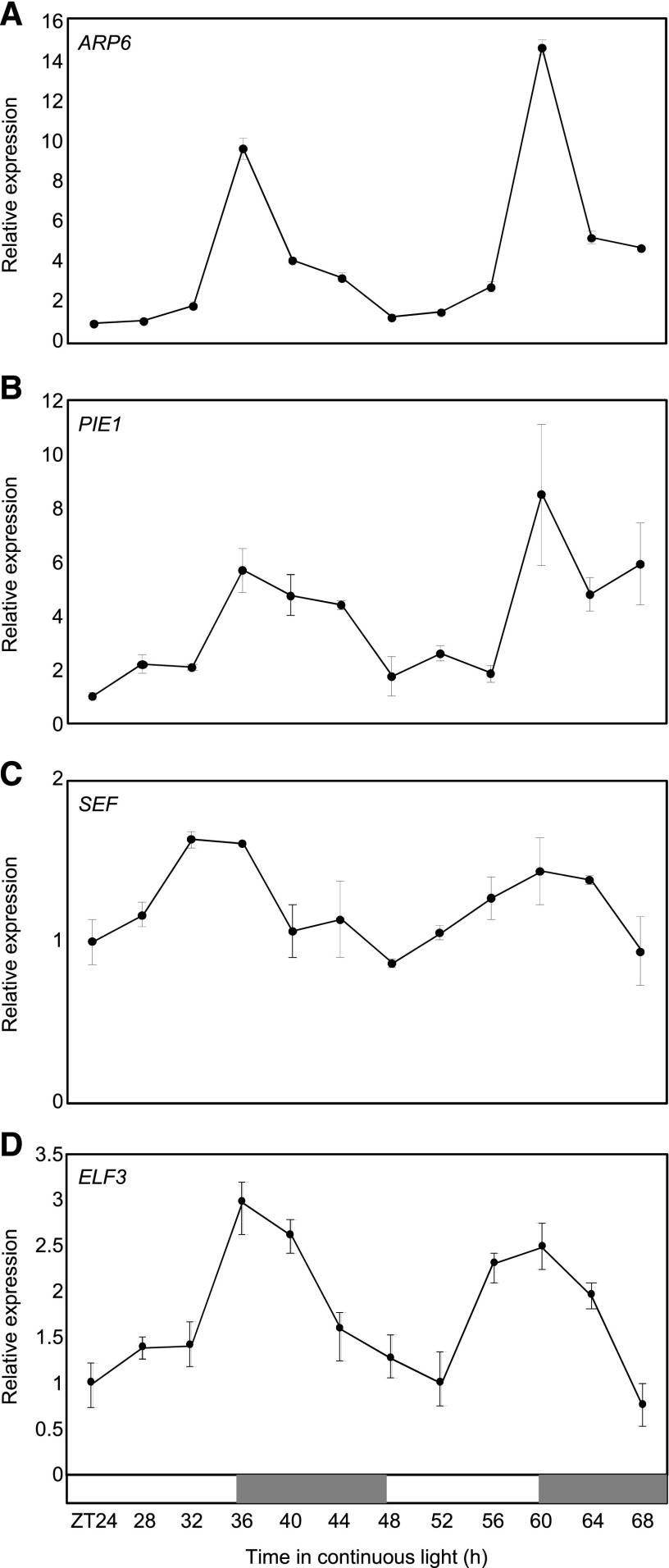
To identify genes mediating the connection between the EC and chromatin architecture, we surveyed the DIURNAL dataset (http://diurnal.mocklerlab.org) for transcripts having a similar expression pattern to the EC. Transcripts encoding histone variants as well as SWR1 components required for H2A.Z deposition were included, and they were regulated in diurnal and circadian patterns (Supplemental Fig. S1A). To confirm these observations, we examined the circadian expression of genes associated with H2A.Z exchange by reverse transcription quantitative PCR (RT-qPCR) analysis. Genes encoding the SWR1 components, including ARP6, PIE1, and SEF, displayed circadian oscillation under free-running conditions with a peak around dusk (Fig. 2, A–C), similar to ELF3 (Fig. 2D). The transcript accumulation of histone variant genes, HTA8, HTA9, and HTA11, however, did not significantly oscillate during the circadian cycle in our conditions (Supplemental Fig. S1B).
Figure 2.
Coexpression of the SWR1 components with EC. A to D, Circadian expression of SWR1 components and ELF3. Seedlings grown under neutral day conditions (12-h light: 12-h dark) for 2 weeks were transferred to continuous light conditions at ZT0. Whole seedlings were harvested from ZT24 to ZT68 to analyze transcript accumulation. Transcript levels of ARP6 (A), PIE1 (B), SEF (C), and ELF3 (D) were determined by RT-qPCR. Gene expression values were normalized to the eIF4A expression. Biological triplicates were averaged. Bars indicate the mean ± se. The white and gray boxes indicate the subjective day and night, respectively.
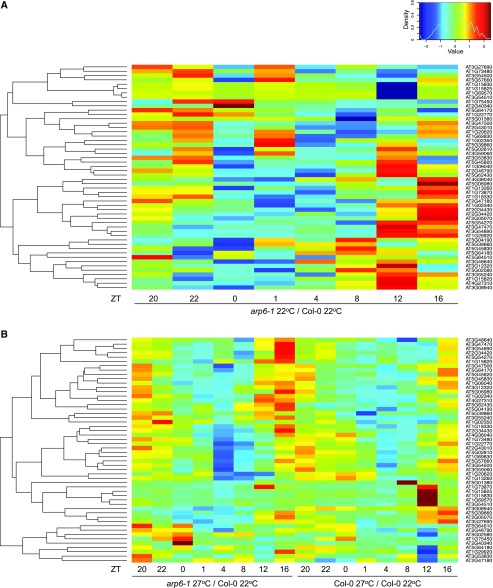
To determine if H2A.Z-nucleosomes are important for EC function, we analyzed whether EC targets are misexpressed in arp6-1, which is compromised in its ability to incorporate H2A.Z-nucleosomes (Kumar and Wigge, 2010). Expression of EC target genes over a diurnal cycle was analyzed using RNA-seq. These genes in this cluster showed a pattern of increased expression at the end of day and early night in arp6-1 compared to wild type (Fig. 3A), a pattern consistent with reduced EC function in arp6-1.
Figure 3.
Elevated expression of EC target genes in arp6-1. Heat map visualization of log fold changes of the expression levels of EC target genes in arp6-1 at 22°C compared with Col-0 at 22°C grown under long day conditions (A) and in arp6-1 at 27°C compared with Col-0 at 22°C grown under short days (B). Values in (A) and (B) represent log2 (TPM in arp6-1 22°C/TMP in Col-0 22°C) and log2 (TPM in arp6-1 27°C/TMP in Col-0 22°C), respectively. The log fold change of the expression level of each gene was calculated by Z-score (mean = 0, sd = 1). The heat map was generated using the heatmap.2 function in R (v3.2.5).
Because warm temperature reduces the ability of the EC to associate with target promoters (Box et al., 2015; Ezer et al., 2017), we investigated how these genes respond to 27°C. As expected, EC targets were upregulated in wild type at 27°C particularly around evening and nighttime (Fig. 3B), reflecting impaired EC function. This pattern was, however, strongly enhanced by the arp6-1 mutation (Fig. 3B). These results indicate that the EC and H2A.Z-nucleosomes function to repress EC targets.
ELF3 Physically Interacts with SEF
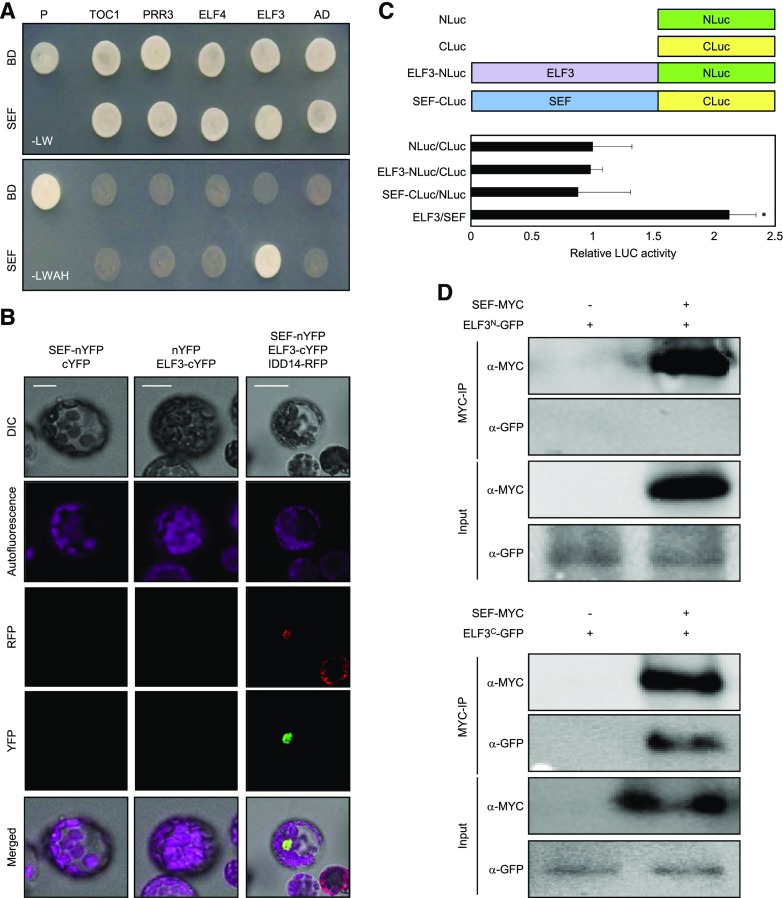
Having established a connection between the EC-dependent gene repression and H2A.Z-nucleosomes, we sought to investigate if there might be a direct physical interaction between the components involved in these processes. To test this possibility, we performed yeast-two-hybrid (Y2H) assays. Constructs of components of the SWR1 complex—ARP4, ARP6, SEF, SWR COMPLEX PROTEIN2, and SWR COMPLEX PROTEIN5, fused with GAL4 DNA-binding domain (BD) and evening-expressed clock components fused with GAL4 activation domain (AD)—were coexpressed in yeast cells. Cell growth on selective medium revealed that SEF interacts specifically with ELF3 (Fig. 4A; Supplemental Fig. S2). Although ELF4 is also a member of the EC (Huang et al., 2016), we did not observe interactions of SEF with ELF4 in yeast cells (Fig. 4A). Because the PIE1 protein is a catalytic core of SWR1 complex, we also examined interactions between PIE1 and EC components. We had difficulties cloning the full-length PIE1 and instead assayed domains of PIE1. Our results showed that the SANT domain of PIE1 was able to interact with ELF3 (Supplemental Fig. S3). Together, the results indicate that PIE1, possibly along with SEF, participate in the physical interactions with ELF3.
Figure 4.
Interactions of the SWR1 complex with EC. A, Y2H assays. Y2H assays were performed with the SEF protein fused to the DNA BD of GAL4 and evening-expressed clock components fused with the transcriptional AD of GAL4 for analysis of interactions. Interactions were examined by cell growth on selective media. -LWHA indicates Leu, Trp, His, and Ade drop-out plates. -LW indicates Leu and Trp drop-out plates. GAL4 was used as a positive control (P). B, BiFC assays. Partial fragments of YFP protein were fused with SEF and ELF3, and coexpressed in Arabidopsis protoplasts. Reconstituted fluorescence was examined by confocal microscopy. IDD14-RFP was used as a nucleus marker. Scale bars = 10 µm. DIC, differential interference contrast. C, Interaction of ELF3-NLuc with SEF-CLuc. Partial fragments of Luc (NLuc and CLuc) were fused with ELF3 or SEF. The fusion constructs were coexpressed in Arabidopsis protoplasts and Luc activities were measured and normalized against total protein. Three independent biological replicates were averaged and statistically analyzed with Student’s t test (*P < 0.05). Bars indicate the mean ± se. D, Co-IP assays. A. tumefaciens cells containing 35S:ELF3N(1-345aa)-GFP, 35S:ELF3C(346-695aa)-GFP, and 35S:SEF-MYC constructs were coinfiltrated to 3-week–old N. benthamiana leaves. Epitope-tagged proteins were detected immunologically using corresponding antibodies.
To confirm the physical interaction in vivo, we carried out bimolecular fluorescence complementation (BiFC) assays using Arabidopsis protoplasts. The SEF complementary DNA (cDNA) sequence was fused in-frame to the 5′-end of a gene sequence encoding the N-terminal half of yellow fluorescent protein (YFP), and the ELF3 gene was fused in-frame to the 5′-end of a sequence encoding the C-terminal half of YFP. The fusion constructs were then transiently coexpressed in Arabidopsis protoplasts. Strong yellow fluorescence was detected in the nucleus of cells coexpressing the combination of SEF-ELF3 (Fig. 4B), while coexpression with empty vectors did not show visible fluorescence (Fig. 4B). To quantify the physical interaction, split Luciferase (Luc) assay was also employed. ELF3 was fused with the amino part of Luc (NLuc), and SEF was fused with carboxyl-part of Luc (CLuc; Fig. 4C). Coexpression of ELF3-NLuc and SEF-CLuc in Arabidopsis protoplasts resulted in 2-fold increase of Luc activity, while coexpression of controls showed background-level Luc activities (Fig. 4C). The in planta interactions of ELF3 and SEF were confirmed by coimmunoprecipitation (Co-IP) assays using Nicotiana benthamiana cells transiently coexpressing 35S:ELF3-GFP and 35S:SEF-MYC fusion constructs. Because the full-length ELF3 protein is a large protein, it is difficult to express transiently in N. benthamiana. As an alternative, we designed fragments of ELF3 fused with GFP and used them to test physical interactions with SEF (Fig. 4D). Co-IP analysis revealed that the C-terminal region of ELF3 was responsible for interactions with SEF in planta (Fig. 4D). These results indicate that the EC directly interacts with the SWR1 complex, and this may facilitate the direct deposition of H2A.Z-nucleosomes at EC target genes.
H2A.Z-Nucleosomes Are Deposited at EC Target Loci
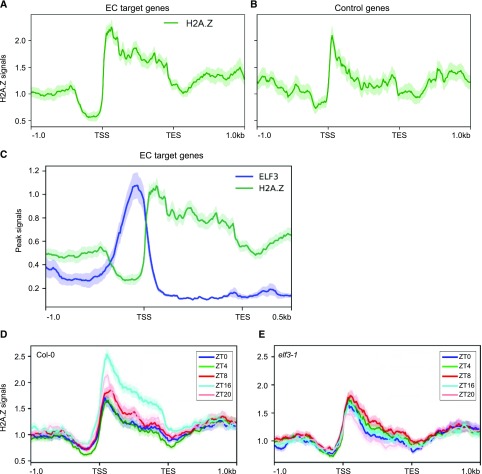
Because EC target genes are misregulated in arp6-1 and ELF3 and SEF interact directly, we investigated if H2A.Z-nucleosomes are enriched at EC target genes. HTA11 occupancy, assayed by chromatin-immunoprecipitation sequencing (ChIP-seq; Cortijo et al., 2017), showed a strong enrichment across the gene body of ELF3 targets (Fig. 5A). The occupancy was particularly high at the region surrounding the TSS at the presumptive +1 nucleosome, but was also markedly higher over the gene body of ELF3 targets (Fig. 5A). By comparison, randomly selected control genes showed H2A.Z enrichment around the TSS only (Fig. 5B; Supplemental Table S2).
Figure 5.
ELF3 and H2A.Z occupancy of EC target genes. A and B, H2A.Z enrichment (normalized HTA11-FLAG ChIP-seq reads) on 52 EC target genes (A) and control genes (B) was analyzed. For the class of “control genes,” a sample of 52 genes was randomly selected to compare their H2A.Z occupancy with EC target genes (see also Supplemental Table S2). (A) and (B) are plotted from 1 kb upstream of the TSS to 1 kb downstream of the TES of the corresponding genes. C, ELF3 and H2A.Z average binding plot on 52 EC target genes. D and E, H2A.Z enrichment in wild type (D) and elf3-1 mutant (E) at various time points under long day conditions.
We then compared the genome-wide association of ELF3 with H2A.Z. Consistent with the fact that the EC is recruited to target sites via LUX binding sites and G-box motifs (Ezer et al., 2017), we observed that these motifs were strongly enriched at ELF3-binding sites (Ezer et al., 2017). Notably, the footprint bound by ELF3 in the promoters of EC target genes, including PRR7, PRR9, and LUX, was devoid of H2A.Z-nucleosome signal (Fig. 5C; Supplemental Fig. S4, A–C), but the adjacent regions including the gene bodies were highly occupied with H2A.Z-nucleosomes (Fig. 5C; Supplemental Fig. S4, A–C). This antiphasing between H2A.Z occupancy and the EC was observed for all the EC targets (Fig. 5C; Supplemental Fig. S4, A–C). H2A.Z-nucleosomes are refractory to transcription (Thakar et al., 2010), suggesting that the initial binding of the EC to the promoters of target genes may assist subsequent recruitment of H2A.Z-nucleosomes to repress EC target gene expression.
We thus further determined whether H2A.Z deposition is dependent on the EC. ChIP-seq assays showed that the H2A.Z occupancy was observed in EC-target genes particularly during night time (Fig. 5D; Supplemental Fig. S4D), whereas enrichment of H2A.Z-nucleosomes at those loci was compromised in the elf3-1 mutant (Fig. 5E; Supplemental Fig. S4D). As a negative control, randomly selected control genes showed no temporal H2A.Z enrichment regardless of genetic background (Supplemental Fig. S4E). Further, we also obtained two additional lists of control genes: (1) genes bound by both ELF3 and LUX, but not upregulated in elf3-1 and lux-4 mutants (Supplemental Table S3); and (2) genes upregulated in elf3-1 and lux-4, but not bound by either ELF3 or LUX (Supplemental Table S4). Again, H2A.Z deposition was not diurnally regulated and influenced by ELF3 in both cases (Supplemental Figs. S5 and S6). These results indicate that the EC-SWR1 complex contributes to H2A.Z deposition at the EC target loci.
Temporal Regulation of H2A.Z Deposition at the PRR7 and PRR9 Promoters Underlies Proper Circadian Oscillation
Given the essential role of the EC in the circadian clock and the direct physical and functional interaction between the EC and the SWR1 complex, SWR1 might be also important for proper circadian clock function. We used genetic mutants affected in the key SWR1 subunits, arp6-1 and sef-1, and analyzed circadian function by monitoring the expression of a circadian output gene, COLD, CIRCADIAN RHYTHM, AND RNA BINDING2 (CCR2). RT-qPCR analysis revealed that the rhythmic amplitude of CCR2 expression was significantly dampened in the two mutants (Supplemental Fig. S7). Similarly, the circadian expression of CCR2 was also reduced in hta9-1 hta11-2 double mutant (Supplemental Fig. S8), in a comparable trend to that of SWR1 mutants. These results suggest that SWR1 activity and proper circadian exchange of H2A.Z-nucleosomes might be important for circadian function.
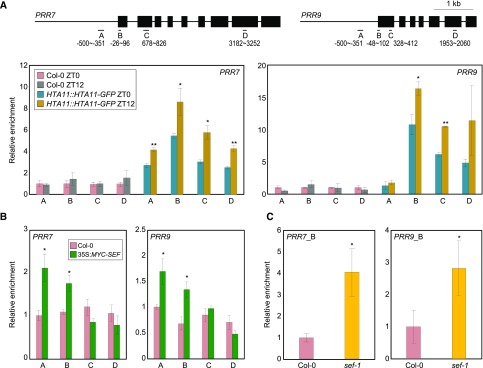
Next we aimed to decipher the clock factors that are direct targets of the SWR1 complex. We thus employed pHTA11:HTA11-GFP transgenic plants and examined H2A.Z deposition at core clock gene promoters by ChIP assays with a GFP-specific antibody. ChIP-qPCR analysis showed that H2A.Z deposition specifically occurs at the gene bodies of the morning-expressed genes PRR7 and PRR9 (Fig. 6A), whereas other core clock genes did not show significant diurnal enrichment of the H2A.Z variant (Supplemental Fig. S9). Furthermore, H2A.Z accumulation was primarily observed around dusk at the PRR7 and PRR9 loci (Fig. 6A), which is consistent with the temporal expression patterns of the ELF3 and SWR1 components (Fig. 2).
Figure 6.
H2A.Z deposition at PRR7 and PRR9 loci by the SWR1 complex. In (A) to (C), fragmented DNA was eluted from the protein–DNA complexes and used for qPCR analysis. Enrichment was normalized relative to eIF4A. Three independent biological replicates were averaged, and the statistical significance of the measurements was determined. Bars indicate the means ± se. A, Accumulation of H2A.Z at clock gene loci. Two-week–old plants grown under neutral day conditions were used for ChIP analysis with anti-GFP antibody. Gene structures are presented (upper representation). Underbars represent the amplified genomic regions. Statistically significant differences between ZT0 and ZT12 samples are indicated by asterisks (Student’s t test, *P < 0.05, **P < 0.01). B, Binding of SEF to clock gene promoters. Two-week–old 35S:MYC-SEF transgenic plants grown under neutral day conditions were harvested at ZT12. Statistically significant differences between wild-type and 35S:MYC-SEF plants are indicated by asterisks (Student’s t test, *P < 0.05). C, Recruitment of Pol II at PRRs in sef-1. Two-week–old plants grown under neutral day conditions were harvested at ZT12 and used for ChIP analysis with an anti-N terminus of Arabidopsis Pol II antibody. qPCR was performed with a primer pair amplifying the B region of each gene promoter (see also Fig. 6A). Statistically significant differences between wild-type and sef-1 plants are indicated by asterisks (Student’s t test, *P < 0.05).
We also generated transgenic plants overexpressing SEF (35S:MYC-SEF), in which the core component of the SWR1 complex, SEF, is fused in-frame to six copies of MYC-coding sequence. qPCR analysis after ChIP assays with an anti-MYC antibody showed that the SWR1 component binds to the PRR7 and PRR9 loci (Fig. 6B). These results support the specific association of SEF with the morning-expressed genes and agree with the pattern of H2A.Z deposition. H2A.Z-nucleosome deposition usually inhibits Pol II accessibility by stimulating closed chromatin formation (Kumar and Wigge, 2010). Consistent with this, Pol II recruitment was significantly increased at PRRs in the sef-1 mutant (Fig. 6C), which has lower catalytic activity of H2A.Z deposition. These results indicate that H2A.Z deposition occurs around dusk at EC target loci such as PRR7 and PRR9 to repress their expression.
ELF3 Is Necessary for H2A.Z Deposition at PRRs in the Control of Circadian Oscillation
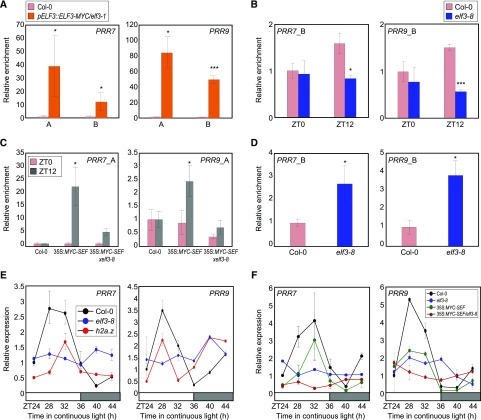
The SEF-ELF3 direct interaction suggests that there may be functional coordination in the temporal regulation of PRR7 and PRR9. To investigate this, we employed pELF3::ELF3-MYC/elf3-1 and 35S:MYC-ELF3 transgenic plants (Jang et al., 2015) and performed ChIP assays using an anti-MYC antibody. ChIP-qPCR analysis confirmed that ELF3 binds to the PRR7 and PRR9 loci (Fig. 7A), and this binding occurs preferentially at dusk (Supplemental Fig. S10). Consistent with the requirement of ELF4 and LUX in a functional EC, overexpression of ELF3 still resulted in rhythmic binding.
Figure 7.
H2A.Z exchange at PRR7 and PRR9 loci by ELF3. A, Binding of ELF3 to PRR promoters. Two-week–old pELF3::ELF3-MYC/elf3-1 seedlings grown under neutral day conditions were harvested at ZT12 and used to conduct ChIP assays. Statistically significant differences between Col-0 and pELF3::ELF3-MYC/elf3-1 plants are indicated by asterisks (Student’s t test, *P < 0.05, ***P < 0.001). B, H2A.Z deposition at clock gene promoters in elf3-8. Two-week–old plants grown under neutral day conditions were used for ChIP analysis with anti-H2A.Z antibody. Statistically significant differences between Col-0 and elf3-8 plants are indicated by asterisks (Student’s t test, *P < 0.05, ***P < 0.001). C, SEF binding to the PRR loci in elf3-8 background. Two-week–old plants grown under neutral day conditions were used for ChIP analysis with anti-MYC antibody. Statistically significant differences between ZT0 and ZT12 samples are indicated by asterisks (Student’s t test, *P < 0.05). D, Recruitment of Pol II at clock gene promoters in elf3-8. Two-week–old plants grown under neutral day conditions were harvested at ZT12 and used for ChIP analysis with an anti-N terminus of Arabidopsis Pol II antibody. Statistically significant differences between ZT0 and ZT12 samples are indicated by asterisks (Student’s t test, *P < 0.05). In (A) to (D), fragmented DNA was eluted from the protein–DNA complexes and used for qPCR analysis. Enrichment was normalized relative to eIF4A. Three independent biological replicates were averaged, and the statistical significance of the measurements was determined. Bars indicate the mean ± se. E, Circadian expression of PRR7 and PRR9 in elf3-8 and h2a.z. F, PRR expression in 35S:MYC-SEF/elf3-8. In (E) and (F), seedlings grown under neutral day conditions for 2 weeks were transferred to continuous light conditions at ZT0.
Chromatin binding of ELF3 is causal for H2A.Z deposition and is independent of H2A.Z occupancy. We genetically crossed pELF3::ELF3-MYC transgenic plants with arp6-1, which lacks genome-wide H2A.Z deposition, and the pELF3::ELF3-MYC x arp6-1 plants were used for ChIP assays using an anti-MYC antibody (Supplemental Fig. S11). As a result, ELF3 binding to PRR loci was comparable in both wild-type and arp6-1 backgrounds (Supplemental Fig. S12), indicating that ELF3 binding is an active process to trigger H2A.Z deposition at cognate regions.
To determine the possible regulation of H2A.Z deposition by ELF3, we examined H2A.Z occupancy by ChIP-qPCR at PRR7 and PRR9 in elf3-8. As expected, H2A.Z deposition was reduced at these loci in elf3-8, particularly around dusk (Fig. 7B; Supplemental Fig. S13), indicating that ELF3 is both present on these promoters and facilitates the insertion or stability of repressive H2A.Z-nucleosomes (Fig. 7B). Consistently, binding of SEF was specifically observed at dusk, and its association to the PRR7 and PRR9 promoters occurred in an ELF3-dependent manner (Fig. 7C).
Consistent with the role of the EC and SWR1 complex in the repression of gene expression, the EC-SWR1 complex contributes to the declining phase of PRR7 and PRR9 expression during the evening and night period, when its expression is returning to basal levels (Farré et al., 2005). As expected, Pol II accessibility of PRR7 and PRR9 was elevated at dusk in elf3-8 (Fig. 7D). In further support of these results, expression of PRR7 and PRR9 was elevated during evening and night-time in elf3-8 and H2A.Z-deficient hta8 hta9 hta11 (hta.z) mutants (Fig. 7E). In addition, consistent with the repressing functions of PRR7 and PRR9 on CCA1 and LHY (Nakamichi et al., 2010), expression of CCA1 and LHY was significantly repressed during subjective morning in elf3-8 and hta.z mutants (Supplemental Fig. S14). Clock-controlled PHYTOCHROME-INTERACTING FACTOR4 expression was also affected in elf3-8 and hta.z mutants, and especially, its expression was derepressed during night period (Supplemental Fig. S14). To confirm the genetic interactions of EC and SWR1, we crossed elf3-8 with 35S:MYC-SEF transgenic plants (Supplemental Fig. S15). 35S:MYC-SEF transgenic plants displayed repressed rhythmic expression of PRR7 and PRR9 compared with wild-type plants, whereas arrhythmic expression was observed in elf3-8 (Fig. 7F). Notably, the 35S:MYC-SEF/elf3-8 plants showed arrhythmicity similar to elf3-8 (Fig. 7F), indicating that the SWR1 complex depends on the EC in the control of circadian oscillation.
In summary, the EC interaction with the SWR1 complex provides a mechanism to recruit transcriptionally repressive H2A.Z-nucleosomes at circadian-regulated genes. In this way, targets may be repressed over the course of the night until the EC abundance declines and/or strong activation signals reactivate gene expression. In the case of circadian regulation, the PRR7 and PRR9 genes are under the temporal regulation of H2A.Z exchange. The EC-SWR1 complex is recruited to PRR7 and PRR9, facilitating H2A.Z deposition to reduce gene expression during evening and night-time. This indicates a role for the EC in establishing circadian waves of gene expression.
DISCUSSION
EC-Dependent Coordination of Chromatin Structures
The ELF3, ELF4, and LUX proteins form the tripartite EC complex and cooperatively regulate a variety of developmental processes. They are functionally intertwined in the gating of day-inducible genes (Hazen et al., 2005; Huang and Nusinow, 2016), and thus their genetic mutants share phenotypic alterations, such as elongated hypocotyls, early flowering, and altered circadian rhythms (Hicks et al., 2001; Hazen et al., 2005; Nusinow et al., 2011). Consistently, their binding sites largely overlap, and the EC controls target gene expression (Ezer et al., 2017). Here, we show that the EC recruits repressive chromatin domains to regulate the evening transcriptome. The EC interacts with the SWR1 complex and establishes transcriptionally repressive H2A.Z-nucleosomes at its target genes. The high occupancy of H2A.Z-nucleosomes across the gene bodies of target genes provides an effective barrier to RNA Pol II, maintaining these genes in a repressed state.
Notably, mutants eliminating EC activity, like elf3 and lux, exhibit an arrhythmic circadian clock. In contrast, SWR1 mutants are rhythmic but with reduced amplitude. Therefore, deposition of H2A.Z by the EC at core clock genes, such as PRR7 and PRR9, is important for correct expression amplitude. However, circadian clock rhythmicity appears not to depend on this mechanism. Additionally, we cannot rule out the possibility that ELF3 may recruit additional chromatin modifiers and/or remodelers. The EC might be an important platform facilitating chromatin reconfiguration with multiple epigenetic modifications.
Temporal exchange of H2A.Z-nucleosomes provides a mechanism for diurnal gating of several physiological processes. For example, a majority of stress-responsive genes are gated primarily during day period (Seo and Mas, 2015), and consistently, a substantial number of stress-inducible genes that are responsive to drought, temperature, or pathogens are under the control of EC-dependent H2A.Z deposition to ensure their suppression during night-time (Ezer et al., 2017). In addition, the circadian clock relies on a series of waves of transcriptional activation and repression. The recruitment of H2A.Z-nucleosomes provides a mechanism for the stable repression of target clock genes over the course of the night, which can be robustly activated the following day. Overall, the EC-SWR1 complex is a global transcriptional regulator that functions in the diurnal gating of many developmental and physiological processes and provides a more stable mechanism for maintaining repressive states during night-time.
Chromatin-Based Regulation at the Core of the Circadian Clock
Time-of-day–dependent accumulation of chromatin marks such as H3ac and H3K4me3 occurs with the circadian transcript abundance of clock genes including CCA1, LHY, TOC1, PRR7, PRR9, and LUX (Hsu et al., 2013; Voss et al., 2015). Mechanistically, H3ac stimulates an open chromatin conformation (Song and Noh, 2012), whereas H3K4me3 inhibits the binding of clock repressor proteins to the core clock gene promoters, avoiding advanced circadian repressor binding (Malapeira et al., 2012). The SDG2/ATXR3 histone methyltransferase contributes to the H3K4me3 accumulation and thus controls the timing of clock gene expression, from activation to repression (Malapeira et al., 2012). Several additional chromatin modifiers, including HISTONE DEACETYLASE6 (HDA6), HDA19, and JUMONJI C DOMAIN-CONTAINING PROTEIN30/JUMONJI DOMAIN CONTAINING5, are also connected with the circadian oscillation (Jones and Harmer, 2011; Lu et al., 2011; Wang et al., 2013), although the mechanisms behind circadian gene regulation remain to be determined.
The epigenetic regulation of core clock genes relies on a complex web of chromatin and clock components. For example, CCA1 facilitates repressive chromatin signatures to regulate TOC1 expression around dawn while HDAs contribute to the declining phase of TOC1 (Perales and Más, 2007). Another MYB-like transcription factor known as REVEILLE8/LHY-CCA1-LIKE5 favors H3 acetylation at the TOC1 promoter, most likely by antagonizing CCA1 function throughout the day (Farinas and Mas, 2011).
In this study, we identify a repressive chromatin state that shapes the rhythmic oscillations in gene expression. Circadian H2A.Z deposition underlies normal circadian oscillation, and PRR7 and PRR9 are primary targets of the SWR1 complex. H2A.Z-nucleosome deposition occurs around dusk, when the SWR1 components are highly expressed, to suppress gene expression. The interaction between the SWR1 complex and ELF3 provides a direct mechanism to facilitate H2A.Z exchange at cognate regions, contributing to precise oscillations in circadian gene expression. Because the PRR7 and PRR9 loci are also subjected to H3ac and H3K4me3 modifications (Malapeira et al., 2012), the higher-order combination of multiple chromatin modifications ultimately shapes the circadian waveforms of gene expression throughout the day–night cycle.
In mammals, rhythmical H2A.Z deposition at the promoters of CLOCK:BMAL1 targets has been observed, although the underlying mechanism is not known (Menet et al., 2014). This suggests that H2A.Z-nucleosomes may have a conserved function in the eukaryotic circadian clock.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana; Columbia-0 ecotype) was used for all experiments unless otherwise specified. Arabidopsis seeds were surface-sterilized and sown on 0.7% (w/v) agar plates containing half-strengthened Murashige and Skoog media. After 3-d stratification at 4°C, the seeds were grown in a reach-in plant growth chamber (Conviron) with 170 µmol m−2 s−1 light intensity and 70% humidity under short day (8-h light/16-h dark), neutral day (12-h light/12-h dark), or long day (16-h light/8-h dark) conditions at 22°C or 27°C, as indicated in figure legends. The arp6-1, arp6-3, elf3-1, elf3-8, hta9-1hta11-2, pHTA11:HTA11-FLAG, pHTA11:HTA11-GFP, and sef-1 plants were previously reported (March-Díaz et al., 2007; Kumar and Wigge, 2010; Coleman-Derr and Zilberman, 2012; Rosa et al., 2013; Nitschke et al., 2016). To produce transgenic plants overexpressing the SEF and ELF3 genes, a full-length cDNA was subcloned into the binary pBA002 vector under the control of the Cauliflower mosaic virus 35S promoter. Agrobacterium tumefaciens-mediated Arabidopsis transformation was then performed.
MNase-Seq and ChIP-Seq Experiments and Analysis
Approximately 1 g of 9-d–old Arabidopsis seedlings was harvested at the time points indicated in figure legends. The harvested seedlings were ground in liquid nitrogen to fine powder. The tissue powder was fixed for 10 min with 1% (v/v) formaldehyde (cat. no. F8775; Sigma-Aldrich) in buffer 1 (0.4 m of Suc, 10 mm of HEPES, 10 mm of MgCl2, 5 mm of β-mercaptoethanol, 0.1 mm of phenylmethylsulfonyl fluoride (PMSF), and 1× protease inhibitor; cat. no., 11836145001; Roche). The reaction was quenched by adding Gly to a final concentration of 127 mM. The homogenate was filtered through Miracloth (EMD Millipore) twice and centrifuged to collect the pellet. The pellet was washed in buffer 2 (0.24 m of Suc, 10 mm of Tris-HCl at pH 8.0, 10 mm of MgCl2, 5 mm of β-mercaptoethanol, 0.1 mm of PMSF, 1× protease inhibitor, and 1% [v/v] Triton) and then spun down in buffer 3 (1.7 m of Suc, 10 mm of Tris-HCl at pH 8.0, 10 mm of MgCl2, 5 mm of β-mercaptoethanol, 0.1 mm of PMSF, 1× protease inhibitor, and 0.15% [v/v] Triton). The nuclei pellet was then resuspended in MNase Digestion buffer (20 mm of Tris-HCl at pH 8.0, 50 mm of NaCl, 1 mm of DTT, 0.5% [v/v] NP-40, 1 mm of CaCl2, 0.5 mm of PMSF, and 1× protease inhibitor) and was flash-frozen in liquid nitrogen twice to break the nuclear envelope. Chromatin was digested by adding MNase (cat. no. N3755; Sigma-Aldrich) to a final concentration of 0.4 U/mL for 12.5 min. The reaction was terminated by adding EDTA to a final concentration of 5 mM. For ChIP-seq samples, H2A.Z in transgenic line pHTA11:HTA11-FLAG was immunoprecipitated by anti-FLAG M2 magnetic beads (cat. no. M8823; Sigma-Aldrich) and then eluted with 3XFLAG peptide (cat. no. B23112; Bimake). After reverse cross linking, the DNA was purified with solid phase reversible immobilization beads. The libraries were constructed using TruSeq ChIP Sample Preparation Kit (cat. no. IP-202-1024; Illumina) according to the manufacturer’s instructions. The libraries were sequenced on a NextSeq 500 (Illumina) platform. The raw reads obtained from the sequencing facilities were analyzed using a combination of publicly available software and in-house scripts. We first assessed the quality of reads using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Potential adaptor contamination and low quality trailing sequences were removed using the tool Trimmomatic (Bolger et al., 2014). Then, the reads were mapped to The Arabidopsis Information Resource 10 reference genome using the software BowTie2 (Langmead and Salzberg, 2012). Duplicates were removed with the tool Picard (https://github.com/broadinstitute/picard) and the read counts was normalized by the sample’s genome-wide reads coverage. Nucleosome positioning and occupancy were determined using the program DANPOS (Chen et al., 2013). Nucleosome and H2A.Z average binding profiles and heatmaps were generated using the program deepTools (Ramírez et al., 2014).
The “randomly selected control genes” were generated using the “sample()” function in R. The indices of the 52 genes were generated by the R code “index ← sample(33557, 52),” where “33,557” is the total number of Arabidopsis genes. The corresponding Arabidopsis Genome Initiative gene names were generated by the R code “random52 ← all_gene_names[index,].” In addition, we also obtained two additional lists of control genes with new filters: genes bound by both ELF3 and LUX, but not upregulated in elf3-1 and lux-4 mutants (Supplemental Table S3); and genes upregulated in elf3-1 and lux-4, but not bound by either ELF3 or LUX (Supplemental Table S4). The filter criteria for the former gene list are: having both ELF3 and LUX binding within 1,000 bp upstream of the gene; and the log fold changes of Transcripts Per Million (TPM) values compared with Col-0 at ZT16 are smaller than 0.5-fold in both elf3-1 and lux-4 mutants. The filter criteria for the latter gene list are: having no ELF3 or LUX binding within 1,000 bp upstream of the gene; and the log fold changes of TPM values compared with Col-0 at ZT16 are larger than 1.5-fold in both elf3-1 and lux-4 mutants.
RNA-Seq Experiment and Analysis
Approximately 30 mg of 7-d–old Arabidopsis seedlings were harvested and their total RNA was extracted using the MagMAX-96 Total RNA Isolation kit (AM1830; Ambion) according to manufacturer’s instructions. Library preparation was performed using 1 μg of high integrity total RNA (RNA integrity number > 8) using the TruSeq Stranded mRNA library preparation kit (cat. no. RS-122-2103; Illumina) according to the manufacturer’s instructions. The libraries were sequenced on a NextSeq 500 platform (Illumina).
For bioinformatics analysis, we first assessed the quality of reads using the program FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Potential adaptor contamination and low quality trailing sequences were removed using the tool Trimmomatic (Bolger et al., 2014), before alignment to The Arabidopsis Information Resource 10 transcriptome using the software TopHat (Trapnell et al., 2009). Potential optical duplicates resulting from library preparation were removed using the Picard tools (https://github.com/broadinstitute/picard), and the read counts were normalized by the sample’s genome-wide reads coverage. Raw counts were determined by HTseq-count (Anders et al., 2015), and the software Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) was utilized to calculate Fragments Per Kilobase Million, which was then converted into TPM.
RT-qPCR Analysis
Total RNA was extracted using TRI reagent (TAKARA Bio) according to the manufacturer’s recommendations. Reverse transcription was performed using Moloney Murine Leukemia Virus reverse transcriptase (Dr. Protein) with oligo(dT18) to synthesize first-strand cDNA from 2 μg of total RNA. Total RNA samples were pretreated with an RNase-free DNase. cDNAs were diluted to 100 μL with TE buffer, and 1 μL of diluted cDNA was used for PCR amplification.
RT-qPCR reactions were performed in 96-well blocks using the Step-One Plus Real-Time PCR System (Applied Biosystems). The PCR primers used are listed in Supplemental Table S5. The values for each set of primers were normalized relative to the EUKARYOTIC TRANSLATION INITIATION FACTOR 4A1 (eIF4A) gene (At3g13920). All RT-qPCR reactions were performed with biological triplicates using total RNA samples extracted from three independent replicate samples. The comparative ΔΔCT method was employed to evaluate relative quantities of each amplified product in the samples. The threshold cycle (CT) was automatically determined for each reaction with the analysis software set using default parameters. The specificity of the RT-qPCR reactions was determined by melting curve analysis of the amplified products using the standard method employed by the software.
Y2H Assays
Y2H assays were performed using the BD Matchmaker system (Clontech). The pGADT7 vector was used for the GAL4 AD fusion, and the pGBKT7 vector was used for GAL4 BD fusion. The yeast strain AH109 harboring the LacZ and His reporter genes was used. PCR products were subcloned into the pGBKT7 and pGADT7 vectors. The expression constructs were cotransformed into yeast AH109 cells and transformed cells were selected by growth on SD/-Leu/-Trp medium and SD/-Leu/-Trp/-His/-Ade. Interactions between proteins were analyzed by measuring β-galactosidase activity using ο-nitrophenyl-β-d-galactopyranoside as a substrate.
BiFC Assays
The ELF3 gene was fused in-frame to the 5′ end of a gene sequence encoding the C-terminal half of EYFP in the pSATN-cEYFP-C1 vector (E3082). The SEF cDNA sequence was fused in-frame to the 5′ end of a gene sequence encoding the N-terminal half of EYFP in the pSATN-nEYFP-C1 vector (E3081). Expression constructs were cotransformed into Arabidopsis protoplasts. Expression of the fusion constructs was monitored by fluorescence microscopy using a model no. LSM510 confocal microscope (Carl Zeiss).
ChIP Assays
The epitope-tagged transgenic plant samples were cross linked with 1% (v/v) formaldehyde, ground to powder in liquid nitrogen, and then sonicated. The sonicated chromatin complexes were bound with corresponding antibodies. Anti-MYC (cat. no. 05-724; Millipore), anti-Pol II (cat. no. sc-33754; Santa Cruz Biotechnology), anti-H2A.Z antibodies (cat. no. ab4174; Abcam), and salmon sperm DNA/protein A agarose beads (cat. no. 16-157; Millipore) were used for ChIP. DNA was purified using phenol/chloroform/isoamyl alcohol and sodium acetate (pH 5.2). The level of precipitated DNA fragments was quantified by qPCR using specific primer sets (Supplemental Table S6). Values were normalized according to input DNA levels. Values for control plants were set to 1 after normalization against eIF4a for qPCR analysis.
Split‐Luc Assays
The coding regions of SEF and ELF3 were cloned into pcFLucC or pcFLucN vector. The recombinant constructs were cotransformed with into Arabidopsis protoplasts by polyethylene glycol-mediated transformation. The pUBQ10::GUS plasmid was also cotransformed as an internal control to normalize the LUC activity.
Co-IP Assays
A. tumefaciens cells containing 35S:SEF-MYC and 35S:ELF3-GFP constructs were infiltrated to 3-week–old Nicotiana benthamiana leaves. N. benthamiana leaves were homogenized in protein extraction buffer (25 mM of Tris-HCl at pH 7.5, 150 mM of NaCl, 5% v/v glycerol, 0.05% w/v Nonidet P-40, 2.5 mM of EDTA, 1 mM of PMSF, and 1× complete cocktail of protease inhibitors). After protein extraction, anti-MYC antibodies (cat. no. 05-724; Millipore) coupled to Protein-A sepharose beads (Sigma-Aldrich) were mixed and incubated for 4 h at 4°C. The precipitated samples were washed at least four times with the protein extraction buffer and then eluted by 1× SDS-PAGE loading buffer to perform SDS-PAGE with anti-MYC (1:2,000 dilution; Millipore) or anti-GFP antibodies (1:1,000 dilution; cat. no. sc-9996; Santa Cruz Biotechnology).
Statistical Analysis
Unless otherwise specified, quantitative data are presented as mean ± sd and significance was assessed by the two-tailed Student’s t test.
Accession Numbers
The raw sequencing data reported in this work have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE109101.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Diurnal expression of H2A.Z-related genes.
Supplemental Figure S2. Y2H assays.
Supplemental Figure S3. Interaction of ELF3 with PIE1.
Supplemental Figure S4. H2A.Z occupancy on EC-target genes.
Supplemental Figure S5. H2A.Z enrichment at genes bound by both ELF3 and LUX, but not differentially expressed in elf3-1 and lux-4 mutants.
Supplemental Figure S6. H2A.Z enrichment at genes upregulated in elf3-1 and lux-4, but not bound by either ELF3 or LUX.
Supplemental Figure S7. Reduced rhythmic amplitude in genetic mutants of SWR1 components.
Supplemental Figure S8. Influences on circadian clock in hta9-1hta11-2.
Supplemental Figure S9. Deposition of H2A.Z at clock gene promoters.
Supplemental Figure S10. Binding of ELF3 to the PRR7 and PRR9 promoters at ZT12.
Supplemental Figure S11. Protein accumulation of ELF3 in pELF3::ELF3-MYC and pELF3::ELF3-MYCxarp6-1.
Supplemental Figure S12. ELF3 binding to the PRR loci in arp6-1 background.
Supplemental Figure S13. H2A.Z deposition in elf3-8 mutant throughout a day.
Supplemental Figure S14. Expression of CCA1, LHY, and PHYTOCHROME-INTERACTING FACTOR4 in h2a.z and elf3-8 mutants.
Supplemental Figure S15. Protein accumulation of SEF in 35S:MYC-SEF and 35S:MYC-SEFxelf3-8.
Supplemental Table S1. EC target gene list.
Supplemental Table S2. Fifty-two randomly selected control genes.
Supplemental Table S3. List of genes bound by both ELF3 and LUX, but not differentially expressed in elf3-1 and lux-4 mutants.
Supplemental Table S4. List of genes upregulated in elf3-1 and lux-4, but not bound by either ELF3 or LUX.
Supplemental Table S5. Primers used in RT-qPCR analysis.
Supplemental Table S6. Primers used in ChIP assays.
Acknowledgments
We thank Dr. Hui Lan for bioinformatics data analysis.
Footnotes
This work was supported by the National Research Foundation of Korea’s Basic Science Research programs (NRF-2019R1A2C2006915), the Basic Research Laboratory (NRF-2017R1A4A1015620), and the Rural Development Administration’s Next-Generation BioGreen 21 Program (PJ01314501 to P.J.S.); the Federación Española de Enfermedades Raras/Spanish Ministry of Economy and Competitiveness, the Ramon Areces Foundation, the Generalitat de Catalunya (Agency for Management of University and Research Grants), the Centres de Recerca de Catalunya Program/Generalitat de Catalunya, and the Spanish Ministry of Economy and Competitiveness “Severo Ochoa Program for Centers of Excellence in R&D” 2016–2019 (SEV-2015-0533 to P.M.); and the Gatsby Foundation (GAT3273/GLB to P.A.W.).
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AAR, et al. (2015) ELF3 controls thermoresponsive growth in Arabidopsis. Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Carré IA, Kim JY (2002) MYB transcription factors in the Arabidopsis circadian clock. J Exp Bot 53: 1551–1557 [DOI] [PubMed] [Google Scholar]
- Chen K, Xi Y, Pan X, Li Z, Kaestner K, Tyler J, Dent S, He X, Li W (2013) DANPOS: Dynamic analysis of nucleosome position and occupancy by sequencing. Genome Res 23: 341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Helfer A, Nusinow DA, Kay SA (2012) ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 7: 170–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D (2012) Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo S, Charoensawan V, Brestovitsky A, Buning R, Ravarani C, Rhodes D, van Noort J, Jaeger KE, Wigge PA (2017) Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Mol Plant 10: 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Kandasamy MK, McKinney EC, Meagher RB (2005) The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017) The Evening Complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, McClung CR (2015) Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610 [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol 3: e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Albertson TM, Wagner DR (2001) EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell 13: 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks KA, Millar AJ, Carré IA, Somers DE, Straume M, Meeks-Wagner DR, Kay SA (1996) Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA (2016) Identification of Evening Complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics 15: 201–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Nusinow DA (2016) Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet 32: 674–686 [DOI] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Jang K, Lee HG, Jung SJ, Paek NC, Seo PJ (2015) The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci Rep 5: 12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Harmer S (2011) JMJD5 functions in concert with TOC1 in the Arabidopsis circadian system. Plant Signal Behav 6: 445–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with BowTie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, Tobin EM (2011) The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol 155: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Webb CJ, Knowles SM, Kim SH, Wang Z, Tobin EM (2012) CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol 158: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira J, Khaitova LC, Mas P (2012) Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc Natl Acad Sci USA 109: 21540–21545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R, García-Domínguez M, Florencio FJ, Reyes JC (2007) SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol 143: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- March-Díaz R, Reyes JC (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M (2014) CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 28: 8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitschke S, Cortleven A, Iven T, Feussner I, Havaux M, Riefler M, Schmülling T (2016) Circadian stress regimes affect the circadian clock and cause jasmonic acid-dependent cell death in cytokinin-deficient Arabidopsis plants. Plant Cell 28: 1616–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh YS, Amasino RM (2003) PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15: 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P (2007) A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell 19: 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Mas P, Millar AJ (2013) Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst Biol 7: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD (2005) Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Madhani HD (2006) Patterning chromatin: Form and function for H2A.Z variant nucleosomes. Curr Opin Genet Dev 16: 119–124 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T (2014) deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res 42: W187–W191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ (2003) Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J 22: 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Von Harder M, Cigliano RA, Schlögelhofer P, Mittelsten Scheid O (2013) The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 25: 1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, McClung CR (2004) The Arabidopsis thaliana clock. J Biol Rhythms 19: 425–435 [DOI] [PubMed] [Google Scholar]
- Salomé PA, Weigel D, McClung CR (2010) The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Mas P (2014) Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Mas P (2015) STRESSing the role of the plant circadian clock. Trends Plant Sci 20: 230–237 [DOI] [PubMed] [Google Scholar]
- Shu H, Gruissem W, Hennig L (2013) Measuring Arabidopsis chromatin accessibility using DNase I-polymerase chain reaction and DNase I-chip assays. Plant Physiol 162: 1794–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HR, Noh YS (2012) Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol Cells 34: 279–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Green R (2011) RNA-based regulation in the plant circadian clock. Trends Plant Sci 16: 517–523 [DOI] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG (2005) The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev 19: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar A, Gupta P, McAllister WT, Zlatanova J (2010) Histone variant H2A.Z inhibits transcription in reconstituted nucleosomes. Biochemistry 49: 4018–4026 [DOI] [PubMed] [Google Scholar]
- To TK, Kim JM (2014) Epigenetic regulation of gene responsiveness in Arabidopsis. Front Plant Sci 4: 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, et al. (2015) The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat Commun 6: 7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S, Yakir E, Green RM (2011) Circadian clocks and adaptation in Arabidopsis. Mol Ecol 20: 1155–1165 [DOI] [PubMed] [Google Scholar]