Covalent mRNA modifications play crucial roles in gene regulation and are required for plant embryonic and postembryonic development.
Abstract
Posttranscriptional control of gene expression is indispensable for the execution of developmental programs and environmental adaptation. Among the many cellular mechanisms that regulate mRNA fate, covalent nucleotide modification has emerged as a major way of controlling the processing, localization, stability, and translatability of mRNAs. This powerful mechanism is conserved across eukaryotes and controls the cellular events that lead to development and growth. As in other eukaryotes, N6-methylation of adenosine is the most abundant and best studied mRNA modification in flowering plants. It is essential for embryonic and postembryonic plant development and it affects growth rate and stress responses, including susceptibility to plant RNA viruses. Although the mRNA modification field is young, the intense interest triggered by its involvement in stem cell differentiation and cancer has led to rapid advances in understanding how mRNA modifications control gene expression in mammalian systems. An equivalent effort from plant molecular biologists has been lagging behind, but recent work in Arabidopsis (Arabidopsis thaliana) and other plant species is starting to give insights into how this essential layer of posttranscriptional regulation works in plants, and both similarities and differences with other eukaryotes are emerging. In this Update, we summarize, connect, and evaluate the experimental work that supports our current knowledge of the biochemistry, molecular mechanisms, and biological functions of mRNA modifications in plants. We devote particular attention to N6-methylation of adenosine and attempt to place the knowledge gained from plant studies within the context of a more general framework derived from studies in other eukaryotes.
Control of gene expression is of paramount importance in biology, and a variety of molecular mechanisms working at the DNA, pre-mRNA, mRNA, and protein levels have evolved to ensure that appropriate levels of activity of gene products operate at any given time. It has long been recognized that chemical modification of the transcription template, either of the DNA itself or of associated histone proteins, is key to the regulation of gene expression. Thanks to rapid progress in the last decade, it has now become clear that chemical modification of mRNA also plays crucial roles in the control of endogenous gene activity and in shaping the outcome of host-virus interactions. This Update provides a brief overview of mRNA modifications identified in plants and their viruses, and an account of what is known of their functional relevance and the molecular mechanisms underlying their functions. We also include a discussion of emerging controversies and outstanding questions in this field.
SEEING MRNA MODIFICATIONS: HIGH-THROUGHPUT SEQUENCING DELIVERS THE BREAKTHROUGH
Although chemically modified nucleotides have been known to exist in RNA, even in mRNA, since more than 40 years (Holley et al., 1965; Desrosiers et al., 1974; Perry and Kelley, 1974; Boccaletto et al., 2018), the mRNA modification field has only taken off with the recent introduction of high-throughput sequencing-based methods to map modified nucleotides in specific mRNAs transcriptome wide (see Box 1 for a historical context of the development of the field). Three main categories of such methods enjoy widespread use (see Linder and Jaffrey [2019] for a detailed review focused on methods). (1) Antibodies specific to a modification are used to immunoprecipitate fragmented mRNA, and RNA sequencing (RNA-seq) is applied to input and immunoprecipitated RNA fractions. Significant enrichment then identifies mRNA intervals likely to contain a modified nucleotide (Dominissini et al., 2012; Meyer et al., 2012; Edelheit et al., 2013; Schwartz et al., 2013; Luo et al., 2014; Delatte et al., 2016; Dominissini et al., 2016; Li et al., 2016; Cui et al., 2017). (2) Modified nucleotides are specifically derivatized using unique reactivity with an appropriate compound, and the presence of the derived nucleotide is read either as a mutation signature or as a stop upon reverse transcription (Squires et al., 2012; Carlile et al., 2014; Lovejoy et al., 2014; Schwartz et al., 2014a; Li et al., 2015; David et al., 2017; Enroth et al., 2019; Sun et al., 2019; Zhang et al., 2019b). The cross-linking immunoprecipitation (CLIP) variant miCLIP constitutes an important special case in this category: a modification-specific antibody is UV cross-linked to purified RNA, allowing mutation-dependent mapping of cross-link sites (Ke et al., 2015; Linder et al., 2015) as in other CLIP techniques (Ule et al., 2018). (3) Differential RNA cleavage at modified versus unmodified nucleotides is used to identify modified sites based on the percentage of sequence read-throughs and stops at each nucleotide (Birkedal et al., 2015; Garcia-Campos et al., 2019; Zhang et al., 2019d). In addition to those three types of methods, direct RNA sequencing by Oxford Nanopore Technology (Garalde et al., 2018) has recently been used successfully to map N6-methyladenosine (m6A) sites based on systematic base-calling errors at modified sites (Liu et al., 2019; Parker et al., 2019; Yang et al., 2019). In general, methods in categories 2 and 3 produce modification maps with nucleotide resolution, while methods in category 1 produce intervals containing a modification. It remains an active field of research to develop methods to detect modifications for two reasons. First, new methods for the detection of known RNA modifications are in high demand, because no single method developed thus far perfectly combines the desired sensitivity and specificity: high-confidence modification sites should ideally rely on results obtained by two orthogonal methods (Zhang et al., 2019b). Indeed, antibody-based methods in categories 1 and 2 have been used since 2012 to detect m6A sites (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2013; Luo et al., 2014; Ke et al., 2015; Linder et al., 2015), but the recent development of RNase cleavage-dependent m6A mapping (Garcia-Campos et al., 2019; Zhang et al., 2019d) shows that the limited sensitivity of antibody-based methods led to substantial underestimation of the number of m6A sites in the transcriptome. Second, it is possible that some mRNA modifications await discovery, thus necessitating the development of methods for their detection.
MRNA MODIFICATIONS IDENTIFIED IN PLANTS AND OTHER EUKARYOTES
In plants, there are now reports on mapping of three distinct covalent nucleotide modifications in the bodies of mRNA: m6A (Li et al., 2014c; Luo et al., 2014; Wan et al., 2015; Shen et al., 2016; Duan et al., 2017; Anderson et al., 2018; Miao et al., 2019; Parker et al., 2019), 5-methylcytidine (m5C; Cui et al., 2017; David et al., 2017; Yang et al., 2019), and pseudouridine (Ψ; Sun et al., 2019). Other types of mRNA modifications, including C-U editing of mitochondrial and plastidial mRNAs (Shikanai, 2006), alternative 5′-caps (Kiledjian, 2018; Wang et al., 2019), and untemplated addition of nucleotides to mRNA 3′-ends (De Almeida et al., 2018), will not be covered here. The discovery of mRNA modifications in plants is lagging behind that in animal cells, in which at least six additional modifications have been mapped and, in some cases, also functionally analyzed (Boccaletto et al., 2018). These modified nucleotides include inosine (I; Bass and Weintraub, 1988; Shevchenko and Morris, 2018), internal (as opposed to cap) 7-methylguanosine (m7G; Zhang et al., 2019b), cap-proximal 2′-O,N6-dimethyladenosine (m6Am; Wei et al., 1975a; Linder et al., 2015; Mauer et al., 2017; Boulias et al., 2019), N1-methyladenosine (m1A; Dominissini et al., 2016; Li et al., 2016, 2017b; Safra et al., 2017), 4-acetylcytidine (ac4C; Arango et al., 2018), 5-hydroxymethylcytidine (hm5C; Delatte et al., 2016), and 2′-O-methylation (any nucleotide [Nm]; Furuichi et al., 1975; Wei et al., 1975b; Dai et al., 2017; Bartoli et al., 2018). For at least one of these modifications, m7G, plant homologs exist of the enzyme responsible for its introduction into tRNA (Alexandrov et al., 2002) and some mRNA sites (Zhang et al., 2019b). For others, such as ac4C and hm5C, the identified modifying enzymes do not have direct counterparts in plants.
Regardless of the existence of close homologs in plant genomes of RNA-modifying enzymes identified in mammalian cells, it is possible that chemical modifications in plant mRNA, not necessarily limited to the ones shown to occur in other eukaryotes, await discovery and functional characterization. Some studies do indeed provide evidence for the presence of additional modified nucleotides in plant mRNA. For example, liquid chromatography-mass spectrometry analysis of total hydrolysates indicates the presence of internal m7G in mRNA of several plant species (Chu et al., 2018). In addition, a thorough examination of sites of recurrent misincorporation by reverse transcriptase in Arabidopsis (Arabidopsis thaliana) RNA-seq data suggested the rare presence of several mRNA modifications with potential to alter Watson-Crick base pairing (Vandivier et al., 2015). These included 3-methylcytidine (m3C), whose presence in some mRNAs was validated by RNA immunoprecipitation with a m3C-specific antibody (Vandivier et al., 2015). It is worth noting, however, that the number of misincorporation sites was orders of magnitude higher in the uncapped mRNA pool undergoing degradation than in intact, capped mRNAs. Thus, rather than resulting from regulated modification, many modified nucleotides identified by this approach may be chemically damaged (e.g. by oxidation) such that they accumulate throughout the life of an mRNA until its degradation. It is also important to note that, despite their clear utility, detection methods of modified nucleotides involving total hydrolysis of poly(A+) fractions such as liquid chromatography-mass spectrometry and thin-layer chromatography may easily include contamination from tRNA and/or rRNA in which the relative, and certainly also the absolute, content of modified nucleotides other than m6A is manyfold higher than in mRNA (Boccaletto et al., 2018). The difficulty in obtaining pure mRNA is highlighted in a study by Legrand et al. (2017), in which total RNA subjected to two rounds of poly(A) selection followed by two consecutive rounds of rRNA depletion still contained 7.1% rRNA.
CARTOGRAPHY OF MRNA MODIFICATIONS IN PLANTS
Cartography of mRNA modifications (i.e. their mapping to specific sites in mRNAs) is very much in its infancy, particularly in plants, in which m6A stands out as the so-far best understood mRNA modification, with only limited knowledge available on Ψ and m5C. A recent report shows that Ψ exists in hundreds of mRNAs in Arabidopsis (Sun et al., 2019), but the functions of Ψ in mRNA or the identities of pseudouridine synthases responsible for mRNA pseudouridylation remain unknown. Three different studies have mapped hundreds of m5C sites in Arabidopsis mRNA, but there is discrepancy in assessments of whether m5C is enriched in coding sequences or in 3′-untranslated regions (UTRs), perhaps as a consequence of the different tissues used or because of actual inconsistencies between results obtained by the two different m5C mapping methods employed (Cui et al., 2017; David et al., 2017). These studies also showed that the tRNA methyltransferase NSUN2/TRM4B may catalyze C5-cytidine methylation in mRNA, because some mRNA-m5C sites are lost in trm4b mutants (Cui et al., 2017; David et al., 2017). In addition, quantitative differences in the extent of m5C modification of specific mRNAs between different tissues were noted, perhaps pointing to regulatory properties of this modification (David et al., 2017). In one case, orthogonal methods were used for m5C mapping, yielding a high-confidence set of m5C sites (Yang et al., 2019). Interestingly, this report also showed that m5C facilitates mRNA long-distance transport through the phloem (Yang et al., 2019), but the underlying mechanisms of this important phenomenon, including the identities of possible m5C-binding proteins, remain unknown. We focus the remainder of this Update on m6A, simply because, at present, its importance is more clearly established and the molecular mechanisms involved are better studied than for any other mRNA modification.
BIOCHEMICAL FRAMEWORK FOR THE FUNCTION OF M6A
Occurrence of m6A in Plant Transcriptomes
In agreement with early biochemical studies of animal, viral, and plant mRNA (Wei et al., 1976; Dimock and Stoltzfus, 1977; Schibler et al., 1977; Wei and Moss, 1977; Canaani et al., 1979; Nichols and Welder, 1981), transcriptome-wide mapping of m6A has identified RR[m6A]CH (R = A/G, H = A/C/U) as the most significantly enriched motif in m6A peaks of all eukaryotes analyzed to date (Dominissini et al., 2012; Meyer et al., 2012; Schwartz et al., 2013; Ke et al., 2015; Linder et al., 2015; Lence et al., 2016; Zhao et al., 2017; Li et al., 2019), including plants (Luo et al., 2014; Wan et al., 2015; Shen et al., 2016; Duan et al., 2017; Wang et al., 2017; Anderson et al., 2018; Miao et al., 2019; Parker et al., 2019). This finding strongly supports the existence of a conserved mechanism of m6A deposition in eukaryotic mRNA and is in perfect agreement with the fact that the adenosine methyltransferase complex is conserved across eukaryotes but not in prokaryotes, in which m6A occurs in a completely different sequence context in mRNA (Box 2; Deng et al., 2015). In addition to its presence in a defined consensus motif, m6A was predominantly found in the 3′-UTR of mRNAs by most of the above-mentioned studies in eukaryotes, again supporting a highly conserved mechanism of adenosine methylation, while it is distributed more or less evenly along prokaryotic transcripts (Deng et al., 2015). Nonetheless, recent reports have raised the question of whether different m6A motifs exist in plants (Li et al., 2014c; Anderson et al., 2018; Wei et al., 2018b; Luo et al., 2019; Miao et al., 2019; Zhang et al., 2019a), a debate that is detailed in Box 2 and illustrated in Figure 1. It is our judgment that the current evidence most strongly supports predominant use of the pan-eukaryotic RR[m6A]CH motif. In spite of this, there does appear to be a difference in the functional categories of m6A-containing mRNAs between plants and cultured animal cells. In plants, many m6A-containing mRNAs encode ribosomal and photosynthesis-related proteins, mitochondrial electron transport factors, and other basic metabolic enzymes (Luo et al., 2014; Wan et al., 2015; Shen et al., 2016; Wang et al., 2017; Anderson et al., 2018), while such housekeeping factors appear to be depleted from sets of m6A-containing mRNAs in yeast and mammalian cell cultures (Schwartz et al., 2014b; Ke et al., 2017). It is a question of outstanding importance to define if and how the labeling of these many housekeeping transcripts by m6A contributes to plant growth and development. Finally, a recent report deposited in bioRxiv shows that primary microRNA transcripts in Arabidopsis also contain m6A and proposes that the modification regulates microRNA biogenesis (Bhat et al., 2019), as previously proposed in animals (Alarcón et al., 2015; Berulava et al., 2015).
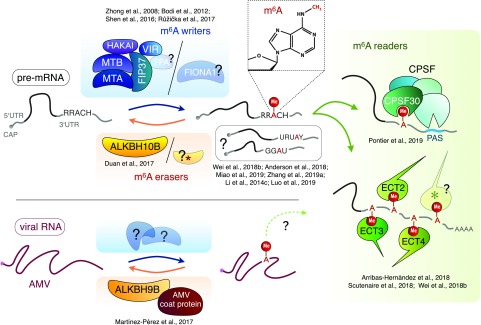
Figure 1.
Schematic representation of the m6A pathway and the functions of its characterized components. m6A writers are depicted on a blue field, readers on green, and erasers on orange. The canonical m6A consensus motif (RRACH) is chosen for the general representation, but alternative motifs (UGUAY and GGAU) recently proposed as plant specific are indicated in a separate box (see Box 2 for details). An endogenous m6A target is depicted as a pre-mRNA to highlight the connection of m6A writing to transcription. YTHDF m6A readers (ECT2, ECT3, ECT4, and probably additional ECTs) are represented as binding to the same transcript for convenience, but there are still no data clarifying whether different ECTs can bind in cis or not. If so, they could compete or have synergistic effects, perhaps interacting with each other as proposed for animal YTHDFs (Shi et al., 2017). Asterisks represent putative additional readers/erasers (listed in the gray boxes of Fig. 2), as many homologs in the plant YTH and ALKBH families remain uncharacterized. AMV, Alfalfa mosaic virus; CPSF, cleavage and polyadenylation specificity factor. PAS, polyadenylation signal.
Writing m6A
mRNA-modifying enzymes are often referred to as writers, by analogy with signal transduction systems in which writers catalyze the formation of a regulatory posttranslational modification, readers act as effectors by binding to the modified amino acid, and erasers remove the modification to reset signaling (Lim and Pawson, 2010). We adopt this nomenclature here but note that its appropriateness is intensely debated, because it is difficult to obtain direct proof that the same mRNA molecules undergo reversible cycles of methylation and demethylation (Ke et al., 2017; Mauer et al., 2017, 2019; Meyer and Jaffrey, 2017; Rosa-Mercado et al., 2017; Darnell et al., 2018; Wei et al., 2018a; Shi et al., 2019). Despite the simple biochemistry of the m6A writer reaction, nucleophilic substitution resulting in transfer of the methyl group from S-adenosylmethionine (SAM) to the N6-amine of adenosine, the major eukaryotic mRNA adenosine methyltransferase turns out to be extraordinarily complicated. Initial attempts at its purification from mammalian cells resulted in the definition of two subcomplexes, ∼200-kD MT-A and ∼875-kD MT-B, both required for full methyltransferase activity, even in vitro (Bokar et al., 1994). While we now know that MT-A contained the catalytic core consisting of a dimer of the two methyltransferase-like proteins METTL3 (Bokar et al., 1997) and METTL14 (Liu et al., 2014), the exact composition of the originally defined MT-B complex is still unclear. Both MT-A subunits have direct homologs in plants: METTL3 corresponds to MTA (Zhong et al., 2008) and METTL14 corresponds to MTB (Růžička et al., 2017). Several additional factors either fully or partly required for m6A deposition in vivo have been identified. Many of these factors are also conserved and include the following proteins, denoted with plant name first and mammalian homolog following a slash (see Figs. 1 and 2 for additional details): the splicing factor FKBP12 Interacting Protein37 (FIP37/WTAP; Vespa et al., 2004; Zhong et al., 2008; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014b; Shen et al., 2016; Růžička et al., 2017); the protein VIRILIZER (VIR/KIAA1429), originally identified in Drosophila melanogaster (Niessen et al., 2001; Ortega et al., 2003; Schwartz et al., 2014b; Růžička et al., 2017); the putative ubiquitin E3 ligase HAKAI/CBLL1 (Horiuchi et al., 2013; Růžička et al., 2017); the Zn finger protein ZC3H13 (Guo et al., 2018; Knuckles et al., 2018; Wen et al., 2018) without orthologs in plants (Balacco and Soller, 2019); and the RNA-binding proteins RBM15A/B (Horiuchi et al., 2013; Yan and Perrimon, 2015; Lence et al., 2016; Patil et al., 2016). Interestingly, the closest plant homolog of RBM15A/B is FLOWERING LOCUS PA (FPA), an RNA-binding protein required for the control of flowering time (Schomburg et al., 2001). However, the homology between FPA and RBM15A/B is confined to the RNA recognition motifs and the overall degree of identity is low. Although the possible involvement of FPA in m6A deposition remains uninvestigated, mutants in FPA and m6A pathway components share at least one molecular phenotype: partial loss of function causes transcriptional read-through and chimeric RNA formation (Hornyik et al., 2010; Duc et al., 2013; Pontier et al., 2019). Nonetheless, it is clear that the requirement of FPA for m6A writer function, if any, cannot be absolute, because in contrast to most knockout mutants in m6A writer components (Vespa et al., 2004; Zhong et al., 2008; Růžička et al., 2017), null alleles of fpa complete embryogenesis (Schomburg et al., 2001).
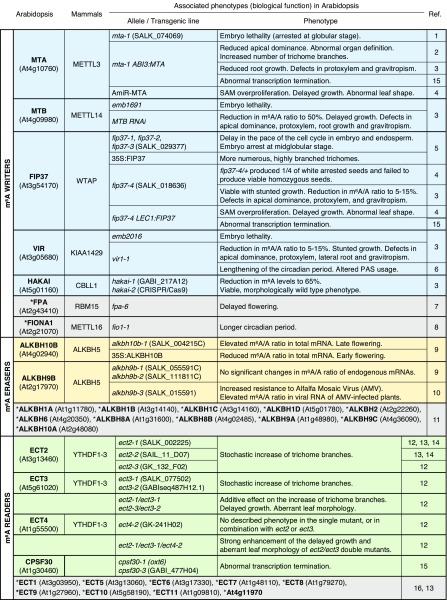
Figure 2.
Biological functions of the m6A pathway in plants as inferred by the mutant phenotype of its characterized components in Arabidopsis. The mammalian homologs of each factor is indicated. Arabidopsis bona fide writers are shaded in blue, erasers in orange, and readers in green. Arabidopsis genes marked with asterisks (as also labeled in Fig. 1) inside gray-shaded boxes are orthologs of m6A pathway components in other organisms or paralogs of such genes in Arabidopsis, but their possible or expected roles as m6A players in plants have not been verified experimentally. For this reason, columns 4 and 5 are omitted in the gray boxes corresponding to putative erasers and readers. Column 3 is also omitted for simplicity as it can be summarized as follows: AtALKBH proteins are homologs of the mammalian ALKBH1 to ALKBH8 and FTO family (Mielecki et al., 2012); all ECTs belong to the YTHDF clade (YTHDF1–YTHDF3 in mammals), while At4g11970 presents homology with the YTHDC clade (YTHDC1 and YTHDC2 in mammals; Scutenaire et al., 2018). In addition to knockout, knockdown, and overexpression lines, transgenic plants expressing point mutants of ECT2 (12–14), ECT3 (12), ECT4 (12), and CPSF30 (15) with impaired ability to bind m6A, or a catalytically inactive ALKBH10b (9), are also described in the indicated references and behave like null mutants for the phenotypes described in all cases. References are as follows: 1, Zhong et al., 2008; 2, Bodi et al., 2012; 3, Růžička et al., 2017; 4, Shen et al., 2016; 5, Vespa et al., 2004; 6, Parker et al., 2019; 7, Schomburg et al., 2001; 8, Kim et al., 2008; 9, Duan et al., 2017; 10, Martínez-Pérez et al., 2017; 11, Mielecki et al., 2012; 12, Arribas-Hernández et al., 2018; 13, Scutenaire et al., 2018; 14, Wei et al., 2018b; 15, Pontier et al., 2019; 16, Li et al., 2014a.
Although the MTA/MTB methyltransferase accounts for the vast majority of m6A sites in Arabidopsis (Zhong et al., 2008; Shen et al., 2016; Růžička et al., 2017; Anderson et al., 2018), additional m6A methyltransferases exist. The essential mammalian enzyme METTL16 methylates structured RNAs containing the nonamer UAC(A)GAGAA (Pendleton et al., 2017). Its few identified targets include the MAT2A mRNA encoding SAM synthetase, and the embryo-lethal phenotype of mettl16 mutants may indeed result from disrupted SAM homeostasis (Pendleton et al., 2017; Warda et al., 2017; Mendel et al., 2018). Interestingly, a clear METTL16 homolog, FIONA1 (FIO1), exists in plants. fio1 loss-of-function mutants are viable and fertile but exhibit increased period length in the circadian clock (Kim et al., 2008), a phenotype shared with mutants homozygous for the hypomorphic vir-1 allele (Růžička et al., 2017; Parker et al., 2019). The biochemical functions of plant FIO1/METTL16, including as an m6A methyltransferase, remain unexplored, however.
It is a relevant question how cells avoid that the specificity of the m6A methyltransferase be undercut by direct incorporation of m6ATP during transcription. After all, degradation of m6A-containing RNA would result in m6AMP that could be converted into an RNA Polymerase II substrate (m6ATP) through salvage pathways. The answer appears to lie in a combination of two factors (Chen et al., 2018): first, the existence of a conserved enzyme with specific m6AMP deaminase activity; second, the fact that adenylate kinase has poor activity toward m6AMP, resulting in poor conversion of m6AMP into m6ADP. Interestingly, knockout of the Arabidopsis m6AMP deaminase MAPDA1 leads to reduced root growth, suggesting that avoidance of m6A recycling may be of importance in vivo (Chen et al., 2018).
Erasing m6A
One of the spectacular early findings in the m6A field was the discovery that enzymes in the ALKBH family have m6A demethylase activity (Jia et al., 2011; Zheng et al., 2013), suggesting the regulatory capacity of the modification for the first time. Several of these enzymes catalyze oxidative dealkylation of N-methylated nucleotides (Ougland et al., 2015). The Arabidopsis genome encodes 13 members of the ALKBH family of oxidases (Mielecki et al., 2012), two of which (ALKBH9B and ALKBH10B) have been shown to have m6A demethylase activity in vitro and have m6A-related functions in vivo (Figs. 1 and 2; Duan et al., 2017; Martínez-Pérez et al., 2017). While such demethylases are important for the control of flowering time (ALKBH10B) and susceptibility to viruses (ALKBH9B) and have an impact on m6A/A ratios in viral RNA (ALKBH9B) and, more subtly, in endogenous mRNA (ALBH10B; Duan et al., 2017; Martínez-Pérez et al., 2017), their exact functions in vivo remain ill defined.
Reading m6A
At least two different properties of RNA may change upon chemical modification of nucleotides. (1) The structure may change as a consequence of altered base pairing properties or rigidity of structure. In turn, this may create or influence the accessibility of binding sites for proteins, or even small molecules, in nearby sequences. (2) The modification itself may create a binding site for an RNA-binding protein with specific affinity for the modified nucleotide.
For m6A, there is evidence for both types of function in mammalian cells (Wang et al., 2014; Liu et al., 2015), although category 2 appears to be more prevalent (Patil et al., 2018). The best studied proteins with specific m6A-binding capacity contain a so-called YT521-B Homology (YTH) domain of ∼140 amino acids (Imai et al., 1998; Hartmann et al., 1999; Stoilov et al., 2002; Zhang et al., 2010) that recognizes the N6-methyl group on adenosine via a highly conserved hydrophobic binding pocket containing three aromatic side chains surrounding the methyl group (the aromatic cage; Li et al., 2014b; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014; Zhu et al., 2014). Two phylogenetic classes of YTH domains can be defined, YTHDF and YTHDC. In mammals, YTHDF-containing proteins are predominantly cytoplasmic, bind to all m6A sites in mRNA, and the three paralogs (YTHDF1–YTHDF3) share high amino acid similarity throughout their entire length, including in their long N-terminal intrinsically disordered regions (IDRs). On the contrary, the YTHDC domain is found in two very different proteins in mammals: YTHDC1 is nuclear and binds to some sites in mRNAs and nuclear non-coding RNAs, whereas YTHDC2 is enriched in perinuclear regions of the cytoplasm and its mRNA-binding profile shows little overlap with m6A sites (Patil et al., 2016; Hsu et al., 2017; Kretschmer et al., 2018; Zaccara et al., 2019). YTHDC2 is specific to mammals and contains several other folded domains in addition to the YTHDC domain (Bailey et al., 2017; Wojtas et al., 2017; Kretschmer et al., 2018), while YTHDC1 has long N- and C-terminal IDRs (Patil et al., 2016). YTHDF- and YTHDC1-type proteins are found in many eukaryotes (Balacco and Soller, 2019), including plants, whose genomes encode more YTH domain proteins than other organisms (Li et al., 2014a; Scutenaire et al., 2018). For example, 13 YTH domain-containing proteins are encoded in Arabidopsis (Fig. 2) compared with five in humans. The 13 Arabidopsis YTH-domain proteins can be divided into 11 YTHDF proteins called EVOLUTIONARILY CONSERVED C-TERMINAL REGION1 (ECT1) to ECT11, one classical YTHDC1-type protein (At4g11970), and one YTHDC protein, unusual in that it also contains additional folded domains: it is CPSF30, the 30-kD subunit of the cleavage and polyadenylation specificity factor involved in pre-mRNA cleavage and 3′-end formation (Zhang et al., 2008; Thomas et al., 2012; Bruggeman et al., 2014; Pontier et al., 2019).
Several lines of evidence suggest that while m6A-binding specificity resides in the YTH domain, the effector function, at least of YTHDF proteins, resides in the IDR. For example, tethering the IDR of YTHDF2 to reporter mRNAs is sufficient to cause their localization to P-bodies in mammalian cells (Wang et al., 2014). The IDR of YTHDF2 also interacts with the deadenylase complex CCR4-NOT (Du et al., 2016) and, via the adaptor protein HRSP12, with the endoribonuclease RNase P/MRP (Park et al., 2019). However, since the affinity of isolated YTH domains for m6A-modified RNA is modest (0.1–5 μm; Li et al., 2014b; Luo and Tong, 2014; Theler et al., 2014; Xu et al., 2014, 2015; Zhu et al., 2014), it is possible that the IDR participates in RNA binding in vivo, as suggested by the loss of mRNA binding in vivo of a mutant in human YTHDF3 containing a deletion in the IDR (Zhang et al., 2019c). In this regard, it is noteworthy that three Arabidopsis YTHDF proteins (ECT5, ECT9, and ECT10) contain an amino acid substitution expected to result in 10-fold higher affinity toward m6A RNA than other YTH domains, based on structural and biochemical analyses of analogous mutants in human YTHDF1 (Xu et al., 2015; Scutenaire et al., 2018). These proteins may, therefore, differ in requirements for their IDRs for RNA interactions compared with other YTH proteins, or they may simply bind with higher affinity. It is an interesting property of both mammalian YTHDFs and plant ECTs that they are able to undergo phase transition in vitro to a condensed liquid or gel-like phase (Arribas-Hernández et al., 2018; Fu and Zhuang, 2019; Gao et al., 2019; Ries et al., 2019). It is of considerable interest to determine whether such phase separation properties underlie biological functions and the subcellular localization of YTHDF proteins. To date, YTH domain proteins are the only characterized class of m6A readers in plants (Figs. 1 and 2). In mammalian cells, several other RNA-binding proteins have been proposed to function as m6A readers (Balacco and Soller, 2019), including the translation initiation factor eIF3 that is recruited to m6A-containing 5′-UTRs to stimulate cap-independent translation initiation (Meyer et al., 2015).
MOLECULAR ROLES OF M6A IN PLANTS
Cytoplasmic Roles
What effect does the presence of m6A in an mRNA have on its stability and translatability? In yeast and mammalian cell culture, the evidence is strong that it accelerates mRNA decay (see e.g. Herzog et al. [2017] or Ke et al. [2017] for animals, Bushkin et al. [2019] for yeast, or Zaccara et al. [2019] for the most recent review), as proposed in early studies (Sommer et al., 1978). Accelerated mRNA decay in mammals involves YTHDF proteins, perhaps in particular YTHDF2 (Wang et al., 2014; Ivanova et al., 2017; Zhang et al., 2017; Zhao et al., 2017; Paris et al., 2019), but whether the mechanism relies mostly on CCR4-NOT deadenylase recruitment (Du et al., 2016), perhaps concurrent with sorting to P-bodies (Wang et al., 2014), enhanced endonucleolysis (Park et al., 2019), or a combination remains unclear. m6A may also stimulate translation, in this case via other YTHDF proteins (Wang et al., 2015; Li et al., 2017a; Shi et al., 2017). In particular, YTHDF1 promotes the translation of m6A targets via interaction with the eukaryotic initiation factor eIF3 (Wang et al., 2015) in a process that may be of particular biological relevance in neurons (Shi et al., 2018; Weng et al., 2018). A cytoplasmic role of METTL3 itself has also been proposed in the activation of translation, also via eIF3 recruitment (Lin et al., 2016; Choe et al., 2018).
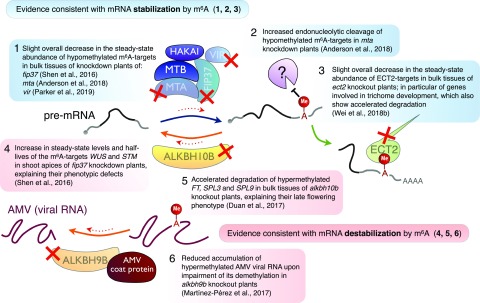
In plants, transcriptomic analyses have also been applied to analyze the effect of m6A on mRNA abundance (Fig. 3). Since knockout of m6A writer subunits is embryonically lethal (see "Functions in Development" below; Zhong et al., 2008), plants expressing either MTA or FIP37 from embryo-specific promoters in the respective knockout backgrounds have been used to obtain postembryonic tissues with 80% to 90% loss of m6A (Bodi et al., 2012; Shen et al., 2016). Studies using either these plants or the partial loss-of-function vir-1 allele (Růžička et al., 2017) showed a slight tendency for m6A targets to be less abundant in tissues of plants with reduced levels of m6A (Shen et al., 2016; Anderson et al., 2018; Parker et al., 2019). The same tendency was observed in knockouts of the m6A reader ect2 (Wei et al., 2018b). Altogether, those observations suggest that m6A may stabilize mRNAs, potentially by protection against endonucleolytic cleavage that occurs in m6A-depleted plants around sites of m6A deposition (Anderson et al., 2018). On the other hand, overaccumulation of m6A-containing mRNAs that encode important developmental regulators has also been found in m6A-depleted mutants (Shen et al., 2016). Moreover, ALKBH10B-mediated m6A demethylation of transcripts encoding key flowering-related genes is associated with their increased abundance and stability (Duan et al., 2017). Thus, the existence of m6A-induced mRNA destabilization in plants should not be ruled out.
Figure 3.
Plant studies reporting on effects of m6A on target RNA accumulation and, in a few cases, decay rates upon global inhibition of transcription.
We note that the interpretation of the transcriptome profiles reported of wild-type and m6A-deficient plants (Fig. 3) suffers from limitations that may preclude clear conclusions to be drawn on whether m6A actually stabilizes or destabilizes mRNA. First, they use mRNA isolated from all cells of seedlings, while analysis of activity of the MTA promoter suggests that m6A deposition may not be active in all cells (Zhong et al., 2008). Thus, in total extracts, the fraction of a particular mRNA target whose abundance can be directly influenced by the loss of methyltransferase activity is diluted by contributions from cells that do not even express the methyltransferase. That is especially relevant for assessing the overall abundance of m6A targets in plants, as they predominantly include ubiquitously expressed housekeeping genes (Anderson et al., 2018). Furthermore, as knockdown of MTA, FIP37, or VIR causes global changes in plant morphogenesis that may affect the expression domains of m6A-containing mRNAs, it compromises the ability of such experiments to yield conclusive results. Second, steady-state measurements comparing a stable mutant with a wild type do not provide kinetic information, as degradation and synthesis rates will inevitably be confounded. In conclusion, it is at present not clear if mRNAs are stabilized or destabilized by m6A (Fig. 3). There is limited direct evidence for either outcome, and the possibility of target- and/or cell-specific outcomes of mRNA methylation simply has not yet been investigated. Ideally, transcriptome-wide pulse-chase analyses that do not disrupt global transcription [e.g. using thiol(SH)-linked alkylation for the metabolic sequencing of RNA; Herzog et al., 2017] should be employed in the relevant cells rather than bulk tissue of the wild type versus mta/mtb/fip37/vir knockdowns or ect knockouts to reveal the effect of m6A deposition and ECT binding on transcript stability. Finally, the possible effects of m6A on translational control in plants remain entirely unexplored.
Nuclear Roles
In addition to the intensely studied effects on mature mRNA, roles of m6A in pre-mRNA processing have also been reported. For example, m6A is required for sex determination in flies, because the sex determination pathway relies on alternative splicing of the sex-lethal (sxl) pre-mRNA, a process that in turn depends critically on m6A deposition around the alternatively spliced sxl exon (Haussmann et al., 2016; Lence et al., 2016). In plants, a recent study has uncovered a special role of m6A in mRNA 3′-end formation and transcription termination at recently duplicated genes: at such loci, read-through transcription into the downstream gene is observed in m6A-deficient plants and, interestingly, in plants containing point mutations to abrogate the aromatic cage in the YTH domain of CPSF30 (Pontier et al., 2019). It is possible that m6A-directed CPSF30 function stimulates pre-mRNA cleavage and RNA Polymerase II termination more generally but that the effect of losing this reinforcement only becomes visible at loci with recent gene duplications (Pontier et al., 2019). In agreement with the idea of m6A-dependent 3′-processing in plants, Parker et al. (2019) recently reported that lack of m6A is associated with a shift to usage of more proximal poly(A) sites and transcriptional read-through, and Luo et al. (2019) observed correlation between the presence of m6A and alternative polyadenylation site usage in maize (Zea mays).
BIOLOGICAL ROLES OF M6A IN PLANTS
Functions in Development
The study of plants provided one of the key discoveries in the development of the mRNA modification field: using Arabidopsis, Fray and coworkers (Zhong et al., 2008) were able to prove in 2008 that m6A is crucial for ontogenesis in a multicellular eukaryote, as embryos defective in the homolog of the m6A methyltransferase identified in yeast and mammals (MTA) arrested at the globular stage. Subsequent phenotypic analyses of hypomorphic alleles or postembryonic knockdowns of mta, mtb, fip37, and vir showed that reduction of m6A results in slower growth, abnormal organ definition, loss of apical dominance, increased number of trichome branches, defective gravitropic responses, and aberrant development of lateral roots and vasculature (Bodi et al., 2012; Růžička et al., 2017; see Fig. 2 for a comprehensive description of knockout and knockdown lines). Crucial additional insights were gained from the work of Shen et al. (2016), who showed that stem cell differentiation in the shoot apical meristem depends on m6A: in plants depleted of FIP37 postembryonically, the shoot apical meristem dramatically increases in size and forms aberrant leaf primordia with a significant delay compared with the wild type. Interestingly, these strong differentiation phenotypes are associated with expansion of the organizing center of the meristem expressing the key transcription factor WUSCHEL (At2g17950), whose mRNA is modified by m6A (Shen et al., 2016). Another m6A target, the mRNA of the meristematic transcription factor SHOOT MERISTEMLESS (At1g62360), also overaccumulates in fip37 knockdown plants, apparently as a consequence of increased mRNA stability (Shen et al., 2016). Thus, m6A in plants is clearly required for meristem function, perhaps in particular for the step of stem cell differentiation, reminiscent of the requirement for m6A for embryonic stem cell differentiation in mammals (Batista et al., 2014; Geula et al., 2015). m6A is also involved in the maintenance of circadian and seasonal plant rhythms, as mutants defective in m6A deposition or removal exhibit lengthening of the circadian period (Parker et al., 2019) and late flowering (Duan et al., 2017), respectively. Finally, recent phenotypic and molecular studies of plants lacking one or several m6A readers provide additional mechanistic insights into the role of m6A in development (Arribas-Hernández et al., 2018; Scutenaire et al., 2018; Wei et al., 2018b). Such advances are discussed in detail below.
Functions in Stress Adaptation
Many mechanisms of posttranscriptional gene regulation, perhaps most prominently small RNA-based gene regulation, are employed to mediate rapid changes in gene expression required for stress adaptation (Sunkar et al., 2007), and it would, therefore, not be surprising to find such roles of the m6A-YTH system. Although early studies based on publicly available microarray data on Arabidopsis and rice (Oryza sativa) YTH domain proteins pointed to their induction by various abiotic and biotic stresses (Li et al., 2014a), relatively little has been done to test their functional implication in such responses. Scutenaire et al. (2018) found that ECT2 relocalizes to cytoplasmic stress granules upon heat stress. Similarly, Arribas-Hernández et al. (2018) observed that ECT2 and ECT4, and to a lesser degree ECT3, relocalize to cytoplasmic foci distinct from P-bodies upon osmotic stress. These studies show that reader proteins respond to stress, but they do not establish whether this is inconsequential or important for the adaptive response. Similar correlative evidence between m6A levels and a biotic stress response in tobacco (Nicotiana tabacum) plants has been reported. In response to tobacco mosaic virus infection, m6A/G ratios and expression of m6A writers decreased whereas mRNA levels of one potential demethylase increased (Li et al., 2018). The available evidence does not, however, allow a distinction between whether such changes are brought about by the virus to promote infection or by the host to limit infection, or even whether they are relevant for the host-virus interaction at all. In animals, examples of both positive and negative regulation of innate and adaptive immunity by m6A have been reported (see Williams et al. [2019] for review), and similar complexity might be expected for different pathogens or cell types in plants.
A role of m6A in selective stabilization of mRNAs responsive to salt stress has recently been suggested (Anderson et al., 2018). This study showed that transcripts involved in salt and osmotic stress were hypermethylated under salt treatment and more highly expressed in treated versus nontreated samples. On the contrary, mRNAs with a tendency to lose m6A marks in response to the treatment were related to more general processes like photosynthesis, and their abundance decreased upon salt stress. Nevertheless, normalization of relative m6A contents in treated versus untreated samples is not trivial when considering on/off genes such as salt stress-response genes. In addition, since m6A peaks are more easily mapped in abundant transcripts with antibody-based mapping approaches (Shen et al., 2016; Garcia-Campos et al., 2019), a bias toward the detection of de novo methylation of salt-responsive transcripts highly expressed after treatment cannot be excluded. Thus, orthogonal mapping approaches with the ability to accurately quantify m6A stoichiometry (e.g. MAZTER-seq; Garcia-Campos et al., 2019) will be valuable to address the dynamics of m6A marks in response to stress. In addition, genetic studies, ideally using sophisticated conditional loss-of-function systems of m6A writers and readers, will be required to evaluate the physiological relevance of the m6A regulatory pathway in stress adaptation.
Methylation of Viral RNA as an Immune Response in Plants
Modification of viral RNA by m6A can modulate the outcome of host-virus interactions in mammals. In some examples, such as the cytoplasmic (+)-strand RNA virus Zika, m6A attenuates viral infectivity (Gokhale et al., 2016; Lichinchi et al., 2016). In other cases, including some m6A sites in HIV, the presence of m6A on viral RNA enhances viral gene expression (Kennedy et al., 2017), making it clear that there is no general outcome of N6-adenosine methylation in host-virus interactions (Williams et al., 2019). In plants, RNA of Alfalfa mosaic virus (AMV) has been shown to contain m6A (Martínez-Pérez et al., 2017). In this case, the multifunctional AMV coat protein recruits the m6A eraser ALKBH9B, resulting in decreased m6A levels in viral RNA (Fig. 1). This m6A erasure dramatically increases systemic infection, indicating that m6A has strong antiviral effects in the case of AMV (Martínez-Pérez et al., 2017). It is not yet clear at which precise points in the infection cycle m6A acts (e.g. initial translation of viral RNA, transcription, cell-to-cell movement, or stability of viral RNA), and the molecular mechanisms underlying its clear antiviral effect remain unexplored. This includes important questions concerning possible links between nonself RNA recognition and N6-adenosine methylation, the identity of the m6A writer, whether reader proteins are involved, and if so how.
MOLECULAR MECHANISMS UNDERLYING BIOLOGICAL ROLES OF M6A IN PLANTS
The clear biological importance of m6A in plants immediately begs mechanistic questions. Given the biochemical framework established for at least some m6A-dependent functions, we can now ask these questions in more precise terms. These include the following. (1) How much of the observed m6A effects is mediated by readers? (2) How many readers are involved? (3) How much is explained by YTH domain proteins? (4) Which mRNAs are targeted, and what are the effects of reader binding to the relevant targets?
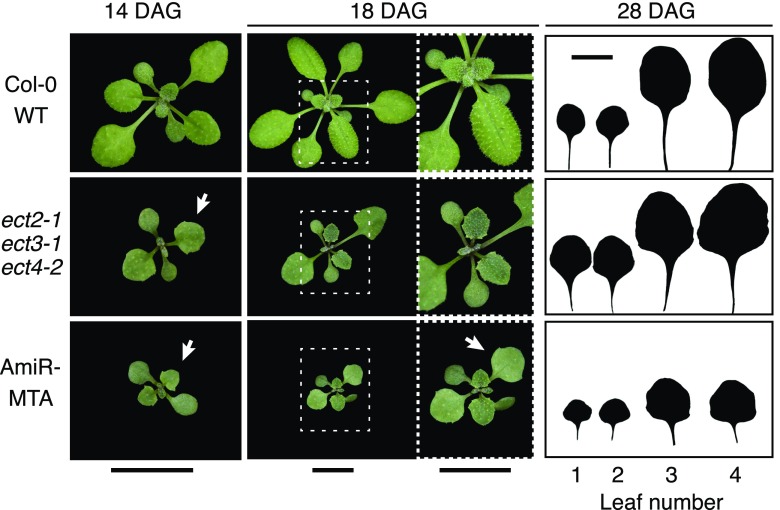
A few recent studies have started to wrestle with the considerable problem of doing genetics with 11 different, potentially redundant, ECT/YTHDF proteins (Li et al., 2014a). Two studies showed that single knockouts of ECT2 led to weak and stochastic increases in trichome branching (Scutenaire et al., 2018; Wei et al., 2018b), resembling one of the defects previously described in postembryonic mta knockdown mutants (Bodi et al., 2012). No other developmental phenotypes could be observed, suggesting that YTHDF reader function is partly involved in specifying branch numbers on trichomes but failing to make a case for reader function in the processes of clear biological importance unveiled by the studies of m6A-deficient plants (Fig 2; Vespa et al., 2004; Zhong et al., 2008; Bodi et al., 2012; Shen et al., 2016; Růžička et al., 2017). In contrast, the simultaneous knockout of three YTHDF proteins, ECT2, ECT3, and ECT4, offered substantial additional insights (Arribas-Hernández et al., 2018). This work demonstrated that the slow development and aberrant leaf morphology in postembryonic mta knockdown plants can be largely recapitulated upon knockout or inactivation of m6A binding of ECT2/3/4, strongly suggesting that the control of these important developmental processes by m6A requires this specific set of YTH domain proteins (Figs. 2 and 4). This study also revealed that both ECT2 and ECT3 play similar roles in the definition of trichome branch numbers and that the combined ect2/ect3 knockout has a much stronger defect than either single mutant, even exceeding the severity observed upon knockdown of m6A (Bodi et al., 2012; Arribas-Hernández et al., 2018). Thus, at least for the phenotypes of slow postembryonic growth, defective leaf morphogenesis, and definition of trichome branch numbers, the YTHDF proteins ECT2 and ECT3, in some cases also ECT4, are responsible for a considerable part of the effects dictated by m6A deposition. For the effects of m6A in other developmental processes (see Fig. 2 for a compilation), we do not have the answers yet.
Figure 4.
Phenotypic comparison between mta knockdown plants with low levels of m6A (AmiR-MTA; Shen et al., 2016) and the triple mutant ect2/ect3/ect4 defective in m6A reader function (Arribas-Hernández et al., 2018). Notice the similarity in the developmental delay (number of leaves at each stage), reduced stature (although aggravated in AmiR-MTA plants), and identical leaf shape (white arrows on photographs and silhouettes of the first true four leaves): triangular blade with more serrations than in wild-type (Col-0 WT) leaves. Plants were grown side by side in Percival growth chambers at 21°C/18°C (day/night) and with a long-day (16 h) light regime. DAG, Days after germination. Bars = 1 cm.
Only a single study has attempted to answer our question 4. Wei et al. (2018b) used formaldehyde cross-linking followed by ECT2 immunoprecipitation and sequencing to identify ECT2 mRNA targets. Nearly one-third of all expressed genes were identified as mRNA targets using this approach. These targets also included the known trichome regulators TRANSPARENT TESTA GLABRA1 (At5g24520), IRREGULAR TRICHOME BRANCH1 (At2g38440), and DISTORTED TRICHOME2 (At1g30825), whose mRNAs were less stable in ect2 mutants than in the wild type, perhaps accounting for their ∼0.6 to 0.8 times lower levels, as measured by quantitative reverse transcription PCR from RNA of whole seedlings. Hence, a model was proposed in which ECT2 stabilizes these mRNAs and their decreased expression in ect2 mutants underlies the increased trichome branch number phenotype (Wei et al., 2018b). As appealing as this model may seem at first sight, some problems are apparent with the evidence supporting it. First, the study did not recover the consensus RR[m6A]CH motif identified in multiple m6A-seq studies (Box 2) as enriched around ECT2 cross-linking sites. This question is more pertinent because formaldehyde, not UV light, was used as a cross-linker. Upon reaction with Lys side chains, formaldehyde forms a Schiff base intermediate that readily reacts with nucleophiles in either proteins or RNA, thereby facilitating indirect cross-links to RNA via other proteins (Hoffman et al., 2015). Thus, many targets identified by formaldehyde-CLIP may not be direct. Second, the transcriptomic analysis measuring decay rates and steady-state abundance were done with entire seedlings, not necessarily informative for gene expression changes in the few cells that matter for the described phenotype: trichomes at an early stage of development. Third, the function of the genes whose decreased abundance in ect2 mutants is proposed to underlie the stochastic increase in trichome branches does not match with the morphological and molecular phenotypes observed (Box 3; Koornneeff et al., 1982; Hülskamp, 2004; Saedler et al., 2004; Zhang et al., 2005; Pattanaik et al., 2014; Arribas-Hernández et al., 2018; Scutenaire et al., 2018; Wei et al., 2018b). For these reasons, we consider it fair to conclude that the evidence is strong that the YTHDF proteins ECT2, ECT3, and ECT4 mediate some m6A-dependent effects on growth and development in a manner that requires m6A binding. On the other hand, how precisely they do so remains much less well defined at present.
CONCLUDING REMARKS
We are just starting to elucidate how mRNA modifications and m6A in particular are at the core of plant development (see Advances Box), response to abiotic stress, and antiviral defense. Although it has not been addressed yet, modulation of growth in response to other phytopathogens and herbivory is likely to be another function. With m6A-YTH axes as the fundamental regulatory units, we now have the guidelines and the tools to find precise answers to how m6A exerts its regulatory functions. Plants might be particularly adept at exploiting the regulatory capacity of m6A, since they grow and develop throughout their life span in the face of a changing environment. They adapt their growth pace in different organs to shape their bodies in unique ways according to environmental cues: root- and stem-branching patterns, leaf shape and size, number of reproductive organs, and overall architecture are dynamically remodeled according to light, water, and nutrient availability, herbivory, and pathogen attack. The expanded families of YTHDF and ALKBH members in plants (Fig. 2) may reflect this necessity (see Outstanding Questions Box). By tissue- or stimulus-dependent expression of these growth regulators, plants may be able to balance the growth rate and final size of different organs in response to different conditions. Potentially, additional mRNA modifications could be combined with m6A to establish a sophisticated network for growth control (see Outstanding Questions Box). A challenge of the next few years will be to clearly define the molecular workings of these potent gene regulatory systems to enable precise tests of this hypothesis of dynamic plant growth control via mRNA modification systems.
Footnotes
This work was supported by the Det Frie Forskningsråd (Danish Council for Independent Research; grant no. 9040-00409B), by the European Research Council (ERC-CoG 726417 “PATHORISC”), and by the Novo Nordisk Foundation (grant no. NNF16OC0021712).
Articles can be viewed without a subscription.
References
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015) N6-Methyladenosine marks primary microRNAs for processing. Nature 519: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov A, Martzen MR, Phizicky EM (2002) Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson ADL, Anderson ZD, Beilstein MA, Fray RG, Lyons E, et al. (2018) N6-Methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Rep 25: 1146–1157 [DOI] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. (2018) Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175: 1872–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribas-Hernández L, Bressendorff S, Hansen MH, Poulsen C, Erdmann S, Brodersen P (2018) An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30: 952–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AS, Batista PJ, Gold RS, Chen YG, de Rooij DG, Chang HY, Fuller MT (2017) The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline. eLife 6: e26116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balacco DL, Soller M (2019) The m6A writer: Rise of a machine for growing tasks. Biochemistry 58: 363–378 [DOI] [PubMed] [Google Scholar]
- Bartoli KM, Schaening C, Carlile TM, Gilbert WV (2018) Conserved methyltransferase Spb1 targets mRNAs for regulated modification with 2′-O-methyl ribose. bioRxiv 271916 [Google Scholar]
- Bass BL, Weintraub H (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. (2014) m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B (2015) N6-Adenosine methylation in miRNAs. PLoS ONE 10: e0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat SS, Bielewicz D, Grzelak N, Gulanicz T, Bodi Z, Szewc L, Bajczyk M, Dolata J, Smolinski DJ, Fray RG, et al. (2019) mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. bioRxiv 557900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H (2015) Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Ed Engl 54: 451–455 [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piątkowski P, Bagiński B, Wirecki TK, de Crécy-Lagard V, Ross R, Limbach PA, Kotter A, et al. (2018) MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG (2012) Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front Plant Sci 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F (1994) Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei: Internal mRNA methylation requires a multisubunit complex. J Biol Chem 269: 17697–17704 [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM (1997) Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3: 1233–1247 [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, Guez T, Vasseur JJ, Debart F, Aravind L, et al. (2019) Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol Cell 75: 631–643.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q, Garmier M, de Bont L, Soubigou-Taconnat L, Mazubert C, Benhamed M, Raynaud C, Bergounioux C, Delarue M (2014) The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol 165: 732–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushkin GG, Pincus D, Morgan JT, Richardson K, Lewis C, Chan SH, Bartel DP, Fink GR (2019) m6A modification of a 3′ UTR site reduces RME1 mRNA levels to promote meiosis. Nat Commun 10: 3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani D, Kahana C, Lavi S, Groner Y (1979) Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res 6: 2879–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Urs MJ, Sánchez-González I, Olayioye MA, Herde M, Witte CP (2018) m6A RNA degradation products are catabolized by an evolutionarily conserved N6-methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30: 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. (2018) mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 561: 556–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JM, Ye TT, Ma CJ, Lan MD, Liu T, Yuan BF, Feng YQ (2018) Existence of internal N7-methylguanosine modification in mRNA determined by differential enzyme treatment coupled with mass spectrometry analysis. ACS Chem Biol 13: 3243–3250 [DOI] [PubMed] [Google Scholar]
- Cui X, Liang Z, Shen L, Zhang Q, Bao S, Geng Y, Zhang B, Leo V, Vardy LA, Lu T, et al. (2017) 5-Methylcytosine RNA methylation in Arabidopsis thaliana. Mol Plant 10: 1387–1399 [DOI] [PubMed] [Google Scholar]
- Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, Dominissini D, He C (2017) Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods 14: 695–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB, Ke S, Darnell JE Jr. (2018) Pre-mRNA processing includes N6 methylation of adenosine residues that are retained in mRNA exons and the fallacy of “RNA epigenetics.”. RNA 24: 262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Burgess A, Parker B, Li J, Pulsford K, Sibbritt T, Preiss T, Searle IR (2017) Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell 29: 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida C, Scheer H, Zuber H, Gagliardi D (2018) RNA uridylation: A key posttranscriptional modification shaping the coding and noncoding transcriptome. Wiley Interdiscip Rev RNA 9: e1440. [DOI] [PubMed] [Google Scholar]
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. (2016) RNA biochemistry: Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351: 282–285 [DOI] [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C (2015) Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res 43: 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F (1974) Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 71: 3971–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimock K, Stoltzfus CM (1977) Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry 16: 471–478 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. (2016) The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L (2016) YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 7: 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan HC, Wei LH, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G (2017) ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29: 2995–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duc C, Sherstnev A, Cole C, Barton GJ, Simpson GG (2013) Transcription termination and chimeric RNA formation controlled by Arabidopsis thaliana FPA. PLoS Genet 9: e1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R (2013) Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9: e1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J (2019) Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res 47: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhuang X (2019) m6A-binding YTHDF proteins promote stress granule formation by modulating phase separation of stress granule proteins. bioRxiv 694455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE (1975) Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci USA 72: 1904–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, Li P (2019) Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res 29: 767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. (2018) Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206 [DOI] [PubMed] [Google Scholar]
- Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S, Winkler R, Nir R, Lasman L, Brandis A, et al. (2019) Deciphering the “m6A code” via antibody-independent quantitative profiling. Cell 178: 731–747 [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. (2015) Stem cells: m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. (2016) N6-Methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20: 654–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Tang HW, Li J, Perrimon N, Yan D (2018) Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc Natl Acad Sci USA 115: 3674–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S (1999) The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell 10: 3909–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M (2016) m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304 [DOI] [PubMed] [Google Scholar]
- Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, Ameres SL (2017) Thiol-linked alkylation of RNA to assess expression dynamics. Nat Methods 14: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EA, Frey BL, Smith LM, Auble DT (2015) Formaldehyde crosslinking: A tool for the study of chromatin complexes. J Biol Chem 290: 26404–26411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A (1965) Structure of a ribonucleic acid. Science 147: 1462–1465 [DOI] [PubMed] [Google Scholar]
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T (2013) Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem 288: 33292–33302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. (2017) Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülskamp M. (2004) Plant trichomes: A model for cell differentiation. Nat Rev Mol Cell Biol 5: 471–480 [DOI] [PubMed] [Google Scholar]
- Imai Y, Matsuo N, Ogawa S, Tohyama M, Takagi T (1998) Cloning of a gene, YT521, for a novel RNA splicing-related protein induced by hypoxia/reoxygenation. Brain Res Mol Brain Res 53: 33–40 [DOI] [PubMed] [Google Scholar]
- Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, Monahan J, Carrieri C, Enright AJ, O’Carroll D (2017) The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell 67: 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011) N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. (2015) A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., Darnell RB (2017) m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31: 990–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AVR, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. (2017) Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 22: 830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M. (2018) Eukaryotic RNA 5′-end NAD+ capping and DeNADding. Trends Cell Biol 28: 454–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et al. (2018) Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev 32: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneeff M, Dellaert LWM, van der Veen JH (1982) EMS- and relation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res Fundam Mol Mech Mutagen 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Kretschmer J, Rao H, Hackert P, Sloan KE, Höbartner C, Bohnsack MT (2018) The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA 24: 1339–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand C, Tuorto F, Hartmann M, Liebers R, Jacob D, Helm M, Lyko F (2017) Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res 27: 1589–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. (2016) m6A modulates neuronal functions and sex determination in Drosophila. Nature 540: 242–247 [DOI] [PubMed] [Google Scholar]
- Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al. (2017a) Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 27: 444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang X, Li Z, Lu C, Zhang Q, Chang L, Li W, Cheng T, Xia Q, Zhao P (2019) Transcriptome-wide analysis of N6-methyladenosine uncovers its regulatory role in gene expression in the lepidopteran Bombyx mori. Insect Mol Biol 28: 703–715 [DOI] [PubMed] [Google Scholar]
- Li D, Zhang H, Hong Y, Huang L, Li X, Zhang Y, Ouyang Z, Song F (2014a) Genome-wide identification, biochemical characterization, and expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol Biol Rep 32: 1169–1186 [Google Scholar]
- Li F, Zhao D, Wu J, Shi Y (2014b) Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res 24: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C (2016) Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 12: 311–316 [DOI] [PubMed] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al. (2017b) Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C (2015) Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597 [DOI] [PubMed] [Google Scholar]
- Li Y, Wang X, Li C, Hu S, Yu J, Song S (2014c) Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol 11: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Shi J, Yu L, Zhao X, Ran L, Hu D, Song B (2018) N6-Methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol J 15: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM (2016) Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20: 666–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Pawson T (2010) Phosphotyrosine signaling: Evolving a new cellular communication system. Cell 142: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR (2015) Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Jaffrey SR (2019) Discovering and mapping the modified nucleotides that comprise the epitranscriptome of mRNA. Cold Spring Harb Perspect Biol 11: a032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Begik O, Lucas MC, Ramirez JM, Mason CE, Wiener D, Schwartz S, Mattick JS, Smith MA, Novoa EM (2019) Accurate detection of m6A RNA modifications in native RNA sequences. Nat Commun 10: 4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014) A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T (2015) N6-Methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO (2014) Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 9: e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. (2014) Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Wang Y, Wang M, Zhang L, Peng H, Zhou Y, Jia G, He Y (2019) Natural variation in RNA m6A methylation and its relationship with translational status. Plant Physiol 182: 332–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Tong L (2014) Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc Natl Acad Sci USA 111: 13834–13839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V (2017) Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci USA 114: 10755–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017) Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur JJ, Rentmeister A, Gross SS, Pellizzoni L, Debart F, et al. (2019) FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol 15: 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, Pillai RS (2018) Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol Cell 71: 986–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR (2017) Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol 33: 319–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR (2015) 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Zhang T, Qi Y, Song J, Han Z, Ma C (2019) Evolution of the RNA N6-methyladenosine methylome mediated by genomic duplication. Plant Physiol 182: 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielecki D, Zugaj D, Muszewska A, Piwowarski J, Chojnacka A, Mielecki M, Nieminuszczy J, Grynberg M, Grzesiuk E (2012) Novel AlkB dioxygenases: Alternative models for in silico and in vivo studies. PLoS ONE 7: e30588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JL, Welder L (1981) Nucleotides adjacent to N6-methyladenosine in maize poly(A)-containing RNA. Plant Sci Lett 21: 75–81 [Google Scholar]
- Niessen M, Schneiter R, Nothiger R (2001) Molecular identification of virilizer, a gene required for the expression of the sex-determining gene Sex-lethal in Drosophila melanogaster. Genetics 157: 679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, Niksic M, Bachi A, Wilm M, Sánchez L, Hastie N, Valcárcel J (2003) Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem 278: 3040–3047 [DOI] [PubMed] [Google Scholar]
- Ougland R, Rognes T, Klungland A, Larsen E (2015) Non-homologous functions of the AlkB homologs. J Mol Cell Biol 7: 494–504 [DOI] [PubMed] [Google Scholar]
- Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. (2019) Targeting the RNA m6A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 25: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, Kim YK (2019) Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell 74: 494–507 [DOI] [PubMed] [Google Scholar]
- Parker MT, Knop K, Sherwood AV, Schurch NJ, Mackinnon K, Gould PD, Hall A, Barton GJ, Simpson GG (2019) Nanopore direct RNA sequencing maps an Arabidopsis N6-methyladenosine epitranscriptome. bioRxiv 706002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR (2016) m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537: 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Pickering BF, Jaffrey SR (2018) Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol 28: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik S, Patra B, Singh SK, Yuan L (2014) An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front Plant Sci 5: 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK (2017) The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169: 824–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP, Kelley DE (1974) Existence of methylated messenger RNA in mouse L cells. Cell 1: 37–42 [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier D, Picart C, El Baidouri M, Roudier F, Xu T, Lahmy S, Llauro C, Azevedo J, Laudié M, Attina A, et al. (2019) The m6A pathway protects the transcriptome integrity by restricting RNA chimera formation in plants. Life Sci Alliance 2: e201900393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR (2019) m6A enhances the phase separation potential of mRNA. Nature 571: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Mercado NA, Withers JB, Steitz JA (2017) Settling the m6A debate: Methylation of mature mRNA is not dynamic but accelerates turnover. Genes Dev 31: 957–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, Eeckhout D, El-Showk S, Li H, Zhong S, et al. (2017) Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol 215: 157–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R, Mathur N, Srinivas BP, Kernebeck B, Hülskamp M, Mathur J (2004) Actin control over microtubules suggested by DISTORTED2 encoding the Arabidopsis ARPC2 subunit homolog. Plant Cell Physiol 45: 813–822 [DOI] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S (2017) The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255 [DOI] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP (1977) Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol 115: 695–714 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. (2013) High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. (2014a) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. (2014b) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep 8: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutenaire J, Deragon JM, Jean V, Benhamed M, Raynaud C, Favory JJ, Merret R, Bousquet-Antonelli C (2018) The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30: 986–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H (2016) N6-Methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell 38: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko G, Morris KV (2018) All I’s on the RADAR: Role of ADAR in gene regulation. FEBS Lett 592: 2860–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C (2017) YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27: 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wei J, He C (2019) Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell 74: 640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, Li J, Hao P, Zhang Y, Zhang F, et al. (2018) m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563: 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T. (2006) RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol Life Sci 63: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S, Lavi U, Darnell JE Jr. (1978) The absolute frequency of labeled N-6-methyladenosine in HeLa cell messenger RNA decreases with label time. J Mol Biol 124: 487–499 [DOI] [PubMed] [Google Scholar]
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T (2012) Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40: 5023–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Rafalska I, Stamm S (2002) YTH: A new domain in nuclear proteins. Trends Biochem Sci 27: 495–497 [DOI] [PubMed] [Google Scholar]
- Sun L, Xu Y, Bai S, Bai X, Zhu H, Dong H, Wang W, Zhu X, Hao F, Song CP (2019) Transcriptome-wide analysis of pseudouridylation of mRNA and non-coding RNAs in Arabidopsis. J Exp Bot 70: 5089–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Theler D, Dominguez C, Blatter M, Boudet J, Allain FH (2014) Solution structure of the YTH domain in complex with N6-methyladenosine RNA: A reader of methylated RNA. Nucleic Acids Res 42: 13911–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PE, Wu X, Liu M, Gaffney B, Ji G, Li QQ, Hunt AG (2012) Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24: 4376–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Hwang HW, Darnell RB (2018) The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb Perspect Biol 10: a032243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandivier LE, Campos R, Kuksa PP, Silverman IM, Wang LS, Gregory BD (2015) Chemical modifications mark alternatively spliced and uncapped messenger RNAs in Arabidopsis. Plant Cell 27: 3024–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M (2004) The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol 134: 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, Lang Z (2015) Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol 16: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014) N6-Methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N6-Methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li S, Zhao Y, You C, Le B, Gong Z, Mo B, Xia Y, Chen X (2019) NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc Natl Acad Sci USA 116: 12094–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Tang K, Zhang D, Wan Y, Wen Y, Lu Q, Wang L (2017) High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS ONE 12: e0185612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, Bohnsack MT (2017) Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep 18: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Gershowitz A, Moss B (1975a) N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257: 251–253 [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B (1975b) Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4: 379–386 [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B (1976) 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15: 397–401 [DOI] [PubMed] [Google Scholar]
- Wei CM, Moss B (1977) Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 16: 1672–1676 [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018a) Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell 71: 973–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, Xiao Y, Zhang X, Duan HC, Jia G (2018b) The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30: 968–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, et al. (2018) Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell 69: 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, Liu Y, Xu T, Wang X, Wong SZH, et al. (2018) Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97: 313–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GD, Gokhale NS, Horner SM (2019) Regulation of viral infection by the RNA modification N6-methyladenosine. Annu Rev Virol 6: 235–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS (2017) Regulation of m6A transcripts by the 3ʹ→5ʹ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell 68: 374–387 [DOI] [PubMed] [Google Scholar]
- Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J (2015) Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem 290: 24902–24913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J (2014) Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10: 927–929 [DOI] [PubMed] [Google Scholar]
- Yan D, Perrimon N (2015) spenito is required for sex determination in Drosophila melanogaster. Proc Natl Acad Sci USA 112: 11606–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Perrera V, Saplaoura E, Apelt F, Bahin M, Kramdi A, Olas J, Mueller-Roeber B, Sokolowska E, Zhang W, et al. (2019) m5C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr Biol 29: 2465–2476 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR (2019) Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20: 608–624 [DOI] [PubMed] [Google Scholar]
- Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. (2017) m6A modulates haematopoietic stem and progenitor cell specification. Nature 549: 273–276 [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhang YC, Liao JY, Yu Y, Zhou YF, Feng YZ, Yang YW, Lei MQ, Bai M, Wu H, et al. (2019a) The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet 15: e1008120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Addepalli B, Yun KY, Hunt AG, Xu R, Rao S, Li QQ, Falcone DL (2008) A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE 3: e2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LS, Liu C, Ma H, Dai Q, Sun HL, Luo G, Zhang Z, Zhang L, Hu L, Dong X, et al. (2019b) Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol Cell 74: 1304–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dyachok J, Krishnakumar S, Smith LG, Oppenheimer DG (2005) IRREGULAR TRICHOME BRANCH1 in Arabidopsis encodes a plant homolog of the actin-related protein2/3 complex activator Scar/WAVE that regulates actin and microtubule organization. Plant Cell 17: 2314–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, Cao X (2019c) RNA-binding protein YTHDF3 suppresses interferon-dependent antiviral responses by promoting FOXO3 translation. Proc Natl Acad Sci USA 116: 976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen LQ, Zhao YL, Yang CG, Roundtree IA, Zhang Z, Ren J, Xie W, He C, Luo GZ (2019d) Single-base mapping of m6A by an antibody-independent method. Sci Adv 5: eaax0250. [DOI] [PMC free article] [PubMed]
- Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, Rafalska I, Heinrich B, Bujnicki JM, Allain FHT, et al. (2010) The YTH domain is a novel RNA binding domain. J Biol Chem 285: 14701–14710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C (2017) m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature 542: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Roundtree IA, Wang P, Wang X, Wang L, Sun C, Tian Y, Li J, He C, Xu Y (2014) Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine. Cell Res 24: 1493–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]