Multifaceted effects of N6-methyladenosine modification of messenger RNA on translational status in maize.
Abstract
N6-methyladenosine (m6A) is the most abundant modification of eukaryotic mRNA. Although m6A has been demonstrated to affect almost all aspects of RNA metabolism, its global contribution to the post-transcriptional balancing of translational efficiency remains elusive in plants. In this study, we performed a parallel analysis of the transcriptome-wide mRNA m6A distribution and polysome profiling in two maize (Zea mays) inbred lines to assess the global correlation of m6A modification with translational status. m6A sites are widely distributed in thousands of protein-coding genes, confined to a consensus motif and primarily enriched in the 3′ untranslated regions, and highly coordinated with alternative polyadenylation usage, suggesting a role of m6A modification in regulating alternative polyadenylation site choice. More importantly, we identified that the m6A modification shows multifaceted correlations with the translational status depending on its strength and genic location. Moreover, we observed a substantial intraspecies variation in m6A modification, and this natural variation was shown to be partly driven by gene-specific expression and alternative splicing. Together, these findings provide an invaluable resource for ascertaining transcripts that are subject to m6A modification in maize and pave the way to a better understanding of natural m6A variation in mediating gene expression regulation.
N6-methyladenosine (m6A) is the most prevalent and physiologically relevant mRNA modification and is currently the best example of a complete epitranscriptomic system with known writer (Zhong et al., 2008; Liu et al., 2014; Ping et al., 2014; Schwartz et al., 2014; Shen et al., 2016; Mendel et al., 2018; Yue et al., 2018, 2019), reader (Wang et al., 2014a, 2015; Xiao et al., 2016; Li et al., 2017; Shi et al., 2017; Arribas-Hernández et al., 2018; Huang et al., 2018; Scutenaire et al., 2018; Wei et al., 2018; Wu et al., 2018), and eraser proteins (Jia et al., 2011; Zheng et al., 2013; Duan et al., 2017) in both plants and mammals. m6A has been demonstrated to be essential for the earliest stages of cell fate determination and cell differentiation in plants (Bodi et al., 2012; Shen et al., 2016), metazoans (Yue et al., 2015; Haussmann et al., 2016; Lence et al., 2016), and mammals (Wang et al., 2014b; Geula et al., 2015; Hsu et al., 2017), and is linked with diseases in humans (Homo sapiens) and other mammalian species (Jia et al., 2011; Zheng et al., 2013; Lin et al., 2016; Cui et al., 2017; Yoon et al., 2017; Zhang et al., 2017a; Wang et al., 2018) as well as required for yeast (Saccharomyces cerevisiae) and mice (mus musculus) meiosis (Zheng et al., 2013; Bodi et al., 2015; Xu et al., 2017). Reduced levels of m6A also affect the circadian period in mice (Fustin et al., 2013), and lead to partial infertility in Drosophila (Lence et al., 2016; Kan et al., 2017).
m6A mediates its physiological effects by influencing the fate of mRNA, and has been connected with a wide range of mRNA metabolism, including nuclear export (Fustin et al., 2013; Roundtree et al., 2017), secondary structure (Liu et al., 2015; Liu et al., 2017), pre-mRNA splicing (Zhao et al., 2014; Haussmann et al., 2016; Xiao et al., 2016), alternative polyadenylation (APA) site choice (Ke et al., 2015; Molinie et al., 2016; Kasowitz et al., 2018; Yue et al., 2018), stability (Wang et al., 2014a; Shi et al., 2017; Huang et al., 2018), translatability (Meyer et al., 2015; Wang et al., 2015; Zhou et al., 2015; Li et al., 2017; Shi et al., 2017; Slobodin et al., 2017; Zhou et al., 2018), pri-microRNA processing (Alarcón et al., 2015), and other mechanisms accompanying RNA maturation (Meyer and Jaffrey, 2014; Yue et al., 2015; Yang et al., 2017).
The impact of m6A on translation has been subjected to substantial examinations in recent years (Meyer, 2018). Several studies have reported stimulatory effects of m6A on translation (Meyer et al., 2015; Wang et al., 2015; Coots et al., 2017; Li et al., 2017; Shi et al., 2017), whereas other studies have shown inhibitory effects (Choi et al., 2016; Qi et al., 2016; Slobodin et al., 2017). Current research has found that diverse effects of m6A on translation regulation are dictated by many factors, including its effect on RNA structures (Wang et al., 2014b; Liu et al., 2015; Roost et al., 2015; Spitale et al., 2015; Liu et al., 2017), the location within a transcript (Meyer et al., 2015; Qi et al., 2016), the proteins (readers) that recognize it (Wang et al., 2015; Li et al., 2017; Shi et al., 2017), and the cellular environment (Zhou et al., 2015; Zhou et al., 2018), among other factors (Han et al., 2017; Roignant and Soller, 2017; Meyer, 2018).
How m6A affects translation has not yet been studied in any plant species. In this study, we illustrated the patterns and features of mRNA m6A marks in two maize (Zea mays) inbred lines, B73 and Mo17, and examined the global extent of m6A correlating with translational status. By combining a parallel transcriptome-wide profiling of m6A distribution and polysome occupancy, we showed that m6A modification conferred a negative correlation with the translational status at a global scale. Meanwhile, the incidence of m6A modification near the start codon tended to enhance the translational status. These results indicate that the involvement of m6A in affecting translational status is multifaceted, and varied in the context of its strength and genic location. Furthermore, we identified that thousands of genes were subject to natural variation in the specific modification of m6A, which is at least partly associated with the intraspecies variations in alternative splicing.
RESULTS
Features of the m6A Methylome in Maize
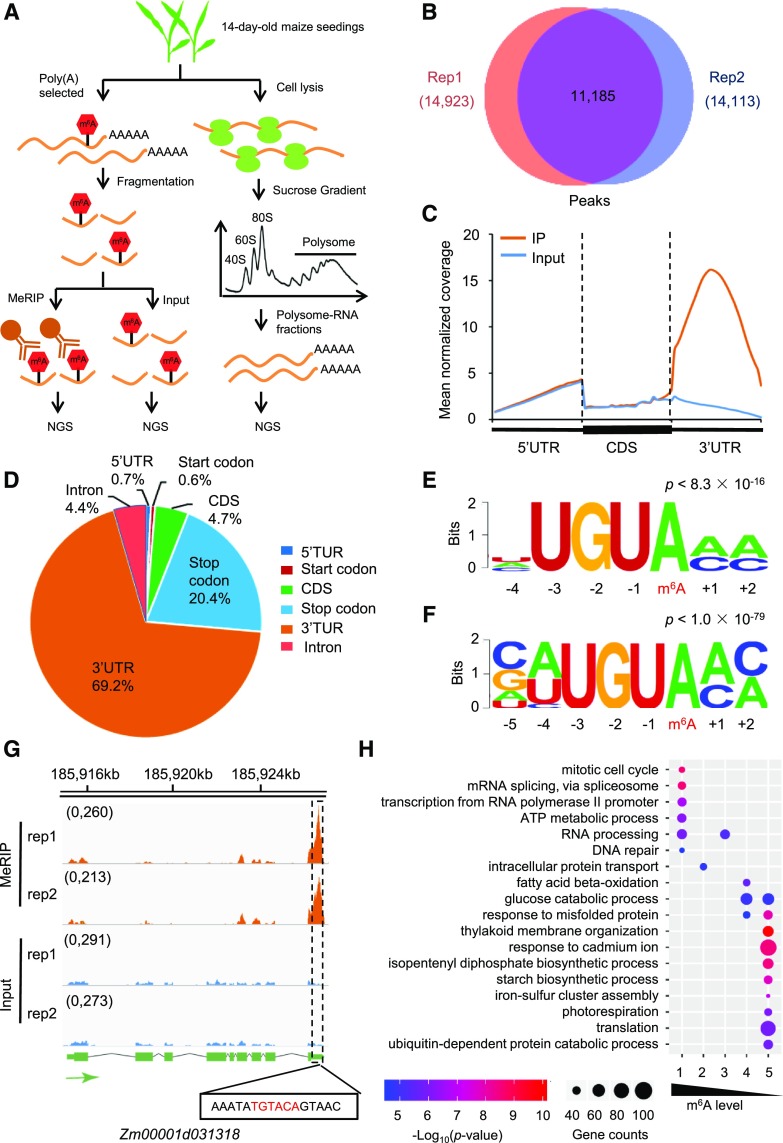
To obtain insight into the roles of m6A in affecting translational status, we conducted m6A RNA immunoprecipitation sequencing (m6A-seq; Dominissini et al., 2012; Meyer et al., 2012), polysome profiling (Juntawong et al., 2014; Zhang et al., 2017b), and input RNA sequencing (RNA-seq) for the same samples collected from two maize inbred lines, B73 and Mo17 (Fig. 1A). To determine the locations of m6A modifications throughout the transcriptome, we adapted an analytical algorithm for identifying m6A peaks as described elsewhere (Dominissini et al., 2013; Yoon et al., 2017). m6A peaks were highly overlapped between two biologically independent replicates (Fig. 1B), and the concordant peaks from two replicates were used for subsequent bioinformatics analyses. In B73, m6A-seq revealed a total of 11,185 peaks, including 8,265 protein-coding mRNAs (Supplemental Table S1), 76 long non-coding RNAs (Supplemental Table S2) and 140 transposable element transcripts (Supplemental Table S3). The majority of protein-coding genes (>90%) contain a single m6A residue (Supplemental Fig. S1). Using m6A reverse transcription quantitative PCR (RT-qPCR), all of the nine randomly selected m6A peak-containing genes were verified (Supplemental Fig. S2), implying a high authenticity of our data.
Figure 1.
The m6A methylome in the maize inbred line B73. A, Schematic diagram of m6A-seq and polysome profiling in parallel. NGS, next generation sequencing. B, Overlap of m6A peaks between two biological replicates (Rep1 and Rep2). C, Metagene profile of m6A peak distribution along a normalized transcript composed of three rescaled nonoverlapping segments, 5′ UTR, CDS, and 3′ UTR. D, Pie chart depicting the percentage of m6A peaks within six transcript segments. E, Sequence motif identified from the top 2,000 most significant m6A peaks by the software MEME. F, Sequence motif identified from the top 2,000 most significant m6A peaks by the software HOMER. G, A representative Integrative Genomics Viewer (http://www.igv.org/) plot showing a m6A motif sequence (highlighted in red) identified in (E) and (F) at the center of a peak within the 3′ UTR of Zm00001d031318. The peak summit is indicated with a dashed rectangle box. H, GO enrichment analysis using the software FuncAssociate 3.0 for five groups of genes ranked by m6A levels. The color bar stands for the −log10(P value) of each GO term. The size of the circle indicates the number of genes in each GO term.
Consistent with previous studies in both mammals and plants (Meyer et al., 2012; Li et al., 2014; Luo et al., 2014; Wan et al., 2015), m6A peaks in protein-coding genes were primarily enriched in the 3′ untranslated region (UTR; ∼69.2%) and in the vicinity of the stop codon (∼20.4%; defined as a 200-nt window centered on the stop codon), while less present in coding sequences (CDS; ∼4.7%), near start codons (∼0.6%; defined as a 200-nt window centered on the start codon), in the 5′UTR (∼0.7%), and in spliced intronic regions (∼4.4%; Fig. 1, C and D; Supplemental Fig. S3). De novo motif analysis of m6A peaks using both the MEME (Bailey et al., 2009) and the HOMER software programs (Heinz et al., 2010) identified a UGUAMM sequence motif (M = A or C; Fig. 1, E–G) that is exactly the same as the motif previously identified from a set of m6A-methylated genes in rice (Oryza sativa; Li et al., 2014), and similar to a URUAY motif recently considered as a plant-specific m6A motif proven to be bound by the m6A reader ECT2 in Arabidopsis (Arabidopsis thaliana; Wei et al., 2018); however, it is distinct from the canonical RRACH motif reported in other organisms (Dominissini et al., 2012; Meyer et al., 2012).
To predict the biological functions associated with m6A-modified genes, we performed gene ontology (GO) enrichment analysis and found that proteins encoded by genes with the highest level of m6A modification (top 20%; see “Materials and Methods”) were involved in a variety of cellular functions, including RNA processing, ATP metabolism, transcription regulation, etc. Transcripts encoding mitotic cell cycle control were identified as the most significantly enriched group (Fig. 1H). Collectively, these results indicate that the high level of m6A modification tends to mark transcripts with regulatory functions.
We performed all the analyses in Mo17 (Supplemental Figs. S4–S6; Supplemental Tables S4 and S5), and all the results about the m6A methylome in Mo17 were consistent with the findings in B73. Taken together, our analysis revealed that thousands of transcripts were post-transcriptionally modified by m6A in maize, and the overall topology of the m6A methylome showed both conserved and unique features compared to other organisms such as mammals (Dominissini et al., 2012; Meyer et al., 2012).
Possible Relation of m6A to the Choice of Poly(A) Sites
In mammals, the m6A modification has been demonstrated to play a role in choosing APA sites (Ke et al., 2015; Molinie et al., 2016; Kasowitz et al., 2018; Yue et al., 2018). The strong enrichment of m6A peaks in 3′UTRs prompted us to investigate whether m6A marks are correlated with APA usage in maize. From a total of 18,676 expressed genes detected in our B73 RNA-seq data set (Fig. 2A; Supplemental Table S6; and see “Materials and Methods”), we found that 67.6% of genes (n = 8,549) containing at least two poly(A) sites were m6A-methylated, which was remarkably higher than 24.8% of genes (n = 10,127) without APA sites (Fisher’s exact test, P value < 2.2 × 10−16; Fig. 2, A, B, and D). Vice versa, 69.7% of m6A-modified genes (n = 8,291) were identified to harbor APA events, which was significantly higher than 26.7% of nonmethylated genes (n = 10,385; Fisher’s exact test, P value < 2.2 × 10−16; Fig. 2, A, C, and D). Moreover, the intimate association of m6A marks with APA usage was also consistently observed in Mo17 (Supplemental Fig. S7; Supplemental Table S7). These results clearly indicate that the m6A modification may be associated with the decision to choose poly(A) sites in maize.
Figure 2.
Association of m6A with APA usage in B73. A, The number of genes defined in each category in the corresponding analysis. B, Proportion of m6A-modifed transcripts within transcripts with (left) or without APA usage (right). P values were calculated using the Fisher’s exact test. C, Proportion of transcripts containing APA usage within m6A-methylated (left), and nonmethylated transcripts (right). P values were calculated using the Fisher’s exact test. D, Integrative Genomics Viewer plots of two examples representing m6A located in the proximal (left) or distal (right) APA. The arrows indicate the gene direction from the 5′ to 3′ end.
To further ascertain whether the effect of m6A on APA usage is dependent on its location on 3′UTRs, we divided m6A-methylated genes into six categories according to m6A sites on different genic segments. Surprisingly, we found that besides m6A-methylated sites on 3′UTRs, it was evident that genes with m6A marks on any other segments also exhibited a significant correlation with APA usage than genes without m6A modification (Supplemental Fig. S8), suggesting that the effect of m6A modification on APA usage may be a general output regardless of its genic location.
Effect of the m6A Modification on Translation
It has been reported in various species that the m6A strength is negatively correlated with the transcript abundance, possibly by affecting mRNA decay (Li et al., 2014; Wang et al., 2014a; Wan et al., 2015; Martínez-Pérez et al., 2017; Anderson et al., 2018). Consistently, we revealed a significantly negative correlation (r = −0.66, P value < 2.2 × 10−16 for B73 and r = −0.65, P value < 2.2 × 10−16 for Mo17) between m6A and the mRNA level in maize (Supplemental Fig. S9). Then, we performed transcriptome-wide polysome profiling and calculated the translational status of each expressed gene (ratio of the polysome-bound fraction to total mRNA) to assess the effect of m6A on translation at a global scale (Supplemental Fig. S10). As shown in Figure 3A, although m6A-modified transcripts displayed a tendency of a higher level of translational status than transcripts without m6A marks, we observed that hypermethylated transcripts had the lowest degree of translational status after ranking the genes into five groups based on the m6A strength, suggesting that the excessive extent of m6A modification may likely attenuate the translational status. In contrast, the level of gene transcription showed a positive correlation with translational status (Supplemental Fig. S11).
Figure 3.
Effect of the m6A modification on translation in B73. A, The m6A level and translational status shows a negative correlation. m6A-depleted and m6A-modified transcripts are displayed in blue and deep red, respectively, and the P value was calculated using the Wilcoxon rank-sum test. The P value among groups with different m6A levels was calculated using the Kruskal–Wallis test. B, Transcripts with m6A residues near the start codon (deep red) exhibit the highest level of translational status. *P < 0.05, **P < 0.01, and ***P < 0.001. C, K-means clustering analysis of all the m6A-modified transcripts based on m6A intensity and translational status. Each level of m6A methylation and translational status was converted to percentiles using the empirical cumulative distribution function. The percentile indicates converted m6A level and translational status. The color indicates the relative m6A methylation and translational status. A total of four clusters were identified and the number of genes in each cluster is shown. D, GO enrichment analysis for Cluster 1 (left) and Cluster 3 (right) using the software FuncAssociate 3.0 (permutation-based corrected P value < 0.05). All significantly enriched GO terms are listed in Supplemental Table S8. Each node indicates an enriched GO term and the node size is proportional to the total number of genes in each pathway.
We next examined the relationship between m6A methylation and translational status by plotting the fraction of genes with m6A peaks in each genic segment. Surprisingly, we found that although the overall level of m6A near the start codon was low (Fig. 1C), transcripts with m6A marks in the vicinity of the start codon showed the highest translational status than any other segments (Fig. 3B), raising an intriguing possibility that the occurrence of m6A nearby the start codon may have the ability to enhance mRNA translation. Indeed, after we performed m6A methylated RNA immunoprecipitation (MeRIP) toward the polysome-mRNA fractions, we found that two randomly selected genes with m6A sites near the start codon exhibited aggregated m6A strength relative to that detected from the total mRNA. In contrast, two genes with m6A sites in the 3′UTR did not show such regularity (Supplemental Fig. S12). These results suggest that at least for the genes tested, the presence of the m6A mark near the start codon in mRNA may facilitate the loading of mRNA onto the ribosomes. Moreover, we performed GO term enrichment analysis for m6A-methylated genes grouped according to m6A sites on different genic segments. Interestingly, we found that proteins encoded by transcripts with the m6A mark in the vicinity of the start codon were enriched in the category of nucleosome (Supplemental Fig. S13). Meanwhile, the functional category of histone binding was enriched in proteins encoded by transcripts with m6A sites in the CDS (Supplemental Fig. S13). In contrast, we could not detect any GO enrichment for all the other groups of genes.
To further investigate whether m6A may coordinate translational control in the context of distinct biological pathways, we performed the k-means clustering analysis to group all the m6A-modified genes into four classes based on the levels of m6A and translational status (Fig. 3C; Supplemental Fig. S14; see “Materials and Methods”). We then conducted GO term enrichment analysis across the different clusters (Supplemental Table S8). In Cluster 1, which was signified by the low level of m6A methylation but the high level of translation efficiency, two of the most significant enriched groups were translation and RNA methylation processes (Fig. 3, C and D).
Interestingly, the Cluster 1 group also showed characteristics as the lowest proportion of APA usages (Supplemental Fig. S15A) and the highest proportion of transcripts with m6A near the start codon (Supplemental Fig. S15B). In contrast, genes involved in purine metabolism were exceedingly enriched in Cluster 3, which exhibited the highest level of m6A modification but the lowest level of translational status (Fig. 3, C and D), as well as the highest proportion of APA usages (Supplemental Fig. S15A) and the lowest proportion of transcripts with m6A near the start codon (Supplemental Fig. S15B). As the specific functional enrichment was identified for all four clusters, it clearly indicates that genes participating in distinct biological pathways may be subject to m6A-mediated translational control in different manners.
Notably, after investigating the correlation of m6A modification and translational status in Mo17 (Supplemental Figs. S16–S19; Supplemental Table S9), we saw the same pattern as we identified in B73. Altogether, these results suggest that m6A modification may play a role in bridging the transcription and translation, hierarchically organized by its strength and genic location in maize.
Natural Variation in m6A Modification between B73 and Mo17
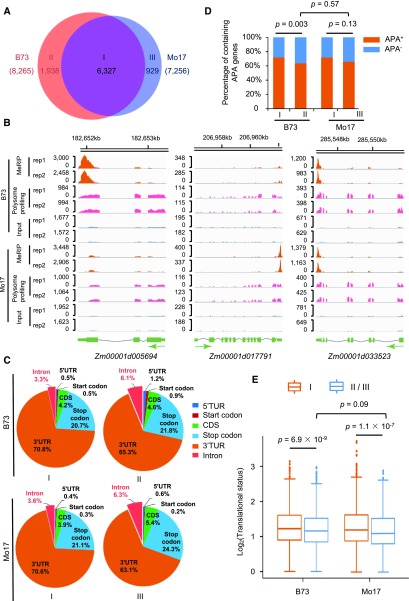
We next examined the extent of natural intraspecies variation in m6A modification. At the gene level, we found that 6,237 genes (type I) were commonly modified by m6A in both B73 and Mo17, while there were 1,938 genes (type II) and 929 genes (type III) that were only modified in B73 or Mo17, respectively (Fig. 4, A and B). As a post-transcriptional modification, the specific appearance of m6A sites is conceivably decided by gene-specific expression. However, we found that the inbred-specific expression could only explain a small proportion of genes within both the type II and III groups (Supplemental Fig. S20). In addition, we identified hundreds of genes with differential levels of m6A modification between B73 and Mo17 (Supplemental Fig. S21). These results indicate that although the overall topology of the m6A methylome is largely conserved between B73 and Mo17, there is a substantial number of genes with natural variation in m6A modification.
Figure 4.
Natural variation in the m6A modification between B73 and Mo17. A, Venn diagram showing the number of genes commonly (type I) and specifically modified by m6A in B73 (type II) and Mo17 (type III). B, Representative Integrative Genomics Viewer plots showing B73-specific (left), Mo17-specific (middle), and common (right) m6A modification. C, Pie charts depicting the percentage of m6A peaks within six transcript segments for three types of genes as indicated in (A). D, Proportion of transcripts containing APA usage for three types of genes as indicated in (A). P values were calculated using the Fisher’s exact test. E, Comparison of translational status among three types of genes as indicated in (A). P values were calculated using the Wilcoxon rank-sum test.
We then searched for the relevant features associated with or possibly responsible for inbred-specific m6A modification. Interestingly, relative to type I, m6A peaks in type II or type III showed a marked increase in the proportion of the spliced intronic segment (Fisher’s exact test, P value < 0.001; Fig. 4C), suggesting that the alternative splicing of mRNA between two inbred lines may at least in part be attributed to the inbred-specific m6A modification. In contrast, the proportion of genes with APA usage in type II and type III turned out to be less compared to that in type I (Fig. 4D), suggesting that the specific m6A-modification in type II or type III was not driven by the alteration in APA usage. Moreover, the specific modification in type II occurred with higher probability in the vicinity of the start codon (Fisher’s exact test, P value = 0.03), but not for other genic segments, when compared with the specific modification in type III (Fig. 4C). However, the proportions of genes with APA usage were comparable between type II and type III (Fig. 4D). Furthermore, we did not observe the enrichment of any significant GO terms for genes belonging to either type II or type III.
To address whether such natural variation in m6A modification had effects in gene expression regulation, we assessed the levels of translational status for genes in type II and type III, and made comparisons to type I. The results showed that the translational status was statistically lower for type II (Wilcoxon rank-sum test, P value = 6.9 × 10−9) and type III (Wilcoxon rank-sum test, P value = 1.1 × 10−7) compared to type I (Fig. 4E), indicating that genes commonly modified by m6A in both B73 and Mo17 may possess higher translational status than genes specifically modified in either B73 or Mo17. In contrast, there was no significant difference of the translational status between type II and type III at a global scale (Fig. 4E), or across each of the genic segments (Supplemental Fig. S22). Taken together, these results indicate that the natural variation in m6A actively occurs, and thereby may confer the other layer of gene expression regulation within species.
DISCUSSION
Topology and Features of m6A Modification in Maize
Our transcriptome-wide m6A mapping revealed an extremely asymmetric distribution of mRNA m6A methylation with the majority of m6A sites enriched in the 3′UTR in maize. In fact, a marked bias of m6A sites in 3′UTRs has been observed in nearly all species examined to date, including rice (Li et al., 2014) and Arabidopsis (Luo et al., 2014; Wan et al., 2015), suggesting that although the degree of this skewness seems variable, an evolutionary constraint may target the m6A deposition to the 3′UTR of genes regardless of gene structure or coding potential. This raises an intriguing question of what the underlying mechanisms of methylation specificity are. In this study, we found that m6A-methylated genes are highly likely to harbor APA events in maize. Meanwhile, we extended the investigation of the relationship between m6A and APA site selection in other plant species, including Arabidopsis (Col-0, Can-0, Hen-16) and rice, using published data (Li et al., 2014; Luo et al., 2014; Duan et al., 2017). The results revealed that the effect of m6A modification on APA usage was conserved in all plant species investigated (Supplemental Fig. S23). APA is a widespread phenomenon in eukaryotes, generating mRNAs with alternative 3′ends (Elkon et al., 2013; Tian and Manley 2017). The causative relation between m6A and APA was demonstrated by a recent study in mouse, showing that a nuclear m6A reader YTHDC1 plays a critical role in m6A-dependent processing of pre-mRNA transcripts, and the loss of YTHDC1 altered the APA usage for more than 800 genes (Kasowitz et al., 2018). In addition, it is likely that the involvement of m6A in APA usage may operate the other way around in maize, i.e. the occurrence of APA in 3′UTR marking transcripts with m6A sites. Alternatively, the bias of m6A methylation in the 3′UTR may be achieved by preferential recruitment of m6A methylase to the 3′UTR of genes by interacting with 3′UTR-binding proteins (Yue et al., 2018), or microRNAs, which were recently demonstrated to regulate the binding of m6A methylase to mRNA in a sequence-dependent manner (Chen et al., 2015).
Early studies from numerous organisms have confirmed a RRACH sequence as the canonical m6A consensus motif (Dominissini et al., 2012, 2013; Meyer et al., 2012), whereas we found that maize may utilize a distinct motif sequence (UGUAMM). The search using the UGUAMM sequence as the inquiry against all the expressed genes yielded a total of 34,173 sites, indicating that the majority of UGUAMM motifs in mRNA lack the m6A modification. Therefore, it remains unclear how the methylation machinery selectively targets a subset of consensus motifs in the 3′UTR. We suspect that cis-elements such as the neighboring RNA sequences or secondary structures may likely have accessory roles in methylation specificity.
m6A-Mediated Translation Control in Maize
Although not explored in plants yet, the impact of m6A on translation has been extensively studied in many other organisms (Meyer et al., 2015; Wang et al., 2015; Choi et al., 2016; Qi et al., 2016; Coots et al., 2017; Li et al., 2017; Slobodin et al., 2017), leading to a paradoxical conclusion that m6A marks enable both stimulatory and inhibitory effects on translation. In this study, after integrating the analyses from the mRNA transcriptome, m6A profiling, and polysome occupancy, we uncovered a general theme illustrating how m6A marks affect translational status in maize (Fig. 5): Transcripts with low m6A levels exhibit relatively high translational status; however, the excessive deposition of m6A marks on transcripts caused negative effects on translational status, indicating that the hypermethylated state of transcripts may inhibit the accretion of ribosomes and lead to the decrease in the translational status. Compared to other genic segments, the presence of m6A marks in the vicinity of the start codon may have the greatest effect on enhancing the translational status. Based on these findings, we conclude that dependent on its strength and genic location, the cotranscriptional m6A modification on mRNAs has multidimensional effects on translational status in maize.
Figure 5.
A general theme describing the multidimensional correlations of m6A marks on translational status. A, Transcripts with low m6A levels exhibit high translational status. B, Moderate m6A marks are associated with the median level of translational status. C, Excessive deposition of m6A marks decreases the translational status. D, The deposition of m6A marks in the vicinity of the start codon may promote the translational status.
As an epitranscriptome mark, m6A would be primarily recognized by reader proteins to fulfill its biological functions. This raises an attractive question of what the mechanistic connection between readers and m6A is, that influences the translational status. Although the translational regulation of m6A-modified mRNAs by m6A readers has not been reported in plants yet, this fascinating field has emerged to be unveiled recently in mammalian cells. The YTHDF2 protein, for instance, decreases the amount of m6A-methylated mRNA in translatable fractions by sequestering m6A-containing mRNAs to processing bodies and eventually facilitating their degradation (Wang et al., 2014a). In contrast, YTHDF1 and YTHDF3 recognizes m6A residues in 3′UTRs and promotes translation through interaction with initiation factors or ribosomal subunit proteins (Wang et al., 2014a, 2015; Li et al., 2017; Shi et al., 2017). Moreover, besides being conveyed by reader proteins, the effect of m6A residues on translation could also be accomplished by its direct impact on secondary and tertiary RNA structures (Wang et al., 2014b; Roost et al., 2015; Spitale et al., 2015; Liu et al., 2015, 2017; ), or its impairment on ribosome stalling or tRNA accommodation at methylation-affecting codons, leading to the reduced translation kinetics (Choi et al., 2016).
How m6A sites near start codons enhance the translational status is also an important area of future study. In principle, only the translating ribosomes can detect start codons, and this event occurs in the cytoplasm. However, m6A modification should take place primarily in the nucleus. Therefore, the start codon itself is unlikely to be the cause of the m6A enrichment near the start codon. This is distinct from m6A-mediated regulation of cap-dependent or -independent translation, both scenarios of which are related to m6A residues located in the 5′UTR (Meyer et al., 2015).
In sum, we conducted a parallel analysis of the transcriptome-wide m6A profiling and polysome occupancy in two maize inbred lines. We found many conserved and unique features of m6A localization in maize when compared to other organisms, and demonstrated that m6A modification is involved in orchestrating transcription and translation at a global scale in maize. We further characterized the mode of m6A methylation correlated with translational status by its strength and location in transcripts. Lastly, we found that thousands of genes exhibit distinctly inbred-specific methylation, highlighting that m6A modification confers a new dimension of natural variance in posttranscriptional gene regulation.
MATERIALS AND METHODS
Plant Materials
Seeds of maize (Zea mays) inbred lines B73 and Mo17 were sterilized by 70% ethanol for 1 min followed by 5% sodium hypochlorite solution for 5 min and rinsed five times with sterile water. Then seeds were sowed in pots containing a mixture of vermiculite and soil (1:1, v/v) and grown in the growth chamber at 28°C for 16 h in the light and 25°C for 8 h in the dark. After 14 d, aerial tissues were harvested, immediately frozen in liquid nitrogen, and stored at −80°C.
m6A MeRIP
Total RNA was extracted using TRIzol reagent (cat. no. 15596-018; Ambion Life Technologies) according to the manufacturer’s protocol. Polyadenylated RNA was isolated using the GenElute mRNA Miniprep Kit (Sigma-Aldrich) following the manufacturer’s instructions. Immunoprecipitation of m6A was adapted from the protocol of the Magna MeRIP m6A Kit (Millipore). Briefly, mRNA was adjusted to 27 μL with the concentration of ∼1 μg/μL, followed by adding 3 μL of 10× RNA fragmentation buffer (CS220011) for 4 min at 94°C and then adding 3 μL of EDTA (0.5 m) to terminate the reaction. The fragmented RNA was purified by ethanol precipitation. Then, 30 μL of magnetic A/G beads (CS203152) was preincubated with 10 μg of anti-m6A antibody (MABE1006) in 1× immunoprecipitation (IP) buffer for 30 min at room temperature. A total of 0.5-μg fragmented RNA was saved for RNA-seq as input control, and 20-μg fragmented mRNA was incubated with the antibody-beads mixture to a final volume of 500 μL for 2 h at 4°C with constant rotating. After washing 3 times with 1× IP buffer, RNA was eluted with 100 μL of elution buffer two times and all elutes from the same samples were combined, and subsequently purified using the RNA Clean & Concentrator Kit (ZYMO). Purified m6A-IP samples and input RNAs were subjected to library construction.
Polysome Profiling
Polysome isolation by differential centrifugation was performed as described in Zhang et al. (2017b). In brief, 2 g of tissue powder was homogenized in 5 mL of polysome extraction buffer (200 mm of Tris-HCl at pH 9.0, 200 mm of KCl, 35 mm of MgCl2, 25 mm of EGTA, 1% [v/v] Triton X-100, 1% [v/v] TWEEN 20, 2% [v/v] polyoxyethylene, 5 mm of dithiothreitol, 0.5 mg/mL of heparin, 100 μg/mL of chloramphenicol, and 25 μg/mL of cycloheximide). Crude cell lysate was centrifuged at 13,200 g for 15 min at 4°C. The supernatants were loaded on the top of a 1.7 m of Suc cushion and centrifuged at 45,000 rpm (model no. SW 55 Ti Swinging Bucket Rotor in a model no. l-100XP Ultracentrifuge; Beckman Coulter) for 3 h at 4°C. The ribosome pellet was resuspended in 200 μL of resuspension buffer (200 mm of Tris-HCl at pH 9.0, 200 mm of KCl, 35 mm of MgCl2, 25 mm of EGTA, 100 μg/mL of chloramphenicol, and 25 μg/mL of cycloheximide). Then the solution was layered over a 20% to 60% Suc density gradient and centrifuged at 41,000 rpm (model no. SW 55 Ti Swinging Bucket rotor; Beckman Coulter) for 2 h at 4°C. After ultracentrifugation, the gradients were monitored and fractionated into 14 fractions at an absorbance of 254 nm using a Gradient Fractionator (Biocomp) including a UV detector. The polysome-RNA fractions were pooled and treated by 5% (w/v) SDS/0.2 m of EDTA, and then extracted twice with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1; v/v/v). The mixture was centrifuged at 12,000 rpm for 5 min at 10°C and RNA was precipitated by isopropanol followed by washed using 70% (v/v) ethanol, and eventually resuspended for the library construction.
Library Construction and Sequencing
Libraries of RNA-seq, m6A-seq, and polysome profiling were generated using the NEBNext Ultra II RNA Library Prep Kit (model no. E7770S; New England Biolabs) according to the manufacturer’s instructions. The libraries were sequenced with 150-bp paired reads on a model no. HiSeq X Ten platform (Illumina).
m6A Peak Calling
Raw paired-end reads of m6A-seq and input RNA-seq were filtered and adapter sequences were trimmed out by the tool Trimmomatic v0.35 (Bolger et al., 2014) with the parameters ILLUMINACLIP TruSeq3-PE-2.fa MINLEN 30. Cleaned reads from B73 samples were aligned to the maize B73 reference genome, AGPv4.38 (Jiao et al., 2017) and cleaned reads from Mo17 samples were mapped to the maize Mo17 reference genome, CAU-1.0 (Sun et al., 2018) using the software Hisat2 v2.1.0 (Kim et al., 2015) with the parameters −5 1 −3 1–dta. Reads matching to less than five places were retained for further analysis. m6A peaks were identified using the MACS2 peak calling algorithm (Zhang et al., 2008) with the input as background and the parameter of effective genome size was adjusted to the transcriptome size (417272442) and the q value was set to 0.01. Peaks that overlapped in at least 50% of their length between two biological replicates were designated as high confidence m6A peaks using the software package BedTools v2.17.0 (Quinlan and Hall 2010). The m6A intensity was defined as fold changes of m6A peaks from MACS2 output.
The analysis of m6A peak enrichment based on six nonoverlapping transcript segments was performed as follows: 5′UTR [transcription start site, CDS start codon − 101 bp], start codon segment [CDS start codon − 100 bp, CDS start codon + 100 bp], CDS [CDS start codon + 101 bp, CDS stop codon − 101 bp], stop codon segment [CDS stop codon − 100 bp, CDS stop codon + 100 bp], 3′UTR [CDS stop codon + 101 bp, transcription termination site], and intron segment. Each of the m6A sites is represented only once in the analysis. For genes with multiple mRNA transcripts, the longest one was selected. Each high confidence peak was annotated to one of these regions using BedTools (Quinlan and Hall 2010).
m6A Motif Analysis
All m6A peaks were sorted according to fold change and the top 2,000 peaks were chosen for the de novo motif analysis using the both of the two programs MEME v4.10.2 (Bailey et al., 2009) and HOMER v4.9 (Heinz et al., 2010). The 101-nt–long sequences derived from the sense strand and centered around the peak summit were extracted using the “fastaFromBed” function in BedTools (Quinlan and Hall 2010) and used as input for MEME (Bailey et al., 2009) and HOMER (Heinz et al., 2010). The meme script in MEME (Bailey et al., 2009) and the findMotifs.pl script in HOMER (Heinz et al., 2010) were used for the de novo motif analysis.
APA Analysis
Genes with multiple poly(A) sites were defined according to maize B73 gene annotations (AGPv4.38; www.maizegdb.org).
RNA-Seq Analysis and Translational Status Calculation
Raw paired-end reads of polysome profiling and RNA-seq were processed and aligned the same as described above for m6A peak calling. Gene transcriptional and translational levels were estimated by calculating fragments per kilobase of transcript per million fragments (FPKM) by the software StringTie v1.3.3 (Pertea et al., 2015) with default parameters. The translational status was calculated by “FPKM(translational level)/FPKM(transcriptional level)” as described in Lei et al. (2015). Inbred-specific expression was defined as genes with the FPKM ≥ 1 of mRNA abundance in one in-bred, but FPKM < 1 in the other.
GO Analysis
K-means clustering was used to explore genes coordinated in the levels of m6A methylation and translational status. On the basis of both the Elbow (the factoextra function in the R package; https://www.r-project.org/) and Average silhouette (the factoextra function in the R package) methods, four clusters were determined as the optimal number of clusters. To avoid skewing distance calculation due to difference in scale and variance of the expression measurements, each level of m6A methylation and translational status was firstly converted to percentiles using the empirical cumulative distribution function. The software FuncAssociate v3.0 (http://llama.mshri.on.ca/funcassociate/; Berriz et al., 2009) was used to assess enrichment of GO terms for each cluster. The gene list for background input was explicitly defined as the set of genes included in all clusters. We defined significant enrichment as GO terms with adjusted P value < 0.05. Adjusted P value was the fraction (as percent) of 1,000 null-hypothesis simulations having attributes with this single-hypothesis P value or smaller.
Enriched GO terms were visualized with the software Cytoscape 3.3.0 (Shannon et al., 2003) installed with the Enrichment Map plugin (Merico et al., 2010). Within the enrichment maps, each node indicates an enriched GO pathway and the node size is proportional to the total number of genes in each pathway.
RT-qPCR
RT-qPCR was performed as described in Duan et al. (2017). Briefly, 0.5-μg RNA from m6A-IP, input, and polysome profiling were used for reverse transcription using the Maxima H Minus cDNA Synthesis Master Mix with ds-DNase (Thermo Fisher Scientific). RT-qPCR was performed using TB Green Premix Ex Taq (TaKaRa). Zm00001d019684 was used as an internal control gene for the normalization. All primers used in this study are listed in Supplemental Table S10.
Analyses in Mo17
To reduce mapping bias, we built a Mo17 pseudogenome by substituting single nucleotide polymorphisms in the maize B73 reference genome (AGPv4.38) to Mo17 nucleotides. After that, reads of m6A-seq, RNA-seq, and polysome profiling from Mo17 samples were remapped to the Mo17 pseudogenomes, and all the related analyses in Mo17 were conducted the same as for B73, as described above.
Accession Numbers
The high-throughput sequencing data generated in this study have been deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus database repository (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE124543. All raw data in this study also were converted to bigWig files (https://genome.ucsc.edu/goldenpath/help/bigWig.html) and deposited in the web-based tool Comparative Genomics (https://genomevolution.org/coge/) for visualization. The accession numbers for these bigwig files are listed in Supplemental Table S11.
Supplemental Material
The following supplemental materials are available.
Supplemental Figure S1. The number of methylated transcripts containing different m6A sites in B73.
Supplemental Figure S2. RT-qPCR validation for nine randomly selected genes containing m6A methylation.
Supplemental Figure S3. The number of methylated transcripts containing various combinations of m6A sites within six transcript segments in B73.
Supplemental Figure S4. Distribution pattern of m6A sites in Mo17.
Supplemental Figure S5. The conserved m6A motif sequence in Mo17.
Supplemental Figure S6. GO enrichment analysis for m6A-methylated genes in Mo17.
Supplemental Figure S7. Association of m6A with APA usage in Mo17.
Supplemental Figure S8. The proportion of genes with APA usage according to the genic location of m6A in B73 (top) and Mo17 (bottom).
Supplemental Figure S9. Correlation between m6A strength and mRNA abundance in B73 and Mo17.
Supplemental Figure S10. The repeatability between two biological replicates for both RNA-seq data (left) and polysome profiling data (right) in B73 (top) and Mo17 (bottom).
Supplemental Figure S11. Positive effect of RNA abundance on translational status in B73.
Supplemental Figure S12. m6A-RT-qPCR of polysome-associated mRNA and total mRNA.
Supplemental Figure S13. GO analysis of genes with m6A marks at different transcript segments in B73.
Supplemental Figure S14. “Elbow” and “Average silhouette” method for the identification of the optimal number of clusters in B73.
Supplemental Figure S15. The relationship between clusters and m6A-related characteristics in B73.
Supplemental Figure S16. Positive effect of RNA abundance on the translational status in Mo17.
Supplemental Figure S17. Effect of the m6A modification on translation in Mo17.
Supplemental Figure S18. “Elbow” and “Average silhouette” method for the identification of the optimal number of clusters in Mo17.
Supplemental Figure S19. Effect of the m6A modification on translation in Mo17.
Supplemental Figure S20. The specific expressed and modified genes in B73 and Mo17.
Supplemental Figure S21. Hundreds of genes with differential levels of m6A modification between B73 and Mo17.
Supplemental Figure S22. Translational status between Type II and Type III across the different mRNA segments.
Supplemental Figure S23. Proportion of m6A-modified genes within transcripts with or without APA usage among different species.
Supplemental Table S1. The list of m6A-containing protein-coding genes showing peak summit locations, m6A level, and translational status in B73.
Supplemental Table S2. The list of m6A-containing long non-coding RNAs showing peak summit locations and m6A level in B73.
Supplemental Table S3. The list of m6A-containing transposable elements showing peak summit locations and m6A level in B73.
Supplemental Table S4. The list of m6A-containing protein-coding genes showing peak summit locations, m6A level, and translational status in Mo17.
Supplemental Table S5. The list of m6A-containing transposable elements showing peak summit locations and m6A level in Mo17.
Supplemental Table S6. mRNA abundance of all expressed genes in two independent biological replicates in B73.
Supplemental Table S7. mRNA abundance of all expressed genes in two independent biological replicates in Mo17.
Supplemental Table S8. GO term enrichments for the clusters identified in Figure 3C.
Supplemental Table S9. GO term enrichments for the clusters identified in Supplemental Figure S19A.
Supplemental Table S10. The list of primers used in the study.
Supplemental Table S11. The list of accession numbers for “bigWig” files in this study.
ACKNOWLEDGMENTS
We thank all the members of our laboratories for helpful discussions and assistance during this project.
Footnotes
This work was supported by the National Key Research and Development Program of China (2016YFD0101201 and 2017YFD0101104 to Y.H.).
Articles can be viewed without a subscription.
References
- Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015) N6-methyladenosine marks primary microRNAs for processing. Nature 519: 482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SJ, Kramer MC, Gosai SJ, Yu X, Vandivier LE, Nelson ADL, Anderson ZD, Beilstein MA, Fray RG, Lyons E, et al. (2018) N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Reports 25: 1146–1157.e3 [DOI] [PubMed] [Google Scholar]
- Arribas-Hernández L, Bressendorff S, Hansen MH, Poulsen C, Erdmann S, Brodersen P (2018) An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30: 952–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP (2009) Next generation software for functional trend analysis. Bioinformatics 25: 3043–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Bottley A, Archer N, May ST, Fray RG (2015) Yeast m6A methylated mRNAs are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PLoS One 10: e0132090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, May S, Fray RG (2012) Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front Plant Sci 3: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. (2015) m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16: 289–301 [DOI] [PubMed] [Google Scholar]
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O’Leary SE, Dominissini D, Rechavi G, Soltis SM, et al. (2016) N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol 23: 110–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coots RA, Liu XM, Mao Y, Dong L, Zhou J, Wan J, Zhang X, Qian SB (2017) m6A facilitates eIF4F-independent mRNA translation. Mol Cell 68: 504–514.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. (2017) m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Reports 18: 2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G (2013) Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8: 176–189 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206 [DOI] [PubMed] [Google Scholar]
- Duan HC, Wei LH, Zhang C, Wang Y, Chen L, Lu Z, Chen PR, He C, Jia G (2017) ALKBH10B is an RNA N6-methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29: 2995–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R (2013) Alternative cleavage and polyadenylation: Extent, regulation and function. Nat Rev Genet 14: 496–506 [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806 [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. (2015) Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347: 1002–1006 [DOI] [PubMed] [Google Scholar]
- Han R, Slobodin B, Agami R (2017) The methylated way to translation. Oncotarget 8: 93313–93314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann IU, Bodi Z, Sanchez-Moran E, Mongan NP, Archer N, Fray RG, Soller M (2016) m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 540: 301–304 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. (2017) Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res 27: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018) Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 20: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011) N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J (2014) Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 111: E203–E212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L, Grozhik AV, Vedanayagam J, Patil DP, Pang N, Lim KS, Huang YC, Joseph B, Lin CJ, Despic V, et al. (2017) The m6A pathway facilitates sex determination in Drosophila. Nat Commun 8: 15737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM, Wang PJ (2018) Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet 14: e1007412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, Haripal B, Zucker-Scharff I, Moore MJ, Park CY, et al. (2015) A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev 29: 2037–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: A fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, et al. (2015) Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J 84: 1206–1218 [DOI] [PubMed] [Google Scholar]
- Lence T, Akhtar J, Bayer M, Schmid K, Spindler L, Ho CH, Kreim N, Andrade-Navarro MA, Poeck B, Helm M, et al. (2016) m6A modulates neuronal functions and sex determination in Drosophila. Nature 540: 242–247 [DOI] [PubMed] [Google Scholar]
- Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al. (2017) Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 27: 444–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang X, Li C, Hu S, Yu J, Song S (2014) Transcriptome-wide N6-methyladenosine profiling of rice callus and leaf reveals the presence of tissue-specific competitors involved in selective mRNA modification. RNA Biol 11: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Choe J, Du P, Triboulet R, Gregory RI (2016) The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 62: 335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. (2014) A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T (2015) N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T (2017) N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 45: 6051–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. (2014) Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez M, Aparicio F, López-Gresa MP, Bellés JM, Sánchez-Navarro JA, Pallás V (2017) Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc Natl Acad Sci USA 114: 10755–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel M, Chen KM, Homolka D, Gos P, Pandey RR, McCarthy AA, Pillai RS (2018) Methylation of structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol Cell 71: 986–1000.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico D, Isserlin R, Stueker O, Emili A, Bader GD (2010) Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One 5: e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD. (2018) m6A-mediated translation regulation. Biochim Biophys Acta Gene Regul Mech 1862: 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR (2014) The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR (2015) 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (2012) Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, Howard BD, Daneshvar K, Mullen AC, Dedon P, et al. (2016) m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat Methods 13: 692–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014) Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res 24: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi ST, Ma JY, Wang ZB, Guo L, Hou Y, Sun QY (2016) N6-methyladenosine sequencing highlights the involvement of mRNA methylation in oocyte meiotic maturation and embryo development by regulating translation in Xenopus laevis. J Biol Chem 291: 23020–23026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Soller M (2017) m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet 33: 380–390 [DOI] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET (2015) Structure and thermodynamics of N6-methyladenosine in RNA: A spring-loaded base modification. J Am Chem Soc 137: 2107–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017) YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6: e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al. (2014) Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports 8: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutenaire J, Deragon JM, Jean V, Benhamed M, Raynaud C, Favory JJ, Merret R, Bousquet-Antonelli C (2018) The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell 30: 986–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Liang Z, Gu X, Chen Y, Teo ZW, Hou X, Cai WM, Dedon PC, Liu L, Yu H (2016) N6-methyladenosine RNA modification regulates shoot stem cell fate in Arabidopsis. Dev Cell 38: 186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, He C (2017) YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 27: 315–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B, Han R, Calderone V, Vrielink J, Loayza-Puch F, Elkon R, Agami R (2017) Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell 169: 326–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhou Y, Chen J, Shi J, Zhao H, Zhao H, Song W, Zhang M, Cui Y, Dong X, et al. (2018) Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat Genet 50: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Tian B, Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Tang K, Zhang D, Xie S, Zhu X, Wang Z, Lang Z (2015) Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol 16: 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, Liu B, Xiao F, Wang W, Huang G, et al. (2018) Loss of YTHDF2-mediated m6A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res 28: 1035–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014a) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C (2015) N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC (2014b) N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol 16: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, Xiao Y, Zhang X, Duan HC, Jia G (2018) The m6A Reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30: 968–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, Chen Y, Xiao Y, Gao Y, Zhang Q, et al. (2018) A novel m6A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res 29: 23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. (2016) Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol Cell 61: 507–519 [DOI] [PubMed] [Google Scholar]
- Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li YF, Chen YS, Zhang XX, Wang CX, Jiang LY, et al. (2017) Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res 27: 1100–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017) Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L et al. (2017) Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell 171: 877–889 e817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Nie X, Yan Z, Weining S (2019) N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol J 17: 1194–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, et al. (2018) VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C (2015) RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29: 1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, et al. (2017a) m6A modulates haematopoietic stem and progenitor cell specification. Nature 549: 273–276 [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, et al. (2017b) Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell 29: 1952–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. (2014) FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 24: 1403–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. (2013) ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB (2015) Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Shu XE, Mao Y, Liu XM, Yuan X, Zhang X, Hess ME, Brüning JC, Qian SB (2018) N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell 69: 636–647.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]