Abstract
Rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) colonization status facilitates isolation and decolonization and reduces MRSA infections. Liquid but not dry swabs allow fully automated detection methods. However, the accuracy of culture and polymerase chain reaction (PCR) using liquid and dry swabs has not been analyzed. We compared different swab collection systems for routine nasal–throat MRSA screening in patients admitted to a tertiary care trauma center in Germany. Over 3 consecutive months, dry swabs (month 1), ESwabs (month 2), or MSwabs (month 3) were processed using Cepheid GeneXpert, Roche cobas and BD-MAX™ MRSA tests compared to chromogenic culture. Among 1680 subjects, the MRSA detection rate using PCR methods did not differ significantly between dry swabs, ESwab, and MSwab (6.0%, 6.2%, and 5.3%, respectively). Detection rates using chromogenic culture were 2.9%, 3.9%, and 1.9%, using dry, ESwab, and MSwab, respectively. Using chromogenic culture as the “gold standard”, negative predictive values for the PCR tests ranged from 99.2–100%, and positive predictive values from 33.3–54.8%. Thus, efficient and accurate MRSA screening can be achieved using dry, as well as liquid E- or MSwab, collection systems. Specimen collection using ESwab or MSwab facilitates efficient processing for chromogenic culture in full laboratory automation while also allowing molecular testing in automated PCR systems.
Keywords: Staphylococcus aureus, MRSA, screening, PCR, chromogenic culture, dry swab, MSwab, ESwab
Introduction
Staphylococcus aureus (SA) and methicillin-resistant SA (MRSA) are major causes of healthcare-acquired infections and have been responsible for outbreaks in healthcare settings worldwide [1–3]. SA and MRSA infections are associated with significant healthcare costs [4]. Guidelines and recommendations [5], as well as hospital standard procedures, recommend active screening and isolation and/or decolonization of patients as measures to control the spread of MRSA and SA [6, 7].
The German MRSA screening guidelines [8] recommend culture-based methods as the basis of MRSA screening. Due to the high negative predictive value (NPV) and rapid turnaround time, PCR-based methods are recommended to complement culture-based testing; swabs from at least the 2 nares should be used for MRSA screening. Applying culture- and PCR-based technologies to MRSA screening requires the use of appropriate collection media suitable for accurate performance of both technologies.
Screening of potential MRSA-positive patients has been shown to be effective in most studies in reducing the frequency of MRSA transmissions in hospitals [3, 7, 9–11]. Over the last years, multiple medium-to-high throughput automated PCR systems have become commercially available [12–14]. Using these MRSA screening tests, the value of rapid detection of colonization with MRSA and SA and consequent implementation of appropriate barrier precautions have been repeatedly demonstrated [15, 16].
The introduction of PCR screening tests is associated with investment and operational costs, which proved inaccessible for some organizations. A possibility of reducing costs for hospitals is to reach a high degree of automation, which, however, requires sample collection with specific swabs that allow liquid sample processing [17, 18]. Advances with liquid and flocked swabs have demonstrated improved pathogen detection [19]. The ESwab system (Copan Diagnostics Inc., Murrieta, CA) is a nylon-flocked swab liquid-based system for collection and transport; the organism inoculum is released into 1 mL of Amies liquid, allowing to perform PCR assays and cultures [20, 21]. Copan's MSwab system is validated for use with MRSA/SA and HSV 1 and 2 assays on a Roche's cobas 4800 system and for limited mostly Gram-positive aerobic bacterial cultures.
While different swab types for MRSA screening had previously been evaluated for individual automated techniques, we are not aware of a study investigating the accuracy of dry and liquid swab collection systems using culture and different automated PCR systems.
We therefore compared the performance of conventional dry swabs with 2 different liquid swabs (ESwab and MSwab) using chromogenic culture and three automated PCR systems for the detection of MRSA colonization.
Materials and Methods
Study Design
This study was performed over three consecutive months from September to November 2016 using routine admission patient samples prospectively collected at the Unfallkrankenhaus Berlin (ukb), a tertiary care trauma center in Berlin, Germany. The ukb serves 600 hospital beds with 25 specialized departments. All samples were tested onsite at the Satellite Laboratory in ukb and subsequently at the Central Laboratory, Labor Berlin. On site, swabs were tested with Cepheid GeneXpert MRSA (GeneXpert), and subsequently, swabs were transported to the central laboratory for chromogenic culture and additional PCR-based testing.
The study design is illustrated in Figure 1. Inclusion criteria for the study were as follows: patients eligible for routine admission screening following the collection site's screening policy (systematic screening of all patients at admission to the trauma center emergency room).
Figure 1.
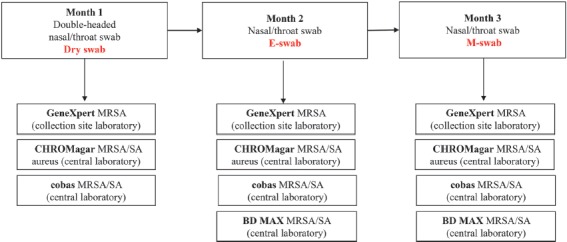
Overview of study design
Collection of the nasal/throat swabs from patients as part of routine MRSA admission screening were as follows: month 1 a double-headed dry swab (Swab: Cotton, Applicator: Wood, Copan, Brescia, Italy), month 2 ESwab (Copan), and month 3 MSwab (Copan). Testing on the GeneXpert system was performed 24/7 immediately after sample receipt. Specimens were transported (3 times daily Monday–Friday and 2 times daily Sa/So) to the centralized laboratory for further analysis. Dry swabs (month 1) were suspended in a 1 mL MSwab medium upon arrival at the Labor Berlin.
Routine culture-based testing using chromogenic media for MRSA and PCR testing was performed from Monday to Sunday. The residual patient materials in E- or MSwab media were stored at 4–8 °C over the weekend until testing resumed.
In the first month, the dry swab designated for molecular testing was suspended in 2 mL of elution buffer medium (GeneXpert, Cepheid) at the ukb satellite laboratory and tested using a GeneXpert. In the second and third month, at the ukb laboratory, 100 μL sample volume from each specimen (nasal and throat swabs) was removed, pooled (200 μL total sample) in 2 mL of elution buffer medium (GeneXpert, Cepheid), and tested using GeneXpert. The residual 900 μL per swab were transported to the Labor Berlin central lab for further processing. All incoming swab samples were streaked onto a bi-plate chromogenic media (chromID MRSA/chromID S. aureus, bioMerieux, France). Dry swabs in MSwab medium, ESwab, and MSwab media were loaded onto the cobas 4800 and BD MAX instruments and processed, following the manufacturer's instructions (see below). All leftover specimens were stored at –80 °C.
A total of 3376 samples were collected from 1689 subjects enrolled between September and November 2016, of which 1680 were evaluable subjects for nasal–throat MRSA colonization analyses (548 for Dry swab, 566 for ESwab, and 566 for MSwab). Overall result interpretation at the ukb for routine MRSA admission screening was as follows: the GeneXpert result was reported as preliminary in all cases. In cases where the GeneXpert result was positive but the chromogenic culture was negative, the culture was repeated from the original collection swab/medium. Positive GeneXpert results were reported immediately with the preliminary report, and the result of the repeated chromogenic culture was reported as the final result.
Chromogenic Culture for MRSA
Direct culture was performed using MRSA and SA chromogenic bi-plates (bioMerieux, Lyon, FR, and chromID MRSA/chromID S. aureus). Presumptive culture isolates were sub-cultured to 5% sheep blood agar and confirmed as SA using Gram staining, followed by latex agglutination testing using Stapaurex Plus Latex Agglutination Test (Remel, Dartford, UK).
Molecular Tests for the Detection of MRSA and SA
The GeneXpert MRSA test is validated for ESwabs and was performed by pooling dry swabs into the elution buffer vial and resuspending thoroughly. The entire elution buffer was transferred into the chamber S of the GeneXpert cartridge and incubated for 15 min before starting the MRSA test.
The cobas MRSA/SA test on the cobas 4800 system (cobas) is validated for use with MSwabs [12]. The testing was performed according to the manufacturer's instructions with the exception of the other swab types used.
The BD MAX MRSA XT test (Becton Dickinson, New Jersey, USA) (BD MAX) is validated for use with Liquid Stuart swabs and was performed on samples collected during months 2 and 3 following the instructions of the manufacturer [22].
Statistical Analysis
The agreements between molecular and chromogenic culture tests were summarized using Venn diagrams for dry swab, ESwabs, and MSwabs. With respect to chromogenic culture, the sensitivity, specificity, positive predicted value, and negative predicted value were calculated for each assay by swab type. The 95% score confidence intervals were provided for the sensitivity and specificity estimates. A generalized linear mixed model (GLMM) was used to estimate the proportions (positivity rate or sensitivity estimate) across assays and to perform statistical hypothesis testing. The model included terms for swab, assay, and their interaction as fixed effects and was used with the SAS GLIMMIX procedure to account for the multiple observations on the same sample by different assays.
Ethics
Ethics approval (EA2_069_16) was obtained from the institutional review board of the Charité University Medical Center.
Results
To evaluate collection devices for universal admission screening of nasal–throat MRSA colonization, we collected 3 different swab types (one month for each swab type) —dry swabs (month 1), MSwabs (month 2), and ESwabs (month 3) processed with different PCR-based and culture-based tests (Figure 1).
First, we compared the performance of dry swabs routinely used for MRSA admission screening to the performance of liquid (ESwab and MSwab) swab collection systems to enable high-throughput processing of swabs on automated liquid microbiology platforms plus on automated PCR-based systems. The positivity rate for chromogenic culture and the PCR-based tests is shown in Figure 2A. Among nasal–throat swabs with positive results obtained from dry swabs during month 1 of testing, there were 15 samples in which MRSA was detected by GeneXpert and cobas and by chromogenic culture (Figure 2B); in 10 samples, GeneXpert and cobas both detected MRSA, whereas chromogenic culture was negative. In addition, GeneXpert and cobas test detected an additional 8 and 7 MRSA isolates, respectively, which were not detected by any other method. Chromogenic culture detected only one additional MRSA isolate that was not detected by either GeneXpert or cobas. Second, among nasal–throat swabs with positive results obtained from ESwabs during month 2 of testing, there were 14 samples in which MRSA was detected by GeneXpert, cobas, and BD MAX, as well as by chromogenic culture (Figure 2C); in an additional 7 samples, GeneXpert, cobas, and BD MAX test detected MRSA, whereas chromogenic culture was negative. Individual PCR-based tests detected between 3 and 8 MRSA isolates without matching results in any other test. Chromogenic culture detected only 1 MRSA isolate that was not detected by any PCR-based test. Lastly, among nasal–throat swabs with positive results obtained from MSwabs during month 3 of testing, there were 10 samples in which MRSA was detected by GeneXpert, cobas, and BD MAX, as well as by chromogenic culture (Figure 2D); in an additional 10 samples, GeneXpert, cobas, and BD MAX detected MRSA whereas chromogenic culture was negative. Of interest, there were 8 positive results in cobas and BD MAX that were not confirmed by the GeneXpert or chromogenic culture. Individual PCR-based tests detected between 3 and 4 MRSA isolates without matching results in any other test.
Figure 2.
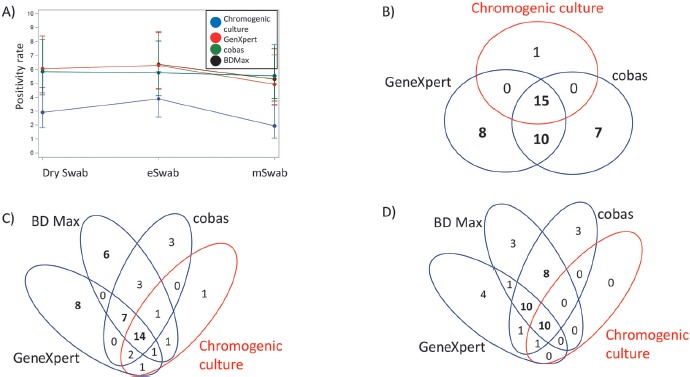
Agreement of results between PCR-based MRSA tests and chromogenic culture using dry swabs, ESwabs, and MSwabs. (A) MRSA positivity rate for PCR-based tests and chromogenic culture (in red) using different swab types and collection media. Numbers of positive results for chromogenic culture vs. cobas and GenXpert in dry swabs (B), ESwabs (C), and MSwabs (D). Agreement in numbers of equal to or above 5 are shown in bold
Compared to chromogenic culture all PCR-based tests demonstrated a high sensitivity; a trend towards higher sensitivity was observed for the dry swab and MSwab compared to the ESwab (Table 1). While this finding was not statistically significant (p = 0.359), it was found with all methods. Of note, we observed a relatively low number of MRSA-positive specimens, which limited the statistical power of the study. These sensitivities translated into very high negative predictive values ranging from 99.2–100%, based on a relatively low prevalence of 5–6% by PCR and 2–4% by culture. However, positive predictive values (PPVs) not surprisingly were markedly lower ranging from 33.3 to 54.8%.
Table 1.
Sensitivity, specificity, positive, and negative predictive values for the detection of MRSA using PCR-based methods compared to chromogenic culture in nasal–throat specimens obtained using dry, E-, or MSwabs collection media
| GeneXpert | cobas | BD MAX | ||
|---|---|---|---|---|
| Dry swab | Sensitivity | 93.8% (15/16) | 93.8% (15/16) | n.p. |
| (CI) | (71.7–99.7) | (71.7–99.7) | ||
| Specificity | 96.6% (508/526) | 96.8% (514/531) | n.p. | |
| (CI) | (94.7–97.8) | (94.9–98.0) | n.p. | |
| PPV | 45.5% (15/33) | 46.9% (15/32) | n.p. | |
| NPV | 99.8% (508/509) | 99.8% (514/515) | n.p. | |
| ESwab | Sensitivity | 86.4% (19/22) | 81.0% (17/21) | 81.8% (18/22) |
| (CI) | (66.7–95.3) | (60.0–92.3) | (61.5–92.7%) | |
| Specificity | 97.0% (525/541) | 97.4% (516/530) | 96.7% (526/544) | |
| (CI) | (95.3–98.2) | (95.6–98.4) | (94.8–97.9) | |
| PPV | 54.3% (19/35) a | 54.8% (17/31) b | 50.0% (18/36) c | |
| NPV | 99.4% (525/528) | 99.2% (516/520) | 99.2% (526/530) | |
| MSwab | Sensitivity | 100% (11/11) | 100% (11/11) | 90.9% (10/11) |
| (CI) | (74.1–100) | (74.1–100) | (62.3–99.5) | |
| Specificity | 96.9% (538/555) | 96.3% (526/546) | 96.4% (535/555) | |
| (CI) | (95.1–98.1) | (94.4–97.6) | (94.5–97.7) | |
| PPV | 39.3% (11/28)a | 35.5% (11/31) b | 33.3% (10/30) c | |
| NPV | 100.0% (538/538) | 100.0% (526/526) | 99.8% (535/536) | |
We also determined the sensitivity of each of the three PCR-based methods applying a rule of majority for nasal–throat swabs in which at least two PCR-based tests had given a positive result along with a positive chromogenic culture. Sensitivities for the GeneXpert performed from dry, ESwabs, or MSwabs, respectively, were 93.8 (95% CI: 71.7–99.7%), 86.4 (95% CI 66.7–95.3%), and 100% (95% CI 74.1–100%); for cobas, sensitivities were 93.8 (95% CI 71.7–99.7%), 81.0 (95% CI 60.0–92.3%), and 100% (95% CI 74.1–100%). For BD Max, sensitivities for ESwab and MSwab were 81.8 (95% CI 61.5–92.7%) and 90.9% (95% CI 62.3–99.5%).
Since PCR assays may be considerably more sensitive than culture, the designation “gold standard” for chromogenic culture is at least questionable. Therefore, we determined the sensitivity of chromogenic culture compared to a reference standard composed of at least two positive PCR test results. The positive percent agreement was low between chromogenic culture and the reference PCR standard, 60% (15/25) for dry swabs, 63.3% (19/30) for ESwabs, and 37.9% (11/29) for MSwabs.
Since results presented above could be interpreted as higher sensitivity of PCR-based tests compared to chromogenic culture, we also analyzed the threshold cycle (Ct) values of test results obtained in PCR-based tests. Ct results are shown for the cobas test results (Table 2). The Ct values obtained in cobas did not differ significantly in dry swab samples that were positive by PCR-based tests plus chromogenic culture vs. those swabs that were discordant (positive by 1 or 2 PCR-based tests and negative by chromogenic culture).
Table 2.
Ranges of cobas MRSA Ct values in patient samples with discordant results in PCR-based tests and chromogenic culture obtained from (a) dry swab, (b) ESwab, and (c) MSwab collection media
| a) | ||||||
|---|---|---|---|---|---|---|
| Swab | Chromogenic culture | GeneXpert | cobas | N | Mean Ct value (SD) | Ct value range |
| Dry swab | Negative | Negative | Negative | 500 | n.a. | n.a. |
| Negative | Positive | 7 | 37.93 (3.22) | 31.0–40.1 | ||
| Positive | Negative | 8 | n.a. | n.a. | ||
| Positive | Positive | Positive | 10 | 35.67 (5.63) | 24.0–40.7 | |
| Negative | Negative | 1a | ||||
| Positive | Positive | 15 | 33.15 (3.11) | 27.0–38.0 | ||
| b) | |||||||
|---|---|---|---|---|---|---|---|
| Swab | Chromogenic culture | GeneXpert | BD Max | cobas | N | Mean Ct value (SD) | Ct value range |
| ESwab | Negative | Negative | Negative | Negative | 500 | n.a. | n.a. |
| Negative | Positive | 3 | 29.47 (9.36) | 22.5–40.1 | |||
| Positive | Negative | 6 | n.a. | n.a. | |||
| Positive | Positive | 3 | 35.93 (3.27) | 32.2–38.3 | |||
| Positive | Negative | Negative | 8 | n.a. | n.a. | ||
| Positive | Positive | 7 | 33.63 (3.04) | 28.3–37.1 | |||
| Positive | Negative | Negative | 1 | n.a. | n.a. | ||
| Negative | Positive | Negative | 1 | n.a. | n.a. | ||
| Positive | Positive | 1 | 29.9 (0) | 29.9–29.9 | |||
| Positive | Negative | Negative | 1 | n.a. | n.a. | ||
| Positive | 2 | 37.45 (1.2) | 26.6–38.3 | ||||
| Positive | Negative | 1 | n.a. | n.a. | |||
| Positive | 14 | 32.31 (3.46) | 25.9–38.3 | ||||
| c) | |||||||
|---|---|---|---|---|---|---|---|
| Swab | Chromogenic culture | GeneXpert | BD Max | cobas | N | Mean Ct value (SD) | Ct value range |
| Mswab | Negative | Negative | Negative | Negative | 518 | n.a. | n.a. |
| Negative | Positive | 3 | 34.27 (5.01) | 28.6–38.1 | |||
| Positive | Negative | 3 | n.a. | n.a. | |||
| Positive | Positive | 6 | 33.83 (3.18) | 27.7–36.1 | |||
| Positive | Negative | Negative | 4 | n.a. | n.a. | ||
| Negative | Positive | 1 | 36.5 (0) | 36.5 | |||
| Positive | Negative | 1 | n.a. | n.a. | |||
| Positive | Positive | 10 | 34.1 (2.01) | 31.2–37.1 | |||
| Positive | Positive | Negative Positive |
Positive Positive |
1 10 |
38.1 (0) 31.14 (4.51) |
38.1 24.7–37.0 |
|
aCultural growth on chromogenic media was notable for sporadic growth.
Discussion
This study was performed to assess the accuracy of dry and liquid swab types for high-throughput MRSA screening using automated PCR systems and chromogenic culture.
The most important result of our study is the finding that dry swabs did not perform significantly different from liquid E- and MSwab collection systems regardless of the detection method being chromogenic culture or PCR. Detection rates for MRSA ranged from 5.3 to 6.0% for the different swab collection systems and did not differ significantly between the PCR-based tests (GeneXpert, cobas, and BD MAX). Detection rates using chromogenic culture with dry, ESwab, and MSwab collection systems compared to PCR systems were markedly lower at 2.9%, 3.9%, and 1.9%, respectively; this finding of lower detection rates using culture compared to PCR-based methods has been reported in other studies [12, 23]. The admission prevalence reported in our tertiary care trauma center falls within the reported ranges of 3–7% for Europe [11, 14, 24–27]. Within the PCR-based assays, no significant difference in performance was noted when using dry or liquid swab collection despite the fact that these PCR-based tests are validated for ESwab (GeneXpert), MSwab (cobas), or Liquid Stuart swabs (BDMAX). The ability to utilize liquid swab collection devices for high volume screening will allow flexibility with total laboratory automation systems and may help to improve standardization of sample collection and processing.
Our finding that liquid swabs did not yield significantly higher detection rates compared to dry swabs came as a surprise. Manufacturers of PCR-based testing systems have validated liquid swab collection systems (E-or MSwab) to allow for high-throughput testing using both fully automated culture systems (liquid microbiology) or fully automated PCR systems with high accuracies. Dry swabs, which were used routinely at the collection site for processing on the GeneXpert test, had not been validated by any of the PCR manufacturers but come at a significantly lower price. Of interest, two independent studies also reported that the BDMAX MRSA test, in addition to the approved Liquid Stuart swabs, can be performed with other swab types, e.g., dry and ESwab with equal performance [21, 28]. However, the lack of statistically different detection rates between dry and liquid swabs in our study may have been caused by the relatively low overall detection rate for MRSA despite the large number of patients screened as part of routine admission screening.
A critical issue for the choice of testing methods used in MRSA screening is the negative predictive value. The PCR methods used in our study (GeneXpert, cobas 4800, BDMAX) demonstrated very high negative predictive values (>99%). The very few false-negative results could be partly attributed to variant SCCmec types that may not be detected by some PCR assays. While such results can occur with any PCR assay, it is important to note that in the present study, high overlap of MRSA positives between different PCR tests was observed, and high Ct values (suggestive of low bacterial counts) were noted in the majority of specimens with discordant results in PCR tests. The U.S. Department of Veterans Affairs hospitals recently analyzed the negative predictive values of routine MRSA admission and transfer screening as a stewardship tool for de-escalation, as well as avoidance of anti-MRSA therapy in a cohort of almost 250,000 patients over a period of 11 years [29]. The NPV of MRSA nares screening for ruling out MRSA infection was 96.5%, thus very similar to the one reported in the present study, and the MRSA positivity rate was 22.9%.
Positive predictive values as an indicator of specificity ranged from 33–54% compared to direct chromogenic culture and were lowest in liquid swabs, especially in those optimized for molecular testing (MSwab: 27–28%; ESwab 37–42%; dry swabs 41–49%), but did not vary between PCR methods; the calculated positive predictive values were lower than previously described in other studies; possible reasons would be the use of direct culture in the present study vs. direct and enrichment culture in other studies, i.e., those for FDA clearance of assays [12, 30]. Other reasons for this finding could be the low MRSA prevalence and the presence of nonviable organisms (which are detected by PCR but not culture methods), as well as the overall lower sensitivity of the culture method used. Furthermore, the impact of differences in sensitivity/bacterial load is further supported by the Ct values observed in the samples with discordant results between culture and cobas; the majority of Ct values were >34 or near the lower range of detection for this assay. The reduced specificity of PCR assays could theoretically be attributed to detection of mecA dropout strains of SA [13, 23]. However, previous large multi-center evaluations of the specificity of PCR assays for MRSA in nasal swabs demonstrated specificities of ≥95% [12, 13, 23]. In this regard, since no absolute gold standard has been established, these results should be designated as “discordants” rather than “false-positives”.
Diagnostic microbiology laboratories now have several commercial options for implementing rapid, automated, and sensitive high-throughput screening for SA and MRSA using fully automated culture-based and PCR-based technologies in parallel. The German KRINKO guidelines [8] recommend culture-based methods as the basis of MRSA screening. Due to the high negative predictive value and rapid turnaround time, PCR-based methods are recommended as an add-on to culture-based testing; however, rapid PCR test results should be labeled as preliminary until confirmed by culture. The requirement for rapid PCR-based testing plus culture-based confirmation–while only applied in a minority of laboratories–can easily be accomplished by the methods described in this study. Both the instrumentation for PCR testing, as well as the introduction of automated plating instruments (liquid microbiology) allow high-throughput screening and timely reporting of results, so the isolation of suspected MRSA carriers can be limited significantly.
The introduction of screening programs with PCR tests is often inhibited by the high investment and operational cost that nosocomial infections in itself cause the healthcare system due to prolonged duration of hospital stay, additional medical complications, increased morbidity, and therefore higher overall costs. Another key consideration for laboratories is the rate of invalid tests for molecular assays. Particularly in a routine admission screening setting with high-throughput needs, invalid results markedly reduce the efficiency of the testing set-up. In this regard, of the 1680 nasal–throat samples, 1655 (98.5%) yielded valid cobas results and 25 (1.5%) yielded invalid results; for the GenXpert, 9/1680 (0.5%) gave invalid results, while the BDMAX did not yield invalid results (0/1132 (0%)).
More recently, PCR-based MRSA tests with vastly reduced turnaround times and POC usability have been introduced [31]. These systems allow testing in less than 30 min and can be placed in the admission areas (emergency room) and operated by personnel untrained in laboratory techniques. However, these systems–while FDA-cleared based on large multicenter trials with performance equal to laboratory-based PCR systems–have not been tested in interventional settings to demonstrate their overall clinical utility.
Despite the large number of patients consecutively enrolled into our study, the study design has limitations. First, the testing of the 3 different swab systems in separate months rather than simultaneously may have introduced bias to the comparability. However, we are not aware of changes in the admission screening policy or patient population attended at the trauma center. We did not perform further analysis on samples that revealed discordant results between culture and PCR or between different PCR tests.
In summary, liquid and dry swab systems performed similarly in PCR- and culture-based MRSA admission screening. The PCR-based test systems identified 2–3-fold more MRSA isolates compared to chromogenic culture regardless of dry or liquid swab collection devices utilized. Implementation of highly efficient and accurate testing workflows will enable efficient routine screening for MRSA to reduce the rate of MRSA infections. Implementation of highly efficient and accurate testing workflows will enable efficient routine screening for MRSA and SA to reduce the rate of MRSA and SA infections.
Acknowledgements
We greatly appreciate the support of all personnel involved in the sample management and testing.
Footnotes
Abbreviations
PCR - polymerase chain reaction
Funding Sources
This work was supported by Roche Molecular Systems, Pleasanton, USA.
Authors' Contributions
NvA and OL planned the study. NvA, KG, TH, and BH managed the study. KG prepared and tested the sample. NvA collected the data. NvA, TH, BH, OL, MN, and JD analyzed the data. NvA, TH, BH, MW, OL, EMM, MN, and JD interpreted the data. NvA, AK, BH, MW, OL, EMM, MN, and JD prepared the manuscript.
Conflict of Interest
EMM, MN, and JD are current employees of Roche Molecular Systems, and OL was an employee of Roche Molecular Systems.
References
- 1.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, Roosendaal R, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med. 2010;362(1):9–17. Epub 2010/01/08. doi: 10.1056/NEJMoa0808939. PubMed PMID: 20054045. [DOI] [PubMed] [Google Scholar]
- 2.Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001;32(Suppl 2):S114–32. Epub 2001/04/26. doi: 10.1086/320184. PubMed PMID: 11320452. [DOI] [PubMed] [Google Scholar]
- 3.Kock R, Friedrich A, On Behalf Of The Original Author Group C Systematic literature analysis and review of targeted preventive measures to limit healthcare-associated infections by meticillin-resistant Staphylococcus aureus. Euro Surveill. 2014;19(37). Epub 2014/09/27. PubMed PMID: 25259535. [DOI] [PubMed] [Google Scholar]
- 4.Reed SD, Friedman JY, Engemann JJ, Griffiths RI, Anstrom KJ, Kaye KS, et al. Costs and outcomes among hemodialysis-dependent patients with methicillin-resistant or methicillin-susceptible Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol. 2005;26(2):175–83. Epub 2005/03/11. doi: 10.1086/502523. PubMed PMID: 15756889. [DOI] [PubMed] [Google Scholar]
- 5.Muto CA, Jernigan JA, Ostrowsky BE, Richet HM, Jarvis WR, Boyce JM, et al. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect Control Hosp Epidemiol. 2003;24(5):362–86. Epub 2003/06/06. doi: 10.1086/502213. PubMed PMID: 12785411. [DOI] [PubMed] [Google Scholar]
- 6.Huang SS, Singh R, McKinnell JA, Park S, Gombosev A, Eells SJ, et al. Decolonization to Reduce Postdischarge Infection Risk among MRSA Carriers. N Engl J Med. 2019;380(7):638–50. Epub 2019/02/15. doi: 10.1056/NEJMoa1716771. PubMed PMID: 30763195; PubMed Central PMCID: PMCPMC6475519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SS, Yokoe DS, Hinrichsen VL, Spurchise LS, Datta R, Miroshnik I, et al. Impact of routine intensive care unit surveillance cultures and resultant barrier precautions on hospital-wide methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis. 2006;43(8):971–8. Epub 2006/09/20. doi: 10.1086/507636. PubMed PMID: 16983607. [DOI] [PubMed] [Google Scholar]
- 8.Ruscher C. Recommendations for prevention and control of methicillinresistant Staphylococcus aureus (MRSA) in medical and nursing facilities. Bundesgesundheitsblatt, Gesundheitsforschung, Gesundheitsschutz. 2014;57(6):696–732. Epub 2014/07/06. PubMed PMID: 24987771. [PubMed] [Google Scholar]
- 9.Cunningham R, Jenks P, Northwood J, Wallis M, Ferguson S, Hunt S. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J Hosp Infect. 2007;65(1):24–8. Epub 2006/12/06. doi: 10.1016/j.jhin.2006.09.019. PubMed PMID: 17145100. [DOI] [PubMed] [Google Scholar]
- 10.French GL. Methods for screening for methicillin-resistant Staphylococcus aureus carriage. Clin Microbiol Infect. 2009;15(Suppl 7):10–6. Epub 2009/12/03. doi: 10.1111/j.1469-0691.2009.03092.x. PubMed PMID: 19951329. [DOI] [PubMed] [Google Scholar]
- 11.Hardy K, Price C, Szczepura A, Gossain S, Davies R, Stallard N, et al. Reduction in the rate of methicillin-resistant Staphylococcus aureus acquisition in surgical wards by rapid screening for colonization: a prospective, cross-over study. Clin Microbiol Infect. 2010;16(4):333–9. Epub 2009/07/23. doi: 10.1111/j.1469–0691.2009.02899.x. PubMed PMID: 19622077. [DOI] [PubMed] [Google Scholar]
- 12.Peterson LR, Woods CW, Davis TEJr., Wang ZX, Young SA, Osiecki JC, et al. Performance of the cobas MRSA/SA Test for Simultaneous Detection of Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus From Nasal Swabs. Am J Clin Pathol. 2017;148(2):119–27. Epub 2017/09/14. doi: 10.1093/ajcp/aqx040. PubMed PMID: 28898981. [DOI] [PubMed] [Google Scholar]
- 13.Mehta SR, Estrada J, Ybarra J, Fierer J. Comparison of the BD MAX MRSA XT to the Cepheid Xpert(R) MRSA assay for the molecular detection of methicillin-resistant Staphylococcus aureus from nasal swabs. Diagn Microbiol Infect Dis. 2017;87(4):308–10. Epub 2017/01/18. doi: 10.1016/j.diagmicrobio.2016.12.011. PubMed PMID: 28094151. [DOI] [PubMed] [Google Scholar]
- 14.Bulliard E, Grandbastien B, Senn L, Greub G, Blanc DS, et al. Evaluation of three consecutive versions of a commercial rapid PCR test to screen for methicillin-resistant Staphylococcus aureus. Clin Microbiol infect. 2019. Epub 2019/04/14. doi: 10.1016/j.cmi.2019.03.029. PubMed PMID: 30980926. [DOI] [PubMed] [Google Scholar]
- 15.Patel PA, Robicsek A, Grayes A, Schora DM, Peterson KE, Wright MO, et al. Evaluation of multiple real-time PCR tests on nasal samples in a large MRSA surveillance program. Am J Clin Pathol. 2015;143(5):652–8. Epub 2015/04/16. doi: 10.1309/ajcpmdy32ztdxpfc. PubMed PMID: 25873498. [DOI] [PubMed] [Google Scholar]
- 16.Roisin S, Laurent C, Nonhoff C, Deplano A, Hallin M, Byl B, et al. Positive predictive value of the Xpert MRSA assay diagnostic for universal patient screening at hospital admission: influence of the local ecology. Eur J Clin Microbiol Infect Dis. 2012;31(5):873–80. Epub 2011/08/30. doi: 10.1007/s10096-011-1387-7. PubMed PMID: 21874398. [DOI] [PubMed] [Google Scholar]
- 17.De Silva S, Wood G, Quek T, Parrott C, Bennett CM. Comparison of Flocked and Rayon Swabs for Detection of Nasal Carriage of Staphylococcus aureus among Pathology Staff Members. J Clin Microbiol. 2010;48(8):2963–4 doi: 10.1128/Jcm.01617-09. PubMed PMID: WOS:000280550500050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silbert S, Kubasek C, Uy D, Widen R. Comparison of ESwab with Traditional Swabs for Detection of Methicillin-Resistant Staphylococcus aureus Using Two Different Walk-Away Commercial Real-Time PCR Methods. J Clin Microbiol. 2014;52(7):2641–3. doi: 10.1128/Jcm.00315-14. PubMed PMID: WOS:000339279700052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhoeven P, Grattard F, Carricajo A, Pozzetto B, Berthelot P. Better detection of Staphylococcus aureus nasal carriage by use of nylon flocked swabs. J Clin Microbiol. 2010;48(11):4242–4. Epub 2010/09/17. doi: 10.1128/jcm.01425-10. PubMed PMID: 20844232; PubMed Central PMCID: PMCPMC3020835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Silva S, Wood G, Quek T, Parrott C, Bennett CM. Comparison of flocked and rayon swabs for detection of nasal carriage of Staphylococcus aureus among pathology staff members. J Clin Microbiol. 2010;48(8):2963–4. Epub 2010/05/28. doi: 10.1128/jcm.01617-09. PubMed PMID: 20504992; PubMed Central PMCID: PMCPMC2916558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silbert S, Kubasek C, Uy D, Widen R. Comparison of ESwab with traditional swabs for detection of methicillin-resistant Staphylococcus aureus using two different walk-away commercial real-time PCR methods. J Clin Microbiol. 2014;52(7):2641–3. Epub 2014/04/25. doi: 10.1128/jcm.00315-14. PubMed PMID: 24759722; PubMed Central PMCID: PMCPMC4097676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen XC, Madsen TV, Engberg J. Evaluation of Xpert MRSA Gen 3 and BD MAX MRSA XT for meticillin-resistant Staphylococcusaureus screening in a routine diagnostic setting in a low-prevalence area. J Med Microbiol. 2017;66(1):90–5. Epub 2016/12/21. doi: 10.1099/jmm.0.000411. PubMed PMID: 27995869. [DOI] [PubMed] [Google Scholar]
- 23.Snyder JW, Munier GK, Johnson CL. Comparison of the BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR assay to culture by use of BBL CHROMagar MRSA for detection of MRSA in nasal surveillance cultures from intensive care unit patients. J Clin Microbiol. 2010;48(4):1305–9. Epub 2010/02/26. doi: 10.1128/jcm.01326-09. PubMed PMID: 20181916; PubMed Central PMCID: PMCPMC2849557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harbarth S, Fankhauser C, Schrenzel J, Christenson J, Gervaz P, Bandiera-Clerc C, et al. Universal screening for methicillin-resistant Staphylococcus aureus at hospital admission and nosocomial infection in surgical patients. JAMA. 2008;299(10):1149–57. Epub 2008/03/13. doi: 10.1001/jama.299.10.1149. PubMed PMID: 18334690. [DOI] [PubMed] [Google Scholar]
- 25.Jeyaratnam D, Whitty CJ, Phillips K, Liu D, Orezzi C, Ajoku U, et al. Impact of rapid screening tests on acquisition of meticillin resistant Staphylococcus aureus: cluster randomised crossover trial. Br Med J. 2008;336(7650):927–30. Epub 2008/04/18. doi: 10.1136/bmj.39525.579063. BE. PubMed PMID: 18417521; PubMed Central PMCID: PMCPMC2335244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gopal Rao G, Michalczyk P, Nayeem N, Walker G, Wigmore L. Prevalence and risk factors for meticillin-resistant Staphylococcus aureus in adult emergency admissions–a case for screening all patients? J Hosp Infect. 2007;66(1):15–21. Epub 2007/03/23. doi: 10.1016/j.jhin.2007.01.013. PubMed PMID: 17376560. [DOI] [PubMed] [Google Scholar]
- 27.Rioux C, Armand-Lefevre L, Guerinot W, Andremont A, Lucet JC. Acquisition of methicillin-resistant Staphylococcus aureus in the acute care setting: incidence and risk factors. Infect Control Hosp Epidemiol. 2007;28(6):733–6. Epub 2007/05/24. doi: 10.1086/516664. PubMed PMID: 17520551. [DOI] [PubMed] [Google Scholar]
- 28.Dalpke AH, Hofko M, Stock C, Zimmermann S. Evaluation of the BD Max MRSA XT assay for use with different swab types. J Clin Microbiol. 2014;52(12):4343–6. Epub 2014/09/19. doi: 10.1128/jcm.02306-14. PubMed PMID: 25232162; PubMed Central PMCID: PMCPMC4313310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mergenhagen KA, Starr KE, Wattengel BA, Lesse AJ, Sumon Z, Sellick JA. Determining the utility of methicillin-resistant Staphylococcus aureus nares screening in antimicrobial stewardship. Clin Infect Dis. 2019. Epub 2019/10/02. doi: 10.1093/cid/ciz974. PubMed PMID: 31573026. [DOI] [PubMed] [Google Scholar]
- 30.Peterson LR, Schora DM. Methicillin-Resistant Staphylococcus aureus Control in the 21st Century: Laboratory Involvement Affecting Disease Impact and Economic Benefit from Large Population Studies. J Clin Microbiol. 2016;54(11):2647–54. Epub 2016/06/17. doi: 10.1128/jcm.00698-16. PubMed PMID: 27307459; PubMed Central PMCID: PMCPMC5078538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg SK, Lu K, Duncan J, Peterson LR, Liesenfeld O. Equivalent Performance of the Cobas((R)) Cdiff Test for Use on the Cobas((R)) Liat((R)) System and the Cobas((R)) 4800 System. Eur J Microbiol Immunol. 2017;7(4):310–8. Epub 2018/02/07. doi: 10.1556/1886.2017.00034. PubMed PMID: 29403660; PubMed Central PMCID: PMCPMC5793701. [DOI] [PMC free article] [PubMed] [Google Scholar]


