Genome-wide association studies were used to analyse potassium use efficiency in rice. Novel associations were found along with a role for sodium replacement via the OsHKT2;1 sodium transporter.
Keywords: Fertilizer use, GWAS, HKT2;1, potassium, potassium use efficiency, rice, sustainable agriculture, sodium
Abstract
Increasing the potassium use efficiency (KUE) of crops is important for agricultural sustainability. However, a greater understanding of this complex trait is required to develop new, high-KUE cultivars. To this end, a genome-wide association study (GWAS) was applied to diverse rice (Oryza sativa L.) genotypes grown under potassium-stressed and -replete conditions. Using high-stringency criteria, the genetic architecture of KUE was uncovered, together with the breadth of physiological responses to low-potassium stress. Specifically, three quantitative trait loci (QTLs) were identified, which contained >90 candidate genes. Of these, the sodium transporter gene OsHKT2;1 emerged as a key factor that impacts on KUE based on (i) the correlation between shoot Na+ and KUE, and (ii) higher levels of HKT2;1 expression in high-KUE lines.
Introduction
Potassium (K+) is the most abundant cation in most plants. It is an essential cofactor for many enzymes and has a dominant role in turgor provision and water homeostasis (Maathuis, 2009). The large amounts of K+ that are required by plants is typically sustained by application of K+ fertilizer in agronomic contexts. Global demand for K+ fertilizers is currently >30 Mt annually and steadily increasing (FAO, 2017). Though there are ample K+ reserves, production and application of K+ fertilizer has important environmental influences: potash fertilizers contribute to agricultural energy use and greenhouse gas emissions (Brentup and Pallière, 2008; Camargo et al., 2013). In 2016, >95% of potash was produced in the northern hemisphere (USGS, 2017), exacerbating deleterious environmental consequences through transportation-related emissions. Agriculture is also implicated in adding to atmospheric K+ deposition (Allen et al., 2010). Taken together, judicious use of potash fertilizers clearly forms an important part of future sustainable agriculture.
At the same time, deficiency for K+ in agricultural soils is widespread, and rapidly increasing in areas such as the Australian wheat belt and Chinese rice paddies (Römheld and Kirkby, 2010). Underfertilization sometimes results from agricultural malpractice, but is more commonly due to economic considerations, with the cost of K+ fertilizer purchase and application proving insurmountable. A sustainable solution to mitigate the economic and environmental consequences of growing K+ demand, while meeting food demand, is to develop crops with higher K+ use efficiency (KUE).
In order to increase crop KUE, knowledge of its genetic underpinnings is important to inform targeted improvement. Studies have been conducted with a range of species and have led to the identification of quantitative trait loci (QTLs) associated with plant responses to K+ deficiency (e.g. Wu et al., 1998; Prinzenberg et al., 2010; Kong et al., 2013; Zhao et al., 2014). Similarly, transcriptomics studies (e.g. Armengaud et al., 2004; Wang et al., 2012) in low-K+ conditions point to genes that encode membrane proteins involved in transport and other proteins for transcriptional regulation. Genes for such proteins can therefore be seen as putative targets for crop improvements (Shin, 2014; Wang and Wu, 2015), but a more complete understanding of the genetic underpinnings of KUE is still required.
In rice, QTLs for several traits, including K+ uptake and tissue K+ concentration in salt- and non-stressed plants, have been reported (Koyama et al., 2001; Lin et al., 2004; Garcia-Oliveira et al., 2009). Furthermore, QTLs in the context of K+ deficiency have been published (Wu et al., 1998; Miyamoto et al., 2012; Fang et al., 2015), although little overlap in the identified regions was apparent. However, both Miyamoto et al. (2012) and Fang et al. (2015) described associations in a large (~7 Mb) QTL on chromosome 6 that were linked with shoot sodium (Na+), K+, and calcium concentrations.
The detection of QTLs and genes related to agriculturally important traits in rice has been aided in recent years by genome-wide association studies (GWAS) which typically yield much higher resolution than conventional QTL mapping approaches. Studies have examined abiotic stresses such as aluminium (Famoso et al., 2011) and salt (Kumar et al., 2015; Campbell et al., 2017; Patishtan et al., 2018), and were able to detect novel loci as well as gene candidates. However, the response of rice to K+ deficiency has yet to be examined using GWAS. In this study, the genetic architecture of low K+ stress was explored using the Rice Diversity Panel 1 (Zhao et al., 2011; Eizenga et al., 2014) and, in doing so, novel QTLs were detected as well as some which co-localized with those in the previous literature. From this, putative targets for crop improvement were proposed.
Materials and methods
Plant growth and germplasm
Five seeds from each of 324 rice (Oryza sativa) cultivars (see Supplementary Table S1 at JXB online for a full list of accessions) were germinated in sand flooded with distilled water for 2 weeks prior to transfer to hydroponic treatments. Seedlings were placed in 9 litre boxes which contained a nutrient solution adapted from Yoshida et al. (1976) consisting of: (in mM) 1.4 NH4NO3, 0.3 NaH2PO4, 1 CaCl2, 1.6 MgSO4·7H2O, and 0.2 Na2O3Si; and (in µM) 9.5 MnCl2, 0.07 (NH4)6Mo7O24, 18 H3BO3, 0.15 ZnSO4, 0.16 CuSO4, 71 citric acid monohydrate. K+ was added as KCl to a final concentration of 0.1 (low-K+ or LK treatment) or 1 mM (high-K+ or HK treatment). Nutrient solutions were changed weekly. One seedling from each cultivar was placed in each treatment, and growth trials were replicated five times. Plant were grown in a glasshouse for 4 weeks (or as indicated in the text) with 12 h day and night periods with temperatures of 32 °C and 28 °C in the day and night, respectively. The relative humidity was maintained between 50% and 60%. For detailed growth experiments on IR64, plants were grown as described above in the presence of 0.01, 0.1, 0.5, 1, or 5 mM K+ (added as KCl) and a total amount of 3 mM Na+.
Tissue cation analysis
Sampled plants were separated into roots and shoots, and their fresh weights were recorded before being oven dried at 80 °C for 3 d. Tissues were then re-weighed before K+ and Na+ concentrations were determined after extraction in 20 mM CaCl2 for 24 h. Cation concentrations were measured using a flame photometer (Sherwood Scientific, Cambridge, Cambridgeshire, UK).
Trait measurement
Briefly, each rice genotype was grown in K+-deficient (0.1 mM) and -replete (1 mM) nutrient solutions (see above). Relative growth rate (RGR) was calculated as [ln(FWend)−ln(FWstart)]/(tend−tstart), where FW is the whole-plant fresh weight. K+ and Na+ tissue concentrations were measured as described above. Phenotype data (Supplementary Table S3) were based on five biological replicates, and least squares means were calculated from raw data. Cultivars with fewer than three replicates were excluded from the analysis. Two different KUE metrics were used: KUE-RGR (defined as the percentage reduction in RGR between LK and HK conditions) and KUE-K (defined as RGR at LK treatment divided by shoot K+ concentration at LK treatment). The latter trait examines the K+ utilization, while KUE-RGR can be influenced by both the uptake and utilization of K+.
Genome-wide association studies
GWAS were carried out using R 3.3.3 and the GenABEL R package (Aulchenko et al., 2007) for KUE metrics, RGR, and tissue cation concentrations. Single nucleotide polymorphisms (SNPs) with a minor allele frequency <0.05 and a call rate <0.9 were excluded from analyses to minimize the risk of spurious associations. Mixed linear models were used for analyses to control for the population structure present in rice (Zhao et al., 2011) which can also induce spurious associations between traits and genetic loci. The top three principal components for population structure were included as fixed effects if this resulted in a model with a genomic inflation factor (Devlin and Roeder, 1999) nearer unity. Previous work has found the use of mixed models with principal components as covariates to be successful in limiting the occurrence of false signals (Zhao et al., 2011; Kumar et al., 2015; Patishtan et al., 2018). Associations between SNPs and genotypes were declared significant if their P-value was <1×10–5 (Crowell et al., 2016) and the false discovery rate (Benjamini and Hochberg, 1995) was <10%.
Identification of quantitative trait loci and candidate genes
A minimum of two significant associations within a 200 kbp window was required for a significant association to be considered as a QTL to minimize the risk of false positives. This genomic region window size was chosen because linkage disequilibrium in rice declines rapidly over this distance (Zhao et al., 2011; McCouch et al., 2016) and genes that are proximal to associations can be considered more credible candidates for influencing the trait in question. QTLs which overlapped were grouped into a single QTL. Genes within QTLs were sourced from those found using the the Rice Genome Annotation Project website (http://rice.plantbiology.msu.edu). Candidate genes were found among these genes, with those with products relating to transport, signalling, and transcription considered to be more credible candidates. Co-localization of significantly associated SNPs and genes within QTLs was examined using the Rice Diversity Allele Finder (http://rs-bt-mccouch4.biotech.cornell.edu/AF/). Such co-localization with a gene could indicate relevance to the trait, and non-synonymous SNPs could lead to changes that ultimately alter KUE.
HKT2;1 expression analysis
Seeds for the following cultivars germinated (where ‘L’ indicates low KUE and ‘H’ indicates high KUE): Cybonnet (L), Dom Sufid (L), Edith (L), Padi Kasalle (L), Tox 782-20-1 (L), 116 (H), Sathi (H), Saturn (H), Ghati Kamma Nangarhar (H), and Wanica (H). Plants were grown a described above on an adapted Yoshida nutrient solution containing (in mM) 2.9 NH4NO3, 0.3 H3PO4, 0.01 KCl, 1 CaCl2·2H2O, and 1.6 MgSO4 (with micronutrients as described above). Medium was adjusted to pH 5.6 using methyl glucamine and supplemented with either 0 mM or 1 mM NaCl. Plants were grown for 4 weeks, after which roots from the three plants of each cultivar were pooled and frozen in liquid nitrogen. The root samples were ground to a powder in liquid nitrogen and total RNA was extracted using a Nucleospin RNA Plant and Fungi kit (Macherey-Nagel Bioanalysis). cDNA was synthesized using a Superscript II reverse transcriptase kit (Invitrogen) with oligo(dT) primers. Quantitative PCRs (qPCRs) were performed using the QuantStudio 3 (Thermo Fisher) system and Fast SYBR green master mix (Thermo Fisher) with 5ʹCTCCATCGACTGCTCACTCA3ʹ and 5ʹGGACAGTGCAAATGTTGTCG3ʹ as forward and reverse HKT2;1-specific primers. The expression of elongation factor 1α was used as an internal control with 5ʹCACATTGCCGTCAAGTTTGC3ʹ and 5ʹCCATACCAGCATCACCGTTC3ʹ forward and revers primers, respectively. Data are presented as the average of three biological replications.
Results and discussion
Influence of potassium stress on growth and tissue cation concentrations
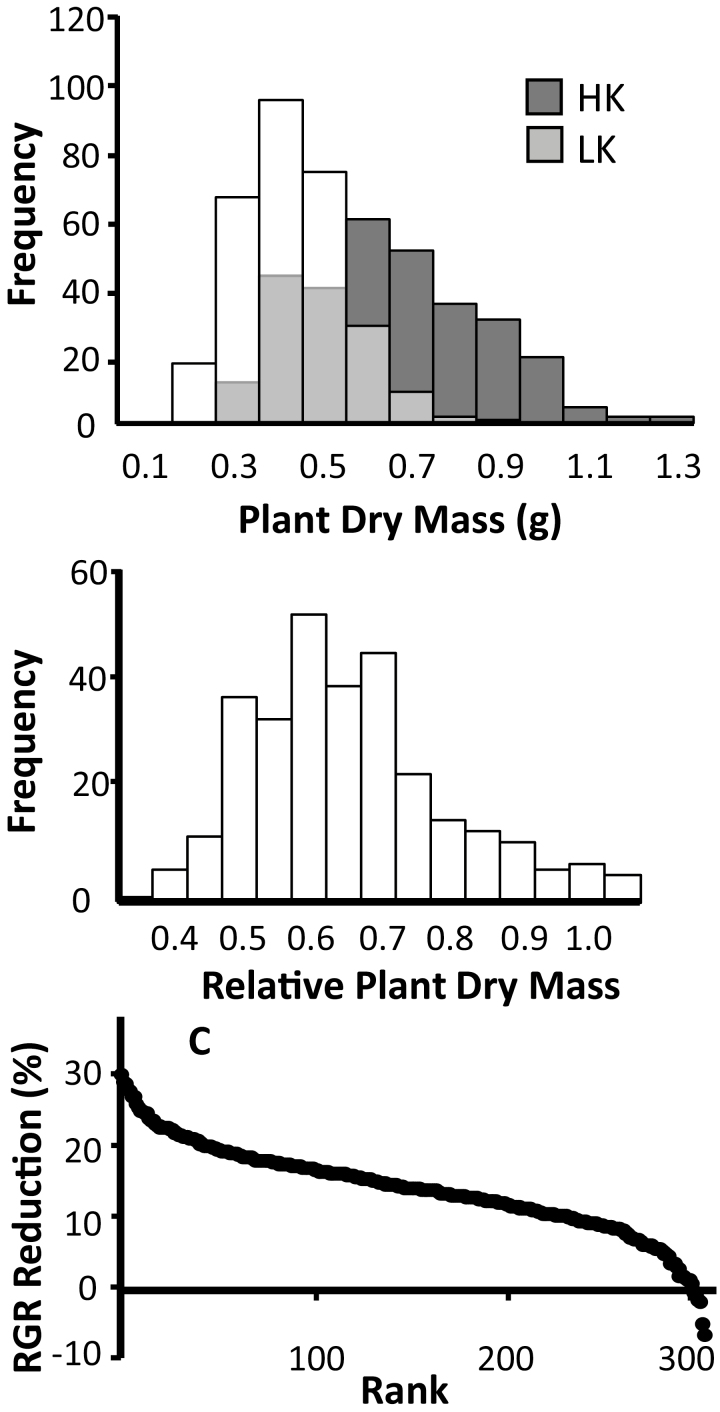
Lowering the medium K+ concentration from 1 mM (HK) to 0.1 (LK) mM had a substantial effect on growth and tissue cation levels. Figure 1a shows that the mean final mass of LK plants was ~40% of that achieved by HK plants. However, at the tissue level, plant growth was not affected uniformly. For example, root to shoot mass ratio was significantly higher in the LK treatment compared with the HK treatment (data not shown). Furthermore, Fig. 1b shows that rice cultivars vary greatly in their growth response to LK. The RGR reduction ranged from 30% to –5% when comparing LK and HK growth data. In other words, the RGR of some lines declines by nearly a third between LK and HK conditions, while others were not at all or only little affected, irrespective of a 10-fold change in medium K+ concentration.
Fig. 1.
Responses of rice genotypes to potassium stress. (a) Mean plant dry mass of cultivars when grown in the presence of 0.1 mM (LK) and 1 (HK) mM potassium. (b) Relative plant dry mass (dry mass LK/dry mass HK). (c) Reduction in relative growth rate (RGR) in LK compared with HK conditions.
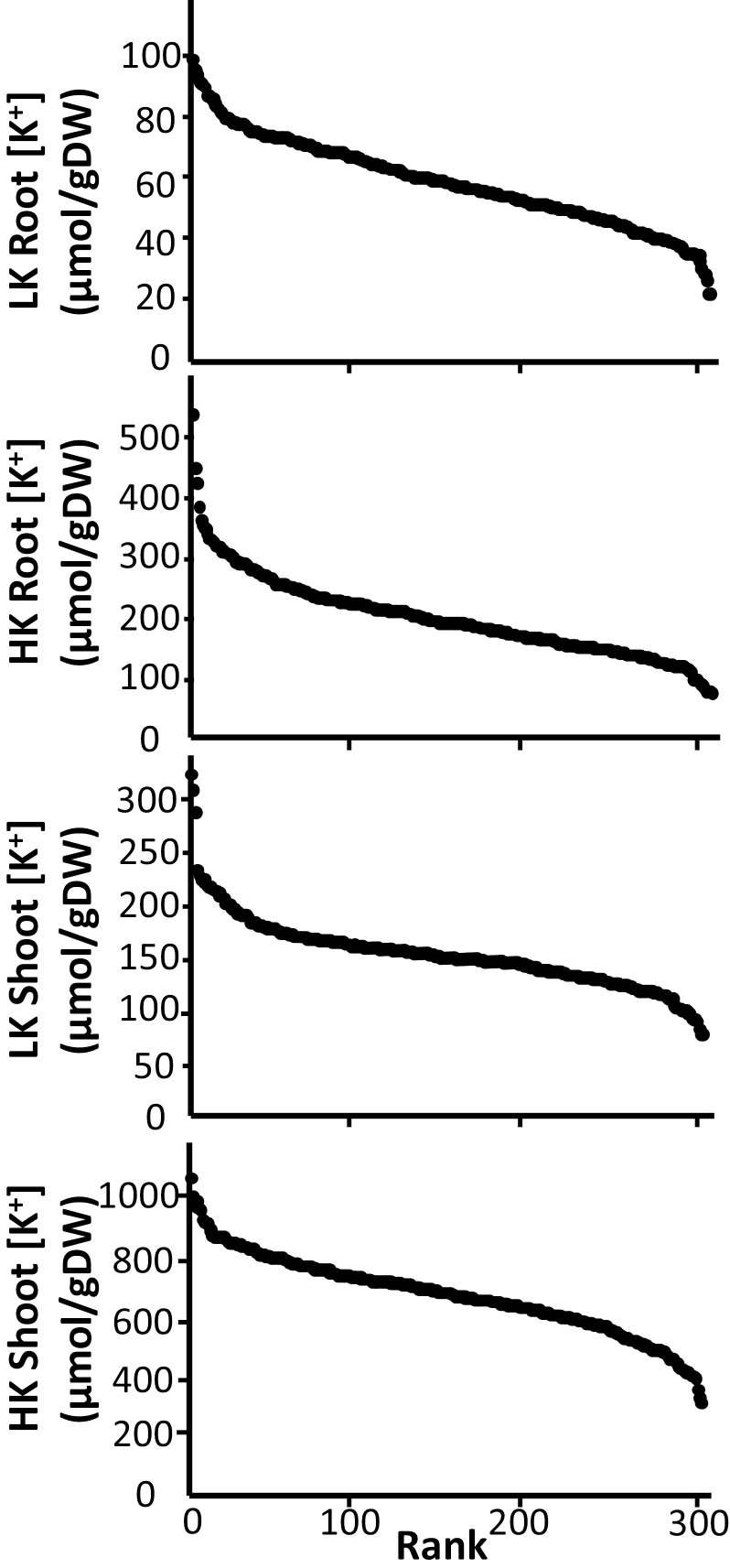
As expected, both root and shoot K+ concentrations were lower in the LK treatment. Across the cultivars, the average shoot K+ concentration declined from 686 μmol gDW–1 to 154 μmol gDW–1 between the HK and LK conditions, while the root concentrations declined from 198 μmol gDW–1 to 59 μmol gDW–1 (Fig. 2). Shoot K+ concentrations were consistently greater than those of roots. In combination, the growth and tissue K+ data show that the LK conditions were effective in causing stress which reduced rice growth, probably arising from insufficient tissue K+ levels. Indeed, many previous studies have shown a strong link between tissue K+ and growth across several plant species (e.g. Asher and Ozanne, 1967; Fageria, 1976; Spear et al., 1978).
Fig. 2.
Distribution of root (top two panels) and shoot (bottom two panels) K+ concentration across the diversity panel for plants grown on LK (0.1 mM) and HK (1 mM) K+ medium.
While low tissue K+ is strongly linked with reduced RGR between treatments, the association is less clear within a specific treatment: in both LK and HK treatments, only weak non-significant correlations were derived between tissue K+ and growth. Such seemingly contradictory outcomes can be explained by the existence of considerable (genetic) variation in the sensitivity of cultivars when exposed to declining levels of tissue K+.
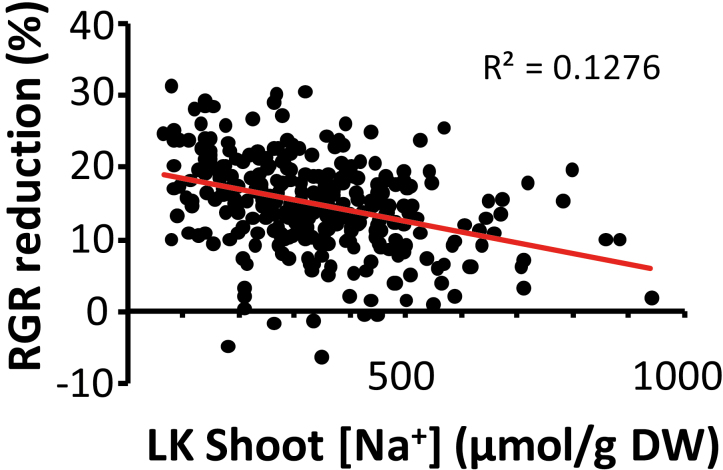
Table 1 shows growth and tissue cation data for the 10 highest and lowest ranking rice cultivars for KUE. KUE-RGR is a measure for the relative growth reduction when changing from HK to LK conditions (RGR_LK/RGR_HK) and differed significantly between cultivars (one-way ANOVA, P<0.01). KUE-K denotes the utilization of K+ (amount of growth per unit K+; RGR_LK/shoot K_LK), and this too varied significantly between cultivars (one-way ANOVA, P<0.001) with a 5- to 6-fold difference between the lowest and highest values (Supplementary Table S4). Interestingly, KUE_RGR and LK shoot [Na+] showed a highly significant negative correlation (r= –0.385, P<0.001; Fig. 3) and similar, but weaker, negative correlations were found between KUE-RGR and HK shoot [Na+], LK root [Na+], and HK root [Na+], respectively (Supplementary Fig. S1). Such evidence points to a potential beneficial effect of Na+ in rice shoots when K+ is limiting, and this may be the result of replacement of K+ by Na+. However, in contrast to KUE-RGR, KUE-K did not correlate significantly with either root or shoot levels of Na+. Indeed, very little overlap between the KUE-K and KUE-RGR was apparent, with only two cultivars (GSOR 117 and 142) emerging as high-KUE lines irrespective of the KUE definition (see Supplementary Table S4). The lack of similarity between KUE-RGR and KUE-K emphasizes the different phenomena these metrics describe: while KUE-K is determined by high growth rates and low shoot [K+] (e.g. ~90 mM and ~250 mM in high- and low-KUE-K lines, respectively, see Table 1), KUE-RGR expresses how well growth is maintained by cultivars in the face of a shortage of K+. Though both approaches are valuable in an agronomic context, one may be more suitable for optimizing local requirements such as soil nutrient status or availability of K fertilizer. The wide variability in either parameter suggests that there is a large scope to enhance these traits.
Table 1.
Growth and tissue cation concentrations for high- and low-KUE accessions
| KUE_K | KUE_RGR | ||||
|---|---|---|---|---|---|
| Low KUE | High KUE | Low KUE | High KUE | ||
| RGR | 0.088 | 0.11 | RGR | ND | ND |
| DW HK (g) | 0.45 | 0.79 | DW HK (g) | 0.72 | 0.42 |
| DW LK (g) | 0.26 | 0.47 | DW LK (g) | 0.29 | 0.34 |
| ShootK HK (mM) | 656 | 646 | ShootK HK (mM) | 713 | 626 |
| ShootK LK (mM) | 244 | 86 | ShootK LK (mM) | 136 | 135 |
| ShootNa HK (mM) | 47 | 40 | ShootNa HK (mM) | 39 | 66 |
| ShootNa LK (mM) | 352 | 232 | ShootNa LK (mM) | 197 | 369 |
| RootK HK (mM) | 253 | 184 | RootK HK (mM) | 236 | 170 |
| RootK LK (mM) | 57 | 52 | RootK LK (mM) | 53 | 59 |
| RootNa HK (mM) | 92 | 67 | RootNa HK (mM) | 79 | 91 |
| RootNa LK (mM) | 104 | 148 | RootNa LK (mM) | 140 | 200 |
| ShootK:Na ratio (HK) | 14 | 16.2 | ShootK:Na ratio (HK) | 18.3 | 9.5 |
| ShootK:Na ratio (LK) | 0.69 | 0.37 | ShootK:Na ratio (LK) | 0.69 | 0.37 |
Fig. 3.
Significant (P<0.05) correlation between RGR reduction and shoot tissue Na+ concentration of plants grown on LK medium.
Genome-wide association studies of potassium stress
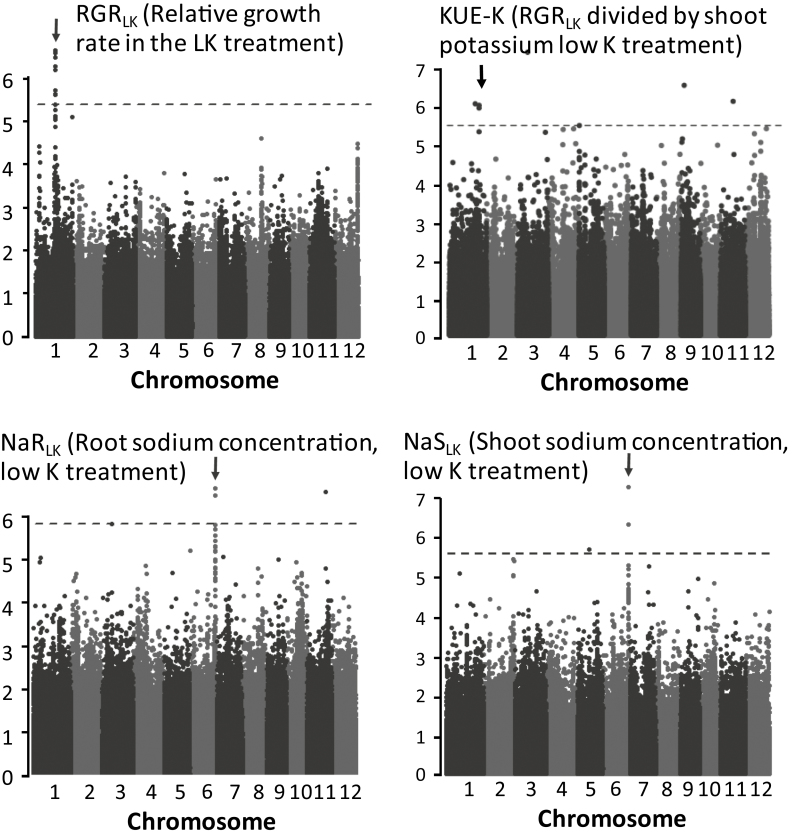
In order to better understand which mechanisms contribute to KUE, GWAS was applied to the growth, cation, and KUE data (Supplementary Table S2). Based on the stringency criteria outlined in the Materials and methods, a total of four association signals were detected; one each for KUE-K (defined as RGR/shoot K), RGR at LK treatment, shoot [Na+], and root [Na+] at LK treatment (Fig. 4; Table 2). Furthermore, the two Na+-related signals co-localized at a position ~29.5 Mbp along chromosome 6 and had the same significantly associated SNPs.
Fig. 4.
Manhattan plots for traits (RGR at LK, KUE-K, root [Na+] at LK, and shoot [Na+] at LK) that generated significant association signals (arrows) using criteria as explained in the Materials and methods. Note that ‘shoot Na’ and ‘root Na’ trait data associate with the same locus on chromosome 6.
Table 2.
Summary of quantitative trait loci identified in GWAS
| Trait | Description | Chr | Position | Significant SNP positions |
|---|---|---|---|---|
| RGR LK | Relative growth rate at low K treatment | 1 | 22,260,180–22,463,799 | 22,360,180; 22,361,410; 22,361,482; 22,363,799 |
| RGR_K | K use efficiency defined as RGR/shoot K concentration at LK treatment | 1 | 34,344,598–34,563,159 | 34,444,598; 34,463,159 |
| NaR_LK | Root Na concentration at low K treatment | 6 | 29,440,164–29,640,591 | 29,540,164; 29,540,591 |
| NaS_LK | Shoot Na concentration at low K treatment | 6 | 29,440,164–29,640,591 | 29,540,164; 29,540,591 |
The three independent QTLs subsumed a total of 86 unique genes (Supplementary Table S5) and 8 significantly associated SNPs (Table 2). Interrogation of the Rice Diversity Allele Finder (http://rs-bt-mccouch4.biotech.cornell.edu/AF/) showed that the two SNPs belonging to the RGR_LK association were synonymous and were located in the coding region of a putative retrotransposon protein (LOC_Os01g39640). One of the KUE-K associations was a synonymous SNP in the intron of another putative retrotransposon protein (LOC_Os01g59580), and both SNPs in the Na+-related signal were synonymous and located in the coding region of the gene for OsHKT2;1 (LOC_Os06g48810), an Na+ transporter.
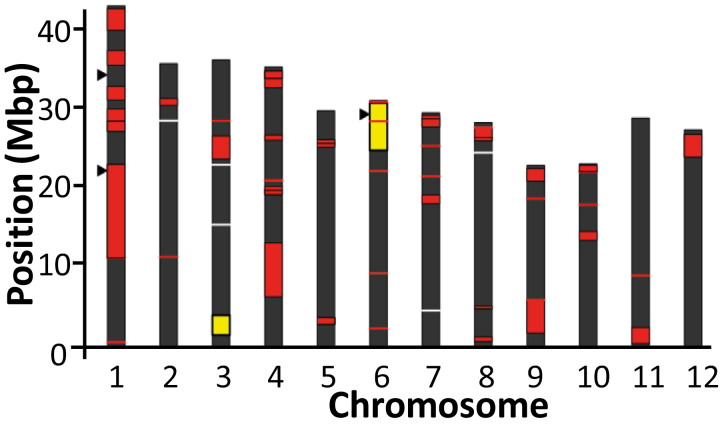
QTLs repeatedly found across different studies can help to identify robust candidates for crop improvement. The positions of QTLs identified in this study were therefore compared against those previously reported (Fig. 5). Though it is noted that many previous studies had relatively low resolution, leading to QTLs that span many megabase pairs (e.g. Fang et al., 2015), an overlap was found for the chromosome 1 RGR-K signal which is positioned at the beginning of an ~10 Mbp QTL described by Fang et al. (2015). The tissue Na+-associated signals on chromosome 6 found in this study were previously described by Miyamoto et al. (2012) who identified a 6.4 Mbp region on chromosome 6 related to Na+ uptake and, using a map-based cloning strategy, isolated a 100 kb chromosomal region that contained HKT2;1.
Fig. 5.
Co-incidence of previously described QTLs and loci identified in this study related to low K+ growth in the rice genome. Each bar represents a chromosome, and previously reported QTLs are marked in white (Wu et al., 1998), yellow (Miyamoto et al., 2012), or red (Fang et al., 2015). Triangles indicate the position of QTLs derived from this study.
Putative drivers of KUE
Out of the 86 genes covered by the significant association signals, the 42 annotated genes were further evaluated to identify potential drivers of KUE. Gene Ontology analysis is problematic with a sample of this size and it is therefore not surprising that no enriched functional class was discovered. In addition to HKT2;1, three further genes (OsCML1, calmodulin-related calcium sensor protein; OsSub52, putative subtilisin homologue; and OsHKT2;4, Na+ transporter) were previously shown to respond transcriptionally to low-K+ conditions (Shankar et al., 2013), suggesting that they may play a role in K+ homeostasis. Furthermore, on the basis of functional annotations, the list contains a large proportion (>10%) of genes that are involved in ‘disease resistance’ (n=7) and in ‘RNA translation’ (n=5), pointing to a potential role for these processes in establishing KUE. There is a well-documented link between K+ deficiency and disease (e.g. Davis et al., 2018); rice diseases such as brown leaf spot, scab, and stem rot are generally not problematic in K+-replete fields, but can easily overwhelm K+-deficient rice. It is not directly obvious how disease impacts on KUE, but LK treatment could (transcriptionally or otherwise) prime plants and thus make them more disease resilient. Improved resilience could alter KUE via generic growth effects. Ribosomal functioning is frequently mentioned as an example process that requires high levels (>100 mM) of K+ (e.g. Maathuis, 2009). Similar to disease resistance, the link between RNA translation and KUE may be convoluted, but more efficient ribosomal constituents, and enzymes involved in translation could improve growth and/or allow plants to synthesize proteins adequately at lower cytoplasmic K+ levels. In contrast to the above, the connection between Na+ and K+ (and hence between Na+ and KUE) is well established (e.g. Maathuis and Amtmann, 1999). Thus the appearance of two putative Na+ transporters, in combination with significant signals in the root Na+ and shoot Na+ traits, strongly suggest that Na+ transport is an important contributing factor in KUE.
HKT2;1 plays a role in KUE via shoot sodium
The cation transport category contains two ‘high-affinity K transporters’. HKT2;1 and HKT2;4 are part of significant association signals when either root or shoot Na+ concentration was used as the trait. HKT2;4 (Os06g48800) is located in the plasma membrane and expressed in the peripheral layers of rice roots and in the shoot vasculature (Sassi et al., 2012). Members of subgroup II HKTs typically perform K:Na co-transport, but in heterologous systems HKT2;4 was shown to move K+ without the need for Na+ (Horie et al., 2011). Thus, HKT2;4 could be involved in K+ (re)distribution, for example between root and shoot. However, its loss of function did not generate a K+-dependent phenotype, though this could be due to functional redundancy with, for example, the very similar HKT2;3 (Horie et al., 2011).
In contrast to HKT2;4, HKT2;1 strongly discriminates against K+ and, in a physiological context, is believed to function exclusively as an Na+ transporter (Horie et al., 2007; Miyamoto et al., 2012). This would fit in with the observation that HKT2;1 is associated with tissue Na+ phenotypes (Supplementary Table S5). Earlier work by Horie et al. (2007) showed that HKT2;1 is mostly expressed in rice roots and that expression is induced during low-K+ conditions. Furthermore, HKT2;1 was previously identified in a QTL associated with high Na+ accumulation in K-deficient rice plants (Miyamoto et al., 2012). Thus, HKT2;1 has been identified in multiple QTL studies and is transcriptionally regulated in a K+-dependent manner. It therefore forms a high confidence candidate that impacts on KUE via the replacement of non-essential K+ by the physicochemically similar monovalent Na+.
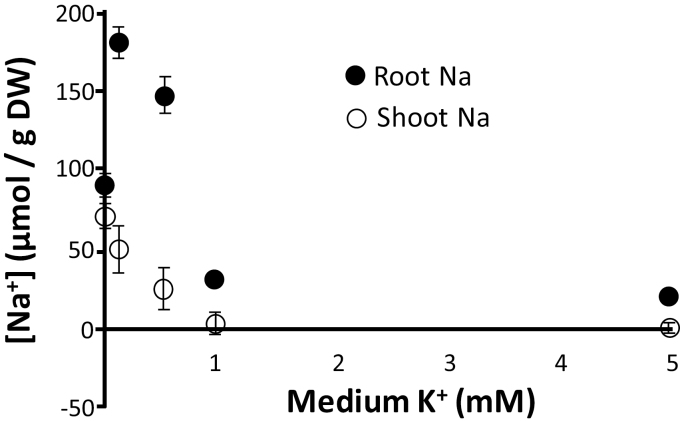
Na+ behaves as a beneficial nutrient for K+-starved glycophytes when present at moderate concentration (e.g. Maathuis, 2013). Substitution of K+ by Na+ in such conditions could make a valuable contribution to maintaining non-critical functions of K+, such as turgor generation, and thus contribute to KUE. Detailed growth experiments with one of the cultivars (IR64) show that there is a clear negative correlation between external K+ levels and tissue Na+, for both roots and shoots (Fig. 6). In addition, our physiological data suggest that raised root and shoot Na+ has a positive effect on KUE: Fig. 3 shows that both root and shoot levels of Na+ negatively correlate with KUE-RGR but that this is clearly more significant for shoot Na+ in the LK treatment. This phenomenon also becomes clear when overall tissue cation composition is compared between high- and low-KUE lines (Table 1). In HK conditions, shoot K+ (~650 μmol gDW–1) and shoot Na+ (~50 μmol gDW–1) generate a K:Na ratio of ~10–18, and is similar for high- and low-KUE accessions (Table 1), using either KUE definition. However, LK treatment causes a dramatic change in the K:Na ratio to less than one of ~ 0.7 and 0.3 in low- and high-KUE lines, respectively, reflecting the greater capacity of high-KUE cultivars to exploit Na+ as a K+ replacement.
Fig. 6.
Reducing levels of medium K+ drastically increases Na+ concentrations in both roots and shoots of rice cultivar IR64. Plants were grown hydroponically for 7 weeks in the presence of varying K+ levels and 3 mM NaCl. Error bars show the SD of three biological replicates.
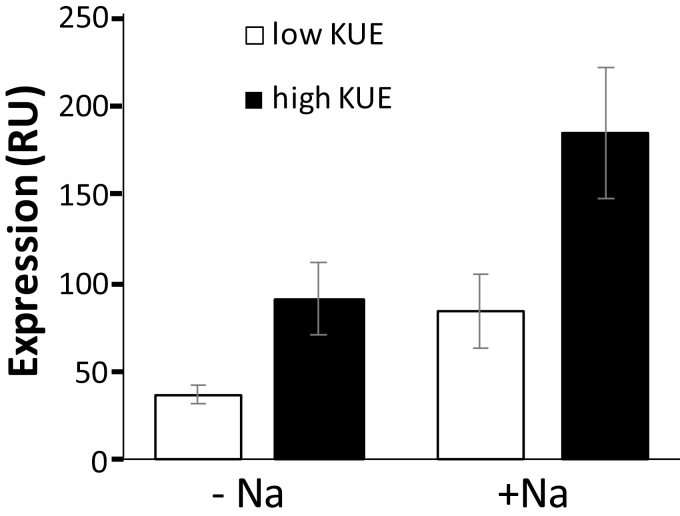
Since there is a clear positive impact of Na+ on KUE-RGR, it is imperative to identify the molecular mechanisms involved. Our GWAS identified HKT2;1 as a potential causative agent for Na+-dependent variation in KEU. There is considerable allelic variation in the HKT2;1 coding sequence which contains five non-synonymous SNPs that are located in the cytoplasmic N-terminus and at the end of the first and sixth transmembrane spans (Oomen et al., 2012). Extensive measurements on oocytes that heterologously express HKT2;1 showed that neither of the amino acid substitutions has a significant effect on HKT2;1 functional properties (Oomen et al., 2012). However, the HKT2;1 promoter region contains a large number (>50) of polymorphisms (e.g http://snp-seek.irri.org/), many of which are located in transcription factor-binding domains (e.g. PlantPan2; http://plantpan2.itps.ncku.edu.tw/) and consequently could affect expression levels. We therefore tested whether HKT2;1 expression levels differed between five high- and five low-KUE lines grown on 0.01 mM K+ and with or without 1 mM Na+. Figure 7 shows that in these very low K+-grown plants, the average expression level of HKT2;1 in both low- and high-KUE lines is induced in the presence of Na+ (1 mM) as was reported previously (Horie et al., 2007). However, in both conditions, HKT2;1 expression levels were >2-fold higher in high-KUE lines, a difference that was highly significant in the minus NaCl condition (P=0.015) but less so in the plus NaCl treatment (P=0.066).
Fig. 7.
qPCR analysis of HKT2;1 expression in roots of five high-KUE cultivars (GSOR 54, 109, 133, 357, and 366; see Supplementary Table S1) and five low-KUE rice cultivars (GSOR 42, 115, 276, 377, and 401). Plants were grown for 4 weeks in medium containing 0.01 mM K+ supplemented with 0 mM or 1 mM NaCl. Data are means for three biological replicates, with error bars denoting the SD.
Although no significant association signals were detected, further Na+ transporters may be involved in tissue K+ substitution by Na+; for example, OsHKT1;5 is involved in shoot Na+ exclusion by retrieving Na+ from the xylem stream and via phloem recirculation (Kobayashi et al., 2017). Down-regulation of this mechanism during low-K+ conditions could therefore augment K+ substitution. Other HKTs such as OsHKT2;2, which is primarily root located and could mediate uptake of both K+ and Na+ (Oomen et al., 2012), is another potential contributor.
Conclusions
A clearer picture of the physiological and molecular underpinnings of KUE variability would be extremely useful in developing high-KUE crops. Differences in KUE can be achieved through various mechanisms including: an altered cellular K+ distribution, especially between the vacuole and cytoplasm; tissue K+ distribution, namely preferential allocation of K+ to the most sensitive tissue such as translocation to the shoot; changes in K+ uptake capacity, especially at low external K+; changes in K+ supply such as enhancing available soil K+ via root exudation; and the functional replacement of K+ with other ions such as Na+ and Ca2+. The relative contribution of these mechanisms is largely unknown and may depend on plant species, developmental stage, and soil properties.
In this study, KUE was explored using a rice diversity panel. Variation in KUE was found to be considerable, and the underlying genetic architecture was examined. By deliberately applying high-stringency criteria, KUE-related high-resolution QTLs were discovered that identified K+ substitution by Na+ as a likely component of KUE in low-K+ conditions. Although it is likely that multiple Na+ and K+ transporters play a role in this process, OsHKT2;1 emerged as the prime suspect responsible for increased Na+ uptake. This transporter and other identified candidates could serve as breeding targets to improve crop performance during low-K+ conditions.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Diversty panel rice accessions.
Table S2. Rice accession trait means by genotype.
Table S3. Phenotypic parameters included in GWAS analysis.
Table S4. High- and low-KUE accessions.
Table S5. QTLs and candidate genes from GWAS.
Fig. S1. Correlations between growth and tissue Na+ concentrations.
Fig. S2. All Manhattan plots of GWAS analyses
Acknowledgments
This work was supported by a BBSRC doctoral grant to TNH.
Glossary
Abbreviations:
- GWAS
genome-wide association study
- KUE
potassium use efficiency
- QTL
quantitative trait locus
- SNP
single nucleotide polymorphism
References
- Allen AG, Cardoso AA, Wiatr AG, Machado CMD, Paterlini WC, Baker J. 2010. Influence of intensive agriculture on dry deposition of aerosol nutrients. Journal of the Brazilian Chemical Society 21, 87–97. [Google Scholar]
- Armengaud P, Breitling R, Amtmann A. 2004. The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiology 136, 2556–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher CJ, Ozanne PG. 1967. Growth and potassium content of plants in potassium cultures maintained at constant potassium concentrations. Soil Science 103, 155–161. [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23, 1294–1296. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 57, 289–300. [Google Scholar]
- Brentup F, Pallière C. 2008. Energy efficiency and greenhouse gas emissions in European nitrogen fertilizer production and use. York: International Fertiliser Society. [Google Scholar]
- Camargo GGT, Ryan MR, Richard TL. 2013. Energy use and greenhouse gas emissions from crop production using the Farm Energy Analysis Tool. BioScience 63, 263–273. [Google Scholar]
- Campbell MT, Bandillo N, Al Shiblawi FRA, et al. 2017. Allelic variants of OsHKT1;1 underlie the divergence between indica and japonica subspecies of rice (Oryza sativa) for root sodium content. PLoS Genetics 13, e1006823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell S, Korniliev P, Falcão A, Ismail A, Gregorio G, Mezey J, McCouch S. 2016. Genome-wide association and high-resolution phenotyping link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nature Communications 7, 10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JL, Armengaud P, Larson TR, Graham IA, White PJ, Newton AC, Amtmann A. 2018. Contrasting nutrient–disease relationships: potassium gradients in barley leaves have opposite effects on two fungal pathogens with different sensitivities to jasmonic acid. Plant, Cell & Environment 41, 2357–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. 1999. Genomic control for association studies. Biometrics 55, 997–1004. [DOI] [PubMed] [Google Scholar]
- Eizenga GC, Ali ML, Bryant RJ, Yeater KM, McClung AM, McCouch SR. 2014. Registration of the rice diversity panel 1 for genomewide association studies. Journal of Plant Registrations 8, doi: 10.3198/jpr2013.03.0013crmp. [DOI] [Google Scholar]
- Fageria NK. 1976. Influence of potassium concentration on growth and potassium uptake by rice plants. Plant and Soil 44, 567–573. [Google Scholar]
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR. 2011. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genetics 7, e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Wu W, Zhang X, Jiang H, Lu W, Pan J, Hu J, Guo L, Zeng D, Xue D. 2015. Identification of quantitative trait loci associated with tolerance to low potassium and related ions concentrations at seedling stage in rice (Oryza sativa L.). Plant Growth Regulation 77, 157–166. [Google Scholar]
- FAO. 2017. World fertilizer trends and outlook to 2020. Rome: FAO. [Google Scholar]
- Garcia-Oliveira AL, Tan L, Fu Y, Sun C. 2009. Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. Journal of Integrative Plant Biology 51, 84–92. [DOI] [PubMed] [Google Scholar]
- Horie T, Brodsky DE, Costa A, Kaneko T, Lo Schiavo F, Katsuhara M, Schroeder JI. 2011. K+ transport by the OsHKT2;4 transporter from rice with atypical Na+ transport properties and competition in permeation of K+ over Mg2+ and Ca2+ ions. Plant Physiology 156, 1493–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. 2007. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. The EMBO Journal 26, 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi NI, Yamaji N, Yamamoto H, et al. 2017. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. The Plant Journal 91, 657–670. [DOI] [PubMed] [Google Scholar]
- Kong F-M, Guo Y, Liang X, Wu C-H, Wang Y-Y, Zhao Y, Li S-S. 2013. Potassium (K) effects and QTL mapping for K efficiency traits at seedling and adult stages in wheat. Plant and Soil 373, 877–892. [Google Scholar]
- Koyama ML, Levesley A, Koebner RM, Flowers TJ, Yeo AR. 2001. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiology 125, 406–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Singh A, Mithra SV, et al. 2015. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Research 22, 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, Hu XH, Ren ZH, Chao DY. 2004. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theoretical and Applied Genetics 108, 253–260. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. 2009. Physiological functions of mineral macronutrients. Current Opinion Plant Biology 12, 250–258. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM. 2013. Signalling and regulation of sodium fluxes in plants. Journal Experimental Botany 65, 849–858. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Amtmann A. 1999. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Annual Botany 84, 123–133. [Google Scholar]
- McCouch SR, Wright MH, Tung CW, et al. 2016. Open access resources for genome-wide association mapping in rice. Nature Communications 7, 10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Ochiai K, Takeshita S, Matoh T. 2012. Identification of quantitative trait loci associated with shoot sodium accumulation under low potassium conditions in rice plants. Soil Science and Plant Nutrition 58, 728–736. [Google Scholar]
- Oomen RJ, Benito B, Sentenac H, Rodríguez-Navarro A, Talón M, Véry AA, Domingo C. 2012. HKT2;2/1, a K+-permeable transporter identified in a salt-tolerant rice cultivar through surveys of natural genetic polymorphism. The Plant Journal 71, 750–762. [DOI] [PubMed] [Google Scholar]
- Patishtan J, Hartley TN, Fonseca de Carvalho R, Maathuis FJM. 2018. Genome-wide association studies to identify rice salt-tolerance markers. Plant, Cell & Environment 41, 970–982. [DOI] [PubMed] [Google Scholar]
- Prinzenberg AE, Barbier H, Salt DE, Stich B, Reymond M. 2010. Relationships between growth, growth response to nutrient supply, and ion content using a recombinant inbred line population in Arabidopsis. Plant Physiology 154, 1361–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Kirkby EA. 2010. Research on potassium in agriculture: needs and prospects. Plant and Soil 335, 155–180. [Google Scholar]
- Sassi A, Mieulet D, Khan I, Moreau B, Gaillard I, Sentenac H, Véry AA. 2012. The rice monovalent cation transporter OsHKT2;4: revisited ionic selectivity. Plant Physiology 160, 498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar A, Singh A, Kanwar P, Srivastava AK, Pandey A, Suprasanna P, Kapoor S, Pandey GK. 2013. Gene expression analysis of rice seedling under potassium deprivation reveals major changes in metabolism and signaling components. PLoS One 8, e70321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R. 2014. Strategies for improving potassium use efficiency in plants. Molecules and Cells 37, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear SN, Edwards DG, Asher CJ. 1978. Response of cassava, sunflower, and maize to potassium concentration in solution I. Growth and plant potassium concentration. Field Crops Research 1, 375–389. [Google Scholar]
- USGS. 2017. Mineral commodity summaries 2017. Washington DC: USGS. [Google Scholar]
- Wang C, Chen HF, Hao QN, Sha AH, Shan ZH, Chen LM, Zhou R, Zhi HJ, Zhou XA. 2012. Transcript profile of the response of two soybean genotypes to potassium deficiency. PLoS One 7, e39856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu WH. 2015. Genetic approaches for improvement of the crop potassium acquisition and utilization efficiency. Current Opinion in Plant Biology 25, 46–52. [DOI] [PubMed] [Google Scholar]
- Wu P, Ni JJ, Luo AC. 1998. QTLs underlying rice tolerance to low-potassium stress in rice seedlings. Crop Science 38, 1458–1462. [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory manual for physiological studies of rice. Los Banos: The International Rice Research Institute. [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, et al. 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nature Communications 2, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Li X, Zhang S, Wang J, Yang X, Tian J, Hai Y, Yang X. 2014. Mapping QTLs for potassium-deficiency tolerance at the seedling stage in wheat (Triticum aestivum L.). Euphytica 198, 185–198. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.