The metabolic-oxidative stress profile of the rice flag leaf during drought stress in the reproductive stage is highly predictive for grain yield loss sensitivity of 292 accessions at harvest time.
Keywords: Drought, leaf oxidative stress status, leaf primary metabolism, Oryza sativa, PLSR, reproductive stage
Abstract
Crop yield stability requires an attenuation of the reduction of yield losses caused by environmental stresses such as drought. Using a combination of metabolomics and high-throughput colorimetric assays, we analysed central metabolism and oxidative stress status in the flag leaf of 292 indica rice (Oryza sativa) accessions. Plants were grown in the field and were, at the reproductive stage, exposed to either well-watered or drought conditions to identify the metabolic processes associated with drought-induced grain yield loss. Photorespiration, protein degradation, and nitrogen recycling were the main processes involved in the drought-induced leaf metabolic reprogramming. Molecular markers of drought tolerance and sensitivity in terms of grain yield were identified using a multivariate model based on the values of the metabolites and enzyme activities across the population. The model highlights the central role of the ascorbate–glutathione cycle, particularly dehydroascorbate reductase, in minimizing drought-induced grain yield loss. In contrast, malondialdehyde was an accurate biomarker for grain yield loss, suggesting that drought-induced lipid peroxidation is the major constraint under these conditions. These findings highlight new breeding targets for improved rice grain yield stability under drought.
Introduction
Drought-induced closure of stomata not only reduces water loss, but also limits CO2 diffusion into the leaf intercellular spaces and thus decreases photosynthetic carbon assimilation. This alteration results in the disruption of cellular homeostasis and leads to an enhanced generation of reactive oxygen species (ROS), mainly in the peroxisomes and chloroplasts (Suzuki et al., 2012; Noctor et al., 2014). In leaf tissues of C3 plants exposed to light and under drought stress, peroxisomes are considered the major production site of hydrogen peroxide (H2O2), primarily because of the enhanced activity of the photorespiratory pathway (Noctor, 2002). In chloroplasts, ROS production arises when excitation energy exceeds the level required for CO2 assimilation (Asada, 2006), a condition that is favoured by drought-induced stomatal closure (Miller et al., 2010). Under persistent water-limited conditions, the cellular ROS-scavenging capacity is exceeded by ROS production that, in turn, leads to oxidative damage that drives the cell into senescence (Halliwell, 2006) and, in extreme cases, cell death (Van Breusegem and Dat, 2006). To reduce oxidative damage, plants employ a complex enzymatic and non-enzymatic antioxidative system, which is triggered by ROS (Mittler et al., 2011; Baxter et al., 2014; You and Chan, 2015).
The decreased photosynthetic carbon assimilation, caused by stomatal closure, results in a reprogramming of plant central metabolism and growth to try to maintain the activity of essential metabolic pathways while simultaneously adapting to the stressful conditions (Obata and Fernie, 2012; Claeys and Inzé, 2013). Under drought stress, accumulation of leaf metabolites thought to have protective functions (e.g. fructose, glucose, raffinose, proline, and glycine betaine) or to accumulate as a consequence of the stress (e.g. protein breakdown causing the overall increase in free amino acids) has been reported for a number of plant species (Verslues and Juenger, 2011; Krasensky and Jonak, 2012; Obata and Fernie, 2012; Fàbregas and Fernie, 2019). This metabolic response to drought varies during different developmental stages (Hummel et al., 2010; Skirycz et al., 2010). Among them, the reproductive stage is considered the most drought sensitive in plants (Passioura, 2012) and particularly in rice, which, among the cereal crops, shows greatest sensitivity to water limitation (Venuprasad et al., 2007). The top leaves of rice, and primarily the flag leaf, are the most important source of assimilates for the developing panicles (Yoshida, 1972). Drought-induced alteration of metabolism and redox state in these leaves during the reproductive stage was shown to be linked with a reduction in grain yield (Biswal and Kohli, 2013; Sandhu et al., 2014).
In the last decade, an increasing number of studies used metabolomics as a large-scale screening tool to identify markers for plant trait improvement (Fernandez et al., 2016; Kumar et al., 2017). This interest mainly relies on the fact that metabolic markers showed equal or even higher predictive power for plant traits than traditional genetic markers (Fernandez et al., 2016). Metabolomics-based prediction of biomass was used in Arabidopsis recombinant inbred lines and accessions grown under optimal (Meyer et al., 2007; Sulpice et al., 2009; Steinfath et al., 2010) and suboptimal (Sulpice et al., 2013) conditions. These studies were performed under controlled conditions, thus reducing environmental effects and increasing the likelihood of finding strong relationships between metabolite levels and the trait of interest (Fernandez et al., 2016). In crop species, trait prediction based on metabolic profiling of large field-grown populations of genetically diverse accessions remains rare, and even more rare in experiments simultaneously conducted under optimal and non-optimal conditions (Riedelsheimer et al., 2012, 2013; Xu et al., 2016).
Here we present a large field study aimed at improving our understanding of the drought-induced reprogramming of metabolism and oxidative stress status in rice and its effect on grain yield. The flag leaf central metabolome together with a range of oxidative stress markers and redox state-related enzymes were analysed in a collection of 292 phenotypically and genetically diverse rice lines that were exposed to well-watered and drought conditions during the reproductive stage. The data set was used to analyse the relationship between oxidative stress and central metabolism under the two different treatments, to generate multivariate models that predict grain yield loss under drought stress, and to identify metabolic and oxidative stress markers of tolerance and sensitivity to drought. The combination of oxidative stress markers and enzyme activities with the metabolomics data set for the prediction, and the large number of field-grown accessions, make this study an extensive and valuable source of information to identify biomarkers for improved grain yield stability of rice under drought.
Materials and methods
Genetic resources and plant growth
Two-hundred and ninety-two accessions of Oryza sativa subsp. indica were used in a field experiment at the International Rice Research Institute (IRRI), Los Baños, Philippines during the 2013 dry season. The accessions are largely the same as those in the PRAY-indica panel (http://ricephenonetwork.irri.org) including traditional and improved indica lines, from tropical and subtropical regions. The experiment comprised a control field and drought stress field, each with three replicates of the population arranged in a serpentine design (see Supplementary Fig. S1 at JXB online). To synchronize flowering, the accessions were divided into six groups according to days required to flower (previously collected data), and progressively sown and transplanted with intervals of 10 d between each group. Drought stress consisted of 14 consecutive days of water withholding applied only to the stress field at the reproductive stage (targeting 50% flowering). At the end of stress, the field was re-watered until all the accessions reached maturity for harvest (further details in Kadam et al., 2018).
Phenotyping
Percentage grain yield loss (GY loss) of each accession was calculated (GYcontrol–GYdrought)/(GYcontrol×100) as the mean values of the GY loss of all replicates (three for drought and two for control). A variable, Sam-Flow, was calculated as the date of leaf sampling (Sam), minus the date of 50% flowering (Flow) for every genotype, under control and drought treatment, separately. The genotypes together with their GY loss, Flow, and Sam-Flow values are shown in Supplementary Table S1.
Leaf sampling
Eight flag/top leaves from the main tiller of eight plants per plot (that were not used for yield determination) were sampled and immediately frozen in liquid nitrogen. Three drought field replicates of all accessions were collected (09.30–11.00 h) on day 14 of the stress treatment. Two control field replicates of the entire population were collected, 2 d later, during the same time window. Samples were ground in liquid nitrogen, shipped to the Netherlands on dry ice, and stored at –80 °C until further analysis in Germany and Belgium, to where samples were also shipped on dry ice.
Metabolite profiling
Metabolite profiling was performed as described by Riewe et al. (2012, 2016). For each accession and treatment, equal amounts of replicates (three for drought and two for control) were pooled, resulting in 584 samples (292 each for drought and control). Samples were extracted in MeOH/H2O (15.0±1.0 mg FW), dried, and in-line derivatized (MPS2 autosampler, GERSTEL) prior to GC-MS analysis (AGILENT/LECO). Metabolites were identified using ChromaTOF software (LECO) and the library provided by the Golm Metabolome Database (GMD; http://gmd.mpimp-golm.mpg.de/download/). Peak intensities were determined using the R package TargetSearch (Cuadros-Inostroza et al., 2009) and normalized against an internal standard (D8-valine), fresh weight, and detector response variation.
Glucose, fructose, and sucrose detection
Glucose, fructose, and sucrose were quantified spectrophotometrically (Riewe et al., 2008). In brief, NADPH production at 340 nm was converted to saccharide content after the sequential addition of hexokinase (glucose), phosphoglucoisomerase (fructose), and invertase (sucrose) into a reaction mix, containing extract, glucose-6-phosphate dehydrogenase, ATP, and NADP+.
Oxidative stress markers
Malondialdehyde (MDA) is a by-product of lipid peroxidation. Its content was assayed according to Hodges et al. (1999). Leaf material (50 mg FW) was homogenized (80% v/v ethanol), using a MagNA Lyser (Roche, Vilvoorde, Belgium). After centrifugation, the supernatant was allowed to react with thiobarbituric acid (TBA) to produce the chromogen, TBA–MDA. Absorbance of TBA–MDA was measured at 440, 532, and 600 nm, using a micro-plate reader (Synergy Mx, Biotek Instruments Inc., Vermont, VT, USA). Protein oxidation (ProtOx) was estimated through measuring the protein carbonyl content, utilizing dinitrophenylhydrazine (DNPH) derivatization (Levine et al., 1994).
Molecular antioxidants
Total non-enzymatic antioxidant capacity (TAC) was assayed after homogenizing and extracting leaf tissue (50 mg FW) in 80% ethanol (v/v). The extract was centrifuged, and the tripyridyltriazine (TPTZ) assay reagent was mixed with the extract (Benzie and Strain, 1999). Absorbance change at 600 nm was measured using a microplate reader. Trolox (0–650 µM) was used as standard. Total polyphenol content (Poly) was assayed after the same homogenizing, extracting, and centrifugation steps, using a Folin–Ciocalteu reagent for phenol detection (Zhang et al., 2006), with gallic acid as standard.
Antioxidant enzymes
Soluble proteins were extracted according to Murshed et al. (2008) and quantified by the Lowry method (Lowry et al., 1951). Enzyme activities were determined in a semi-high-throughput set-up (Zinta et al., 2014; AbdElgawad et al., 2016). Ascorbate peroxidase (APX), dehydroascorbate reductase (DHAR), monodehydroascorbate reductase (MDHAR), and glutathione reductase (GR) activities were measured in extracts obtained from 100 mg of frozen tissue, in 1 ml of extraction buffer: 50 mM MES/KOH (pH 6.0) containing 0.04 M KCl, 2 mM CaCl2, and 1 mM ASC, homogenized by MagNALyser (Roche). APX, MDHAR, DHAR, and GR activities were determined in microplates according to the method of Murshed et al. (2008). Their activities were assayed in 50 mM K-phosphate, 50 mM HEPES pH 7.6, 50 mM HEPES pH 7, and 50 mM HEPES pH 8, respectively. APX, MDHAR, and GR activities were measured by monitoring the decrease in ASC (ɛ290=2.8 mM–1 cm–1), NADH (ɛ340=6.22 mM–1 cm–1), and NADPH (ɛ340=6.22 mM–1 cm–1). Peroxidase (POX) activity was determined by the oxidation of pyrogallol in 100 mM phosphate buffer (ɛ430=2.46 mM–1 cm–1) (Kumar and Khan, 1983). Superoxide dismutase (SOD) activity was analysed by measuring the inhibition of nitro-blue tetrazolium (NBT) reduction (ɛ550=12.8 mM–1 cm–1) (Dhindsa et al., 1982). Glutathione peroxidase (GPX) activity was measured as described by Drotar et al. (1985), in a coupled enzyme assay with GR, measuring the decrease in NADPH absorption. Catalase (CAT) activity was assayed by monitoring the H2O2 decomposition at 240 nm (ɛ240=39.4 M−1 cm−1) (Aebi, 1984). Ascorbate oxidase (AO) activity was measured as the rate of decrease of ascorbate (absorbance at 290 nm; Yoshimura et al., 1998). Glutathione S-transferase (GST) activity was determined by measuring conjugation of GSH to 1-chloro-2,4-dinitrobenzene (CDNB) at 340 nm (Habig et al., 1974). Peroxiredoxin (PRX) activity was determined according to Horling et al. (2003), by measuring the decrease in H2O2 concentration in the reaction mixture. Glutaredoxin (Grx) activity was determined by measuring the reduction of 2-hydroxy-ethyl-disulfide by GSH in the presence of NADPH and yeast GR (Lundberg et al., 2001). Thioredoxin (Trx) activity was determined by measuring NADPH oxidation (Wolosiuk et al., 1979) at 340 nm. Ferredoxin-NADP(H) reductase (Frx) activity was determined as the reduction of potassium ferricyanide at 420 nm (Rodriguez et al., 2007).
Photorespiration enzymes
Glycolate oxidase (GOX) activity was determined by the formation of a glyoxylate complex with phenylhydrazine (ε324=17 mM–1 cm–1; Feierabend and Beevers, 1972). Hydroxypyruvate reductase (HPR) activity was determined according to Schwitzguebel and Siegenthaler (1984) as the oxidation of NADH that was followed at 340 nm upon hydroxypyruvate addition.
Data analysis
Results of all metabolites and oxidative stress markers/enzymes were log10 transformed to improve normality. Metabolites showing >5% missing values among the samples were excluded from the analysis. A list of the results of metabolites and oxidative stress markers/enzymes used for statistical analyses is included in Supplementary Table S2.
Statistical analyses and graphical representations were performed using R (version 3.4.3; The R Foundation for Statistical Computing). Fold change (FC) was calculated (on non-log10-transformed data) dividing drought values by control values. Hierarchical clustering analysis was conducted using the hclust function (stats package) and based on complete linkage analysis of pairwise dissimilarity, calculated as 1–rs (rs, Spearman rank correlation coefficient). Dendrograms were created by using the dendextend package, and heat maps of correlations were created using the heatmap.2 function (gplot package). Imputation of missing values, prior to principal componenet analysis (PCA) and partial least squares regression (PLSR) analysis, was performed by the knnImputation function in the DMwR package. PCA was performed using the prcomp function (stats package), and the value of each variable was centred (mean subtraction) and scaled (standard deviation division) before analysis.
To identify the variables predictive for grain yield loss, a cross-validated PLSR (pls package) was used (Mevik and Wehrens, 2007; Mumm et al., 2016). Observations were auto-scaled in the PLSR procedure. The number of latent variables to include in the model was selected by testing the predictability value (Q2) using an increasing number of latent variables from 1 to 10. The relative importance of the metabolites in the models was summarized using rank-products.
Results
Relationship between differences in flowering time and grain yield loss
The rice population was grown in a field trial as part of a study aimed at collecting information on phenotypic trait performance under well-watered and drought stress conditions (Kadam et al., 2018). To evaluate the impact of drought stress on grain yield stability, the percentage of grain yield loss (GY loss) under drought versus control conditions was calculated across the 292 genotypes as an indicator of stress tolerance/susceptibility (Supplementary Table S1).
Even though the accessions were sown and transplanted on different dates to minimize flowering time differences, flowering synchronization was not perfect, and could represent a confounding effect on yield results under drought (Kadam et al., 2018). Correlation analysis was performed to evaluate the influence of flowering time differences (Supplementary Table S1) on the drought-induced GY loss performance of the 292 accessions (Supplementary Fig. S2). Flowering (Flow) under drought significantly and negatively correlated (P-value <0.001) with GY loss (rs= –0.35) but only 12% (R2=0.12) of the variation was explained by the corresponding linear model. In general, the correlation trend shows that accessions that already flowered before stress imposition (<10% of the total) displayed a relatively higher severity of GY loss than those that had nearly or already flowered (booting stage for 60% and heading stage for 30% of the total) during stress imposition. Interestingly, a significant (P-value <0.001) and almost identical negative correlation was observed between Flow under control (rs= –0.37; R2=0.13) and GY loss. This similarity is determined by the almost perfect correlation (rs=0.96; R2=0.94; P-value <0.001) observed between Flow under control and drought (Supplementary Fig. S2). Nevertheless, drought stress significantly affected (paired t-test’s P-value <0.001) the date of 50% flowering with a delay of ~3 d (mean ±SD: 83.9±10.6) compared with control (81.0±10.3). The almost perfect correlation between Flow under the two treatments indicates that the flowering delay under drought is virtually identical in the 292 accessions.
Drought induces accumulation of amino acids and affects the level of antioxidant enzymes and organic acids
Leaf samples of the 292 accessions were analysed by untargeted GC-MS-based metabolite profiling to assess the variation in polar metabolites under well-watered and drought conditions. A total of 88 metabolites were identified, predominantly primary metabolites (amino acids, sugars, and organic acids). The amount of the three most abundant sugars (sucrose, fructose, and glucose) was determined spectrophotometrically (glucose also by MS). The same leaf materials were analysed for the oxidative stress status of the different accessions under the two treatments. For this, the level of molecular antioxidants (2) and oxidative stress markers (2), and the activity of enzymes (16) involved in ROS-scavenging mechanisms and photorespiration were quantified. The 111 variables, metabolites, and oxidative stress markers/enzymes considered in this study (Supplementary Table S3) are hereafter referred to as MetabOxi.
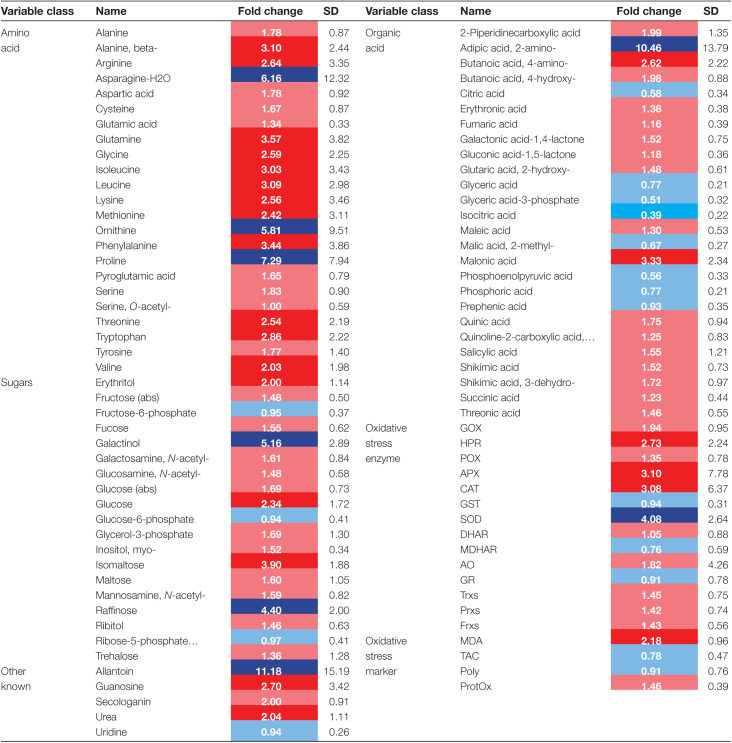
PCA was used to gain insight into the overall effect of drought on the MetabOxi profiles of the accessions. Among the first three principal components (PCs), PC1 explained 29.5% of the total variation, and almost completely separated the control and drought-stressed samples (Supplementary Fig. S3), suggesting a strong influence of drought on the MetabOxi profiles of the accessions. To evaluate the treatment effect on the level of the individual MetabOxi variables, one-way ANOVA was conducted. This showed that drought significantly influenced (Bonferroni-corrected P-value <0.05) most (91 out of 111) of the MetabOxi variables across the 292 genotypes (Supplementary Table S4). To quantify the magnitude of these alterations, we conducted an FC analysis (stress over control values) for the 91 MetabOxi variables that changed significantly between treatments (Table 1). The majority (75 out of 91) displayed an FC increase (FC >1), with allantoin and 2-aminoadipic acid showing the largest increase (FC >10). Interestingly, the level of all amino acids significantly increased under drought, with the highest FC values for Pro, Asn-H2O, and Orn. Organic acids, sugars, oxidative stress markers, and enzymes displayed a more diverse response. In particular, most of the organic acids showed a decrease (FC <1), with two tricarboxylic acid cycle (TCA) intermediates, isocitric and citric acid, and two glycolysis intermediates, glyceric acid-3-phosphate and phosphoenolpyruvic acid, displaying the largest decrease (FC <0.6). Of the sugars, galactinol and raffinose displayed a strong increase (FC >4), as did the ROS-scavenging enzymes SOD, APX, and CAT (3>FC>4). Collectively, these results highlight the strong influence of drought stress on the metabolome and oxidative stress status of the population, mainly characterized by an increase in the level of amino acids and activity of specific antioxidant enzymes, and by a decrease in the level of organic acids.
Table 1.
The effect of drought on the MetabOxi variables showing a significant response to stress
Drought stress increases the correlations within and between metabolite classes
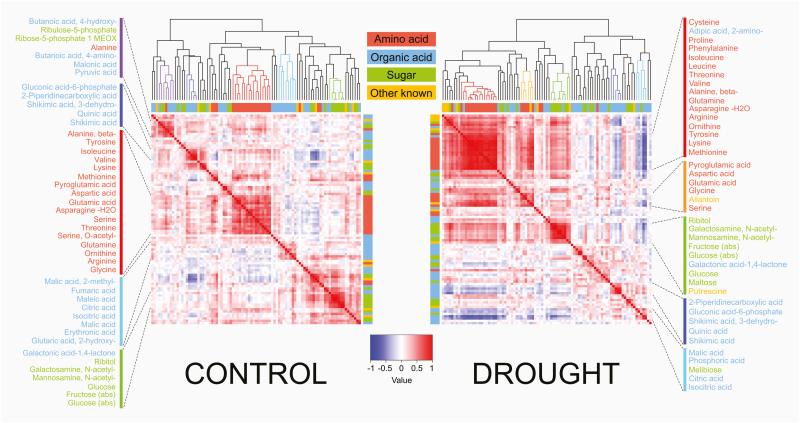
Correlation analysis was performed to assess the nature and strength of the associations between flag leaf metabolites under control and drought conditions (Supplementary Table S5, S6). Figure 1 presents the degree of correlation among metabolites, together with their hierarchical clustering under the two treatments (see Supplementary Fig. S4 for a detailed representation of the dendrograms). By comparing the two heat maps, similarities as well as stress-induced population-wide differences in the correlations between leaf metabolites can be identified. Metabolites of the same class (mainly amino acids, sugars, and organic acids) clustered together in both treatments (clusters of the same colour in Fig. 1). Only a single drought-specific cluster (yellow), including five amino acids (Asp, Glu, Gly, Ser, and pyroglutamic acid) and allantoin, was identified. Drought stress resulted in stronger correlations between metabolites within each cluster than for control conditions, as evidenced, particularly, by the amino acid cluster (red). Drought stress also increased the correlations (positive or negative) between clusters representing different metabolite classes. For example, under drought, the amino acid (red) and sugar cluster (green) displayed an increased correlation, with the latter also showing an increased negative correlation with the organic acid clusters (light blue and dark blue). In particular, between the two organic acid clusters, the one containing three TCA intermediates, citric, isocitric, and malic acid (light blue), showed the strongest negative correlation with the amino acid (red) and sugar (green) clusters. Weaker correlations with the same clusters were displayed by the other organic acid cluster (dark blue), enriched in metabolites of the shikimate pathway (shikimic acid, quinic acid, and 3-dehydroshikimic acid). Interestingly, the drought-specific amino acid-enriched cluster (yellow) displayed a correlation pattern with the other metabolite clusters similar to the major amino acid cluster (red) to which it was strongly positively correlated. Under control conditions, the above-mentioned clusters showed very low correlations with each other. In summary, drought stress resulted in a generally stronger correlation within and between all the leaf metabolite classes than under control conditions and induced one new (stress-specific) cluster, containing Asp, Glu, Gly, Ser, pyroglutamic acid, and allantoin.
Fig. 1.
Hierarchical clustering and heatmap of the metabolites versus metabolites pairwise correlations under the two treatments for the 292 rice accessions. Spearman correlations under control (left) and drought stress (right) conditions. Colour bars on the top and side of the heatmaps represent the four main classes of metabolites (amino acid, organic acid, sugar, and other known). In the dendrograms, main clusters are coloured according to the class of the majority of the metabolites they included: red (amino acids), light blue (TCA cycle organic acids), blue (shikimic acid pathway), green (sugars), purple (mixed cluster), and yellow (mainly amino acid). All the other minor clusters are coloured in black. The metabolites included in each cluster are displayed at the side of the two heatmaps. Metabolite names are coloured according to the four main classes of known metabolites mentioned above.
Leaf amino acid metabolism is strongly linked with stress-induced photorespiratory and antioxidative enzymes
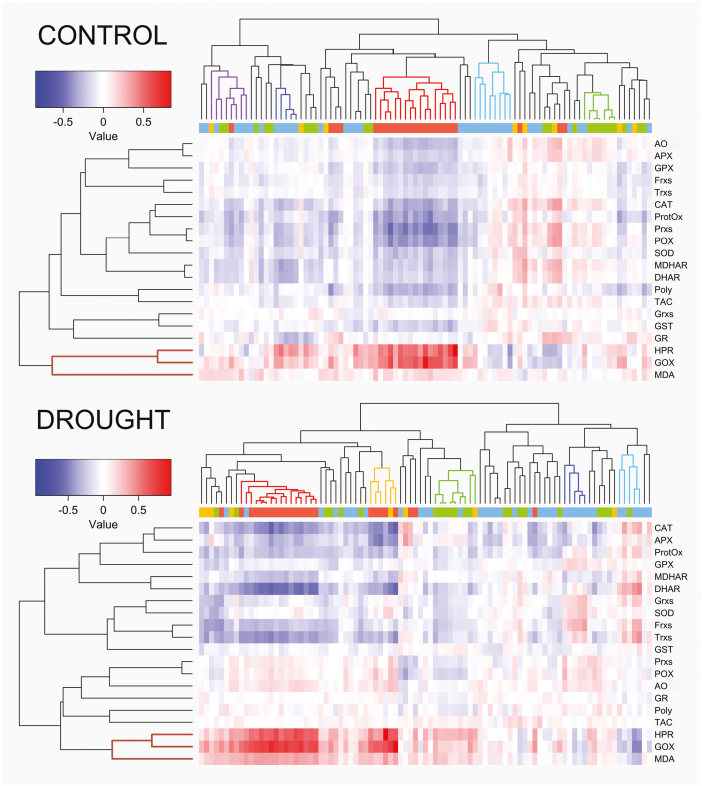
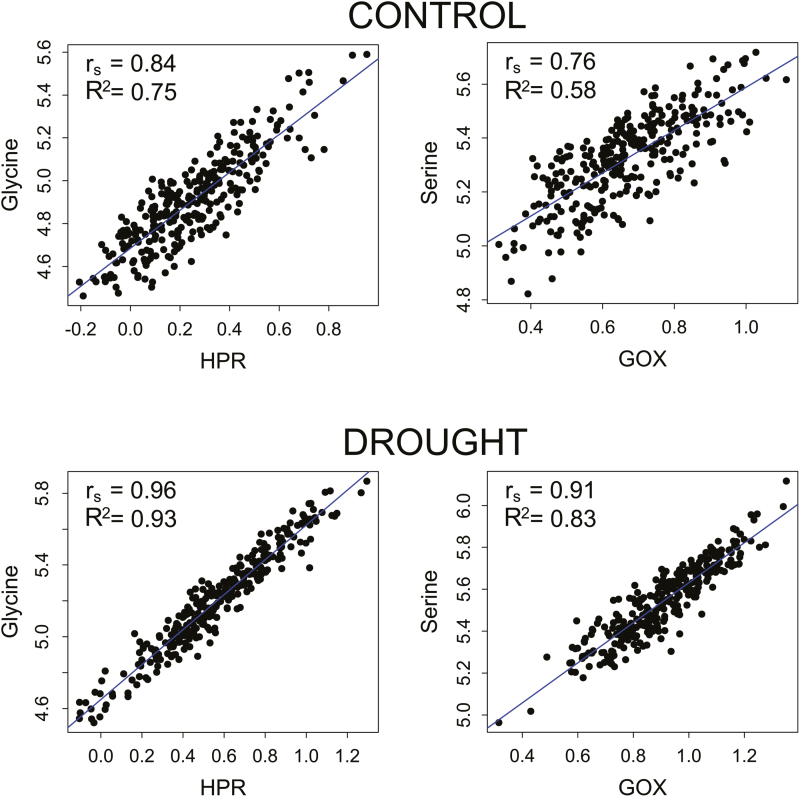
To investigate the relationships between leaf oxidative stress status and metabolism, correlation analysis and hierarchical clustering were conducted on the combined data sets (Fig. 2; Supplementary Tables S7, S8). Strikingly, a cluster formed by two photorespiratory enzymes, HPR and GOX, and the lipid peroxidation product MDA was present under both control and drought conditions (brown). HPR and GOX showed the strongest positive correlation with a number of metabolites under both treatments, and, especially, with the main cluster of amino acids (red). The two enzymes also displayed a strong positive correlation with the drought-specific cluster of amino acids (yellow), which contains Gly and Ser, known to be produced by the photorespiratory pathway. MDA showed low correlations with all other metabolites under control conditions, whereas, under stress, these correlations became stronger, with an overall pattern very similar to that of HPR and GOX (Fig. 2). Under drought stress, the HPR–GOX–MDA cluster also showed a stronger negative correlation with the TCA cycle cluster (light blue) than under control conditions. Considering the other oxidative stress markers/enzymes, almost all (15 out of 17) displayed negative correlations with the single amino acid cluster (red) under control conditions. Under drought, a more limited number (7 out of 17) showed a negative correlation with the two amino acid clusters (red and yellow). However, among them, specific ROS scavenger enzymes such as DHAR, CAT, and APX showed very strong negative correlations with the amino acid clusters (red and yellow), stronger than in control conditions. In summary, these results show the presence of a strong correlation between the flag leaf oxidative stress status and metabolome in both treatments, but with the strongest and more specific associations under drought, particularly between oxidative stress enzyme activities/markers and amino acids. The best correlations between the variables of the two data sets were the same under drought and control conditions: HPR with Gly and GOX with Ser (Fig. 3). For GOX and Ser, the correlation value (rs) was 0.76 under control conditions and 0.91 under drought stress, with the two linear models able to explain 58% and 83% of the variation, respectively. For HPR and Gly, the correlation was even stronger, with rs=0.84 under control and rs=0.96 under drought conditions, and with 75% and 93% of the variation, respectively, explained by the linear models.
Fig. 2.
Hierarchical clustering and heatmap of the oxidative stress markers/enzymes versus metabolites pairwise correlations under the two treatments for the 292 rice accessions. Spearman correlations under control (top) and drought stress (bottom) conditions. The metabolite colour code of the bar below the dendrograms as well as the colour of the main clusters are the same as in Fig. 1. Malondialdehyde (MDA), polyphenols (Poly), protein oxidation (ProtOx), total antioxidant capacity (TAC), ascorbate oxidase (AO), ascorbate peroxidase (APX), catalase (CAT), dehydroascorbate reductase (DHAR), ferredoxins (Frxs), glycolate oxidase (GOX), glutathione peroxidase (GPX), glutathione reductase (GR), glutaredoxins (Grxs), glutathione S-transferase (GST), hydroxypyruvate reductase (HPR), monodehydroascorbate reductase (MDHAR), peroxidase (POX), peroxiredoxins (Prxs), superoxide dismutase (SOD), and thioredoxins (Trxs).
Fig. 3.
Linear models between the photorespiratory enzymes and hydroxypyruvate reductase, and glycolate oxidase and the amino acids glycine and serine under the two treatments. Linear models of the two best correlations between MetabOxi variables under the two treatments. Axes are expressed as log10-transformed detector response values for the metabolites glycine and serine and log10 enzyme activity for hydroxypyruvate reductase (HPR) and glycolate oxidase (GOX). In each plot are reported the Spearman correlation value (rs) between the two variables and the variation explained by the specific linear model (R2). (This figure is available in colour at JXB online.)
Single MetabOxi variables are highly correlated with the genotypic variation in grain yield stability
Next, a correlation analysis was carried out on the control and drought values of the 111 MetabOxi variables and GY loss (Supplementary Table S9) to assess if single variables could be associated with yield stability of the 292 rice accessions.
A higher number of significant correlations (Bonferroni-corrected P-value <0.05) with GY loss were found using drought values of the MetabOxi variables (53) than the control (25) values (Supplementary Table S9). Under both treatments, positive correlations outnumbered the negative ones (23 out of 25 positive correlations under control and 48 out of 53 under drought). The variables that displayed the strongest correlations differed between the two treatments. Under control conditions (Table 2), erythritol showed the best correlation (rs=0.41) with GY loss, followed by a number of amino acids (Phe, Leu, and β-Ala as the top ones) and 2-amino-adipic acid, all displaying similar and positive values (rs ~0.30). The percentage of GY loss variance explained by the linear models (R2) under control conditions was low (<20%), with erythritol showing the highest contribution (17%).
Table 2.
MetabOxi variables displaying the best correlations between their control values and grain yield loss
| Variable | Control | |||||
|---|---|---|---|---|---|---|
| GY loss | Sam-Flow | |||||
| Spearman’s rho | Bonferroni-corrected P-value | R 2 | Spearman’s rho | Bonferroni-corrected P-value | R 2 | |
| Erythritol | 0.41 | 4.28E-11 | 0.17 | 0.75 | 5.17E-34 | 0.42* |
| Phenylalanine | 0.36 | 4.69E-08 | 0.13 | 0.57 | 7.20E-16 | 0.23* |
| Alanine, beta- | 0.33 | 1.75E-07 | 0.12 | 0.04 | 1 | 0.00 |
| Adipic acid, 2-amino- | 0.31 | 5.48E-07 | 0.11 | 0.11 | 1 | 0.01 |
| Leucine | 0.33 | 1.62E-06 | 0.11 | 0.27 | 0.0313 | 0.05* |
| Maltose | 0.31 | 1.65E-05 | 0.09 | 0.17 | 1 | 0.01 |
| Proline | 0.31 | 2.83E-05 | 0.09 | 0.21 | 1 | 0.00 |
| Lysine | 0.26 | 0.0005 | 0.07 | –0.06 | 1 | 0.02 |
| Glucosamine, N-acetyl- | 0.30 | 0.0010 | 0.07 | 0.76 | 7.95E-43 | 0.50* |
| Isoleucine | 0.25 | 0.0019 | 0.06 | 0.00 | 1 | 0.00 |
| Ribitol | 0.25 | 0.0021 | 0.06 | 0.43 | 5.03E-10 | 0.15* |
| Mannosamine, N-acetyl- | 0.26 | 0.0029 | 0.06 | 0.41 | 1.55E-08 | 0.13* |
| Ornithine | 0.23 | 0.0061 | 0.06 | –0.10 | 1 | 0.00 |
| Tyramine | 0.26 | 0.0063 | 0.05 | 0.44 | 2.96E-09 | 0.14* |
| Poly | –0.24 | 0.0092 | 0.05 | –0.01 | 1 | 0.00 |
List of the 15 best (highest significance) Spearman correlations between control values of the MetabOxi variables and grain yield loss (GY loss). Spearman rank correlation values and significance between the same variables and flowering at sampling (Sam-Flow) are displayed. R2, variance explained by the linear model created between the trait and each variable. An asterisk indicates a MetabOxi variable significantly correlated with both grain yield loss and flowering at sampling.
Under drought, correlation values strongly increased (Table 3; Supplementary Table S10), with 17 MetabOxi variables displaying higher values than the top one under control conditions. The two best correlations with GY loss under drought were displayed by the lipid peroxidation product MDA (rs=0.63) and the antioxidant enzyme DHAR (rs= –0.56), both showing a similar percentage of GY loss variance explained by their respective linear models (R2=0.38 and 0.37). Interestingly, DHAR was the only top ranked variable to show a negative correlation with GY loss. Under drought, many more amino acids than under control conditions ranked among the top correlated variables with GY loss (Thr, Arg, and Val, as the top ones). Again, all these amino acids displayed similar correlation values with GY loss (rs ~0.45), but the correlations under drought, as well as the R2 values of their linear models (~20%), were substantially higher than under control conditions.
Table 3.
MetabOxi variables displaying the best correlations between their drought values and grain yield loss
| Variable | Drought | |||||
|---|---|---|---|---|---|---|
| GY loss | Sam-Flow | |||||
| Spearman’s rho | Bonferroni-corrected P-value | R 2 | Spearman’s rho | Bonferroni-corrected P-value | R 2 | |
| MDA | 0.63 | 3.65E-30 | 0.38 | 0.23 | 0.1106 | 0.04 |
| DHAR | –0.56 | 5.81E-29 | 0.37 | –0.06 | 1 | 0.00 |
| Threonine | 0.48 | 6.17E-18 | 0.25 | 0.33 | 1.21E-06 | 0.11* |
| Arginine | 0.48 | 6.75E-18 | 0.25 | 0.36 | 1.73E-06 | 0.11* |
| Valine | 0.47 | 9.99E-17 | 0.24 | 0.39 | 2.40E-09 | 0.14* |
| Ornithine | 0.46 | 2.44E-16 | 0.23 | 0.35 | 1.79E-07 | 0.12* |
| Isoleucine | 0.46 | 1.13E-15 | 0.22 | 0.41 | 3.79E-12 | 0.18* |
| Serine | 0.46 | 2.03E-15 | 0.22 | 0.25 | 0.0248 | 0.05* |
| Methionine | 0.45 | 2.54E-15 | 0.22 | 0.41 | 1.34E-11 | 0.17* |
| Leucine | 0.45 | 7.23E-15 | 0.21 | 0.40 | 1.20E-12 | 0.19* |
| Glutamine | 0.44 | 8.17E-15 | 0.21 | 0.38 | 6.60E-11 | 0.16* |
| Phenylalanine | 0.44 | 1.96E-14 | 0.21 | 0.45 | 1.38E-16 | 0.24* |
| Tyrosine | 0.44 | 5.84E-14 | 0.20 | 0.39 | 1.65E-12 | 0.19* |
| Alanine, beta- | 0.43 | 3.82E-13 | 0.19 | 0.32 | 3.48E-08 | 0.13* |
| Lysine | 0.41 | 9.76E-13 | 0.19 | 0.35 | 1.39E-08 | 0.13* |
List of the 15 best (highest significance) Spearman correlations between drought values of the MetabOxi variables and grain yield loss (GY loss). Spearman rank correlation values and significance between the same variables and flowering at sampling (Sam-Flow) are displayed. R2, variance explained by the linear model created between the trait and each variable. An asterisk indicates a MetabOxi variable significantly correlated with both grain yield loss and flowering at sampling.
Considering the previously described significant and similar correlation values between GY loss and flowering under both treatments (Supplementary Fig. S2), we also decided to investigate if values of the MetabOxi variables were associated with differences in flowering among the accessions at sampling time. For this purpose, a new flowering variable (Sam-Flow) was created by subtracting the date of 50% flowering from the date of leaf sampling for every genotype (Supplementary Table S1), and correlation analysis was then performed between Sam-Flow and the control and drought values of the 111 MetabOxi variables (Supplementary Table S10). Interestingly, the top two correlated metabolites with GY loss under control conditions, erythritol and Phe, displayed significant and particularly high correlations (rs=0.75 and 0.57, respectively) with Sam-Flow. In contrast to the control conditions, the two top correlated variables with GY loss, MDA and DHAR, showed no significant correlation with Sam-Flow (Table 3).
Collectively, these results suggest that only drought values of the MetabOxi variables, and primarily of MDA and DHAR, are highly associated with GY loss performance across the 292 genotypes. In addition, the non-significant correlation of MDA and DHAR with Sam-Flow further validates their true association with GY loss only. Nevertheless, the trait prediction accuracy of these two variables remains below 40% of the GY loss phenotypic variance.
Prediction of grain yield loss by multivariate partial least squares regression models reveals the importance of leaf antioxidant system for yield stability
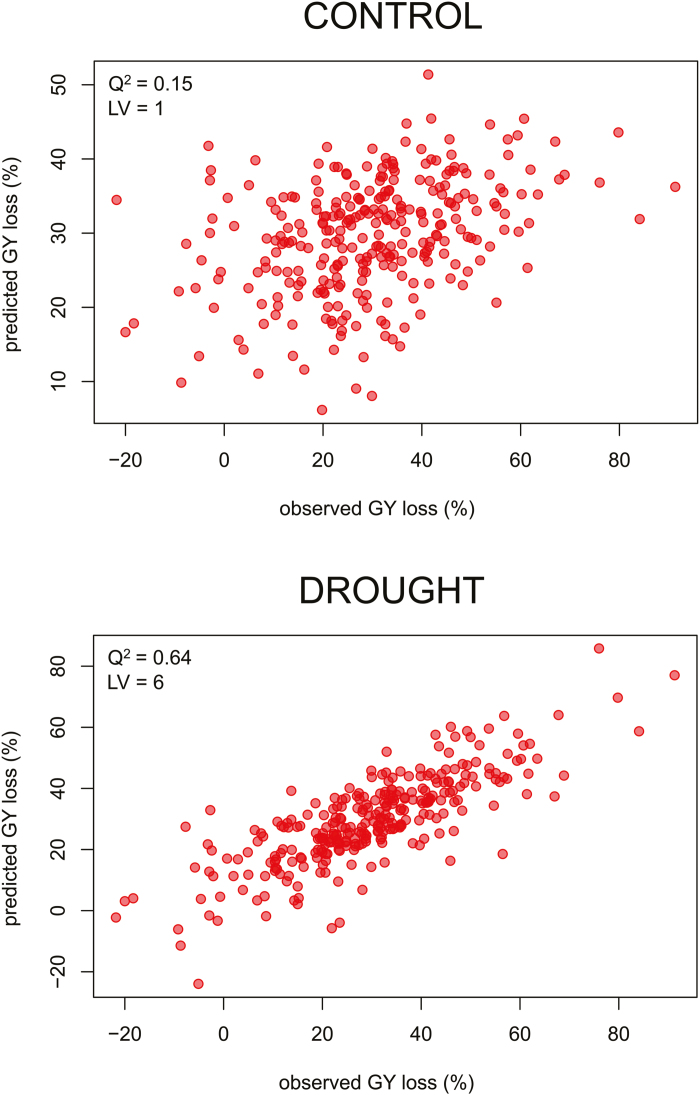
Two models based on multivariate PLSR were generated to predict GY loss performance of the population considering the 111 MetabOxi variables. In the first model, control values of variables were used for prediction of GY loss under drought. The best model (1LV; see Supplementary Fig. S5) showed a low predictability (Q2=0.15) (Fig. 4). This suggests that non-stressed values of flag leaf MetabOxi variables are not very accurate markers of tolerance/sensitivity for drought-induced GY loss across the population. In contrast, the best PLSR model based on values of the MetabOxi variables under drought conditions (6LVs; see Supplementary Fig. S5) displayed a high predictability (Q2=0.64) (Fig. 4), indicating a strong association between stress-induced changes in leaf metabolism/oxidative stress and GY loss.
Fig. 4.
Predicted versus observed values of the control and drought PLSR models for the prediction of grain yield loss in the 292 rice accessions. PLSR plot of the cross-validated models for GY loss based on the control (top) and drought (bottom) values of the 111 MetabOxi variables. Predictability (Q2) and linear latent variables (LVs) of the two models are reported. (This figure is available in colour at JXB online.)
Both the PLSR models were based on 10 different single submodels generated by the cross-validating procedure. By multiplying the 10 ranks of each MetabOxi variable in the single submodels, their overall ranking was calculated (Supplementary Tables S11, S12). A low rank-product implies that the variable has a high importance for the model. Among the top five predicting variables of the control model (Table 4), galactaric acid and erythritol ranked first and second, both showing a similarly low rank-product value (32 and 64, respectively). Interestingly, galactaric acid poorly correlates with GY loss, whereas erythritol, as mentioned before, showed the highest correlation with the trait under control conditions. Additionally, erythritol showed a strong correlation with Sam-Flow whereas galactaric acid did not (Table 4). The next highest ranking predictors of the model, 2-aminoadipic acid, Trp, and allantoin, showed a much lower importance than the first two, as represented by their high rank-product values. Interestingly, Trp, like erythritol, showed a strong positive correlation with Sam-Flow.
Table 4.
Best predicting variables of the control and drought PLSR models for the prediction of grain yield loss in the 292 rice accessions
| Control PLSR model | Drought PLSR model | ||||||
|---|---|---|---|---|---|---|---|
| Variable | Rank-product | GY loss | Sam-Flow | Variable | Rank-product | GY loss | Sam-Flow |
| r s | r s | r s | r s | ||||
| Galactaric acid | 32 | 0.09 | 0.00 | DHAR | 1 | –0.56 * | –0.06 |
| Erythritol | 64 | 0.41 * | 0.75 * | MDA | 1024 | 0.63 * | 0.23 |
| Adipic acid, 2-amino- | 9.841E+04 | 0.31 * | 0.11 | MDHAR | 5.905E+04 | –0.03 | 0.17 |
| Tryptophan | 7.680E+05 | 0.27 * | 0.73 * | TAC | 1.806E+07 | 0.08 | 0.07 |
| Allantoin | 2.540E+07 | 0.18 * | 0.18 | AO | 3.931E+07 | 0.22 * | –0.17 * |
Top five ranked predicting variables of the double cross-validated PLSR models for grain yield loss prediction based on control (left) and drought (right) values. Variables are ranked based on their rank-product value. Variables with the lower rank-product value are the ones with the larger discriminative power. rs, Spearman’s rho of correlation with grain yield loss (GY loss) and flowering at sampling (Sam-Flow). An asterisk indicates a significantly correlated variable.
In the drought-based PLSR model of GY loss, all the five top predicting variables are oxidative stress enzymes or markers (Table 4). The most important predicting variable of the model, by far, is the antioxidant enzyme DHAR that ranked first in all the 10 single submodels (rank-product=1). The second highest ranking predicting variable of the model is the lipid peroxidation product MDA that displayed a still relatively low rank-product value (1024) albeit already considerably higher than that of DHAR. DHAR and MDA are also the two variables that showed the highest correlation coefficients with GY loss, the first negative and the second positive. The model selected the antioxidant enzyme MDHAR and the level of TAC as third and fourth best predicting variables even if both poorly correlate with GY loss. The fifth most important predicting variable of the PLSR drought model, AO, is, like DHAR and MDHAR, an enzyme involved in the ascorbate–glutathione redox cycle. The high rank-product value displayed by MDHAR, TAC, and AO indicates that they have a substantially lower importance in the model than DHAR and MDA. Differently from the control model, the top predictors of the drought model did not show significant correlations with Sam-Flow (Table 4), except for AO that displayed a weak correlation.
PLSR prediction models of grain yield stability in the population highlighted that only the drought-stressed values of the MetabOxi variables could provide an accurate prediction of the trait. In addition, only antioxidant enzymes and oxidative stress markers ranked among the top ranked predicting variables of the drought model and they showed low correlations with Sam-Flow. This indicates that stress-induced alteration in the leaf oxidative stress status—much more than the metabolome—is tightly linked to the prevention of GY loss under drought.
Discussion
Drought affects flag leaf central metabolism and induces leaf senescence, protein degradation, and nitrogen recycling
Drought-induced stomatal closure reduces leaf photosynthetic activity, which induces metabolic reprogramming aimed to simultaneously adapt to the stressful condition and maintain active essential metabolic pathways (Obata and Fernie, 2012; Claeys and Inzé, 2013). In this study, the imposition of drought stress during flowering resulted in a population-wide alteration of flag leaf central metabolism and oxidative stress status. The stress induced an increase in the level of most metabolites, with a marked increase in the level of all the free amino acids and a, slightly less marked, increase of almost all the sugars, and decrease in the organic acids (Table 1).
The increase in amino acid levels is a response to water limitation that has been observed before in leaves of many plant species (Obata and Fernie, 2012; Krasensky and Jonak, 2012; Obata et al., 2015; Fàbregas and Fernie, 2019) when exposed to severe drought stress in the vegetative stage. This accumulation has been associated with protein catabolism induced by premature stress-induced leaf senescence (Araújo et al., 2011; Watanabe et al., 2013; Hildebrandt et al., 2015). The presence of a strong drought-induced catabolic activity is further supported by the large increase under drought in the level of two metabolites, allantoin and 2-aminoadipic acid (Table 1). Allantoin is an intermediate in purine catabolism known to be important for nitrogen remobilization and more recently postulated to have a role in stress tolerance by activating the production of abscisic acid (Watanabe et al. 2014). 2-Aminoadipic acid is the central metabolite involved in the plant catabolic pathway of lysine (Zhu et al., 2000).
Accumulation of specialized metabolites able to induce water retention and positive turgor pressure and to counteract the enhanced generation of ROS helps to protect cellular functions against the damage caused by drought-induced dehydration (Verslues and Juenger, 2011; Krasensky and Jonak, 2012; Nakabayashi and Saito, 2015). We found that the amino acid showing the highest population-wide increase was Pro, widely reported in the literature to accumulate under stressful conditions and considered as an osmolyte, ROS scavenger, and stabilizer of protein structure (Hare and Cress, 1997; Verslues and Juenger, 2011). Similarly, the highest increase in the sugars was displayed by two members of the raffinose family oligosaccharides (RFO), galactinol and raffinose. These two sugar alcohols have been reported to accumulate, like Pro, in leaves exposed to environmental stress and to exert an osmoprotective action and scavenging activity against hydroxyl radicals (Nishizawa et al., 2008; Van den Ende, 2013; Fàbregas and Fernie, 2019). Interestingly and contrary to the literature, in our study the strong increase in Pro, galactinol, and raffinose seemed not to be associated with a signature of stress tolerance. Indeed, drought values of these three metabolites neither showed negative correlations with GY loss (Supplementary Table S9) nor ranked as good model predictors for GY loss (Supplementary Table S12).
Only a few metabolite levels decreased under drought, and most of them belonged to the class of organic acids, with TCA cycle (isocitric and citric acid) and glycolysis (glyceric acid-3-phosphate and phosphoenolpyruvic acid) intermediates displaying the highest decrease (Table 1). The TCA cycle and glycolysis are two interconnected pathways involved in the production of metabolic intermediates used in biosynthesis elsewhere in the cell (Araujo et al., 2012). Under drought, the strong reduction in the levels of metabolites of these two pathways could be due to an overall reduced biosynthetic activity of stressed leaves. This indirectly supports the fact that the increased levels of all the amino acids must be due to stress-induced protein degradation.
Drought stress alters the relationships between central metabolic pathways
Consistent with previous studies, hierarchical clustering analysis (Fig. 1) displayed the tendency of metabolites of the same class (amino acids, organic acids, and sugars) to cluster together as evidence of commonly shared biosynthetic or catabolic pathways (Carreno-Quintero et al., 2012; Obata and Fernie, 2012; Riedelsheimer et al., 2012; Obata et al., 2015). Drought induced a unique stress-specific cluster including allantoin and four amino acids, Asp, Glu, Gly, and Ser (yellow in Fig. 1) that separated from the main cluster of the amino acids (red). Above, we have already discussed the role of allantoin in nitrogen remobilization as an intermediate product of purine catabolism. A similar role is exerted by two amino acids of the cluster, Asp and Glu, both involved in nitrogen remobilization as major long-distance phloem transport nitrogen forms (after their conversion to Asn and Gln) in senescent leaves (Watanabe et al., 2013; Avila-Ospina et al., 2014). The other two amino acids of the stress-specific cluster, Gly and Ser, are well known markers of photorespiration, as an increased activity of that process results in higher production of these compounds (Bauwe et al., 2010; Maurino and Peterhansel, 2010). The presence of this cluster, under drought conditions only, further supports the idea that physiological processes such as stress-induced senescence, nutrient recycling, and photorespiration are co-ordinately enhanced by drought and have an impact on leaf central metabolism (Avila-Ospina et al., 2014; Hildebrandt et al., 2015; Hodges et al., 2016). Confirming this hypothesis are the increased negative correlations that we found under drought between the amino acid clusters (red and yellow in Fig. 1) and the TCA cycle cluster (light blue). These negative correlations link high levels of photorespiration and presence of leaf senescence to low levels of mitochondrial respiration under drought stress (Atkin and Macherel, 2009).
Photorespiration and ROS scavenging activity are linked with metabolic and oxidative damage induced by drought
In addition to metabolic alterations, the drought-induced decrease in carbon assimilation results in enhanced generation of ROS in leaf cells (Suzuki et al., 2012; Noctor et al., 2014). Overall, we found that the activity of two photorespiratory enzymes, HPR and GOX, strongly positively correlated with the main amino acid clusters (Fig. 2). More specifically, the amounts of Gly and Ser in the flag leaf of the 292 genotypes almost perfectly correlated with the activity of the photorespiratory enzymes HPR and GOX under stress and showed very high correlations also under control conditions (Fig. 3). These results support the hypothesis of a direct interaction between photorespiration and amino acid metabolism (Fernie et al., 2013; Hodges et al., 2016), and they help to shed light on the contribution of the photorespiratory pathway to the overall supply of Gly and Ser in rice leaves under stressed and non-stressed conditions (Ros et al., 2013). The strength of these correlations (Fig. 3) highlights the importance of photorespiration in rice leaf central metabolism and suggests that its reduction to improve crop yield (Betti et al., 2016; Walker et al., 2016; South et al., 2019; Weber and Bar-Even, 2019) should be approached with great care as this could lead to leaf metabolic impairments resulting in even higher yield losses under drought.
In the presence of limited CO2 supply, the photorespiratory pathway has often been considered as a protective mechanism able to reduce ROS generation, and the consequent oxidative damage, by consuming the excess of energy produced in the chloroplast (Wingler et al., 2000; Takahashi and Badger, 2011; Hodges et al., 2016). However, the MDA values detected in this study do not support the ROS protective action of photorespiration. MDA is a lipid peroxidation product widely accepted as a marker of membrane oxidative damage (Møller et al., 2007). In the present study, under stress conditions, MDA values display a correlation pattern with the main metabolic clusters very similar to that of GOX and HPR (Fig. 2). This suggests that during enhanced photorespiration, a high rate of H2O2 production in the peroxisomes results in oxidation of cellular membrane lipids as previously described in maize leaves exposed to drought (Avramova et al., 2017).
Plants have evolved a complex enzymatic and non-enzymatic antioxidative system to protect cells from the enhanced generation of ROS and their oxidative action (You and Chan, 2015). Under drought stress, we found that only the activity of some antioxidant enzymes, DHAR, CAT, and APX, showed a strong negative correlation with the metabolic clusters associated with stress (Fig. 2), suggesting a specialized role for these enzymes in counteracting the adverse effects induced by drought. CAT and APX are considered the main antioxidant enzymes involved in H2O2 removal in leaves (Noctor et al., 2014). In particular, CAT directly converts H2O2 to water and oxygen (Mhamdi et al., 2010) and exerts most of its activity in the peroxisomes, where this enzyme counteracts H2O2 generation by GOX during photorespiration (Bauwe et al., 2010). APX protects chloroplast membranes by reducing H2O2 to water through the oxidation of ascorbate (Das and Roychoudhury, 2014). An efficient regeneration of reduced ascorbate is therefore essential for H2O2 scavenging. DHAR regenerates the oxidized ascorbate by using reduced glutathione as electron donor (Das and Roychoudhury, 2014), and the enzyme participates, like APX, in the ascorbate–glutathione pathway, the main redox hub in plants (Foyer and Noctor, 2011). CAT and APX activity displayed a strong population-wide increase under drought stress, whereas, surprisingly, DHAR activity did not (Table 1). This could indicate that DHAR is not as ubiquitously important as CAT and APX, which are up-regulated across the whole population, or, alternatively, other regulatory mechanisms such as post-translational modification or allosteric interactions could control DHAR in vivo, but are not discriminated in the in vitro assay (Sulpice et al., 2010). Nevertheless, differences in in vitro—analysed—DHAR activity in the vegetative stage between rice genotypes differing in drought tolerance have been described before (Selote and Khanna-Chopra, 2004). Conversely, and even more surprisingly, SOD, the enzyme showing the highest drought-induced increase (Table 1), did not show any strong positive or negative association with the metabolic clusters representative of stress (Fig. 2). In plants, SODs, localized in chloroplasts, mitochondria, and cytosol, catalyse the conversion of the highly oxidative anion superoxide (O2–) to the less harmful H2O2 (Halliwell, 2006). Apparently, under drought stress, SOD activity increases independent of the genotype, thus representing a commonly shared mechanism to generate high amounts of H2O2 that, in turn, can be detoxified by more genotype-dependent, localized, and effective antioxidative responses.
Multivariate modelling of GY loss reveals that the enzymes of the ascorbate–glutathione cycle are essential for GY stability under drought
In contrast to univariate statistics, multivariate analysis considers the simultaneous relationships between all the variables of a given data set, thus increasing its predictive power, and it was used before in metabolomics-based plant trait prediction (Meyer et al., 2007; Sulpice et al., 2009, 2013; Steinfath et al., 2010; Mumm et al., 2016). The PLSR model based on control values of the MetabOxi variables showed low predictability for GY loss (Fig. 4). This observation suggests that basal levels of flag leaf primary metabolites, oxidative stress markers/enzymes, and their interactions have little influence in determining the genotypic GY loss sensitivity upon the introduction of drought stress. Despite its low predictability, the PLSR control model suggested the highest importance for erythritol and galactaric acid. Consistent with the literature (Obata and Fernie, 2012; Fàbregas and Fernie, 2019), we found increased levels of erythritol under drought (Table 1), but only its control values displayed a positive correlation with drought-induced GY loss and an even stronger correlation with flowering time differences (Tables 2, 4). Considering the almost perfect correlation between flowering time differences between the two treatments and its influence on GY loss (Supplementary Fig. S2), it seems possible that the high ranking of erythritol in the control model reflects the relative importance of flowering differences at the time of stress imposition on GY loss performance of the 292 accessions. On the other hand, control values of the other equally important predictor of the model, galactaric acid, were not significantly correlated with GY loss, or with flowering differences (Tables 2, 4). In a previous study on 21 rice genotypes, galactaric acid was described to correlate positively with plant growth, under both control and drought stress conditions (Degenkolbe et al., 2013). Its presence as top predictor in the control PLSR model might reveal a hidden link between genotypic-induced differences in plant growth rate under optimal conditions and GY loss tolerance under drought stress.
In contrast to the control model, the PLSR model based on drought values of the MetabOxi variables showed high predictability of GY loss (Fig. 4). This clearly indicates that the stress-induced interaction between metabolites and oxidative stress markers/enzymes is tightly linked to the GY loss performance of the population. The top predictors of the drought model highlighted the importance of the enzymatic reduction of oxidized ascorbate as a key mechanism to reduce the negative effect of oxidative stress on GY loss (Table 4). In particular, DHAR outclassed all the other predictors in terms of model contribution. Above, we have already discussed the role of this enzyme in counteracting the adverse metabolic changes induced by drought (Fig. 2; Table 3). Interestingly, another antioxidant enzyme, MDHAR, ranked third in the model. MDHAR, just like DHAR, regenerates reduced ascorbate so that it can be used again by APX in the scavenging of H2O2 (Das and Roychoudhury, 2014). MDHAR, by the direct conversion of monodehydroascorbate to ascorbate before it spontaneously converts to dehydroascorbate (Smirnoff, 2000), might reduce the DHAR workload, thus increasing the efficiency of ascorbate reduction. For this reason, MDHAR activity under drought might have been selected as a good predictor by the PLSR model despite its null correlation with GY loss (Table 4), and this demonstrates the value of this modelling approach above simple correlation analysis. The presence of MDA among the top three predictors of the drought model reinforces the link between leaf oxidative damage and GY loss. MDA values under drought showed the highest positive correlation with GY loss (Tables 3, 4) and displayed a strong correlation with photorespiratory activity and leaf senescence (Fig. 2). All these findings suggest that MDA represents the best biomarker of GY loss sensitivity under drought and that oxidative damage of leaf membrane lipids is among the most damaging processes caused by drought.
Conclusions
The metabolic and oxidative stress profiles of the rice flag leaf changed dramatically during drought stress in the reproductive stage. These changes proved to be highly informative for the grain yield loss sensitivity of the different rice accessions at harvest time. Multivariate modelling of grain yield loss revealed that the co-ordinated activity of enzymes involved in the ascorbate–glutathione cycle, and among them primarily DHAR, is an essential mechanism of drought tolerance in rice. Our study suggests that the co-expression of specific antioxidant enzymes of the ascorbate–glutathione cycle (DHAR and MDHAR) could represent a robust mechanism of tolerance that can minimize grain yield losses under drought. Finally, the genetic diversity of the 292 rice accessions used in this study offers the possibility to find genomic associations for the identified key enzymatic and metabolic determinants of grain yield loss under drought. These associations could be developed into genetic markers to be used in breeding for grain yield stability under drought in rice.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Experimental design of the field trial.
Fig. S2. Correlations between flowering differences and grain yield loss.
Fig. S3. Principal component analysis based on the 111 MetabOxi variables.
Fig. S4. Hierarchical clustering results for the 91 metabolites under the two treatments.
Fig. S5. Predictability of the PLSR models with increasing LVs
Table S1. Percentage of grain yield loss (GY loss %), days to 50% flowering (Flow), and leaf sampling date minus date of 50% flowering (Sam-Flow) under control (CON) and drought stress (DRO) conditions, for the 292 genotypes.
Table S2. Leaf values (log10 transformed) of the 111 MetabOxi variables in the 292 genotypes of the population under control and drought stress conditions.
Table S3. List of the 111 MetabOxi variables considered in this study.
Table S4. Effect of treatment on the 111 MetabOxi variables.
Table S5. Spearman rank correlation matrix of the 91 metabolites under control conditions.
Table S6. Spearman rank correlation matrix of the 91 metabolites under drought conditions.
Table S7. Spearman rank correlation matrix of the 21 oxidative stress markers/enzymes and the 91 metabolites under control conditions.
Table S8. Spearman rank correlation matrix of the 21 oxidative stress markers/enzymes and the 91 metabolites under drought conditions.
Table S9. List of the 111 MetabOxi variables ranked by significance of their Spearman rank correlation with grain yield loss (GY loss) under control and drought conditions.
Table S10. List of the 111 MetabOxi variables ranked by significance of their Spearman rank correlation with flowering at sampling (Sam-Flow) under control and drought conditions.
Table S11. Ranking of the cross-validated PLSR model based on control values of the variables.
Table S12. Ranking of the cross-validated PLSR model based on drought values of the variables.
Acknowledgements
This work is part of the Growing Rice like Wheat research programme financially supported by an anonymous private donor, via Wageningen University Fund, for the first author’s PhD fellowship. GC-MS analysis was enabled by the Transnational Access capacities of the European Plant Phenotyping Network (EPPN, grant agreement no. 284443) funded by the FP7 Research Infrastructures Programme of the European Union. The authors wish to thank Andrea Apelt (IPK-Gatersleben) for excellent technical assistance during the GC-MS analysis.
References
- AbdElgawad H, Zinta G, Hegab MM, Pandey R, Asard H, Abuelsoud W. 2016. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Frontiers in Plant Science 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi H. 1984. Catalase in vitro. Methods in Enzymology 105, 121–126. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Nunes-Nesi A, Nikoloski Z, Sweetlove LJ, Fernie AR. 2012. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant, Cell & Environment 35, 1–21. [DOI] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. 2011. Protein degradation—an alternative respiratory substrate for stressed plants. Trends in Plant Science 16, 489–498. [DOI] [PubMed] [Google Scholar]
- Asada K. 2006. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiology 141, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin OK, Macherel D. 2009. The crucial role of plant mitochondria in orchestrating drought tolerance. Annals of Botany 103, 581–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. 2014. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany 65, 3799–3811. [DOI] [PubMed] [Google Scholar]
- Avramova V, AbdElgawad H, Vasileva I, Petrova AS, Holek A, Mariën J, Asard H, Beemster GT. 2017. High antioxidant activity facilitates maintenance of cell division in leaves of drought tolerant maize hybrids. Frontiers in Plant Science 8, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. 2010. Photorespiration: players, partners and origin. Trends in Plant Science 15, 330–336. [DOI] [PubMed] [Google Scholar]
- Baxter A, Mittler R, Suzuki N. 2014. ROS as key players in plant stress signalling. Journal of Experimental Botany 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Benzie IF, Strain JJ. 1999. Ferric reducing/antioxidant power assay: direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration. Methods in Enzymology 299, 15–27. [DOI] [PubMed] [Google Scholar]
- Betti M, Bauwe H, Busch FA, et al.. 2016. Manipulating photorespiration to increase plant productivity: recent advances and perspectives for crop improvement. Journal of Experimental Botany 67, 2977–2988. [DOI] [PubMed] [Google Scholar]
- Biswal AK, Kohli A. 2013. Cereal flag leaf adaptations for grain yield under drought: knowledge status and gaps. Molecular Breeding 31, 749–766. [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CW, Mumm R, Bouwmeester H, Visser RG, Keurentjes JJ. 2012. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiology 158, 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H, Inzé D. 2013. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiology 162, 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA. 2009. TargetSearch—a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K, Roychoudhury A. 2014. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Frontiers in Environmental Science 2, 53. [Google Scholar]
- Degenkolbe T, Do PT, Kopka J, Zuther E, Hincha DK, Köhl KI. 2013. Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS One 8, e63637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhindsa RS, Plumb-Dhindsa PL, Reid DM. 1982. Leaf senescence and lipid peroxidation: effects of some phytohormones, and scavengers of free radicals and singlet oxygen. Physiologia Plantarum 56, 453–457. [Google Scholar]
- Drotar A, Phelps P, Fall R. 1985. Evidence for glutathione peroxidase activities in cultured plant cells. Plant Science 42, 35–40. [Google Scholar]
- Fàbregas N, Fernie AR. 2019. The metabolic response to drought. Journal of Experimental Botany 33, 585–588. [DOI] [PubMed] [Google Scholar]
- Feierabend J, Beevers H. 1972. Developmental studies on microbodies in wheat leaves: I. Conditions influencing enzyme development. Plant Physiology 49, 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez O, Urrutia M, Bernillon S, Giauffret C, Tardieu F, Le Gouis J, Langlade N, Charcosset A, Moing A, Gibon Y. 2016. Fortune telling: metabolic markers of plant performance. Metabolomics 12, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Bauwe H, Eisenhut M, et al. 2013. Perspectives on plant photorespiratory metabolism. Plant Biology 15, 748–753. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology 155, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. 1974. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. Journal of Biological Chemistry 249, 7130–7139. [PubMed] [Google Scholar]
- Halliwell B. 2006. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiology 141, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA. 1997. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regulation 21, 79–102. [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araújo WL, Braun HP. 2015. Amino acid catabolism in plants. Molecular Plant 8, 1563–1579. [DOI] [PubMed] [Google Scholar]
- Hodges M, Dellero Y, Keech O, Betti M, Raghavendra AS, Sage R, Zhu XG, Allen DK, Weber AP. 2016. Perspectives for a better understanding of the metabolic integration of photorespiration within a complex plant primary metabolism network. Journal of Experimental Botany 67, 3015–3026. [DOI] [PubMed] [Google Scholar]
- Hodges DM, Delong JM, Forney CF, Prange RK. 1999. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. [DOI] [PubMed] [Google Scholar]
- Horling F, Lamkemeyer P, König J, Finkemeier I, Kandlbinder A, Baier M, Dietz KJ. 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiology 131, 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel I, Pantin F, Sulpice R, et al.. 2010. Arabidopsis plants acclimate to water deficit at low cost through changes of carbon usage: an integrated perspective using growth, metabolite, enzyme, and gene expression analysis. Plant Physiology 154, 357–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam NN, Struik PC, Rebolledo MC, Yin X, Jagadish SVK. 2018. Genome-wide association reveals novel genomic loci controlling rice grain yield and its component traits under water-deficit stress during the reproductive stage. Journal of Experimental Botany 69, 4017–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasensky J, Jonak C. 2012. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. Journal of Experimental Botany 63, 1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Bohra A, Pandey AK, Pandey MK, Kumar A. 2017. Metabolomics for plant improvement: status and prospects. Frontiers in Plant Science 8, 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar KB, Khan PA. 1983. Age-related changes in catalase and peroxidase activities in the excised leaves of Eleusine coracana Gaertin. cv PR 202 during senescence. Experimental Gerontology 18, 409–417. [DOI] [PubMed] [Google Scholar]
- Levine RL, Williams JA, Stadtman ER, Shacter E. 1994. Carbonyl assays for determination of oxidatively modified proteins. Methods in Enzymology 233, 346–357. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry 193, 265–275. [PubMed] [Google Scholar]
- Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, Johansson M, Holmgren A. 2001. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. Journal of Biological Chemistry 276, 26269–26275. [DOI] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. 2010. Photorespiration: current status and approaches for metabolic engineering. Current Opinion in Plant Biology 13, 249–256. [DOI] [PubMed] [Google Scholar]
- Mevik B-H, Wehrens R. 2007. The pls package: principle component and partial least squares regression in R. Journal of Statistical Software 18, 1–24. [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, et al. 2007. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G. 2010. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. Journal of Experimental Botany 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. 2010. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell & Environment 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. 2011. ROS signaling: the new wave? Trends in Plant Science 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. 2007. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology 58, 459–481. [DOI] [PubMed] [Google Scholar]
- Mumm R, Hageman JA, Calingacion MN, et al.. 2016. Multi-platform metabolomics analyses of a broad collection of fragrant and non-fragrant rice varieties reveals the high complexity of grain quality characteristics. Metabolomics 12, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshed R, Lopez-Lauri F, Sallanon H. 2008. Microplate quantification of enzymes of the plant ascorbate–glutathione cycle. Analytical Biochemistry 383, 320–322. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Saito K. 2015. Integrated metabolomics for abiotic stress responses in plants. Current Opinion in Plant Biology 24, 10–16. [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Shigeoka S. 2008. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiology 147, 1251–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Veljovic-Jovanovic S, Driscoll S, Novitskaya L, Foyer CH. 2002. Drought and oxidative load in the leaves of C3 plants: a predominant role for photorespiration? Annals of Botany 89, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH. 2014. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiology 164, 1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Fernie AR. 2012. The use of metabolomics to dissect plant responses to abiotic stresses. Cellular and Molecular Life Sciences 69, 3225–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Witt S, Lisec J, Palacios-Rojas N, Florez-Sarasa I, Yousfi S, Araus JL, Cairns JE, Fernie AR. 2015. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiology 169, 2665–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura JB. 2012. Phenotyping for drought tolerance in grain crops: when is it useful to breeders? Functional Plant Biology 39, 851. [DOI] [PubMed] [Google Scholar]
- Riedelsheimer C, Brotman Y, Méret M, Melchinger AE, Willmitzer L. 2013. The maize leaf lipidome shows multilevel genetic control and high predictive value for agronomic traits. Scientific Reports 3, 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE. 2012. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nature Genetics 44, 217–220. [DOI] [PubMed] [Google Scholar]
- Riewe D, Grosman L, Zauber H, Wucke C, Fernie AR, Geigenberger P. 2008. Metabolic and developmental adaptations of growing potato tubers in response to specific manipulations of the adenylate energy status. Plant Physiology 146, 1579–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Jeon HJ, Lisec J, Heuermann MC, Schmeichel J, Seyfarth M, Meyer RC, Willmitzer L, Altmann T. 2016. A naturally occurring promoter polymorphism of the Arabidopsis FUM2 gene causes expression variation, and is associated with metabolic and growth traits. The Plant Journal 88, 826–838. [DOI] [PubMed] [Google Scholar]
- Riewe D, Koohi M, Lisec J, Pfeiffer M, Lippmann R, Schmeichel J, Willmitzer L, Altmann T. 2012. A tyrosine aminotransferase involved in tocopherol synthesis in Arabidopsis. The Plant Journal 71, 850–859. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Lodeyro A, Poli HO, et al.. 2007. Transgenic tobacco plants overexpressing chloroplastic ferredoxin-NADP(H) reductase display normal rates of photosynthesis and increased tolerance to oxidative stress. Plant Physiology 143, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Cascales-Miñana B, Segura J, Anoman AD, Toujani W, Flores-Tornero M, Rosa-Tellez S, Muñoz-Bertomeu J. 2013. Serine biosynthesis by photorespiratory and non-photorespiratory pathways: an interesting interplay with unknown regulatory networks. Plant Biology 15, 707–712. [DOI] [PubMed] [Google Scholar]
- Sandhu N, Singh A, Dixit S, Sta Cruz MT, Maturan PC, Jain RK, Kumar A. 2014. Identification and mapping of stable QTL with main and epistasis effect on rice grain yield under upland drought stress. BMC Genetics 15, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitzguebel JP, Siegenthaler PA. 1984. Purification of peroxisomes and mitochondria from spinach leaf by Percoll gradient centrifugation. Plant Physiology 75, 670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote DS, Khanna-Chopra R. 2004. Drought-induced spikelet sterility is associated with an inefficient antioxidant defence in rice panicles. Physiologia Plantarum 121, 462–471. [Google Scholar]
- Skirycz A, De Bodt S, Obata T, et al.. 2010. Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiology 152, 226–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnoff N. 2000. Ascorbic acid: metabolism and functions of a multi-facetted molecule. Current Opinion in Plant Biology 3, 229–235. [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR. 2019. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363, eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinfath M, Gärtner T, Lisec J, Meyer RC, Altmann T, Willmitzer L, Selbig J. 2010. Prediction of hybrid biomass in Arabidopsis thaliana by selected parental SNP and metabolic markers. Theoretical and Applied Genetics 120, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Nikoloski Z, Tschoep H, et al. 2013. Impact of the carbon and nitrogen supply on relationships and connectivity between metabolism and biomass in a broad panel of Arabidopsis accessions. Plant Physiology 162, 347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, et al. 2009. Starch as a major integrator in the regulation of plant growth. Proceedings of the National Academy of Sciences, USA 106, 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, et al.. 2010. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. The Plant Cell 22, 2872–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G. 2012. ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell & Environment 35, 259–270. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Badger MR. 2011. Photoprotection in plants: a new light on photosystem II damage. Trends in Plant Science 16, 53–60. [DOI] [PubMed] [Google Scholar]
- Van Breusegem F, Dat JF. 2006. Reactive oxygen species in plant cell death. Plant Physiology 141, 384–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W. 2013. Multifunctional fructans and raffinose family oligosaccharides. Frontiers in Plant Science 4, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad R, Lafitte HR, Atlin GN. 2007. Response to direct selection for grain yield under drought stress in rice. Crop Science 47, 285–293. [Google Scholar]
- Verslues PE, Juenger TE. 2011. Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Current Opinion in Plant Biology 14, 240–245. [DOI] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR. 2016. The costs of photorespiration to food production now and in the future. Annual Review of Plant Biology 67, 107–129. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Balazadeh S, Tohge T, Erban A, Giavalisco P, Kopka J, Mueller-Roeber B, Fernie AR, Hoefgen R. 2013. Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiology 162, 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Matsumoto M, Hakomori Y, Takagi H, Shimada H, Sakamoto A. 2014. The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant, Cell & Environment 37, 1022–1036. [DOI] [PubMed] [Google Scholar]
- Weber APM, Bar-Even A. 2019. Update: improving the efficiency of photosynthetic carbon reactions. Plant Physiology 179, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC. 2000. Photorespiration: metabolic pathways and their role in stress protection. Philosophical Transactions of the Royal Society B: Biological Sciences 355, 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolosiuk RA, Crawford NA, Yee BC, Buchanan BB. 1979. Isolation of three thioredoxins from spinach leaves. Journal of Biological Chemistry 254, 1627–1632. [PubMed] [Google Scholar]
- Xu S, Xu Y, Gong L, Zhang Q. 2016. Metabolomic prediction of yield in hybrid rice. The Plant Journal 88, 219–227. [DOI] [PubMed] [Google Scholar]
- Yoshida S. 1972. Physiological aspects of grain yield. Annual Review of Plant Physiology 23, 437–464. [Google Scholar]
- Yoshimura K, Ishikawa T, Nakamura Y, Tamoi M, Takeda T, Tada T, Nishimura K, Shigeoka S. 1998. Comparative study on recombinant chloroplastic and cytosolic ascorbate peroxidase isozymes of spinach. Archives of Biochemistry and Biophysics 353, 55–63. [DOI] [PubMed] [Google Scholar]
- You J, Chan Z. 2015. ROS regulation during abiotic stress responses in crop plants. Frontiers in Plant Science 6, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhang J, Shen J, Silva A, Dennis DA, Barrow CJ. 2006. A simple 96-well microplate method for estimation of total polyphenol content in seaweeds. Journal of Applied Phycology 18, 445–450. [Google Scholar]
- Zhu X, Tang G, Galili G. 2000. The catabolic function of the alpha-aminoadipic acid pathway in plants is associated with unidirectional activity of lysine-oxoglutarate reductase, but not saccharopine dehydrogenase. Biochemical Journal 351, 215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinta G, AbdElgawad H, Domagalska MA, Vergauwen L, Knapen D, Nijs I, Janssens IA, Beemster GT, Asard H. 2014. Physiological, biochemical, and genome-wide transcriptional analysis reveals that elevated CO2 mitigates the impact of combined heat wave and drought stress in Arabidopsis thaliana at multiple organizational levels. Global Change Biology 20, 3670–3685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.