Summary
Mechanisms that control aging are important yet poorly defined. To discover longevity control genes, we performed a forward genetic screen for delayed reproductive aging in C. elegans. Here we show that am117 is a nonsense mutation in the phm-2 gene, which encodes a protein homologous to human scaffold attachment factor B. phm-2(lf) mutant worms have an abnormal pharynx grinder, which allows live bacteria to accumulate in the intestine. This defect shortens lifespan on highly pathogenic bacteria but extends lifespan and health span on the standard E. coli diet by activating innate immunity pathways that lead to bacterial avoidance behavior and dietary restriction. eat-2(lf) mutants displayed a similar phenotype, indicating accumulation of live bacteria also triggers extended longevity in this mutant. The analysis of phm-2 elucidates connections between pathogen response and aging by defining a mechanism of longevity extension in C. elegans - bacterial colonization, innate immune activation and bacterial avoidance behavior.
Keywords: phm-2, eat-2, Innate immunity, Bacterial avoidance, Dietary restriction, C. elegans, Aging and lifespan
Graphical Abstract
eTOC Blurb
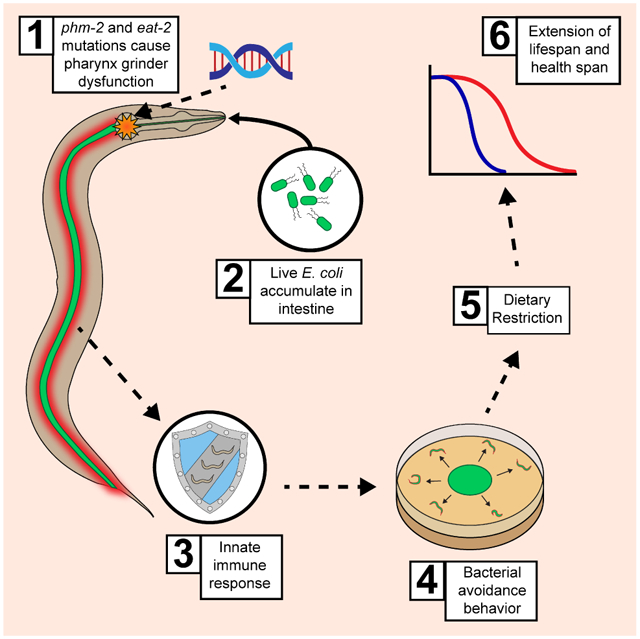
Kumar et al. link immunity and aging in C. elegans. The authors characterize phm-2 and eat-2, mutants that allow live bacteria to accumulate in the intestine, causing delayed reproductive and somatic aging. The mechanism combines molecular immune activation and behavioral food avoidance, leading to dietary restriction and extended lifespan.
Introduction
Studies of the microbiome of mammals have begun to elucidate the complex interactions between animal hosts and bacteria, ranging from beneficial commensals to deleterious pathogens. There is currently little information about how these interactions affect aging, which is characterized by progressive degenerative changes of tissue structure and function that impair physiology and ultimately lead to death. Caenorhabditis elegans is a terrestrial, non-parasitic nematode that is a powerful genetic system for studies of aging. During its 16-day lifespan, C. elegans displays a wide range of age-related changes such as reproductive and musculoskeletal degeneration (Brenner, 1974; Johnson and Wood, 1982; Klass, 1977). Tests of environmental, genetic, and pharmacologic interventions have led to the discovery and characterization of a relatively small number of endogenous pathways that can be modified to extend lifespan and health span. Dietary restriction is the original lifespan extending intervention; first described in rodents (McCay et al., 1935), it is now established to work in a wide range of organisms including C. elegans. Pathways regulated by dietary restriction are beginning to be defined and may include the target of rapamycin (TOR) energy sensing pathway that controls longevity (Kapahi et al., 2010). The insulin/insulin-like growth factor-1 signaling (IIS) pathway plays an evolutionarily conserved role in C. elegans and other animals (Guarente and Kenyon, 2000; Kenyon, 2010). Diminished mitochondrial function can extend lifespan, suggesting that wild-type (WT) levels of mitochondrial activity promote rapid aging (Hansen et al., 2005; Lakowski and Hekimi, 1996; Lee et al., 2003). The identification of additional mechanisms that extend lifespan is an important goal.
Here we identify phm-2 as a gene that influences C. elegans aging and demonstrate that it functions by a surprising mechanism: phm-2(lf) mutants are colonized by bacteria, which triggers an innate immune response that includes behavioral avoidance of bacterial food. Hughes et. al. (2011) identified the am117 mutation in a forward genetic screen for delayed reproductive aging, and here we demonstrated that am117 also delayed somatic aging, displayed a scrawny body morphology, and avoided the bacterial lawn. Interaction studies with other aging mutations revealed a strong interaction with eat-2, which has been interpreted as a genetic model of dietary restriction. We used positional cloning approaches to identify the affected gene as phm-2, previously defined by two mutations that were not molecularly identified (Avery, 1993a). PHM-2 contains two highly conserved protein domains: a SAP domain and a RRM_SAF domain. Both of these domains are also present in the human protein scaffold attachment factor B (SAFB). phm-2 mutants have a defective pharynx grinder (Avery, 1993a) and are hypersensitive to pathogenic bacteria because the grinder defect allows live bacteria to enter the intestinal lumen (Darby et al., 2002; Labrousse et al., 2000; Portal-Celhay et al., 2012; Smith et al., 2002). Thus, we hypothesized that in standard culture conditions, phm-2 mutants allow live E. coli to enter the intestinal lumen, resulting in mild pathogenicity and bacterial avoidance behavior. Consistent with this model, we demonstrated that live E. coli accumulated in the intestine of phm-2(lf) mutant animals, and phm-2(lf) mutants cultured on live E. coli displayed transcriptional changes and molecular events typical of bacterial infections. This response is important for the delayed aging phenotype, since culture on non-pathogenic bacteria abrogated the scrawny body morphology and aging phenotypes.
eat-2(lf) mutants have been used frequently to analyze dietary restriction in worms since pioneering studies by Lakowski and Hekimi (1998). Here we demonstrate eat-2(lf) mutants accumulated live E. coli in the intestine, indicating that they have a defect in pharynx grinder function, and eat-2(lf) mutants displayed activation of innate immunity and bacterial avoidance behavior. Furthermore, when eat-2(lf) mutants were cultured on nonpathogenic bacteria, the phenotypes were suppressed. Thus, eat-2 and phm-2 mutants are not examples of simple dietary restriction, but are a combination of bacterial colonization, innate immune activation, bacterial avoidance behavior and dietary restriction – an unexpected mechanism of lifespan extension in C. elegans.
Results
am117 mutant hermaphrodites displayed delayed reproductive and somatic aging, bacterial avoidance behavior, decreased body size, and increased stress resistance.
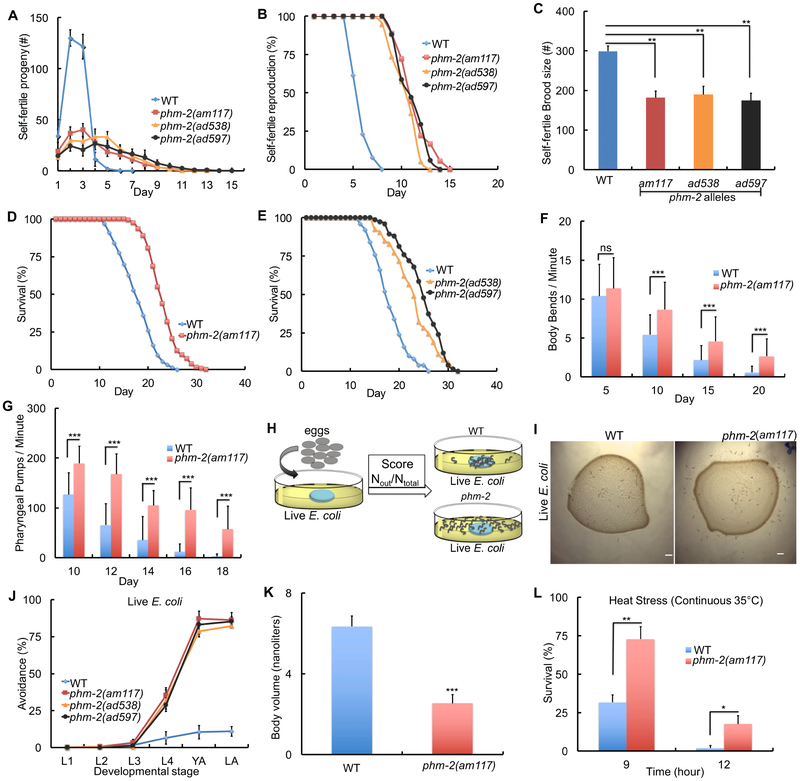
The am117 mutation was identified in a forward genetic screen for mated hermaphrodites that display extended reproductive spans (Hughes et al., 2011). The screen was conducted using standard culture conditions: a small lawn of live E. coli OP50 on NGM dishes at 20°C. Similar to mated hermaphrodites, self-fertile am117 hermaphrodites displayed reduced early progeny production and increased late progeny production, resulting in a reduced total brood size and an extended reproductive span (Figure 1A-C). To investigate somatic aging, we analyzed adult lifespan. am117 mutant animals displayed a significant extension of mean (34%) and maximum (24%) lifespan compared to wild type (Figure 1D, Table S1). We define maximum adult lifespan as the average lifespan of the 10% of the population that are longest lived. The am117 extended lifespan phenotype was also observed at 15°C and 25°C, demonstrating that the effect is not temperature dependent (Figure S1A-B, Table S1).
Figure 1. The am117 mutation delayed reproductive and somatic aging, caused bacterial avoidance behavior, reduced body size and increased stress resistance.
(A-K) Animals were cultured at 20°C on NGM dishes with a small lawn of live E. coli OP50 bacteria. (A-C) L4 stage hermaphrodites were cultured individually and monitored daily for self-progeny production. Values are the average (+/− S.D.). (N=6-12 animals, Tukey post hoc HSD; **, P <0.01). (D-E) Survival curves - see Table S1 for summary statistics. (F-G) Bars represent average (+/−S.D.) body bends per minute (F) and pharyngeal pumps per minute (G) (N=30-60 animals analyzed in each of 2-3 biological replicates; ***, P < 0.0001, not significant (ns), P > 0.05 by Student’s t-test). (H) Schematic of method used to quantify bacterial avoidance behavior. (I) Bright field photographs of dishes with a lawn of live E. coli OP50 (dark circle) and the surrounding medium without bacteria; wild-type animals are mostly inside the bacterial lawn, whereas am117 mutant animals are mostly outside the bacterial lawn. Scale bar = 100 μm. (J) “Avoidance” is the average percent of animals (+/− S.D.) outside the bacterial lawn at the larval stages L1, L2, L3, L4, day 1 adult (YA) and day 2 adult (LA). See Table S2. (K) Bars represent the average volume of individual worms (+/−S.D.) four days after the L4 stage determined by analyzing dissecting microscope images with the wormsizer algorithm (N=35-66 animals analyzed); ***, P < 0.0001 by Student’s t-test). (L) Bars represent average (+/−S.D.) fractional survival. Animals were cultured at 20°C on NGM dishes until day 1 of adulthood, shifted to 35°C, and scored for survival after 9 and 12 hours (N=106-149 animals in each of 3 biological replicates; *, P < 0.05; **, P < 0.005 by Student’s t-test).
See also Figures S1-2 and Tables S1-3.
To investigate the effect of am117 on age-related degenerative changes (Collins et al., 2008a; Huang et al., 2004), we analyzed neuromuscular processes that can be monitored non-invasively. Wild-type hermaphrodites display rapid and highly coordinated sinusoidal body movement as young adults, and the speed and coordination of body movements display age-related declines. To analyze body movement quantitatively, we counted body bends of worms on solid medium using a dissecting microscope. am117 hermaphrodites displayed a significantly higher rate of body movement on days 10, 15, and 20 compared to wild type (Figure 1F). The rate of pharyngeal pumping can be analyzed quantitatively using a dissecting microscope and displays age-related decline. am117 hermaphrodites displayed significantly higher rates of pharyngeal pumping on days 10–18 compared to wild type (Figure 1G). Thus, the gene affected by am117 is necessary to promote rapid somatic aging, including age-related declines of neuromuscular activity and survival probability.
We noticed that am117 mutant animals were frequently outside the lawn of bacteria, a phenotype called lawn leaving behavior or bacterial avoidance behavior (Zhang et al., 2005) (Figure 1H-I). Wild-type animals display this behavior when cultured with pathogenic bacteria or RNAi bacteria that cause toxicity (Melo and Ruvkun, 2012). To quantify this behavior, we counted the number of animals inside and outside the bacterial lawn. Eggs were placed in the center of the bacterial lawn, and animals were scored at the L1, L2, L3, L4, day 1 adult, and day 2 adult stages. am117 mutant animals displayed bacterial avoidance behavior beginning at the L4 stage and peaking during adulthood with more than 85% of animals outside the bacterial lawn. By contrast, wild-type animals displayed fewer than 15% of animals outside the bacterial lawn (Figure 1J, Table S2). Sensory perception is important for worms to detect and chemotax towards food. There are 60 ciliated neurons in C. elegans that function in perception of the external environment (Perkins et al., 1986; Starich et al., 1995). Because defects in these neurons might cause animals to wander away from food, we investigated the morphology of amphid neurons in am117 mutant animals using DiO staining. am117 mutant animals displayed morphology similar to wild type, indicating that the bacterial avoidance behavior is not likely to be caused by abnormal sensory perception (Figure S1J-L). Bacterial avoidance behavior might reduce food ingestion, leading to dietary restriction. Indeed, am117 mutant animals displayed a scrawny phenotype. To quantify body volume, we used dissecting microscope images and the worm sizer algorithm to analyze live animals 4 days after the L4 stage. Body volume of am117 animals was reduced 60% compared to wild type (Figure 1K, S2A-B). These results indicate that the am117 mutation causes bacterial avoidance behavior that may result in dietary restriction and a scrawny body morphology.
Many C. elegans mutations that delay aging also increase stress resistance (Johnson et al., 2000; Lithgow et al., 1995). To analyze the effect of am117 on stress resistance, we monitored heat resistance at 35°C. am117 mutant animals displayed significantly increased survival compared to wild type (Figure 1L). Thus, the gene affected by am117 is necessary to promote the levels of sensitivity to stress observed in wild-type animals.
Lifespan extension caused by am117 interacted genetically with eat-2.
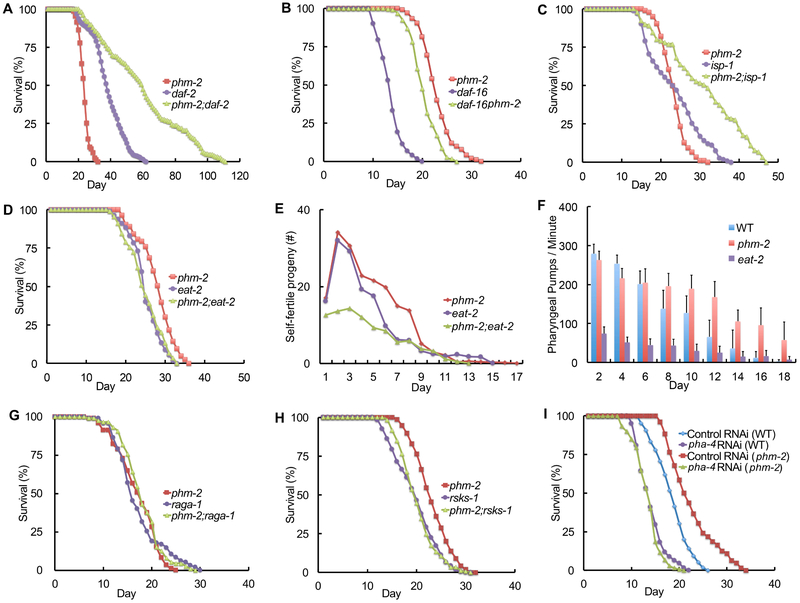
To elucidate the mechanism of action of am117, we generated and analyzed double mutant strains with additional mutations that influence lifespan. Multiple genes in the IIS pathway influence C. elegans lifespan (Friedman and Johnson, 1988; Kenyon et al., 1993; Kimura et al., 1997; Morris et al., 1996). Mutations that partially reduce the activity of daf-2, which encodes a protein homologous to the vertebrate insulin/IGF-1 receptor, extend lifespan. This signaling pathway controls the activity of a FOXO transcription factor encoded by daf-16, and daf-16 activity is necessary for the lifespan extension caused by mutations in upstream signaling genes (Lin et al., 1997). am117; daf-2(lf) double mutant animals displayed an extremely long mean lifespan of 58 days that was significantly longer than either single mutant strain (Figure 2A, Table S1). daf-16(lf) am117 double mutant animals displayed a significant extension of mean (58%) and maximum (43%) lifespan compared to daf-16(lf) single mutant animals (Figure 2B, Table S1). Thus, the am117 lifespan extension was additive with the daf-2(lf) lifespan extension and did not require daf-16 activity. Furthermore, am117 extended lifespan and self-fertile reproductive span in the daf-16(lf); daf-2(lf) genetic background (Figure S1C-D, Table S1). The IIS pathway mediates entry into an alternative third larval stage called dauer, a diapause state that is stress resistant (Kimura et al., 1997; Malone and Thomas, 1994; Ogg et al., 1997). am117 mutant animals did not display an obvious Daf-c phenotype. To test am117 in a more sensitive assay, we analyzed am117;daf-2(lf) double mutant animals. The am117 mutation did not affect the temperature sensitive Daf-c phenotype caused by daf-2(lf), indicating the gene affected by am117 is not necessary to inhibit formation of dauer larvae (Figure S1E).
Figure 2. Genetic interactions between phm-2(am117) and genes involved in longevity.
(A-D, G-I) Survival curves - see Tables S1 and S4 for summary statistics. Hermaphrodites had the following genotypes: wild type, phm-2(am117), daf-2(e1370), daf-16(mu86), isp-1(qm150), eat-2(ad1116), raga-1(ok386), and rsks-1(ok1255). (E) Self-progeny production as in Figure 1 (N=8-10 animals). (F) Pharyngeal pumping as in Figure 1 (N=15-55 animals analyzed in each of 2 biological replicates). See Table S3 for summary statistics. (I) Wild-type or phm-2(am117) hermaphrodites were cultured on pha-4 RNAi bacteria or control RNAi bacteria.
Reducing the activity of multiple genes that are critical for mitochondrial activity extends C. elegans lifespan. The isp-1 gene encodes an iron sulfur cluster containing protein that is essential for the function of complex III, and isp-1(lf) mutations extend lifespan (Feng et al., 2001; Lakowski and Hekimi, 1996). The am117; isp-1(lf) double mutant animals displayed an extended lifespan that was significantly longer than isp-1(lf) single mutant animals (Figure 2C, Table S1). Thus, the lifespan extensions caused by reducing mitochondrial function and the am117 mutation were additive.
Energy metabolism genes can influence lifespan. The AMP-activated protein kinase (AMPK) alpha subunit AAK-2 promotes longevity by controlling energy metabolism, since an aak-2(lf) mutation reduces lifespan (Apfeld et al., 2004; Curtis et al., 2006). am117; aak-2(lf) double mutant animals displayed significantly extended mean (42%) and maximum (38%) lifespans compared to aak-2(lf) single mutant animals (Figure S1F, Table S1). The target of rapamycin (TOR) signaling network plays a critical role in nutrient homeostasis and influences adult longevity (Kapahi et al., 2010). rict-1(lf) mutations affect TOR signaling and reduce lifespan. am117; rict-l(lf) double mutant animals displayed significantly extended mean (93%) and maximum (70%) lifespans compared to rict-1(lf) single mutant animals (Figure S1G, Table S1). Thus, the activities of rict-1 and aak-2 were not necessary for the lifespan extension caused by am117. Dietary restriction influences the lifespan of many organisms, demonstrating that ad libitum feeding during laboratory culture promotes a rapid lifespan. Mutations of the eat-2 gene reduce the pharyngeal pumping rate and cause a lifespan extension (Lakowski and Hekimi, 1998; McKay et al., 2004). The lifespan of am117; eat-2(lf) double mutant animals was not significantly longer than eat-2(lf) single mutant animals (Figure 2D, Table S1). Furthermore, these double mutant animals did not display an additive extension of reproductive span (Figure 2E). These results indicate that eat-2(lf) and am117 may extend lifespan by a similar mechanism. By contrast to eat-2(lf) mutant animals, which display a substantial reduction of pharyngeal pumping rate, the am117 mutant animals displayed a pharyngeal pumping rate similar to wild-type in young adults and higher than wild-type in older adults (Figure 2F, Table S3). To investigate the possibility that am117 impairs food ingestion even though the pharyngeal pumping rate is normal, we measured feeding by exposing worms to fluorescent beads for 15 minutes and monitoring fluorescence in the intestine. Wild type and am117 mutant animals displayed strong fluorescence, indicating robust ingestion, whereas eat-2(lf) animals displayed significantly less fluorescence, indicating a reduced ingestion rate (Figure S1I).
To further characterize genetic interactions, we analyzed the raga-1, rsks-1 and pha-4 genes. The C. elegans raga-1 gene encodes a highly conserved ras-related GTPase, and raga-1(lf) mutations delay aging and age-related changes (Schreiber et al., 2010). The C. elegans rsks-1 gene encodes a ribosomal S6 kinase that is evolutionarily conserved and promotes development, metabolism, and autophagy; rsks-1(lf) mutant animals display delayed aging and age-related changes, as well as reduced fertility (Hansen et al., 2007; Kapahi et al., 2010; Pan et al., 2007; Selman et al., 2009; Vellai et al., 2003; Wullschleger et al., 2006). The lifespan of am117; raga-1(lf) or am117; rsks-1(lf) double mutant animals was not significantly longer than raga-1(lf) or rsks-1(lf) single mutant animals, respectively, indicating that the lifespan extensions are not additive (Figure 2G-H, Table S1). C. elegans pha-4 encodes an ortholog of a mammalian FOXA transcription factor, and pha-4 is essential for foregut development and necessary for the eat-2(lf) lifespan extension phenotype (Hansen et al., 2008; Panowski et al., 2007). When pha-4 activity was reduced by feeding RNAi beginning at L1 stage larvae, the lifespan of wild type and am117 mutant animals was reduced similarly (Figure 2I, Table S4). Thus, the lifespan extension caused by the am117 mutation requires pha-4 activity. As a control for specificity, we demonstrated that pha-4 feeding RNAi did not suppress the daf-2(lf) lifespan extension phenotype (Figure S1H, Table S4). Based on these genetic studies, we hypothesized that the am117 mutation causes bacterial avoidance behavior, which results in dietary restriction characterized by a scrawny body morphology and delayed aging phenotypes that interact genetically with eat-2, raga-1, rsks-1 and pha-4.
Identification of phm-2 as the gene affected by the am117 mutation.
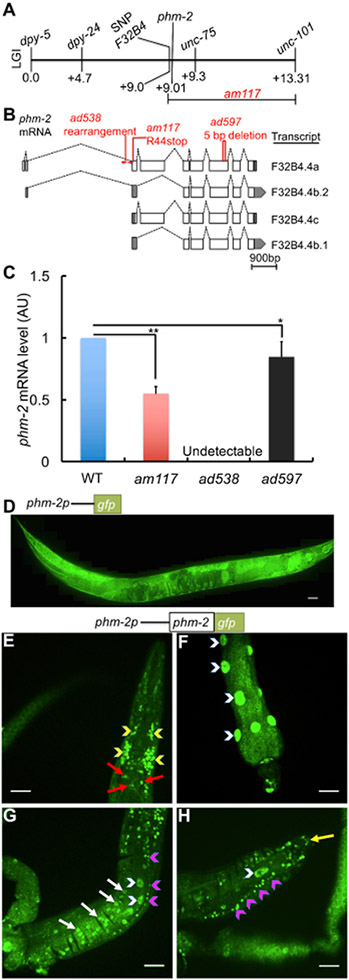
To identify the gene affected by the am117 mutation, we used a positional cloning approach. Hughes et al. (2011) used the readily-scored scrawny body morphology phenotype to position am117 on the right arm of chromosome I by linkage to single nucleotide polymorphism (SNP) markers. Multi-factor mapping experiments indicated am117 is positioned in a 4.3 map unit interval between a SNP in cosmid F32B4 at +9.0 and unc-101 at +13.3 (Figure 3A). We performed whole-genome sequencing using genomic DNA from the am117 mutant strain. Candidate mutations in the mapping interval were identified by comparison to the wild-type reference sequence and confirmed by standard DNA sequencing (Hillier et al., 2008). One candidate mutation caused a nonsense change in the predicted open reading frame F32B4.4. Gene structure predictions indicate that the F32B4.4 locus generates four transcripts; F32B4.4a is the longest and contains nine exons, whereas F32B4.4b.2, F32B4.4c, and F32B4.4b.1 transcripts contain 7, 7, and 6 exons, respectively (Figure 3B). The am117 mutation is a C to T transition in codon 44 of the F32B4.4a transcript that changes an arginine to a stop codon (Figure 3B).
Figure 3. The am117 mutation is an allele of phm-2.
(A) A genetic map of the right arm of C. elegans linkage group (LG) I. Loci defined by SNPs and mutations that cause visible phenotypes are named above, and map units are shown below. Multi-factor mapping experiments positioned am117 between a SNP in cosmid F32B4 and unc-101, an interval that includes phm-2. (B) Diagrams of four phm-2 transcripts. Boxes represent exons, shaded regions are untranslated, and dotted lines indicate introns. Position and nature of mutations is indicated above. (C) Bars represent average phm-2 mRNA levels (+/−S.D.) in arbitrary units (AU) determined by qPCR in wild-type, phm-2(am117), phm-2(ad538) and phm-2(ad597) animals. The WT value was set equal to 1.0, and mutant values are expressed as fold change over WT (N=4 biological replicates, Tukey post hoc HSD; *, P < 0.05; **, P < 0.01). (D-H) Diagrams of the upstream phm-2 promoter region (not to scale) extending ~475 bp upstream of the translation start site of F32B4.4a fused to the GFP coding region (D) or the phm-2 and GFP coding regions (E-H). Panel D shows a fluorescence confocal microscope image of a live transgenic animal (genotype amEx315) at the adult stage displaying expression in most tissues from head to tail including pharynx, intestine, hypodermis, muscle and tail side neurons. Scale bar = 50 μm. (E-H) Fluorescence images show PHM-2::GFP fusion protein detected by immunostaining of fixed transgenic animals (genotype amEx320) at the adult stage. Panel E shows pharyngeal nuclei (red arrows) and nerve ring (yellow arrowheads) in the head region; panel F shows intestinal nuclei (white arrowheads); panel G shows ventral nerve cord (pink arrowheads), hypodermis (white arrows) and intestinal nuclei (white arrowheads) in the middle region of the worm; panel H shows tail neurons (yellow arrow), ventral nerve cord (pink arrowheads) and intestinal nuclei (white arrowhead) in the tail region. Scale bar = 20 μm.
See also Figures S3-4.
We noticed that the phm-2 gene, which has not been molecularly identified, is positioned on the genetic map in the range of 0.08 – 0.35 map units left of unc-75, which is consistent with the position of F32B4.4. Avery (1993a) used X-ray mutagenesis to generate two phm-2 mutations, ad538 and ad597, that were recovered in a screen for eating defective mutants. phm-2 mutants display morphological defects in the pharyngeal grinder - it is unable to come to its full forward position, the muscle fibers of the terminal bulb appear to be shortened, and the space posterior to the grinder is expanded (Avery, 1993a). To test the model that phm-2 mutations affect F32B4.4, we determined the DNA sequence of the F32B4.4 locus using genomic DNA from ad538 and ad597 mutant animals. The phm-2(ad597) allele contains a 5 base pair deletion that eliminates a portion of exon 7 that is common to all four transcripts, and the phm-2(ad538) allele displayed abnormal PCR amplification suggestive of a rearrangement of intron 2 and exon 3 (Figure 3B).
To determine how these mutations affect F32B4.4 transcripts, we analyzed mRNA levels by quantitative PCR (qPCR). phm-2(ad538) mutants lacked detectable mRNA, consistent with a gross rearrangement of the locus, whereas phm-2(ad597) and am117 mutant animals displayed significantly reduced transcript levels that were nonetheless readily detectable, consistent with a nonsense mutation and a small deletion that may reduce mRNA stability (Fig. 3C). These results indicate that the phm-2 gene corresponds to F32B4.4 and am117, ad538 and ad597 are each alleles of phm-2. The phm-2(ad597) mutation is likely to be a strong loss-of function or null allele based on the lack of detectable mRNA.
To analyze the effect of phm-2 mutations ad538 and ad597 on reproduction, we monitored progeny production of self-fertile hermaphrodites. Thesephm-2(lf) mutations caused a decrease of early progeny production, resulting in a smaller self-fertile brood size, and an increase in late progeny production, resulting in an extended self-fertile reproductive span (Figure 1A-C). phm-2(ad538) and phm-2(ad597) mutant animals displayed significant extensions of mean (30%, 33%) and maximum (28%, 27%) lifespan, respectively (Figure 1E, Table S1). These phm-2(lf) mutations caused bacterial avoidance behavior (Figure 1J). Thus, the phm-2(ad538) and phm-2(ad597) mutations that were identified based on eating defects (Avery, 1993a) caused the same reproductive and somatic aging phenotypes used to identify the am117 mutation (Hughes et al., 2011).
To confirm that the am117 mutation inphm-2 causes these phenotypes, we tested a wild-type version of phm-2, a cDNA of the F32B4.4a transcript, for rescue ability. We generated five independently derived transgenic strains containing extrachromosomal arrays with wild-type copies of phm-2 in the background of phm-2 (am 117). All these transgenic strains displayed a significant decrease in lifespan compared to non-transgenic phm-2(am117) animals (Figure S3A, Table S1). Furthermore, expression of wild-type phm-2 significantly increased the body volume by 42% (Figure S3B). Thus, the phm-2 locus was sufficient to partially rescue the extended lifespan and scrawny body morphology phenotypes, consistent with the model that phm-2 is the gene affected by the am117 mutation.
To analyze the predicted PHM-2 protein, we performed a BLAST search to identify related proteins. An alignment of C. elegans PHM-2 revealed 19.9%, 20.5%, and 16.7% overall identity to human, mouse, and Drosophila proteins, respectively, demonstrating that PHM-2 is an evolutionarily conserved member of the scaffold attachment factor B (SAFB) protein family (Figure S4). C. elegans PHM-2 protein includes two highly conserved motifs, the SAP domain that may be involved in DNA binding and the RRM_SAF domain that may be involved in RNA recognition (Garee and Oesterreich, 2010). These results suggest that C. elegans phm-2 and human SAFB are descended from a common ancestral gene.
phm-2 is broadly expressed, and the PHM-2 protein localizes to the nucleus.
To determine the expression pattern of phm-2, we generated a plasmid that expresses green fluorescent protein (GFP) under the control of the upstream predicted phm-2 promoter, injected the plasmid into wild-type animals to generate transgenic animals, and monitored fluorescence. The upstream phm-2 promoter region consisted of ~475 bp of genomic DNA that extends from the start codon of F32B4.4a to the 3′ end of the adjacent gene (Figure 3B). These transgenic animals displayed GFP expression in multiple tissues including the hypodermis, muscles, neuron, vulva, and intestine, indicating that phm-2 is expressed in most if not all tissues (Figure 3D).
The PHM-2 protein encoded by the F32B4.4a transcript contains 1073 amino acids. To determine the sub-cellular localization of this protein, we generated plasmids that express PHM-2 fused to GFP under the control of the upstream predicted phm-2 promoter, injected these into phm-2(am117) animals to generate transgenic animals, and used immunostaining to detect the fusion protein. PHM-2::GFP was detected in a wide range of cells including the pharynx, nerve ring, intestine, neurons, and ventral nerve cord. The protein displayed punctate staining suggestive of nuclear localization (Figure 3E-H).
phm-2(lf) mutants displayed abnormal pharynx morphology, live bacteria accumulation in the intestine, and an activated innate immune response.
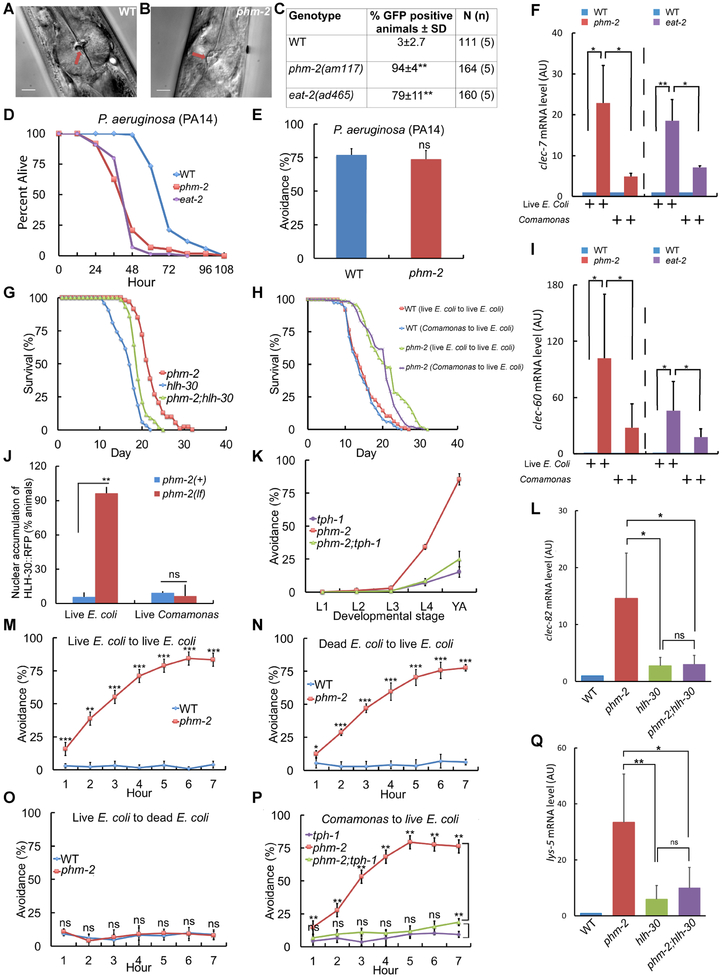
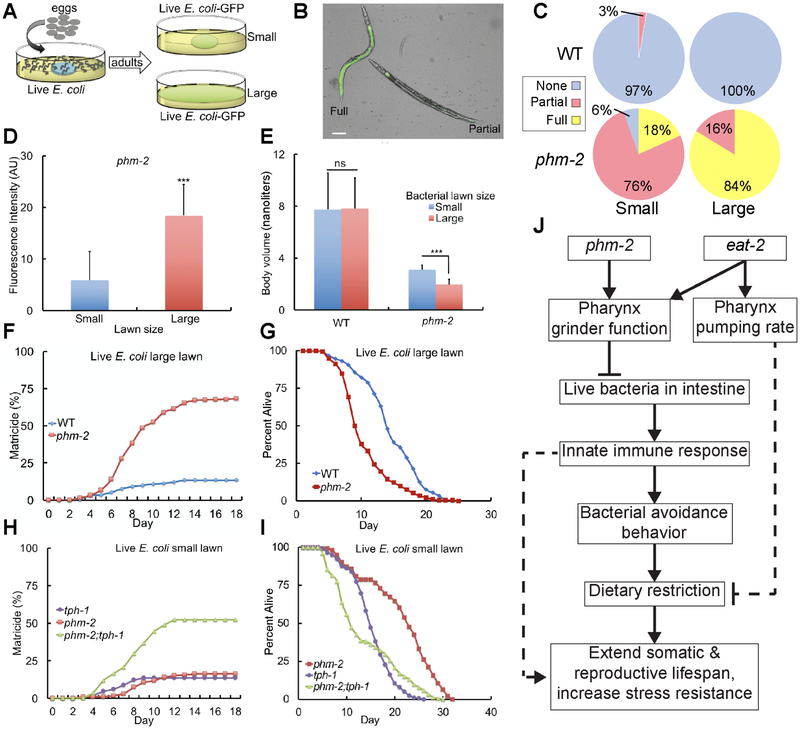
A critical function of the C. elegans pharynx is to crush live bacteria before they enter the intestine, thus preventing bacterial colonization (Avery, 1993b; Doncaster, 1962). phm-2(am117) mutant animals exhibited abnormal pharynx grinder morphology (Figure 4A-B), consistent with the defects observed in thephm-2(ad538) and phm-2(ad597) mutants identified by Avery (1993a). To determine if this defect allows live bacteria to accumulate in the intestine, we cultured adult animals on E. coli OP50-GFP for 24 hours. phm-2(am117) mutant animals displayed significantly more fluorescent animals than wild type, demonstrating that live E. coli accumulate in the intestine of this mutant strain (Figure 4C, S5G). Reducing the activity of rsks-1 or pha-4 suppressed the lifespan extension caused by phm-2(lf) but did not block bacterial accumulation, indicating these genes function downstream of bacterial accumulation to influence lifespan (Figure S5H-I). phm-2(lf) mutants have been reported to be hypersensitive to pathogenic bacteria, since the pharynx grinder defect makes them more susceptible to colonization by live bacteria (Labrousse et al., 2000; Portal-Celhay et al., 2012; Smith et al., 2002). Consistent with this observation, phm-2(am117) mutant animals were more susceptible than wild-type animals to killing by the pathogenic bacteria P. auruginosa strain PA14 (Figure 4D). Wild-type and phm-2(am117) mutants displayed robust avoidance of this pathogenic strain (Figure 4E).
Figure 4. phm-2(lf) animals displayed abnormal pharynx morphology, live bacteria accumulation and an innate immune response.
(A,B) Representative bright field photographs of the terminal bulb of the pharynx illustrate an open, abnormal grinder in phm-2(am117) mutant animals (red arrow). Scale bar = 10 μm. (C) Quantification of number of fluorescent animals for wild-type, phm-2(am117), and eat-2(ad465) adult animals cultured with green E. coli OP50-GFP for 24 hours. N(n) indicates total number of animals and number of independent trials. Tukey post hoc HSD; **, P < 0.01 compared to WT. (D) Wild-type, phm-2(am117), and eat-2(ad465) animals were cultured on live E. coli OP50 bacteria from egg until the L4 stage, transferred to pathogenic P. aeruginosa PA14 bacteria in NGM dishes with FudR at 25°C, and analyzed every 12 hours for survival. phm-2(am117) and eat-2(ad465) animals had a significantly shorter mean lifespan compared to WT (Five biological experiments, N=98-106 animals, P < 0.001 by ANOVA). (E) Wild-type and phm-2(am117) animals were cultured on live E. coli OP50 bacteria from egg until the L4 stage, transferred to pathogenic P. aeruginosa PA14 bacteria, and scored after 24 hours. Values are the average (+/− SD) of three biological replicates, N≥100 animals per replicate. Both strains displayed similar, robust avoidance (ns, not significant, P > 0.05 by Student’s t-test). (F, I, L, Q) Bars represent mRNA levels (+/−S.D) for clec-7, clec-60, clec-82, and lys-5 pathogen response genes determined by qPCR in wild-type, phm-2(am117), eat-2(ad465), hlh-30(tm1978), and phm-2;hlh-30 animals cultured on live E. coli OP50 (F, I as indicated and L, Q) or live Comamonas bacteria (F, I as indicated). Values in arbitrary units (AU) are the average of three to five biological replicates. ANOVA; *, **, P < 0.05, P < 0.005; ns, not significant, P > 0.05. (G) Survival curves for phm-2(am117), hlh-30(tm1978), and phm-2(am117); hlh-30(tm1978). See Table S1 for summary statistics. (H) Survival curves of wild type or phm-2(am117) animals grown from embryo to adult on either live E. coli OP50 or live Comamonas DA1877 and then transferred to live E. coli OP50 at the adult stage for the remainder of their lifespan. See Table S6 for summary statistics. (J) Quantification of percent of animals that displayed nuclear localized fluorescence for phm-2(+) and phm-2(am117) animals that express HLH-30::RFP cultured on live E. coli OP50 or live Comamonas (N≥100 animals). (K) phm-2(am117);tph-1(mg280) animals did not display bacterial avoidance behavior on live E. coli compared to phm-2(lf). See Table S5 for summary statistics. (M-O) Avoidance was measured hourly after wild-type or phm-2(am117) adults were transferred to the test dish. Values are the average of five biological replicates, N=148-162 animals (M) and three to four biological replicates, N=77-142 animals (N,O). ns, not significant, P > 0.05 by Student’s t-test. (P) Animals were cultured on Comamonas bacteria from embryo to young adult and then tested on live E. coli OP50. phm-2(am117);tph-1(mg280) animals displayed less bacterial avoidance behavior compared to phm-2(lf) and were similar to tph-1(lf) (N=108-146 animals, 4 biological replicates, Tukey post hoc HSD; n.s., not significant, P > 0.05).
See also Figures S5-8 and Tables S1, S5 and S6.
C. elegans has an innate immune system and responds to bacterial pathogens by expression of protective genes (Irazoqui et al., 2010; Sifri et al., 2003). C. elegans HLH-30 is the ortholog of vertebrate transcription factor EB (TFEB); hlh-30 can modulate longevity and plays an important role in autophagy, lysosomal biogenesis, and regulation of innate immune response genes (Lapierre et al., 2013; Settembre et al., 2011). Exposure to highly pathogenic bacteria induces nuclear accumulation of HLH-30::RFP (Visvikis et al., 2014). To test the hypothesis that live E. coli in the intestine of phm-2(lf) mutant animals activate the innate immune response, we analyzed the transcription of response genes and the nuclear accumulation of HLH-30. The transcript levels of multiple genes involved in the pathogen response were dramatically higher in phm-2(lf) mutant animals compared to wild-type animals when cultured on live E. coli, including the C-type lectin genes clec-7, clec-60, and clec-82, the antimicrobial peptide gene F53A9.8, and the lysozyme gene lys-5 (Figure 4F, I, S5M-O). This difference in transcript levels between wild type and mutant was largely or completely abrogated when animals were cultured on live Comamonas, a non-pathogenic bacteria (Figure 4F, I, S5M-O). The phm-2 promoter was not induced by exposure to highly pathogenic bacteria (Figure S5A-F). The HLH-30 transcription factor accumulates in the nucleus when it is activated. Two day old adult phm-2(lf) mutant animals displayed nuclear accumulation of HLH-30::RFP compared to wild-type animals when cultured on live E. coli, and this difference was not observed when cultured on live Comamonas (Figure 4J, S5J). To investigate the functional consequences of hlh-30 activation, we analyzed the expression levels of innate immune genes and the lifespan of phm-2(lf); hlh-30(lf) double mutants. hlh-30 was necessary for the increased transcript levels of clec-82, lys-5, and clec-7, although it was not necessary in the case of clec-60 (Figure 4L, Q, S5K-L). Thus, hlh-30 appears to mediate at least part of the transcriptional response to bacterial accumulation in phm-2(lf) mutant animals. phm-2(lf); hlh-30(lf) double mutant animals displayed a lifespan that was intermediate between phm-2(lf) and hlh-30(lf) single mutants (Figure 4G, Table S1). Therefore, hlh-30 appears to be partially required for the phm-2(lf) lifespan extension. Thus, live E. coli accumulate in the intestine of phm-2(lf) mutant animals which appears to activate the innate immune response.
Analysis of phm-2(lf) bacterial avoidance behavior.
We observed that phm-2(lf) mutant animals did not display bacterial avoidance behavior when cultured on UV-killed E. coli or non-pathogenic live Comamonas bacteria. This raises the question: how do phm-2(lf) mutant animals sense that live E. coli are pathogenic and implement the bacterial avoidance behavior? To test the possibility that the worms sense a secreted metabolite released by live E. coli, we analyzed filtered supernatant from overnight bacterial cultures. Adding supernatant from live E. coli OP50 cultures to Comamonas bacteria was not sufficient to provoke the bacterial avoidance behavior (Figure S6C), and adding supernatant from live Comamonas bacteria to E. coli bacteria was not sufficient to inhibit bacterial avoidance behavior (Figure S6D-E). Furthermore, adding a block of live E. coli OP50 to the cover of the dish was not sufficient to provoke the bacterial avoidance behavior (Figure S6F-G). Mixtures of live E. coli OP50 and Comamonas resulted in intermediate levels of bacterial avoidance behavior (Figure S6H-I). These results do not support the model that a secreted metabolite caused bacterial avoidance behavior.
We investigated whether phm-2(lf) animals learn to avoid live E. coli. In the first learning paradigm, animals were trained by culture from egg to adult on one type of bacteria, and then these adults were tested on another type of bacteria (Figure S6L). If worms learn avoidance behavior, then the training experience is predicted to influence their behavior during the test. phm-2(lf) mutant animals trained on dead E. coli, which they do not avoid, and tested on live E. coli, displayed robust avoidance behavior that was measurable in one hour and similar to animals trained on live E. coli (Figure 4M-N). phm-2(lf) mutant animals trained on live E. coli, which they avoid, and tested on dead E. coli did not display avoidance behavior (Figure 4O). Similar results were obtained when Comamonas was used instead of dead E. coli (Figure S6J-K). Thus, in this learning paradigm prior experience had no measurable effect on avoidance behavior, indicating the behavior is not learned.
The second learning paradigm was a binary choice assay; animals were trained by culture from egg to adult on one type of bacteria, adults were tested on dishes with two small lawns with different types of bacteria, and the number of worms in each lawn was scored. Wild type preferred live E. coli over Comamonas and live E. coli over dead E. coli, and these preferences were not affected by the training bacteria (Figure S7). phm-2(lf) mutants preferred Comamonas over live E. coli and dead E. coli over live E. coli, and these preferences were not affected by the training bacteria (Figure S7). Thus, the phm-2(lf) mutants displayed strikingly different bacterial preferences than wild type, consistent with avoidance of live E. coli. However, in this learning paradigm prior experience had no measurable effect on avoidance behavior, indicating the behavior is not learned. These experiments indicate that avoidance behavior is a rapid response of phm-2(lf) mutant animals to live E. coli, since it can be observed after one hour, and it involves a continuous sensing mechanism rather than learning and memory.
To explore the mechanism of bacterial avoidance behavior, we used a candidate gene approach by analyzing double mutant strains. Most of the genes involved in longevity did not strongly influence the bacterial avoidance behavior of phm-2(lf) mutant animals, including daf-16, isp-1, aak-2, rict-1, and rsks-1. However, daf-2(lf) caused a modest reduction of avoidance (Figure S8, Table S5). We investigated C. elegans pathways reported to be involved in avoiding pathogenic bacteria (Kao et al., 2011; Kim et al., 2002; Pradel et al., 2007; Pujol et al., 2001; Reddy et al., 2009; Troemel et al., 2006). Mutations of pmk-1, mlk-1, hlh-30, and npr-1 did not strongly influence the bacterial avoidance behavior of phm-2(lf) mutant animals (Figure S8, Table S5). The serotonin biosynthesis enzyme tryptophan hydroxylase is encoded by the tph-1 gene, which is necessary for avoidance of pathogenic bacteria (Melo and Ruvkun, 2012; Zhang et al., 2005). The analysis of double mutant phm-2(lf); tph-1(lf) animals demonstrated that the tph-1 mutation suppressed the bacterial avoidance behavior caused by phm-2(lf) (Figure 4K, P, Table S5). A detailed time course revealed that the suppression is partial and most effective in young and old adults, whereas it was less effective in day 2 adults (Figure S8L). These results indicate that tph-1 is necessary to mediate this bacterial avoidance behavior.
Live E. coli are pathogenic to phm-2(lf) mutant animals, and bacterial avoidance behavior is adaptive.
To test the model that accumulation of live E. coli in phm-2(lf) mutant animals is deleterious and bacterial avoidance behavior is adaptive, we spread live E. coli OP50 bacteria over the entire surface of the NGM dish so that animals could not avoid the bacteria; we refer to such dishes as having a large lawn. By contrast, standard culture dishes have a central spot of bacteria, and we refer to such dishes as having a small lawn. When cultured with live E. coli OP50-GFP, phm-2(lf) mutant animals displayed significantly increased fluorescence intensity when cultured on the large lawn compared to the small lawn (Figure 5A, D). Furthermore, a greater fraction of the animals displayed strong fluorescence (Figure 5B-C). Thus, the bacterial avoidance behavior limits the accumulation of live E. coli in the intestine.
Figure 5. Bacterial avoidance behavior by phm-2(lf) animals limits bacterial accumulation in the intestine and increases survival.
(A) Schematic of method. (B) Representative bright field photographs and (C) Quantification of categories of phenotype severity. Animals were categorized as Full (E. coli OP50-GFP throughout the intestine), Partial (E. coli OP50-GFP in part of the intestine) or None (no detectable fluorescence in the intestine). Five biological replicates with N≥112 animals were analyzed. Scale bar = 50 μm. (D) Quantification of whole animal fluorescence. phm-2(am117) adult animals were cultured with a small (blue) or large (red) lawn of E. coli OP50-GFP for 24 hours. Values in arbitrary units (AU) are the average (+/−S.D), N≥13 animals. ***, P < 0.001, by Student’s t-test. (E) Worm volume as in Figure 1 (N=40-55 animals analyzed; ns, not significant, P > 0.05 by Student’s t-test). (F, H) Values are the cumulative fraction of animals that displayed matricidal hatching versus days of adulthood. Genotypes were wild type, phm-2(am117), and tph-1(mg280). Panel F is based on six to eight biological replicates, N=295-342 animals, and panel H is based on three biological replicates, N≥168 animals. (G) Percent alive for wild type and phm-2(am117) animals cultured on a large lawn of live E. coli, and (I) for phm-2(am117), tph-1(mg280), and phm-2;tph-1 animals cultured on a small lawn of live E. coli. Note that animals that died of matricidal hatching were not censored from these data. See Table S6 for summary statistics. (J) Model of the mechanism of phm-2(lf) and eat-2(lf) lifespan extensions. Dotted lines indicate uncertainty.
See also Table S6.
To investigate the consequences of increased bacterial accumulation, we monitored the fate of animals cultured on the dishes with large bacterial lawns. phm-2(lf) mutant animals displayed a high frequency of early death due to matricidal hatching and a shorter mean lifespan compared to wild type (Figure 5F-G, Table S6). Consistent with this finding, phm-2(lf); tph-1(lf) double mutant animals displayed a high frequency of matricidal hatching and a shorter mean lifespan when cultured on a small lawn of live E. coli OP50, since they do not avoid the bacteria (Figure 5H-I, Table S6). phm-2(lf) mutant animals cultured on the large lawn of bacteria displayed an enhanced scrawny body morphology compared to animals cultured with a small lawn of bacteria (Figure 5E). Reduced body size is a non-specific phenotype that might result from a variety of defects. We interpret that the very small body size of animals cultured on the large lawn results from strong pathogenesis, whereas we interpret that the slightly bigger body size of animals cultured on the small lawn results from mild pathogenesis and dietary restriction caused by food avoidance behavior. Thus, bacterial avoidance behavior of phm-2(lf) animals promotes growth and prevents matricidal hatching, and forced exposure to live E. coli is indeed deleterious.
Colonization of phm-2(lf) mutants by pathogenic E. coli causes delayed aging.
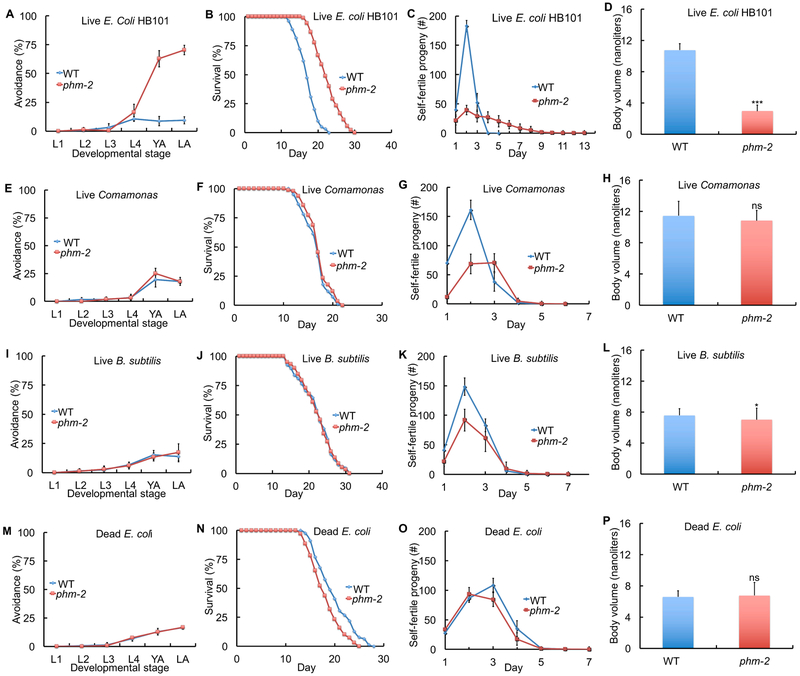
We hypothesized that live bacterial accumulation in the intestine causes bacterial avoidance behavior, resulting in dietary restriction that is the cause of the scrawny body morphology and contributes to the delayed aging phenotypes (Figure 5J). To test this model, we tried to identify bacterial food sources that would not trigger the avoidance behavior. When phm-2(lf) mutant animals were cultured on another strain of live E. coli, the B/K12 hybrid strain HB101, bacterial avoidance behavior, scrawny body morphology and delayed aging phenotypes were still observed (Figure 6A-D, S2C-D, Table S2, S6).
Figure 6. Live E. coli accumulation in phm-2(lf) mutants caused avoidance behavior, scrawny body morphology, and delayed aging.
WT or phm-2(am117) hermaphrodites were cultured with a small bacterial lawn as indicated. (A, E, I, M) Avoidance as in Figure 1 - see Table S2. (B, F, J, N) Survival curves - see Table S6 for summary statistics. (C, G, K, O) Self progeny production as in Figure 1 (N=9-16 animals). (D, H, L, P) Worm volume as in Figure 1 (N=37-58 animals). ***, P < 0.0001; *, P < 0.05; ns, not significant, P > 0.05 by Student’s t-test.
E. coli is not a natural food source for C. elegans, a soil-dwelling animal (Félix and Duveau, 2012; Schulenburg and Ewbank, 2007). Furthermore, E. coli is a mild pathogen for older adult wild-type C. elegans (Garigan et al., 2002). We analyzed the soil bacteria Comamonas (strain DA1877) and Bacillus subtilis, because they are not pathogenic for C. elegans. Strikingly, phm-2(lf) mutant animals did not avoid these bacterial lawns and did not display the scrawny body morphology and delayed aging phenotypes (Figure 6E-L, S2E-H, Table S2, S6). Furthermore, phm-2(lf) mutant animals cultured on dead E. coli OP50 that was killed by UV light did not avoid the bacterial lawn or display the scrawny body morphology and delayed aging phenotypes (Figure 6M-P, S2I-J, Table S2, S6). Similar results were obtained when E. coli OP50 was killed by antibiotic treatment (Figure S6A-B). These results suggest that live E. coli accumulation is the cause of multiple phenotypes including avoidance behavior, scrawny body morphology and delayed somatic and reproductive aging.
To investigate whether the extended lifespan of phm-2(lf) mutant animals was caused by an effect on development or an effect on adult animals, we compared phm-2(am117) animals cultured from embryo to adult on Comamonas or live E. coli and then transferred to live E. coli at adulthood. phm-2(lf) mutant animals displayed a similar lifespan in both cases, indicating that the bacterial food source during development does not strongly influence lifespan, whereas the bacterial food source during adulthood is critical (Figure 4H, Table S6).
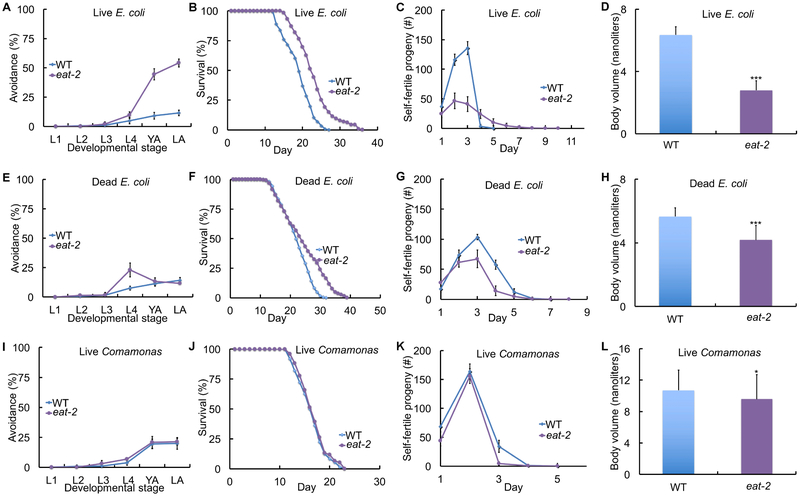
eat-2(lf) animals displayed live E. coli accumulation that contributed to the extended lifespan phenotype.
eat-2(lf) mutants were identified by Avery (1993a) based on feeding defects and observed to have slow, strong, often regular pharyngeal pumping. Lakowski and Hekimi (1998) observed that eat-2(lf) mutants display extended longevity. These results led to the model that the reduced pumping rate reduces food ingestion, causing caloric restriction and delayed aging. We confirmed that eat-2(lf) mutant animals displayed a reduced rate of pharyngeal pumping and a reduced rate of ingestion as measured by uptake of fluorescent beads compared to wild-type and phm-2(lf animals (Figure 2F, S1I). Surprisingly, we observed that eat-2(lf) mutant animals accumulated live E. coli in the intestine, similar to phm-2(lf) mutants (Figure 4C, S5G). Based on these results, we hypothesized that eat-2(lf) mutant animals have defective pharynx grinder function that allows live bacteria to enter the intestine. This model predicts that eat-2(lf) mutant animals will display activation of innate immune genes and hypersensitivity to highly pathogenic bacteria. Indeed, eat-2(lf mutants displayed higher levels of clec-7, clec-60, clec-82, F53A9.8, and lys-5 transcripts compared to wild type, and in four cases these levels were reduced by culture on Comamonas (Figure 4F, I, S5M-O). Furthermore, eat-2(lf) mutant animals were hypersensitive to highly pathogenic P. auruginosa strain PA14 (Figure 4D). To test additional predictions of this model, we measured bacterial avoidance behavior and analyzed the ability of non-pathogenic food sources to rescue the eat-2(lf) aging phenotypes. When cultured on live E. coli, eat-2(ad465) animals displayed strong bacterial avoidance behavior as well as extended self-fertile reproduction, extended lifespan, and scrawny body morphology (Figure 7A-D; Table S2, S6). eat-2(ad465) animals cultured on dead E. coli displayed mild food avoidance behavior, a small decrease in reproduction, and a small extension of lifespan compared to wild type (Figure 7E-H; Table S2, S6). eat-2(ad465) animals cultured on Comamonas were almost indistinguishable from wild type, since they displayed no bacterial avoidance behavior and reproduction and lifespan returned to wild-type levels; they still displayed a slightly reduced body volume (Figure 7I-L; Table S2, S6). These results indicate that eat-2(lf) mutations cause live E. coli accumulation in the intestine, which leads to activation of an innate immune response including bacterial avoidance behavior. Live E. coli accumulation appears to be the primary cause of eat-2(lf) aging phenotypes, since culture on nonpathogenic bacteria abrogated these defects even though it did not abrogate the slow pharyngeal pumping rate.
Figure 7. Live E. coli accumulation in eat-2(lf) mutants caused avoidance behavior, scrawny body morphology, and delayed aging.
WT or eat-2(ad465) hermaphrodites were cultured with a small bacterial lawn as indicated. (A, E, I) Avoidance as in Figure 1 - see Table S2. (B, F, J) Survival curves - see Table S6 for summary statistics. (C, G, K) Self progeny production as in Figure 1 (N=9-12 animals). (D, H, L) Worm volume as in Figure 1 (N=30-50 animals). ***, P < 0.0001; *, P < 0.05, by Student’s t-test.
Discussion
Identification of phm-2, which encodes a SAFB protein conserved in mammals, in a forward genetic screen for delayed reproductive aging.
The main emphasis of aging research has been lifespan, which is determined by age-related degeneration of life-support systems. However, there is increasing appreciation that extending health span as well as lifespan is an important objective of medical research (Bansal et al., 2015; Peña et al., 2014; Rockwood et al., 2005). Reproductive aging is an important component of health span; in human females, age-related degenerative changes increase the rate of birth defects and cause a progressive loss of fertility that culminate in menopause (Hartge, 2009; Te Velde and Pearson, 2002). Reproductive aging has been well documented in C. elegans and is characterized by a progressive decline in the rate of egg laying and increased disorganization of gonad morphology (Hughes et al., 2007). Whereas many genetic interventions have been reported to extend lifespan, only a subset influence reproductive aging (Kumar et al., 2014; Luo et al., 2009; Wang et al., 2014).
To identify genes that influence reproductive aging, Hughes et al. (2011) performed a forward genetic screen for mated hermaphrodites that display an extended reproductive span. The mutation am117 caused a substantial extension of mated reproductive span, increased the number of progeny generated late in the reproductive period, and delayed age-related degenerative changes in the morphology of the hermaphrodite gonad (Hughes et al., 2011). Here we demonstrate that the am117 mutation also influenced somatic aging, since it significantly extended mean and maximum lifespan. Furthermore, am117 mutant animals displayed significantly increased stress resistance, a phenotype frequently associated with enhanced longevity (Lithgow et al., 1995). The am117 mutation delayed the age-related decline of body movement and pharyngeal pumping significantly, indicating the mutation affects somatic aging.
We used positional cloning approaches to identify am117 as a nonsense mutation in F32B4.4. The phm-2 gene was discovered and named by Avery (1993a), who identified the ad538 and ad597 alleles based on pharyngeal pumping defects and positioned these mutations on the right arm of chromosome I. Here we demonstrate that ad597 is a 5 base pair deletion and ad538 appears to be a gross rearrangement of F32B4.4, and thus we refer to this gene as phm-2. Consistent with this model, both ad597 and ad538 caused delayed reproductive and somatic aging, similar to the am117 mutation. These results provide a molecular characterization of phm-2, and identify am117, ad597, and ad538 as strong loss-of-function or null alleles based on the molecular analysis and the similar phenotypes.
PHM-2 protein contains two highly conserved domains: a SAP domain and a RRM_SAF domain. The SAP domain is a putative DNA binding domain, whereas the RRM_SAF domain is a putative RNA recognition motif. Both domains are also contained in the human protein scaffold attachment factor B (SAFB). In humans, the SAFB family of proteins has three members: SAFB1, SAFB2, and SALMT. Human SAFB proteins are ubiquitously expressed in most tissues and localized in the nucleus. Similarly, we demonstrated that C. elegans PHM-2 is expressed in most or all C. elegans tissues and nuclear localized. The function of human SAFB is beginning to be revealed, but many important questions remain. SAFB appears to directly bind promoter regions and regulate transcription of many genes, including hsp27 and repression of the estrogen receptor (Aravind and Koonin, 2000; Oesterreich, 2003; Oesterreich et al., 2000; Townson et al., 2004; Weighardt et al., 1999). These data indicate that human SAFB and C. elegans phm-2 are homologous genes that are derived from a common ancestral gene. phm-2 is the only SAFB homolog in C. elegans, as opposed to three proteins in humans that may have redundant functions. Therefore, C. elegans is a relevant and powerful model system to investigate this important gene family.
phm-2 mutations cause a pharyngeal grinder defect that allows live bacteria to accumulate in the intestine, leading to an innate immune response that includes bacterial avoidance behavior.
The identification of phm-2 as the affected gene provided two key insights: (1) Avery (1993a) showed that phm-2 mutants have a defective pharynx grinder; (2) subsequent studies of pathogenicity showed phm-2 mutants are hypersensitive to pathogenic bacteria, because the grinder defect allows live bacteria to enter the intestinal lumen (Köthe et al., 2003; Labrousse et al., 2000; Portal-Celhay et al., 2012; Smith et al., 2002). Thus, we hypothesized that in standard culture conditions, phm-2 mutants allow live E. coli to enter the intestinal lumen, resulting in colonization by a weak pathogen that triggers an innate immune response that includes bacterial avoidance behavior.
Consistent with this model, we demonstrated that (1) Live E. coli accumulated in the intestine of phm-2(lf) mutant animals, as determined by fluorescently labeled bacteria; (2) phm-2(lf) mutants cultured on live E. coli displayed transcriptional changes typical of bacterial infections. Innate immune response genes such as C-type lectins, antimicrobial, and lysozyme genes were activated by live E. coli in phm-2(lf) mutant animals; (3) The HLH-30 transcription factor is the C. elegans homolog of mammalian TFEB, the master transcriptional regulator of the autophagy, and in worms HLH-30 protein accumulates in the nucleus in response to pathogenic bacteria (Visvikis et al., 2014). Here we demonstrated that phm-2(lf) mutant animals displayed nuclear localization of HLH-30. A hlh-30(lf) mutation suppressed the increased transcript levels of multiple innate immune genes, although it did not suppress the bacterial avoidance behavior, indicating that hlh-30 mediates part but not all of the immune response. A hlh-30(lf) mutation partially suppressed the phm-2(lf) lifespan extension, indicating that the immune response may contribute to the delayed aging phenotypes. An alternative interpretation is that hlh-30 activity is necessary for the extended lifespan of phm-2(lf) mutants because it promotes autophagy in response to dietary restriction. (4) Culture on non-pathogenic bacteria such as killed E. coli, live B. subtilis or live Comamonas abrogated the scrawny body morphology and aging phenotypes. Remarkably, on these food sources phm-2(lf) mutants closely resembled wild type; (5) Forced culture on live E. coli caused a shortened lifespan, suggestive of pathogenicity, consistent with previous studies (Portal-Celhay et al., 2012). Forced bacterial exposure was accomplished by using a large lawn that cannot be avoided or creating a double mutant with tph-1, which abrogates the bacterial avoidance behavior. These results indicate that the pharyngeal grinder defect allows live E. coli to enter the intestine and accumulate, triggering an innate immune response and bacterial avoidance behavior. If the lawn is small and can be avoided, then phm-2(lf) mutants display delayed somatic and reproductive aging. If the lawn is large and cannot be avoided, then phm-2(lf) mutants displayed a shortened lifespan due to bacterial pathogenicity. If the lawn consists of non-pathogenic bacteria, then animals do not display bacterial avoidance behavior, and the body morphology and lifespan are similar to wild type. Thus, the lifespan phenotype is conditional upon the level of bacterial pathogenicity: highly pathogenic bacteria result in a shortened lifespan, weakly pathogenic bacteria result in an extended lifespan, and non-pathogenic bacteria result in a wild-type lifespan.
The effects of pathogenic bacteria on phm-2(lf) mutants represent an interesting contrast to the effects on wild-type animals. Live E. coli are mildly pathogenic for older wild-type animals, which display age-related accumulation of bacteria in the intestine. This contributes to senescent death, since wild-type animals cultured on killed E. coli live several days longer than animals cultured on live E. coli (Garigan et al., 2002; Garsin et al., 2003; Gems and Riddle, 2000). By contrast, phm-2(lf) animals cultured on killed E. coli live several days shorter than animals cultured on live E. coli. In phm-2(lf) mutants, live E. coli enter the intestine of young animals, triggering bacterial avoidance behavior and an extended lifespan. In wild-type animals, live E. coli only enter the intestine of older adults that have developed pharyngeal dysfunction; these older adults are mobility impaired and do not avoid bacteria, and the accumulation of live bacteria shortens the lifespan.
A role for innate immunity in promoting longevity is beginning to emerge, and our analysis of phm-2 contributes to this area. daf-2(lf) mutants are long lived and resistant to bacterial pathogens, suggesting these two phenotypes might be related (Garsin et al., 2003). Furthermore, Troemel et al., (2006) showed that p38 MAPK pathway is important for innate immunity and contributes to the daf-2(lf) longevity phenotype. Our results show that colonization with pathogenic bacteria cause the lifespan extension of phm-2 mutants, directly implicating the innate immune response in enhanced longevity. Part of the response is transcriptional activation of innate immune genes and nuclear localization of HLH-30, and part is bacterial avoidance behavior that leads to dietary restriction (Figure 5J). An alternative interpretation that is consistent with the data is that live bacteria in the intestine lead to dietary restriction by impairing nutrient acquisition or otherwise inhibiting growth. Further work is necessary to establish the relative contributions of the damage caused by bacterial colonization versus the innate immune response that consists of molecular and behavioral changes triggered by bacterial colonization and/or damage.
Bacterial lawn avoidance behavior reflects ongoing monitoring of bacterial pathogenicity and requires serotonin signaling.
Bacterial avoidance behavior is a fascinating phenomenon that must involve sensing the pathogenicity of the bacteria and mounting a locomotion response. The behavior was first described in wild-type C. elegans in response to highly pathogenic bacteria such as S. marcescens and P. aeruginosa (Pujol et al., 2001; Zhang et al., 2005). In addition, the behavior has been documented in response to RNAi bacteria that reduce the activity of many different critical genes (Melo and Ruvkun, 2012). To characterize this behavior, we determined if prior exposure influences subsequent behavior, which might suggest that worms learn to avoid the bacteria. However, phm-2(lf) mutant animals initially cultured on pathogenic live E. coli or non-pathogenic killed E. coli displayed similar behavior, indicating that bacterial avoidance behavior involves continuous sensing rather than learning. This sensing can occur quickly, since phm-2(lf) mutant animals transferred to live E. coli displayed measurable avoidance behavior within one hour.
Several innate immune genes and pathways have been linked to bacterial avoidance behavior, such as FSHR-1 (Powell et al., 2009) the NSY-1/SEK-1/PMK-1 p38 family MAP kinase (Kim et al., 2002), Toll-like receptor (Pujol et al., 2001; Tenor and Aballay, 2008), and DAF-7/TGF-b (Reddy et al., 2009). Neuropeptide receptor 1 (npr-1) animals fail to avoid pathogen lawns of P. aeruginosa (Reddy et al., 2009). To investigate the role of these pathways in the phm-2(lf) bacterial avoidance behavior, we analyzed genetic interactions with pmk-1, mlk- 1, npr-1, daf-2, daf-16, and several other signaling pathways. The results indicate these genes are not necessary for the phm-2(lf) bacterial avoidance behavior. Serotonin is a highly conserved neurotransmitter that is used in the gastrointestinal tract and implicated in the response to enteric pathogens. For example, serotonin signaling mediates the Garcia effect, where rats and mammals learn to avoid with activation of nausea (Endo et al., 2000). To investigate the role of serotonin signaling, we used a tph-1(lf) mutation. tph-1 encodes a tryptophan hydrolase enzyme critical for serotonin synthesis, and tph-1(lf) mutants are defective in serotonin signaling (Szø et al., 2000). tph-1 activity was necessary for the phm-2(lf) bacterial avoidance behavior, indicating serotonin signaling plays a critical role in sensing pathogenic bacteria or mounting the locomotion response. This observation is consistent with studies of RNAi bacterial avoidance, which also requires tph-1 (Melo and Ruvkun, 2012). Furthermore, avoiding the highly pathogenic bacteria P. aeruginosa depends on tph-1, and continuous exposure to pathogenic bacteria increased transcription of tph-1 and the biosynthesis of serotonin in ADF neurons that promote avoidance behavior (Zhang et al., 2005). Together, these results indicate that phm-2(lf) bacterial avoidance behavior requires serotonin signaling but not several other innate immune response genes.
eat-2(lf) mutants displayed live bacterial accumulation in the intestine, activation of innate immunity, and bacterial avoidance that contribute to delayed aging.
Mutations in eat-2 were identified by Avery (1993a) based on a reduced rate of pharyngeal pumping. Lakowski and Hekimi (1998) observed that mutations in multiple eat genes cause an extended lifespan and focused on eat-2 because of the magnitude of the effect. They hypothesized that the reduced pharyngeal pumping rate leads to reduced bacterial ingestion and thereby dietary restriction. eat-2(lf) mutants have been used frequently to analyze dietary restriction, resulting in about 70 publications to date. There are alternative methods to achieve dietary restriction in worms, such as dilution of the bacterial food (Greer et al., 2007). A puzzling observation that has emerged is that different methods of dietary restriction cause somewhat different phenotypes (Greer and Brunet, 2009).
Here we demonstrate that eat-2(lf) mutants accumulate live E. coli in the intestine, indicating they have a defect in pharynx grinder function as well as a reduced rate of pumping. Consistent with this observation, eat-2(lf) mutants displayed activation of innate immune genes, bacterial avoidance behavior, and hypersensitivity to highly pathogenic bacteria. Thus, we hypothesized that the eat-2(lf) aging phenotype is caused by a mechanism similar to the phm-2(lf) phenotype. Indeed, when eat-2(lf) mutants were cultured on nonpathogenic bacteria (dead E. coli or Comamonas), the bacterial avoidance, scrawny body morphology, and longevity phenotypes were all suppressed. Similarly, Kauffman et al., (2010) previously observed that culture on Comamonas suppressed the eat-2 scrawny and extended lifespan phenotypes. Thus, although eat-2(lf) mutants have a reduced rate of pharyngeal pumping, the rate is still adequate for full nutrition when these animals do not avoid the bacterial lawn. These observations have implications for the mechanism of eat-2 longevity extension and the interpretation of studies that used this reagent. Both phm-2 and eat-2 mutants ingest live E. coli, which appears to be the root cause of the aging phenotypes. In principle, the aging phenotypes could be caused by damage from live bacteria, the innate immune response, the bacterial avoidance behavior that leads to dietary restriction, or a combination of these effects. Thus, eat-2 and phm-2 mutants are not examples of simple dietary restriction (Figure 5J). These observations may explain why eat-2 mutants have been shown to behave differently from dietary restriction achieved by dilution of the bacterial food.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kerry Kornfeld (kornfeld@wustl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans and Bacterial strains
C. elegans hermaphrodites were cultured on 6 cm Petri dishes containing NGM agar. To produce a small lawn of live bacteria, we pipetted 200 μl of an overnight culture of Escherichia coli strains OP50 or HB101, Baccillus subtilis, or Comamonas DA1877 and incubated overnight. To produce a small lawn of dead bacteria, we seeded NGM dishes with live E. coli OP50, cultured for 24 hours, and exposed to ultraviolet light by placing dishes in a UV Stratalinker 2400 for 15 minutes at maximum power. Bacterial killing was evaluated by inoculating LB medium with UV-treated bacteria; lack of growth at 37°C confirmed effective killing (Gems and Riddle, 2000). The wild-type strain was N2 Bristol. The identification and initial characterization of phm-2(am117) was previously described (Hughes et al., 2011). phm-2(am117) was outcrossed to WT up to 16 times. The phm-2 alleles ad538 and ad597 were outcrossed to WT 3 times. The genotypes of all double and triple mutant strains generated were confirmed by PCR and/or DNA pyrosequencing (Qiagen PSQ96). Additional alleles and strains used in the study are listed in the Key Resources Table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Primary polyclonal rabbit anti-GFP | Laboratory of Swathi Arur | Anti-GFP antibody |
| Secondary goat anti-rabbit Alexa-594 antibody | Life Technologies | A11012 |
| Bacterial and Virus Strains | ||
| Escherichia coli OP50 | CGC | OP50 |
| Escherichia coli OP50-GFP | CGC | E. coli OP50-GFP |
| Pseudomonas aeruginosa PA14 | Laboratory of Dennis Kim | PA14 |
| Comamonas DA1877 | Laboratory of Michael Nonet | DA1877 |
| Escherichia coli HB101 | Author’s laboratory | HB101 |
| Baccillus subtilis | Author’s laboratory | PY79 |
| Ahringer RNAi Libraries in E. coli HT115 (D3) | Source BioScience | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| DiO dye | Thermofisher | DiO |
| latex microspheres | Polysciences Inc. | Catalog #18242 |
| DAPI | Vector Laboratories | H-1200 |
| Fluorodeoxyuridine (FUdR) | Sigma-Aldrich | F0503 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| C. elegans: WT wild-type | CGC | WT |
| C. elegans: phm-2(am117) I | Author’s laboratory | WU740 |
| C. elegans: phm-2(ad538) I | CGC | DA538 |
| C. elegans: phm-2(ad597) I | CGC | DA597 |
| C. elegans: daf-16(mu86) I | CGC | CF1038 |
| C. elegans: daf-2(e1370) III | CGC | CB1370 |
| C. elegans: isp-1(qm150) IV | CGC | MQ887 |
| C. elegans: eat-2(ad465) II | CGC | DA465 |
| C. elegans: eat-2(ad1116) II | CGC | DA1116 |
| C. elegans: aak-2(ok524) X | CGC | RB754 |
| C. elegans: rict-1(mg360) II | CGC | KQ6 |
| C. elegans: rsks-1(ok1255) III | CGC | RB1206 |
| C. elegans: hlh-30(tm1978) IV | Laboratory of Mitani | TM1978 |
| C. elegans: tph-1(mg280) II | CGC | MT15434 |
| C. elegans: pmk-1(km25) IV | CGC | KU25 |
| C. elegans: mlk-1(ok2471) V | CGC | RB1908 |
| C. elegans: npr-1(ur89) X | CGC | IM222 |
| C. elegans: raga-1(ok386) II | CGC | VC222 |
| C. elegans: daf-16(mgDf47);daf-2(e1370) | CGC | GR1309 |
| C. elegans: acIs101[(F35E12.5:: gfp);pRF4(rol-6(su1006))] | CGC | AY101 |
| C. elegans: agIs17[irg-1::GFP; myo-2::mCherry] | CGC | AU133 |
| C. elegans: phm-2(am117) I (16 times) | This study | WU1686 |
| C. elegans: phm-2(ad538) I (3 times) | This study | WU1678 |
| C. elegans: phm-2(ad597) I (3 times) | This study | WU1679 |
| C. elegans: daf-16(mu86) phm-2(am117) | This study | WU1528 |
| C. elegans: phm-2(am117);daf-2 | This study | WU1683 |
| C. elegans: daf-16 phm-2(am117);daf-2 | This study | WU1526 |
| C. elegans: phm-2(am117);isp-1 | This study | WU1542 |
| C. elegans: phm-2(am117);eat-2(ad1116) | This study | WU1543 |
| C. elegans: phm-2(am117);aak-2 | This study | WU1734 |
| C. elegans: phm-2(am117);rict-1 | This study | WU1642 |
| C. elegans: phm-2(am117);rsks-1 | This study | WU1643 |
| C. elegans: phm-2(am117);hlh-30 | This study | WU1527 |
| C. elegans: phm-2(am117);pmk-1 | This study | WU1678 |
| C. elegans: phm-2(am117);mlk-1 | This study | WU1688 |
| C. elegans: phm-2(am117);npr-1 | This study | WU1668 |
| C. elegans: phm-2(am117);tph-1 | This study | WU1669 |
| C. elegans: phm-2(am117);raga-1 | This study | WU1855 |
| C. elegans: phm-2(am117); acIs101 | This study | WU1677 |
| C. elegans: phm-2(am117); agIs17 | This study | WU1676 |
| C. elegans: phm-2(am117);(hlh-30p::hlh-30::RFP);pRF4(rol-6)] | This study | WU1798 |
| C. elegans: amEx315[(phm-2p::gfp); pRF4(rol-6)], Genetic background WT | This study | WU1594 |
| C. elegans: amEx321[(phm-2p::phm-2::stop codon::gfp);pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1684 |
| C. elegans: amEx322[(phm-2p::phm-2::stop codon::gfp);pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1685 |
| C. elegans: amEx324[(phm-2p::phm-2::stop codon::gfp);pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1696 |
| C. elegans: amEx328[(phm-2p::phm-2::stop codon::gfp);pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1700 |
| C. elegans: amEx331[(phm-2p::phm-2::stop codon::gfp);pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1703 |
| C. elegans: amEx320[(phm-2p::phm-2::gfp); pRF4(rol-6)], Genetic background phm-2(am117) | This study | WU1671 |
| C. elegans: amIs13(hlh-30p::hlh-30::RFP);pRF4(rol-6)], Genetic background hlh-30(tm1978) | This study | WU1796 |
| Oligonucleotides | ||
| ama-1 Forward: 5′ATCGGAGCAGCCAGGAACTT | This study | N/A |
| ama-1 Reverse: 5′ GACTGTATGATGGTGAAGCTGG | This study | N/A |
| phm-2 Forward: 5′ CCATCTCGTCCAGAGTTGATAC | This study | N/A |
| phm-2 Reverse: 5′ GAGCTCCGAAGTGCTAATGT | This study | N/A |
| clec-7 Forward: 5′ GGCCGGCTTCAAATGTTTATC | This study | N/A |
| clec-7 Reverse: 5′ TAGTGGACATTACCATGCAGTC | This study | N/A |
| clec-60 Forward: 5′ CTGAGCCAAGAACCACAAGA | This study | N/A |
| clec-60 Reverse: 5′ GAAGTGCTGACTGACGAAAGA | This study | N/A |
| clec-82 Forward: 5′ TTCCGCCGTTGTCTGTTT | This study | N/A |
| clec-82 Reverse: 5′ CACTTGAGCTGGCTAGATTGA | This study | N/A |
| F53A9.8 Forward: 5′GTTCACCATGCAGGAGATCA | This study | N/A |
| F53A9.8 Reverse: 5′ TCTCCATCTTGGTGTTGAGTTT | This study | N/A |
| lys-5 Forward: 5′ CGGGAAGTGTAGATACTGTTGG | This study | N/A |
| lys-7 Reverse: 5′ AGAGACGCCTTAACTTGGTTAG | This study | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| Graph, Result and Statistical analysis | Microsoft Office | Excel, Powerpoint |
| Result and Statistical analysis | Astatsa | http://astatsa.com |
| Result and Statistical analysis | OASIS | https://sbi.postech.ac.kr/oasis/ |
| Image editing | Adobe | Photoshop and illustrator |
| Image editing | Fiji ImageJ | https://fiji.sc |
| WormSizer | Laboratory of L. Baugh | https://github.com/bradtmoore/wormsizer |
| Other | ||
METHOD DETAILS
RNA interference (RNAi)
RNA interference was performed by feeding bacteria that express dsRNA (RNAi bacteria), as described by Kamath et al. (2003). Briefly, E. coli HT115 bacteria with the control plasmid (L4440) or a plasmid encoding pha-4 were obtained from the Ahringer library (Kamath et al., 2001). The identity of the clones was confirmed by DNA sequencing. Single colonies of RNAi bacteria were isolated from LB dishes containing 50μg/ml ampicillin and used to inoculate overnight starter cultures in LB medium containing 50μg/ml ampicillin. The starter cultures were used to inoculate larger cultures at 1:100 dilution in LB medium containing 50μg/ml ampicillin and grown for 6 hours at 37°C. RNAi bacteria were seeded onto NGM agar dishes containing 1mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 50μg/ml carbenicillin. L1 stage larvae were transferred to RNAi dishes and analyzed after reaching adulthood.
Identification of am117 by whole genome sequencing
To identify the am117 molecular lesion, we generated genomic DNA from the am117 strain, performed whole genome sequencing, and compared the results to the WT sequence. In the region close to unc-75 (+9.3 on chromosome I) where am117 was positioned by Hughes et al. (2011), three mutations were detected that affect the coding sequence of F32B4.4, Y53C10.5 and F56H6.9.
Plasmid construction and transgenic strain generation
To analyze transcription of phm-2 isoform a (F32B4.4a), we PCR amplified the 496bp region between the start codon of phm-2 and the 3’ end of the adjacent upstream gene from genomic DNA of wild-type animals. This promoter region was inserted into plasmid pDG218 to generate pSK1 [phm-2p::gfp::unc-54 3’UTR]. Briefly, pDG218 was derived from pBluescript SK+ (Stratagene) by inserting the coding region for green fluorescent protein (GFP) and the unc-54 3′-UTR, both amplified from pPD95.77, a gift from A. Fire (Stanford University, Palo Alto, CA).
To analyze expression of PHM-2 protein, we PCR-amplified full length, wild-type phm-2 coding sequence (3222 bp) from cDNA and ligated into pSK1 to generate pSK2 [phm-2p::phm-2::stop codon::gfp::unc-54 3’UTR]. pSK2 encodes PHM-2 protein but the gfp coding sequence is out-of-frame following the stop codon of phm-2. The stop codon in pSK2 was removed by site directed mutagenesis (New England Biolabs) to generate pSK3 [phm-2p::phm-2::gfp::unc-54 3’UTR]. pSK3 encodes a PHM-2::GFP fusion protein.
Plasmid DNA was prepared using a Miniprep column (Qiagen) and quantified using a Nanodrop (Thermo).
Transgenic animals were generated by injecting pSK1 into wild-type hermaphrodites and pSK2 or pSK3 into phm-2(am117) hermaphrodites. All injections were performed with the dominant Rol marker pRF4 (Mello et al., 1991). We selected independently derived Rol self-progeny that transmitted the Rol phenotype. These transgenes formed extrachromosomal arrays, since the Rol phenotype was transmitted to only a sub-set of the self-progeny.
To measure the expression pattern of HLH-30, we PCR amplified a 1497bp fragment containing the coding sequence for W02C12.3a from pDONR201 (Dharmacon) and a 2000bp fragment containing W02C12.3a promoter (Dharmacon) and subcloned these into the pBlueScript SK+ vector pDG219 to create pJM18 [hlh-30p::hlh-30::rfp]. To generate transgenic animals with an extrachromosomal array, we injected pJM18 and pRF4 into hlh-30(tm1978) worms. The extrachromosomal array was integrated into the genome by UV irradiation (UV Stratalinker 2400), and we selected RFP positive worms with a roller phenotype that segregated only transgenic animals.
We used standard genetic techniques to construct strains with phm-2(am117); HLH-30::RFP. Eggs were allowed to develop into two day old adults on live E. coli OP50 and live Comamonas bacteria, and animals were paralyzed with 1% NaN3 and mounted on a 2% agarose pad for imaging. Images were captured using the 10X objective of a Zeiss Axioplan fluorescent compound microscope equipped with a Zeiss AxioCam MRm digital camera. More than 100 animals were analyzed in three biological replicates.
Measurement of lifespan, reproduction, pharyngeal pumping, and body movement
Studies of lifespan were begun on day zero by placing L4 stage hermaphrodites on a Petri dish. Hermaphrodites were transferred to a fresh Petri dish daily during the reproductive period (approximately the first ten days) to eliminate self-progeny and every 2-3 days thereafter. Each hermaphrodite was examined daily using a dissecting microscope for survival, determined by spontaneous movement or movement in response to prodding with a platinum wire. Dead worms that displayed matricidal hatching, vulval extrusion, or desiccation due to crawling off the agar were excluded from the data analysis. One exception to this data analysis approach was that animals that displayed matricidal hatching were not excluded for experiments shown in Table S6 rows 21-25. Mean lifespan was calculated as the number of days from the L4 stage to the last day a worm was observed to be alive.
For the experiment described in Figure 4H, WT or phm-2(am117) embryos were hatched on NGM agar dishes seeded with 200 μL of either E. coli OP50 or Comamonas DA1877. At adulthood, worms were transferred to plates seeded with E. coli OP50 for the remainder of their lifespan.
Daily reproduction, brood size, pharyngeal pumping, and body movement were determined as described in Kumar et al., (2016). Briefly, to analyze progeny production, one L4 stage hermaphrodite was placed on a Petri dish (day one) and transferred to a fresh dish daily until at least 4 days without progeny production. Progeny were counted after two days. Pharyngeal pumping was counted during a 10 second interval using a dissecting microscope. Body movement was assayed by observation for 20 seconds using a dissecting microscope. Petri dishes were tapped to stimulate animals to move before scoring.
Measurement of heat stress resistance and dauer formation
Thermotolerance assays were performed as described by McColl et al., (2010). Briefly, WT and phm-2(am117) L4 stage hermaphrodites were cultured at 20°C for one day, adults were transferred to 35°C and scored as alive or dead based on responding to prodding with a platinum wire after either 9 or 12 hours.
Dauer formation was assayed as described by Kimura et al.,(1997). To analyze dauer formation of daf-2(e1370) and phm-2(am117); daf-2(e1370) animals, we transferred eggs to 20°C, 22.5°C and 25°C and scored dauer larvae after culturing 3-5 days.
Measurement of worm size
To measure animal size, we captured images of live adult animals four days after the L4 stage on unseeded NGM dishes using a Leica M80 microscope with a 1.25X objective and an IC80 HD camera. Images were batch-processed with IrfanView 4.37 or ImageJ, and worm volume was automatically measured using the WormSizer 1.2.0 plugin for Fiji ImageJ 1.49n, as described in Moore et al., (2013). Roller transgenic animals were paralyzed in a drop of sodium azide on unseeded NGM dishes to prevent the head and tail from touching.
Measurement of bacterial avoidance behavior
We prepared small, uniform circular lawns by pipetting 100 μL of overnight cultures of various bacteria (live E. coli OP50 or HB101, live B. subtillis live Comamonas DA1877,) on the center of NGM dishes and allowing at least one day to dry. To produce a small lawn of dead bacteria, we seeded NGM dishes with 100 μl of live E. coli OP50, cultured for 24 hours, then exposed to ultraviolet light. The next day 50-80 eggs were transferred on top of the bacterial lawn without disturbing or spreading the lawn. Each animal was scored as inside or outside the lawn (Avoidance=Nout/Nttotal) at larval stages (L1 to L4) and one (YA) and two (LA) day old adults. For antibiotic dishes, live E. coli OP50 bacteria was seeded onto NGM agar dishes containing 12.5 μg/ml tetracycline or 50 μg/ml kanamycin. To generate lawns with a mixture of bacteria, we grew E. coli OP50 and Comamonas DA1877 overnight, created mixed cultures of various ratios (100:0, 10:90, 50:50, 90:10 and 0:100), and seeded the mixed culture on NGM dishes. To determine whether bacterial secondary metabolites modulate avoidance behavior, we grew E. coli OP50 and Comamonas DA1877 overnight, pelleted the bacteria at 10,000 RPM for 20 minutes, collected the supernatant, and filtered the supernatant using a 0.22 μm syringe filter. To generate odorant signals, we cut a square NGM block with either E. coli OP50 or Comamonas DA1877, attached the block to the inside of a lid, placed the lid over dishes seeded with E. coli OP50 or Comamonas DA1877, and wrapped the dishes with parafilm. For studies of the pathogenic bacteria P. aeruginosa strain PA14, WT and phm-2(am117) eggs were allowed to develop to the L4 stage on live E. coli OP50, washed 2-3 times in S basal and transferred to a P. aeruginosa strain PA14 lawn on NGM dishes. Animals were scored for avoidance after 24 hours.
To determine if experience influences bacterial lawn avoidance behavior, we placed 50-80 eggs on a “training dish” containing a small bacterial lawn (live E. coli OP50, live Comamonas DA1877, or UV-killed E. coli OP50). Young adults (one day after L4 stage) were washed 2-3 times in S basal, transferred to a “testing dish” with a small bacterial lawn and scored as inside or outside the lawn every hour.
Measurement of bacterial lawn choice behavior
To analyze the preference between two bacterial lawn options, we generated dishes with two small lawns by pipetting 90 μl of overnight cultures on either side of a NGM dish. 50-60 eggs were placed on a “training” dish with a small bacterial lawn (live E. coli OP50, live Comamonas DA1877, or dead E. coli OP50). Young adults (one day after L4 stage) were washed 2-3 times in S basal, transferred to a “testing dish” with two small bacterial lawns, and scored as inside lawn 1, inside lawn 2 or outside both lawns after 24 hours.
Measurement of matricidal hatching
We prepared NGM dishes with a large lawn (400 μl of live E. coli OP50 spread over the entire surface) or a small lawn (100 μl of live E. coli OP50 spotted in the center); dishes were allowed to dry at least one day. To quantify the prevalence of matricidal hatching, we placed 40-60 eggs (WT or phm-2(am117)) on these dishes and allowed them to develop to the L4 stage (defined as day 0). Hermaphrodites were transferred to a fresh dish daily during the reproductive period (approximately the first fifteen days) to eliminate self-progeny. Using a dissecting microscope, each hermaphrodite was examined daily for survival, determined by spontaneous movement or movement in response to prodding with a platinum wire. Dead hermaphrodites that displayed live progeny inside the body were scored as matricidal hatching.
Measurement of survival and induction of reporter genes on P. aeruginosa
Slow killing was assayed as described (Estes et al., 2010; Powell and Ausubel, 2008). Briefly, the P. aeruginosa PA14 strain was cultured in LB medium, seeded on 6 cm Petri dishes with 50 μM FUdR, incubated for 24 hours at 37°C and then for 24 hours at room temperature. 20-30 L4 stage hermaphrodites were transferred to these dishes, incubated at 25°C, and scored for survival every 12 hours. Experiments were repeated at least five times.
Two transgenic strains had pathogen responsive promoters driving GFP: F35E12.5 (acIs101[(F35E12.5::gfp);pRF4(rol-6(su1006))], and irg-1 (agIs17[irg-1::GFP; myo-2::mCherry]) (Estes et al., 2010; Troemel et al., 2006). One transgenic strain had the phm-2 promoter driving GFP (amEx315[(phm-2p::gfp); pRF4(rol-6)]. To measure GFP induction, we placed 30–40 young adult hermaphrodites (one day past the L4 stage) on SK dishes with P. aeruginosa PA14 and cultured for 24 hours. Animals were washed 3 times with M9 and placed on a large pre-made 1.5% agarose pad with a 20ul drop of 1M sodium azide. A rectangular coverslip was applied, and slides were imaged on a Zeiss Axioplan fluorescent compound microscope equipped with a Zeiss AxioCam MRm digital camera using a 10x objective.
Measurement of fluorescent microsphere ingestion
The protocol was modified from Kiyama et al., (2012). Briefly, Fluoresbrite Plain YG 45.58um +/−0.80um solid latex microspheres in water (Polysciences Inc. 9003-53-6, catalog #18242, lot# 589756) were dissolved in 90% ethanol at a concentration of 3.64×10^10 and diluted 1:1000 in S-basal. 10^8 microspheres were applied to 60mm NGM dishes and spread with a flame-sterilized glass spreader. The S-basal was allowed to soak into the NGM overnight and protected from light. Worms were washed 3-4 times in a tabletop microcentrifuge with 1ml of S-basal + 0.1% Tween-20 (SBTw) to remove bacteria, and then applied to a dish. Dishes were incubated with open lids for 15 minutes in the dark, and then 1ml of 50mM sodium azide in SBTw was added to immobilize the worms. Worms were transferred to an Eppendorf tube and pelleted by centrifugation; the supernatant was removed, and ice-cold methanol was added for 10 minutes. Worms were washed once with SBTw and applied to large pre-made 1.5% agarose pads on glass slides with a rectangular coverslip. Slides were imaged on a Zeiss Axioplan fluorescent compound microscope equipped with a Zeiss AxioCam MRm digital camera using a 5x objective.
Measurement of GFP-labeled E. coli in the intestinal lumen
To measure intact bacteria in the intestine, we utilized a strain of E. coli labeled with GFP obtained from the CGC that was cultured overnight with 50μg/ml ampicillin at 37 °C with shaking (Labrousse et al., 2000). Feeding experiments were performed as described above for microsphere ingestion, except that live bacteria were used to seed NGM dishes (100 ul in the center of a dish, or 400 ul spread across the entire surface of a dish), and worms were allowed to feed for 24 hours. Based on the fluorescent images, we defined three patterns: GFP fluorescence throughout the intestine (full), in some but not all the intestine (partial), and not strongly in the intestine (none).
To measure bacterial ingestion in worms treated with pha-4 RNAi, we allowed worms to hatch in S-basal + 0.1% cholesterol. L1 animals were transferred to NGM agar dishes containing 1mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 50μg/ml carbenicillin and seeded with E. coli HT115 bacteria with either the control plasmid (L4440) or a plasmid encoding pha-4. At adulthood, worms were transferred to RNAi dishes seeded with a bacterial solution containing 25% by volume GFP expressing E. coli OP50 and 75% by volume control (L4440) or pha-4 RNAi E. coli bacteria. Worms were cultured for 24 hours before being washed and visualized as previously described.
Measurement of DiO staining
To stain animals with DiO, we followed the protocol of Collins et al., (2008a) with slight modifications. Briefly, ~2mg DiO was dissolved in 1ml dimethyl formamide and then diluted in M9 to a final concentration of 20ug/ml. L4 stage animals were picked into M9, washed once with M9, incubated in DiO solution for 4 hours, washed 3 times with M9, and applied to a large pre-made 1.5% agarose pad with a 20ul drop of 1M sodium azide. After applying a rectangular coverslip, the slides were imaged on a Zeiss Axioplan florescent compound microscope equipped with a Zeiss AxioCam MRm digital camera using a 40x objective. osm-3(p802) and N2 animals were used as positive and negative controls, respectively, for the Dyf-d phenotype.
Protein imaging in vivo
To perform live imaging of fluorescent proteins, we picked >20 animals into S-basal + 4mM levamisole and placed the animals onto a large pre-made 1.5% agarose pad.
To image intact worms with antibodies, we picked >20 animals into PBS + 0.1% Tween-20 (PBSTw), washed with PBSTw, fixed in 3% PFA (EM grade) for 30-60 minutes, washed twice with PBSTw, incubated with DAPI (1:1000; 0.1mg/ml) for 5-10 minutes, and mounted the animals on a large pre-made 2.5% agarose pad in DABCO in 90% glycerol. The slide was allowed to dry overnight and then imaged using a PerkinElmer spinning-disc confocal microscope with a 40x or 63x oil-immersion lens.
To image dissected worms with antibodies, we picked >50 animals into PBS, washed twice with PBS, immobilized with levamisole, and dissected by cutting at the pharynx or at the tail with two 25G needles. Worms were fixed in 3% PFA for 10 minutes, post-fixed in ice-cold (−20°C) high-grade methanol (Fisher) for 2 hours, and washed 3 times in PBSTw. Primary polyclonal rabbit anti-GFP antibody (gift from Swathi Arur) was applied at 1:100 in 30% Goat Serum overnight at room temperature. After 3 PBSTw washes, secondary goat anti-rabbit Alexa- 594 antibody was applied at 1:400 in 30% Goat Serum overnight at 4°C. After 3 PBSTw washes and a 5 minute DAPI incubation, worms were applied to a large pre-made 2.5% agarose pad in DABCO in 90% glycerol. The slide was allowed to dry overnight and then imaged with a Perkin Elmer spinning-disc confocal microscope using a 40x, 63x, or 100x oil-immersion lens.
Pharynx morphology
To image pharynx morphology, we washed WT and phm-2(am117) young adult animals 2-3 times in S basal, paralyzed the animals with 1% sodium azide, mounted the animals on 2% agarose pads and covered with a coverslip. DIC images were acquired using a Zeiss Axioplan fluorescent compound microscope equipped with a Zeiss AxioCam MRm digital camera.
Quantitative real-time PCR (RT-PCR)
To generate synchronous populations of worms for RNA extraction, we allowed WT, phm-2(am117), eat-2(ad465), hlh-30(tm1978) and phm-2(am117);hlh-30(tm1978) adults to lay eggs for 4-6 hours at 20°C on NGM dishes seeded with live E. coli OP50. Eggs were allowed to develop until 12 hours after the L4 stage. These synchronized young adults were washed and collected for RNA isolation. RNA analysis was performed as previously described with slight modifications (Davis et al., 2009). Briefly, RNA was isolated using a Trizol (Invitrogen) and chloroform extraction (GlycoBlue was used to aid precipitation in isopropanol), treated with DNAseI, and further purified using an RNeasy spin column (Quiagen). RNA was quantified using a NanoDrop and diluted in nuclease-free water. cDNA was synthesized using either High Capacity cDNA Reverse Transcription kit (Applied Biosystems) or iScript cDNA Synthesis Kit (BioRad Laboratories, Hercules, CA). Quantitative, real-time PCR was performed using an Applied Biosystems Step One Plus Real-Time PCR system and either SYBR green master mix (Applied Biosystems) or iTaq Universal SYBR Green Supermix (BioRad Laboratories, Hercules, CA). mRNA fold change was calculated using the comparative CT method (Schmittgen and Livak, 2008) by comparing mRNA levels of the gene of interest with mRNA levels of the reference gene ama-1. A minimum of three biological replicates, each with three technical replicates, were performed.
Statistical Analysis
All data from RT-PCR experiments and measurement of brood size, dauer formation, heat stress resistance, lifespan and bacterial avoidance behavior were analyzed using the two-tailed Student’s t-test for samples with unequal variances by using Excel. For comparisons of more than two groups, an ANOVA (http://astatsa.com) was used. P-values less than 0.05 were considered statistically significant. To determine whether the choice of a statistical test for lifespan affected the conclusions, we used the log-rank (Mantel-Cox) method to analyze a subset of the lifespan experiments https://sbi.postech.ac.kr/oasis/ (Yang et al., 2011). Both tests produced similar P-values.
Supplementary Material
Highlights.
phm-2 encodes a SAFB protein conserved in mammals
phm-2 and eat-2 mutants have a defective pharynx and accumulate bacteria in the gut
Bacterial accumulation activates innate immunity and bacterial avoidance behavior
Bacterial avoidance leads to dietary restriction and delays aging
Acknowledgments
We thank the Caenorhabditis Genetics Center for providing stains, Michael Nonet for assistance with cloning phm-2, Kenneth Wong for regent generation, and Elaine Mardis and David Larson for sharing their knowledge, expertise, and facilities. This work was supported by NIH grant R01 AG02656106A1 to KK.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apfeld J, O’Connor G, McDonagh T, DiStefano PS, and Curtis R (2004). The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 18, 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, and Koonin EV (2000). SAP - A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci 25, 112–114. [DOI] [PubMed] [Google Scholar]
- Avery L (1993a). The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L (1993b). Motor neuron M3 controls pharyngeal muscle relaxation timing in Caenorhabditis elegans. J. Exp. Biol 175, 283–297. [DOI] [PubMed] [Google Scholar]
- Bansal A, Zhu LJ, Yen K, and Tissenbaum HA (2015). Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc. Natl. Acad. Sci 112, E277–E286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Huang C, Hughes S, and Kornfeld K (2008a). The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JJ, Evason K, Pickett CL, Schneider DL, and Kornfeld K (2008b). The anticonvulsant ethosuximide disrupts sensory function to extend C. elegans lifespan. PLoS Genet. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis R, O’Connor G, and DiStefano PS (2006). Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell 5, 119–126. [DOI] [PubMed] [Google Scholar]
- Darby C, Hsu JW, Ghori N, and Falkow S (2002). Caenorhabditis elegans: Plague bacteria biofilm blocks food intake. Nature 417, 243–244. [DOI] [PubMed] [Google Scholar]
- Davis DE, Hyun CR, Deshmukh K, Bruinsma JJ, Schneider DL, Guthrie J, Robertson JD, and Kornfeld K (2009). The cation diffusion facilitator gene cdf-2 mediates zinc metabolism in Caenorhabditis elegans. Genetics 182, 1015–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doncaster CC (1962). Nematode feeding mechanisms. 1. observations on rhabditis and pelodera. Nematologica 8, 313–320. [Google Scholar]
- Endo T, Minami M, Hirafuji M, Ogawa T, Akita K, Nemoto M, Saito H, Yoshioka M, and Parvez SH (2000). Neurochemistry and neuropharmacology of emesis - The role of serotonin. Toxicology 153, 189–201. [DOI] [PubMed] [Google Scholar]
- Estes K. a, Dunbar TL, Powell JR, Ausubel FM and Troemel ER (2010). bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 107, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, and Duveau F (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussière F, and Hekimi S (2001). Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Friedman DB, and Johnson TE (1988). A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garee JP, and Oesterreich S (2010). SAFB1’s multiple functions in biological control-lots still to be done! J. Cell. Biochem 109, 312–319. [DOI] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Abringet J, and Kenyon C (2002). Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, and Ausubel FM (2003). Long-lived C. elegans daf-2 Mutants are resistant to bacterial pathogens. Science (80-. ). 300, 1921. [DOI] [PubMed] [Google Scholar]
- Gems D, and Riddle DL (2000). Genetic, behavioral and environmental determinants of male longevity in Caenorhabditis elegans. Genetics 154, 1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, and Brunet A (2009). Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, and Brunet A (2007). An AMPK-FOXO Pathway Mediates Longevity Induced by a Novel Method of Dietary Restriction in C. elegans. Curr. Biol 17, 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, and Kenyon C (2000). Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262. [DOI] [PubMed] [Google Scholar]
- Hansen M, Hsu AL, Dillin A, and Kenyon C (2005). New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 0119–0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, and Kenyon C (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, and Kenyon C (2008). A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartge P (2009). Genetics of reproductive lifespan. Nat. Genet 41, 637–638. [DOI] [PubMed] [Google Scholar]
- Hillier LDW, Marth GT, Quinlan AR, Dooling D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang W, et al. (2008). Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods 5, 183–188. [DOI] [PubMed] [Google Scholar]
- Huang C, Xiong C, and Kornfeld K (2004). Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl. Acad. Sci 101, 8084–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Evason K, Xiong C, and Kornfeld K (2007). Genetic and pharmacological factors that influence reproductive aging in nematodes. PLoS Genet. 3, 0254–0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SE, Huang C, and Kornfeld K (2011). Identification of mutations that delay somatic or reproductive aging of Caenorhabditis elegans. Genetics 189, 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui JE, Troemel ER, Feinbaum RL, Luhachack LG, Cezairliyan BO, and Ausubel FM (2010). Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, and Wood WB (1982). Genetic analysis of life-span in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 79, 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, Cypser J, De Castro E, De Castro S, Henderson S, Murakami S, Rikke B, Tedesco P, and Link C (2000). Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. In Experimental Gerontology, pp. 687–694. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, and Ahringer J (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kao CY, Los FCO, Huffman DL, Wachi S, Kloft N, Husmann M, Karabrahimi V, Schwartz JL, Bellier A, Ha C, et al. (2011). Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PWL, Thomas EL, and Kockel L (2010). With TOR, less is more: A key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, orces-Zimmerman MR, Landis JN, and Murphy CT (2010). Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ (2010). The genetics of ageing. Nature 464, 504–512. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, and Tabtiang R (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M-W, et al. (2002). A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, and Ruvkun G (1997). Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science (80-. ). 277, 942–946. [DOI] [PubMed] [Google Scholar]
- Kiyama Y, Miyahara K, and Ohshima Y (2012). Active uptake of artificial particles in the nematode Caenorhabditis elegans. J. Exp. Biol 215, 1178–1183. [DOI] [PubMed] [Google Scholar]
- Klass MR (1977). Aging in the nematode Caenorhabditis elegans: Major biological and environmental factors influencing life span. Mech. Ageing Dev 6, 413–429. [DOI] [PubMed] [Google Scholar]
- Köthe M, Antl M, Huber B, Stoecker K, Ebrecht D, Steinmetz I, and Eberl L (2003). Killing of Caenorhabditis elegans by Burkholderia cepacia is controlled by the cep quorum-sensing system. Cell. Microbiol 5, 343–351. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kocsisova Z, and Kornfeld K (2014). Keep on Laying Eggs Mama, RNAi My Reproductive Aging Blues Away. PLoS Genet. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dietrich N, and Kornfeld K (2016). Angiotensin Converting Enzyme (ACE) Inhibitor Extends Caenorhabditis elegans Life Span. PLoS Genet. 12, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrousse a, Chauvet S, Couillault C, Kurz CL, and Ewbank JJ (2000). Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol 10, 1543–1545. [DOI] [PubMed] [Google Scholar]
- Lakowski B, and Hekimi S (1996). Determination of Life-Span in Caenorhabditis elegans by Four Clock Genes. Sci. New Ser 272, 1010–1013. [DOI] [PubMed] [Google Scholar]
- Lakowski B, and Hekimi S (1998). The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci 95, 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, et al. (2013). The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat. Commun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RYN, Fraser AG, Kamath RS, Ahringer J, and Ruvkun G (2003). A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 33, 40–48. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, and Kenyon C (1997). daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science (80-. ). 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, and Johnson TE (1995). Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc. Natl. Acad. Sci 92, 7540–7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Shaw WM, Ashraf J, and Murphy CT (2009). TGF-ß Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genet. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, and Thomas JH (1994). A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics 136, 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, and Maynard LA (1935). The effect of retarded growth upon the length of life span and upon the ultimate body size. J. Nutr 5, 63–79. DOI: [PubMed] [Google Scholar]
- McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, and Lithgow GJ (2010). Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 12, 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JP, Raizen DM, Gottschalk A, Schafer WR, and Avery L (2004). eat-2 and eat-18 are Required for Nicotinic Neurotransmission in the Caenorhabditis elegans Pharynx. Genetics 166, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, and Ambros V (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, and Ruvkun G (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BT, Jordan JM, and Baugh LR (2013). WormSizer: High-throughput Analysis of Nematode Size and Shape. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, and Ruvkun G (1996). A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536–539. [DOI] [PubMed] [Google Scholar]
- Oesterreich S (2003). Scaffold Attachment Factors SAFB1 and SAFB2: Innocent Bystanders or Critical Players in Breast Tumorigenesis? J. Cell. Biochem 90, 653–661. [DOI] [PubMed] [Google Scholar]
- Oesterreich S, Zhang Q, Hopp T, Fuqua S. a, Michaelis M, Zhao HH, Davie JR, Osborne CK, and Lee a V (2000). Tamoxifen-bound estrogen receptor (ER) strongly interacts with the nuclear matrix protein HET/SAF-B, a novel inhibitor of ER-mediated transactivation. Mol. Endocrinol 14, 369–381. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, and Ruvkun G (1997). The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, and Kapahi P (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, and Dillin A (2007). PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447, 550–555. [DOI] [PubMed] [Google Scholar]
- Peña FG, Theou O, Wallace L, Brothers TD, Gill TM, Gahbauer EA, Kirkland S, Mitnitski A, and Rockwood K (2014). Comparison of alternate scoring of variables on the performance of the frailty index. BMC Geriatr. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, and Culotti JG (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol 117, 456–487. [DOI] [PubMed] [Google Scholar]
- Portal-Celhay C, Bradley ER, and Blaser MJ (2012). Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Microbiol. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, and Ausubel FM (2008). Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. Methods Mol. Biol 415, 403–427. [DOI] [PubMed] [Google Scholar]
- Powell JR, Kim DH, and Ausubel FM (2009). The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc. Natl. Acad. Sci. U. S. A 106, 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, and Ewbank JJ (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A 104, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol N, Link EM, Liu LX, Kurz CL, Alloing G, Tan MW, Ray KP, Solari R, Johnson CD, and Ewbank JJ (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol 11, 809–821. [DOI] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, and Kim DH (2009). A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science (80-. ). 323, 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwood K, Song X, MacKnight C, Bergman H, Hogan D, McDowell I, and Al E (2005). A global clinical measure of fitness and frailty in elderly people. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, and McIntire SL (2010). Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H, and Ewbank JJ (2007). The genetics of pathogen avoidance in Caenorhabditis elegans. Mol. Microbiol 66, 563–570. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JMA, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. (2009). Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. (2011). TFEB links autophagy to lysosomal biogenesis. Science (80-. ). 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifri CD, Begun J, Ausubel FM, and Calderwood SB (2003). Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun 71, 2208–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MP, Laws TR, Atkins TP, Oyston PCF, De Pomerai DI, and Titball RW (2002). A liquid-based method for the assessment of bacterial pathogenicity using the nematode Caenorhabditis elegans. FEMS Microbiol. Lett 210, 181–185. [DOI] [PubMed] [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, and Riddle DL (1995). Mutations affecting in chemosensory neurons of Caenorhabditis elegans. Genetics 139, 171–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szø JY, Victor M, Loer C, Shi Y, and Ruvkun G (2000). Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564. [DOI] [PubMed] [Google Scholar]
- Tenor JL, and Aballay A (2008). A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townson SM, Kang K, Lee AV, and Oesterreich S (2004). Structure-function analysis of the estrogen receptor α corepressor scaffold attachment factor-B1: Identification of a potent transcriptional repression domain. J. Biol. Chem 279, 26074–26081. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, and Kim DH (2006). p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2, 1725–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velde ER, and Pearson PL (2002). The variability of female reproductive ageing. Hum. Reprod Update 8, 141–154. [DOI] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, and Müller F (2003). Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 426, 620–620. [DOI] [PubMed] [Google Scholar]
- Visvikis O, Ihuegbu N, Labed SA, Luhachack LG, Alves AMF, Wollenberg AC, Stuart LM, Stormo GD, and Irazoqui JE (2014). Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity 40, 896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Oakley HD, Carr CE, Sowa JN, and Ruvkun G (2014). Gene Pathways That Delay Caenorhabditis elegans Reproductive Senescence. PLoS Genet. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Cobianchi F, Cartegni L, Chiodi I, Villa a, Riva S, and Biamonti G (1999). A novel hnRNP protein (HAP/SAF-B) enters a subset of hnRNP complexes and relocates in nuclear granules in response to heat shock. J. Cell Sci 112 ( Pt 1, 1465–1476. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, and Hall MN (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. [DOI] [PubMed] [Google Scholar]
- Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, and Kim S (2011). OASIS: Online application for the survival analysis of lifespan assays performed in aging research. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, and Bargmann CI (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.